Abstract
Background
Primary sarcopenia is usually known as age-related skeletal muscle loss; however, other factors like endocrine, lifestyle and inflammation can also cause muscle loss, known as secondary sarcopenia. Although many studies have used different sarcopenia animal models for exploring the underlying mechanism and therapeutic approaches for sarcopenia, limited study has provided evidence of the relevance of these animal models. This study aims to investigate the similarity and difference in muscle qualities between primary and secondary sarcopenia mice models, using naturally aged mice and dexamethasone-induced mice.
Methods
21-month-old mice were used as naturally aged primary sarcopenia mice and 3-month-old mice received daily intraperitoneal injection of dexamethasone (20 mg/ kg body weight) for 10 days were used as secondary sarcopenia model. This study provided measurements for muscle mass and functions, including Dual-energy X-ray absorptiometry (DXA) scanning, handgrip strength test and treadmill running to exhaustion test. Besides, muscle contraction, muscle fibre type measurements and gene expression were also performed to provide additional information on muscle qualities.
Results
The results suggest two sarcopenia animal models shared a comparable decrease in forelimb lean mass, muscle fibre size, grip strength and muscle contraction ability. Besides, the upregulation of protein degradation genes was also observed in two sarcopenia animal models. However, only primary sarcopenia mice were identified with an early stage of mtDNA deletion.
Conclusion
Collectively, this study evaluated that the dexamethasone-induced mouse model could be served as an alternative model for primary sarcopenia, according to the comparable muscle mass and functional changes. However, whether dexamethasone-induced mice can be used as an animal model when studying the molecular mechanisms of sarcopenia needs to be carefully evaluated.
The translational potential of this article
The purpose of sarcopenia research is to investigate appropriate treatments for reversing the loss of skeletal muscle mass and functions. Using animal models for the preclinical study could predict the safety and efficacy of the treatments. This study compared the typical age-related sarcopenia mice model and dexamethasone-induced secondary sarcopenia mice to provide evidence of the pathological and functional changes in the mice models.
Keywords: Sarcopenia, Dexamethasone, Animal model, Skeletal muscle function, Skeletal muscle mass
1. Background
“Sarcopenia” was first used to describe the loss of skeletal muscle mass by Irwin Rosenberg in 1989 [1]. According to the five main organisations of sarcopenia research, the European Working Group of Sarcopenia in Older People (EWGSOP), the Sarcopenia Definition and Outcomes Consortium (SDOC), the Asian Working Group for Sarcopenia (AWGS), the International Working Group on Sarcopenia (IWGS), and the Foundation for the National Institutes of Health (FNIH) Sarcopenia Project, clinical sarcopenia diagnosis included low muscle strength, low muscle mass and decreased in physical performance [[2], [3], [4]]. With the global growing elderly population, the prevalence of sarcopenia increased. The statistics for the majority of sarcopenia in different countries suggest a high percentage of older people diagnosed with sarcopenia nowadays [5].
It is generally believed that the imbalance of protein degradation and protein synthesis level results in the loss of skeletal muscle mass. One of the most widely studied signalling pathways to control protein degradation and synthesis is the PI3K-Akt signalling pathway, which could be activated by various factors with the aim to maintain muscle mass. The inhibition of the PI3K-Akt signalling pathway activates the forkhead box transcription factors (FOXO), which are the key factors for the activation of two muscle-specific E3 ubiquitin ligases, namely muscle specific ring finger protein 1 (MuRF-1) and muscle atrophy F-box (Atrogin-1), resulting in protein degradation and loss of muscle mass [6,7]. Previous studies have indicated that these atrogenes were highly expressed and playing important roles in ageing related sarcopenia and various types of muscle atrophy associated with corticoid administration and diabetes [[8], [9], [10], [11]]. Meanwhile, inhibiting the PI3K-Akt signalling pathway downregulates rapamycin (mTOR) expression and leads to the inhibition of the protein synthesis process [6,12]. Besides the imbalance of protein degradation and synthesis, other factors such as satellite cell dysfunction, cellular senescence, chronic inflammation and mitochondrial dysfunction have been reported to impair the maintenance of muscle mass [13]. It is evidenced that Wnt-3a, TGF-b and FGF could affect cellular senescence process and induce senescent cells for expressing senescence-associated secretory phenotypes (SASPs), which affect the nearby healthy satellite cells and skeletal muscle cells. The senescent cells with upregulated mitochondrial oxidation were found to exhibit increased oxidative stress and mitochondrial dysfunction [14]. Chronic-inflammation is well known as an age-associated phenotypic change [15], and there are more evidence suggested the corelation between chronic-inflammation and sarcopenia. Tumour necrosis factor α (TNF-α) and interleukin-6 (IL-6) are the most studied inflammatory cytokines which were increased in age-related sarcopenia and further affect muscle development [16]. Recently, there are more cytokines were identified as sarcopenia biomarkers, including GDF-15 and FGF-21. GDF-15 was recently listed as a sensitive biomarker for age-related sarcopenia and found to be independently and negatively associated with muscle functions [17,18].
To understand the underlying pathomechanism and development of effective treatments, various animal models of sarcopenia have been established [19]. Sarcopenia research investigates the changes in skeletal muscle qualities, including muscle mass and functions. Clinical relevant assessments have been employed in animal research, including body composition can be measured by Dual-energy X-ray absorptiometry (DXA) scanning machine [20], grip strength test for forelimb muscles strength in rodents [21], treadmill running test for measuring the physical performance such as the endurance of rodents [22]. In addition, animal models allow for more in-depth study of the changes in muscle mass. Specific skeletal muscles can be isolated for calculating muscle wet weight, which directly provides the changes in skeletal muscles tissues weight. Muscle tissue histology results can evaluate skeletal muscle fibre size distribution. Improving muscle functions is the main purpose of current treatment for sarcopenia. Skeletal muscle contraction is used to maintain body movements, an ex-vivo muscle functional test system can be used for analyzing the contraction of skeletal muscle after electronic stimulation in animal models [23,24].
In general, currently used animal models can be divided into age-related primary sarcopenia and secondary sarcopenia induced by other factors [25]. Glucocorticoid is one of the common factors that can cause muscle wasting and have been observed in human patients that long-term or high dosage of glucocorticoid treatment will result in the loss of skeletal muscle qualities [26]. Besides, glucocorticoid-induced mice were used as an in vivo animal model in numerous studies related to the loss of skeletal muscles [[27], [28], [29]]. The main purpose of current research for treating sarcopenia is to improve muscle functions and physical performance. Although different research teams have investigated the different molecular changes in naturally aged and glucocorticoid-induced mice [30,31], exploring the similarities in muscle function between two animal models is still valued. The purpose of this study is to compare natural ageing and dexamethasone induced muscle wasting models at functional, histological and molecular levels, and to evaluate if the dexamethasone-induced model could be served as alternative model for primary sarcopenia.
2. Methods
2.1. Animals
C57BL/6 female mice were used for this study. 3-Month-old mice were used as the young healthy control group (CON), 21-month-old mice were used as age-related sarcopenia mice (AGED) and 3-month-old mice received daily intraperitoneal injection of dexamethasone (20 mg/kg body weight, Sigma #D4902) for 10 days were used as secondary sarcopenia model (DEX) [32]. Animals were housed in 12 h light/12 h dark cycles at 22–28°C with standard chow diet and water. All experiment animals used in this study were under the license approved by the Animal Experimentation Ethics Committee (18/256/MIS-5-B).
2.2. Body composition
Whole-body, hindlimb and forelimb lean mass, fat mass and bone mineral density (BMD) were measured by small animal DXA (UltraFocusDXA, Faxitron Bioptics, Tucson, Arizona, USA). Dedicated Bioptics Vision software was used for further data analysis.
2.3. qPCR
Freshly isolated gastrocnemius muscles were frozen in liquid nitrogen immediately and stored in liquid nitrogen before RNA extraction. TRIzol (Invitrogen #15596018) was used for total RNA purification and High-Capacity cDNA Reverse Transcription Kit (Applied BiosystemsTM #4368813) was used to obtain cDNA. Gene expression quantification was performed by using the QuantStudioTM 7 Flex Real-Time PCR System (Applied BiosystemsTM). Myostatin (Mstn), atrogin-1and muscle RING-finger protein-1 (MuRF-1) were used for protein degradation measurement. The eukaryotic translation initiation factor 4 gamma 1 (e-IF4G-1) and the p70 ribosomal S6 kinase (p70s6k) were used for protein synthesis measurement. All gene expression results were normalised using the housekeeping gene Gapdh.
2.4. Muscle function tests
For forelimb grip strength measurement, mice were placed on the grip strength meter with its tail gently pulled back. Each mouse was allowed to perform the test five times and 5 min of rest was given between each test. To avoid inaccuracy, the highest and lowest results were excluded. The remainder values were recorded and normalised to the whole-body weight. A rodent treadmill (Ugo basile #47303) was used for the running fatigue test. The training protocol and running to fatigue test were performed as previously described [33]. In brief, a 3-day-training protocol was given to the mice before starting the fatigue test. These mice were then placed on the running channel at a speed of 12 m/min to analyse the running to fatigue distance. By automatically recording the running distance and time after the mice remained in the fatigue zone for 5 s. The running channel was stopped after the mice remained in the fatigue zone for five times. For the ex vivo muscle functional test, mice were anesthetised and the hindlimb gastrocnemius (GA) muscles were isolated carefully and placed in the Krebs buffer with 95% O2 and 5% CO2. Achilles's tendon, as the terminal of GA muscle, was tied to mount on the Dynamic Muscle Control system (DMC v5.4; Aurora Scientific, Inc.). A single 150 Hz stimulus was given to GA muscle three times and recorded the responses for twitch force calculation. A continuous 150 Hz stimulus was given three times and recorded the responses for tetanic force calculation. Both twitch and tetanic force were normalised to GA muscle cross-sectional area (CSA). Max rate of contraction and relaxation, half relaxation time and time to max force were recorded during the whole process. After 5 min of resting, GA muscles were followed with repeated stimulus every 5 s for a total of 300 s and the contraction response was recorded for fatigability measurement. 5 min and 10 min after the repeated stimulus, a 150 Hz stimulus was given to GA muscle and the tetanic forces were recorded for recovery rate calculation. The results were analysed using Dynamic Muscle Analysis system (DMA v3.2; Aurora Scientific, Inc.).
2.5. Immunostaining
Freshly isolated gastrocnemius muscles were placed in pre-cool-isopentane then stored in −80 °C for further processing. Frozen muscles were embedded in OCT embedding medium and sectioned at 10 for further staining. For muscle fibre type staining (Table 1), samples were fixed in pre-cool methanol, blocked with 2% horse serum for 1 h at room temperature, followed by primary antibodies cocktail incubation overnight at 4 °C. Secondary antibodies were incubated at room temperature for 1 h. Hematoxylin and eosin (H&E) staining was used to obtain the cross-sectional morphology of muscle fibre. All images were acquired by IX83 Inverted Fluorescence Microscope (Olympus corporation, Tokyo, Japan). ImageJ software was used for image data analysis.
Table 1.
Antibody list.
| Antibody | Brand | Cat.# | Dilution |
|---|---|---|---|
| Myosin Heavy Chain Type IIA | DSHB | #SC-71 | 1:100 |
| Myosin Heavy Chain Type I | DSHB | # BA-F8 | 1:50 |
| Myosin Heavy Chain Type IIB | DSHB | #BF–F3 | 1:100 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | #A21121 | 1:500 |
| Goat anti-Mouse IgG2b Cross-Adsorbed Secondary Antibody, Alexa Fluor 350 | Invitrogen | #A21140 | 1:300 |
| Goat anti-Mouse IgM (Heavy chain) Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen | # A-21426 | 1:500 |
2.6. Genomic DNA extraction
Gastrocnemius muscles were placed in NTES buffer with proteinase K (20 mg/mL) at 55 °C overnight for tissue lysis. Phenol was added to the liaised tissues and centrifuge at 12,000 rpm for 10 min to extract genomic DNA extraction. Supernatants were moved to new tubes with chloroform and centrifuge at 12,000 rpm for 10 min. Supernatants were transferred to new tubes with 100% ethanol, gently mixed the samples and stored at room temperature for 3 min. DNA became visible as white cloudy precipitate, then centrifuge the samples at 7500 rpm for 3 min. Remove the supernatants and resuspend the tissue pallet with 75% ethanol. Centrifuge the samples at 7500 rpm for 3 min. Remove the ethanol and resuspend the tissue pallet in water. The isolated genomic DNA was then used to detect mtDNA long PCR (13.6 kb) and D-17 deletion [34].
2.7. Statistical analysis
The sample size for each experiment was estimated by G-power software (Version 3.1.9.4.). All statistical analyses were performed using SPSS V22.0 software, and statistical significance was determined using one-way ANOVA followed by Tukey's Post Hoc test (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005, #P < 0.001). At least three independent experiments were performed.
3. Results
3.1. Changes of body composition
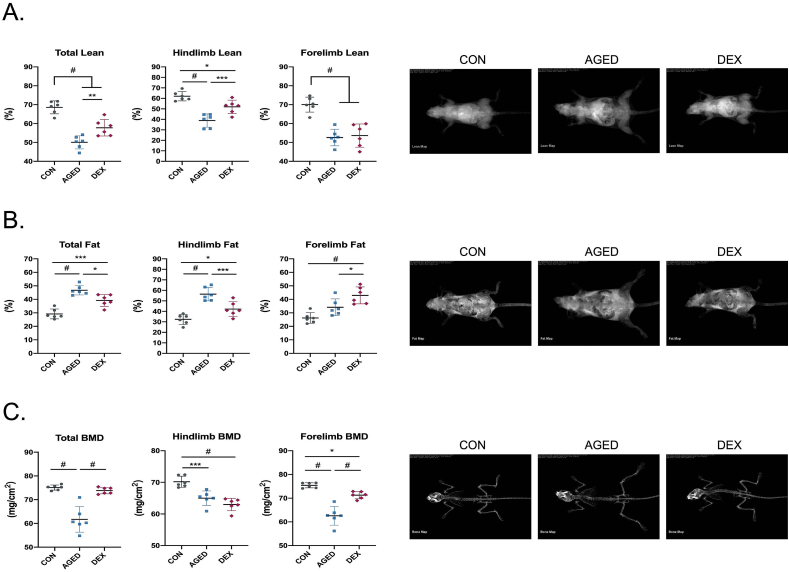
The loss of skeletal muscle mass is commonly observed with ageing and is a parameter for sarcopenia diagnosis. In this study, body composition was measured by animal DXA (Fig. 1A-C). DXA results show a dramatic decrease in whole-body lean mass, hindlimb lean mass and forelimb lean mass by 27%, 37% and 24% respectively in aged mice comparing with healthy young mice (Fig. 1A). In dexamethasone-treated mice, a significant decrease in lean mass (15.7% for whole body, 16% for hindlimb and 24% for forelimb lean mass) was observed compared with healthy young mice. The decrease of forelimb lean mass was comparable in these two types of sarcopenia models. The presence of skeletal muscle loss and increased fat tissue or loss of bone mass is relevant to clinical phenomena of age-related sarcopenic obesity and osteosarcopenia. As expected, the percentage of fat tissue raised significantly in both aged mice and DEX-induced mice (Fig. 1B). Of note, a higher increase level of whole-body fat tissue and hindlimb fat tissue was shown in aged mice, while a higher level of forelimb fat tissue was found in dexamethasone-treated mice. Bone mineral density (BMD) significantly decreased in aged mice, including whole-body, hindlimb and forelimb (Fig. 1C). In clinical, femur BMD is more relevant to the diagnosis of bone loss. The results show that the levels of hindlimb BMD in two animal models are similar.
Fig 1.
The assessment of body composition in sarcopenia animal models.
(A-C) Whole body, hindlimb and forelimb composition including lean mass, fat mass and bone mineral density (BMD) measured by DXA machine (A) Lean mass in whole body, hindlimb and forelimb (B) Fat mass in whole body, hindlimb and forelimb (C) BMD in whole body, hindlimb and forelimb. n = 6 per group. Quantitative data are presented as mean ± SD. Statistical analysis are performed using one-way ANOVA test, with significance set at P < 0.05 (∗:P < 0.05, ∗∗:P < 0.01, ∗∗∗:P < 0.005, #:P < 0.001).
3.2. The decline in skeletal muscle mass
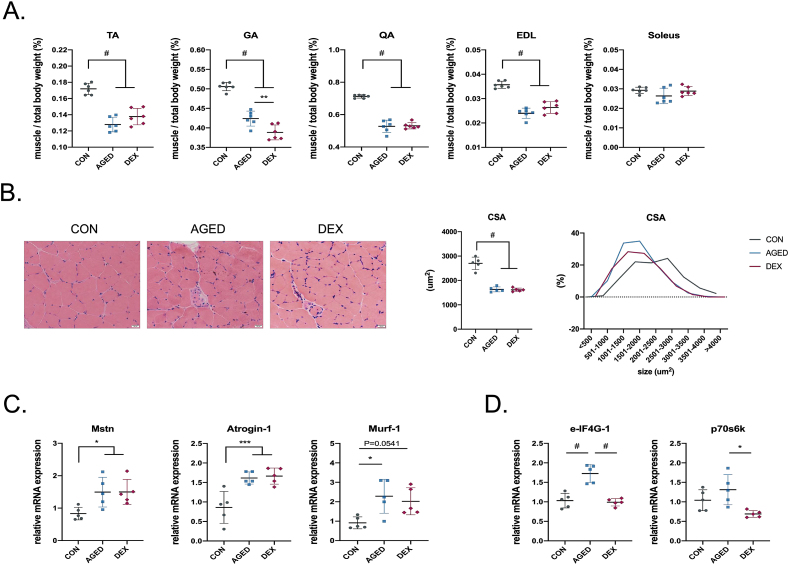
During the ageing process, a severe decline in lower extremity muscle was observed in human patients [35,36]. We isolated the hindlimb muscles for measuring the wet muscle weight. The results ascertain that in age-related and DEX-induced sarcopenia animal models, there are 25.6% and 19.9% decreased in tibialis anterior (TA), 16.2% and 23.2% decreased in gastrocnemius (GA), 25.9% and 25.4% decreased in quadriceps (QA), 32.8% and 26.2% decreased in extensor digitorum longus (EDL) muscle (Fig. 2A). A similarly reduced level of TA, QA and EDL muscle wet weight in both models was observed. No significant changes were found in soleus muscle, probably due to the subtle effect on slow-twitch muscle fibre with ageing [37]. Next, we used H&E staining with GA muscle to show the changes in muscle fibre size. About 40% reduction in GA muscle fibre size was found in both models compared with healthy young mice (Fig. 2B). Evidence of change toward smaller fibre size can be observed in both mouse models, which is relevant to clinical observation.
Fig 2.
The effects on muscle mass and gene expression in sarcopenia animal models.
(A) Quantification of tibialis anterior (TA), gastrocnemius (GA), quadriceps femoris muscle (QA) extensor digitorum longus (EDL) and soleus muscle wet weight. n = 6 per group (B) Quantification of muscle cross-sectional area in H&E stained GA muscle (C-D) Analysis of protein degradation-related genes (Mstn, Atrogin-1 and Murf-1) and protein synthesis-related genes (e-IF4g-1 and p70s6k) expression. n = 5 per group. Quantitative data are presented as mean ± SD. Statistical analysis are performed using one-way ANOVA test, with significance set at P < 0.05 (∗:P < 0.05, ∗∗:P < 0.01, ∗∗∗:P < 0.005, #:P < 0.001).
3.3. Expression of protein degradation and protein synthesis-related genes
The balance of protein degradation and protein synthesis is the key for maintaining skeletal muscle mass [38]. The qPCR results suggest that protein degradation-related genes were upregulated in both aged mice and DEX-induced mice (Fig. 2C). Myostatin and atrogin-1 were increased at the same level in two sarcopenia models (∼1.9 and ∼2.3 times higher than the healthy control group). Surprisingly, we found the up-regulation of protein synthesis-related genes in aged mice (increased ∼1.3 for p70s6k and ∼1.7 for e-IF4G-1, compared with the healthy control group). In contrast, no changes were found in dexamethasone-induced mice compared to healthy young mice (Fig. 2D). The results indicated the dramatic decrease of skeletal muscle mass in two sarcopenia animal models is comparable, which also comes with the upregulation of protein degradation genes.
3.4. Decreased of muscle functions and physical performance
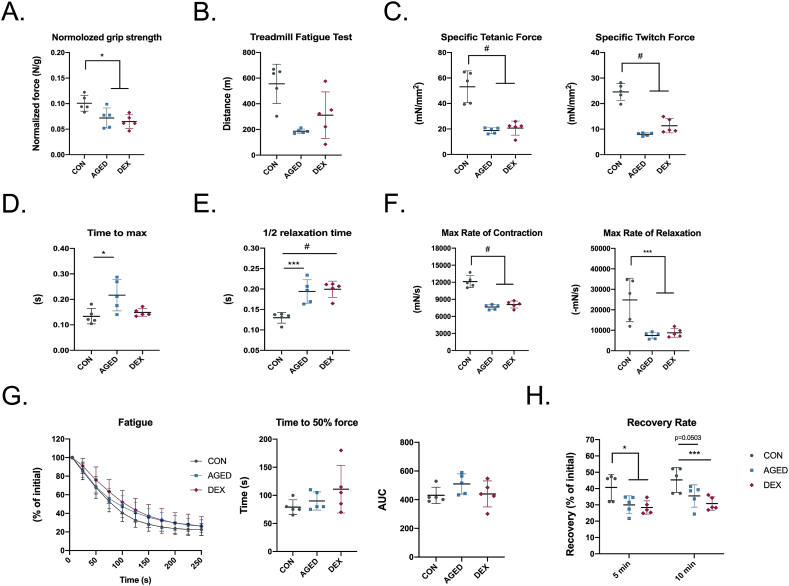
Muscle function is the most important indicator for the diagnosis of sarcopenia. In this study, we first test the forelimb grip strength, which is significantly lower in two sarcopenia models (Fig. 3A). The treadmill running fatigue test shows a dramatic decrease in running distance for the aged mice; meanwhile, a reduction in the dexamethasone-induced mice was observed (Fig. 3B). To test the skeletal muscle contractile properties, freshly isolated gastrocnemius muscles were used for the ex-vivo functional test (Fig. 3C-H). Direct stimulation of gastrocnemius muscles elicited ∼60% lower specific tetanic contractions and twitched contractions in two sarcopenia models, suggesting that the gastrocnemius muscles from two sarcopenia models were weaker compared to those from healthy mice (Fig. 3C). In aged mice, the time to reach maximum twitch force was 62% faster compared with healthy young mice and 45% faster than dexamethasone-induced mice; meanwhile, no significant difference was found between dexamethasone-induced mice and healthy young mice (Fig. 3D). Two sarcopenia models took the equivalent time to relax, which was 50% longer than healthy young mice (Fig. 3E). In aged mice, there is a 36% decrease in maximal rate of contraction and a 70% decrease in maximal rate of relaxation (Fig. 3F). A 33% decrease and 65% decrease in maximal rate of contraction and relaxation were found in dexamethasone-induced mice, which is comparable with the aged mice (Fig. 3F). No difference was found after receiving continued stimulation in three groups (Fig. 3G). The time taken to decrease 50% force and the area under curve analysis also show no difference in the three groups. The recovery ability of aged mice and dexamethasone-induced mice after 5- and 10- minutes of rest were lower than the healthy young control mice (Fig. 3H). Taken together, two sarcopenia models have compatible muscle function levels, which are both significantly worse than the healthy mice.
Fig 3.
The reduction of muscle functions in sarcopenia animal models.
(A) Hindlimb grip strength was measured and normalized by whole body weight (B) Treadmill fatigue test was measured and running distance was recorded (C–H) Ex-vivo GA muscle contraction test (C) Peak tetanic fore and twitch fore normalised by GA muscle cross-sectional area (D) Time to reach maximum twitch force (E) Half-relaxation time (F) Max rate of contraction and relaxation (G) GA muscle fatigability normalised by prefatigued developed tension (H) The recovery rate was measured by the contraction force of GA muscle after 5 min and 10 min rest from fatigue test. n = 5 per group Quantitative data are presented as mean ± SD. Statistical analyses are performed using one-way ANOVA test, with significance set at P < 0.05 (∗:P < 0.05, ∗∗:P < 0.01, ∗∗∗:P < 0.005, #:P < 0.001).
3.5. Changes in skeletal muscle fibre types and mitochondrial DNA damage
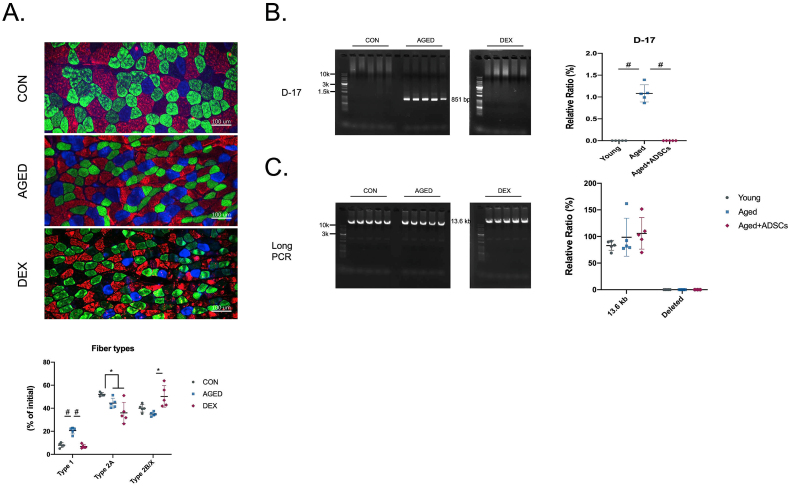
Muscle fibre type was believed to have the ability in controlling muscle functions; thus, we measured the fibre type distribution in three groups of animals—the results showed a completely different pattern between the two sarcopenia models (Fig. 4A). Previous studies have reported that type II muscle fibre will be affected during ageing, which is similar to our finding. However, there was no difference in type I and IIB/X muscle fibre percentages in DEX-induced mice compared with healthy young control mice. A similar change in two sarcopenia models' fibre type compared with the healthy mice is the decrease of type IIA muscle fibre. Compared to the healthy young mice, only the number of type I and type IIA muscle fibres was significantly changed in aged or DEX-induced mice. Type I and type IIA muscle fibres have higher oxidative capacitive; therefore, we next check the mitochondrial quality, which is strongly related to the oxidative property. PCR result shows that only samples from aged mice showed the positive D-17 band at 851 bp, indicating that the presence of mtDNA deletion in aged mice only but not in the young mice nor Dex-induced young mice (Fig. 4B). Long-range PCR was used to detect the damaged mtDNA [39]. Our results showed complete mtDNA at 13.6 kb for three samples, and no deleted band was observed (Fig. 4C). Thus, only the aged mice were identified as early stage of mtDNA deletion.
Fig 4.
The effect on skeletal muscle fibre type switching and mitochondrial damages in sarcopenia animal models.
(A) Immunofluorescence staining and quantification for fibre types in GA muscle sections. Blue: type I; green: type IIA; red: type IIB; black: type IIX staining (B–C) Mitochondrial DNA damage detection. n = 5 per group Quantitative data are presented as mean ± SD. Statistical analyses are performed using one-way ANOVA test, with significance set at P < 0.05 (∗:P < 0.05, ∗∗:P < 0.01, ∗∗∗:P < 0.005, #:P < 0.001).
4. Discussion
The high prevalence of sarcopenia, particularly in ageing society comes with an increasing burden on the healthcare system and socioeconomic. Consequently, there are growing numbers of sarcopenia research using animal models to explore novel therapeutic approaches. For in vivo study, naturally aged mice are the most widely used animal model due to their relevance to primary sarcopenia, but not commonly used in research because it is costly and time-consuming. Therefore, the development of alternative models with phenotype and functional changes similar to naturally aged mice is of research interest. Previous studies have suggested various animal models for sarcopenia research, including naturally aged, diet-induced, gene knockout/overexpression, mtDNA mutations, drug-induced and hindlimb suspension mice [40]; however, each of them have their limitations. Gene-edited and mtDNA mutation mice are costly, and their progressions were not observed in human ageing progression. Although hindlimb suspension can mimic the immobilisation of patients, long-time suspension caused trauma to the tail. Therefore, more user-friendly and less invasive approaches are sought after [41]. With its anti-inflammatory property, dexamethasone (DEX), one common type of glucocorticoids, is widely used for treating various diseases [[42], [43], [44], [45]], especially patients with congenital adrenal hyperplasia and Addison's disease may require lifelong DEX treatment [[46], [47], [48]]. However, long-term or high dosage of DEX treatment often results in multiple side effects, including muscle atrophy [49]. With these, DEX-induced mice have been suggested as a sarcopenia-like model [50,51]. However, there is yet poorly understood how similar these two models are in the context of muscle properties and if this drug-induced muscle atrophy model could serve as alternative sarcopenia for in vivo drug screening. Our findings clearly show that animal models of age-related sarcopenia and DEX-induced muscle atrophy share comparable phenotype changes, especially in terms of body composition and physical performance, but mRNA levels of genes related to protein degradation and synthesis are not identical.
Clinical diagnosis for sarcopenia relies on the measurement of skeletal muscle function and mass [52]. For skeletal muscle mass measurement, the clinical assessment includes dual-energy X-ray absorptiometry (DXA), computed tomography (CT), ultrasound and magnetic resonance imaging (MRI) [53]. For the clinical muscle functional test, handgrip test, gait speed, 6-m walking and 5-time chair stand are widely used diagnostic tests. In this study, DXA scanning, forelimb grip strength test and a running to exhaustion test were performed to mimic the clinical assessments in order to evaluate the muscle qualities in the two tested animal models. The changes in muscle mass and strength in our naturally aged mice are in agreement with previous animal studies. Compared with other naturally aged animal studies on muscle functions, ∼35% decrease in grip strength and ∼65% decrease in treadmill running distance were found similar in our study and others [[54], [55], [56], [57], [58], [59], [60]]. These in vivo results are also relevant to the clinical findings. According to the international sarcopenia research groups, the criteria for low handgrip strength is lower than 20 kg [61], which is around 40% lower than the healthy young adult. For the muscle mass diagnosis, clinical studies suggest that 2 standard deviations (SD) below the mean of young healthy adults will be recognized as low muscle mass [62]. Our naturally aged model results in an over 6 SD lower muscle mass compared with the healthy young mice. Clinical studies also indicate that sarcopenia is often associated with decreased bone qualities, also known as osteosarcopenia [63,64]. The advantage of animal models is the available of additional information in the context of muscle quality assessment. Hindlimb skeletal muscles were isolated for measuring muscle fibre size and muscle contractile force. According to previous studies, around 20–30% reduction of skeletal muscle CSA was found in aged mice, which was also found in our naturally aged animal model [65,66]. Moreover, ex vivo muscle functional test is allowable to analyse the response of skeletal muscle to electronic stimulation directly. There is limited report on the muscle contraction changes in sarcopenia animal models, therefore, our study provides additional evidence on how aged skeletal muscle responds to the stimuli. In human diagnosis, femur BMD is more relevant to the diagnosis of bone loss [59]. We also found lower BMD in the hindlimb of our naturally aged animal models, which is consistent with the typical clinical phenomenon of sarcopenia and the in vivo studies on age-related sarcopenia animal models.
Our study provides evidence showing similar changes in terms of histological and functional aspects, however, according to recent study, the molecular profiles between these two models have significant discrepancies. RNA sequencing and proteomic analysis with quantitative mass spectrometry suggest the likelihood of different underlying molecular mechanisms [30]. Only 34 overlapping genes out of 263 genes were found between the skeletal muscle tissues of age-induced and DEX-induced mice. Besides, the proteomic analysis also indicated a largely divergent between the two animal models. There were only 18 proteins were found could be induced by both age-induced and DEX-induced mice. The downregulation of myoblast differentiation-related protein expression was specifically shown in age-induced mice, which is correlated to the muscle cell ageing process [61]. This result indicates that cell ageing is commonly observed in aged skeletal muscle tissue, however, dexamethasone did not affect the skeletal muscle cell ageing process. Besides, the muscle atrophy-related gene, Atrogin1 and MuRF-1, expression was only shown to increase in response to dexamethasone and their protein levels were not significantly changed, suggesting these ubiquitin ligases may not be good biological markers for age-related muscle atrophy. On the contrary, up-regulation of these muscle atrophy-related genes was observed in our naturally aged and DEX-induced mice showed, which is consistent with other studies [62,63]. The discrepancies might be due to the muscle chosen for analysis. Gastrocnemius muscle was used in our study and other studies, which shows the up-regulation of muscle atrophy-related gene expression, which is different from tibialis anterior muscle used in Hunt's study [30]. The composition dissimilarity between the two types of muscle could result in different profiles in response to ageing.
Even though most molecular changes remain different, some similarities were still found. The similar gene changes of two sarcopenia animal models mostly related to inflammatory cells, including S100a9, IL1b, and Serpina3n, which correlated to predict muscle atrophy, showed upregulation in two sarcopenia animal models [30]. S100a9 was reported to sustain chronic inflammation during ageing [65] and the gene level could also be increased during glucocorticoid induction [66]. Besides, the RNA sequencing results found a similar trend on mitochondrial-associated genes in two animal models. The mitochondrial damage has been known as one of the ageing phenotypic changes in human and can be measured by multiple assays. In this study, we also provide mtDNA deletion data by detecting the D-17 and long-rang PCR. The mtDNA has known as a close circle DNA, and there are numerous pairs of exact or inexact repeats in the mtDNA genome. D-17 is one of the DNA repeats used for detecting mtDNA deletion and is an inducer of mtDNA deletions [34,[67], [68], [69]]. Long-range PCR, also known as long PCR, is commonly used for amplifying longer DNA fragments. For the complete mtDNA, there will only be a clear band located at 16.6 kb. Some bands will be found at a smaller size if there is any deletion on the mtDNA [34,70]. Our results confirmed that the mtDNA deletion inducer, D-17, expression was observed in aged mice. However, the long-range PCR did not show deletion as observed in D-17 Meanwhile, we have noticed that some previous study had found the individual difference in mouse samples, which indicated that long-range PCR deletion would not serve in all aged mice [71]. Also, a similar result, showing the deletion in D-17 but not long-range PCR, was found in ageing mice [72]. These articles provided evidence that there are multiple stages of mtDNA deletion, and two of our animal models are identified as different stages.
Age-related sarcopenia patients are commonly accompanied by complicated age-related diseases, which is challenging to mimic in vivo animal model. Alternative animal models induced by chemicals to produce consistent sarcopenia-like phenotypes provide beneficial options to improve experimental reproducibility. Besides, the purpose of most studying sarcopenia therapeutic approach is to improve muscle functions and physical performance. Thus, this study provides evidence for using DEX-induced mice, an easy-induced and highly reproducible animal model for future studies to improve the loss of muscle functions and muscle mass. To conclude, this study directly compared the skeletal muscle pathological and functional changes
In two sarcopenia animal models. The results suggest that even two animal models might share different molecular mechanisms, despite comparable changes in skeletal muscle mass and functions. Cautions should be taken when using DEX-induced animal model for sarcopenia mechanistic study.
Declaration of competing interest
Dr Belle Yu-Hsuan Wang, Mr Allen Wei-Ting, Dr Nicodemus Wong, Dr Yi-Fan Chen, Dr Chien-Wei Lee and Dr Wayne Yuk Wai Lee declare that they have no conflict of interest.
Acknowledgements
This work was supported by Start-up grant from Chinese University of Hong Kong (Ref. Nos. 4930991 and 4930992); General Research Fund (Ref. No., 14104620); Research Matching Grant Scheme; Area of Excellence (Ref. No. AoE/M-402/20), Research Grants Council, University Grants Committee; Center for Neuromusculoskeletal Restorative Medicine (Ref. No. CT1.1), Health@InnoHK program, Innovation Technology Commission, Hong Kong SAR, China; Health and Medical Research Fund, The Food and Health Bureau, HKSAR (Ref No. 06170546); 2020 Rising Star Award provided by American Society for Bone and Mineral Research.
Contributor Information
Chien-Wei Lee, Email: icehikki@gmail.com.
Wayne Yuk Wai Lee, Email: waynelee@ort.cuhk.edu.hk.
References
- 1.Rosenburg I. Summary comments: epidemiological and methodological problems in determining nutritional status of older persons. Am J Clin Nutr. 1989;50(5):1231–1233. [PubMed] [Google Scholar]
- 2.Cruz-Jentoft A.J., Baeyens J.P., Bauer J.M., Boirie Y., Cederholm T., Landi F., et al. Sarcopenia: European consensus on definition and diagnosis: report of the European working group on sarcopenia in older people. Age Ageing. 2010;39(4):412–423. doi: 10.1093/ageing/afq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cawthon P.M., Manini T., Patel S.M., Newman A., Travison T., Kiel D.P., et al. Putative cut-points in sarcopenia components and incident adverse health outcomes: an SDOC analysis. J Am Geriatr Soc. 2020;68(7):1429–1437. doi: 10.1111/jgs.16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen L.-K., Woo J., Assantachai P., Auyeung T.-W., Chou M.-Y., Iijima K., et al. Asian Working Group for Sarcopenia: 2019 consensus update on sarcopenia diagnosis and treatment. J Am Med Dir Assoc. 2020;21(3):300. doi: 10.1016/j.jamda.2019.12.012. 07. e2. [DOI] [PubMed] [Google Scholar]
- 5.Diz J.B.M., Queiroz BZd, Tavares L.B., Pereira L.S.M. Prevalence of sarcopenia among the elderly: findings from broad cross-sectional studies in a range of countries. Revista Brasileira de Geriatria e Gerontologia. 2015;18:665–678. [Google Scholar]
- 6.Lee J.H., Jeon J.H., Lee M.J. Docosahexaenoic acid, a potential treatment for sarcopenia, modulates the ubiquitin–proteasome and the autophagy–lysosome systems. Nutrients. 2020;12(9):2597. doi: 10.3390/nu12092597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rom O., Reznick A.Z. The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radic Biol Med. 2016;98:218–230. doi: 10.1016/j.freeradbiomed.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Fernando R., Drescher C., Nowotny K., Grune T., Castro J.P. Impaired proteostasis during skeletal muscle aging. Free Radic Biol Med. 2019;132:58–66. doi: 10.1016/j.freeradbiomed.2018.08.037. [DOI] [PubMed] [Google Scholar]
- 9.Larsson L., Degens H., Li M., Salviati L., Lee Y.I., Thompson W., et al. Sarcopenia: aging-related loss of muscle mass and function. Physiol Rev. 2019;99(1):427–511. doi: 10.1152/physrev.00061.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schakman O., Gilson H., Thissen J.-P. Mechanisms of glucocorticoid-induced myopathy. J Endocrinol. 2008;197(1):1–10. doi: 10.1677/JOE-07-0606. [DOI] [PubMed] [Google Scholar]
- 11.Perry B.D., Caldow M.K., Brennan-Speranza T.C., Sbaraglia M., Jerums G., Garnham A., et al. Muscle atrophy in patients with Type 2 Diabetes Mellitus: roles of inflammatory pathways, physical activity and exercise. Exerc Immunol Rev. 2016;22:94. [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon M.-S. mTOR as a key regulator in maintaining skeletal muscle mass. Front Physiol. 2017;8:788. doi: 10.3389/fphys.2017.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mankhong S., Kim S., Moon S., Kwak H.-B., Park D.-H., Kang J.-H. Experimental models of sarcopenia: bridging molecular mechanism and therapeutic strategy. Cells. 2020;9(6):1385. doi: 10.3390/cells9061385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Passos J.F., Nelson G., Wang C., Richter T., Simillion C., Proctor C.J., et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6(1):347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrucci L., Fabbri E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat Rev Cardiol. 2018;15(9):505–522. doi: 10.1038/s41569-018-0064-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bian A.-L., Hu H.-Y., Rong Y.-D., Wang J., Wang J.-X., Zhou X.-Z. A study on relationship between elderly sarcopenia and inflammatory factors IL-6 and TNF-α. Eur J Med Res. 2017;22(1):1–8. doi: 10.1186/s40001-017-0266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H., Kim K.M., Kang M.J., Lim S. Growth differentiation factor-15 as a biomarker for sarcopenia in aging humans and mice. Exp Gerontol. 2020;142 doi: 10.1016/j.exger.2020.111115. [DOI] [PubMed] [Google Scholar]
- 18.Alcazar J., Frandsen U., Prokhorova T., Kamper R.S., Haddock B., Aagaard P., et al. Changes in systemic GDF15 across the adult lifespan and their impact on maximal muscle power: the Copenhagen Sarcopenia Study. Journal of Cachexia, Sarcopenia and Muscle. 2021;12(6):1418–1427. doi: 10.1002/jcsm.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christian C.J., Benian G.M. Animal models of sarcopenia. Aging Cell. 2020;19(10) doi: 10.1111/acel.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shepherd J.A., Ng B.K., Sommer M.J., Heymsfield S.B. Body composition by DXA. Bone. 2017;104:101–105. doi: 10.1016/j.bone.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonetto A, Andersson DC, Waning DL. Assessment of muscle mass and strength in mice. BoneKEy reports. 2015;4 doi: 10.1038/bonekey.2015.101. 732–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graber T.G., Maroto R., Fry C.S., Brightwell C.R., Rasmussen B.B. Measuring exercise capacity and physical function in adult and older mice. J Gerontol: Series A. 2021;76(5):819–824. doi: 10.1093/gerona/glaa205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bondulich M.K., Jolinon N., Osborne G.F., Smith E.J., Rattray I., Neueder A., et al. Myostatin inhibition prevents skeletal muscle pathophysiology in Huntington's disease mice. Sci Rep. 2017;7(1):1–14. doi: 10.1038/s41598-017-14290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biltz N.K., Collins K.H., Shen K.C., Schwartz K., Harris C.A., Meyer G.A. Infiltration of intramuscular adipose tissue impairs skeletal muscle contraction. J Physiol. 2020;598(13):2669–2683. doi: 10.1113/JP279595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz-Jentoft A.J., Bahat G., Bauer J., Boirie Y., Bruyère O., Cederholm T., et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing. 2019;48(1):16–31. doi: 10.1093/ageing/afy169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oray M., Abu Samra K., Ebrahimiadib N., Meese H., Foster C.S. Long-term side effects of glucocorticoids. Expet Opin Drug Saf. 2016;15(4):457–465. doi: 10.1517/14740338.2016.1140743. [DOI] [PubMed] [Google Scholar]
- 27.Ma K., Mallidis C., Bhasin S., Mahabadi V., Artaza J., Gonzalez-Cadavid N., et al. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am J Physiol Endocrinol Metabol. 2003;285(2):E363–E371. doi: 10.1152/ajpendo.00487.2002. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser G., Gerst F., Michael D., Berchtold S., Friedrich B., Strutz-Seebohm N., et al. Regulation of forkhead box O1 (FOXO1) by protein kinase B and glucocorticoids: different mechanisms of induction of beta cell death in vitro. Diabetologia. 2013;56(7):1587–1595. doi: 10.1007/s00125-013-2863-7. [DOI] [PubMed] [Google Scholar]
- 29.Seto J.T., Roeszler K.N., Meehan L.R., Wood H.D., Tiong C., Bek L., et al. ACTN3 genotype influences skeletal muscle mass regulation and response to dexamethasone. Sci Adv. 2021;7(27) doi: 10.1126/sciadv.abg0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt L.C., Graca F.A., Pagala V., Wang Y.-D., Li Y., Yuan Z.-F., et al. Integrated genomic and proteomic analyses identify stimulus-dependent molecular changes associated with distinct modes of skeletal muscle atrophy. Cell Rep. 2021;37(6) doi: 10.1016/j.celrep.2021.109971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato A.Y., Richardson D., Cregor M., Davis H.M., Au E.D., McAndrews K., et al. Glucocorticoids induce bone and muscle atrophy by tissue-specific mechanisms upstream of E3 ubiquitin ligases. Endocrinology. 2017;158(3):664–677. doi: 10.1210/en.2016-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seo E., Truong C.-S., Jun H.-S. Psoralea corylifolia L. seed extract attenuates dexamethasone-induced muscle atrophy in mice by inhibition of oxidative stress and inflammation. J Ethnopharmacol. 2022;296 doi: 10.1016/j.jep.2022.115490. [DOI] [PubMed] [Google Scholar]
- 33.Dougherty J.P., Springer D.A., Gershengorn M.C. The treadmill fatigue test: a simple, high-throughput assay of fatigue-like behavior for the mouse. JoVE. 2016;(111) doi: 10.3791/54052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu C.-Y., Chen Y.-F., Wang C.-H., Kao C.-H., Zhuang H.-W., Chen C.-C., et al. A persistent level of Cisd2 extends healthy lifespan and delays aging in mice. Hum Mol Genet. 2012;21(18):3956–3968. doi: 10.1093/hmg/dds210. [DOI] [PubMed] [Google Scholar]
- 35.Reid K.F., Pasha E., Doros G., Clark D.J., Patten C., Phillips E.M., et al. Longitudinal decline of lower extremity muscle power in healthy and mobility-limited older adults: influence of muscle mass, strength, composition, neuromuscular activation and single fiber contractile properties. Eur J Appl Physiol. 2014;114(1):29–39. doi: 10.1007/s00421-013-2728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skelton D.A., Kennedy J., Rutherford O.M. Explosive power and asymmetry in leg muscle function in frequent fallers and non-fallers aged over 65. Age Ageing. 2002;31(2):119–125. doi: 10.1093/ageing/31.2.119. [DOI] [PubMed] [Google Scholar]
- 37.Nilwik R., Snijders T., Leenders M., Groen B.B., van Kranenburg J., Verdijk L.B., et al. The decline in skeletal muscle mass with aging is mainly attributed to a reduction in type II muscle fiber size. Exp Gerontol. 2013;48(5):492–498. doi: 10.1016/j.exger.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Sartori R., Romanello V., Sandri M. Mechanisms of muscle atrophy and hypertrophy: implications in health and disease. Nat Commun. 2021;12(1):1–12. doi: 10.1038/s41467-020-20123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao P., Gallagher P., Nedungadi S., Manczak M., Kohama S.G., Ferguson B., et al. Mitochondrial DNA deletions and differential mitochondrial DNA content in Rhesus monkeys: implications for aging. Biochim Biophys Acta, Mol Basis Dis. 2012;1822(2):111–119. doi: 10.1016/j.bbadis.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie Wq, He M., Yu Dj, Wu Yx, Wang Xh, Lv S., et al. Mouse models of sarcopenia: classification and evaluation. Journal of Cachexia, Sarcopenia and Muscle. 2021;12(3):538–554. doi: 10.1002/jcsm.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morey-Holton E.R., Globus R.K. Hindlimb unloading rodent model: technical aspects. J Appl Physiol. 2002;92(4):1367–1377. doi: 10.1152/japplphysiol.00969.2001. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y., Wong R.S., Soo Y.O., Chui C.H., Lau F.Y., Chan N.P., et al. Initial treatment of immune thrombocytopenic purpura with high-dose dexamethasone. N Engl J Med. 2003;349(9):831–836. doi: 10.1056/NEJMoa030254. [DOI] [PubMed] [Google Scholar]
- 43.Barlogie B., Smith L., Alexanian R. Effective treatment of advanced multiple myeloma refractory to alkylating agents. N Engl J Med. 1984;310(21):1353–1356. doi: 10.1056/NEJM198405243102104. [DOI] [PubMed] [Google Scholar]
- 44.Alexanian R., Dimopoulos M.A., Delasalle K., Barlogie B. Primary dexamethasone treatment of multiple myeloma. Blood. 1992;80(4):887–890. [PubMed] [Google Scholar]
- 45.Taboada M, Rodríguez N, Varela PM, Rodríguez MT, Abelleira R, González A, et al. Effect of high versus low dose of dexamethasone on clinical worsening in patients hospitalised with moderate or severe COVID-19 Pneumonia: an open-label, randomised clinical trial. European Respiratory Journal. 2022;60(2) doi: 10.1183/13993003.02518-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta P., Bhatia V. Corticosteroid physiology and principles of therapy. Indian J Pediatr. 2008;75(10):1039–1044. doi: 10.1007/s12098-008-0208-1. [DOI] [PubMed] [Google Scholar]
- 47.Rathbun KM, Nguyen M, Singhal M. StatPearls [internet] StatPearls Publishing; 2020. Addisonian crisis.https://www.ncbi.nlm.nih.gov/books/NBK441933 Available: [Google Scholar]
- 48.Camozzi V., Betterle C., Frigo A.C., Zaccariotto V., Zaninotto M., De Caneva E., et al. Vertebral fractures assessed with dual-energy X-ray absorptiometry in patients with Addison's disease on glucocorticoid and mineralocorticoid replacement therapy. Endocrine. 2018;59(2):319–329. doi: 10.1007/s12020-017-1380-8. [DOI] [PubMed] [Google Scholar]
- 49.Kim S., Kim K., Park J., Jun W. Curcuma longa L. Water extract improves dexamethasone-induced sarcopenia by modulating the muscle-related gene and oxidative stress in mice. Antioxidants. 2021;10(7):1000. doi: 10.3390/antiox10071000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shang Y., Kuang M., Wang Z., Huang Y., Liu L., Zhao X., et al. An ultrashort peptide-based supramolecular hydrogel mimicking IGF-1 to alleviate glucocorticoid-induced sarcopenia. ACS Appl Mater Interfaces. 2020;12(31):34678–34688. doi: 10.1021/acsami.0c09973. [DOI] [PubMed] [Google Scholar]
- 51.Meza-Valderrama D., Marco E., Dávalos-Yerovi V., Muns M.D., Tejero-Sánchez M., Duarte E., et al. Sarcopenia, malnutrition, and cachexia: adapting definitions and terminology of nutritional disorders in older people with cancer. Nutrients. 2021;13(3):761. doi: 10.3390/nu13030761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Messina C., Maffi G., Vitale J.A., Ulivieri F.M., Guglielmi G., Sconfienza L.M. Diagnostic imaging of osteoporosis and sarcopenia: a narrative review. Quant Imag Med Surg. 2018;8(1):86. doi: 10.21037/qims.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goodpaster B.H., Chomentowski P., Ward B.K., Rossi A., Glynn N.W., Delmonico M.J., et al. Effects of physical activity on strength and skeletal muscle fat infiltration in older adults: a randomized controlled trial. J Appl Physiol. 2008;105(5):1498–1503. doi: 10.1152/japplphysiol.90425.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palla A., Ravichandran M., Wang Y., Alexandrova L., Yang A., Kraft P., et al. Inhibition of prostaglandin-degrading enzyme 15-PGDH rejuvenates aged muscle mass and strength. Science. 2021;371(6528) doi: 10.1126/science.abc8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shang G.K., Han L., Wang Z.H., Liu Y.P., Yan S.B., Sai W.W., et al. Sarcopenia is attenuated by TRB3 knockout in aging mice via the alleviation of atrophy and fibrosis of skeletal muscles. Journal of cachexia, sarcopenia and muscle. 2020;11(4):1104–1120. doi: 10.1002/jcsm.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayasaka T., Takehara N., Aonuma T., Kano K., Horiuchi K., Nakagawa N., et al. Sarcopenia-derived exosomal micro-RNA 16-5p disturbs cardio-repair via a pro-apoptotic mechanism in myocardial infarction in mice. Sci Rep. 2021;11(1):1–14. doi: 10.1038/s41598-021-98761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Del Campo A., Contreras-Hernández I., Castro-Sepúlveda M., Campos C.A., Figueroa R., Tevy M.F., et al. Muscle function decline and mitochondria changes in middle age precede sarcopenia in mice. Aging (Albany NY) 2018;10(1):34. doi: 10.18632/aging.101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benz E., Trajanoska K., Lahousse L., Schoufour J.D., Terzikhan N., De Roos E., et al. Sarcopenia in COPD: a systematic review and meta-analysis. Eur Respir Rev. 2019;28(154) doi: 10.1183/16000617.0049-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glaser D.L., Kaplan F.S. Osteoporosis: definition and clinical presentation. Spine. 1997;22(24):12S–16S. doi: 10.1097/00007632-199712151-00003. [DOI] [PubMed] [Google Scholar]
- 60.Clynes M.A., Gregson C.L., Bruyère O., Cooper C., Dennison E.M. Osteosarcopenia: where osteoporosis and sarcopenia collide. Rheumatology. 2021;60(2):529–537. doi: 10.1093/rheumatology/keaa755. [DOI] [PubMed] [Google Scholar]
- 61.Moustogiannis A., Philippou A., Taso O., Zevolis E., Pappa M., Chatzigeorgiou A., et al. The effects of muscle cell aging on myogenesis. Int J Mol Sci. 2021;22(7):3721. doi: 10.3390/ijms22073721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu S., Tian Z., Torigoe D., Zhao J., Xie P., Sugizaki T., et al. Aging-and obesity-related peri-muscular adipose tissue accelerates muscle atrophy. PLoS One. 2019;14(8) doi: 10.1371/journal.pone.0221366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang L., Jiao X.-F., Wu C., Li X.-Q., Sun H.-X., Shen X.-Y., et al. Trimetazidine attenuates dexamethasone-induced muscle atrophy via inhibiting NLRP3/GSDMD pathway-mediated pyroptosis. Cell death discovery. 2021;7(1):1–11. doi: 10.1038/s41420-021-00648-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gueugneau M., d'Hose D., Barbé C., de Barsy M., Lause P., Maiter D., et al. Increased Serpina3n release into circulation during glucocorticoid-mediated muscle atrophy. Journal of cachexia, sarcopenia and muscle. 2018;9(5):929–946. doi: 10.1002/jcsm.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swindell W.R., Johnston A., Xing X., Little A., Robichaud P., Voorhees J.J., et al. Robust shifts in S100a9 expression with aging: a novel mechanism for chronic inflammation. Sci Rep. 2013;3(1):1–13. doi: 10.1038/srep01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu K., Passey R.J., Endoh Y., Rahimi F., Youssef P., Yen T., et al. Regulation of S100A8 by glucocorticoids. J Immunol. 2005;174(4):2318–2326. doi: 10.4049/jimmunol.174.4.2318. [DOI] [PubMed] [Google Scholar]
- 67.Tanhauser S.M., Laipis P.J. Multiple deletions are detectable in mitochondrial DNA of aging mice. J Biol Chem. 1995;270(42):24769–24775. doi: 10.1074/jbc.270.42.24769. [DOI] [PubMed] [Google Scholar]
- 68.Xu S.C., He M.D., Lu Y.H., Li L., Zhong M., Zhang Y.W., et al. Nickel exposure induces oxidative damage to mitochondrial DNA in Neuro2a cells: the neuroprotective roles of melatonin. J Pineal Res. 2011;51(4):426–433. doi: 10.1111/j.1600-079X.2011.00906.x. [DOI] [PubMed] [Google Scholar]
- 69.Kume S., Uzu T., Horiike K., Chin-Kanasaki M., Isshiki K., Araki S-i, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120(4):1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spendiff S., Reza M., Murphy J.L., Gorman G., Blakely E.L., Taylor R.W., et al. Mitochondrial DNA deletions in muscle satellite cells: implications for therapies. Hum Mol Genet. 2013;22(23):4739–4747. doi: 10.1093/hmg/ddt327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wanrooij P.H., Tran P., Thompson L.J., Carvalho G., Sharma S., Kreisel K., et al. Elimination of rNMPs from mitochondrial DNA has no effect on its stability. Proc Natl Acad Sci USA. 2020;117(25):14306–14313. doi: 10.1073/pnas.1916851117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen W.-J., Lin I., Lee C.-W., Yoshioka K., Ono Y., Yan Y.-T., et al. Ribonucleotide reductase M2B in the myofibers modulates stem cell fate in skeletal muscle. NPJ Regenerative medicine. 2022;7(1):1–12. doi: 10.1038/s41536-022-00231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]