Abstract
Background
Coronavirus disease 2019 (COVID-19) is currently a global pandemic. The pathogenesis of severe COVID-19 has been widely investigated, but it is still unclear. Human leukocyte antigen (HLA) plays a central role in immune response, and its variants might be related to COVID-19 progression and severity.
Objective
To investigate the hypothesis that individual HLA variations could alter the course of COVID-19 and might be associated with the severity of COVID-19.
Methods
In this study, we conducted an HLA targeted capture enrichment and sequencing of severe COVID-19 patients matched to mild cases. A total of 16 COVID-19 patients, confirmed by SARS-CoV-2 viral RNA polymerase-chain-reaction (PCR) test and chest computed tomography (CT) scan, were enrolled in this study. The HLA targeted capture enrichment and sequencing were conducted. HLA typing was performed by comparing contigs with IPD-IMGT/HLA Database.
Results
In this study, 139 four-digit resolution HLA alleles were acquired. The results showed that HLA-DRB3*01:01 allele was significantly associated with the severity of COVID-19 (odds ratio [OR] = 27.64, 95% confidence interval [CI] = 1.35–560.50, P = 0.0064). And HLA-K*01:01 might be a potential risk factor for COVID-19 severity (OR = 0.11, 95% CI = 0.017–0.66, P = 0.019), but HLA-K*01:02 might be a protective factor (OR = 7.50, 95% CI = 1.48–37.92, P = 0.019).
Conclusion
Three non-classical HLA alleles, including HLA-DRB3*01:01, HLA-K*01:01, HLA-K*01:02 were identified to be associated with the severity of COVID-19 by comparing mild and severe patients. The current findings would be helpful for exploring the influence of HLA gene polymorphisms on the development and severity of COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13258-022-01358-2.
Keywords: Coronavirus disease 2019 (COVID-19), Human leukocyte antigen (HLA), Allele frequency, Disease association, HLA-DRB3, HLA-K
Introduction
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was first reported at the end of December 2019 in Wuhan, China (Zhu et al. 2020), and it has become a global pandemic (Wang et al. 2020a). According to the reports from the World Health Organization, there were 640,395,651 confirmed cases and 6,618,579 deaths by 2 December 2022. The result of the epidemiological survey showed that the incubation period was 1–14 days, and the median incubation period was only about 5 days (Lauer et al. 2020). Generally, people infected with SARS-CoV-2 were moderate or asymptomatic symptoms (Lai et al. 2020). However, some patients developed severe diseases and even acute respiratory distress syndrome (Zhang et al. 2020). Although the pathogenesis of severe COVID-19 has been widely investigated, the underlying mechanisms are still unclear.
The major histocompatibility complex (MHC), also known as human leukocyte antigen (HLA) in humans, is located in the 6p21.3 region of chromosome 6, including a series of closely linked loci. Human leukocyte antigen (HLA) plays an essential role in immune regulation. The polymorphisms of HLA are significantly related to the susceptibility and severity of many diseases, such as tumors (Sadagopan et al. 2022), autoimmune diseases (Naito and Okada 2022), immunodeficiency diseases (Grifoni et al. 2015), allergic diseases (Daya et al. 2021), infectious diseases (Medhasi and Chantratita 2022), metabolic diseases (Tekola Ayele et al. 2012), and so on. Each person has multiple alleles that make up the individual HLA type. A person’s HLA allele might affect the immune system’s response to the SARS-CoV-2 virus. It would mean that the immune system of people with specific HLA types might be more sensitive than others in detecting the presence of viruses, thus resulting in better or worse immune responses. The first report about HLA genetic variation affecting the susceptibility to SARS-CoV-2 and the severity of infection was an in silico analysis. The result suggested that HLA-A, -B, and -C genes might affect susceptibility and severity of COVID-19 (Nguyen et al. 2020). The study of HLA variability might help us understand the differences in immune responses to SARS-CoV-2 infection and the clinical course of COVID-19. In this study, we conducted an HLA targeted capture enrichment and sequencing of severe COVID-19 patients matched to mild cases to determine the association of HLA allelic variations in the susceptibility and severity of COVID-19.
Materials and methods
Patient involvement and diagnostic standards
The COVID-19 patients, enrolled in this study, were admitted into the First Affiliated Hospital of Bengbu Medical College from January 22 to March 4, 2020. According to “New Coronary Virus Pneumonia Treatment plan (trial version 7)” issued by National Health and Care Commission, the samples of throat swabs and/or lower respiratory tract secretions were detected by real-time fluorescence PCR and chest computed tomography (CT) scan. These participants did not have diabetes, hypertension, cancer, cardiovascular disease, or immune-related diseases. The following criteria were used to character the severe COVID-19 patient: (1) shortness of breath, with a respiratory rate ≥ 30 breaths/min; (2) percutaneous oxygen saturation (SpO2) ≤ 93% on room air at rest; (3) arterial oxygen tension (PaO2)/ inspiratory oxygen fraction (FIO2) ≤ 300 mmHg. This study was approved by the institutional ethics board of the First Affiliated Hospital of Bengbu Medical College (No. 2020KY010).
HLA-targeted sequencing
Peripheral blood samples were collected from mild or severe patients to extract DNA. Genomic DNA was quantified using agarose gel (1%) electrophoresis, and DNA concentration was qualified by Qubit® DNA Assay Kit (cat. no. Q33231) in Qubit®3.0 Fluorometer (Life Technologies, CA, USA). The HLA sequences were efficiently enriched using Roche’s NimbleGen capture technology (Roche Technologies, WI, USA). The library and capture experiments were performed using NimbleGen SeqCap EZ Choice Library Kit (cat. no. 06266304001). Fragments between 180 and 280 bp in length were extracted and sequenced using the Illumina HiSeq X Ten system. The HLA-target sequences were sequenced to 100-fold with a coverage of 98% in this study.
HLA typing
The sequencing error rate, data volume, and mapping rate were calculated to evaluate the quality of the sequencing database. The valid sequencing data were aligned by Burrows-Wheeler Aligner (Li and Durbin 2010) to the reference genome (GRCh37/hg19), and the initial alignment results of the binary alignment map (BAM) format were obtained. HLA typing was performed by comparing contigs with currently known HLA sequences in the IPD-IMGT/HLA Database (Robinson et al. 2020).
The variability of HLA in the general population
A larger national HLA data set was the control group, which comprised 10,689 individuals of the Chinese population (Zhou et al. 2016). This work presented a complete polymorphism map of the MHC region mutation loci and HLA genes in the Chinese population by accurate sequence analysis and genotyping of the MHC region. The average sequence coverage over the whole MHC region was 55×, which was the largest dataset of variants in the MHC region of the Chinese population. The raw sequencing data from samples evaluated in the Han-MHC project could be downloaded from the Sequence Read Archive (SRA) with the accession SRA205317.
Statistical analysis
Statistical analyses were performed using R (ver 3.6.3) software. Continuous variables were expressed as means and standard deviations (SD). Categorical variables were expressed as counts and percentages (%). For continuous variables, the Kruskal Wallis rank-sum test was used to calculate the p-value. The Fisher’s exact probability test was used to compare the distribution of HLA allele frequencies in COVID-19 patients and control individuals. In this study, a total of 10,689 Chinese individuals were used as the control group. P-values were adjusted with the Benjamini–Hochberg method to evaluate the distribution of HLA allele frequencies in COVID-19 patients and a larger Chinese control population. The frequencies of HLA alleles were compared between mild and severe patients using the Fisher’s exact test. The odds ratio (OR) with a confidence interval (CI) of 95% were also calculated. The P-value of less than 0.05 was considered statistically significant in this study.
Results
Clinical presentation
A total of 16 COVID-19 patients (including 8 mild and 8 severe patients) were enrolled in this study. The summary of the clinical features was provided in Table 1. Compared to mild patients, the values of neutrophils, lymphocytes, C-reactive protein, albumin, and neutrophil-to-lymphocyte ratio were significantly different in severe cases.
Table 1.
Clinical characteristics of patients with COVID-19 enrolled in this study
| Characteristic | Mild patients | Severe patients | p-value* |
|---|---|---|---|
| Number | 8 | 8 | |
| Age (year, Mean ± SD) | 43.25 ± 8.61 | 45.88 ± 11.00 | 0.603 |
| Gender (number) | 0.614 | ||
| Female | 4 | 3 | |
| Male | 4 | 5 | |
| Symptoms at admission (number) | |||
| Fever | 8 | 8 | 1.000 |
| Cough | 2 | 5 | 0.131 |
| Chest pain | 0 | 5 | 0.007 |
| Dyspnoea | 0 | 2 | 0.131 |
| Diarrhea | 1 | 4 | 0.106 |
| Laboratory findings (Mean ± SD) | |||
| White-cell count (x109/L) | 4.62 ± 1.58 | 6.64 ± 2.76 | 0.093 |
| Total neutrophils (x109/L) | 2.94 ± 1.42 | 5.50 ± 2.71 | 0.033 |
| Total lymphocytes (x109/L) | 1.22 ± 0.41 | 0.73 ± 0.23 | 0.011 |
| Platelet count (x10/L) | 228.25 ± 75.88 | 243.25 ± 85.60 | 0.716 |
| Alanine Aminotransferase (U/L) | 56.75 ± 66.44 | 57.13 ± 25.33 | 0.988 |
| Interleukin − 6 (pg/mL) | 3.51 ± 4.21 | 38.72 ± 50.94 | 0.075 |
| C-reactive protein (mg/L) | 7.56 ± 7.82 | 67.17 ± 70.92 | 0.033 |
| Creatine kinase (U/L) | 166.25 ± 251.25 | 49.13 ± 21.32 | 0.210 |
| Albumin (g/L) | 41.28 ± 4.42 | 35.48 ± 5.23 | 0.031 |
| Neutrophil to Lymphocyte Ratio | 2.80 ± 1.93 | 8.03 ± 4.42 | 0.008 |
*For continuous variables, the Kruskal Wallis rank-sum test was used to calculate the p-value. For categorical variables, the Fisher’s exact probability test was used to calculate the p-value. The p-value of less than 0.05 was considered statistically significant
HLA -targeted sequencing quality control and typing information
According to the sequencing feature of Illumina platforms, for paired-end sequencing data, Q30 (the percent of base pairs with Phred scores greater than 30) should be above 80%, and the average error rate should be below 0.1%. The sequencing error rate, data volume, and mapping rate were calculated to evaluate the quality of the sequencing database (Supplementary Table 1). The sequence data have been submitted to the NCBI SRA database (Accession: PRJNA641014). According to the degree of differentiation, HLA typing can be divided into four categories: two-digit precision, four-digit precision, six-digit precision, and eight-digit precision. The typing accuracy was generally up to four digits, and the highest precision was eight digits.
HLA allele information
In this study, 29 HLA gene loci were sequenced, including 8 classical genes (HLA-A, -B, -C, -DRB1, -DQA1, -DQB1, -DPA1, and -DPB1), and 21 non-classical genes (HLA-DMA, -DMB, -DOA, -DOB, -DRA, -DRB2, -DRB3, -DRB4, -DRB5, -DRB6, -DRB7, -DRB8, -DRB9, -E, -F, -G, -H, -J, -K, -L, and -V). A total of 139 four-digit resolution HLA alleles were acquired. The complete allele information was provided in Supplementary Table 2.
Distribution of HLA allele frequencies in COVID-19 patients
In this study, the allele distributions of HLA alleles were compared between COVID-19 patients and control individuals (n = 10,689), which were obtained from the Chinese population (Zhou et al. 2016). The resulting odds ratios and P values were presented in Supplementary Table 3. In Fisher’s exact test analysis, HLA-H*02:06, HLA-DRB3*01:01, HLA-DPB1*02:02, and HLA-K*01:03 frequencies were higher in COVID-19 patients than that in control individuals (adjusted P < 0.05) (Fig. 1). The detail information of 4 significantly different HLA alleles between the COVID-19 patients and the Chinese control population (adjusted P < 0.05) was listed in Table 2.
Fig. 1.
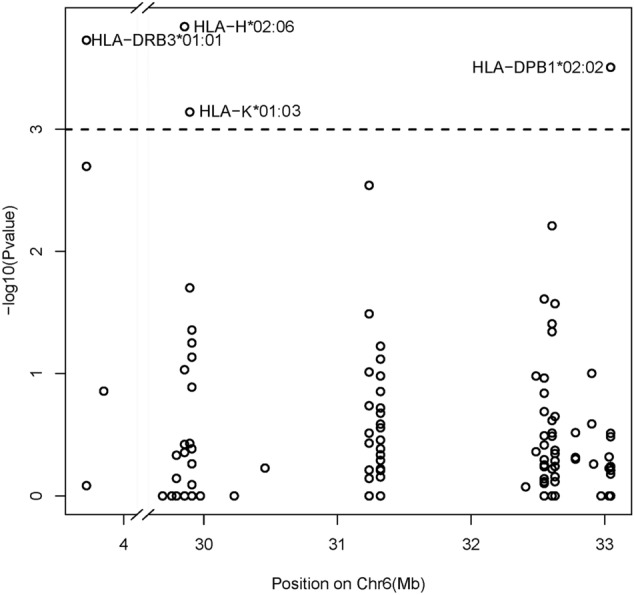
Association tests between all HLA alleles and the occurrence of COVID-19. The –log10(P value) of each HLA was plotted according to the physical location of the HLA region on GRCh37/hg19_chr6. The dashed line indicates the Bonferroni adjusted significance threshold (10− 3)
Table 2.
Distribution of HLA allele frequencies in COVID-19 patients and a larger Chinese control population
| Allele | COVID-19 patients (2 N = 32) no. (%) |
Control population (2 N = 21,378) no. (%) |
Odds ratios | P value | Adjusted P value |
|---|---|---|---|---|---|
| HLA-H*02:06 | 4 (12.5) | 176 (0.8) | 17.21 | 0.0001 | 0.0122 |
| HLA-DRB3*01:01 | 8 (25.0) | 1112 (5.2) | 6.07 | 0.0002 | 0.0122 |
| HLA-DPB1*02:02 | 8 (25.0) | 1199 (5.6) | 5.61 | 0.0003 | 0.0136 |
| HLA-K*01:03 | 4 (12.5) | 271 (1.3) | 11.13 | 0.0007 | 0.0236 |
| HLA-DRB3*03:01 | 9 (28.1) | 1986 (9.3) | 3.82 | 0.0020 | 0.0527 |
| HLA-C*03:03 | 8 (25.0) | 1689 (7.9) | 3.89 | 0.0029 | 0.0628 |
| HLA-DQA1*06:01 | 6 (18.8) | 1133 (5.3) | 4.12 | 0.0062 | 0.1153 |
| HLA-K*01:01 | 10 (31.3) | 11,316 (52.9) | 0.40 | 0.0198 | 0.3246 |
| HLA-DRB1*12:02 | 6 (18.8) | 1531 (7.2) | 2.99 | 0.0245 | 0.3504 |
| HLA-DQB1*03:01 | 12 (37.5) | 4404 (20.6) | 2.31 | 0.0267 | 0.3504 |
| HLA-C*01:06 | 1 (3.1) | 21 (0.1) | 32.81 | 0.0324 | 0.3857 |
| HLA-DQA1*03:02 | 7 (21.9) | 2175 (10.2) | 2.47 | 0.0391 | 0.4241 |
| HLA-A*24:07 | 1 (3.1) | 29 (0.1) | 23.75 | 0.0439 | 0.4241 |
| HLA-DQA1*05:05 | 5 (15.6) | 1322 (6.2) | 2.81 | 0.0453 | 0.4241 |
HLA variants associated with the severity of COVID-19
The Fisher’s exact test results showed that the associations of alleles of HLA-DRB3*01:01, HLA-K*01:01, and HLA-K*01:02 reached a significant difference between COVID-19 patients with mild or severe symptoms (Table 3). HLA-DRB3*01:01 allele was more significantly associated with the severity of COVID-19 (odds ratio [OR] = 27.64, 95% confidence interval [CI] = 1.35–560.50, P = 0.0064). Moreover, HLA-DRB3*01:01 allele was only found in severe patients. The comparative analysis showed that different alleles of HLA-K might play the opposite roles during the process of COVID-19. The frequency of HLA-K*01:01 was 12.5% in severe patients compared with 57.1% in mild ones (OR = 0.11, 95%CI = 0.017–0.66, P = 0.019). Meanwhile, the frequency of HLA-K*01:02 was significantly higher in severe patients (75.0%) than in mild cases (28.6%) (OR = 7.50, 95%CI = 1.48–37.92, P = 0.019).
Table 3.
Association test results for susceptibility HLA Loci with the severity of COVID-19
| Characteristic | Mild patients | Severe patients | p-value* |
|---|---|---|---|
| Number | 8 | 8 | |
| HLA allele frequency | |||
| HLA-DRB3*01:01 | 0.0% | 57.1% | 0.0064 |
| HLA-K*01:01 | 57.1% | 12.5% | 0.0187 |
| HLA-K*01:02 | 28.6% | 75.0% | 0.0261 |
*The frequencies of HLA alleles were compared between mild and severe patients using the Fisher’s exact test. The p-value of less than 0.05 was considered statistically significant
Discussion
COVID-19 is a newly emerging infectious disease, and the susceptibility of the population is generally high. From the response to the SARS-CoV-2 virus, it could be noticed that the response of the human population to infection was very diverse. Some people will be very vulnerable to infection, while others will become very susceptible. Although many researchers have been using genome-wide methods to find susceptibility variants to COVID1-19, others focus on the human leukocyte antigen (HLA) system. As we known, the variation of HLA is an important host genetic factor in determining the outcome of many infectious diseases. For example, HLA-B*07:03 and HLA-DRB1*03:01 were associated with susceptibility and resistance to the development of serve acute respiratory syndrome (SARS) (Ng et al. 2004). And HLA-DRB1*11:01 and DQB1*02:02 might be related to susceptibility to Middle East Respiratory Syndrome (MERS), caused by a beta coronavirus referred to as MERS-CoV (Hajeer et al. 2016).
Up to date, many studies have explored the influence of HLA genotypes on infection susceptibility and mortality of COVID-19. However, the associations between HLA gene polymorphism and SARS-CoV-2 infection or severity risk remain inconsistent. Based on the HLA allelic frequencies from 147 individuals of European descent with variable COVID-19 clinical outcomes, HLA-DRB1*04:01 was found three times as frequently in asymptomatic people, which suggested that people with this allele could protect themselves to some extent from severe symptoms of COVID-19 (Langton et al. 2021). The data from the Iranian population showed that HLA-A*01 and HLA-B*07 might be predominant in the COVID-19 deaths (Saadati et al. 2020). The frequency of HLA-A*01, B*56, and C*01 in Saudi patients was associated with the susceptibility to COVID-19 infection and outcome (Naemi et al. 2021). The data from 137 Japanese patients showed that HLA-A*11:01:01:01, HLA-C*12:02:02:01, and HLA-B*52:01:02:02 were significantly associated with the severity of COVID-19 (Khor et al. 2021). The data from 82 Chinese individuals suggested that B*15:27 alleles may be related to the occurrence of COVID-19 (Wang et al. 2020b). However, a recent report found the classical HLA loci had no significant allele associations with COVID-19 in patients from Italy and Spain (Ellinghaus et al. 2020). Among the Israeli population, no association was found between HLA haplotypes and SARS-CoV-2 infection or severity (Ben Shachar et al. 2021). The results might be biased due to different study designs, ethnic populations, and limited sample sizes.
In this study, we identified strong association between alleles in non-classical HLA genes and the severity of COVID-19. The data in the Table 3 showed that compared the alleles of HLA COVID-19 patients with mild symptoms to individuals with severe symptoms, the associations of alleles of HLA-DRB3*01:01, HLA-K*01:01, and HLA-K*01:02 reached a significant difference by using the Fisher’s exact test. The result suggested that HLA-DRB3*01:01 might be a potential genetic high-risk factor for the severity of COVID-19. HLA-DRB3 is one of the HLA class II beta chain paralogues. It has been identified that HLA-DRB3*01:01 was a potential risk factor for heparin-induced thrombocytopenia, which was an immune-mediated reaction to heparin (Karnes et al. 2017). HLA-K is one kind of pseudogene. There was no report on the association between the polymorphisms of HLA-K and human diseases. Here, we identified 2 HLA-K allelic associations with the severity of COVID-19, HLA-K*01:01 as a risk factor, and HLA-K*01:02 as a protective factor. The results of this study on the breast cancer survivors showed that the decreased HLA-K pseudogene expression was prognostic of poor patient survival and suggested that HLA-K might play a potential function via a pseudogene-gene interaction (Smerekanych et al. 2020).
In this study, the next-generation sequencing method was used to study different alleles of HLA genes. During the first blockade in China, this work was limited to samples from Bengbu city, which reduced the variation in the study group. A major limitation of this approach is the small number of patients. More donors and asymptomatic patients are needed to include in the association analysis.
In conclusion, three non-classical HLA alleles, including HLA-DRB3*01:01, HLA-K*01:01, HLA-K*01:02 were identified to be associated with the severity of COVID-19 by comparing mild and severe patients in this study. The sample size is a huge limitation for this study. Although the current result would need to be confirmed with larger sample sizes and different ethnic groups, the present findings would still be helpful for exploring the influence of HLA gene polymorphisms on the disease progression and severity of COVID-19.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank all the individuals for their participation in this study. This work was supported by the Research Foundation for Advanced Talents of Bengbu Medical College (grant number 15190016).
Declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ben Shachar S, Barda N, Manor S, Israeli S, Dagan N, Carmi S, Balicer R, Zisser B, Louzoun Y. MHC haplotyping of SARS-CoV-2 patients: HLA subtypes are not Associated with the Presence and severity of COVID-19 in the israeli population. J Clin Immunol. 2021;41:1154–1161. doi: 10.1007/s10875-021-01071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daya M, Cox C, Acevedo N, Boorgula MP, Campbell M, Chavan S, Cho MH, David GL, Kachroo P, Lasky-Su J, et al. Multiethnic genome-wide and HLA association study of total serum IgE level. J Allergy Clin Immunol. 2021;148:1589–1595. doi: 10.1016/j.jaci.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Fernandez J, Prati D, Baselli G, Asselta R, et al. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A, Montesano C, Colizzi V, Amicosante M. Key role of human leukocyte antigen in modulating human immunodeficiency virus progression: an overview of the possible applications. World J Virol. 2015;4:124–133. doi: 10.5501/wjv.v4.i2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajeer AH, Balkhy H, Johani S, Yousef MZ, Arabi Y. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Ann Thorac Med. 2016;11:211–213. doi: 10.4103/1817-1737.185756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnes JH, Shaffer CM, Cronin R, Bastarache L, Gaudieri S, James I, Pavlos R, Steiner HE, Mosley JD, Mallal S, et al. Influence of human leukocyte antigen (HLA) alleles and killer cell immunoglobulin-like receptors (KIR) types on heparin-induced thrombocytopenia (HIT) Pharmacotherapy. 2017;37:1164–1171. doi: 10.1002/phar.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor SS, Omae Y, Nishida N, Sugiyama M, Kinoshita N, Suzuki T, Suzuki M, Suzuki S, Izumi S, Hojo M, et al. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, age and sex are associated with severity of japanese COVID-19 with respiratory failure. Front Immunol. 2021;12:658570. doi: 10.3389/fimmu.2021.658570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CC, Liu YH, Wang CY, Wang YH, Hsueh SC, Yen MY, Ko WC, Hsueh PR. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J Microbiol Immunol Infect. 2020;53:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton DJ, Bourke SC, Lie BA, Reiff G, Natu S, Darlay R, Burn J, Echevarria C. The influence of HLA genotype on the severity of COVID-19 infection. HLA. 2021;98:14–22. doi: 10.1111/tan.14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, Azman AS, Reich NG, Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhasi S, Chantratita N. Human leukocyte antigen (HLA) system: genetics and association with bacterial and viral infections. J Immunol Res. 2022;2022:9710376. doi: 10.1155/2022/9710376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naemi FMA, Al-Adwani S, Al-Khatabi H, Al-Nazawi A. Frequency of HLA alleles among COVID-19 infected patients: preliminary data from Saudi Arabia. Virology. 2021;560:1–7. doi: 10.1016/j.virol.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito T, Okada Y. HLA imputation and its application to genetic and molecular fine-mapping of the MHC region in autoimmune diseases. Semin Immunopathol. 2022;44:15–28. doi: 10.1007/s00281-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MH, Lau KM, Li L, Cheng SH, Chan WY, Hui PK, Zee B, Leung CB, Sung JJ. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190:515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, Thompson RF. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J Virol. 2020;94:e00510–00520. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD-IMGT/HLA database. Nucleic Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadati M, Chegni H, Ghaffari AD, Mohammad Hassan Z. The potential association of human leukocyte antigen (HLA)-A and -B with COVID-19 mortality: a neglected risk factor. Iran J Public Health. 2020;49:2433–2434. doi: 10.18502/ijph.v49i12.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadagopan A, Michelakos T, Boyiadzis G, Ferrone C, Ferrone S. Human leukocyte antigen class I antigen-processing machinery upregulation by anticancer therapies in the era of checkpoint inhibitors: a review. JAMA Oncol. 2022;8:462–473. doi: 10.1001/jamaoncol.2021.5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerekanych S, Johnson TS, Huang K, Zhang Y. Pseudogene-gene functional networks are prognostic of patient survival in breast cancer. BMC Med Genomics. 2020;13:51. doi: 10.1186/s12920-020-0687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekola Ayele F, Adeyemo A, Finan C, Hailu E, Sinnott P, Burlinson ND, Aseffa A, Rotimi CN, Newport MJ, Davey G. HLA class II locus and susceptibility to podoconiosis. N Engl J Med. 2012;366:1200–1208. doi: 10.1056/NEJMoa1108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang W, Zhang J, He J, Zhu F. Distribution of HLA allele frequencies in 82 chinese individuals with coronavirus disease-2019 (COVID-19) HLA. 2020;96:194–196. doi: 10.1111/tan.13941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJY, Lee KS, Ang LW, Leo YS, Young BE. Risk factors of severe disease and efficacy of treatment in patients infected with COVID-19: a systematic review, Meta-analysis and Meta-regression analysis. Clin Infect Dis. 2020;71:2199–2206. doi: 10.1093/cid/ciaa576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Cao H, Zuo X, Zhang T, Zhang X, Liu X, Xu R, Chen G, Zhang Y, Zheng X, et al. Deep sequencing of the MHC region in the chinese population contributes to studies of complex disease. Nat Genet. 2016;48:740–746. doi: 10.1038/ng.3576. [DOI] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


