Abstract
New series of benzimidazole ring core conjugated with either dithiocarbamate or thiopropyl linkers, hybridized with different secondary amines were synthesized; 5–15 and 22–31; respectively. The new compounds were characterized by different spectroscopic techniques (1H, 13C 1D & 2D NMR, ESI-MS and IR). They were screened for in vitro anticancer activity against breast cancer using MCF7 cell line. The results obtained revealed that compounds 5, 12, 15 and 25 were the most active among the synthesized series exhibiting IC50 < 10 μg/ml against DOX. To characterize targeting breast cancer on molecular level, binding to 15N-labeled Pin1 enzyme was conducted using state-of-the-art 2D NMR binding experiments. Results showed promising binding between compounds 5, 12, and 25 by chemical shift perturbation (peak shifting or peak disappearance). Molecular docking study were quite valuable to explain the binding mode of active derivatives via hydrogen bonding. Additional contact preferences and surface mapping studies stated the similarity pattern between active candidates which may pave the way for more precise anti breast cancer target optimization.
Keywords: Benzimidazole, Breast cancer, NMR, 15N-1H HSQC, Pin1 inhibitors
1. Introduction
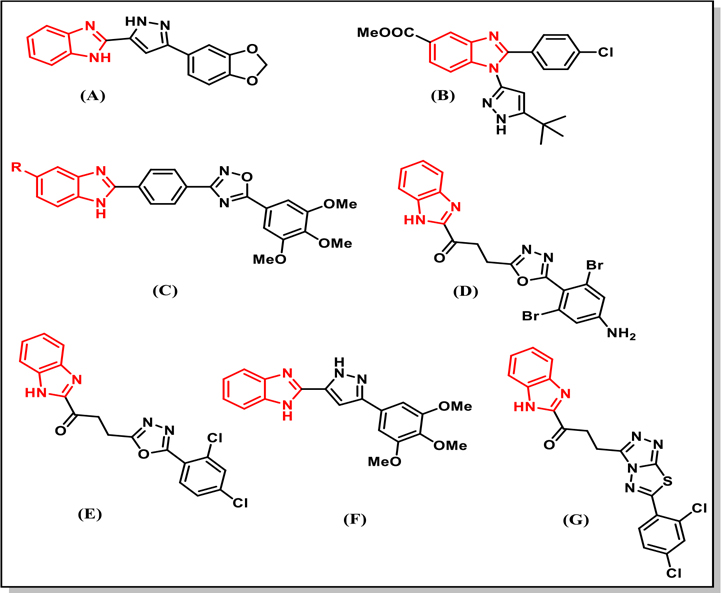
Cancer, which is one of major health problems in the world, is characterized by abnormal cell growth and may have the potential to invade other parts of body (metastasis). One of the most common cancer types is breast cancer which causes death to millions of women every year (it affects one in eight women in the US). So there is an imperative need for discovering more effective drugs to treat breast cancer1. Different classes of heterocyclic compounds have been studied as anticancer agent, one of the most important heterocyclic compounds are benzimidazoles due to their versatile biological activity, such as anti-inflammatory2, antiviral 3,4, antibacterial 5,6, antifungal 7–9, antioxidant 10,11 and anti-proliferative/anticancer activities 12,13. Special attention will be highlighted on benzimidazoles as anticancer agents against breast cancer (Chart 1) 14–19.
Chart 1.
Structures of some literature benzimidazoles with anti-proliferative activity against breast cancer cell lines. LC50 reported for these compounds against MCF7 is > 100 μM.
On the other hand, dithiocarbamates are organosulphrus compounds with variable biological activities, such as antibacterial, antifungal, antiviral including HIV and anticancer. Their antitumor activity due to their metal binding properties resulting from presence of two Sulphur atoms, which have the ability to donate a lone pair of electrons to central metal atom to form stable metal complex20–23. It was reported that linking benzimidazole with dithiocarbamate moieties results in compound with improved anticancer activity24.
Human Pin1 (protein interacting with NIMA “never in mitosis gene a”) is a member of the family of peptidyl-prolyl cis–trans isomerase. Pin1 isomerizes the phosphorylated S/T-P motif, which results in a conformational change in the target protein23. Other members of the same family include FKBP and Cyclophilin. Pin1, regulates a large variety of proteins, especially those involved in mitosis. Downstream effects of substrate isomerization by Pin1 include conformational changes in the phosphorylation state, and stability/turnover of the substrate24,25. The conformational regulation catalyzed by Pin1 has a profound impact on key proteins involved in the regulation of cell growth, genotoxic and other stress responses, the immune response, induction, and maintenance of pluripotency, germ cell development, neuronal differentiation, and survival. Pin1 binds to estrogen receptor where it performs a regulatory effect on the disordered AF1 of estrogen receptors alpha26. It’s also reported that Pin1 increases the DNA binding to ER. Therefore, misregulation and overexpression of Pin1 are common in various cancers, particularly breast cancer.
Molecular targeting protocol for cancer treatment is distinct from conventional chemotherapy in terms of higher tumor specificity. One particular molecular target of high potential is Pin1. Pin1 substrate proteins are important in cell cycle regulation, and it’s up regulation lead to wide variety of human cancers making Pin1 a potential target for anticancer therapy24. It is composed of two domains: N-terminal bounded to ww-domain which is responsible for substrate recognition and a catalytic C-terminal PPIase domain which is responsible for peptidyl-prolyl isomerase activity26.
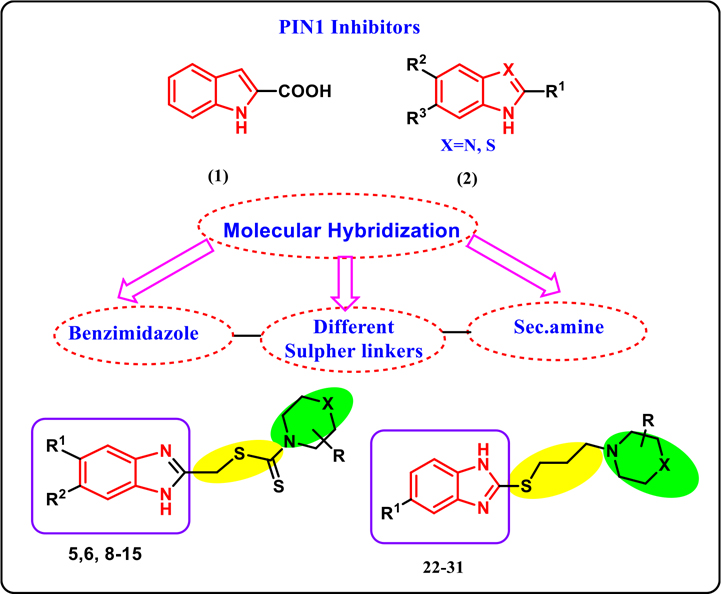
As an important target for cancer, many studies have been pursued to design novel antitumor drugs that specifically inhibit the over-activation of Pin1. NMR-based fragment screening has been reported to target Pin 1 where a large library of small fragments has been tested27. Two competitor ligands had been identified where their binding to the Pin1 active site had been verified by crystallography: The d-peptide inhibitor, and the phenylalaninol phosphate. Moreover, NMR screening identified five competitively binding compounds, of which the indole 2-carboxylic acid was the most potent inhibitor of Pin1 (compound 1).
More Importantly, Potter et al identified a series of new benzimidazole derivatives as inhibitors of PPIase activity of Pin 1 with high activity (compound 2), Chart 228. Moreover benzimidazoles class of Pin1 inhibitors have been characterized as potent non-phosphate, small molecular Pin1 inhibitors (sub-micromolar Ki values) via interactions with Pin1 catalytic site by the hydrogen bonds between the two nitrogen atom (N,NH) of benzimidazole and the cys 113 thiol and the ser154 alcohol, and interaction also between fused ring system of benzimidazole with hydrophobic pocket29.
Chart 2.
Structures of some benzimidazole and its isosteres as Pin1 inhibitors & the target compounds as well as the rational design.
• Why NMR for Drug-Design:
NMR is considered one of the leading applications in drug-Screening because of the high sensitivity, in addition to the possibility to detect the binding site(s) on an atomic level. Many NMR techniques may be applied and are classified in two categories: Ligand-Based and Protein-Based methods.33 It’s one of the known concepts in fragment-based drug design that the compounds to start with are having weak affinities, then they can be optimized using growing or hybridization approaches. Therefore, the use of a sensitive technique like NMR is a must in these cases. In our approach here, we used 15N-1H-HSQC to test the interaction between two novel benzimidazole derivatives and Pin1.30,31 NMR proved very promising to detect binding. The only challenge, in this case, was the solubility of the new compounds and the need to keep the protein in a reasonable amount of aqueous buffer. We tested only three compounds because of the limited solubility of the others.
In summary, the core of our work is based on combining synthetic techniques, computer aided modelling, NMR and cell-based assays to design new benzimidazole derivatives that harbor higher potency against breast cancer. Compounds 1 and 2 have led the way for us for further optimization towards higher efficient benzimidazole derivatives. We synthesized new hybrid compounds that comprised both benzimidazole and amines linked via either dithiocarbamate or thiopropyl linkers and we tested the newly synthesized compounds against breast cancer. In order to look deeper on the molecular level, our work utilize new tool for testing binding affinity depending on NMR methodology technique to study binding to Pin1 as a potential target for antibreast cancer compounds. We believe that our results will pave the way for further studies targeting breast cancer using benzimidazole as a pharmacophore. Moreover, our study will shade the light on using NMR technique in drug design.
2. Results and discussion
2.1. Chemistry
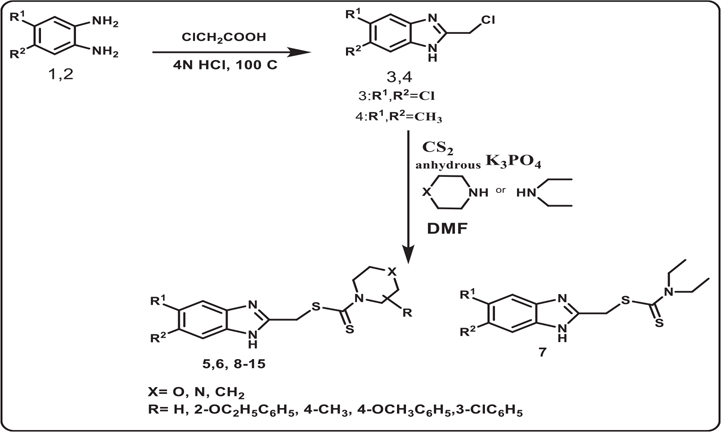
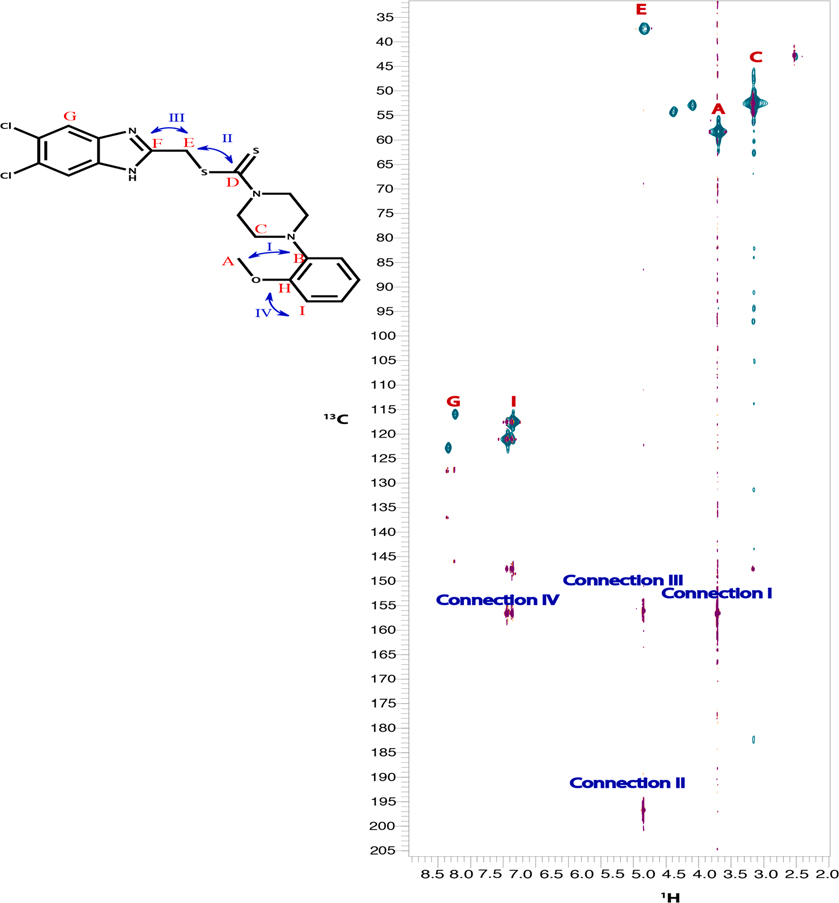
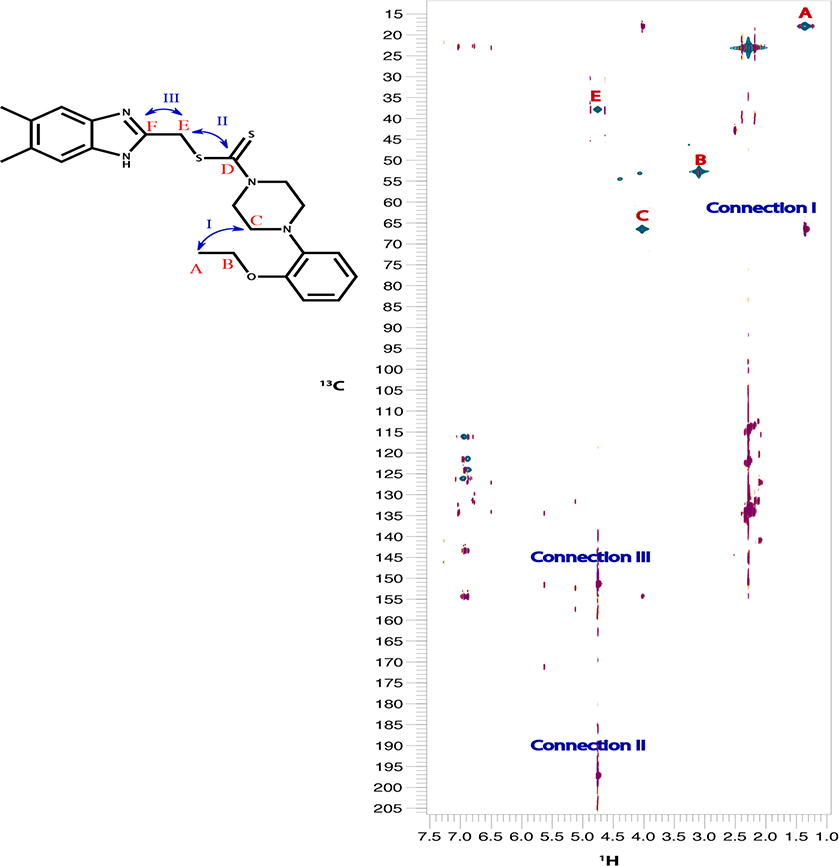
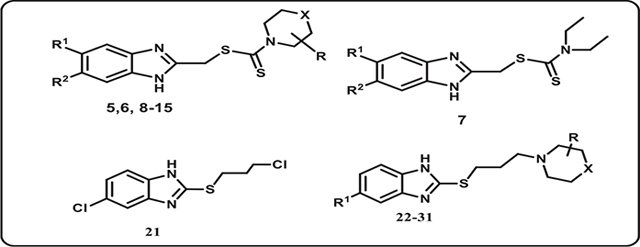
The target compounds in this work were prepared via two schemes. The reaction of phenylene diamine derivatives with chlorocetic acid according to reported procedure to yield intermediate compounds 3, 49,13 (Scheme 1). The newly synthesized benzimidazole-dithiocarbamate-amine conjugates 5–15 were obtained in one pot reaction via two steps. Firstly, different substituted secondary amines were mixed with carbon disulphide in dry dimethyl formamide followed by reaction of the resultant product with intermediates 3 and 4 producing compounds 5–15 (Scheme 1, Table 1). The structure of the synthesized compounds have been confirmed by different spectroscopic techniques including IR, 1H NMR, 13C NMR and MS. Compounds 5–15 have shown characteristic thiocarbonyl band at 950–1050 cm−1 proving the incorporation of DTC group in all the compounds along with the appearance of N-CS band around 1530 cm−1. 1H NMR spectral data for compounds 5–15 clearly proves the incorporation of different amine moieties. Appearance of morpholine protons of compound 5 was shown in the form of a set of multiplet protons at 2.34–3.50 ppm. On the other hand, for compounds 8 & 9 piperazine protons have been appeared as a set of multiplet protons at 2.34–3 ppm, while in compound 10, piperazine protons have been appeared as two sets of singlet protons at 2.4 and 3.1 ppm, the same case was reported for piperidine protons of compound 12 at 3.5 and 4.2 ppm. For compounds 10 &15, the structures were verified using 13C–1H HMBC34 and 13C–1H HSQC35 experiments on BRUKER 600 MHz NMR spectrometer. HMBC shows the long-range bond correlation between 13C and 1H nuclei while HSQC is doing the opposite by showing the direct one bond correlation between the two nuclei. Overlay of HMBC and HSQC spectra verified the structure of both compounds 10 & 15 by showing the one bond and longer-range correlation. The peaks that have been used to verify the structure are annotated in Fig. 1.
Scheme 1.
Synthesis of the target compounds 5–15.
Table 1.
Physicochemical properties of the newly synthesized compounds 5–15 and 21–31.
| |||||||
|---|---|---|---|---|---|---|---|
| Comp | X | R1 | R2 | R | % Yield | Mp, 0C | Molecular Formulae |
|
| |||||||
| 5 | O | Cl | Cl | H | 20 | 171–173 | C13H13Cl2N3OS2 |
| 6 | CH2 | Cl | Cl | H | 48 | 216–218 | C14H15Cl2N3S2 |
| 7 | – | Cl | Cl | NH(C2H5)2 | 45 | 117–119 | C13H15Cl2N3S2 |
| 8 | N | Cl | Cl | 2-OC2H5-C6H4 | 50 | 110–112 | C21H22Cl2N4OS2 |
| 9 | N | Cl | Cl | 4-CH3 | 30 | 205–207 | C14H16Cl2N4S2 |
| 10 | N | Cl | Cl | 4-OCH3-C6H4 | 40 | 220–222 | C20H20Cl2N4OS2 |
| 11 | N | Cl | Cl | 3-Cl-C6H4 | 30 | 135–137 | C19H17Cl3N4S2 |
| 12 | CH2 | CH3 | CH3 | H | 19 | 188–190 | C16H21N3S2 |
| 13 | N | CH3 | CH3 | 4-OCH3 | 39 | 104–106 | C22H26N4OS2 |
| 14 | N | CH3 | CH3 | 3-Cl-C6H4 | 15 | 77–79 | C21H23CIN4S2 |
| 15 | N | CH3 | CH3 | –2-OC2H5-6H4 | 40 | 93–95 | C23H28N4OS2 |
| 21 | Cl | – | – | 36 | 128–130 | C10H10Cl2N2S | |
| 22 | O | H | – | H | 67 | 136–138 | C14H19N3OS |
| 23 | N | H | – | 4-CH3 | 48 | 145–147 | C15H22N4S |
| 24 | N | H | – | H | 41 | 138–140 | C14H20N4S |
| 25 | N | H | – | 4-OCH3-C6H4 | 24 | 110–112 | C21H26N4OS |
| 26 | N | H | – | 2-OC2H5-C6H4 | 23 | 129–130 | C22H28N4OS |
| 27 | N | H | – | 3-Cl-C6H4 | 15 | 133–135 | C20H23CIN4S |
| 28 | N | Cl | – | 4-CH3 | 29 | 158–160 | C15H21CIN4S |
| 29 | N | Cl | 2-OC2H5-C6H4 | 15 | 149–151 | C22H27CIN4OS | |
| 30 | N | Cl | – | 4-OCH3-C6H4 | 39 | 156–158 | C21H25CIN4OS |
| 31 | N | Cl | – | 3-Cl-C6H4 | 19 | 126–128 | C20H22Cl2N4S |
Fig. 1.
An overlay of 13C–1H HMBC (magenta) with 13C–1H HSQC (green) of compound 10 (top) & compound 15 (bottom). Peaks that were of particular interest to verify the structure are annotated on top of each peak. HSQC peaks of interest are highlighted by red letter. HMBC peaks showing long-range connections are highlighted by blue roman numbers.
In Scheme 2, The target compounds 21–31 were prepared. Firstly benzimidazole-2-thiol derivatives 18 & 19 were obtained according to reported procedure14 then they were alkylated by reaction with 1-bromo-3-chloropropane in the presence of potassium carbonate to get the crude intermediates 20, 21 which were further purified by recrystallization to get pure products. The incorporation of S-propyl chloro substituent was carried out via SN2 reaction. The final amine-linked benzimidazole thiopropyl derivatives 22–31 were obtained by reaction of compounds 20 & 21 with a series of amines in DMF. The structure of the synthesized compounds have been confirmed by different spectroscopic techniques including 1H NMR, 13C NMR and MS. 1H NMR spectral data for compounds 22–31 clearly proves the incorporation of different amine moieties, also proves the presence of thiopropyl alkyl chain, in which compounds 21–31 displayed three signals as follows: the S-CH2 group appeared as triplet at 3.75–4.39 ppm, N-CH2 group appeared as triplet at 3.29–3.37 ppm and CH2 group appeared as pentet at 2.3 ppm.
Scheme 2.
Synthesis of the target compounds 21–31.
In compound 22, morpholine protons appeared in the form of a set of multiplet protons at 2.4–3.50 ppm, while in compound 25, 26, piperazine protons appeared in the form of a set of multiplet protons at 2.8–3.14 ppm. In compound 25, the piperazine attached to methoxy phenyl moiety which appeared as singlet proton at 3.67 ppm, while in compound 26 the piperazine attached to ethoxy phenyl moiety displayed two signals as follows: the CH3 group appeared as triplet at 1.32 ppm and OCH2 group appeared as quartet at 4.1 ppm.
2.2. In vitro antitumor activity
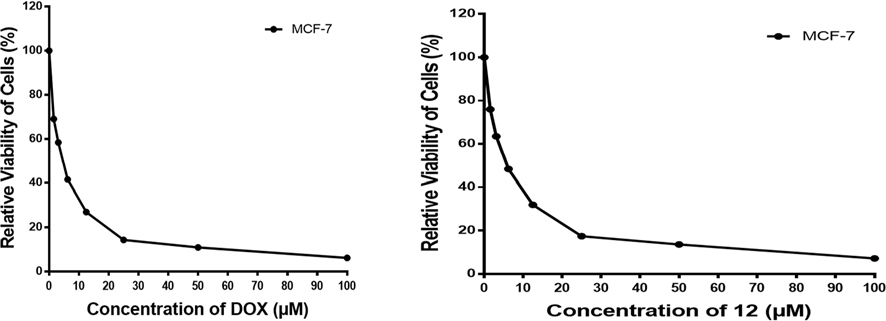
All the newly synthesized compounds were evaluated for their in vitro anticancer effect via the standard MTT method36–37, against a breast cancer cell line (MCF-7). MTT is a calorimetric assay that is based on the ability of live but not dead tumor cells to reduce yellow MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) by mitochondrial dehydrogenases to purple formazan. DMSO is added to dissolve the purple formazan then it’s quantified by measuring absorbance at certain wavelength. The amount of formazan produced is quantified using a dose response curve for compounds-treated vs untreated cells to report on the effectiveness of the compounds. The obtained results revealed that the tested compounds 5, 12, 15, and 25 exhibited very strong potency against tested breast cancer cell line (Fig. 2) in which the compound 15 exhibited the most active candidate with IC50 at 5.58 μg/ml compared to Doxurubucin (reference IC50 at 4.17 μg/ml). Moreover, compounds 7, 8, 10 and 28 have shown strong activity with IC50 ranging from 10 to 20 μg/ml. Compounds 6, 9, 21, 26, 29, and 30 showed moderate activity against the same cell line with IC50 42.75, 30.47, 46.03, 24.07, 32.73, 27.73, 38.46 μg/ml respectively. On the other hand, compounds 22, 23, and 24 showed lowest potency in which 24 exhibited least inhibitory activity with IC50 at 91.96 μg/ml (see Figs. 3 and 4).
Fig. 2.
Viability of MCF-7 cell line at various concentrations.
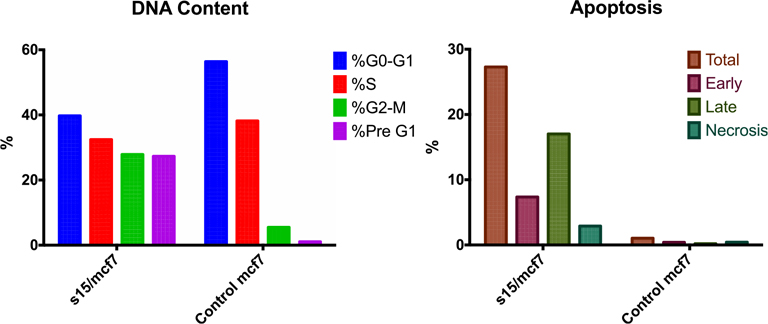
Fig. 3.
Left: Percent of DNA content in compound 15 treated cells vs the control at different cell cycle phases. Right: Percent of apoptotic cells in compound 15 treated cells vs control (total, early, late and necrosis).
Fig. 4.
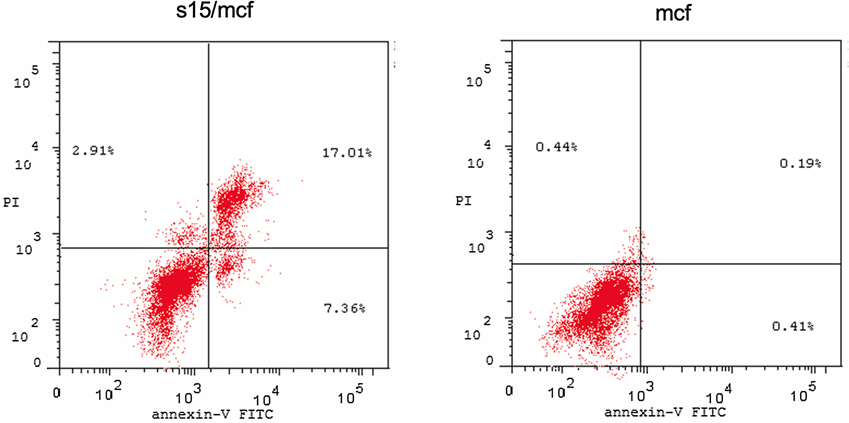
Evaluation of apoptosis induced by compound 15 treatment. Flow cytometry analysis for Annexin-VFITC/PI staining Left: compound 15 treated cells, right: control.
2.3. Apoptosis analysis of compound 15
Based on the results from cell-based assay and NMR binding to Pin1, we chose compound 15 to further assess its effect on the apoptotic process. Mcf7 cells were treated with compound 15 for 24 h and processed for flow cytometry and the presence of Annexin V-FITC/PI was investigated. As shown in Table 3, cells treated with compound 15 are showing higher DNA content at the G2-M phase (27.86% vs 5.47% for the control) indicating cell growth arrest at this phase. Moreover, cells treated with compound 15 showed a high percentage apoptosis rate (27.28% vs 1.04% for the control) corresponding to the sum of the percentage of Annexin V-positive/PI-negative staining and double-positive staining cells (early and late apoptosis, respectively).
Table 3.
Evaluation of DNA content and apoptosis induced by compound 15 (s15) on mcf7 cells lines vs control.
| Sample | Code | MWt | DNA content % |
||||
|---|---|---|---|---|---|---|---|
| %G0-G1 | %S | %G2-M | %Pre G1 | Comment | |||
|
| |||||||
| 1 | s15/mcf7 | 440.62 g/mole | 39.72 | 32.42 | 27.86 | 27.28 | Cell growth arrest at G2/M |
| 2 | Control mcf7 | – | 56.38 | 38.15 | 5.47 | 1.04 | |
| Apoptosis | Necrosis | ||||||
| Total | Early | Late | |||||
| 1 | s15/mcf7 | 440.62 g/mole | 27.28 | 7.36 | 17.01 | 2.91 | |
| 2 | Control mcf7 | – | 1.04 | 0.41 | 0.19 | 0.44 | |
2.4. 2D 15N-1H HSQC NMR
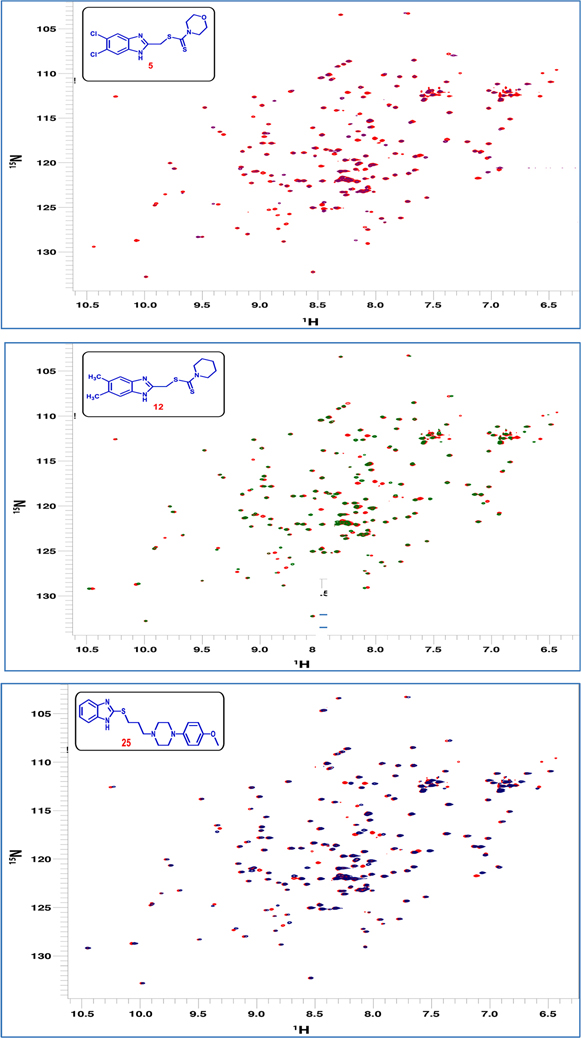
2D 15N-1H HSQC NMR is a very sensitive technique to detect binding between specific biomolecule and a small ligand. Not only it can report about binding or not, HSQC spectra can also report about the time regime of exchange between free and bound form if there’s binding. Slow exchange leads to the appearance of two sets of peaks corresponding to free and bound form, while intermediate exchange causes peaks broadening due to the μs-ms time scale of exchange. On the other hand, the fast exchange leads to averaging and consequently peaks shifting (chemical shift perturbation). Herein, we used 15N-labeled Pin1 enzyme where we recorded HSQC spectra of the protein alone and in the presence of excess amount of three of the newly synthesized compounds. In our research, the main challenge we faced was the solubility of the compounds in a balanced mixture of the protein aqueous buffer. Despite the fact that the compounds were freely soluble in DMSO, it’s inapplicable to use pure DMSO to dissolve the protein since this will result in protein denaturation and probably aggregation. Therefore, only three compounds were tested where we were able to get these compounds to dissolve partially in a mixture of 95% buffer and 5% DMSO. In addition, as a control, the spectrum was recorded only in presence of DMSO and compared to the ones with the ligand. The proportion of Pin1 to the compounds was adjusted to be 1:10. The recorded spectra showed chemical shift perturbation for some of the amino acids in the protein (CSP). Chemical shift perturbation and peak shifting is indicative of fast exchange between free and bound states which results in peaks positions averaging. Moreover, some peaks corresponding to different amino acids showed peak disappearance upon binding. This peak disappearing indicated exchange on the μs-ms time scale (Fig. 5). Furthermore, our NMR results have been verified using cell lines.
Fig. 5.
Overlay of 15N-1H HSQC spectra of free Pin1 enzyme (red) with Pin1 bound to: Compound 5 (violet), compounds 12 (green), and compound 25 (blue). The overlay is showing chemical shift perturbation and peak disappearance for different peaks as indicative of binding on different time scales.
2.5. Structure activity relationship
From the obtained results in Table 2, some points can be highlighted about the synthesized compounds activity. Generally, compounds with dithiocarbamate linkage in Scheme 1 are more potent than compounds with alkyl chain linkage in Scheme 2. Compounds 8, 10, 15, and 25 with bulky amines like methoxyphenylpiperazine and ethoxyphenylpiperazine showed higher cytotoxic activity. Compound 22 with morpholine amine showed low cytotoxic activity while compounds 6 and 12, which have piperidine, showed moderate cytotoxic activity. Changing the substitution at positions 5 and 6 of benzimidazole nucleus did not have a significant effect on the activity of compounds in Schemes 1 And 2.
Table 2.
DNA/methyl green colorimetric assay of the target compounds.
| Compounds | In vitro Cytotoxicity IC50 (μM) · MCF7 |
|---|---|
|
| |
| DOX· · | 4.17 ± 0.2 |
| 5 | 5.70 ± 3.3 |
| 6 | 42.75 ± 2.8 |
| 7 | 12.55 ± 1.1 |
| 8 | 16.23 ± 1.4 |
| 9 | 30.47 ± 2.3 |
| 10 | 19.93 ± 1.7 |
| 11 | 60.20 ± 3.8 |
| 12 | 9.55 ± 0.9 |
| 13 | 46.03 ± 3 |
| 14 | 8.39 ± 0.7 |
| 15 | 5.58 ± 0.3 |
| 21 | 24.07 ± 1.9 |
| 22 | 82.87 ± 4.8 |
| 23 | 77.92 ± 4.1 |
| 24 | 91.69 ± 5.2 |
| 25 | 6.84 ± 0.5 |
| 26 | 32.73 ± 2.4 |
| 27 | 54.63 ± 3.5 |
| 28 | 13.92 ± 1.2 |
| 29 | 27.73 ± 2.1 |
| 30 | 38.46 ± 2.6 |
| 31 | 61.40 ± 3.9 |
IC50 (μM): 1 – 10 (very strong). 11 – 20 (strong). 21 – 50 (moderate) 51 – 100 (weak) and above100 (non-cytotoxic) · · DOX: Doxorubicin
2.6. Molecular modeling study
2.6.1. Conformational analysis
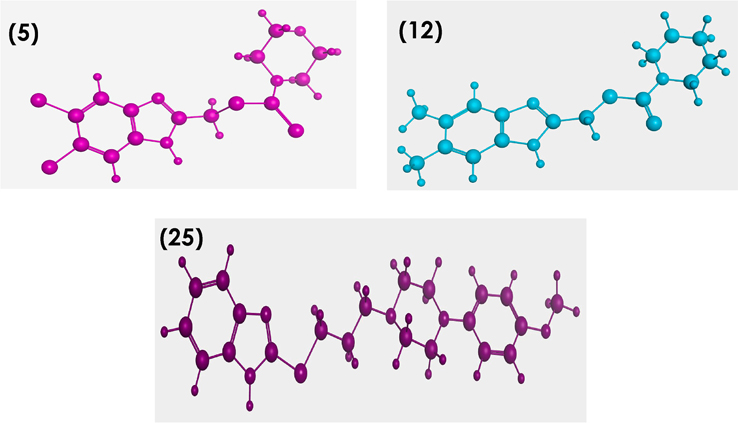
Conformational analysis for compounds 5, 12 and 25 have been performed. The best energy conformers were obtained by conformational searching using multiconformer method (Fig 6). Further in-depth docking study was performed for the aforementioned compounds which showed good binding to Pin1 enzyme via NMR biding technique.
Fig. 6.
Lowest energy conformers of compounds 5, 12 and 25 with balls and cylinders.
2.6.2. Molecular docking study
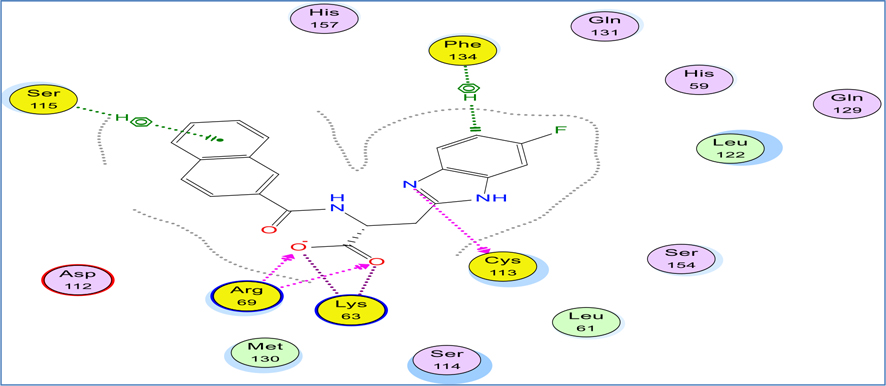
The benzimidazole derivative cocrystallized in Pin1 crystal structure obtained from protein data bank (pdb: 4TYO) was chosen as a reference for our docking study due to the similarity of its benzimidazole ligand core structure to our compounds38. This reference ligand showed a tight binding via the interaction with a network of hydrogen binding with Arg 69, Lys63, and Cys113 in addition to cationic hydrogen binding with both Phe134 and Ser115 residues (Fig. 7). It was reported that the interaction with Lys63 is essential for Pin1 inhibition38.
Fig. 7.
2D binding mode and residues involved in the recognition of reference ligand 3-(6-fluoro-1H-benzimidazol-2-yl)-N-(naphthalen-2-ylcarbonyl)-d-alanine (Arg69, Phe143, Ser115, Lys63, Cys113 (with yellow highlight).
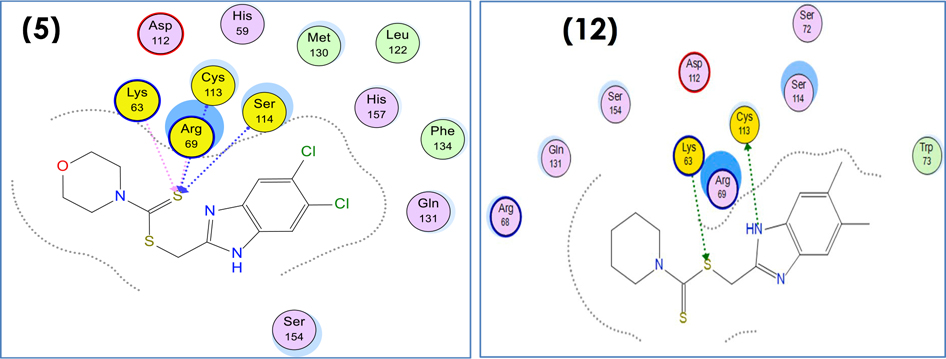
Compound 5 is well-positioned to interact with Lys63, Arg69, Cys 113 and Ser 114 amino acid residues via sulpher atom of thiocarbonyl group (Fig. 8, left side). Also, compound 12 retains good stacking against the shelf-like hydrophobic surface and occupies a similar position to the benzimidazole ring of cocrystallized ligand by binding to Cys 113 and Lys 63 via hydrogen bonding interaction (Fig. 8, right side).
Fig. 8.
2D binding mode and residues involved in the recognition of Compounds 5 and 12 ligands as references at Pin1 enzyme.
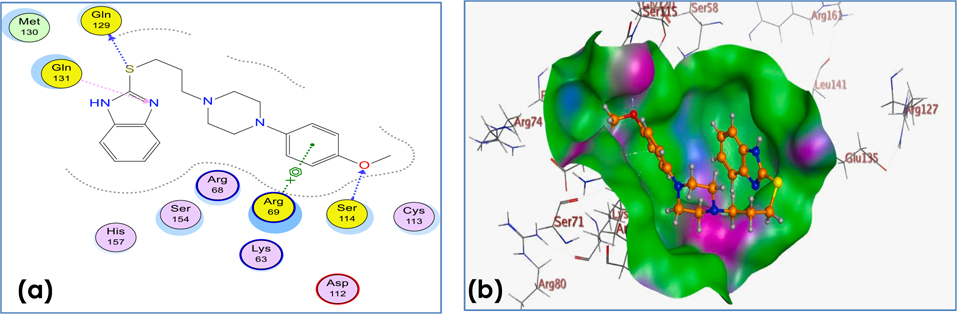
Compound 25 has shown both hydrogen bonding and n-cationic interactions to Ser 114, Gln 131, Gln 129 and Arg69 respectively (Fig. 9a). The aligned conformation of compound 25 in binding site was aligned completely inside the binding site surface map explaining its activity experimentally determined by NMR binding technique (Fig. 9b).
Fig. 9.
(a) 2D binding mode and residues involved in the recognition of active compound 25 at Pin1 enzyme. (b) The aligned conformation of compound 25 (ball and stick) occupying pocket (surface mapping).
2.6.3. Contact preference
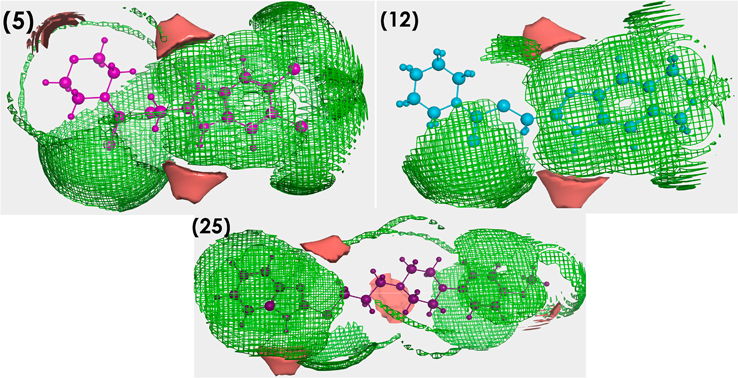
The purpose of the Contact Statistics application is to calculate, from the 3D atomic coordinates of ligand, preferred locations for hydrophobic and hydrophilic ligand atoms. In particular, the aim of this work was to study the interactions between the chemical components of the ligands and the protein microenvironment surrounding them. The results obtained indicate that the information underlying the fragment contacts is valuable as it clearly demonstrates the similarity pattern of distribution of the hydrophobic and hydrophilic sites between our ligands 5, 12 and 25 (shown in Fig. 10) and can also be exploited in understanding results of molecular docking simulations. A better understanding of the interaction patterns of these moieties can lead to improved application of ligand binding prediction, protein function recognition, and drug design tools.
Fig. 10.
Contact statistics map for compounds 5, 12 and 25 (hydrophobic: green, hydrophilic: red).
2.6.4. Surface mapping
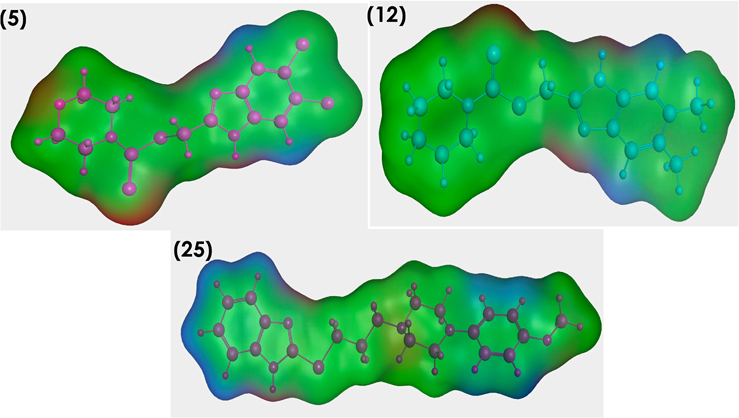
In additional confirmatory investigation step to ensure the similarity between our active candidates’ binders to Pin1 enzyme surface mapping study was performed for the conformers with lowest energy. Fig. 11 showed nearly typical surface mapping contours of selected ligands. Hence, that could be contributing to their good binding to our selected enzymes and put more evidence that their compounds antibreast cancer activity may be attributed to Pin1 inhibition.
Fig. 11.
Surface map for compounds 5, 12 and 25, Pink, hydrogen bond, blue: mild polar, green hydrophobic.
3. Conclusion
A new series of benzimidazoles core compounds with dithiocarbamate and thiopropyl link conjugated with different secondary amines were designed, synthesized and biologically evaluated for their antibreast cancer activity using MCF7 cell line. Pin 1 enzyme was chosen as a new goal for breast cancer targeting. Results revealed that compounds 5, 12, 15 and 25 were the most potent antibreast cancer activity with IC50 5.70 9.55, 5.58 and 6.84 μg/ml respectively. Using 2D 15N-1H HSQC NMR experiments, we were able to show that the three compounds are binding to the purified Pin1 on a fast time scale. The binding has been proved by noticing the chemical shift perturbation and peak disappearance of some peaks in the 2D NMR spectra. This change is some of the peaks upon binding indicates change in the chemical environment of the corresponding residues as a result of binding. Compounds with dithiocarbamate linkage were more potent than compounds with alkyl chain linkage. Molecular modeling study was conducted to understand the compounds binding to Pin 1, docking study proved a similar binding mode of our compounds to reference cocrystallized ligand to Pin 1 enzyme. Contact statistics and surface map studies stated the similarity between active candidates which assures their fitting to active site of Pin 1 enzyme.
4. Experimental section
The synthesis of the designed compounds was performed in Faculty of Pharmacy, Mansoura University, Mansoura, Egypt. The in vitro anticancer screening was conducted in Faculty of Pharmacy, Mansoura University, Mansoura, Egypt. Melting points (°C) were determined on Mettler FP80 melting point apparatus and are uncorrected. All of the new compounds were analyzed for C, H and N and agreed with the proposed structures within ± 0.4% of the theoretical values. 1D 1H- and 13C NMR were recorded on a JOEL 500 MHz & BRUKER 400 MHz FT spectrometers; chemical shifts are expressed in δ ppm with reference to TMS. For samples 10 & 15 13C–1H HMBC and HSQC were measured on a shielded BRUKER 600 MHz FT spectrometer in University of Colorado, Denver, USA. The following parameters were used: 64 scans with 2 s interscan delay and 4096 points in the 1H dimension vs 512 points in the 13C dimension. For samples 23, 24 and 30, 13C-APT NMR (Attached proton test) and 1D-13C NMR were performed on BRUKER 600 MHz – cryoprobe- FT spectrometer in University of Colorado, Denver, USA. For APT experiments, 65,536 points were recorded, with 256 scans and a relaxation delay of 2 s. The HeC J-coupling value was sat to 145 Hz. For the 1D-13C NMR, 65536 points were recorded, with 1024 scans and a relaxation delay of 2 s.
Mass spectral (MS) data were obtained on a Perkin Elmer, Clarus 600 GC/MS and Joel JMS-AX 500 mass spectrometers. Thin layer chromatography was performed on precoated (0.25 mm) silica gel GF254 plates (E. Merck, Germany), compounds were detected with 254 nm UV lamp. Silica gel (60–230 mesh) was employed for routine column chromatography separations. All the fine chemicals and reagents used were purchased from Aldrich Chemicals Co, USA. Molecular docking experiments were performed using ‘Molecular Operating Environment’ software on Core i7 workstation.
4.1. Chemistry
4.1.1. 4.1.1. General procedure for preparation of (5,6-dimethyl-1H-benzo [d]imidazol-2-yl)methyl substituted-1-carbodithioate) or (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl substituted-1-carbodithioate) 5–15
A solution of the appropriate secondary amine (0.05 M) in dry DMF (10 ml) and carbon disulfide (0.1 M) was stirred for 2hr at room temperature, dry K3PO4 (0.05 M) was add to the reaction mixture and stirred for 3hr at room temperature. 2-(chloromethyl)-5,6-dimethyl-1H-benzo[d]imidazole 3 or 5,6-dichloro-2-(chloromethyl)-1H-benzo[d] imidazole 4 (0.05 M) were added to the reaction mixture, and continuously stirred at room temperature for 1–2 days. After completion of the reaction, which was monitored by TLC, the reaction mixture was poured onto crushed ice and the resulting precipitation was collected by filtration and purified by recrystallization from ethanol.
4.1.1.1. (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl morpholine-4-carbodithioate 5.
Puff solid, IR(KBr, cm−1): 800(C-Cl), 1238(C═S), 3400(N—H), 1H NMR (DMSO-d6) δ: 2.21–2.32(m,8H,morphorine-H), 4.7(s,2H,S-CH2), 5.5(s,1H,N—H), 7.3(s, 1H, CH benzimidazole), 7.53(s, 1H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ: 35.6, 49.9, 64.5, 118.3, 130.6, 138.2, 142.5, 198.1, MS m/z(%):362.29, calcd.362.3
4.1.1.1.1. (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl piperidine-1-carbodithioate 6.
Gray solid, MS m/z(%):360.32, 1H NMR(DMSO-d6) δ: 2.49(S,1H,NH), 3.89(S,5H,piperidine-H), 4.2(s,5H,piperidine-H), 4.9(S,2H,CH2), 7.3(s, 1H, CH benzimidazole), 7.53(s, 1H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ: 14.8, 23.4, 26.1, 43.1, 51.3, 64.6, 108.5, 114.5, 119.3, 120.5, 121.8, 123.1, 138.6, 143.7, 147.1, 162.8, MS m/z(%): 160.8(88), 266, 268, calcd.360.32.
4.1.1.2. 4.1.1.3. (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl diethylcarbamodithioate)7.
Puff solid, 1H NMR (DMSO-d6) δ: 1.18(t, 3H, CH3), 1.24(t, J = 7.2 Hz, 3H, CH3), 3.78(q, 2H, CH2), 3.96(q, 2H, CH2), 4.75(s, 2H, S-CH2), 7.79(s, 2H, CH benzimidazole), 13C NMR (100 MHz, DMSO-d6) δ: 15.2, 35.8, 48.7, 116.9, 130.2, 139.1, 140.8, 197.9, MS m/z(%):348.30, calcd.348.3.
4.1.1.2.1. (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl 4-(2-ethoxyphenyl)piperazine-1-carbodithioate 8.
Puff solid, 1H NMR (DMSO-d6) δ: 1.36(t, J = 6.4 Hz, 3H, CH3), 2.7(s, 2H, piperazine-H), 2.89(s, 2H, piperazine-H), 3.1(s, 4H, piperazine-H), 4.07(q, 2H, O-CH2), 4.82(s, 2H,S-CH2), 6.88(s, 1H, CHbenzimidazole), 6.94 (s, 1H, CHbenzimidazole), 7.7–7.8(m, 4H, Ar-H), 12.7(s,1H, NH),13C NMR (100 MHz, DMSO-d6)δ: 16.1, 34.9, 50.1, 51.6, 63.9, 112.7, 117.3, 120.9, 121.2, 121.6, 129.8, 139.2, 141.2, 142.8, 160.9, 197.8 MS m/z (%):481.45, calcd.481.5.
4.1.1.3. (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl 4-methylpiperazine-1-carbodithioate 9.
Brown solid, IR(KBr, cm-1): 865(C-Cl), 1227(C═S), 3447(N—H), 1H NMR (DMSO-d6) δ: 2.39(s, 4H, piperazine-H), 2.49(s, 3H, N-CH3), 3.91(s, 2H, piperazine-H), 4.21(s,2H,piperazine-H), 4.78(s,2H,S-CH2), 7.7(s, 1H, CHbenzimidazole), 7.8(s, 1H, CHbenzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:33.9, 45.9, 50.1, 52.9, 117.1, 127.9, 138.6, 140.9, 196.8, MS m/z(%):375.33, calcd.375.3.
4.1.1.4. (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl 4-(4-methoxyphenyl)piperazine-1-carbodithioate 10.
Brown solid, 1H NMR (DMSO-d6) δ: 3.13(s,4H,piperazine-H), 3.67(s,3H,O-CH3), 4.07 (s,2H,piperazineH), 4.36(s,2H,piperazine-H), 4.8(s,2H,S-CH2), 6.82(d, J = 8 Hz,2H, Ar-H), 6.92(d, J = 8.2 Hz, 2H,Ar-H), 7.72(s, 1H, CHbenzimidazole), 7.82(s, 1H, CHbenzimidazole),12.7(s, 1H, NH), 13C NMR(100 MHz, DMSO-d6) δ:34.8, 50.3, 51.7, 58.2, 116.1, 116.2, 117.3, 129.1, 138.9, 141.3, 144.4, 152.2, 197.9, MS m/z(%): 467.43, calcd.467.4.
4.1.1.5. (5,6-dichloro-1H-benzo[d]imidazol-2-yl)methyl 4-(2-chlorophenyl)piperazine-1-carbodithioate 11.
Brown solid, IR(KBr, cm-1):836(C-Cl), 1224(C═S),3422(N—H), 1H NMR (DMSO-d6) δ: 3.37(s,4H,piperazine-H), 4.07(s,2H,piperazine-H), 4.34(s,2H,piperazine-H), 4.81(s,2H,S-CH2), 6.8 (d, J = 7.5 Hz, 1H, Ar-H), 6.86 (s, 1H, Ar-H), 6.94 (d, J = 8.1 Hz,1H, Ar-H), 7.22(t, J = 8 Hz, 1H, Ar-H), 7.72(s, 1H, CH benzimidazole), 7.82(s, 1H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ: 23.7, 25.1, 34.8, 51.4, 113.5, 126.1, 134.9, 143.8, 192.5, MS m/z(%):471.84, calcd.471.9.
4.1.1.6. (5,6-dimethyl-1H-benzo[d]imidazol-2-yl)methyl piperidine-1-carbodithioate 12.
Puff solid, m.p. = 188°c, yield = 15%, IR(KBr, cm-1): 1234(C═S), 3449(N—H), 1H NMR (DMSO-d6) δ: 1.57(s,3H,CH3), 1.64(S,3H,CH3), 2.49(s,2H,piperidine-H), 3.89(s, 4H,piperidine-H), 4.2(s,4H,piperidine-H),4.08(S,2H,CH2), 6.8(s, 1H, CHbenzimidzole), 7.3(s, 1H, CHbenzimidazole), 12.12(S,1H,NH), 13C NMR(100 MHz, DMSO-d6) δ:17.9, 23.7, 23.9, 33.9, 50.9, 116.8, 128.9, 137.1, 140.5, 199.9 MS m/z(%): 319.49, calcd.319.5.
4.1.1.7. (5,6-dimethyl-1H-benzo[d]imidazol-2-yl)methyl 4-(4-methoxyphenyl)piperazine-1-carbodithioate 13.
Puff solid, m.p. = 104°c, yield = 15%, 1H NMR (DMSO-d6) δ: 1.14(s,3H,CH3), 1.22(s,3H,CH3), 3.12(s,4H,piperazine-H), 3.67(s,3H,O-CH3), 4.02(s,2H,piperazine-H), 4.37(s,2H,piperazine-H), 4.72(s,2H,S-CH2), 6.82(d, J = 8 Hz,2H, Ar-H), 6.92(d, J = 8.2 Hz, 2H, Ar-H), 7.22(s, 1H, CH benzimidazole), 7.27(s, 1H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:18.6, 34.2, 50.9, 51.8, 57.8, 115.8, 116.2, 129.7, 136.9, 140.9, 145.9, 156.1, 198.1, MS m/z(%):426.6, calcd.426.6.
4.1.1.8. (5,6-dimethyl-1H-benzo[d]imidazol-2-yl)methyl 4-(2-chlorophenyl)piperazine-1-carbodithioate 14.
Puff solid, 1H NMR (DMSO-d6) δ: 1.05(s,3H,CH3), 1.17(s,3H,CH3), 2.38 (s,4H,piperazine-H), 4.02(s,2H,piperazine-H), 4.37(s,2H,piperazine-H), 4.729(s,2H,S-CH2), 6.8 (d, J = 7.5 Hz,1H, Ar-H), 6.88(d, J = 8.1, 1H, Ar-H), 6.93(s, 1H, Ar-H), 7.237(t, J = 8 Hz, 1H, Ar-H), 7.27(s, 2H, CH benzimidazole), 12.16(S,1H,NH), 13C NMR(100 MHz, DMSO-d6) δ:18.2, 34,8, 51.9, 52.8, 116.3, 124.1, 125.1, 128.8, 129.9, 130.1, 130.7, 136.7, 140.8, 148.6, 196.2, MS m/z(%):431.01, calcd.431.
4.1.1.9. (5,6-dimethyl-1H-benzo[d]imidazol-2-yl)methyl 4-(2-ethoxyphenyl)piperazine-1-carbodithioate 15.
Yellow solid, IR(KBr, cm-1):1236(C═S), 3421(N—H) 1H NMR (DMSO-d6) δ: 1.34(t, J = 6.75 Hz, 3H,CH3), 2.2(s,3H,CH3), 2.3(s,3H,CH3), 3.09(q,2H,O-CH2), 4.002–4.05 (m,8H,piperazine-H), 4.37(s,1H,NH), 4.7(s,2H,S-CH2), 6.85–6.95(m, 4H, Ar-H), 7.3(s, 1H, CH benzimidazole), 7.4(s, 1H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:16.1, 18.5, 34.5, 52.3, 53.1, 61.2, 112.2, 116.8, 120.7, 121.3, 121.9, 129.9, 138.9, 140.9, 142.1, 160.3, 197.9, MS m/z(%):440.62, calcd.440.6.
4.1.2. General procedure for preparation of 2-((3-chloroprpyl)thio)-1H-benzo[d]imidazole 20 and 5-chloro-2-((3-chloropropyl)thio)-1H-benzo[d] imidazole 21
In 50 ml rounded-bottomed flask the 1H-benzo[d]imidazole-2-thiol 18 or 5-chloro-1H-benzo[d]imidazole-2-thiol 19 (0.005 mol) was added with 1-bromo-2-chloroethane(0.005 mol) and unhydrous K2CO3 (0.0075 mol) in 20 ml dry DMF and stirred at room temprature for 1 hr. After compleation of the reaction which was monitored by TLC, water was added to the reaction mixture and the resulting ppt was collected by filtration to give the titled compounds.
4.1.2.1. (5-chloro-2-((3-chloropropyl)thio)-1H-benzo[d]imidazole) 21.
puff solids, m.p. = 130, 1H NMR (DMSO-d6) δ: 2.16(p, 2H,CH2), 3.37(t, J = 6.75 Hz, 2H, N-CH2), 3.75(t, J = 6.5 Hz, 2H, S-CH2), 7.15(d, J = 8 Hz, 1H, CH benzimidazole), 7.4(d, J = 8 Hz, 2H, CH benzimidazole) 7.49 (s, 1H, CH benzimidazole), MS m/z(%):247.14, calcd.247.
4.1.3. General procedure for preparation of 2-((3-(substituted-1-yl)propyl) thio)-1H-benzo[d]imidazole or 5-chloro-2-((3-(substituted-1-yl)propyl) thio)-1H-benzo[d]imidazole 22–31
A solution of the appropriate secondary amine (0.05 M), 2-((3-chloropropyl)thio)-1H-benzo[d]imidazole 20 or 5-chloro-2-((3-chloropropyl)thio)-1H-benzo[d]imidazole 21(0.05 M) and unhydrous K2CO3(0.1 M) in dry DMF(10 ml) was stirred in room temprature for 3–24 hr. After compleation of reaction which was monitored by TLC, water and EA were added to the reaction mixture, EA layer was separated and dried by Na2SO4 and evaporated in vacuo affording a solid residue
4.1.3.1. 4-(3-((1H-Benzo[d]imidazol-2-yl)thio)propyl)morpholine 22.
Puff solid, 1H NMR (400 MHz, CDCl3) δ: 2.3(p, 2H, CH2), 2.4–2.49(m, 8H, morphorine-H), 3.29(t, J = 5.5 Hz, 2H, N-CH2), 4.2(t, J = 6 Hz, 2H, S-CH2), 7.1–7.4(m, 4H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ: 22.8, 25.9, 39.5, 42.2, 43.1, 116.9, 121.7, 142.3, 146.5, MS m/z(%):277.39, calcd.277.4.
4.1.3.2. 2-((3-(4-Methylpiperazin-1-yl)propyl)thio)-1H-benzo[d] imidazole 23.
Puff solid,m.p. = 140 °c, yield = 48%, 1H NMR (400 MHz, CDCl3) δ: 2.3(p, 2H, CH2), 2.4–2.49(m, 8H, piperazine-H), 3.28(t, J = 5.75 Hz, 2H, N-CH2), 3.3(s, 3H, N-CH2), 4.2(t, J = 5.75 Hz, 2H, S-CH2), 7.1–7.44(m,4H,CHbenzimidazole), 13C NMR(150 MHz, DMSO-d6) δ: 23.3, 25.8, 40.07, 42.63, 109.1 (negative in APT), 117.4 (negative in APT), 121.1 (negative in APT), 122.2 (negative in APT), 136.1, 142.9, 146.9, MS m/z(%):290.43, calcd.290.4.
4.1.3.3. 2-((3-(Piperazin-1-yl)propyl)thio)-1H-benzo[d]imidazole 24.
Puff solid, 1H NMR (400 MHz, CDCl3) δ: 2.32(p, 2H, CH2), 2.4–2.87(m, 8H, piperazine-H), 3.35(t, 2H, N-CH2), 4.39(t, 2H, S-CH2), 7.11–7.45(m, 4H, CHbenzimidazole), 13C NMR(150 MHz, DMSO-d6) δ: 23.3, 25.7, 42.69, 109.1 (negative in APT), 117.5 (negative in APT), 121.3 (negative in APT), 122.2 (negative in APT), 136.1, 142.9, 147, MS m/z(%):276.4, calcd.276.5.
4.1.3.4. 2-((3-(4-(4-Methoxyphenyl)piperazin-1-yl)propyl)thio)-1H-benzo [d]imidazole 25.
Puff solid 1H NMR (400 MHz, CDCl3) δ: 2.34(p,2H,CH2), 2.86–2.99(m,8H, piperazine -H), 3.28(t, J = 5.55 Hz, 2H,N-CH2), 3.673(s,3H,O-CH3), 4.2(t, J = 6 Hz, 2H, S-CH2), 6.82(d, J = 8 Hz, 2H, Ar-H), 6.9(d, J = 8.2 Hz, 2H, Ar-H), 7.1–7.45(m, 4H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:26.7, 34.6, 51.4, 52.8, 54.1, 56.1, 116.4, 118.1, 122.8, 138.9, 145.2, 147.6, 155.2, MS m/z(%):382.53, calcd.382.5.
4.1.3.5. 2-((3-(4-(2-Ethoxyphenyl)piperazin-1-yl)propyl)thio)-1H-benzo [d]imidazole 26.
Puff solid, 1H NMR (400 MHz, CDCl3) δ: 1.32(t, J = 6.75 Hz, 3H, CH3), 2.32 (p, 2H, CH2), 2.89–2.98(m, 8H, piperazine -H), 3.34(t, J = 5.25 Hz, 2H, N-CH2), 4(q, 2H, O-CH2), 4.2(t, J = 5.75 Hz, 2H, S-CH2), 6.85–6.95(m, 4H, Ar-H), 7.1–7.4(m, 4H, CHbenzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:14.8, 23.5, 29.9, 51.6, 53.1, 55.1, 63.6, 112.1, 115.2, 119.5, 120.1, 122.2, 123.4, 137.5, 142.6, 147.1, 162.3, MS m/z(%):396.55, calcd.396.6.
4.1.3.6. 2-((3-(4-(3-Chlorophenyl)piperazin-1-yl)propyl)thio)-1H-benzo [d]imidazole 27.
Puff solid, 1H NMR (400 MHz, CDCl3) δ: 2.3(p, 2H, CH2), 2.49–2.6(m, 8H, piperazine -H), 3.3(t, J = 5.75 Hz, 2H, N-CH2), 4.2(t, J = 5.75 Hz, 2H, S-CH2), 6.82 (d, J = 7.5 Hz, 1H, Ar-H), 6.96(d, J = 8.1, 1H, Ar-H), 7.02(s, 1H, Ar-H), 7.24(t, J = 8 Hz, 1H, Ar-H), 7.3–7.49(m, 4H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:27.2, 33.7, 51.1, 53.2, 55.1, 110.1, 113.5, 117.9, 122.8, 124, 131.6, 136.1, 138.9, 146.1, 147.6, MS m/z(%):386.94, calcd.387.
4.1.3.7. 5-Chloro-2-((3-(4-methylpiperazin-1-yl)propyl)thio)-1H-benzo[d] imidazole 28.
Brown solid, 1H NMR (400 MHz, CDCl3) δ: 2.31 (p, 2H, CH2), 2.49–2.6(m, 8H, piperazine -H), 3.29(t, J = 5.5 Hz, 2H, N-CH2), 3.3(t, 3H, N-CH3), 4.2(t, J = 5.75 Hz, 2H, S-CH2), 7.15(d, J = 8 Hz, 1H, CH benzimidazole), 7.45(s, 1H, CH benzimidazole), 7.56(d, J = 8 Hz, 1H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:26.8, 34.1, 47.9, 51.3, 53.1, 54.9, 116.1, 118.1, 122.2, 129.1, 138.1, 139.9, 146.3, MS m/z(%):324.87, calcd.324.9.
4.1.3.8. 4.1.3.8. 5-Chloro-2-((3-(4-(2-ethoxyphenyl)piperazin-1-yl) propyl)thio)-1H-benzo[d]imidazole 29.
Puff solid, 1H NMR (400 MHz, CDCl3) δ: 1.33(t, J = 6.75 Hz, 3H, CH3), 2.31(p, 2H, CH2), 2.85–2.89(m, 8H, piperazine -H), 3.28(t, J = 5.75 Hz, 2H, N-CH2), 4.01(q, 2H, O-CH2), 4.2(t, J = 5.25 Hz, 2H, S-CH2), 6.83–6.89(m, 4H, Ar-H),7.15(d, J = 8 Hz, 1H, CH benzimidazole), 7.45(s, 1H, CH benzimidazole), 7.5 (d, J = 8 Hz, 1H, CH benzimidazole), 13C NMR (100 MHz, DMSO-d6) δ:15.9, 26.1, 33.9, 51.5, 55.2, 59.8, 112.9, 116.3, 118.2, 120.6, 120.9, 122.1, 125.2, 129.3, 138.4, 140.1, 145.6, 146.2, 147.9, MS m/z(%):431, calcd.431.1.
4.1.3.9. 4.1.3.9. 5-Chloro-2-((3-(4-(4-methoxyphenyl)piperazin-1-yl) propyl)thio)-1H-benzo[d] imidazole 30.
Puff solid, 1H NMR (400 MHz, CDCl3) δ: 2.31(p,2H,CH2), 2.49–2.6(m, 8H, piperazine -H), 3.28(t, J = 5.75 Hz, 2H, N-CH2), 3.6(s, 3H, O-CH3), 4.2(t, J = 6 Hz, 2H, S-CH2), 7.15(d, J = 8 Hz, 2H, Ar-H), 7.173(d, J = 8.2 Hz, 2H, Ar-H), 7.43(s, 1H, CH benzimidazole), 7.48(d, J = 8 Hz, 1H, CH benzimidazole), 7.56(d, J = 8 Hz, 1H, CH benzimidazole), 13C NMR (150 MHz, DMSO-d6) δ: 23.15, 25.69, 40.07, 42.9, 50.07, 55.58 (negative in APT), 109.42 (negative in APT), 110.34 (negative in APT), 114.63 (negative in APT), 116.93 (negative in APT), 118.5 (negative in APT), 121.17 (negative in APT), 122.36 (negative in APT), 126, 126.7, 134.9, 136.8, 141.6, 143.7, 148.4, 149, MS m/z (%):416.97, calcd.417.
4.1.3.10. 4.1.3.10. 5-Chloro-2-((3-(4-(3-chlorophenyl)piperazin-1-yl) propyl)thio)-1H-benzo[d] imidazole 31.
Puff solid, 1H NMR (400 MHz, CDCl3) δ: 2.31(p,2H,CH2), 2.29–3.14(m,8H, piperazine-H), 3.29(t, J = 5.75 Hz, 2H,N-CH2), 4.2(t, J = 5.75 Hz, 2H, S-CH2), 6.76(d, J = 7.5 Hz, 1H, Ar-H), 6.88(d, J = 8.1 Hz, 1H, Ar-H), 6.91(s, 1H, Ar-H),7.15(d, J = 8 Hz, 1H, CH benzimidazole), 7.43(t, J = 8 Hz, 1H, Ar-H), 7.45(s, 1H, CH benzimidazole), 7.57(d, J = 8 Hz, 1H, CH benzimidazole), 13C NMR(100 MHz, DMSO-d6) δ:27.1, 34.3, 51.1, 53.2, 56.7, 112.2, 113.9, 115.7, 116.2, 120.9, 124.1, 129.1, 131.1, 136.1, 139.1, 140.5, 147.5, 149.9, MS m/z(%):421.38, calcd.421.4.
4.2. Experimental NMR for Drug-Design
15N-labeled Pin1 was expressed and purified according to the known protocol 39,40. The sample was concentrated to a final concentration of 200 μM in PBS buffer at pH = 6.5. Compounds number 5, 12 and 25 were dissolved in DMSO and added to the protein to achieve a final concentration of 1 mM in the NMR tube. 15N-1H-HSQC experiments have been performed for the protein alone or with the compound on Varian-900 MHz-cold probe in University of Colorado, Denver. The number of points used in the indirect dimension were 128 and with 32 scans. The spectra were processed using NMR Pipe 41 and figures were created using Sparky 42 software.
4.3. Cytotoxicity assay
Cell line: Mammary gland breast cancer (MCF-7). The cell line was obtained from ATCC via Holding company for biological products and vaccines (VACSERA), Cairo, Egypt. Doxorubicin was used as a standard anticancer drug for comparison. Chemical reagents: the reagents RPMI-1640 medium, MTT and DMSO (sigma co., St. Louis, USA), Fetal Bovine serum (GIBCO, UK) 43. The cell line mentioned above were used to determine the inhibitory effects of compounds on cell growth using the MTT assay. Cell lines were cultured in RPMI-1640 medium with 10% fetal bovine serum. Antibiotics added were 100 units/ml penicillin and 100 μg/ml streptomycin at 37 C in a 5% Co2 incubator. The cell lines were seeded in a 96-well plate at a density of 1.0×104 cells/well at 37 C for 48 h under 5% Co2. After incubation, the cells were treated with different concentration of compounds and incubated for additional 24 h. After 24 h of drug treatment, 20 μl of MTT solution at 5 mg/ml was added and incubated for 4 h. Dimethyl sulfoxide (DMSO) in volume of 100 μl is added into each well to dissolve the purple formazan formed. The absorbance was measured at wavelength 570 nm using a plate reader (EXL 800, USA). The relative cell viability in percentage was calculated as (A570 of treated samples/A570 of untreated sample) X 100 44.
4.4. Apoptosis assay
Apoptosis detection was performed by using Annexin V-FITC and PI apoptosis kit (eBioscienceTM, San Diego, CA, USA). Mcf cells were plated at a 600,000 cells/mL density onto a six well plate. After 24 h of incubation, the cells were treated with compound 15 at 10 Mg/mL. Cells grown in media containing an equivalent amount of DMSO served as the solvent control. After 24 h, the cells were stained with an Annexin V-FITC conjugate and propidium iodide (PI), and the percentage of apoptotic, necrotic, and living cells was determined according to the protocol provided by the Annexin V-FITC and PI apoptosis kit. The cells’ emitted fluorescence was analyzed by flow cytometry (NovoCyte, ACEA Biosciences Inc, San Diego, CA, US) through the NovoExpress 1.3.0 software (ACEA Biosciences Inc, San Diego, CA, US), acquiring 1 × 104 events per sample using the population plot “dot plot”, where each point corresponds to a single event with a specific fluorescence signal in reference to the axes; Annexin V-FITC green fluorescence in abscissa vs. PE red fluorescence in ordinate.
4.5. Molecular modeling
The three-dimensional structures of some selected substituted benzimidazole 5, 12, 25 which represent best anti-breast cancer activity, in their neutral forms, were built by using the MOE of Chemical Computing Group Inc software 2014. The Lowest energy conformers of new analogues ‘global-minima’ was docked into the binding pocket of 4TYO pdb 45 It was obtained from the Protein Data Bank of Brookhaven National Laboratory. The hydrogens were added, then enzyme structure was subjected a refinement protocol where the constraints on the enzyme were gradually removed and minimized until the RMSD gradient was 0.01 kcal/mol Å. Energy minimization was carried out using the molecular mechanics force field ‘AMBER.’ For each benzimidazole derivative, energy minimizations (EM) were performed using 1000 steps of steepest descent, followed by conjugate gradient minimization to a RMSD energy gradient of 0.01 Kcal/mol Å. The active site of the enzyme was detected using a radius of 10.0 Å around MTX. Energy of binding was calculated as the difference between the energy of the complex and individual energies of the enzyme and ligand 46,47. The compounds under study underwent flexible alignment experiment using ‘Molecular Operating Environment’ software (MOE of Chemical Computing Group Inc., on a Core i7 2.3 GHz workstation). The molecules were constructed using the Builder module of MOE. Their geometry was optimized by using the MMFF94 forcefield followed by a flexible alignment using systematic conformational search. The Lowest energy aligned conformers were identified. ADMET Calculations (Absorption, Distribution, Metabolism, Excretion, and Toxicity) were determined using implemented tool in MOE, 2014. Molecular structure of the tested flavonoids was constructed from fragment libraries MOE 48–50. For each analogue, the partial atomic charges were assigned using the semi-empirical mechanical calculation “AM1“ method implemented in the program 51. Conformational search was done. All the conformers were minimized until the RMSD deviation was 0.01 Kcal/mol, then subjected to surface mapping, color coded: pink, hydrogen bond; blue: mild polar; green hydrophobic.
Supplementary Material
Acknowledgements
The authors thank Alexandra Born and Professor Beat Vögeli for providing Pin1 enzyme and the magnet time at University of Colorado, Anschutz Medical Campus, Denver, USA.
Footnotes
Declarations
The authors report no conflict of interest.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmc.2020.115495.
References
- 1.Ramla MM, Omar MA, El-Khamry A-MM, El-Diwani HI. Synthesis and antitumor activity of 1-substituted-2-methyl-5-nitrobenzimidazoles. Bioorgan Med Chem. 2006;14:7324–7332. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Mohsen SA, El-Emary TI, El-Kashef HS. Synthesis, anti-inflammatory and antibacterial activities of novel pyrazolo[4,3-g]pteridines. Chem Pharm Bull (Tokyo). 2016;64:476–482. 10.1248/cpb.c16-00044. [DOI] [PubMed] [Google Scholar]
- 3.Sharma D, Narasimhan B, Kumar P, et al. Synthesis, antimicrobial and antiviral evaluation of substituted imidazole derivatives. Eur J Med Chem. 2009;44:2347–2353. 10.1016/j.ejmech.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Tonelli M, Simone M, Tasso B, et al. Antiviral activity of benzimidazole derivatives. II. Antiviral activity of 2-phenylbenzimidazole derivatives. Bioorgan Med Chem. 2010;18:2937–2953. 10.1016/j.bmc.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 5.Göker H, Özden S, Yıldız S, Boykin DW. Synthesis and potent antibacterial activity against MRSA of some novel 1,2-disubstituted-1H-benzimidazole-N-alkylated-5-carboxamidines. Eur J Med Chem. 2005;40:1062–1069. 10.1016/j.ejmech.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 6.He Y, Yang J, Wu B, Risen L, Swayze EE. Synthesis and biological evaluations of novel benzimidazoles as potential antibacterial agents. Bioorg Med Chem Lett. 2004;14:1217–1220. 10.1016/j.bmcl.2003.12.051. [DOI] [PubMed] [Google Scholar]
- 7.Sharma S, Gangal S, Rauf A. Convenient one-pot synthesis of novel 2-substituted benzimidazoles, tetrahydrobenzimidazoles and imidazoles and evaluation of their in vitro antibacterial and antifungal activities. Eur J Med Chem. 2009;44:1751–1757. 10.1016/j.ejmech.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Kamala G, Srinivasan N, Shankar KR, Suresh R. Synthesis, characterization and antimicrobial evaluation of N-Mannich bases of (2-substituted phenyl) benzimidazole derivatives. AJPR. 2018;8:87–93. [Google Scholar]
- 9.Can NÖ, Acar Çevik U, Sağlık BN, Levent S, Korkut B, Özkay Y et al. Synthesis, molecular docking studies, and antifungal activity evaluation of new benzimidazoletriazoles as potential lanosterol 14α-demethylase inhibitors. J Chem-NY 2017. [Google Scholar]
- 10.Taha M, Mosaddik A, Rahim F, Ali S, Ibrahim M, Almandil NB. Synthesis, antiglycation and antioxidant potentials of benzimidazole derivatives. J King Saud Univ – Sci. 2018. 10.1016/j.jksus.2018.04.003. [DOI] [Google Scholar]
- 11.Özil M, Parlak C, Baltaş N. A simple and efficient synthesis of benzimidazoles containing piperazine or morpholine skeleton at C-6 position as glucosidase inhibitors with antioxidant activity. Bioorg Chem. 2018;76:468–477. 10.1016/j.bioorg.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Kanwal A, Saddique FA, Aslam S, Ahmad M, Zahoor AF. Benzimidazole ring system as a privileged template for anticancer agents. Pharm Chem J. 2018;51(12):1068–1077. 10.1007/s11094-018-1742-4. [DOI] [Google Scholar]
- 13.Cheong JE, Zaffagni M, Chung I, et al. Synthesis and anticancer activity of novel water soluble benzimidazole carbamates. Eur J Med Chem. 2018;144:372–385. 10.1016/j.ejmech.2017.11.037. [DOI] [PubMed] [Google Scholar]
- 14.Abonia R, Cortés E, Insuasty B, Quiroga J, Nogueras M, Cobo J. Synthesis of novel 1,2,5-trisubstituted benzimidazoles as potential antitumor agents. Eur J Med Chem. 2011;46:4062–4070. [DOI] [PubMed] [Google Scholar]
- 15.Kamal A, Reddy TS, Vishnuvardhan MVPS, et al. Synthesis of 2-aryl-1,2,4-oxadiazolo-benzimidazoles: tubulin polymerization inhibitors and apoptosis inducing agents. Bioorgan Med Chem. 2015;23:4608–4623. [DOI] [PubMed] [Google Scholar]
- 16.Kamal A, Shaik AB, Polepalli S, et al. Synthesis of arylpyrazole linked benzimidazole conjugates as potential microtubule disruptors. Bioorgan Med Chem. 2015;23:1082–1095. [DOI] [PubMed] [Google Scholar]
- 17.Husain A, Rashid M, Mishra R, Parveen S, Shin D-S, Kumar D. Benzimidazole bearing oxadiazole and triazolo-thiadiazoles nucleus: design and synthesis as anticancer agents. Bioorg Med Chem Lett. 2012;22:5438–5444. [DOI] [PubMed] [Google Scholar]
- 18.Rashid M, Husain A, Mishra R. Synthesis of benzimidazoles bearing oxadiazole nucleus as anticancer agents. Eur J Med Chem. 2012;54:855–866. [DOI] [PubMed] [Google Scholar]
- 19.Husain A, Rashid M, Shaharyar M, Siddiqui AA, Mishra R. Benzimidazole clubbed with triazolo-thiadiazoles and triazolo-thiadiazines: new anticancer agents. Eur J Med Chem. 2013;62:785–798. [DOI] [PubMed] [Google Scholar]
- 20.Dou Y-H, Xu S-D, Chen Y, Wu X-H. Synthesis, characterization, and anticancer activity of dithiocarbamate ruthenium (II) complexes. Phosphorus Sulfur. 2017;192:1219–1223. [Google Scholar]
- 21.Buac D, Schmitt S, Ventro G, Rani Kona F, Ping Dou Q. Dithiocarbamate-based coordination compounds as potent proteasome inhibitors in human cancer cells. MiniRev Med Chem. 2012;12:1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cvek B Dithiocarbamate complexes with metals in cancer therapy. Mini-Rev Med Chem. 2012;12(12):1173. [DOI] [PubMed] [Google Scholar]
- 23.Bala V, Gupta G, L Sharma V. Chemical and medicinal versatility of dithiocarbamates: an overview. Mini-Rev Med Chem 2014;14:1021–1032. [DOI] [PubMed] [Google Scholar]
- 24.Bacharaju K, Jambula SR, Sivan S, JyostnaTangeda S, Manga V. Design, synthesis, molecular docking and biological evaluation of new dithiocarbamates substituted benzimidazole and chalcones as possible chemotherapeutic agents. Bioorg Med Chem Lett. 2012;22:3274–3277. 10.1016/j.bmcl.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 25.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996:544–547. [DOI] [PubMed] [Google Scholar]
- 26.Shen M, Stukenberg PT, Kirschner MW, Lu KP. The essential mitotic peptidyl-prolyl isomerase pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev. 1998;12(5):706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liou Y-C, Zhou XZ, Lu KP. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci. 2011;36(10):501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajbhandari P, Finn G, Solodin NM, et al. Regulation of estrogen receptor α N-terminus conformation and function by peptidyl prolyl isomerase Pin1. Mol Cell Biol. 2012;32(2):445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baurin Nicolas, #; Aboul-Ela Fareed; Barril Xavier; Davis Ben; Drysdale Martin; Dymock Brian; Finch Harry; Fromont Christophe; Richardson Christine; Simmonite Heather, and; et al. Design and Characterization of Libraries of Molecular Fragments for Use in NMR Screening against Protein Targets. J. Chem. Inf. Comput. Sci. 2004, 44, 6, 2157–2166. [DOI] [PubMed] [Google Scholar]
- 30.Potter AJ, Ray S, Gueritz L, et al. Structure-guided design of α-amino acid-derived Pin1 inhibitors. Bioorg Med Chem Lett. 2010;20(2):586–590. [DOI] [PubMed] [Google Scholar]
- 31.Dong L, Marakovits J, Hou X, Guo C, Greasley S, Dagostino E et al.Bioorg Med Chem Lett 20;2010:2210. [DOI] [PubMed] [Google Scholar]
- 33.Byrne C, Henen MA, Belnou M, et al. A β-turn motif in the steroid hormone receptor’s ligand-binding domains interacts with the peptidyl-prolyl isomerase (PPIase) catalytic site of the immunophilin FKBP52. Biochemistry. 2016;55(38):5366–5376. [DOI] [PubMed] [Google Scholar]
- 34.Cicero DO, Barbato G, Bazzo R. Sensitivity enhancement of a two-dimensional experiment for the measurement of heteronuclear long-range coupling constants, by a new scheme of coherence selection by gradients. J Magn Reson. 2001;148:209–213. 10.1006/jmre.2000.2234). [DOI] [PubMed] [Google Scholar]
- 35.Bax A, Griffey RH, Hawkins BL. Correlation of proton and nitrogen-15 chemical shifts by multiple quantum NMR. J Magn Reson. 1983;55:301–315. 10.1016/0022-2364(83)90241-X). [DOI] [Google Scholar]
- 36.Samir Bondock, Shymaa Adel, Etman Hassan A Badria Farid A. Synthesis and antitumor evaluation of some new 1,3,4-oxadiazole-based Heterocycles EUR. J Med Chem. 2012;48:192e199. [DOI] [PubMed] [Google Scholar]
- 37.Thabrew MI, Hughes RD, McFarlane IG. J Pharm Pharmacol 1997;49:1132e1135. [DOI] [PubMed] [Google Scholar]
- 38.Potter Andrew J., Ray Stuart, Gueritz Louisa, Nunns Claire L., Bryant Christopher J., Scrace Simon F., et al. Moore Structure-guided design of a-amino acid-derived Pin1 inhibitors Bioorg Med Chem Lett 2010;20:586–590. [DOI] [PubMed] [Google Scholar]
- 39.Born A, Henen M, Nichols P, Wang J, Jones D, Vogeli B. Efficient stereospecific Hβ2/3 NMR assignment strategy for mid-size proteins. Magnetochemistry. 2018;4(2):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Born A; Henen M; Nichols P; Wang J; Jones D; Vogeli B Efficient Stereospecific Hβ2/3 NMR Assignment Strategy for Mid-Size Proteins. MAGNETOCHEMISTRY 2018, 4 (2), 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multi-dimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. [DOI] [PubMed] [Google Scholar]
- 42.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31(8):1325–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mosmann T Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 44.Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival. J Immunol Methods. 1986;89:271–277. [DOI] [PubMed] [Google Scholar]
- 45.Guo C, Hou X, Dong L, Marakovits J, Greasley S, Dagostino E et al. Structure-based design of novel human Pin1 inhibitors (III): Optimizing affinity beyond the phosphate recognition pocket. 4TYO (2014) Bioorg Med Chem Lett 2014;24:4187–4191. [DOI] [PubMed] [Google Scholar]
- 46.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliver Rev. 1997;23:3–25. [DOI] [PubMed] [Google Scholar]
- 47.Profeta S, Allinger NL. Molecular mechanics calculations on aliphatic amines. JACS. 1985;107:1907–1918. [Google Scholar]
- 48.Allinger NL. Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms. JACS. 1977;99:8127–8134. [Google Scholar]
- 49.Labute P, Williams C, Feher M, Sourial E, Schmidt JM. Flexible alignment of small molecules. J Med Chem. 2001;44:1483–1490. [DOI] [PubMed] [Google Scholar]
- 50.Kearsley S, Smith GM. An alternative method for the alignment of molecular structures: maximizing electrostatic and steric overlap. Tetrahedron Comput Methodol. 1990;3:615–633. [Google Scholar]
- 51.Cornell WD, Cieplak P, Bayly CI, Gould IR, et al. A second Generation forth field for the simulation of Proteins and Nucleic Acids. JACS. 1995;117:5179–5197. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.