Abstract
Recent discovery of the ribosomal protein (RP) RPL11 interacting with and inhibiting the E3 ubiquitin ligase function of MDM2 established the RP-MDM2-p53 signaling pathway, which is linked to biological events, including ribosomal biogenesis, nutrient availability, and metabolic homeostasis. Mutations in RPs lead to a diverse array of phenotypes known as ribosomopathies in which the role of p53 is implicated. Here, we generated conditional RPL11-deletion mice to investigate in vivo effects of impaired RP expression and its functional connection with p53. While deletion of one Rpl11 allele in germ cells results in embryonic lethality, deletion of one Rpl11 allele in adult mice does not affect viability but leads to acute anemia. Mechanistically, we found RPL11 haploinsufficiency activates p53 in hematopoietic tissues and impedes erythroid precursor differentiation, resulting in insufficient red blood cell development. We demonstrated that reducing p53 dosage by deleting one p53 allele rescues RPL11 haploinsufficiency-induced inhibition of erythropoietic precursor differentiation and restores normal red blood cell levels in mice. Furthermore, blocking the RP-MDM2-p53 pathway by introducing an RP-binding mutation in MDM2 prevents RPL11 haploinsufficiency–caused p53 activation and rescues the anemia in mice. Together, these findings demonstrate that the RP-MDM2-p53 pathway is a critical checkpoint for RP homeostasis and that p53-dependent cell cycle arrest of erythroid precursors is the molecular basis for the anemia phenotype commonly associated with RP deficiency.
Keywords: ribosomal protein, RPL11, MDM2, p53, anemia, ribosomopathy
Abbreviations: BM, bone marrow; BMT, BM transplantation; DBA, Diamond-Blackfan anemia; Hb, hemoglobin; MEF, mouse embryonic fibroblast; Met, methionine; RP, ribosomal protein; Rpl11+/flox, RPL11 heterozygous flox mice
The tumor suppressor p53 is the most commonly mutated gene across many types of cancers (1, 2). p53-mediated cell cycle arrest and apoptosis in response to DNA damage and other cellular stresses are thought to be vital for p53-mediated tumor suppression, a notion that led to p53 being nicknamed the “Guardian of the Genome” (3). p53 is tightly regulated by the E3 ubiquitin ligase MDM2 via direct binding and increased proteosomal degradation via MDM2-mediated p53 polyubiquitination (4). The initial observation that ribosomal protein (RP) RPL11 binds to and inhibits the E3 ligase function of MDM2 led to the discovery of the RP-MDM2-p53 pathway, which involves multiple RPs and has been shown to contribute to tumor suppression, nutrient balance, and metabolic homeostasis (5, 6). Further studies demonstrated that p53 is activated in response to imbalances in RP stoichiometry caused by oncogene activation or RNA interference (7, 8). These studies established a link between p53 and ribosomal biogenesis, a vital and energetically demanding process in all eukaryotic cells (9). Mutations in RPs lead to a class of diseases known as ribosomopathies with wide ranging phenotypes (10), in which the role of p53 has been implicated (11). However, the mechanism underlying RP mutation-dependent p53 activation and subsequent ribosomopathy is not fully understood.
RPL11 was the first RP identified to interact with and inhibit the E3 ligase function of MDM2, leading to p53 stabilization and activation (12, 13). Of the multiple MDM2-binding RPs, RPL11 appears to be particularly important for p53 regulation. RPL11 is protected from proteasomal degradation upon ribosomal biogenesis stress, while many other MDM2-binding RPs, including RPL23, RPL25, and RPS7, are degraded upon ribosomal stress (14). RPS6 deletion impairs ribosome biogenesis and specifically induces expression of RPL11, but not other RPs, to cause p53 activation (15). Upon ribosomal stress, RPL11 interacts with p14ARF (p19Arf in mouse) to augment p53 activation (16). RPL11, but not other RPs, promotes MDM2-mediated MDMX polyubiquitination and degradation (17). Recent studies have shown that upon c-MYC overexpression, the p19Arf-MDM2-p53 oncogenic insult response pathway and the RPL11-MDM2-p53 ribosomal stress response pathway are two nonredundant mechanisms possessing similar capabilities for p53 activation, indicating that, like p19Arf, RPL11 acts as a tumor suppressor (18).
To date, at least 220 distinct mutations in 18 RP genes are reported in Diamond-Blackfan anemia (DBA) patients, and about 50% of all DBA patients can be characterized by mutations in four RPs, the RPL11, RPL5, RPS19, and RPL26 (19). Interestingly, RPS19 and RPS26 activate p53 in an RPL11-dependent manner (20, 21), underscoring a role for RPL11 in linking DBA with the function of p53. Studies in zebrafish have shown that p53 activation is responsible for the morphological deficiencies associated with RPL11 depletion (22). In contrast to the zebrafish study, however, a study in mice indicated that RPL11 deletion decreases p53 activity (23). Whether the discrepancy is due to species difference is unknown. To address this issue, we generated inducible RPL11 deletion mice. Our study recapitulated the anemic phenotype observed in previous RPL11 deletion study; however, contrary to the previous study showing decrease of p53 function, our study showed an increase of p53 function upon RPL11 deletion in mice.
Results
Homozygous deletion of RPL11 in mice inhibits protein synthesis and causes rapid lethality
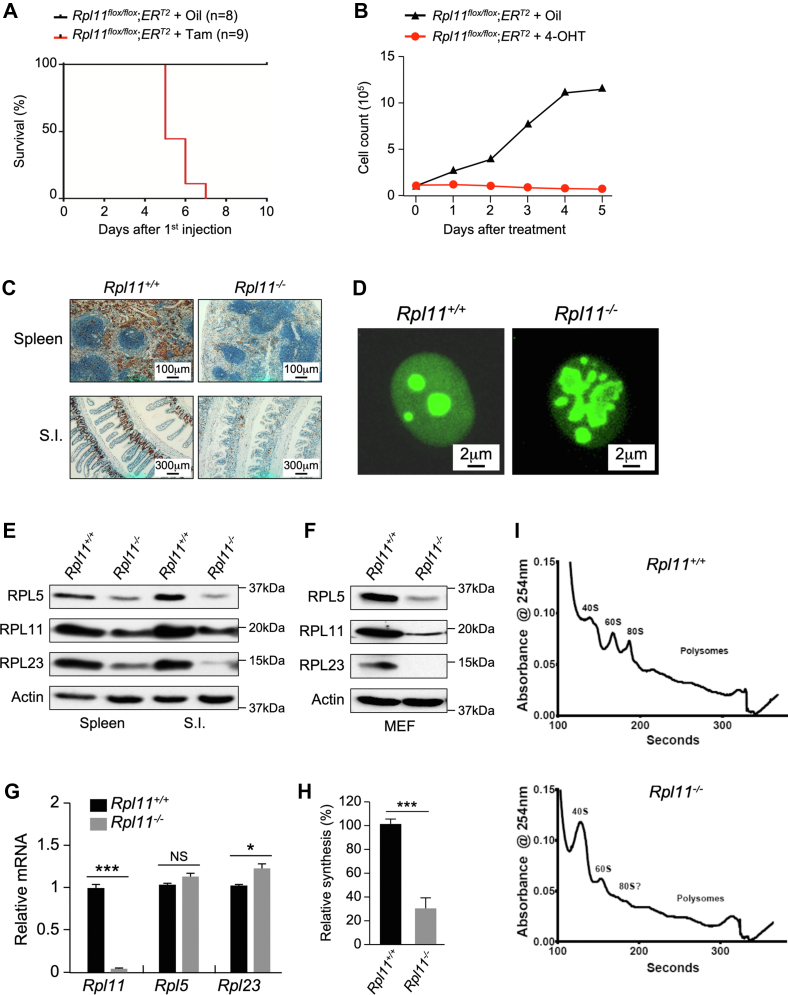
To probe the in vivo function of RPL11, we generated mice expressing a conditional knockout construct, where Rpl11 exon 2 was flanked by two loxP sites. We crossed RPL11 heterozygous flox mice (Rpl11+/flox) with CMV-Cre mice, which constitutively express the Cre transgene, to delete one allele of Rpl11. No Rpl11+/− mice were observed from more than 130 pups generated from the cross, indicating that Rpl11 heterozygosity is embryonic lethal. This is consistent with a previous report using a different gene knockout strategy that also showed Rpl11 heterozygosity is insufficient to support mouse embryonic development (23).
To overcome the embryonic lethality caused by Rpl11 loss, we crossed Rpl11+/flox mice with CreERT2 mice, which express a tamoxifen-inducible Cre recombinase, to generate Rpl11+/flox;ERT2 mice and intercrossing the mice to generate Rpl11flox/flox;ERT2 mice. We injected 6-month old Rpl11flox/flox;ERT2 mice with tamoxifen to delete both Rpl11 alleles. No Rpl11−/− mice survived more than 7 days after tamoxifen injection, indicating that homozygous deletion of RPL11 in adult mice causes rapid lethality (Fig. 1A). Likewise, mouse embryonic fibroblast (MEF) cells immediately ceased proliferation after treatment with 4-hydroxytamoxifen to delete RPL11 (Fig. 1B). In RPL11 homozygous deletion mice, fast-growing tissues, such as spleen and small intestine, showed no staining for the proliferation marker Ki-67 (Fig. 1C). MEF cells with RPL11 deletion showed malformed and heavily fused nucleoli (Fig. 1D). Deletion of RPL11 reduced expression of other RPs in tissues (Fig. 1E) and in MEF cells (Fig. 1F) but had no effect on their mRNA expression (Fig. 1G), indicating the inhibitory effect is posttranscriptional. RPL11 deletion reduced global protein synthesis measured by [35S]-methionine (Met) incorporation (Fig. 1H). The reduction of protein synthesis was likely due to a blockade of translation, as loss of RPL11 prevented maturation of 60S large ribosomal subunit and assemblage of 80S ribosome and polysomes (Fig. 1I). In conclusion, total loss of RPL11 in adult mice inhibits global protein synthesis, causing rapid cessation of cell proliferation and organismal lethality.
Figure 1.
Homozygous deletion of RPL11 in mice inhibits protein synthesis and causes rapid lethality.A, mice bearing RPL11 homozygous deletion exhibit early lethality. Kaplan–Meier curve depicting the survival of Rpl11flox/flox;CreERT2 mice treated with three injections of peanut oil (n = 8) or tamoxifen (n = 9). B, Rpl11flox/flox;CreERT2 MEF cells were seeded into 10-cm tissue culture dishes at 1 × 105 cells/dish. Peanut oil or 4-hydroxitamoxifen (4-OHT) was added to each dish at day-0. Every day, cells were harvested from one dish and counted using a Bio-Rad cell counter. C, representative Ki-67-stained sections from paraffin-embedded WT and Rpl11−/− mouse tissues isolated after treatment with peanut oil or tamoxifen. The normal architecture of the spleen and small intestine (S.I.) and positive Ki-67 signals were seen in the WT mouse tissues. The architectures of the tissues were destroyed, and the Ki-67 signals were markedly reduced in the Rpl11−/− mouse tissues. D, structure of nucleolus of WT and Rpl11−/− cells. Early passages of WT and Rpl11−/− MEF cells were fixed and immune-stained with B23 (nucleophosmin, NPM) antibody to visualize nucleolar structure. (Original magnification 400×). E, deletion of RPL11 inhibits protein expression. Rpl11flox/flox;CreERT2 mouse spleen and small intestine (S.I.) tissues were harvested at day 5 after the first peanut oil or tamoxifen injection. Protein levels in crude tissue extracts were analyzed by Western blot analysis using antibodies against various proteins as indicated. F, WT and Rpl11−/− MEF cells were analyzed by Western blot using antibodies against various proteins as indicated. G, quantitative real-time RT-PCR analysis for relative levels of mRNA of mouse bone marrow tissues. The tissues were harvested from Rpl11flox/flox;CreERT2 mice at day 5 after the first peanut oil or tamoxifen injection, and total RNA were extracted for analysis. (n = 3 per group; error bars represent standard deviation. NS: nonsignificant; ∗p < 0.05; ∗∗∗p < 0.001; Student’s t test. H, protein synthesis rates in WT and Rpl11−/− MEF cells were assessed by [35S]-methionine incorporation. The number of counts was normalized to total protein content. (n = 3 per group; error bars represent standard deviation) ∗∗∗p < 0.001; Student’s t test. I, polysome profiling of the Rpl11+/+ and Rpl11−/− MEF cells. The cells were lysed, and the nuclei, mitochondria, and large cellular debris were removed via centrifugation. The remaining supernatant was loaded onto a linear sucrose gradient, centrifuged at 32,000 rpm for 2 h to separate ribosomal subunits, monosomes, and polysomes. Ribosome profiles were obtained by measuring the absorbance of RNA at 254 nm.
Heterozygous deletion of RPL11 in adult mice impinges on tissues of hematopoiesis
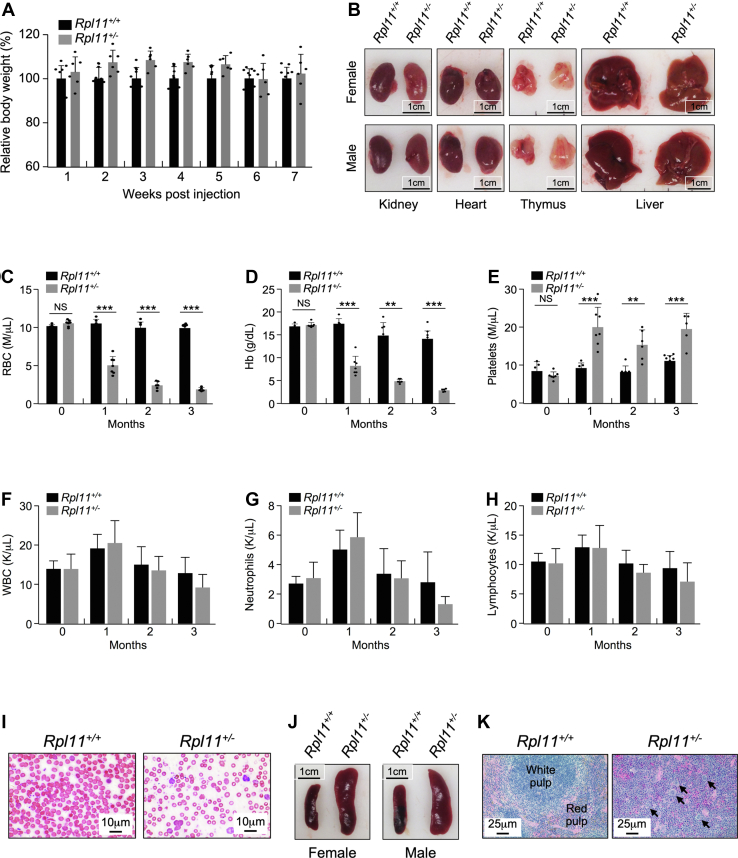
On the other hand, heterozygous RPL11 deletion (Rpl11+/−) generated by treating Rpl11+/flox;ERT2 mice with tamoxifen did not cause lethality nor did it cause visible abnormalities, and mice body weight remains unaffected (Fig. 2A). Consistently, the morphology, wet weight, and gross appearance of multiple organs, including heart, liver, kidney, and thymus were indistinguishable as compared with age-matched littermates (Fig. 2B). We noted that some of the organs from Rpl11+/− mice exhibited decreased coloration (Fig. 2B, kidney, thymus, and liver), indicative of anemia. Analysis of mouse peripheral blood revealed a significant decrease of RBC count soon after tamoxifen treatment (Fig. 2C). Correspondingly, hemoglobin (Hb) content was also decreased (Fig. 2D). In contrast, platelet count was increased in Rpl11+/− mice (Fig. 2E). This increase in platelets is likely a result of thrombocytosis in response to the acute loss of RBC (24). RPL11 haploinsufficiency had no noticeable effect on leukocyte counts in the peripheral blood, which include white blood cells, neutrophils, and lymphocytes (Fig. 2, F–H). Peripheral blood smears exhibited defects in RBC differentiation as demonstrated by irregularities in both shape and size, in addition to reduced quantities, within the Rpl11+/− RBCs (Fig. 2I). Rpl11+/− mice also exhibited severe splenomegaly, as shown by ∼3-fold larger spleens compared to control mice (Fig. 2J). Histological examination of spleen sections showed the white pulp structure was disrupted in the Rpl11+/− mouse spleens, along with a visible increase in megakaryocytes that may contribute to the increased platelet counts (Fig. 2K). Together, these data demonstrated that deletion of one Rpl11 allele in mice causes rapid on set of anemia and splenomegaly.
Figure 2.
Heterozygous deletion of RPL11 in adult mice impinges on tissues of hematopoiesis.A, effect of RPL11 heterozygous deletion on body weight of adult mice. Four-month-old male Rpl11+/+;CreERT2 (n = 11) and Rpl11+/flox;CreERT2 (n = 12) mice were given three injections of tamoxifen every other day. Changes in body weight were recorded every week after the injection. Data were presented as ± standard error of the mean. No significant differences were observed. B, images of kidney, heart, thymus, and liver tissues freshly isolated from female and male Rpl11+/+;CreERT2 and Rpl11+/flox;CreERT2 mice 2 months after treatment with tamoxifen. C–H, Four-month-old male Rpl11+/+;CreERT2 and Rpl11+/flox;CreERT2 mice were treated with tamoxifen injection every other day for 3 times. Peripheral blood cells were collected every month after the treatment for 3 months and measured for red blood cell (RBC) count (C), hemoglobin (Hb) content (D), platelet count (E), white blood cell (WBC) count (F), neutrophil count (G), and lymphocyte count (H). Results are show as mean ± standard deviation from a minimum of five mice per group. (n = 3 per group; error bars represent standard deviation; NS, not significant, ∗∗p < 0.01, ∗∗∗p < 0.001; Student’s t test). I, peripheral blood smears from Rpl11+/+;CreERT2 and Rpl11+/flox;CreERT2 mice 2 months after treatment with tamoxifen. Original magnification (1000×). J, images of representative spleens from 4-month-old female and male Rpl11+/+;CreERT2 and Rpl11+/flox;CreERT2 mice 2 months after treatment with tamoxifen. K, hematoxylin and eosin staining of representative spleen sections 2 months after tamoxifen injection. The images showing disruption of the red/white pulp structures in the Rpl11+/− mice compared to Rpl11+/+ mice. Arrows indicate megakaryocytes. Original magnification (200×). Rpl11+/flox, RPL11 heterozygous flox mice.
RPL11 haploinsufficiency impairs erythropoiesis by causing G1 cell cycle arrest in erythroid cells
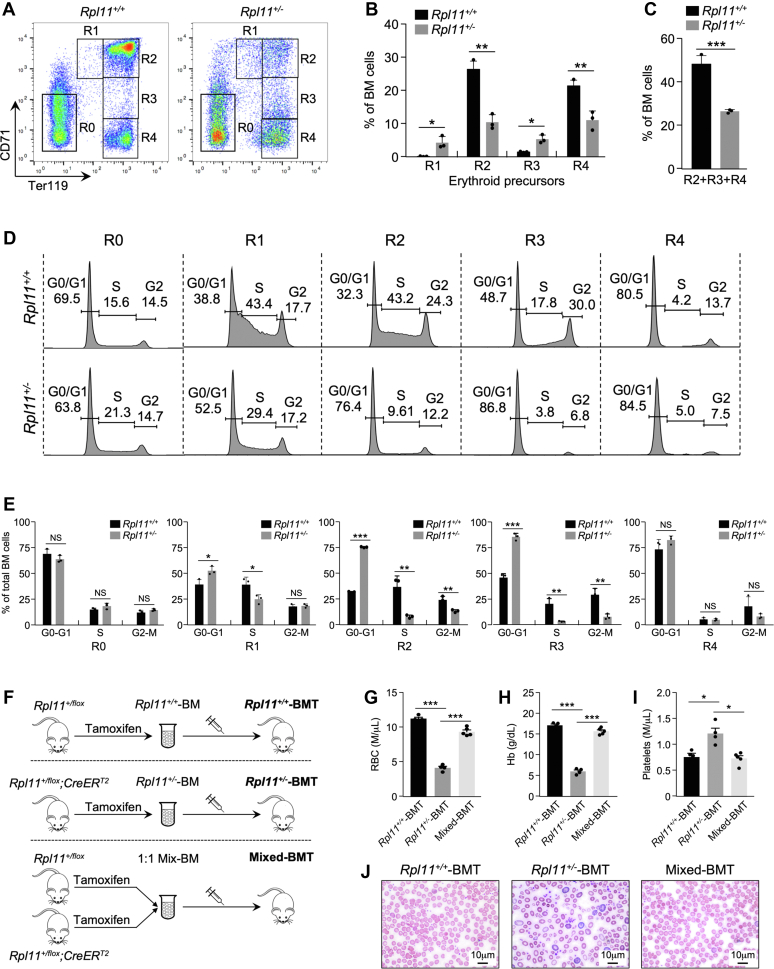
To gain insight into the RPL11 haploinsufficiency-induced anemia, we analyzed mouse bone marrow (BM) for erythroid precursor cell differentiation by flow cytometry. Analysis of BM cells using the cell surface antigens CD71 and Ter119 revealed increase of proerythroblasts (Fig. 3, A and B, R1 area) and polychromatic erythroblasts (R3 area) and decrease of basophilic erythroblasts (R2 area) and orthochromatic erythroblasts (R4 area) in the Rpl11+/− BM, observations that indicate impaired erythroid cell maturation (25, 26). The impaired erythroid maturation in the Rpl11+/− BM was further demonstrated by ∼50% reduction of overall Ter119-positive erythroid cells (Fig. 3C).
Figure 3.
RPL11 haploinsufficiency impairs erythropoiesis by causing G1 cell cycle arrest in erythroid cells.A, representative CD71/Ter119 flow cytometric profiles of BM cells isolated from Rpl11+/+ and Rpl11+/− mice. Gates of different erythroblast populations were according to expression levels of CD71 and Ter119. R0 contains mixed populations of hematopoietic stem cells and early progenitors. R1, R2, R3, and R4 represent proerythroblasts, basophilic, polychromatic, and orthochromatic erythroblasts, respectively. B, bar graphs representing the percent of total cells of the R1-R4 subpopulations. (n = 3 per group; error bars represent standard deviation; ∗p < 0.05; ∗∗p < 0.01; Student’s t test). C, bar graph representing the percentage of Ter119high (R2+R3+R4) cells from total bone marrow. (n = 3 per group; error bars represent standard deviation, ∗∗∗p < 0.001; Student’s t test). D, BM extracted from Rpl11+/+ and Rpl11+/− mice were incubated with CD71 and Ter119 antibodies followed by cell sorting to separate R0-R4 populations. Flow cytometry was applied to analyze the cell cycle distribution of individual erythroid progenitor populations. Quantification of the cell cycle distribution from each phase was shown as a percent of total cells. E, bar graphs represent the average percentage of cells from erythroid progenitor subsets (R0-R4) in each stage of the cell cycle. (n = 3 per group; error bars represent standard deviation; NS: nonsignificant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; Student’s t test). F, schematic representation of the BM transplantation (BMT) procedure. Top: experimental outline of murine BM reconstitution assay with WT (Rpl11+/+) donor mice. Middle: experimental procedure of BM reconstitution assay with heterozygous deletion RPL11 (Rpl11+/−) donor mice. Bottom: experimental procedure of BM reconstitution assay with donor BM isolated from WT and heterozygous deletion RPL11 mice mixed at 1:1 ratio. All recipient mice were treated with 8-Gy γ-irradiation prior to transplantation. G–I, peripheral blood analysis of mice after BMT. Peripheral blood was extracted from BMT-mice 2 months post transplantation for total blood count analysis of RBC (G), hemoglobin (Hb) (H), and platelet (I) from Rpl11+/+-BMT (black bar), Rpl11+/−-BMT (gray bar), and Mixed-BMT (white bar) mice. Results are shown as mean ± standard deviation from a minimum of three mice per group. J, peripheral blood smears from Rpl11+/+-BMT, Rpl11+/−-BMT, and Mixed-BMT mice 2 months after transplantation. Original magnification (1000×). BM, bone marrow.
To explore mechanisms underlying the interruption of erythropoiesis, we analyzed cell cycle status for each erythroid precursor subset. A significant increase of G0/G1-phase and decrease of S-phase were observed in the R1, R2, and R3 cell populations in the Rpl11+/− BMs compared to WT BMs (Fig. 3, D and E, R1, R2, R3 areas), indicating a G1 cell cycle arrest in the Rpl11+/− erythroid cells. The nonerythroid BM cells (CD71low;Ter119low, R0 area) and the orthochromatic erythroblasts (CD71low;Ter119high, R4 area), which are largely postmitotic cells, did not show changes in cell cycle distributions. These data suggested that partial loss of RPL11 induces erythroid lineage-specific cell cycle arrest that blocks erythropoiesis.
To determine whether the hematopoiesis defect in Rpl11+/− mice was intrinsic to BM-derived hematopoietic cells, we carried out BM transplantation (BMT) assays (Fig. 3F). We treated 6-week-old Rpl11+/flox;CreERT2 mice with tamoxifen to delete one Rpl11 allele (Rpl11+/−). We also treated Rpl11+/flox mice with tamoxifen to serve as control (Rpl11+/+). Six weeks after the treatment, 10 million Rpl11+/+ or Rpl11+/− BM cells were harvested and transplanted into γ-irradiated WT mice (designated Rpl11+/+-BMT and Rpl11+/−-BMT, respectively). Compared to control recipients (Rpl11+/+-BMT), the Rpl11+/−-BMT recipient mice exhibited significantly decreased RBC numbers and Hb levels as early as 1 month after transplantation, while their platelet counts were elevated (Fig. 3, G–I, Rpl11+/+-BMT), indicating an intrinsic defect of the Rpl11+/− BM cells. To determine whether RPL11 deficiency affects hematopoietic stem cell self-renewal and reconstitution, we carried out competitive BMT by mixing Rpl11+/− and Rpl11+/+ BM cells at a 1:1 ratio and transplanted the mixture into irradiated WT recipient mice. Interestingly, Mixed-BMT recipient mice exhibited near normal numbers of RBCs, Hb content, and platelet count (Fig. 3, G–I, Mixed-BMT), indicating that the Rpl11+/− BM cells had competitive disadvantage in the mixed BM cell population. The growth disadvantage of Rpl11+/− BM cells was also demonstrated by blood smear, where the erythroid cell irregularity of Rpl11+/− mice was not observed in the Mixed-BMT mice (Fig. 3J). Overall, these data indicated that RPL11 haploinsufficiency compromised the fitness of the BM cells and suggested that the anemia in Rpl11+/− mice is driven by cell intrinsic defects of BM erythroid precursor cells.
RPL11 haploinsufficiency activates p53 in hematopoietic tissues
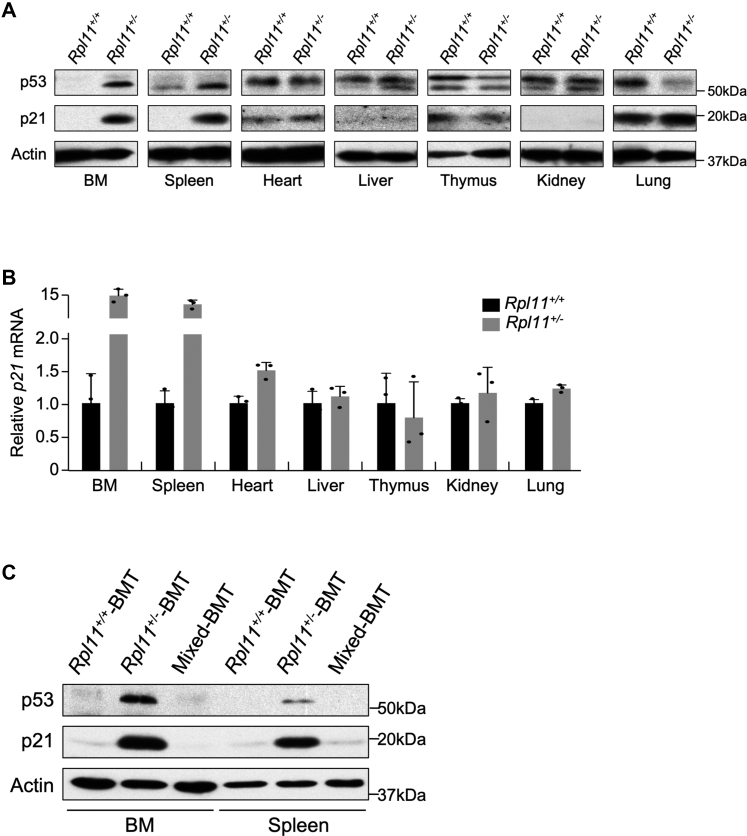
Previous studies showed that RPL11 deficiency in zebrafish leads to p53 activation (22). However, a study in mice suggested that RPL11 deficiency inactivates p53 (23). To clarify the significance of RPL11 deficiency on p53 function, we treated Rpl11+/+;ERT2 and Rpl11+/flox;ERT2 mice with tamoxifen and examined p53 expression in tissues 2 weeks after the treatment, prior to the appearance of anemia. We found that the protein levels of p53 were noticeably increased in the BM and spleen, slightly increased in the liver, and relatively unchanged or nonobservable in other tissues of the Rpl11+/− mice; p21 levels were also increased in the BM and spleen of the Rpl11+/− mice, indicating that RPL11 heterozygous deletion activates p53 in tissues of hematopoiesis (Fig. 4A). The hematopoietic tissue-specific p53 activation was corroborated by changes of p21 (Cdkn1a) mRNA, where it was highly expressed only in BM and spleen tissues of Rpl11+/− mice (Fig. 4B). To determine if the hematopoietic tissue-specific p53 activation in Rpl11+/− mice was BM cell autonomous, we examined p53 in BMT mice. In the BM and spleen tissues of Rpl11+/−-BMT recipient mice, p53 and p21 were elevated, which was not seen in tissues of Mixed-BMT recipient mice (Fig. 4C), demonstrating the RPL11 haploinsufficiency–caused p53 activation was BM cell autonomous.
Figure 4.
RPL11 haploinsufficiency activates p53 in hematopoietic tissues.A, tissue expression pattern of p53 and p21 proteins in Rpl11+/+ and Rpl11+/− mice. Tissues were isolated from adult Rpl11+/+;CreERT2 and Rpl11+/flox;CreERT2 mice after tamoxifen treatment for 2 weeks prior to protein extraction for immunoblot analysis of p53, p21, and actin. B, Rpl11+/+ and Rpl11+/− mice were treated, and tissues were isolated as in (A). Tissue expression pattern of p21 mRNA in Rpl11+/+ and Rpl11+/− mice was analyzed by RT-PCR. C, Bone marrow (BM) and spleen tissues were harvested from BM transplant mice and proteins were extracted for immunoblot analysis of MDM2, p53, p21, and actin. Rpl11+/flox, RPL11 heterozygous flox mice.
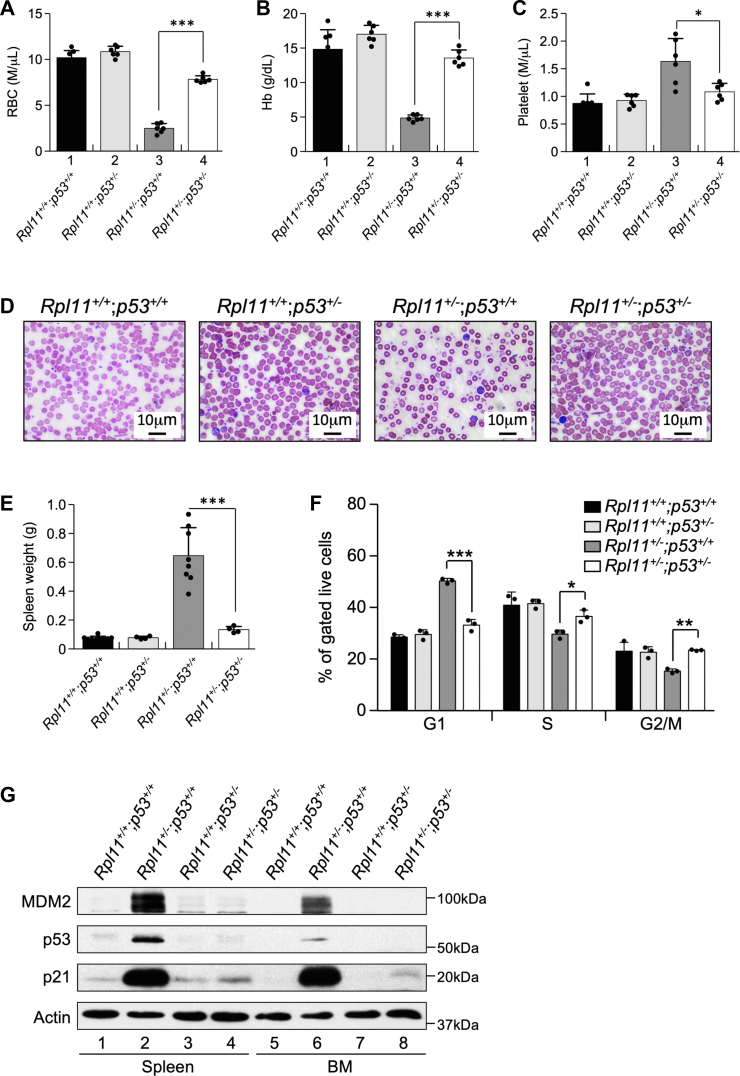
Deletion of one p53 allele rescues RPL11 haploinsufficiency caused anemia and restores cell cycle progression in erythroid cells
To determine whether p53 activation is responsible for the anemic phenotype in the RPL11 haploinsufficiency mice, we generated cohorts of Rpl11+/+;CreERT2;p53+/+, Rpl11+/flox;CreERT2;p53+/+, Rpl11+/+;CreERT2;p53+/−, and Rpl11+/flox;CreERT2;p53+/− mice. These mice were injected with tamoxifen, and their peripheral blood counts were assessed 2 months later. Under WT RPL11 background (Rpl11+/+), deletion of a single p53 allele had no effect on RBC counts, Hb levels, and platelet counts (Fig. 5, A–C, bars 1 and 2). Under heterozygous RPL11 background (Rpl11+/−), deletion of one p53 allele essentially restored RBC counts, Hb levels, and platelet counts (Fig. 5, A–C, bars 3 and 4). Deletion of one p53 allele also restored RBC levels and homogeneity of the Rpl11+/− mice (Fig. 5D). Consistently, deletion of one p53 allele reversed RPL11 heterozygosity-induced splenomegaly (Fig. 5E). Deletion of both p53 alleles similarly reversed RPL11 haploinsufficiency–caused anemic phenotype; however, we did not include these data because complete deletion of p53 results in spontaneous tumor development at early ages in mice (27), making the results unreliable. To understand the effects of p53 deletion on erythropoiesis, we examined the cell cycle status of the erythroid precursor populations in the BM. In contrast to cell cycle arrest found in Rpl11+/−;p53+/+ BM, the cell cycle status from Rpl11+/−;p53+/− BM exhibited similar patterns to that of the WT and Rpl11+/+;p53+/− BMs (Fig. 5F), suggesting that p53 plays a critical role in the development of erythropoietic disorders by driving cell cycle arrest in erythroid precursor populations. To corroborate the hematopoietic defect in the Rpl11+/− mice that is intrinsically related to p53 activation, we isolated BM and spleen tissues from tamoxifen treated mice to analyze p53 expression along with p53 targets p21 and MDM2 by Western blot. Under WT p53 background, heterozygous deletion of RPL11 resulted in p53 stabilization and activation (Fig. 5G, lanes 2 and 6), which was prevented by deletion of a single p53 allele (Fig. 5G, lanes 4 and 8). Together, these data demonstrated that the anemic phenotype in the Rpl11+/− mice is rescued by deletion of a single p53 allele, suggesting that p53 activation is the primary driver of these abnormalities.
Figure 5.
Deletion of one p53 allele rescues RPL11 haploinsufficiency–induced anemia and restores cell cycle progression in erythroid cells.A–C, Six-week old Rpl11+/+;CreERT2;p53+/+, Rpl11+/flox;CreERT2;p53+/+, Rpl11+/+;CreERT2;p53+/−, and Rpl11+/flox;CreERT2;p53+/− male mice were injected with tamoxifen every other day for 3 times to generate Rpl11+/+;p53+/+, Rpl11+/+;p53+/−, Rpl11+/−;p53+/+, and Rpl11+/−;p53+/− mice. Peripheral blood was isolated 2 months after the tamoxifen treatment and analyzed for RBC (A), hemoglobin (Hb) (B), and platelet (C). (n = 3 per group; error bars represent standard deviation; ∗p < 0.05; ∗∗∗p < 0.001; Student’s t test). D, peripheral blood was isolated from abovementioned mice, and blood smears were performed. Original magnification (1000×). E, spleen weight in grams from abovementioned mice. (n = 3 per group; error bars represent standard deviation; ∗∗∗p < 0.001; Student’s t test). F, BM cells were extracted from abovementioned mice and gated for erythroid precursors. The gated cells were analyzed for cell cycle distribution. (n = 3 per group; error bars represent standard deviation; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; Student’s t test). G, BM and spleen tissues were harvested from abovementioned mice, and tissue extracts were analyzed by Western blot for MDM2, p53, p21 and actin. BM, bone marrow; Rpl11+/flox, RPL11 heterozygous flox mice.
RPL11 haploinsufficiency caused anemia involves the RP-MDM2-p53 pathway
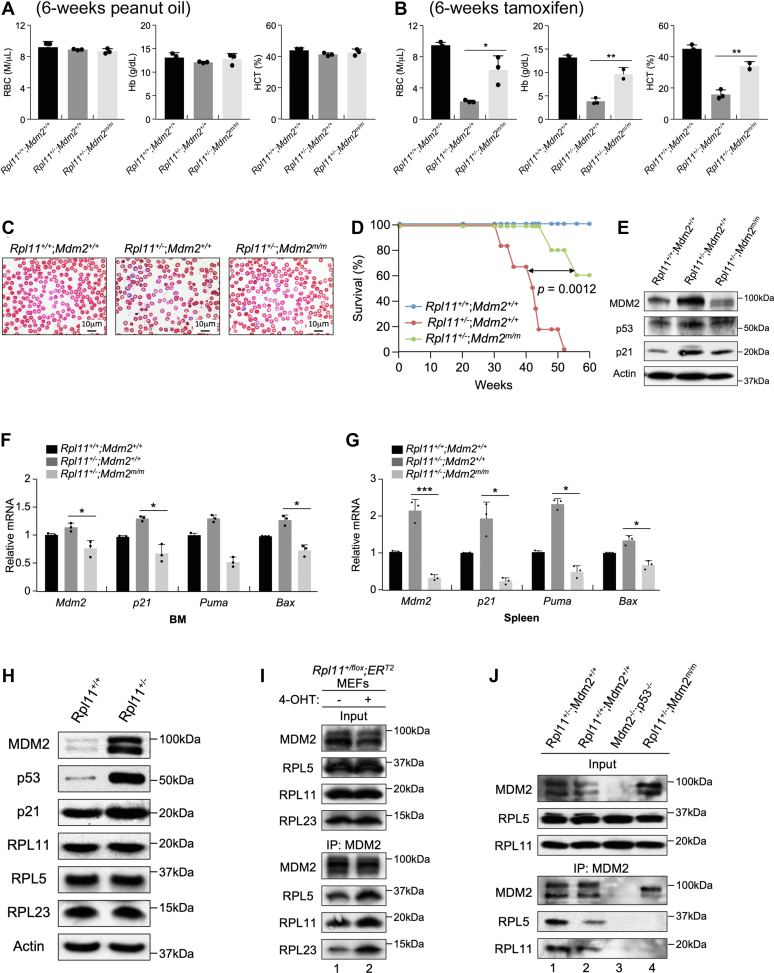
Recent studies uncovered an RP-MDM2-p53 stress response pathway mediated by RP binding to and inhibiting the E3 ligase function of MDM2 (5, 6). Mouse model bearing a single point mutation in the zinc-finger domain of MDM2 (MDM2C305F), which inhibits the binding between MDM2 and RPL11 (28), mitigates p53 response to ribosomal stress (29, 30). To determine whether the RP-MDM2-p53 pathway is involved in RPL11 haploinsufficiency caused anemia, we crossed MDM2C305F-mutant mice (Mdm2m/m) with Rpl11+/flox;CreERT2 mice to generate Rpl11+/flox;CreERT2;Mdm2m/m compound mice. We treated the compound mice with tamoxifen to generate Rpl11+/−;Mdm2m/m mice. The presence of MDM2C305F mutation notably alleviated Rpl11+/−-induced anemia (Fig. 6, A–C) and significantly extended Rpl11+/− mouse survival (Fig. 6D), indicating that an intact RP-MDM2-p53 pathway is necessary for hematopoietic inhibition in response to RPL11 haploinsufficiency. Moreover, the RPL11 haploinsufficiency–induced p53 activation was prevented by the MDM2C305F mutation at both protein (Fig. 6E) and mRNA (Fig. 6, F and G) levels. RPL11 haploinsufficiency did not change general RP expression levels (Fig. 6H) but augmented MDM2 binding to RPL5, RPL23, and RPL11 (Fig. 6I). Using BM lysate, we showed that RPL11 haploinsufficiency would augment MDM2 binding with RPL5 and RPL11 (Fig. 6J, compare lane 1 to lane 2), which was been eliminated by MDM2C305F mutation (Fig. 6J, compare lane 4 to lane 2). Taken together, these data demonstrated that RPL11 haploinsufficiency activates p53 by increasing MDM2 binding to RPs.
Figure 6.
RPL11 haploinsufficiency–induced anemia involves the RP-MDM2-p53 pathway.A–B, Four-month-old Rpl11+/+;CreERT2;Mdm2+/+, Rpl11+/flox;CreERT2;Mdm2+/+, and Rpl11+/flox;CreERT2;Mdm2m/m male mice were treated with peanut oil (A) or tamoxifen (B) for 6 weeks. Peripheral blood was isolated and analyzed for RBC, hemoglobin (Hb), and hematocrit (HCT). (n = 3 per group; error bars represent standard deviation; ∗p < 0.05; ∗∗p < 0.01; Student’s t test). C, peripheral blood was isolated from abovementioned mice, and blood smears were performed. Original magnification (1000×). D, Kaplan–Meier curve depicting the survival of Rpl11+/+;CreERT2;Mdm2+/+ (n = x), Rpl11+/flox;CreERT2;Mdm2+/+ (n = x), and Rpl11+/flox;CreERT2;Mdm2m/m (n = x), male mice treated with three injections of tamoxifen. E, BM tissues were harvested from 4-month-old Rpl11+/+;CreERT2;Mdm2+/+, Rpl11+/flox;CreERT2;Mdm2+/+, and Rpl11+/flox;CreERT2;Mdm2m/m male mice 2 weeks after treated with three injections of tamoxifen. Tissue extracts were analyzed by Western blot for protein expression of MDM2, p53, p21, and actin. F–G, quantitative real-time RT-PCR analyses of Mdm2, p21, Puma, and Bax transcript levels in BM (F) and Spleen (G) tissues isolated from 4-month-old Rpl11+/+;CreERT2;Mdm2+/+, Rpl11+/flox;CreERT2;Mdm2+/+, and Rpl11+/flox;CreERT2;Mdm2m/m male mice 2 weeks after treated with three injections of tamoxifen. GAPDH transcripts were used for normalization. (n = 3 per group; error bars represent standard deviation; ∗p < 0.05; ∗∗∗p < 0.001; Student’s t test). H, WT and Rpl11+/− MEF cells were analyzed by Western blot using antibodies against various proteins as indicated. I, Rpl11+/flox;ERT2 MEFs were exposed to 4-OHT for 20 h prior to immunoprecipitation with anti- MDM2 antibodies and immunoblotting for MDM2, RPL5, RPL11, and RPL23. J, BM tissues were isolated from Rpl11+/+;Mdm2+/+, Rpl11+/−;Mdm2+/+, Rpl11+/+;Mdm2−/−;p53−/−, and Rpl11+/−;Mdm2m/m mice; MDM2 immunoprecipitation was carried out prior to immunoblot analysis for MDM2, RPL5, and RPL11. BM, bone marrow; Rpl11+/flox, RPL11 heterozygous flox mice.
Discussion
Multiple studies have indicated correlations between p53 activation and ribosomopathies such as DBA; the p53 dependency of ribosomopathy-associated phenotypes, however, remains partially clarified (31, 32). Our data showed that RPL11 haploinsufficiency–induced hematopoietic disorder is p53 dependent, but increased p53 activity alone is not sufficient to drive defective hematopoiesis in mice. Previous studies showed that mice with a truncated form of p53, which exhibits increased p53 activity, have early aging phenotype and normal blood chemistry and RBC levels (33). Likewise, the so-called “super-p53” mice, which carry an extra functioning p53 allele, showed a constitutively increased p53 activity, enhanced DNA damage response, and greater resistance to carcinogen-induced tumor formation, yet had no evidence of defective hematopoiesis (34). Therefore, it seems that p53 activation alone is not sufficient to cause hematopoiesis deficiency, and other factors besides p53 activity are involved.
Reports in model organisms and humans have shown that monoallelic inactivation of RP genes contributes development of ribosomopathy and cancer. The mechanistic basis by which RP mutations do so remains poorly understood. Altered signaling in cell stress responsive pathways is consistently observed among diseases caused by RP deficiency. Specifically, deficient in RPs has been shown to increase p53 activity, which seems counterintuitive since multiple ribosomopathies are also associated with cancer predisposition. We speculate that constitutive activation of p53 in the RP-deficient hematopoietic cells leads to elimination of these cells or adds pressure for p53 mutation to facilitate cell survival. Activation of p53 triggers cell cycle checkpoints, apoptosis, senescence, and metabolic deregulation, yet it is unclear which of these processes is important for RP deletion–induced ribosomopathies. Our data indicated that RPL11 haploinsufficiency causes G1 phase cell cycle arrest specifically in in erythroid lineage (Fig. 3), which is likely the common cause behind RP deficiency–mediated anemia. As cells balance RP production to maintain equimolar quantities of ribosome subunits, a tight regulation of RP production is evolved across multiple levels of protein homeostasis, including transcription, translation, and protein degradation (35). Therefore, it seems that obstruction of ribosomal biosynthesis and activation of p53 during RP mutation are two necessary conditions for the development of ribosomopathies. Interestingly, deletion of one Rpl11 allele did not cause observable reduction of RPL11 and other RP protein expression, yet it significantly activated p53 (Fig. 6H). We speculate that in the Rpl11+/− cells RPL11 production is compensated by ramped up transcription and translation of the remaining Rpl11 allele or by tapered off RPL11 degradation. A subtle reduction of RPL11 levels will disturb ribosomal biogenesis, resulting in increased RP-MDM2 interactions and p53 activation.
The work presented here recapitulated many of the observations of a previously published RPL11 deletion mouse study (23). However, the previous study indicated that p53 activity is decreased in the Rpl11+/− mice, leading to increased lymphomagenesis in the Rpl11+/− mice; whereas our results showed that in the absence of cellular stress, basal p53 activity is increased in the Rpl11+/− mice (Fig. 4). We do not have clear explanation about the discrepancy. To validate the increased p53 activity is responsible for RPL11 deficiency–caused anemia, we co-deleted a p53 allele in the Rpl11+/− mice to generate Rpl11+/−;p53+/− double mutant mice. Deletion of one p53 allele nearly corrected all the disease phenotypes of Rpl11+/− mice. Study of the Rpl11+/−;p53+/− double deletion mice established that a full dosage of p53 is required for RPL11 deficiency resulted hematopoiesis inhibition, demonstrating that p53 activation is the cause of the anemia of the Rpl11+/− mice. By generating and studying the Rpl11+/−;Mdm2m/m double mutation mice, where a C305F mutation in MDM2 disrupts RP binding, we further showed that an increased RP binding to MDM2 is necessary for p53 activation and anemia phenotype in the Rpl11+/− mice. The study demonstrated that RPL11 heterozygous deletion triggers a ribosomal stress signal that is transmitted to p53 through RP binding to MDM2, implicating a role for the RP-MDM2-p53 pathway in mediating RP deficiency stress signals to p53. We noticed that the rescue of RPL11 haploinsufficiency–induced anemia by MDM2C305F mutation appears to be incomplete, showing partial recovery of RBC levels in the Rpl11+/−;Mdm2m/m mice and incomplete rescue of the Rpl11+/− mouse late life lethality (Fig. 6). We interpret that this partial rescue is likely because of the MDM2C305F mutation disrupts binding of some, but all MDM2-interacting RPs and those RPs that still bind MDM2C305F could respond to RPL11 deletion caused ribosomal stress to activate p53. Indeed, studies have shown that MDM2C305F mutation disrupts RPL5 and RPL11 binding but not RPL23 binding (28, 29). Given that as many as 16 RPs have been identified to bind MDM2 and regulate p53 activity (36), it is conceivable that some of these RPs that still bind to MDM2C305F mutant could inhibit MDM2C305F function to activate p53.
The specific activation of p53 within hematopoietic tissues in the Rpl11+/− mice is particularly striking, and this phenotype is not specific to RPL11 deletion as downregulation of Rps14 and Rps19 genes induces similar specific p53 activation within the erythroid lineage (37). The precise mechanism of this cell type-specific p53 activation is currently unknown, but we speculate that the highly proliferative nature of erythroid precursors to maintain RBC production requires significant translational capacity, which creates a unique environment that is particularly susceptible to RP imbalance. Furthermore, studies have shown that terminally differentiated and anucleated cells, such as erythroid cells, lack ribosome recycling factors that adds further stresses on the translation machinery within erythroid lineage cells (38). Taken together, erythroid precursors are uniquely positioned to stimulate p53 activation in response to altered RP expression and changed translational capabilities.
Experimental procedures
Mice
C57BL/6 Rpl11+/flox, Rpl11flox/flox, p53+/−, and Rpl11+/flox;p53+/− mice with or without CreERT2 were bred and maintained on a 12 h light and 12 h dark cycle with lights on from 7:00 AM to 7:00 PM. All mice were given standard food pellets (normal chow) and water ad libitum. Cohorts of age-matched male and female mice were used for the study. The Institutional Animal Care and Use Committee at the University of North Carolina Animal Care Facility approved all studies using animals (protocol 16–026).
Tamoxifen treatment
Tamoxifen (T5648, Sigma) was dissolved in a small volume of 100% ethanol at room temperature prior to mixing with peanut oil (P2144, Sigma). The peanut oil, tamoxifen, and ethanol mixture was then rotated at room temperature for 5 h. Mice (6–10 weeks old) were injected intraperitoneally with either vehicle (peanut oil) or 2 mg tamoxifen every other day for a total of three injections. All time-points in the data are started from the initial injection.
Blood testing
Blood was extracted from either the tail vein or behind the eye via capillary tube prior to submitting to the UNC Animal Histopathology & Lab Medicine Core at the University of North Carolina, which is supported in part by an NCI Center Core Support Grant (5P30CA016086–41) to the UNC Lineberger Comprehensive Cancer Center. Blood samples were analyzed using the ProCyte Dx Hematology Analyzer (IDEXX BioResearch) to generate complete blood counts.
Peripheral blood smears
A drop of blood was placed upon a glass slide before being smeared using the edge of a second glass slide. After air dry, Wright-Giemsa (WS16, Sigma) staining of the slides were performed for 1 to 3 min and mounted with resin. Images were acquired using an EVOS AMEX 100 digital scope with a 100× objective.
BM transplant
Rpl11+/flox and Rpl11+/flox;CreERT2 mice were injected with tamoxifen as described previously then allowed to grow for 6 weeks before sacrifice and extraction of BM. WT mice were irradiated (600 cGy) to kill the BM before injected with either Rpl11+/flox (WT-BMT) or Rpl11+/flox;CreERT2 (Rpl11+/−-BMT) or a 1:1 mix of Rpl11+/flox and Rpl11+/flox;CreERT2 (Mix-BMT) from the retro orbital.
Erythroid flow cytometry, cell sorting, and cell cycle analysis
BM cells were isolated and incubated with anti-Ter119 and anti-CD71 antibodies for 1 h at 4 °C with constant rotation. The fluorescently labeled cells were then analyzed using a Beckman Coulter CyAn ADP flow cytometer. Data are shown as a percentage of singlet detected within each gate of the entire BM population. For cell cycle analysis, BM cells were isolated and incubated with anti-Ter119 and anti-CD71 antibodies for 1 h at 4 °C; the cells were then washed and fixed by 5% formalin for 10 min on ice; fixed cells were washed and permeabilized by PBS with 0.1% saponin and further stained by DAPI for 15 min. R1 to R4 erythroid populations were sorted based on the Ter119 and CD71 staining. The DNA content of these selected populations were further analyzed by the DAPI staining for cell cycle progression using an Attune NXT flow cytometer (ThermoFisher).
qRT-PCR analysis of mRNA expression
Total RNA was prepared from mouse tissues using Trizol Reagent (15596–026, Invitrogen). RNA concentration was determined with a NanoDrop spectrophotometer (Thermo Scientific, NanoDrop 2000c), and quality was assessed by agarose gel electrophoresis. cDNA was synthesized using Superscript III reverse transcriptase (18080–051, Invitrogen). qRT-PCR was performed with SYBR Green probes using the Applied Biosystems 7900HT Fast Real-Time PCR system. Results were expressed as the fold-change in transcript levels.
MEF cell culture
Primary MEFs were cultured in a 37 °C incubator with 5% CO2 and 3% O2 in Dulbecco's modified Eagle's medium supplied with 10% fetal bovine serum and penicillin (100 IU/ml)/streptomycin (100 μg/ml). For activation of CreERT2, 1 μM 4-hydroxytamoxifen dissolved in 100% ethanol was added to the culture medium.
Antibodies
Rabbit polyclonal anti-c-MYC (N262; Santa Cruz), mouse anti-p53 (pAB122), and mouse anti-MDM2 (2A10) were purchased commercially. Rabbit polyclonal antibodies to p21 were gifts from Dr Yue Xiong (UNC-Chapel Hill). Rabbit polyclonal antibodies to RPL11 were homemade and previously described (29).
Protein analysis
For Western blotting, proteins were extracted from tissues as previously described (29). Briefly, mouse tissue was homogenized and lysed in 0.5% NP-40 lysis buffer. Results detected by using either Pico, Dura, or Femto enhanced chemiluminescence (ECL) systems (Thermo Scientific, SuperSignal West Dura Substrate) or WesternBright ECL chemiluminescence system (Advansta).
Measurement of protein synthesis by [35S]-methionine labeling
For measuring nascent protein synthesis, cells were washed twice with PBS and starved in Met-free Dulbecco's modified Eagle's medium supplemented with dialyzed 10% FBS for 30 min. The cells were then labeled at 37 °C for 2 h by adding 10 μCi [35S]-Met (Amersham) per 100 mm plate. Cells were then washed thoroughly with PBS, lysed with 0.5% NP-40 buffer, total proteins were precipitated with 20% ice-cold TCA, pelleted, and dissolved in 0.1 N NaOH. Equal amounts of protein were then counted in a scintillation counter for Met incorporation.
Polysome profiling
For sucrose density gradient fractionation of ribosomal subunits and polysomes, cells were scraped and collected after adding cycloheximide (100 μg/ml) in culture for 10 min. The cells were centrifuged at 500g for 5 min and lysed using 1% Triton X-100 lysis buffer containing 140 mM KCl, 5 mM MgCl2, 10 μg/ml cycloheximide, 10 mM DTT, and 20 mM Tris-HCl pH 7.4. Cells were then passed through a 27G needle 5 times to disrupt cell membranes. Nuclear and mitochondrial debris were removed through additional centrifugation (1000g and 13000g, respectively). The remaining supernatant was then loaded onto linear 10 to 50% sucrose gradients and centrifuged at 180,000g for 2 h at 4 °C using a Beckman SW40Ti rotor and fractionated by detecting absorbance at 254 nm using an EM-1 UV monitor (Bio-Rad).
Indirect immunofluorescence staining
MEF cells were washed with PBS two times at room temperature prior to fixation with 10% formalin solution (Sigma) for 10 min at room temperature with gentle shaking. Fixed cells were then rinsed again with PBS to remove residual formaldehyde and permeabilized with PBS/0.2% Triton X-100 for 5 min at 4 °C without shaking. Cells were then incubated with PBS/0.5% BSA before primary antibody incubation for 1 h with gentle shaking. Cells were then washed 3 times with PBS prior to secondary antibody incubation for 1 h at room temperature with gentle shaking. Cells were then washed 3 times with PBS, and DAPI was included with the final wash to stain DNA. Cells were then covered with DAKO mounting solution and a cover slip prior to imaging using an Olympus IX 81 microscope and a SPOT RT3 camera.
Statistical analysis and error bars
Results are represented as mean ± standard deviation. Quantitative PCR data and immunohistochemistry quantifications were evaluated for significance using the two-tailed Student’s t tests. A p value < 0.05 was considered significant for all analyses. Significant differences between experimental groups were ∗p < 0.05, ∗∗p < 0.01, or ∗∗∗p < 0.001.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article. Raw data were generated at UNC Chapel Hill. Derived data supporting the findings of this study are available from the corresponding author Y. Z. on request.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank many people that helped to complete this work through technical expertise and advice. Specifically, we would like to thank Jing Yang, Anqun Tang, and Yong Liu for their helpful advice and technical assistance. We would like to thank Nathan Montgomery and Stephanie Montgomery for their help with mouse peripheral blood analysis and discussions. We would like to thank Janet Dow and the UNC Flow Cytometry Core for technical expertise with flow cytometry and assistance with the bone marrow cell cycle analysis specifically. We would also like to thank Ned Sharpless for his advice and assistance with stem cell biology and flow cytometry experiments.
Author contributions
D. A. F., S. L., and Y. Z. methodology; D. A. F., S. L., J. L., R. L., K. C. A., F. Y., A. J., and Z. G. investigation; D. A. F., S. L., R. L., K. C. A., Z. G., and S. H. formal analysis; F. Y. and A. J. resources; D. A. F, S. H., and Y. Z. writing-original draft; N. M., G. G. W., Y. Y. W., and Y. Z. supervision.
Funding and additional information
This research was supported by grants from the National Institutes of Health (CA100302, CA127770, and CA212407) to Y. Z., a fellowship from the UNC Pharmacology Department Training grant (GM007040-41) to D. F. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ronald Wek
References
- 1.Hollstein M., Sidransky D., Vogelstein B., Harris C.C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 2.Olivier M., Hollstein M., Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor Perspect. Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane D.P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 4.Moll U.M., Petrenko O. The MDM2-p53 interaction. Mol. Cancer Res. 2003;1:1001–1008. [PubMed] [Google Scholar]
- 5.Zhang Y., Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Deisenroth C., Zhang Y. RP-MDM2-p53 pathway: linking ribosomal biogenesis and tumor surveillance. Trends Cancer. 2016;2:191–204. doi: 10.1016/j.trecan.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandori C., Gomez-Roman N., Felton-Edkins Z.A., Ngouenet C., Galloway D.A., Eisenman R.N., et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 8.Nicolas E., Parisot P., Pinto-Monteiro C., de Walque R., De Vleeschouwer C., Lafontaine D.L. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 2016;7 doi: 10.1038/ncomms11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 10.Narla A., Ebert B.L. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fumagalli S., Thomas G. The role of p53 in ribosomopathies. Semin. Hematol. 2011;48:97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Lohrum M.A., Ludwig R.L., Kubbutat M.H., Hanlon M., Vousden K.H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., Wolf G.W., Bhat K., Jin A., Allio T., Burkhart W.A., et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bursac S., Brdovcak M.C., Pfannkuchen M., Orsolic I., Golomb L., Zhu Y., et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fumagalli S., Di Cara A., Neb-Gulati A., Natt F., Schwemberger S., Hall J., et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 2009;11:501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai M.S., Challagundla K.B., Sun X.X., Palam L.R., Zeng S.X., Wek R.C., et al. Physical and functional interaction between ribosomal protein L11 and the tumor suppressor ARF. J. Biol. Chem. 2012;287:17120–17129. doi: 10.1074/jbc.M111.311902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilkes D.M., Chen L., Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng X., Carlson N.R., Dong J., Zhang Y. Oncogenic c-Myc-induced lymphomagenesis is inhibited non-redundantly by the p19Arf-Mdm2-p53 and RP-Mdm2-p53 pathways. Oncogene. 2015;34:5709–5717. doi: 10.1038/onc.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horos R., Ijspeert H., Pospisilova D., Sendtner R., Andrieu-Soler C., Taskesen E., et al. Ribosomal deficiencies in Diamond-Blackfan anemia impair translation of transcripts essential for differentiation of murine and human erythroblasts. Blood. 2012;119:262–272. doi: 10.1182/blood-2011-06-358200. [DOI] [PubMed] [Google Scholar]
- 20.Dutta S., Akey I.V., Dingwall C., Hartman K.L., Laue T., Nolte R.T., et al. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- 21.Cui D., Li L., Lou H., Sun H., Ngai S.M., Shao G., et al. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene. 2014;33:2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty A., Uechi T., Nakajima Y., Gazda H.T., O'Donohue M.F., Gleizes P.E., et al. Cross talk between TP53 and c-Myc in the pathophysiology of Diamond-Blackfan anemia: evidence from RPL11-deficient in vivo and in vitro models. Biochem. Biophys Res. Commun. 2018;495:1839–1845. doi: 10.1016/j.bbrc.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Morgado-Palacin L., Varetti G., Llanos S., Gomez-Lopez G., Martinez D., Serrano M. Partial loss of Rpl11 in adult mice recapitulates Diamond-Blackfan anemia and promotes lymphomagenesis. Cell Rep. 2015;13:712–722. doi: 10.1016/j.celrep.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 24.Rokkam V.R., Kotagiri R. StatPearls. StatPearls Publishing; 2021. Secondary thrombocytosis; pp. 1–7. Treasure Island, FL. [Google Scholar]
- 25.Koulnis M., Pop R., Porpiglia E., Shearstone J.R., Hidalgo D., Socolovsky M. Identification and analysis of mouse erythroid progenitors using the CD71/TER119 flow-cytometric assay. J Vis Exp. 2011;5:2809. doi: 10.3791/2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Socolovsky M., Nam H., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 27.Donehower L.A. The p53-deficient mouse: a model for basic and applied cancer studies. Semin. Cancer Biol. 1996;7:269–278. doi: 10.1006/scbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 28.Lindstrom M.S., Jin A., Deisenroth C., White Wolf G., Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and Attenuate MDM2-induced p53 degradation. Mol. Cell. Biol. 2007;27:1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macias E., Jin A., Deisenroth C., Bhat K., Mao H., Lindstrom M.S., et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., He Y., Jin A., Tikunov A.P., Zhou L., Tollini L.A., et al. Ribosomal protein-Mdm2-P53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2414–E2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaako P., Debnath S., Olsson K., Zhang Y., Flygare J., Lindstrom M.S., et al. Disruption of the 5S RNP-Mdm2 interaction significantly improves the erythroid defect in a mouse model for Diamond-Blackfan anemia. Leukemia. 2015;29:2221–2229. doi: 10.1038/leu.2015.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moniz H., Gastou M., Leblanc T., Hurtaud C., Cretien A., Lecluse Y., et al. Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012;3:e356. doi: 10.1038/cddis.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyner S.D., Venkatachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Cao I., Garcia-Cao M., Martin-Caballero J., Criado L.M., Klatt P., Flores J.M., et al. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perry R.P. Balanced production of ribosomal proteins. Gene. 2007;401:1–3. doi: 10.1016/j.gene.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yong Liu C.D., Zhang Yanping. RP–MDM2–p53 pathway: linking ribosomal biogenesis and tumor surveillance. Trends Cancer. 2016;2:191–204. doi: 10.1016/j.trecan.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutt S., Narla A., Lin K., Mullally A., Abayasekara N., Megerdichian C., et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mills E.W., Wangen J., Green R., Ingolia N.T. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep. 2016;17:1–10. doi: 10.1016/j.celrep.2016.08.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article. Raw data were generated at UNC Chapel Hill. Derived data supporting the findings of this study are available from the corresponding author Y. Z. on request.