Abstract
Objectives:
Cephalometric analysis is essential for diagnosis, treatment planning and outcome assessment of orthodontics and orthognathic surgery. Utilizing artificial intelligence (AI) to achieve automated landmark localization has proved feasible and convenient. However, current systems remain insufficient for clinical application, as patients exhibit various malocclusions in cephalograms produced by different manufacturers while limited cephalograms were applied to train AI in these systems.
Methods:
A robust and clinically applicable AI system was proposed for automatic cephalometric analysis. First, 9870 cephalograms taken by different radiography machines with various malocclusions of patients were collected from 20 medical institutions. Then 30 landmarks of all these cephalogram samples were manually annotated to train an AI system, composed of a two-stage convolutional neural network and a software-as-a-service system. Further, more than 100 orthodontists participated to refine the AI-output landmark localizations and retrain this system.
Results:
The average landmark prediction error of this system was as low as 0.94 ± 0.74 mm and the system achieved an average classification accuracy of 89.33%.
Conclusions:
An automatic cephalometric analysis system based on convolutional neural network was proposed, which can realize automatic landmark location and cephalometric measurements classification. This system showed promise in improving diagnostic efficiency in clinical circumstances.
Keywords: Deep Learning; Orthodontics; Diagnosis; X-Rays; Neural Networks, Computer
Introduction
Landmark localization in the cephalometric analysis is essential for orthodontic and orthognathic diagnosis, treatment planning, and outcome assessment. 1 However, projection from a three-dimensional skull to a two-dimensional cephalogram causes structure superposition and image distortion, inevitably leading to landmark definition disagreement. And even for well-trained orthodontists, it takes more than 0.5 h to manually locate all cephalometric landmarks. 1 Therefore, many efforts have been made to deal with this time-consuming, experience-dependent, and error-prone task.
In recent years, artificial intelligence (AI) has been widely applied in cephalometric analysis, and many traditional machine-learning 2–6 and deep learning methods 7–14 have been developed for AI-based systems. Traditional methods mainly focused on Random Forest combined with geometric or shape information. 6 However, in recent years, convolutional neural networks (CNNs) showed higher accuracy and robustness and have proved more promising for clinical application. 15
To date, many automatic systems for cephalometrics are still at the theoretical stage and haven’t been clinically applied for the incapability of processing different sources of cephalograms and patients with various malocclusions. This may be caused by undersampled, underrepresented data sets and inaccurate annotation of cephalogram training samples. Some researchers collected several hundreds of cephalograms through the Internet or from only one clinical institution, and thus the diversity of malocclusions and cephalogram machines was highly limited. 2,16–18 For instance, Kim et al collected 1000 cephalograms from only one institution, 18 and subjects with mixed dentition or dentofacial deformity were excluded.
Taken together, although considerable efforts have been invested in AI-based automatic landmark localization, an accurate, universal, and clinically applicable system still awaits to be developed. In this research, CephNet was developed based on a substantial amount of cephalograms for automatic cephalometric analysis. CephNet was constructed by two-layer cascaded CNNs and performed accurate cephalometric analysis. Furthermore, an interactive system was developed to refine CephNet through feedback from clinical applications.
Methods and materials
This study was approved by the Institutional Review Board (approval number WCHSIRB-D-2019–120). All procedures were carried out in accordance with approved guidelines, and the personal information of subjects was deleted.
Data collection and preprocessing
A total of 9870 digital cephalograms (568 ~ 2144 pixels wide and 570 ~ 2600 pixels high) were collected from 20 medical institutions in China (details about images and cephalography machines were in Table 1). To better simulate clinical situations, patients aged from 6 to 50 years old with all kinds of malocclusions and skeletal patterns were included. Cephalograms with insufficient resolutions to display soft and hard tissues were excluded.
Table 1.
Medical institutions and the cephalography machines
| Medical institutions | Number of images | Manufacture | Image size (pixels) |
|---|---|---|---|
| Clinic 1 | 1063 | VATECH | 1720*1080 |
| Clinic 2 | 3155 | J.MORITA | 1752*1537 |
| Clinic 3 | 960 | NEW TOM | 793*751 |
| Clinic 4 | 456 | NEW TOM | 2426*2339 |
| Clinic 5 | 233 | MEYER | 2685*2232 |
| Clinic 6 | 342 | VATECH | 2256*2304 |
| Clinic 7 | 221 | FUSSEN | 1201*779 |
| Clinic 8 | 110 | LARGEV | 2703*2040 |
| Clinic 9 | 109 | LARGEV2 | 1167*675 |
| Clinic 10 | 179 | MyRay | 792*749 |
| Clinic 11 | 334 | POINTNIX | 2648*2100 |
| Clinic 12 | 567 | RAYSCAN | 2000*2250 |
| Clinic 13 | 173 | VATECH | 2260*2304 |
| Clinic 14 | 300 | MeiYa | 3400*2800 |
| Clinic 15 | 121 | Orthopantomograph | 2832*2304 |
| Clinic 16 | 232 | CRANFX | 2144*2304 |
| Clinic 17 | 349 | VATECH | 2600*2000 |
| Clinic 18 | 167 | HDX WILL | 2836*2040 |
| Clinic 19 | 334 | Green Smart | 696*806 |
| Clinic 20 | 465 | MEYER | 1216*827 |
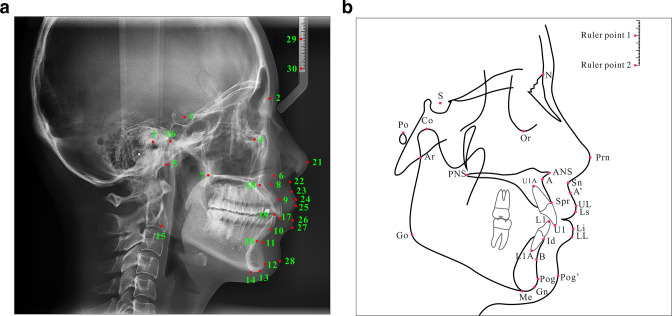
30 cephalometric landmarks (Figure 1, Table 2) in 9870 cephalograms were manually annotated by five experienced orthodontists as ground truth. The 30 landmarks included skeletal, dental, and soft-tissue landmarks and two ruler landmarks. Skeletal landmarks included S, N, Po, Or, Ar, ANS, PNS, A, SPr, B, Pog, Gn, Me, Go, Co. Dental landmarks included U1, UlA, L1, Id, L1A. Soft tissue landmarks included Prn, Sn, A', UL, Ls, Li, LL, Pog'.
Figure 1.
Location instructions and definition of the 30 cephalometric landmarks on cephalogram (A) and schematic (B), and the Ruler point one and Ruler point two were not trained.
Table 2.
Description of cephalometric landmarks
| No. | Landmark description | No. | Landmark description | |
|---|---|---|---|---|
| 1 | Sella (S) | 16 | Condylion (Co) | |
| 2 | Nasion (N) | 17 | Upper incisor (U1) | |
| 3 | Porion (Po) | 18 | Root apex of upper central incisor (U1A) | |
| 4 | Orbitale (Or) | 19 | Lower incisor (L1) | |
| 5 | Articulare (Ar) | 20 | Root apex of lower central incisor (L1A) | |
| 6 | Anterior nasal spine (ANS) | 21 | Pronasale (Prn) | |
| 7 | Posterior nasal spine (PNS) | 22 | Subnasale (Sn) | |
| 8 | Subspinale (A) | 23 | superior labial sulcus (A’) | |
| 9 | Superior prosthion (Spr) | 24 | Upper lip (UL) | |
| 10 | Infradentale (Id) | 25 | Labrale superius (Ls) | |
| 11 | Supramental (B) | 26 | Lower lip (LL) | |
| 12 | Pogonion (Pog) | 27 | Labrale inferius (Li) | |
| 13 | Gnathion (Gn) | 28 | Pogonion of soft tissue (Pog’) | |
| 14 | Menton (Me) | 29 | Ruler point 1 | |
| 15 | Gonion (Go) | 30 | Ruler point 2 |
To improve the accuracy and efficiency of annotation, a gold-standard data set was established to train a CNN model. Initially, 1063 cephalograms were annotated by an orthodontist with 15 years of experience (LJ) as a gold-standard data set to train and unify the location standard of the other four orthodontists. Both the intraclass consistency of the four orthodontists and the interclass consistency of them and LJ reached over 80% and has been shown in Table 3.
Table 3.
Reliability of manually annotated landmarks by four fellow orthodontists and the principal investigator
| Landmarks | Intraclass coefficient correlation (95% confidence interval) | Interclass coefficient correlation (95% confidence interval) | p |
|---|---|---|---|
| Sella | 0.912 (0.901–0.923) | 0.883 (0.849–0.976) | <0.05 |
| Nasion | 0.905 (0.802–0.998) | 0.873 (0.826–0.987) | |
| Porion | 0.836 (0.791–0.972) | 0.914 (0.892–0.976) | |
| Orbitale | 0.867 (0.801–0.992) | 0.803 (0.781–0.842) | |
| Articulare | 0.888 (0.859–0.961) | 0.893 (0.834–0.932) | |
| ANS | 0.854 (0.801–0.921) | 0.912 (0.881–0.945) | |
| PNS | 0.874 (0.821–0.923) | 0.893 (0.863–0.924) | |
| A | 0.852 (0.809–0.912) | 0.912 (0.871–0.967) | |
| Superior prosthion (SPr) | 0.882 (0.829–0.935) | 0.903 (0.867–0.949) | |
| B | 0.867 (0.819–0.949) | 0.883 (0.830–0.917) | |
| Pogonion | 0.843 (0.805–0.925) | 0.912 (0.867–0.959) | |
| Gn | 0.866 (0.798–0.942) | 0.892 (0.823–0.967) | |
| Me | 0.912 (0.871–0.967) | 0.873 (0.832–0.938) | |
| Go | 0.886 (0.823–0.961) | 0.912 (0.848–0.937) | |
| Co | 0.863 (0.831–0.949) | 0.894 (0.861–0.961) | |
| U1 | 0.914 (0.882–0.973) | 0.895 (0.872–0.942) | |
| U1A | 0.875 (0.810–0.928) | 0.854 (0.843–0.892) | |
| L1 | 0.906 (0.878–0.937) | 0.913 (0.867–0.967) | |
| Id | 0.886 (0.827–0.934) | 0.892 (0.803–0.958) | |
| L1A | 0.899 (0.865–0.949) | 0.874 (0.832–0.935) | |
| Prn | 0.873 (0.841–0.938) | 0.793 (0.781–0.834) | |
| Sn | 0.893 (0.869–0.923) | 0.885 (0.841–0.927) | |
| A’ | 0.872 (0.840–0.934) | 0.869 (0.821–0.903) | |
| UL | 0.891 (0.867–0.934) | 0.854 (0.830–0.871) | |
| Ls | 0.912 (0.863–0.957) | 0.852 (0.829–0.876) | |
| Li | 0.904 (0.878–0.967) | 0.877 (0.843–0.891) | |
| LL | 0.888 (0.847–0.936) | 0.865 (0.832–0.901) | |
| Pog’ | 0.873 (0.851–0.930) | 0.894 (0.834–0.935) |
Based on the manually annotated data set, a CNN model was trained to roughly annotate the remained cephalograms. Subsequently, all annotated data sets including the gold-standard data set were mixed and each landmark was refined by a corresponding orthodontist with one landmark a round using Colabeler (v. 2.0.4, Hangzhou kuaiyi Technology Co., Ltd. Hangzhou, China). In the next stage, 100 orthodontists were recruited and a SaaS system was developed for further fine adjustment of landmark locations. The SaaS system was built using the Java web framework and run on Alibaba Cloud servers in China (https://www.aliyun.com). Each orthodontist was assigned a unique system account with specific authority to part of the data, and they adjusted the landmark locations through web browsers.
Considering actual clinical situations, image augmentation was used to improve the model performance. In this study, selected geometric and photometric transformations were applied according to clinical conditions, 18 including rotation, translation, zoom, random color manipulation, noise injection, etc. The transformations were applied randomly within the maximum transformation value, and data sets increased 10–15 times to approximately 150,000.
Neural network architecture
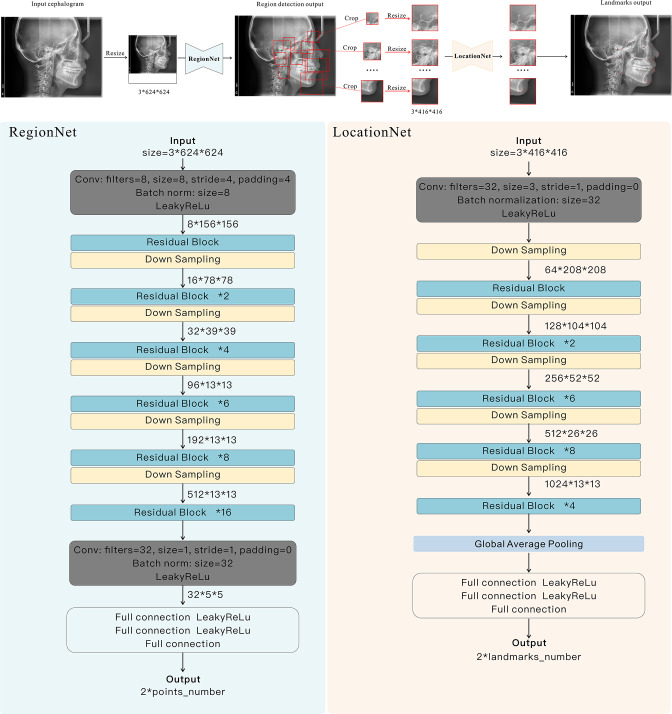
A cascade framework “CephNet” was built for landmark localization on digital cephalograms. The CephNet consisted of two-stage neural networks, the RegionNet and the LocationNet (Figure 2). In the first stage, RegionNet aimed to detect 10 regions of interest (ROIs), each containing 1–9 landmarks. In the second stage, LocationNet was proposed to accurately locate landmarks in the ROIs.
Figure 2.
Structure of CephNet. The network took lateral cephalograms as inputs, and adopted two connected convolutional streams (i.e. Region-stream and Location-stream) to learn global and local discriminative structural representations. RegionNet was constructed to locate the center points of ROIs, and local subimages were used for the accurate landmark localization by LocationNet.
The RegionNet was built to detect ROIs by predicting one representative point (center point) for each ROI. The network primarily comprised convolution layers, residual blocks, downsampling blocks, and fully connected layers. In each Convolutional layer, there were Batch Normalization (BN) and LeakyReLU operations.
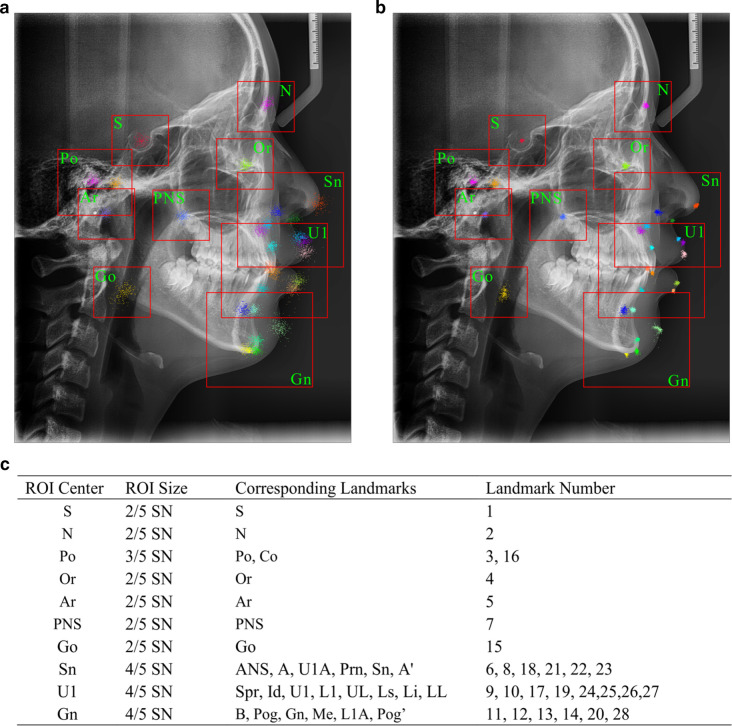
For each center point, square boxes with the different side lengths referring to SN (the distance between Sella and Nasion, which is easy to locate and stable in craniofacial growth) were captured as ROIs to analyze local contextual information. Closely connected landmarks were included in the same ROI (Figure 2). In total, 10 ROIs centered in S, N, Or, Po, PNS, Ar, Go, Gn, U1, and Sn were detected, and the landmarks in each ROI were shown in Figure 3.
Figure 3.
The centers, sizes, and corresponding landmarks of ROIs. (A) ROIs boxes drawn in cephalogram with the prediction error of one-stage network (RegionNet), (B) ROIs boxes drawn in cephalogram with the prediction error of CephNet, (C) the sizes and corresponding landmarks of ROIs. ROIs, regions of interest
Based on the result of RegionNet, subimages were cropped and resized to 416 × 416 pixels. Then, the LocationNet took these images as input to predict the coordinate offsets of certain number of landmarks. The LocationNet used an architecture similar to RegionNet, but shallower and simpler than RegionNet in order to limit model complexity because of the small image size.
Implementation and network training
259 cephalograms were randomly assigned to the test data set and the remaining 9611 samples to the training data set. 1000 cephalograms (10.40%) were randomly selected from the training data set as the validation set. The loss function was defined as follows:
Where,
n indicated the center points or landmarks in RegionNet and LocationNet; indicated the groundtruth x-axis coordinate value, indicated the predicted x-axis coordinate value; indicated the ground-truth y-axis coordinate value, indicated the predicted y-axis coordinate value, while and indicated the width and height of the cephalogram.
Model evaluation and accuracy assessment
An ablation experiment was conducted to validate the necessity of our network components. First, a one-stage network with the same architecture as RegionNet was built to locate all the landmarks, which was named as cNet. The RegionNet and cNet were trained and tested on the same data set. The landmark localization results were drawn relative to ground-truth positions of the respective image based on the error vectors from test data set. 6 The error vectors were normalized referring to SN in direction and ruler points in the distance.
To evaluate the landmark localization accuracy, the successful detection rates (SDR) of 28 landmarks (ruler points excluded) were assessed. Detection was regarded successful when the absolute deviation between the AI-predicted landmark and ground-truth landmark was less than α mm. α was set as 1 mm, 2 mm, and 3 mm, as studies have recognized landmark deviation of 2–3 mm as clinically acceptable. 15,19 Further, 11 cephalometric measurements were conducted based on landmark locations, including skeletal measurements (SNA, SNB, ANB, MP-SN, FMA), dental measurements (U1-NA, L1-NB, U1-SN, L1-MP), and soft-tissue measurements (UL-EP, LL-EP). For clinical cephalometric analysis, absolute values of measurements were less important than the classification results, e.g. ANB value was used to classify skeletal patterns of patients of Class I, II, III. Therefore, the accuracy of the cephalometric analysis was tested by the success classification rates (SCRs). 5 And the standard values wildly used in China were applied to classify the Lower, Normal, and Higher groups.
Results
Evaluation of the two-stage network
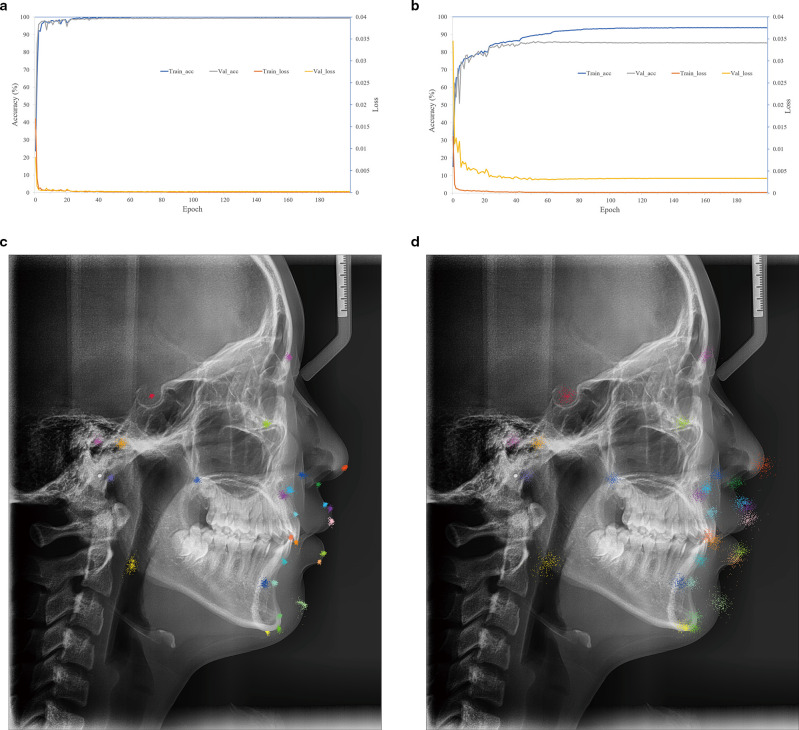
CephNet was a two-stage cascade CNN, in which the first stage (RegionNet) detected ROIs, and the second stage (LocationNet) located landmarks. As shown in Figure 4A, the train and validation loss steeply decreased; meanwhile, the accuracy rapidly increased during the first 30 epochs. After 200 epochs, the training accuracy reached 99.99%, and the validation reached 99.29%. Landmark prediction errors vectors of 259 test images concentrated around the ground-truth landmark (Figure 4C), indicating the LocationNet successfully learned local features from the ROIs to accurately locate landmarks.
Figure 4.
Performance comparison between the two-stage CephNet and one-stage cNet. (A) Loss and accuracy of LocationNet on training and validation data set for S (Sella); (B) loss and accuracy of the cNet on training and validation data set for 28 landmarks; (C) qualitative results for 28 landmarks on test data set by two-stage network, and all error vectors referring to SN were drawn relative to the ground-truth landmark positions of the respective cephalogram; (D) qualitative results for 28 landmarks on test dataset by cNet. For the accuracy, and the landmark prediction error ≤2 mm was defined as true perdition. Train_acc: training accuracy; Train_loss: training loss; Val_ acc: validation accuracy; Val_ loss: validation loss.
The ablation study that applied cNet to locate all the 28 landmarks showed that the training and validation accuracy only reached 93.88 and 85.33% respectively after 200 epochs (Figure 4B), and the prediction errors were not clinically acceptable (Figure 4D). It illustrated that one-step network (such as RegionNet) was insufficient to accurately locate landmarks, and the ROIs-based LocationNet was necessary.
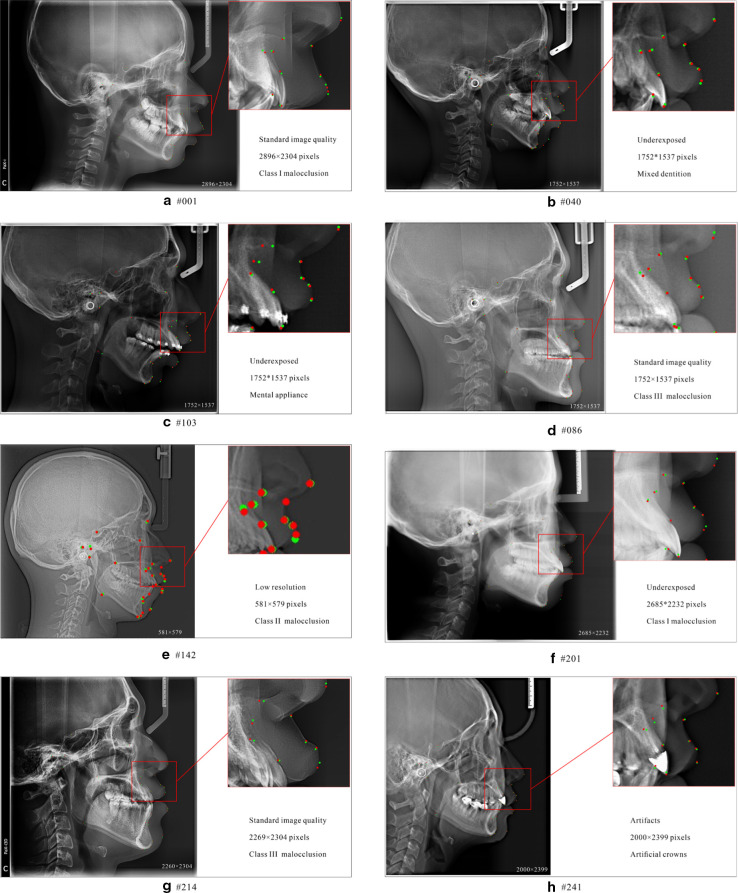
To further verify the generalization and stability of CephNet, more complicated cases were tested. As showed in Figure 5, CephNet showed robust performance in cephalograms with various malocclusions (Angle Class I, II or III), oral conditions (mixed and permanent dentition, restorations, orthodontic appliance) or imaging quality (different brightness, exposure, or artifacts).
Figure 5.
Cephalometric landmark localization samples on test data set. Green dots represent groundtruth landmarks and red dots represent predicted landmarks, and A ~ H showed the performance of CephNet in cephalograms with different quality, resolution, and malocclusions.
Accuracy of automatic landmark localization
Statistical prediction errors and success detection rates (SDR) of 28 landmarks were presented in Table 4. The average prediction error was 0.94 ± 0.74 mm. Except for Go (1.85 ± 1.16 mm) and Pog' (1.69 ± 1.28 mm), deviation of other 26 landmarks was less than 1.5 mm. And the least error was Sn (0.50 ± 0.29 mm).
Table 4.
The result of success detection rates for 28 landmarks
| Number | Landmarks | Success detection rate (%) | Mean (mm) |
SD (mm) |
||
|---|---|---|---|---|---|---|
| 1 mm | 2 mm | 3 mm | ||||
| 1 | S | 95.75 | 99.61 | 100.00 | 0.50 | 0.31 |
| 2 | N | 65.64 | 96.14 | 100.00 | 0.88 | 0.54 |
| 3 | Po | 45.95 | 83.40 | 93.05 | 1.32 | 0.95 |
| 4 | Or | 54.83 | 94.98 | 99.23 | 1.02 | 0.63 |
| 5 | Ar | 58.69 | 94.59 | 98.07 | 1.00 | 0.69 |
| 6 | ANS | 57.53 | 87.64 | 97.68 | 1.09 | 0.75 |
| 7 | PNS | 68.73 | 97.30 | 100.00 | 0.85 | 0.49 |
| 8 | A | 52.12 | 91.12 | 98.07 | 1.08 | 0.63 |
| 9 | Spr | 90.35 | 97.68 | 100.00 | 0.56 | 0.46 |
| 10 | Id | 76.06 | 97.30 | 99.61 | 0.73 | 0.52 |
| 11 | B | 54.05 | 94.98 | 99.23 | 1.00 | 0.59 |
| 12 | Pog | 75.68 | 96.14 | 99.61 | 0.72 | 0.55 |
| 13 | Gn | 73.75 | 96.53 | 100.00 | 0.77 | 0.51 |
| 14 | Me | 72.59 | 98.46 | 99.61 | 0.85 | 0.51 |
| 15 | Go | 28.57 | 57.14 | 86.10 | 1.85 | 1.16 |
| 16 | Co | 35.14 | 79.15 | 95.37 | 1.44 | 0.84 |
| 17 | U1 | 92.28 | 98.07 | 98.84 | 0.57 | 0.49 |
| 18 | U1A | 43.63 | 86.49 | 98.46 | 1.24 | 0.69 |
| 19 | L1 | 83.78 | 97.68 | 99.61 | 0.69 | 0.44 |
| 20 | L1A | 33.59 | 73.75 | 94.98 | 1.48 | 0.88 |
| 21 | Prn | 45.95 | 91.12 | 100.00 | 1.12 | 0.59 |
| 22 | Sn | 93.44 | 100.00 | 100.00 | 0.50 | 0.29 |
| 23 | A' | 90.35 | 99.23 | 100.00 | 0.55 | 0.33 |
| 24 | UL | 81.08 | 98.84 | 100.00 | 0.68 | 0.43 |
| 25 | Ls | 65.25 | 93.44 | 99.23 | 0.94 | 0.63 |
| 26 | Li | 83.78 | 99.23 | 100.00 | 0.68 | 0.40 |
| 27 | LL | 97.68 | 100.00 | 100.00 | 0.43 | 0.24 |
| 28 | Pog’ | 35.91 | 68.34 | 86.87 | 1.69 | 1.28 |
| Average | 66.15 | 91.73 | 97.99 | 0.94 | 0.74 | |
The average SDR within 1 mm reached 66.15%, and within 2 mm reached 91.73% respectively. The most accurate landmarks (S, Spr, U1, Sn, A', and LL) showed more than 90% SDR within 1 mm, and the least accurate point (Go) was 57.14% SDR within 2 mm, and 86.10% within 3 mm.
Landmarks surrounding stable and clear anatomical structures exhibited higher SDR than landmarks interfered by overlapped anatomical structures. Besides, as landmark definitions of “the most front” or “the most convex” point was difficult to unify, larger error was shown in Prn and Pog’. Further, the gathered error vectors revealed the error distribution and influence factors (Figure 4C and D). The error vectors of Go were distributed around the mandibular angle along the airway, indicating the location was disturbed by the overlap of bilateral mandibular angles and low-density shadows of the airway.
Accuracy of cephalometric analysis
To verify the clinical value of this system, the diagnostic accuracy of eleven clinical measurements was tested. The comparison between CephNet and manual measurement in terms of SCR were shown in Table 5. The SCRs of all measurements exceeded 84%, and the average SCR of CephNet was 89.33%. Among the three subgroups, the Normal group showed more than 85% of SCR, but the SCR of Lower and Higher groups varied from 63.64 to 100.00%, which might be related to insufficient test samples.
Table 5.
The success classification rates of 11 cephalometric measurements
| Measurements | Ground truth | AI prediction | Accuracy | Total (%) | ||
|---|---|---|---|---|---|---|
| n | True | False | ||||
| SNA (°) | 88.03% | |||||
| Lower (<78.8) | 66 | 42 | 24 | 63.64% | ||
| Normal (78.8–86.8) | 177 | 173 | 4 | 97.74% | ||
| Higher (>86.8) | 16 | 13 | 3 | 81.25% | ||
| SNB (°) | 88.80% | |||||
| Lower (<76.2) | 94 | 76 | 18 | 80.85% | ||
| Normal (76.2–84) | 149 | 141 | 8 | 94.63% | ||
| Higher (>84) | 16 | 13 | 3 | 81.25% | ||
| ANB (°) | 84.56% | |||||
| Lower (<0.7) | 53 | 41 | 12 | 77.36% | ||
| Normal (0.7–4.7) | 129 | 110 | 19 | 85.27% | ||
| Higher (>4.7) | 77 | 68 | 9 | 88.31% | ||
| MP-SN (°) | 90.35% | |||||
| Lower (<31.0) | 69 | 69 | 0 | 100.00% | ||
| Normal (31.0–39.0) | 136 | 122 | 14 | 89.71% | ||
| Higher (>39.0) | 54 | 43 | 11 | 79.63% | ||
| FMA (°) | 88.03% | |||||
| Lower (<21.2) | 56 | 52 | 4 | 92.86% | ||
| Normal (21.2–33.4) | 177 | 156 | 21 | 88.14% | ||
| Higher (>33.4) | 26 | 20 | 6 | 76.92% | ||
| U1-NA (mm) | 84.17% | |||||
| Lower (<2.7) | 45 | 34 | 11 | 75.56% | ||
| Normal (2.7–7.5) | 147 | 130 | 17 | 88.44% | ||
| Higher (>7.5) | 67 | 54 | 13 | 80.60% | ||
| L1-NB (mm) | 89.96% | |||||
| Lower (<4.6) | 88 | 80 | 8 | 90.91% | ||
| Normal (4.6–8.8) | 139 | 128 | 11 | 92.09% | ||
| Higher (>8.8) | 32 | 25 | 7 | 78.13% | ||
| U1-SN (°) | 87.64% | |||||
| Lower (<99.4) | 61 | 53 | 8 | 86.89% | ||
| Normal (99.4–112) | 132 | 123 | 9 | 93.18% | ||
| Higher (>112) | 66 | 51 | 15 | 77.27% | ||
| L1-MP (°) | 93.44% | |||||
| Lower (<89.4) | 67 | 66 | 1 | 98.51% | ||
| Normal (89.4–103.6) | 155 | 142 | 13 | 91.61% | ||
| Higher (>103.6) | 37 | 34 | 3 | 91.89% | ||
| UL-EP (mm) | 92.28% | |||||
| Lower (<−2.0) | 85 | 72 | 13 | 84.71% | ||
| Normal (-2.0–2.0) | 166 | 161 | 5 | 96.99% | ||
| Higher (>2.0) | 8 | 6 | 2 | 75.00% | ||
| LL-EP (mm) | 95.37% | |||||
| Lower (<0.0) | 86 | 82 | 4 | 95.35% | ||
| Normal (0.0–6.0) | 167 | 160 | 7 | 95.81% | ||
| Higher (>6.0) | 6 | 5 | 1 | 83.33% | ||
| Average | 89.33% | |||||
Discussion
In this research, a novel and accurate system for automatic cephalometric landmark location and analysis were developed based on a two-stage CephNet. This system was capable of identifying different cephalograms regardless of the cephalography machines or craniomaxillofacial variability of patients. In the multicenter test data set, the average location error of this system was 0.94 ± 0.74 mm, outperforming most previous studies with an average location error of about 2 mm. 16 A study using the SOTA method reached a similar level of average location error of 0.79 ± 0.91 mm with this study, but the researchers only included subjects over 14 years old without dentofacial deformity, which didn’t completely apply to clinical practice. 18 Based on cephalometric landmark localization, this system showed high SCR (89.33%) in 11 commonly used cephalometric measurements 19 and was sufficient for clinical application.
The accuracy and robustness of CephNet was mainly attributed to the highly qualified data sets and advanced network architecture. The data set of this study contained 9870 cephalograms taken by different cephalography machines with various image sizes and quality, and the craniofacial deformities of patients were highly diverse. Previous studies included much fewer cephalograms than this study, and the cephalograms were of identical size from a sole source. 5,16,18,20 Also, the high consistency of manual landmark annotation among the five orthodontists also guaranteed the quality of training samples for this system.
On the other hand, a staged procedure was performed in this study for high-quality landmark annotation. Similar to this study, some automatic cephalometric systems split the landmark localization process into two steps: first detection of ROIs and then landmark localization within ROIs. This method significantly increased the signal-to-noise ratio. 12–16 However, previous studies ignored the spatial and anatomical relationship between ROIs, and the specific size of the ROIs was not clearly defined. Some researchers assigned each landmark to one ROI, 12,14,16,18,20 while this study included anatomically related landmarks in the same ROI. This initiative largely decreased the computation work and simplified the ROI detection process. Also, the sizes of ROIs of different landmarks in previous studies were identical, 20 while this study set three different ROI sizes for landmark groups. As anatomical structures around each landmark were different, the shape and size of ROIs also need to be adaptive. On the other hand, as cephalogram samples were of different magnifications and resolutions, ROI sizes based on pixels would produce different content for one landmark and compromise the results of landmark localization training. In this study, as S and N points in cephalograms have long been recognized as stable in growth and also clear enough for localization, 21,22 SN distance was applied as a reference to set ROIs sizes. This initiative significantly increased the consistency of sizes and locations of ROIs of different cephalograms. On one hand, the pixel distance of SN was proportionate with cephalograms regardless of image magnification. On the other hand, as actual SN distance was relatively stable among populations, sizes of ROIs of the same landmark in different cephalograms could be unified.
Conclusion
In conclusion, this automatic cephalometric analysis system supported by ample training samples and an innovative algorithm showed high accuracy and applicability. This system is expected to facilitate orthodontists and significantly enhanced their efficiency at work.
Footnotes
Acknowledgments: The authors thank the clinics and medical institution for providing the cephalograms and the orthodontists who use our SaaS system and provide clinical evaluation and feedback. The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Conflicts of Interest: The authors declare that they have no competing interests.
Funding: This research was funded by Research and Develop Program, West China Hospital of Stomatology Sichuan University, NO. LCYJ2019-22; National Natural Science Foundation of China, No. 31971240; Chengdu artificial intelligence application and development industrial technology basic public service platform, No. 2021-0166-1-2, and Major Special Science and Technology Project of Sichuan Province, No. 2022ZDZX0031.
Ethics approval and consent to participate: Informed consent was obtained from all subjects involved in the study.
Consent for publication: Written informed consent has been obtained from the patients to publish this paper.
Fulin Jiang and Yutong Guo have contributed equally to this study and should be considered as co-first authors.
Availability of data and materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
Contributor Information
Fulin Jiang, Email: 875668697@qq.com.
Yutong Guo, Email: guoyutong6@126.com.
Cai Yang, Email: 404265070@qq.com.
Yimei Zhou, Email: 478600113@qq.com.
Yucheng Lin, Email: 719104558@qq.com.
Fangyuan Cheng, Email: 986587808@qq.com.
Shuqi Quan, Email: 552119979@qq.com.
Qingchen Feng, Email: 784597463@qq.com.
Juan Li, Email: lijuan@scu.edu.cn.
REFERENCES
- 1. Broadbent BH. A new x-ray technique and its application to orthodontia. The Angle Orthodontist. 1931;1(2):45-66, [Google Scholar]
- 2. Wang C-W, Huang C-T, Lee J-H, Li C-H, Chang S-W, Siao M-J, et al. A benchmark for comparison of dental radiography analysis algorithms. Med Image Anal 2016; 31: 63–76. doi: 10.1016/j.media.2016.02.004 [DOI] [PubMed] [Google Scholar]
- 3. El-Fegh I, Galhood M, Sid-Ahmed M, Ahmadi M. Automated 2-D cephalometric analysis of X-ray by image registration approach based on least square approximator. Annu Int Conf IEEE Eng Med Biol Soc 2008; 2008: 3949–52. doi: 10.1109/IEMBS.2008.4650074 [DOI] [PubMed] [Google Scholar]
- 4. Ibragimov B, Likar B, Pernus F, editors . Vrtovec T. Computerized cephalometry by game theory with shape-and appearance-based landmark refinement Proceedings of International Symposium on Biomedical imaging (ISBI). ; 2015. [Google Scholar]
- 5. Lindner C, Wang C-W, Huang C-T, Li C-H, Chang S-W, Cootes TF. Fully automatic system for accurate localisation and analysis of cephalometric landmarks in lateral cephalograms. Sci Rep 2016; 6(1): 33581. doi: 10.1038/srep33581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Urschler M, Ebner T, Štern D. Integrating geometric configuration and appearance information into a unified framework for anatomical landmark localization. Med Image Anal 2018; 43: 23–36: S1361-8415(17)30134-2. doi: 10.1016/j.media.2017.09.003 [DOI] [PubMed] [Google Scholar]
- 7. Arık SÖ, Ibragimov B, Xing L. Fully automated quantitative cephalometry using convolutional neural networks. J Med Imaging (Bellingham) 2017; 4: 014501. doi: 10.1117/1.JMI.4.1.014501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Armato SG, Petrick NA, Lee H, Park M, Kim J. Cephalometric landmark detection in dental x-ray images using convolutional neural networks. In: Lee H, Park M, Kim J, eds. SPIE Medical Imaging; Orlando, Florida, United States. ; 3 March 2017. doi: 10.1117/12.2255870 [DOI] [Google Scholar]
- 9. Lee J-H, Yu H-J, Kim M-J, Kim J-W, Choi J. Automated cephalometric landmark detection with confidence regions using Bayesian convolutional neural networks. BMC Oral Health 2020; 20(1): 270. doi: 10.1186/s12903-020-01256-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noothout JMH, De Vos BD, Wolterink JM, Postma EM, Smeets PAM, Takx RAP, et al. Deep learning-based regression and classification for automatic landmark localization in medical images. IEEE Trans Med Imaging 2020; 39: 4011–22: 12. doi: 10.1109/TMI.2020.3009002 [DOI] [PubMed] [Google Scholar]
- 11. Oh K, Oh I-S, Le VNT, Lee D-W. Deep anatomical context feature learning for cephalometric landmark detection. IEEE J Biomed Health Inform 2021; 25: 806–17. doi: 10.1109/JBHI.2020.3002582 [DOI] [PubMed] [Google Scholar]
- 12. Payer C, Štern D, Bischof H, Urschler M. Integrating spatial configuration into heatmap regression based cnns for landmark localization. Med Image Anal 2019; 54: 207–19: S1361-8415(18)30578-4. doi: 10.1016/j.media.2019.03.007 [DOI] [PubMed] [Google Scholar]
- 13. Zeng M, Yan Z, Liu S, Zhou Y, Qiu L. Cascaded convolutional networks for automatic cephalometric landmark detection. Med Image Anal 2021; 68: 101904: S1361-8415(20)30268-1. doi: 10.1016/j.media.2020.101904 [DOI] [PubMed] [Google Scholar]
- 14. Zhang J, Liu M, Shen D. Detecting anatomical landmarks from limited medical imaging data using two-stage task-oriented deep neural networks. IEEE Trans Image Process 2017; 26: 4753–64. doi: 10.1109/TIP.2017.2721106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park J-H, Hwang H-W, Moon J-H, Yu Y, Kim H, Her S-B, et al. Automated identification of cephalometric landmarks: part 1-comparisons between the latest deep-learning methods YOLOV3 and SSD Angle Orthod 2019; 89: 903–9. doi: 10.2319/022019-127.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J, Kim I, Kim Y-J, Kim M, Cho J-H, Hong M, et al. Accuracy of automated identification of lateral cephalometric landmarks using cascade convolutional neural networks on lateral cephalograms from nationwide multi-centres. Orthod Craniofac Res 2021; 24 Suppl 2: 59–67. doi: 10.1111/ocr.12493 [DOI] [PubMed] [Google Scholar]
- 17. Nishimoto S, Sotsuka Y, Kawai K, Ishise H, Kakibuchi M. Personal computer-based cephalometric landmark detection with deep learning, using cephalograms on the Internet. J Craniofac Surg 2019; 30: 91–95. doi: 10.1097/SCS.0000000000004901 [DOI] [PubMed] [Google Scholar]
- 18. Kim IH, Kim YG, Kim S, Park JW, Kim N. Comparing intra-observer variation and external variations of a fully automated cephalometric analysis with a cascade convolutional neural net. Sci Rep 2021; 11(1): 7925. doi: 10.1038/s41598-021-87261-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hwang HW, Moon JH, Kim MG, Donatelli RE, Lee SJ. Evaluation of automated cephalometric analysis based on the latest deep learning method. Angle Orthod 2021; 91: 329–35. doi: 10.2319/021220-100.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim YH, Lee C, Ha EG, Choi YJ, Han SS. A fully deep learning model for the automatic identification of cephalometric landmarks. Imaging Sci Dent 2021; 51: 299–306. doi: 10.5624/isd.20210077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Afrand M, Ling CP, Khosrotehrani S, Flores-Mir C, Lagravère-Vich MO. Anterior cranial-base time-related changes: a systematic review. Am J Orthod Dentofacial Orthop 2014; 146: 21–32. doi: 10.1016/j.ajodo.2014.03.019 [DOI] [PubMed] [Google Scholar]
- 22. Bastir M, Rosas A, O’higgins P. Craniofacial levels and the morphological maturation of the human skull. J Anat 2006; 209: 637–54. doi: 10.1111/j.1469-7580.2006.00644.x [DOI] [PMC free article] [PubMed] [Google Scholar]