Abstract
Fibrillar collagen–integrin interactions in the extracellular matrix (ECM) regulate a multitude of cellular processes and cell signalling. Collagen I fibrils serve as the molecular scaffolding for connective tissues throughout the human body and are the most abundant protein building blocks in the ECM. The ECM environment is diverse, made up of several ECM proteins, enzymes, and proteoglycans. In particular, glycosaminoglycans (GAGs), anionic polysaccharides that decorate proteoglycans, become depleted in the ECM with natural aging and their mis‐regulation has been linked to cancers and other diseases. The impact of GAG depletion in the ECM environment on collagen I protein interactions and on mechanical properties is not well understood. Here, we integrate ELISA protein binding assays with liquid high‐resolution atomic force microscopy (AFM) to assess the effects of GAG depletion on the interaction of collagen I fibrils with the integrin α2I domain using separate rat tails. ELISA binding assays demonstrate that α2I preferentially binds to GAG‐depleted collagen I fibrils in comparison to native fibrils. By amplitude modulated AFM in air and in solution, we find that GAG‐depleted collagen I fibrils retain structural features of the native fibrils, including their characteristic D‐banding pattern, a key structural motif. AFM fast force mapping in solution shows that GAG depletion reduces the stiffness of individual fibrils, lowering the indentation modulus by half compared to native fibrils. Together these results shed new light on how GAGs influence collagen I fibril–integrin interactions and may aid in strategies to treat diseases that result from GAG mis‐regulation.
Keywords: atomic force microscopy, collagen, collagen interactions, extracellular matrix, fast force mapping AFM, fibril, glycosaminoglycans, integrin, solution AFM
Abbreviations
- AFM
atomic force microscopy
- AM‐AFM
amplitude‐modulation AFM
- ANOVA
analysis of variance
- BSA
bovine serum albumin
- CMP
collagen model peptide
- DMMB
dimethylmethylene blue
- ECM
extracellular matrix
- EDTA
ethylenediaminetetraacetic acid
- ELISA
enzyme‐linked immunosorbent assay
- FFM‐AFM
fast force mapping AFM
- GAG
glycosaminoglycan
- InVOLS
inverse optical lever sensitivity
- PG
proteoglycan
- α2I
I‐domain of integrin α2 subunit
1. INTRODUCTION
Intermolecular interactions between fibrillar collagens and collagen‐binding integrins in the extracellular matrix (ECM) drive numerous cellular processes, including cell adhesion and motility, platelet aggregation, cell development, differentiation, and hemostasis. 1 , 2 , 3 While much knowledge has been reaped from studying integrin binding to triple helical collagen model peptides, the ECM is quite complex, composed of collagen fibril assemblies and also other ECM proteins, matrix metalloproteinases, and proteoglycans (PGs), which contain glycosaminoglycan (GAG) polysaccharides 4 (Figure 1a). As part of the natural aging process, the ECM environment will be modified and the GAG content altered. 5 , 6 In this study, we probe how GAG depletion impacts the mechanical properties of collagen I and its critical interactions with the integrin α2I domain.
FIGURE 1.
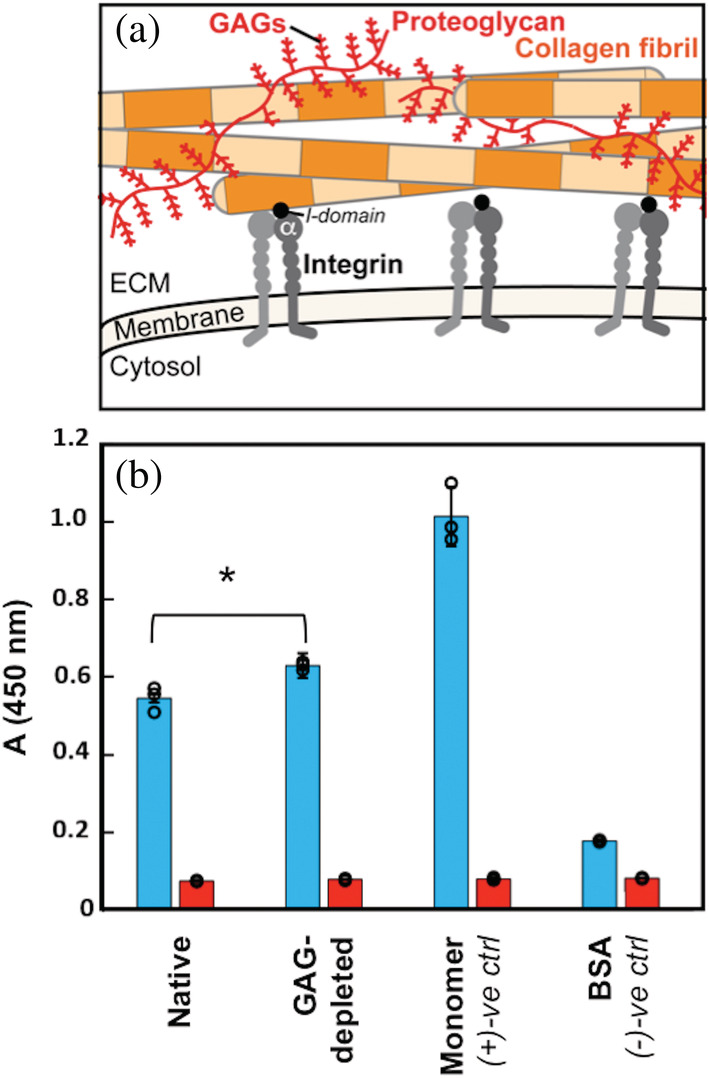
Collagen I fibril–integrin α2I interactions in the native ECM environment. (a) Cartoon highlighting multiple components of the ECM. In this study, we focus on collagen I fibrils (orange); integrin cell surface receptors (gray), specifically the I‐domain (black) of the α‐subunit (αI); and glycosaminoglycans (GAGs), which are polysaccharide chains displayed on proteoglycans (red). Integrin αI domains and GAGs interact with individual collagen I fibrils in the ECM. (b) ELISA binding assay of integrin α2I (10 μg/ml) binding to collagen I in varying contexts (native: collagen I fibrils from rat tail tendon ECM in phosophate buffered saline (PBS), GAG‐depleted: collagen I fibrils from ECM with an estimated 85% GAG depletion in PBS, monomer: purified collagen I from rat tail tendon in 10 mM acetic acid, and BSA: 50 mg/mL bovine‐serum albumin in PBS). A one‐way ANOVA analysis confirmed that there is a significant difference between α2I binding to native and GAG‐depleted collagen I fibrils (15% increase for GAG‐depleted collagen I fibrils) with a p‐value of <0.05 (*). Each binding condition was performed in the presence of either 5 mM MgCl2 (blue) or 5 mM EDTA (red). Error bars indicate the standard deviation of the measurements, which were taken in triplicate. Individual measurements are shown as circles. ECM, extracellular matrix
The most abundant fibrillar collagen in the human body is collagen I, primarily found in tendons, bone, and skin. 7 , 8 Collagen I self‐assembles into fibrillar structures that are up to hundreds of nanometers in diameter and hundreds of micrometers long. 9 , 10 , 11 , 12 These fibrils are made up of triple helical monomer units, heterotrimers of three protomer chains. 9 , 10 , 11 , 12 The uniform staggered arrangement of triple helical monomers within the fibrils gives the fibrils a banded topography defined by regions of low‐ and high‐protein density, referred to as the gap and overlap regions. 9 , 10 , 11 , 12 This repeating pattern, called D‐banding, has a characteristic periodicity of 67 nm that can be observed by high‐resolution microscopy methods. 9 , 10 , 11 , 12 These fibril units serve as the main building blocks of collagen in the ECM and act as the molecular scaffolding for cells that make up connective tissues. 9 , 10 , 11 , 12 Collagen I is highly biologically active, interacting with numerous ECM biomolecules (Figure 1a) to enable cell signaling, regulate collagen turnover, and facilitate many cellular processes. 7 , 8 , 13 , 14
Integrins are an important class of collagen interaction partners for cell signaling. 1 One of the most widely distributed collagen‐binding integrins, integrin α2β1 is primarily found on platelets and epithelial cells. 1 , 2 , 3 , 15 Integrin α2β1 interacts with collagen I via a divalent metal coordination of its metal ion‐dependent adhesion site (MIDAS) on the inserted (I) domain of the α2 subunit (α2I) with a glutamate in a GXX'GEX” motif in collagen I, where X' is often hydroxyproline (O) and X” is R or N. 16 , 17 , 18 , 19 The α2I domain retains its structure and interaction affinity for collagen I in isolation. 17 Thus, it can be used as a biologically active mimic for collagen I–integrin α2β1 interactions. Previously, our lab and others have studied collagen–integrin αI interactions in the context of triple helical collagen model peptides (CMPs). 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 Farndale et al. have developed a library of CMPs to determine the specificity of integrin αI domains for collagen binding motifs. 28 The GFOGER sequence was identified as a high‐affinity motif in collagen I, 18 and a number of lower affinity sites were identified with a canonical sequence of GXOGER/N. 16 , 26 We have investigated how collagen–αI interactions are perturbed upon modulation of these peptide sequences or in the presence of disease‐affiliated substitutions. 20 , 21 , 22 , 23 However, in the ECM, collagen I triple helical monomers assemble into fibrils. In these CMPs and collagen monomers, all possible integrin binding sites are fully exposed. Yet, upon assembling into fibrils, many binding sites become buried within the fibril core and seemingly inaccessible from the fibril surface. We have previously proposed that molecular motions on the collagen I fibril surface may facilitate increased surface accessibility to some integrin recognition sites that are otherwise hidden by the complex fibril architecture. 29
In the ECM, the highly conserved collagen assembly is regulated by PGs, in which protein cores are bound to anionic polysaccharide chains, called GAGs. 5 , 30 A large diversity of PGs and GAGs are distributed across connective tissues for proper ECM activity. 5 , 30 In all tissues, PGs recognize a number of specific amino acid sequence motifs on collagen fibrils 5 to act as part of the ECM scaffolding. In addition to their structural roles, GAGs have been shown to be essential for wound healing, cell adhesion, and cell motility. 5 As humans age, GAGs become depleted in the ECM 5 , 6 and the mis‐regulation of GAGs is associated with various cancers, diabetes, and fibrosis. 5 , 6 , 31 , 32 , 33 , 34 , 35 It has previously been proposed that mis‐regulation of GAGs perturbs the mechanical properties of the ECM, leading to a loss of biological function and aberrant cell activity. 36 The mechanical properties of collagen impact its molecular interactions, and collagen stiffness has been shown to direct cell movement and determine cell fate. 37 , 38 However, the mechanism and extent to which GAGs alter the mechanical properties of individual collagen I fibrils and its association with fibril function has not been elucidated.
Given that collagenous tissues function in a fully hydrated environment, it is important to investigate the fibrillar form of collagen I in solution. In this study, we probe the influence of GAG depletion on the (1) collagen–integrin α2I binding activity using an enzyme‐linked immunosorbent assay (ELISA) and (2) structure and mechanical properties of collagen I fibrils through advanced AFM techniques in solution. Together, these results provide a better understanding of how GAG depletion, an inevitable result of aging, impacts the interactions of collagen I fibrils with its cell receptor, integrin α2I, in its native environment.
2. RESULTS
2.1. Assessing the impact of GAG depletion on collagen–integrin α2I domain interactions
To determine the impact of GAG depletion on collagen–α2I interactions, we compared ELISA binding assays of recombinant α2I to native collagen I fibrils and to chondroitinase treated collagen I fibrils. For these experiments, fibrils were derived from the same rat tail (Figure 1b). As the collagen I–α2I interaction is dependent on coordination of a divalent metal cation, the assay is performed in the presence of Mg2+ or ethylenediaminetetraacetic acid (EDTA) as a negative control. As expected, minimal α2I binding is observed in the presence of EDTA, where metal ions are depleted. Bovine serum albumin (BSA) was used as a negative control, as no specific α2I interaction is expected. Acid solubilized, purified collagen I triple helical monomer serves as a positive control for collagen I–α2I binding. We see a high level of α2I binding to this triple helical collagen I monomer, consistent with all integrin binding sites being fully exposed. The integrin α2I domain shows a lower affinity to native collagen fibrils relative to collagen triple helical monomers (Figure 1b). This has been shown previously by other labs, which attribute this reduced affinity to the reduced accessibility of integrin binding sites, 39 many of which are bundled into the fibril core upon fibril assembly. GAG‐depleted fibrils, relative to native fibrils, show slightly higher binding to integrin α2I (Figure 1b). The affinity of α2I to GAG‐depleted fibrils is trending toward the fully exposed triple helical monomer. This may suggest that integrin binding sites within the GAG‐depleted collagen I fibrils are becoming more accessible upon removal of GAGs. Both native and GAG‐depleted collagen I fibrils display dose‐dependent binding to α2I in ELISA assays (Figure 2). For this second set of ELISA assays, fibrils were derived from a rat tail separate from that used in Figure 1. For all concentrations of α2I, GAG‐depleted fibrils consistently have an increased response to α2I relative to native fibrils, reinforcing the stronger affinity of α2I to GAG‐depleted collagen I fibrils. Thus, the ability of integrin α2I to interact with collagen I fibrils is modulated by GAGs in the ECM environment, and the regulation of GAG concentration may be imperative for proper collagen activity. In order to better understand how the presence of GAGs modulates collagen I fibril–α2I interactions, we probed how GAG content modulates the structure and mechanical properties of the individual collagen I fibrils.
FIGURE 2.
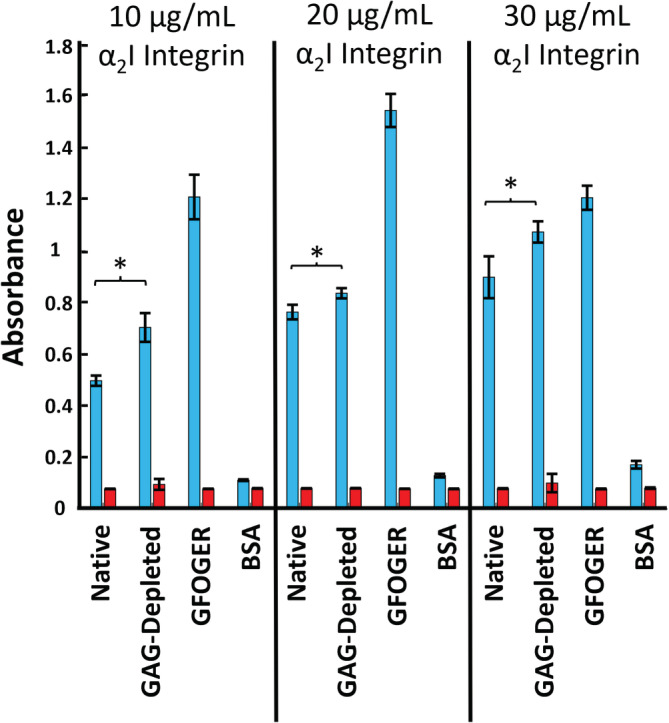
Dose‐dependent binding of integrin α2I to collagen I fibrils. ELISA binding assay of integrin α2I to collagen I fibrils with increasing concentrations of α2I. (Native: collagen I fibrils from rat tail tendon ECM in PBS, GAG‐depleted: collagen I fibrils from ECM with an estimated 70% GAG depletion in PBS, GFOGER: synthetic triple helical collagen mimetic peptide (Ac‐GPC‐GPP5‐GFOGER‐GPP5‐GPC‐GY‐NH2) containing the high affinity α2I binding sequence, GFOGER, in 10 mM acetic acid, and BSA: 50 mg/mL bovine‐serum albumin in PBS.) Each binding condition was performed in the presence of either 5 mM MgCl2 (blue) or 5 mM EDTA (red). Error bars indicate the standard deviation of the measurements, which were taken in triplicate. “*” indicates significant differences with a p‐value of <0.05. ECM, extracellular matrix; GAG, glycosaminoglycans
2.2. Characterizing native collagen I fibrils in solution
The emergence of sophisticated AFM imaging modes has enabled direct, high‐resolution images of structure or mechanical/surface properties of biological systems in situ under physiological aqueous conditions. 40 , 41 We characterized the topography of intact collagen I fibrils extracted from rat tail tendon in air and in solution by amplitude‐modulation (AM‐) AFM. These experiments were performed using collagen derived from a rat tail separate from the ELISA experiments. Collagen I fibrils are expected to display a canonical repeating 67‐nm banding pattern that is produced by the staggered arrangement of triple helical units, referred to as D‐banding. 42 AM‐AFM confirms such banding is present in rat tail tendon collagen I fibrils adsorbed to a glass slide in air and when immersed in phosphate buffered saline (PBS) buffer solution (Figure 3). Here, the sinusoidal periodicity in the height along the main fibril axis is observed in both imaging environments, with alternating high (“overlap,” yellow), and low (“gap,” red) regions. Height profiles extracted from the apex of the collagen I fibrils in air reveal a distinct sinusoidal oscillation of overlap and gap regions (Figure S1a,c). Although this periodicity in the height profiles is not as clear from images in PBS, it is discernible in the phase channel (Figure S2a,b). Two‐dimensional Fourier transforms of the phase channel from imaging native collagen I fibrils revealed an average periodicity of 65.6 ± 1.0 nm (N = 5) in air and 65.8 ± 0.8 nm (N = 5) in PBS (Table 1). Thus, the fibril periodicity, a structural hallmark of fibrillar collagens, is maintained in an aqueous environment.
FIGURE 3.
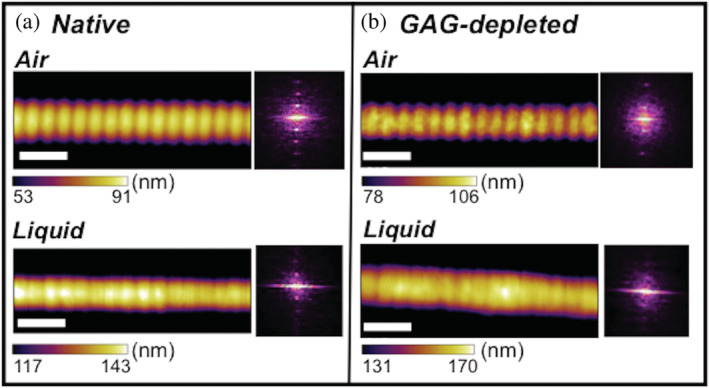
AM‐AFM imaging of collagen I fibrils. Collagen I fibrils bear a regular, characteristic D‐banding pattern that is produced by the staggered arrangement of triple helical collagen monomers in the fibril. This D‐banding is resolved here by AM‐AFM experiments for both (a) native and (b) GAG‐depleted collagen I fibrils (with an estimated 76% GAG depletion) imaged in air and liquid environments. Fibril heights of five fibrils from the same rat tail for all conditions ranged from ≈55–160 nm (Avg. 117.1 ± 47.4 nm) for native fibrils and ≈100–250 nm (Avg. 156.6 ± 59.6 nm) for GAG‐depleted fibrils in air and ≈100–285 nm (Avg. 209.5 ± 85.3 nm) for native fibrils and ≈165–430 (Avg. 286.6 ± 96.5 nm) for GAG‐depleted fibrils in liquid. It is important to note that although the native and GAG‐depleted fibrils may differ in height, fibril height has been shown to not correlate with indentation modulus. 53 The white scale bars are 200 nm in each image. The Fourier transform insets show the periodicity of each fibril and scale bar ranges were chosen to accentuate the D‐banding in the images. Uncropped height, amplitude, and phase images are shown in the SI (Figures S2, S3). AM‐AFM amplitude modulation‐atomic force microscopy; ECM, extracellular matrix; GAG, glycosaminoglycans
TABLE 1.
Overview of results for the control and chondroitinase treated fibrils
| Native | GAG‐depleted | |
|---|---|---|
| Periodicity (air) | 65.6 ± 1.0 nm | 66.0 ± 0.2 nm |
| Periodicity (liquid) | 65.8 ± 0.8 nm | 66.2 ± 1.0 nm |
| Overlap‐gap height difference (air) | 3.6 ± 2.4 nm | 3.6 ± 2.7 nm |
| Modulus (liquid) | 6.4 ± 1.4 MPa | 2.9 ± 1.0 MPa |
Note: Visualization of the distribution of these values are shown in the SI (Figure S7).
Abbreviation: GAG, glycosaminoglycans.
2.3. The fibril nanoscale structure is maintained upon depletion of GAGs
We then asked if this characteristic structure of individual collagen I fibrils is impacted by the concentration of GAGs in their environment. We used AM‐AFM to probe perturbations to the topography of collagen I fibrils in ECM with an estimated 76% reduced GAG concentration upon treatment with chondroitinase. GAG‐depleted collagen I fibrils display D‐band periodicities of 66.0 ± 0.2 nm in air (N = 5) and 66.2 ± 1.1 nm in PBS (N = 4), as measured from 2D Fourier transforms of the phase channel (Table 1). This is consistent with the native fibril data and with previously reported AFM results of chondroitinase‐treated tendons in air 43 .
Another assessment of fibril surface topography is the overlap–gap step height. This is the difference between the maximum height of the overlap and the minimum height of the gap (Figure S4). Upon GAG depletion, the overlap–gap step height also remained unchanged, as chondroitinase‐treated collagen I fibrils exhibit an overlap–gap step height of 3.6 ± 2.7 nm in air compared to 3.6 ± 2.4 nm in native collagen I fibrils (Figure S7a). Step‐heights in liquid were unable to be determined as gap and overlap regions were indistinguishable across height profiles (Figure S1b,d). Overall, the data show that GAG dissociation does not significantly impact fibril periodicity or topography. This indicates that the collagen I fibril nanoscale surface structure is preserved upon GAG depletion in aqueous solution.
2.4. AFM force mapping reveals a reduction in stiffness of individual collagen I fibrils upon GAG depletion relative to native fibrils
Although GAG depletion does not significantly affect the surface structure of collagen I fibrils, we asked if the mechanical properties of the fibrils were perturbed upon GAG depletion. We used fast force mapping (FFM‐) AFM to determine the indentation modulus, a measure of local nanoscale stiffness, of individual collagen I fibrils in solution in native and GAG‐depleted states. Figure 4 presents a 1 × 1 μm force map of native and GAG‐depleted collagen I fibrils, respectively. Unlike in Figure 3, which is an amplitude‐modulated height image, it is important to emphasize that these images are 2D maps displaying the indentation modulus of each pixel (Figure S6). This enables mechanical data to be assessed at different locations within the D‐bands along the fibrils.
FIGURE 4.
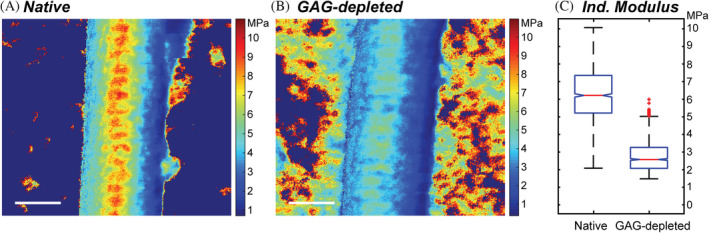
Nanoscale indentation modulus maps of collagen I fibrils in solution. (a,b) Representative 1 × 1 μm fast force maps of individual (a) native and (b) GAG‐depleted collagen fibrils from FFM‐AFM experiments. The color at each pixel shows the local nanoscale indentation moduli (1 pixel/every ~15 nm2). The FFM images are spatial reconstructions of 65,536 discrete force–distance curves that were fitted using standard indentation protocols 53 and subsequently replotted in Cartesian space using in‐house MATLAB code. A large scan size was selected to capture more than 10 D‐bands per image, enabling meaningful comparisons between internal regions, fibrils, and GAG concentration. White scale bars indicate 200 nm. (c) Box and whisker plots compare a subset of indentation moduli data in (a) and (b) for force curves acquired only at the apex down the fibril long axes. In (c), data for five fibrils in each condition (derived from a native or GAG‐depleted tendon taken from the same rat tail) are presented for increased statistical confidence. A one‐way ANOVA analysis confirmed that there is a significant difference between the conditions with a p‐value of <0.01. (Box and whisker plot statistics, Native: IQR = 2.22 MPa, Median = 6.39 MPa, Q1/Q3 = 5.31/7.53 MPa, Outlier cutoffs = [1.98–10.85 MPa], GAG depleted: IQR = 1.13 MPa, Median = 2.64 MPa, Q1/Q3 = 2.13/3.27 MPa Outlier cutoffs = [0.44–4.96 MPa]). ECM, extracellular matrix; FFM, fast force mapping; FFM‐AFM fast force mapping‐atomic force microscopy; GAG, glycosaminoglycans
Notably, D‐banding is evident in both force maps; the overlap region systematically has a larger indentation modulus than the gap regions, leading to a distinct light/dark patterning every ~67 nm down the long axis. While the demarcation between gap/overlap is less well defined than for AM‐AFM data, the results show that even in solution, collagen mechanical properties oscillate as dictated by the underlying D‐banded structure. The indentation modulus is influenced by the distinct molecular packing in the gap and overlap regions. In the collagen I fibril assembly, the gap regions are devoid of one triple helical monomer per minimal fibril unit (five collagen I monomers per minimal fibril unit in the overlap region compared to four monomers per minimal fibril unit in the gap). These gap and overlap structures are maintained and repeated from the surface layer to the fibril core. Figure S8 provides a more comprehensive explanation of the collagen fibril structure. As a result of the fibril structure, the overlap regions will be stiffer in comparison to the gap regions. Previous studies have reported a 20% increase in stiffness of the overlap region relative to the gap region as measured by nanoindentation and attributed it to the 20% denser monomer packing in the overlap region relative to the gap region. 44
Native and GAG‐depleted collagen I fibrils display important differences in the forces measured. A significant reduction in average local indentation modulus is evident upon GAG depletion relative to the native fibril, while preserving the native D‐banded structure. In Figure 4a, overlap regions of the native fibril have moduli of ~9 MPa (red) compared to ~5 MPa (light blue) for GAG‐depleted fibrils. Likewise, corresponding gap regions shift from ~7 MPa (yellow) to ~2 MPa (dark blue) upon GAG depletion (Figure 4b). To better understand these differences, we focused on the 256 force curves along the apex of each fibril (Figure 4c). The native collagen I fibrils exhibit a mean indentation modulus of 6.4 ± 1.4 MPa with a broad distribution of values over the fibrils measured, using an indentation speed of 16 μm/s (Figure 4a,c, S7c). This is consistent with reported indentation moduli of collagen I fibrils with similar indentation speeds in solution. 44 For GAG‐depleted fibrils, both the average indentation modulus (2.9 ± 1.0 MPa) and distribution is reduced (Figure 4b,c), indicating that GAG depletion reduces fibril stiffness relative to the native fibrils.
3. DISCUSSION
With an increasingly aging population, it is important to understand the impact of GAG depletion on interaction processes in the ECM of connective tissues. Our data, using ELISA assays and liquid high‐resolution AFM, indicate that GAG depletion within collagen I fibrils derived from rat tail tendon impacts on both the stiffness of the collagen I fibrils as well as the binding to the integrin α2I domain relative to the native fibril. We show that GAG depletion reduces the indentation modulus by a factor of two‐to‐three relative to the native fibril and that the reduction is apparent across the length of the entire fibril, indicating that the stiffness of the entire collagen I fibril is reduced. The breadth of the effect on the fibril stiffness is consistent with the identification of PG binding sites across broad regions of both the gap and overlap regions within the fibril structure. 45 , 46 , 47 , 48 , 49
In order to probe the impact of GAG depletion on collagen I fibril activity, we monitored the change in integrin α2I binding to collagen I fibrils in a GAG depleted state relative to a native environment. We find that α2I binding to collagen I fibrils is increased upon GAG depletion of the fibrils, trending toward the higher affinity of α2I to fully exposed, monomeric collagen I or triple helical collagen I peptides. Extensive studies using triple helical model peptides have identified unique α2I binding sites on fibrillar collagens. 16 , 18 , 19 , 25 These binding sites are fully exposed in triple helical peptides or the full‐length collagen I monomer. However, when the monomers become incorporated into the fibril, many of these integrin binding sites become buried within the fibril core. Studies of collagen fibrils from different tissues such as cartilage and tendon support the view that the fibril environment and the types of collagen that constitute the fibril are important for integrin binding. 39 , 50 For instance, cartilage is composed primarily of collagen II, IX, and XI, whereas tendon is composed of type I and in this context, studies shows that integrin α2β1 binds differently under these different tissue contexts. 50 Here, we investigate collagen I–integrin interactions in their native context of the ECM from rat tail tendons to understand how environmental factors influence these interactions.
We propose that enhanced integrin binding to collagen I fibrils in response to GAG depletion may arise from two different factors or a combination thereof. One proposal is that GAG depletion on the surface of the collagen fibrils results in increased accessibility of α2I to collagen binding sites as a result of increased fibril exposure. It has been shown via detailed mapping of interaction sites onto the fibril structure that some PG binding regions overlap with integrin binding sites. 46 Therefore, it is possible that the interaction of GAGs with the collagen I fibril may interfere or compete with binding by integrins. A second proposal is that the two to threefold reduction in indentation modulus and reduced stiffness of the GAG‐depleted collagen I fibrils relative to the native state may modify the inherent binding affinity of α2I to collagen. One aspect by which inherent α2I binding may be modulated is that reduced indentation may promote a higher propensity for deformation within the collagen fibril. Despite our observation that nanoscale topography of collagen I fibrils does not change upon GAG‐depletion, this deformation propensity may allow rearrangement of monomers within the fibril and grant α2I access to binding sites within the core that are otherwise inaccessible. Integrin binding sites that are exposed in the collagen model peptides are proposed to be buried and inaccessible from the fibril surface in the complex collagen fibril architecture. 39 , 51 We have previously proposed, through molecular dynamics simulations on the nanosecond timescale that molecular rearrangements may facilitate access to otherwise hidden binding sites of integrin 29 and that monomer fluctuations within the fibril may play a role in regulating collagen–integrin interactions.
In conclusion, we have shown that under physiological solution conditions, GAG depletion enhances collagen I fibril–integrin α2I domain interactions and decreases the indentation modulus of individual collagen I fibrils relative to native fibrils while maintaining their native nanoscale topography. Integrin α2I domain interactions and physical and mechanical properties of collagen I fibrils were separately characterized in tendons from different rat tails. It is increasingly clear that alteration of GAG concentration is intimately associated with the progression of aging, 5 , 6 which leads to disruptions in the functional landscape of the ECM. Still, further research will be required to make a direct association between GAG depletion and cell activity or how the effect propagates to longer length scales beyond the fibril building block. The current study provides new knowledge of how GAG depletion affects collagen I fibril–integrin interactions and how it modulates collagen I fibril structure and mechanics. This expands on the understanding of how aging processes, namely alterations in GAG levels, manifest in the ECM and may aid in the development of new treatment strategies.
4. MATERIALS AND METHODS
4.1. Chondroitinase treatment of rat tail tendons
Rat tail tendons were harvested from frozen–thawed rat tails and only three individual tails were used for all experiments: one tail was used for AFM samples while a second tail and third tail were used for ELISA assay preparation (one tail was used for the ELISA assay in Figure 1 while another tail was used for Figure 2). Flowcharts depicting the preparation of rat tail tendons for AFM and ELISA assays are shown in Figures S9 and S10. In short, eight sections of tendon several centimeters in length were cut and weighed (≈3–6 mg in weight each) from the exposed tail tendons of a singular rat tail and then subdivided into two groups with four sections being used for the native samples and four sections being used for the GAG‐depleted samples. Chondroitinase treatment of tendons was performed in a similar manner to previously published methods. 43 For chondroitinase treatment, the tail tendon sections were placed into a 4 ml solution of 0.15 U/ml chondroitinase ABC (Sigma‐Aldrich) in 0.1 M sodium acetate and 0.1 M Tris–HCl, pH 8. For the native, tendons were placed in the same buffer in the absence of chondroitinase. The tendons were incubated in their respective solutions at 37°C overnight. After incubation, tendon sections would then be used for AFM imaging, GAG quantification, or the ELISA assay.
4.2. Quantification of GAG depletion
GAG depletion was quantified by using the dimethylmethylene‐blue (DMMB) assay. 52 Three native and three GAG depleted tendon sections, from the same tail used for either the AFM or ELISA experiments, following the incubation were then rinsed with water and placed into 1 ml of a papain solution (500 μg papain, 0.1 M dibasic sodium phosphate, 0.01 M EDTA, 0.0144 M L‐cysteine) at 60°C for 24 hr to digest the tendons. The resulting solution was stained with DMMB and absorbance was measured at 525 nm. A standard curve of varying chondroitin sulfate concentrations (0.25–1.5 μg/ml) was used to quantify the GAG concentration in the GAG‐depleted and native samples. The DMMB assay could not be performed on the actual tendon sections that were used for the AFM study or ELISA binding assays as this would compromise the samples. However, in light of the very low variability in GAG content for the native and GAG‐depleted samples (e.g., AFM: native = 1.9–2.2 μg GAG/mg tendon, GAG depleted = 0.12–0.55 μg GAG/mg tendon) and the fact that the GAG reduction was performed on all four sections simultaneously and identically, we estimate that the average GAG content is the average of the values in the other three sections. Results of the DMMB assay for all experiments are shown in Figure S11.
4.3. Integrin α2I domain expression and purification
The integrin α2I domain (residues 142–336) was recombinantly expressed in Escherichia coli BL21(DE3) cells with an N‐terminal His6 tag by induction with 1 mM of IPTG for 16 hr at 25°C to stimulate protein production and then harvested. The cells were lysed with 20% sucrose TES buffer. The His6‐α2I domain was purified using a Ni2+‐charged HisTrap HP column (GE Healthcare Life) and buffer exchanged to PBS, pH 7.4 with a PD‐10 desalting column (GE Healthcare Life). Protein concentration was determined by measuring A 280 with a molar extinction coefficient of 20,400 M−1 cm−1.
4.4. ELISA assay
Following incubation, one native and one GAG‐depleted tendon section (between 3 and 6 mg each) of similar weight (±0.1 mg) from the same tail were placed into 1.5 ml of PBS and subjected to 5 min of tearing by tweezers and sonicated three times for 10 s at 50% amplitude. The tendons were then manually pulled apart again by tweezers for two and a half minutes and sonicated for 15 s at 50% amplitude to create a solution of dispersed fibrils. For the dose‐dependent ELISA binding assay, three native and three GAG‐depleted tendon sections (between 3 and 6 mg each) of similar combined weight (±0.1 mg) from another tail were placed into 5 ml of PBS and were then subject to the same protocol to create a larger solution of dispersed fibrils. Another three native and three GAG‐depleted fibrils from this other tail were also prepared for the DMMB GAG quantification.
Immulon 2HB 96‐well plates (Thermo Scientific) were coated with triplicate samples of native and GAG‐depleted rat tail tendon collagen dispersions as prepared above and purified collagen I monomer (Corning, 10 μg/ml in 20 mM acetic acid, positive control) or synthetic triple helical collagen peptide (Ac‐GPC‐GPP5‐GFOGER‐GPP5‐GPC‐GY‐NH2; LifeTein, LLC) overnight at 4°C. The following day the solutions that did not adhere to the microplates were discarded. The wells were then blocked for 1 hr with 200 μl of 50 mg/ml BSA in PBS at room temperature. Three blank wells were also coated with 200 μl of 50 mg/ml BSA in PBS for 1 hr at room temperature as a negative control. The wells were then washed three times with a solution of PBS with 200 μl of 1 mg/ml BSA containing either 5 mM MgCl2 or 5 mM EDTA. Recombinant integrin α2I domain was then added to bind to the plated collagens or BSA for 1 hr at room temperature. After washing three times with the MgCl2 or EDTA buffer, 100 μl of mouse anti‐integrin α2I antibody was added to the wells at a 1:2,000 dilution for 45 min followed by binding 100 μl of a goat HRP‐conjugated anti‐mouse antibody at a 1:5,000 dilution for 30 min. Washing steps were performed between each binding addition. A TMB Substrate Kit (Pierce) was then used according to the manufacturer's protocol to measure integrin α2I binding activity. Absorbance was measured at 450 nm.
4.5. Atomic force microscopy
For atomic force microscopy (AFM) sample preparation, following incubation, one native and one GAG‐depleted tendon section from the same tail were rinsed with fresh PBS buffer solution (pH 7.4) and smeared onto separate microscope glass slides in order to deposit collagen fibrils onto the surfaces. 53 Prior to experiments, the microscope glass surfaces were sonicated in ultrapure water, rinsed with ethanol, and then dried with ultra‐high purity nitrogen. The samples were then washed with ultrapure water and allowed to dry for 1 hr in a laminar flow hood (1,300 Series A2, Thermo Scientific). Once dry, a stereoscope was used to locate and assess fibril deposition prior to AFM experiments.
Amplitude modulation (AM−) imaging and fast force mapping (FFM−) experiments were performed on a Cypher ES AFM (Asylum Research, Oxford Instruments). Silicon nitride cantilevers AC240 and Biolever‐mini BL‐AC40TS sourced from Oxford Instruments were used for AM imaging in air and liquid, respectively. These cantilevers have a nominal spring constant of k c = 2.0 and 0.1 N m−1, and radii of r = 7 and 8 nm, respectively. Image and force measurements were acquired at 25°C. The glass microscope slide surface with deposited collagen fibrils was mounted on a steel puck within the AFM box (a sealed enclosure). Samples were first imaged in air to locate and characterize the collagen fibrils. Before liquid experiments, the spring constant of the cantilever was first determined in air using the GetReal function in the Asylum Research AFM software, which is a combination of the Sader 54 and thermal noise method. The inverse optical lever sensitivity (InVOLS) was then determined by averaging the results of 10 force curves on a sapphire surface in PBS buffer. Liquid experiments were completed in a droplet of approximately 0.25 ml of fresh 10 mM PBS buffer, pH 7.4 solution on the glass microscope slide surface at 25°C. To minimize thermal drift, this setup was allowed to equilibrate for at least 1 hr in the AFM prior to imaging. To facilitate obtaining high‐quality AM images in liquid conditions, the tip was photothermally excited at its resonance frequency using blueDrive (Asylum Research, Oxford Instruments).
4.6. Fast force mapping
Fast force mapping (FFM) experiments were performed immediately following AM‐AFM imaging on the same fibrils characterized in air and liquid. The same BL‐AC40TS AFM probe as referenced above was used for all nanoindentation experiments. Force maps were acquired at a scan size of 1 × 1 μm with a pixel resolution of 256 × 256 points, Z‐rate of 20 Hz, indentation speed of 16 μm/s, and a setpoint of 2 nN. This setpoint was used specifically to ensure that the indentation did not exceed 15% of the fibrils' total height. The indentation moduli of the fibrils were then determined from the force maps produced using the analysis of the matrix of force curves as described below.
4.7. Image analysis
4.7.1. Periodicity
2D Fourier Transforms were performed on the phase channel of the AM images of collagen fibrils in both air and liquid using the Gwyddion software. 55 Profiles were then drawn on the transformed images, which then revealed the periodicity of the fibrils.
4.7.2. Overlap‐gap step height
For determination of the overlap–gap step height from the AM images, sine curves bearing the periodicity of the respective fibrils were fitted to height profiles along the apex of the corresponding fibril. The maxima/minima values of the sine curve were strictly used to determine the corresponding overlap/gap height values used along the fibrils. Adjacent overlap and gap values were then subtracted to obtain the step heights, and this was performed across an entire height profile (Figure S4). Furthermore, some chondroitinase treated fibrils imaged in air showed signs of debris (aggregates from chondroitinase stock solution) occurring along the height profiles, these areas were not taken into consideration for determining the average height of the fibril or step height (Figure S5). When the treated fibrils were immersed in PBS, visual inspection revealed that there was no evidence that the debris remained on the fibrils.
4.8. Post‐processing analysis
FFM data sets were analyzed using in‐house Matlab (R2018b) scripts. In brief, indentation moduli were calculated for each force curve iteratively using the methods detailed by Andriotis and coworkers. 53 The resulting 256 × 256 matrices of FFM indentation moduli were then reconstructed in Cartesian space as a scaled colormap (Figure 4a,b). Box and whisker plots were derived from a subset of five FFM images for each treatment. Here, force curve data at the height apex along the fibril longitudinal axis was extracted (256 moduli) and pooled among treatments. For this study, the apex corresponds to the values along a straight line created between two of the highest values at opposite ends of the fibril. A one‐way analysis of variance (ANOVA) was performed, and the moduli data from each treatment condition are shown as a box and whisker in the main text Figure 4c.
AUTHOR CONTRIBUTIONS
Jonathan Andrew Roth: Conceptualization (supporting); data curation (lead); formal analysis (lead); investigation (lead); writing – original draft (supporting); writing – review and editing (supporting). Cody Hoop: Conceptualization (supporting); data curation (supporting); visualization (supporting); writing – original draft (supporting); writing – review and editing (supporting). Jonathan Williams: data curation (supporting); formal analysis (supporting); investigation (supporting); methodology (supporting); writing – review and editing (supporting). Robert Hayes: Conceptualization (supporting); funding acquisition (supporting); methodology (supporting)); supervision (supporting); writing ‐ review and editing (supporting). Jean Baum: Conceptualization (lead); funding acquisition (lead); methodology (supporting); project administration (lead); supervision (lead); writing ‐ review and editing (lead).
Supporting information
Figure S1. Full apex height traces from AM images along representative regions of (a, b) native and (c, d) chondroitinase‐treated rat tail collagen fibrils in (a, c) air and (b, d) PBS, pH 7.4. In air, the fibrils have a clear repeating structure with distinct overlap and gap regions while in liquid this clarity is diminished and overlap and gap distinctions cannot be made. Visually there are still regions that resemble distinct overlap and gap regions albeit not throughout the entire profile.
Figure S2. Uncropped AM images of native (a, b) and chondroitinase‐treated (c, d) collagen fibrils in both air (a, c) and liquid (b, d) conditions. The height images are the same data as presented in Figure 3 in the main text. The corresponding amplitude and phase channels are displayed for each fibril in each condition.
Figure S3. Height profile of a native collagen fibril in air. Heights were calculated by averaging the height values along the apex of the fibril. The calculated height of this fibril is 82.1 nm.
Figure S4. Cartoon schematic of the alternating heights between overlap and gap regions along the collagen fibril and a height profile along the apex of a native collagen fibril in air (blue curve) along with a sine curve fitted to the periodicity of the fibril (orange). In order to determine the overlap‐gap step height, the height value corresponding to the maxima of the sine curve is considered the overlap height (letter “O”) is subtracted by the neighboring height value corresponding to the minima of the sine curve, which is considered to be the gap height (letter “G”). This is repeated across the entire fibril and performed for all fibrils in air.
Figure S5. Height image of a chondroitinase treated fibril in air (left) and its height profile fitted with a sine curve similar to Figure S4 (right). In the image, there are signs of debris on the fibril as a result of the chondroitinase treatment (arrows). The debris can be distinguished on the fibril image as well as on the corresponding height profile. Since the debris disallows for the distinction of certain overlap and gap regions and does not represent the true topography of the fibril, these areas (marked by “x”) were not accounted for step‐height.
Figure S6. Fast force mapping. (a) Once a fibril is imaged and visualized in standard tapping mode the same exact area is divided into a 256 × 256 pixel grid, and a force curve is performed at each pixel. A representative force curve from a native fibril in liquid is shown in (b). The contact force for each coordinate is extracted and local modulus calculated. These moduli are then replotted in space to yield force maps such as that presented in Figure 4 of the main text.
Figure S7. Boxplots showing the distribution of overlap‐gap step height differences (a), and modulus values (b) between the control and chondroitinase treated fibrils. The step heights are not significantly different indicating no structural differences while the modulus values are significantly different. Red symbols indicate outliers of the boxplot.
Figure S8. Fibril structure. A collagen monomer (a) consists of five distinct regions D1–D5. D1–D4 have a length of 67 nm while D5 has a length of 30 nm. When monomers aggregate together to form a fibril, they stack together as depicted in (b). Notably, since the D5 is only 30 nm in length, it does not fully cover the D4. Thus, there is a region which has five stacked monomers (overlap) and a region which only have four stacked monomers (gap). This structure is repeated in the z‐direction towards the core of the fibril. The black box on the collagen fibril encapsulates not only the surface layer, but also the inner layer right below it. As is shown, the overlap region of the surface layer is perfectly aligned with the overlap region of the inner layer (likewise for the gap region). This alignment continues with all consecutive layers towards the core of the fibril, resulting in the gap region being less dense with monomers in comparison to the overlap region and thus less stiff in comparison. For a more in‐depth analysis of the fibril structure, the reader is referred to here. 1 , 2 , 3 , 4
Figure S9. For AFM sample preparation, tendons from a single rat tail were cut into eight segments several cm in length. The segments were then separated into two distinct groups, native and GAG‐depleted. GAG‐depleted tendon sections were incubated with chondroitinase to digest the GAGs while the native tendon sections were incubated in the same buffer without the chondroitinase. One tendon section from each group was then smeared onto a glass surface to deposit individual fibrils on the surface. The surfaces were then washed, dried and underwent AFM imaging. Additionally, three tendon sections from each group were used to assess GAG content by a DMMB assay and quantitate the GAG depletion.
Figure S10. For adhesion assay sample preparation, tendons from a single rat tail (different from the rat tail used for the AFM sample prep) were cut into eight segments several cm in length. The segments were then separated into two distinct groups, native and GAG depleted. GAG depleted tendon sections were incubated with chondroitinase to digest the GAGs while the native tendon sections were incubated in the same buffer without the chondroitinase. One tendon section from each condition of similar weight (±0.1 mg) was submitted to sonication and being pulled apart by tweezers to create a suspension of collagen fibrils. These suspensions for both treatments were then used to coat the wells of a microplate to perform the ELISA binding assay. Three wells were coated for each condition. Additionally, three tendon sections from each group were used to assess GAG content by a DMMB assay to quantitate the GAG depletion.
Figure S11. GAG Quantification. DMMB results from the native and GAG‐depleted tendon sections derived from the three rat tails used for experiments in this study. Average GAG content (μg GAG/mg tendon): Rat Tail #1‐ Native = 1.89, GAG Depleted = 0.45; Rat Tail #2‐ Native = 2.10, GAG Depleted = 0.32; Rat Tail #3‐ Native = 2.84, GAG Depleted = 0.82. Average GAG depletion: Rat Tail #1 = 76% depletion, Rat Tail #2 = 85% depletion, Rat Tail #3 = 70% Depletion.
ACKNOWLEDGEMENTS
The authors acknowledge Dr. Joseph Freeman and Michael Pellegrini of Rutgers University Dept. of Biomedical Engineering for assistance with rat tail tendon procedures and Jordan Elliott in the Baum lab for assisting with integrin α2I expression. This work was supported by an NIH T32 Postdocotral Training Program in Translational Research in Regenerative Medicine under award number T32EB005583, NIH grant GM136431 to Jean Baum, and an Exploratory Research Seed Grant from Rutgers University to Robert Hayes.
Roth J, Hoop CL, Williams JK, Hayes R, Baum J. Probing the effect of glycosaminoglycan depletion on integrin interactions with collagen I fibrils in the native extracellular matrix environment. Protein Science. 2023;32(1):e4508. 10.1002/pro.4508
Review Editor: Aitziber L. Cortajarena
Funding information NIH, National Institutes of Health, Grant/Award Numbers: GM136431, T32EB005583; Rutgers University Exploratory Research Seed Grant
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. [DOI] [PubMed] [Google Scholar]
- 2. Springer TA, Wang JH. The three‐dimensional structure of integrins and their ligands, and conformational regulation of cell adhesion. Adv Protein Chem. 2004;68:29–63. [DOI] [PubMed] [Google Scholar]
- 3. Barczyk M, Carracedo S, Gullberg D. Integrins. Cell Tissue Res. 2010;339:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yue B. Biology of the extracellular matrix: An overview. J Glaucoma. 2014;23:S20–S23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ryan CNM, Sorushanova A, Lomas AJ, Mullen AM, Pandit A, Zeugolis DI. Glycosaminoglycans in tendon physiology, pathophysiology, and therapy. Bioconjug Chem. 2015;26:1237–1251. [DOI] [PubMed] [Google Scholar]
- 6. Li Y, Liu Y, Xia W, Lei D, Voorhees JJ, Fisher GJ. Age‐dependent alterations of decorin glycosaminoglycans in human skin. Sci Rep. 2013;3:2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kadler KE, Baldock C, Bella J, Boot‐Handford RP. Collagens at a glance. J Cell Sci. 2007;120:1955–1958. [DOI] [PubMed] [Google Scholar]
- 9. Kadler KE. Fell Muir lecture: Collagen fibril formation in vitro and in vivo. Int J Exp Pathol. 2017;98:4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holmes DF, Lu Y, Starborg T, Kadler KE. Collagen fibril assembly and function. Curr Top Dev Biol. 2018;130:107–142. [DOI] [PubMed] [Google Scholar]
- 11. Kadler KE, Holmes DF, Trotter JA, Chapman JA. Collagen fibril formation. Biochem J. 1996;316(Pt 1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Musiime M, Chang J, Hansen U, Kadler KE, Zeltz C, Gullberg D. Collagen assembly at the cell surface: Dogmas revisited. Cells. 2021;10:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heino J. The collagen family members as cell adhesion proteins. BioEssays : news and reviews in molecular, cellular and developmental biology. 2007;29:1001–1010. [DOI] [PubMed] [Google Scholar]
- 14. Sweeney SM, Orgel JP, Fertala A, et al. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J Biol Chem. 2008;283:21187–21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adorno‐Cruz V, Liu H. Regulation and functions of integrin α2 in cell adhesion and disease. Genes Dis. 2019;6:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hamaia S, Farndale RW. Integrin recognition motifs in the human collagens. Adv Exp Med Biol. 2014;819:127–142. [DOI] [PubMed] [Google Scholar]
- 17. Knight CG, Morton LF, Onley DJ, et al. Collagen‐platelet interaction: Gly‐Pro‐Hyp is uniquely specific for platelet Gp VI and mediates platelet activation by collagen. Cardiovasc Res. 1999;41:450–457. [DOI] [PubMed] [Google Scholar]
- 18. Knight CG, Morton LF, Peachey AR, Tuckwell DS, Farndale RW, Barnes MJ. The collagen‐binding A‐domains of integrins alpha(1)beta(1) and alpha(2)beta(1) recognize the same specific amino acid sequence, GFOGER, in native (triple‐helical) collagens. J Biol Chem. 2000;275:35–40. [DOI] [PubMed] [Google Scholar]
- 19. Raynal N, Hamaia SW, Siljander PR, et al. Use of synthetic peptides to locate novel integrin alpha2beta1‐binding motifs in human collagen III. J Biol Chem. 2006;281:3821–3831. [DOI] [PubMed] [Google Scholar]
- 20. An B, Chang S‐W, Hoop C, Baum J, Buehler MJ, Kaplan DL. Structural insights into the glycine pair motifs in type III collagen. ACS Biomater Sci Eng. 2017;3:269–278. [DOI] [PubMed] [Google Scholar]
- 21. Nunes AM, Zhu J, Jezioro J, et al. Intrinsic local destabilization of the C‐terminus predisposes integrin alpha1 I domain to a conformational switch induced by collagen binding. Protein Sci. 2016;25:1672–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao J, Sun X, Madhan B, Brodsky B, Baum J. NMR studies demonstrate a unique AAB composition and chain register for a heterotrimeric type IV collagen model peptide containing a natural interruption site. J Biol Chem. 2015;290:24201–24209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parmar AS, Nunes AM, Baum J, Brodsky B. A peptide study of the relationship between the collagen triple‐helix and amyloid. Biopolymers. 2012;97:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Knight CG, Morton LF, Onley DJ, et al. Identification in collagen type I of an integrin alpha2 beta1‐binding site containing an essential GER sequence. J Biol Chem. 1998;273:33287–33294. [DOI] [PubMed] [Google Scholar]
- 25. Siljander PRM, Hamaia S, Peachey AR, et al. Integrin activation state determines selectivity for novel recognition sites in fibrillar collagens. J Biol Chem. 2004;279:47763–47772. [DOI] [PubMed] [Google Scholar]
- 26. Munnix IC, Gilio K, Siljander PR, et al. Collagen‐mimetic peptides mediate flow‐dependent thrombus formation by high‐ or low‐affinity binding of integrin alpha2beta1 and glycoprotein VI. J Thromb Haemost. 2008;6:2132–2142. [DOI] [PubMed] [Google Scholar]
- 27. Brodsky B, Thiagarajan G, Madhan B, Kar K. Triple‐helical peptides: An approach to collagen conformation, stability, and self‐association. Biopolymers. 2008;89:345–353. [DOI] [PubMed] [Google Scholar]
- 28. Farndale RW, Lisman T, Bihan D, et al. Cell‐collagen interactions: The use of peptide toolkits to investigate collagen‐receptor interactions. Biochem Soc Trans. 2008;36:241–250. [DOI] [PubMed] [Google Scholar]
- 29. Zhu J, Hoop CL, Case DA, Baum J. Cryptic binding sites become accessible through surface reconstruction of the type I collagen fibril. Sci Rep. 2018;8:16646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gandhi NS, Mancera RL. The structure of glycosaminoglycans and their interactions with proteins. Chem Biol Drug Des. 2008;72:455–482. [DOI] [PubMed] [Google Scholar]
- 31. Lu P, Weaver VM, Werb Z. The extracellular matrix: A dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levental KR, Yu H, Kass L, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Walker C, Mojares E, Del Rio Hernandez A. Role of extracellular matrix in development and cancer progression. Int J Mol Sci. 2018;19:3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Law B, Fowlkes V, Goldsmith JG, Carver W, Goldsmith EC. Diabetes‐induced alterations in the extracellular matrix and their impact on myocardial function. Microsc Microanal. 2012;18:22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vogel V. Unraveling the mechanobiology of extracellular matrix. Annu Rev Physiol. 2018;80:353–387. [DOI] [PubMed] [Google Scholar]
- 37. Lachowski D, Cortes E, Pink D, et al. Substrate rigidity controls activation and Durotaxis in pancreatic stellate cells. Sci Rep. 2017;7:2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. [DOI] [PubMed] [Google Scholar]
- 39. Jokinen J, Dadu E, Nykvist P, et al. Integrin‐mediated cell adhesion to type I collagen fibrils. J Biol Chem. 2004;279:31956–31963. [DOI] [PubMed] [Google Scholar]
- 40. Dufrene YF, Ando T, Garcia R, et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat Nanotechnol. 2017;12:295–307. [DOI] [PubMed] [Google Scholar]
- 41. Krieg M, Fläschner G, Alsteens D, et al. Atomic force microscopy‐based mechanobiology. Nat Rev Phys. 2019;1:41–57. [Google Scholar]
- 42. Petruska JA, Hodge AJ. A subunit model for the tropocollagen macromolecule. Proc Natl Acad Sci U S A. 1964;51:871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rigozzi S, Muller R, Stemmer A, Snedeker JG. Tendon glycosaminoglycan proteoglycan sidechains promote collagen fibril sliding‐AFM observations at the nanoscale. J Biomech. 2013;46:813–818. [DOI] [PubMed] [Google Scholar]
- 44. Baldwin SJ, Quigley AS, Clegg C, Kreplak L. Nanomechanical mapping of hydrated rat tail tendon collagen I fibrils. Biophys J. 2014;107:1794–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Orgel JP, Eid A, Antipova O, Bella J, Scott JE. Decorin core protein (decoron) shape complements collagen fibril surface structure and mediates its binding. PLoS One. 2009;4:e7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sweeney SM, Guy CA, Fields GB, San Antonio JD. Defining the domains of type I collagen involved in heparin‐ binding and endothelial tube formation. Proc Natl Acad Sci U S A. 1998;95:7275–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. San Antonio JD, Karnovsky MJ, Gay S, Sanderson RD, Lander AD. Interactions of syndecan‐1 and heparin with human collagens. Glycobiology. 1994;4:327–332. [DOI] [PubMed] [Google Scholar]
- 48. Scott JE, Glanville RW. Homologous sequences in fibrillar collagens may be proteoglycan binding sites. Biochem Soc Trans. 1993;21:123 S. [DOI] [PubMed] [Google Scholar]
- 49. Scott JE, Orford CR. Dermatan sulphate‐rich proteoglycan associates with rat tail‐tendon collagen at the d band in the gap region. Biochem J. 1981;197:213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Woltersdorf C, Bonk M, Leitinger B, et al. The binding capacity of α1β1‐, α2β1‐ and α10β1‐integrins depends on non‐collagenous surface macromolecules rather than the collagens in cartilage fibrils. Matrix Biol. 2017;63:91–105. [DOI] [PubMed] [Google Scholar]
- 51. Hoop CL, Zhu J, Nunes AM, Case DA, Baum J. Revealing accessibility of cryptic protein binding sites within the functional collagen fibril. Biomolecules. 2017;7:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Coulson‐Thomas V, Gesteira T. Dimethylmethylene blue assay (DMMB). Bio‐Protocol. 2014;4:e1236. [Google Scholar]
- 53. Andriotis OG, Manuyakorn W, Zekonyte J, et al. Nanomechanical assessment of human and murine collagen fibrils via atomic force microscopy cantilever‐based nanoindentation. J Mech Behav Biomed Mater. 2014;39:9–26. [DOI] [PubMed] [Google Scholar]
- 54. Sader JE, Chon JWM, Mulvaney P. Calibration of rectangular atomic force microscope cantilevers. Rev Sci Instrum. 1999;70:3967–3969. [Google Scholar]
- 55. Nečas D, Klapetek P. Gwyddion: An open‐source software for SPM data analysis. Cent Eur J Phys. 2012;10:181–188. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Full apex height traces from AM images along representative regions of (a, b) native and (c, d) chondroitinase‐treated rat tail collagen fibrils in (a, c) air and (b, d) PBS, pH 7.4. In air, the fibrils have a clear repeating structure with distinct overlap and gap regions while in liquid this clarity is diminished and overlap and gap distinctions cannot be made. Visually there are still regions that resemble distinct overlap and gap regions albeit not throughout the entire profile.
Figure S2. Uncropped AM images of native (a, b) and chondroitinase‐treated (c, d) collagen fibrils in both air (a, c) and liquid (b, d) conditions. The height images are the same data as presented in Figure 3 in the main text. The corresponding amplitude and phase channels are displayed for each fibril in each condition.
Figure S3. Height profile of a native collagen fibril in air. Heights were calculated by averaging the height values along the apex of the fibril. The calculated height of this fibril is 82.1 nm.
Figure S4. Cartoon schematic of the alternating heights between overlap and gap regions along the collagen fibril and a height profile along the apex of a native collagen fibril in air (blue curve) along with a sine curve fitted to the periodicity of the fibril (orange). In order to determine the overlap‐gap step height, the height value corresponding to the maxima of the sine curve is considered the overlap height (letter “O”) is subtracted by the neighboring height value corresponding to the minima of the sine curve, which is considered to be the gap height (letter “G”). This is repeated across the entire fibril and performed for all fibrils in air.
Figure S5. Height image of a chondroitinase treated fibril in air (left) and its height profile fitted with a sine curve similar to Figure S4 (right). In the image, there are signs of debris on the fibril as a result of the chondroitinase treatment (arrows). The debris can be distinguished on the fibril image as well as on the corresponding height profile. Since the debris disallows for the distinction of certain overlap and gap regions and does not represent the true topography of the fibril, these areas (marked by “x”) were not accounted for step‐height.
Figure S6. Fast force mapping. (a) Once a fibril is imaged and visualized in standard tapping mode the same exact area is divided into a 256 × 256 pixel grid, and a force curve is performed at each pixel. A representative force curve from a native fibril in liquid is shown in (b). The contact force for each coordinate is extracted and local modulus calculated. These moduli are then replotted in space to yield force maps such as that presented in Figure 4 of the main text.
Figure S7. Boxplots showing the distribution of overlap‐gap step height differences (a), and modulus values (b) between the control and chondroitinase treated fibrils. The step heights are not significantly different indicating no structural differences while the modulus values are significantly different. Red symbols indicate outliers of the boxplot.
Figure S8. Fibril structure. A collagen monomer (a) consists of five distinct regions D1–D5. D1–D4 have a length of 67 nm while D5 has a length of 30 nm. When monomers aggregate together to form a fibril, they stack together as depicted in (b). Notably, since the D5 is only 30 nm in length, it does not fully cover the D4. Thus, there is a region which has five stacked monomers (overlap) and a region which only have four stacked monomers (gap). This structure is repeated in the z‐direction towards the core of the fibril. The black box on the collagen fibril encapsulates not only the surface layer, but also the inner layer right below it. As is shown, the overlap region of the surface layer is perfectly aligned with the overlap region of the inner layer (likewise for the gap region). This alignment continues with all consecutive layers towards the core of the fibril, resulting in the gap region being less dense with monomers in comparison to the overlap region and thus less stiff in comparison. For a more in‐depth analysis of the fibril structure, the reader is referred to here. 1 , 2 , 3 , 4
Figure S9. For AFM sample preparation, tendons from a single rat tail were cut into eight segments several cm in length. The segments were then separated into two distinct groups, native and GAG‐depleted. GAG‐depleted tendon sections were incubated with chondroitinase to digest the GAGs while the native tendon sections were incubated in the same buffer without the chondroitinase. One tendon section from each group was then smeared onto a glass surface to deposit individual fibrils on the surface. The surfaces were then washed, dried and underwent AFM imaging. Additionally, three tendon sections from each group were used to assess GAG content by a DMMB assay and quantitate the GAG depletion.
Figure S10. For adhesion assay sample preparation, tendons from a single rat tail (different from the rat tail used for the AFM sample prep) were cut into eight segments several cm in length. The segments were then separated into two distinct groups, native and GAG depleted. GAG depleted tendon sections were incubated with chondroitinase to digest the GAGs while the native tendon sections were incubated in the same buffer without the chondroitinase. One tendon section from each condition of similar weight (±0.1 mg) was submitted to sonication and being pulled apart by tweezers to create a suspension of collagen fibrils. These suspensions for both treatments were then used to coat the wells of a microplate to perform the ELISA binding assay. Three wells were coated for each condition. Additionally, three tendon sections from each group were used to assess GAG content by a DMMB assay to quantitate the GAG depletion.
Figure S11. GAG Quantification. DMMB results from the native and GAG‐depleted tendon sections derived from the three rat tails used for experiments in this study. Average GAG content (μg GAG/mg tendon): Rat Tail #1‐ Native = 1.89, GAG Depleted = 0.45; Rat Tail #2‐ Native = 2.10, GAG Depleted = 0.32; Rat Tail #3‐ Native = 2.84, GAG Depleted = 0.82. Average GAG depletion: Rat Tail #1 = 76% depletion, Rat Tail #2 = 85% depletion, Rat Tail #3 = 70% Depletion.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


