ABSTRACT
Coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, has affected millions of individuals with various implications. Consistent with the crucial role of the microbiome in determining health and disease in humans, various studies have investigated the gut and respiratory microbiome effect on the COVID-19. Microbiota dysbiosis might support the entry, replication, and establishment of SARS-CoV-2 infection by modulating various mechanisms. One of the main mechanisms that the modulation of respiratory microbiota composition during the COVID-19 infection affects the magnitude of the disease is changes in innate and acquired immune responses, including inflammatory markers and cytokines and B- and T-cells. The diversity of respiratory microbiota in COVID-19 patients is controversial; some studies reported low microbial diversity, while others found high diversity, suggesting the role of respiratory microbiota in this disease. Modulating microbiota diversity and profile by supplementations and nutrients can be applied prophylactic and therapeutic in combating COVID-19. Here, we discussed the lung microbiome dysbiosis during various lung diseases and its interaction with immune cells, focusing on COVID-19.
KEYWORDS: COVID-19, microbiome, respiratory microbiome, microbiome dysbiosis, immune responses
Introduction
Although severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) was first reported in China’s Hubei province in November 2019, it rapidly spread globally and caused Coronavirus Disease 2019 (COVID-19) pandemic disease [1]. Despite the approval of some preventive vaccines and the availability of vaccinization programs, COVID-19 has affected around 311 million people worldwide by now (11 January 2022). Mild fever, fatigue, dry cough, sore throat, diarrhoea, and anosmia are the most common symptoms in SARS-CoV-2-infected patients [2]. Many infected people remain asymptomatic or have only mild upper respiratory tract symptoms, but others exhibit acute respiratory distress syndrome (ARDS) as well as pneumonia, necessitating intubation that can experience fatal complications [3]. Alongside respiratory complications, infection with SARS-CoV-2 can cause acute kidney injury, vascular thrombosis, endothelial sloughing, and shock [4]. Thus, multi-organ failures and complications imposition great stress for healthcare systems and prolonged hospitalization [5].
The microbiome is an aggregate of microorganisms, including bacteria, fungi, viruses, and protozoans, that reside in a particular microenvironment on or within the human body and are involved actively in cellular metabolisms and functions [6]. There is evidence that the interaction between microbes and humans plays a decisive role in determining the health or disease status in the human body owing to the contribution of microbiomes to the improvement or impairment of immune and metabolic functions [7]. Thus, understanding host-microbe interactions and dysbiosis of the microbiome could be useful in diagnosing and introducing appropriate treatment for diseases. Due to the high expression of the angiotensin-converting enzyme 2 (ACE2), a receptor of the SARS-CoV-2 virus, in respiratory and gastrointestinal tracts [8], some emerging scientific and clinical investigations have indicated the dysbiosis of the gut and respiratory microbiome during COVID-19 infection [9,10]. For example, the analyses of stool microbiome from 15 patients during hospitalization in Hong Kong revealed that COVID-19 patients exhibited remarkably reduced bacterial diversity and increased abundance of opportunistic ones, such as Rothia, Streptococcus, Actinomyces, and Veillonella [11]. On the other hand, the invasion of the SARS-CoV-2 virus towards the lungs for beginning COVID-19 disease is related to the lung infection and subsequent immune responses in which the lung microbiome might play an essential role in the initiation, development, and response to therapeutic agents. This review will summarize the lung microbiome dysbiosis during various lung diseases and its interaction with immune cells and responses. We focused on the alterations in the lung microbiome in patients infected with COVID-19 and how these alterations can be used to improve treatment outcomes.
Lung microbiome dysbiosis during diseases
The studies based on the traditional culture systems have reported that the bronchial tree in healthy individuals is sterile [12,13]. Nowadays, the advent of high-throughput sequencing technologies and culture-independent techniques, such as sequencing the 16S rRNA gene, revealed that the bronchial tree is not sterile, even in healthy people. The lungs are a microbial ecosystem with a collection of microorganisms, both viable and non-viable, colonizing in the bronchial tree and parenchymal tissues, and are essential for health, including immunoregulation, epithelial integrity, and colonization resistance [14]. The main genera have been identified in healthy lungs are Streptococcus, Prevotella, Fusobacterium, Haemophilus, and Veillonella [15,16]. Compared to the gastrointestinal (GI) tract, which contains 1011-1012 bacteria per gram of tissue, the microbial population in the lungs of healthy individuals is relatively lower (about 103-105 bacteria per gram of tissue) [17]. It is reported that there are 10–100 bacteria per 1000 human cells in the lung tissue samples [18], and three factors determine its composition: 1) microbial immigration, such as direct mucosal dispersion, bacteria inhalation, and microaspiration; 2) microbial elimination, such as host defence responses, mucociliary clearance, and cough; and 3) regional growth conditions, such as pH, temperature, and nutrient availability [19,20].
It has been shown that both acute and chronic respiratory diseases dramatically alter the lung microbiome composition (Table 1). For instance, patients with idiopathic pulmonary fibrosis (IPF) [35], cystic fibrosis (CF) [36] and bronchiectasis [37] have exhibited an increase in bacterial burden in their lower airways. In addition to changes in the load of bacteria, some specific bacteria are frequently identified in the airways of unhealthy people, such as Streptococcus, Veillonella, Neisseria, and Haemophilus (in IPF); Staphylococcus aureus, Pseudomonas aeruginosa, and Burkholderia spp. (in CF); and Haemophilus, P. aeruginosa, Prevotella, and Veillonella (in bronchiectasis) [38]. In asthma, bacterial composition and dysbiosis in the lung airway contribute to the severity of the disease and its pathogenesis. Using 16S rRNA-based methods, Huang et al. indicated airway dysbiosis in patients with the severe asthmatic condition compared with milder asthma which promotes inflammatory responses by recruiting neutrophils in a Th17-dependent manner. They also found that the lungs of patients with severe asthma were remarkably enriched with Actinobacteria and Klebsiella [39]. In patients with chronic obstructive pulmonary disease (COPD), the microbiome analysis revealed a reduction in microbial diversity associated with remodelling of the alveolar and bronchiolar tissue, the infiltration of CD4+ T-cells, and emphysematous destruction. The COPD patients exhibited an increase in Actinobacteria and Proteobacteria and reduced Bacteroidetes and Firmicutes. This alteration in microbiota composition was associated with the infiltration of neutrophils, B-cells, and eosinophils, suggesting the correlation between host immune responses and lung dysbiosis [40]. Furthermore, numerous studies showed that the gut microbiota composition in lung cancer patients is significantly different from healthy control ones, and the respiratory microbiome dysbiosis contributes to inflammation responses and tumorigenesis of lung cancer [41]. A systematic review conducted on 41 studies indicated that pre-existing Mycobacterium tuberculosis is associated with the risk of lung cancer [42]. The examination of bronchoalveolar lavage fluid (BALF) samples from 28 patients (20 with lung cancer and 8 at the benign stage) revealed the increase of genera Megasphaera and Veillonella in patients with lung cancer and TM7 and Firmicutes phyla compared with patients at the benign stage [31].
Table 1.
Alteration of lung microbiome during respiratory diseases.
| Disease | Sample type | Sample size | Microbiome alteration | Ref | |
|---|---|---|---|---|---|
| CF | BAL | 95 | ↑Streptococcus, Staphylococcus, Pseudomonas | [21] | |
| CF | Sputum | 17 | ↑Staphylococcus, Pseudomonas, Achromobacter, Stenotrophomonas | [22] | |
| CF | BAL | 12 | ↑Burkholderia, Streptococcus, Prevotella, Porphyromonas, Haemophilus, Veillonella | [23] | |
| COPD | Sputum | 16 | ↑Pseudomonas, Moraxella, Corynebacterium | [24] | |
| COPD | Lung tissue | 24 | ↑Firmicutes, Lactobacillus | [18] | |
| COPD | Sputum | 8 | ↑Fusobacterium, Bacteroidetes, Actinobacteria, Firmicutes, Streptococcus, ↓Proteobacteria | [25] | |
| Asthma | Nasopharyngeal | 234 | ↑Proteobacteria, Streptococcus | [26] | |
| Asthma | Bronchoscopy | 42 | ↑Neisseria, Haemophilus, Porphyromonas, Fusobacterium | [27] | |
| Asthma | BAL and endobronchial brushing | 58 | ↑Pseudomonas, Lactobacillus, Rickettsia | [28] | |
| Lung cancer | Saliva | 30 | ↑Selenomonas, Veillonella, Capnocytophaga, ↓Streptococcus, Neisseria | [29] | |
| Lung cancer | Lung tissues and bronchoscopy | 42 | ↑Streptococcus, ↓Staphylococcus | [30] | |
| Lung cancer | BAL | 28 | ↑Veillonella, Megasphaera | [31] | |
| IPF | BAL | 55 | ↑Streptococcus, Staphylococcus | [32] | |
| IPF | BAL | 65 | ↑Streptococcus, Veillonella, Haemophilus, Neisseria | [33] | |
| IPF | Bronchoscopy | 35 | ↑Pseudomonas, Streptococcus, Haemophilus, Prevotella, Veillonella | [34] | |
CF, cystic fibrosis; BAL, bronchoalveolar lavage; COPD, chronic obstructive pulmonary disease; IPF, idiopathic pulmonary fibrosis.
In addition to the mentioned pathological complications, there is evidence that smoking is another condition that affects the lung microbiota. Smoking and exposure to tobacco could increase the risk of infection with bacteria by increasing the microbial diversity at the lower respiratory tract [43]. Cigarette smoking increases the risk of ARDS in patients following severe trauma. The lung of these patients with smoking status was enriched with pathogenic bacteria, including Haemophilus, Fusobacterium, Streptococcus, Prevotella, and Treponema [44]. Also, cigarette smoking could alter virus populations in the lungs. Gregory et al. indicated that the abundance of bacteriophages infecting Actinomyces, Haemophilus, Xanthomonas, Rodoferax, Aeromonas, Prevotella, and Capnocytophaga was increased in smokers, whereas Morganella and Enhydrobacter bacteriophages were prevailing in non-smokers [45].
Immune responses against SARS-CoV-2 infection
The intracellular adherence and tight junctions act as connectors of adjacent cells as well as barriers to regulate paracellular permeability. The impairment of barrier function during diseases increases epithelial permeability and, subsequently, pathogen entry [46]. During infection with viruses, they can bind to their cognate cellular receptors and induce the stimulation of pattern recognition receptors (PRRs) in the epithelial cells, resulting in the secretion of molecules, enzymes, peptides, reactive oxygen species (ROSs), and chemokines with anti-microbial activities. Collectively, the released molecules recruit immune cells and facilitate their communication with each other, contributing to the immune responses that are essential for controlling the infection [47]. In the case of COVID-19, the infection of alveolar type II cells with higher expression of ACE2 receptor through SARS-CoV-2 disrupts the critical functions of these cells, including stabilization of airway epithelial barrier, airway regeneration following injury, and production of pulmonary surfactant [48]. These cells also are involved in the immune responses against pathogens and alveolar damage by producing cytokines to stimulate the recruitment and activation of immune cells, especially macrophages, in defence of the alveolus [49]. Following the binding of the S1 subunit in the viral spike (S) protein, through its receptor-binding domain (RBD), to the receptor, the S protein undergoes protease cleavage by serine protease transmembrane protease serine 2 (TMPRSS2), resulting in uncovering the S2 site and subsequent membrane fusion between the virus and alveolar type II cells, endocytosis of the virus, and release of viral compartments into the cell cytoplasm [50,51]. Neuropilin 1 (NRP1) and Furin also have been identified as co-factors of virus internalization [51,52]. The infected cells release damage-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs), for-example viral RNA, which are recognized by various intracellular PRRs, like RIG-I/MDA5/MAVS/TRAF3/IRF3/IRF7/type I IFNs and TLRs/TRIF/MyD88/TRAF6/p65/p50/TNF pathways (Figure 1). Certainly, targeting these pathways could be beneficial therapeutic strategies in virus-induced diseases. For example, targeting TNF- and IFN-mediated responses during the respiratory syncytial virus infection protects lung cells against the harmful effects of the viral infection [53,54].
Figure 1.
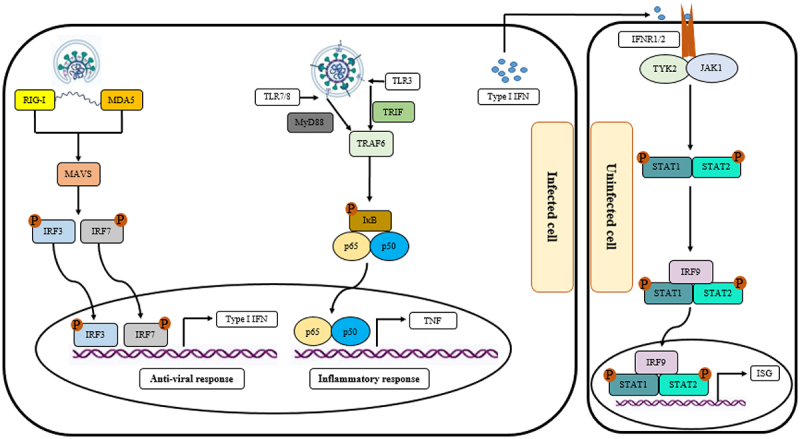
The effect of infection with SARS-CoV-2 on the innate immune response. Following infecting the target cells, the RNA of the virus was recognized by the cytosolic pattern recognition receptors (PRRs), such as the melanoma differentiation-associated protein (MDA5) and the retinoic acid-inducible gene I (RIG-I), which recruit the mitochondrial antiviral signaling protein (MAVS), followed by the phosphorylation and activation of interferon regulatory factor 3 and 7 (IRF3 and IRF7). The phosphorylated form of IRF3 and IRF7 upregulates the expression of type I interferons, as the antiviral response. In addition to the cytosolic receptors, the infection with SARS-CoV-2 is recognized by the endosomal receptors, such as toll-like receptors 3, 7, and 8 (TLR3, TLR7, and TLR8), leading to trigger a cascade to upregulate inflammatory responses. The secreted type I IFNs from the infected cells are recognized through their cognate receptors on the uninfected cells, resulting in the activation of IRF9, STAT1, and STAT2, and, subsequently, upregulation of interferon-stimulated gene (ISG).
The involvement of the host immune system is characterized by the contribution of the innate immune system in the first phase. A typical alveolar immune landscape in the healthy lung is composed mainly of alveolar macrophages located at the air-liquid interface and a few granulocytes, including eosinophils and basophils [55]. Triggering PRR signalling within cells involved in the innate immune responses elevates the levels of pro-inflammatory factors, including tumour necrosis factor (TNF)-α and granulocyte macrophage-colony stimulating factor (GM-CSF) in the plasma of COVID-19 patients as well as interleukin (IL)-1β, IL-6, and IL-8, in their BALF and plasma [56–59]. Additionally, PRR signalling could promote the recruitment and infiltration of neutrophils, monocytes, and T-cells to the infection site by upregulating CXCL8, CCL2, and CCL7 [57,60]. Importantly, the signalling of PRR activates the production of anti-viral type I IFNs within plasmacytoid dendritic cells (DCs) [61]. IFNs could stimulate phagocytosis in macrophages and the Janus kinase signal transducer and activator of transcription (JAK-STAT) pathway, which directs the polarization of anti-viral helper T-cells [62].
Besides the innate immune system, adaptive immune responses in COVID-19 patients contribute to managing and controlling viral infection. The lysed and damaged epithelium cells in the lung and subepithelial DCs present virus antigens to CD8+ and CD4+ T-cells, leading to cytotoxicity activation against virus-infected cells and induction of apoptosis as well as promoting differentiation of CD4+ T-cells towards Th1, Th2, Th17, and T follicular helper (FH) [63]. Recently, Pavel et al. indicated that the imbalance in Th2/Th1 and Th17/Th1 cytokines could affect the outcomes in patients with SARS-CoV2 infection and is related to their mortality risk [64]. Also, TFH cells help the development of B-cells into plasma cells, promoting the production of virus-specific IgG, IgM, and IgA [63]. Mazzoni et al. exhibited that asymptomatic patients showed lower proportions of the viral-specific CD4+ T-cells and lower humoral response, suggesting that the frequency of SARS-CoV-2-reactive T-cells is associated with the disease severity. Furthermore, they revealed that asymptomatic individuals show lower multifunctionality of antigen-specific T-cells and lower expression of TIGIT and PD-1 immune-checkpoints compared with symptomatic patients. This indicates that immune responses in symptomatic patients against SARS-CoV-2 can not eliminate the virus rapidly, thus, resulting in repeated activation of immune system cells [65]. In another study, Qin et al. found that the population of memory and regulatory T-cells were lessened in COVID-19 patients, whereas naïve T-cell counts were increased compared to healthy ones [66]. Single-cell analysis of T-cells from COVID-19 patients revealed that CD4+ T-cells were activated with high expression of regulatory responses and CD25 as well as suppression of FOXP3 expression in severe COVID-19. These cells showed unique differentiation pathways in the patient’s lungs with both Th1 and Th2 characteristics. In addition, highly activated CD25-expressing CD4+ T-cells facilitated SARS-CoV-2 entry by producing the protease Furin [67].
Infection with SARS-CoV-2 and microbiome dysbiosis
Since super-infection or co-infection with bacteria partially attributed to the worth outcome and mortality of virus pandemics, including the 1918 H1N1 and 2009 H1N1 influenza, various studies examined this in the infection with COVID-19. The microbiome in humans differs across sex, age, ethnicity, and race, suggesting the unique and specific profiles of the microbial population [68]. The infection of the GI tract by SARS-CoV-2 could disrupt gut microbiota and lead to dysbiosis, GI symptoms, and intestinal inflammation. This imbalance can not be fully restored after even three months of recovery [69]. Also, there is evidence that gut microbiota dysbiosis during infection with SARS-CoV-2 could affect the severity of COVID-19 [70]. It has been shown that gut microbiota could regulate immune responses and modulation of their composition had preventive and therapeutic benefits [71]. For instance, beneficial bacteria Bifidobacteria and Lactobacilli and butyrate-producing bacteria Eubacterium rectale and Faecalibacterium prausnitzii were decreased in patients with COVID-19 [72,73], whereas the population of opportunistic bacteria such as Clostridium hathewayi and Clostrium ramosum were increased and positively correlated with COVID-19 severity [73]. Zuo et al. indicated that the faecal samples from patients with COVID-19 were enriched with Collinsella tanakaei, Collinsella aerofaciens, Morganella morganii, and Streptococcus infantis, whereas the samples from none-to-low SARS-CoV-2 infectivity displayed a higher abundance of Lachnospiraceae bacterium 1_1_57FAA, Alistipes onderdonkii, and Bacteroides stercoris [74]. Furthermore, there is a negative correlation between the diversity of gut microbiota and COVID-19 severity levels; the diversity decreases with increasing the symptoms of severity in which the “severe” patients show the least diversity [75]. Khan et al. demonstrated that the severely COVID-19 patients had elevated levels of IL-21 in comparison with mildly ill and healthy participants, while there were no remarkable differences in the levels of INF-γ and TNF-α among the groups [75]. The investigation of alterations in the gut microbiota of 115 patients and its influence on COVID-19 severity revealed that moderate and severe patients were enriched with Proteobacteria compared with mild ones. In contrast, the abundance of butyrate-producing bacteria, including Lachnospira and Roseburia, and the Firmicutes/Bacteroidetes ratio was lower in moderate and severe patients [76]. The produced butyrate, short-chain fatty acids (SCFAs), and propionate with gut microbiota could increase the expression of Tc17 cells and CD8+ cytotoxic T lymphocytes (CTLs). For instance, butyrate could increase granzyme B and IFN-γ expression on CTLs as well as promote the Tc17 cells switch towards cytotoxic phenotypes [77]. In a study, Zhou et al. compared the gut microbiota composition and immune responses in moderate COVID-19 patients with and without fever. They found that opportunistic pathogens, including Saccharomyces cerevisiae and Enterococcus faecalis, were enriched in COVID-19 patients with fever. S. cerevisiae was positively associated with diarrhoea symptoms in these patients, whereas E. faecalis was positively associated with D-dimer and lactate dehydrogenase (LDH) and negatively correlated with IL-4 and CD8+ T-cells. On the other hand, the proportion of species with protective and anti-inflammatory effects, including Eubacterium ramulus and Bacteroides fragilis, was increased in non-fever patients. Also, there was a negative correlation between enrichment with E. ramulus with aspartate aminotransferase (AST), LDH, and IL-6. This study suggested that the gut microbiota contributes to the induction of fever in COVID-19 patients by increasing the pathogenic bacteria in the GI tract, which stimulate the secretion of inflammatory cytokines, including IL-6 [78]. Tao et al. found that the changes in the gut microbiota composition is positively correlated with the higher expression of the pro-inflammatory factor IL-18 [79]. Thus, gut microbiota diversity and composition could be act as prognostic biomarkers for COVID-19 severity and immune responses.
Profiling of the respiratory microbiome in COVID-19 patients
Besides the gut microbiota, infection with SARS-CoV-2 could affect the population and metabolism of respiratory microbiota. The profiling microbiomes of the COVID-19 respiratory tract by Haiminen et al. revealed that metabolic pathways are modulated, including a decrease in lipid metabolism (such as sphingolipid metabolism) and glycan metabolism pathways (such as glycan degradation), and an increase in carbohydrate metabolism pathways (such as glycolysis and gluconeogenesis) [80]. It has been shown that sphingosine, as a part of sphingolipids, hinders the interaction of the S subunit of SARS-CoV-2 with ACE2 and reduces the virus infectivity [81]. Also, the changes in glucose metabolism in both innate immune cells and pulmonary epithelial cells contribute to cytokine synthesis and inflammatory responses [82]. Under high glucose conditions, viral replication and cytokine production increase, leading to lung epithelial cell death [83]. Thus, lung microbiome alterations could affect the metabolism in immune cells and lung epithelial cells, affecting the SARS-CoV-2 life cycle. Because bacterial dysbiosis is discussed in detail throughout the article, Table 2. summarizes the dysbiosis of other respiratory microbiota in COVID-19 patients. There is evidence that respiratory viral co-infection in respiratory diseases is 3–68.0% [84]. For instance, Lin et al. stated that viral co-infections in SARS-CoV-2 patients were 3.2% [85]. Furthermore, a metagenomics analysis revealed that in addition to betacoronavirus, COVID-19 samples were associated with other viral co-infections, such as Tombusvirus, Partitivirus, Victorivirus, Totivirus, and Chrysovirus [86].
Table 2.
Dysbiosis of respiratory microbiota in COVID-19 patients.
| Microorganism | Sample type | Sample size | Dysbiosis | Ref |
|---|---|---|---|---|
| Fungi | BAL | 26 | - ↑Candida spp. - ↑Ascomycota in patients with COVID-19 not colonized with Candida spp. |
[87] |
| Fungi | BAS, BAL | 90 | ↑Candida albicans | [88] |
| Fungi | Sputum, EA | 99 | - ↑Candida glabrata - ↑Candida albicans |
[3] |
| Virus | Throat swab, Nasal swab, Sputum | 23 | - ↑Human alphaherpesvirus 1 - ↑Rhinovirus B - ↑ Human orthopneumovirus |
[89] |
| Virus | Nasopharyngeal swabs | 8 | - ↑Betacoronavirus - ↓Alphacoronavirus - ↓Cystovirus - ↑Macavirus - ↓Myovirus |
[90] |
| Virus | Nasopharyngeal swabs | 10 | - ↑Betacoronavirus - ↓Siphovirus - ↓Alphapapillomavirus - ↓Myovirus |
[86] |
| Archaea | Nasopharyngeal swabs | 8 | - ↑Halogeometricum - ↑Haloquadratum - ↑Natrialba - ↓Methanospirillum - ↓Methanoregula - ↓Methanocaldococcus |
[90] |
| Archaea | Nasopharyngeal swabs | 10 | - ↑Methanosarcina - ↑Methanocaldococcus - ↑Thermococcus - ↑Haloarcula - ↓Methanobrevibacter - ↓Methanococcus - ↓Methanocorpusculum |
[86] |
BAL, bronchoalveolar lavage; BAS, bronchial aspirates; EA, endotracheal aspirates.
It has been stated that the lung microbiota composition of healthy individuals is mainly enriched with commensal bacteria, including Tropheryma whipplei, Prevotella spp., Streptococcus spp., and Veillonella spp., that maintain the immune homoeostasis [91]. The bacterial diversity in COVID-19 patients is controversial; some studies found low microbial diversity [92,93], while others reported a high diversity [94]. These paradoxical results could be owing to the patients’ severity, treatment, disease stage, and differences in sampling location of the respiratory tract. Figure 2 represents the alteration in the respiratory bacteria during infection with COVID-19. To identify the association between upper and lower respiratory tract microbiota and the COVID-19 severity, Lloréns-Rico et al. used nasopharyngeal swabs and BAL samples. They explained that the microbiome variation within the upper respiratory tract could be affected by the type of oxygen support (predominantly mechanical ventilation), time in ICU, and treatments (such as antibiotics), while viral load showed a reduced impact [95]. For instance, COVID-19 patients who develop ventilator-associated pneumonia have an impaired innate immune defence that leads to their susceptibility to secondary infection as well as impaired clearance of SARS-CoV-2 infection [96]. Otherwise, there is evidence that the lung microbiome composition between COVID-19 negative and COVID-19 positive patients who developed ventilator-associated pneumonia was similar [97]. Another study revealed that COVID-19 symptomatic patients exhibited low nasopharyngeal bacteria diversity compared with asymptomatic and COVID-19 negative ones. They explained that running sneezing and nose could wash off the nasal microbiota of the symptomatic patients. In the symptomatic patients, nasal microbiota dysbiosis led to high levels of Cutibacterium and Lentimonas, while reducing the abundance of Prevotellaceae, Flectobacillus, Luminiphilus, Jannaschia, and Comamonas [98]. Liu et al. conducted an investigation to identify metabolome features and nasopharyngeal bacteria of COVID-19 patients. The sera examination of COVID-19 patients revealed that the levels of chlorogenic acid methyl ester (CME), L-proline, and lactic acid were notably reduced compared with COVID-19 negative individuals. Also, the pharynges of these patients were depleted from Gemella haemolysans, Gemella morbillorum, and Leptotrichia hofstadii. In contrast, the population of Prevotella histicola, Veillonella dispar, and Streptococcus sanguinis were increased. They found that the abundance of L. hofstadii and G. haemolysans were remarkably associated with the serum levels of CME [99]. CME could reduce the expression of inflammatory factors and alleviate the pathological impairment of lung tissue [100]. Gaibani et al. characterized the diversity of different bacteria in critically ill COVID-19 patients and healthy subjects using a 16S rRNA profiling on BAL samples. They found that the lung of COVID-19 patients enriched with Pseudomonas alcaligenes, Acinetobacter schindleri, Acinetobacter spp., Sphingobacterium spp., and Enterobacteriaceae. In contrast, Veillonella dispar, Haemophilus influenzae, Granulicatella spp., Streptococcus spp., and Porphyromonas spp., characterized in the lung of COVID-19-negative participants [101]. In another study, Merenstein et al. indicated the lower abundance of Actinomyces, Hemophilus, and Neisseria in the oropharyngeal of severe COVID-19 patients in comparison with normal ones. They also showed that the lower lymphocyte/neutrophil ratio was associated with the lower microbiome composition and diversity and inversely associated with the disease severity [102]. Iebba et al. identified Veillonella infantium and Prevotella salivae as predominant in patients suffering COVID-19, while Rothia mucilaginosa and Neisseria perflava were abundant in controls. Furthermore, the levels of cytokines IL-2, IL-5, IL-6, INF-γ, G-CSF, GM-CSF, and TNF-α were augmented in the COVID-19 patients, whereas only IL-12p70 was elevated in control subjects [103]. In a cross-sectional study, Soffritti et al. demonstrated that oral microbiome dysbiosis is inversely correlated with the COVID-19 severity. They reported the higher abundance of Streptococcus, Prevotella, Veillonella, Lactobacillus, Porphyromonas, Capnocytophaga, Aggregatibacter, Abiotrophia, and Atopobium in COVID-19 patients, whereas Haemophilus, Rothia, Fusobacterium, Parvimonas, and Gemella spp. showed lower frequency. Interestingly, the oral fungal and virome in COVID-19 patients were increased in comparison with controls. While Saccharomyces spp. and Candida were more abundant fungi in control participants, Nakaseomyces, Aspergillus, and Malassezia spp. were high in the COVID-19 patients. Bacteriophages targeted towards Streptococcus (phage PH10 and phage EJ-1), Lactobacillus (phage phiadh), and Staphylococcus (phage ROSA) as well as herpes simplex virus type 1 (HSV-1) were abundant in the COVID-19 patients. Furthermore, a reduction in mucosal sIgA responses was exhibited in more severely COVID-19 patients, suggesting the importance of local immune response in controlling virus infection at the early phase. Among the pro-inflammatory cytokines/chemokines, the levels of IL-6 and IL-17 were meaningfully higher in the oral of COVID-19 patients, without remarkable changes in GM-CSF and TNF-α [104].
Figure 2.
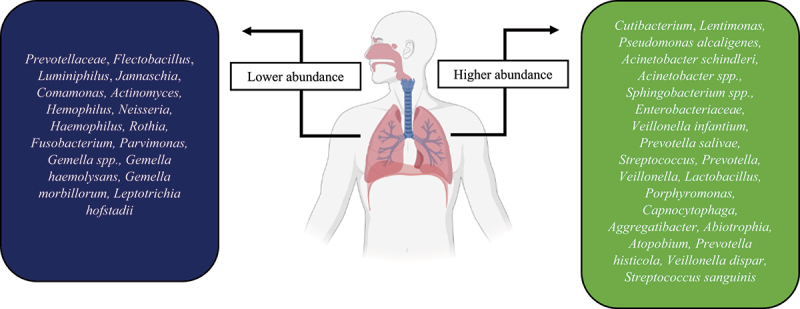
The higher and lower abundance of bacteria in the respiratory tract of COVID-19 patients.
There is evidence reporting bacterial, fungal, and viral co-infection with COVID-19. Two meta-analysis studies showed bacterial co-infection in 7% and 14% of hospitalized and critically ill patients, respectively [105,106]. The most commonly reported bacterial co-infection with COVID-19 patients are Enterobacteriaceae, H. influenza, and P. aeruginosa [106,107]. Gaibani et al. found that infection with carbapenem-resistant Acinetobacter baumannii elevated during COVID-19 [101]. In addition, Redondoviridae and Anelloviridae, airway commensal DNA viruses, exhibited more colonization frequency in COVID-19 patients and were completely linked to the severity of the disease [102].
The microbiome and treatments for COVID-19
Due to the undeniable role of microbiota in the COVID-19 disease, management of microbiota to shift its composition towards a healthy state could be effective in controlling SARS-CoV-2 infection and preventing the disease severity. Patients suffering from asymptomatic or mild COVID-19 infections are advised to eat a balanced, healthy, and anti-inflammatory diet, including legumes, fruits, grains, and vegetables. It has been reported that there is a reverse correlation between the amount of fibre intake with inflammatory markers, for example TNF-α, C-reactive protein, IL-6, and IL-18. Furthermore, high-fibre diets are associated with higher adiponectin and lower glycaemia, which have positive anti-inflammatory effects. Also, the anti-inflammatory activities of dietary fibre could support the functions of immunosuppressive and anti-viral drugs [108]. Various studies, including clinical trials, have been directed to investigate the preventive and therapeutic efficacy of microbiota modulation on COVID-19 using probiotics, synbiotics, prebiotics, nutraceuticals, and trace elements. For example, probiotics such as Paenibacillus and Lactobacilli produce peptides with the binding ability to ACE2, inhibiting the binding of the SARS-CoV-2 to its targeted cells [109,110]. Ceccarelli et al. concluded that treatment of patients suffering severe COVID-19 pneumonia with probiotics was correlated with a reduced risk of death [111]. Probiotics also promote anti-viral immune responses by activating toll-like receptor 4 (TLR4) signalling and inducing the secretion of type I and II IFNs [112,113]. Liu et al. indicated that orally receiving capsules as faecal microbiota transplantation (FMT) in COVID-19 patients could improve GI symptoms and alter B-cell populations, in which naïve B-cells were decreased and memory and non-switched B-cells were augmented. In discharged patients after FMT administration, the abundance of Actinobacteria was increased from 4% to 15%, and the proportion of Proteobacteria was reduced from 9.2% to 2.8%. However, they reported abdominal pain and diarrhoea as side effects of FMT administration [114]. In a case report study with two patients, Biliński et al. showed that FMT administration to treat concomitant COVID-19 and C. difficile infection led to rapid resolution of COVID-19 [115]. A clinical trial is investigating the impact of FMT, as immunomodulatory intervention, on reduced risk of progression of COVID-19 disease with increased inflammatory parameters and cytokine storm (NCT04824222). A meta-analysis study demonstrated that vitamin D deficiency is associated with COVID-19 severity; lower levels in poor prognosis patients [116]. It has been reported that vitamin D could increase the production of butyrate, which improves the gut barrier, exerts anti-inflammatory activities, and promotes anti-viral effects [71]. Table 3. summarizes the clinical trials conducted to evaluate microbiota modulation’s efficacy on COVID-19.
Table 3.
The effect of nutrients and probiotics in microbiota modulation of COVID-19 under clinical trials.
| Intervention/treatment | Participants (n) | Goal | ClinicalTrials.gov Identifier |
|---|---|---|---|
| Probiorinse of Lactococcus Lactis W136 | 23 | Reduction of the severity of COVID-19 symptoms | NCT04458519 |
| NSS using Saccharomyces bourllardii | 80 | Reduction of complications in COVID-19 patients and comorbidities | NCT04507867 |
| Combination of Pediococcus acidilactici CECT7483, Lactobacillus plantarum CECT7484, L. plantarum CECT30292, and L. plantarum CECT7485 | 300 | How does the combination of the probiotics reduce the risk of progression of COVID-19 to moderate or severe | NCT04517422 |
| Lactobacillus | 201 | Effects on the incidence and severity of COVID-19 in the elderly population living in a nursing home | NCT04756466 |
| NBT-NM108 | 100 | Evaluating the effectiveness and feasibility of NBT-NM108, as a treatment, in modulating gut microbiota of COVID-19. | NCT04540406 |
| Dry extract of polyphenols (tannins) from quebracho and chestnut | 124 | Evaluating the efficacy of Tannin against COVID-19 infection | NCT04403646 |
| Lactobacillus Coryniformis K8 | 314 | Evaluating the protective effect against COVID-19 in healthcare workers | NCT04366180 |
| Probiotic strains with maltodextrin as excipient | 41 | Improvement of symptoms in COVID-19 patients and reduction of hospitalization days | NCT04390477 |
| Lactobacillus salivarius with Vit D and Zinc | 60 | Effects on immune responses in COVID-19 patients | NCT04937556 |
| Lactobaciltus rhamnosus GG | 182 | Effects on the microbiome of household contacts exposed to COVID-19 | NCT04399252 |
| Lactobacilli and Bifidobacteria | 300 | Facilitating faster recovery from COVID-19 and enhancing immune responses | NCT04907877 |
NSS, Nutritional support system.
Conclusion
Human microbiota can affect immune responses, thus influencing disease progression and prevention. In the case of COVID-19, respiratory microbiota dysbiosis could be associated with underactive and overactive immune responses, which results in various clinical complications. Currently, the respiratory microbiome alteration is effect or cause of the COVID-19 is not clearly distinguished. However, the composition of microbiota differs from person to person and may explain the inter-individual variation in response to COVID-19. Due to the indispensable roles of microbiota in response to COVID-19, the assumption of inexpensive and safe supplementations and nutrients, such as probiotics, can be considered a preventive strategy for COVID-19 negative individuals or an adjunctive for treatment strategies to limit the progression of COVID-19 infection in the suffering patients. Furthermore, personalized diet regimens could direct the therapeutic strategies towards personalized medicine. In the sampling, it should be considered that sampling the lungs for microbial flora sequencing is technically demanding due to the relatively low biomass. In addition, lower respiratory tract sampling by bronchoscopy requires the instrument to be passed through the oral or nasal route, which may lead to the contaminations of samples.
Funding Statement
This work was supported by the Zhejiang Provincial Science and Technology Projects (No. LGF19H030001 to YJC), and the General Project Funds from the Health Department of Zhejiang Province (No. 2018KY269, and 2021KY481 to MHC).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Heymann DL, Shindo N.. COVID-19: what is next for public health? Lancet. 2020;395:542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ferreira-Santos D, Maranhão P, Monteiro-Soares M.. Identifying common baseline clinical features of COVID-19: a scoping review. BMJ Open. 2020;10:e041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Piva S, Filippini M, Turla F, et al. Clinical presentation and initial management critically ill patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in Brescia, Italy. J Crit Care. 2020;58:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Quah P, Li A, Phua J. Mortality rates of patients with COVID-19 in the intensive care unit: a systematic review of the emerging literature. Crit Care. 2020;24:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gilbert JA, Blaser MJ, Caporaso JG, et al. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ogunrinola GA, Oyewale JO, Oshamika OO, et al. The human microbiome and its impacts on health. Int J Microbiol 2020. 2020;2020:7. 10.1155/2020/8045646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Segal JP, Mak JWY, Mullish BH, et al. The gut microbiome: an under-recognised contributor to the COVID-19 pandemic? Therap Adv Gastroenterol. 2020;13:1756284820974914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Srinath BS, Shastry RP, Kumar SB. Role of gut-lung microbiome crosstalk in COVID-19. Res Biomed Eng. 2020;38:181–191. 10.1007/s42600-020-00113-4 [DOI] [Google Scholar]
- [11].Gu S, Chen Y, Wu Z, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020;71:2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rosell A, Monsó E, Soler N, et al. Microbiologic determinants of exacerbation in chronic obstructive pulmonary disease. Arch Intern Med. 2005;165:891–897. [DOI] [PubMed] [Google Scholar]
- [13].Thorpe JE, Baughman RP, Frame PT, et al. Bronchoalveolar lavage for diagnosing acute bacterial pneumonia. J Infect Dis. 1987;155:855–861. [DOI] [PubMed] [Google Scholar]
- [14].Wang L, Hao K, Yang T, et al. Role of the lung microbiome in the pathogenesis of chronic obstructive pulmonary disease. Chin Med J (Engl). 2017;130:2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huffnagle GB, Dickson RP, Lukacs NW. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 2017;10:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mathieu E, Escribano-Vazquez U, Descamps D, et al. Paradigms of lung microbiota functions in health and disease, particularly, in asthma. Front Physiol. 2018;9:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yagi K, Huffnagle GB, Lukacs NW, et al. The lung microbiome during health and disease. Int J Mol Sci. 2021;22:10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sze MA, Dimitriu PA, Hayashi S, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185:1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dickson RP, Huffnagle GB, Goldman WE. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 2015;11(7):e1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Frayman KB, Armstrong DS, Carzino R, et al. The lower airway microbiota in early cystic fibrosis lung disease: a longitudinal analysis. Thorax. 2017;72:1104–1112. [DOI] [PubMed] [Google Scholar]
- [22].Feigelman R, Kahlert CR, Baty F, et al. Sputum DNA sequencing in cystic fibrosis: non-invasive access to the lung microbiome and to pathogen details. Microbiome. 2017;5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Laguna TA, Wagner BD, Williams CB, et al. Airway microbiota in bronchoalveolar lavage fluid from clinically well infants with cystic fibrosis. PLoS One. 2016;11:e0167649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Millares L, Ferrari R, Gallego M, et al. Bronchial microbiome of severe COPD patients colonised by pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 2014;33:1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee S-W, Kuan C-S, Ls-H W, et al. Metagenome and metatranscriptome profiling of moderate and severe COPD sputum in Taiwanese Han males. PLoS One. 2016;11:e0159066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Durack J, Lynch SV, Nariya S, et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. 2017;140:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Denner DR, Sangwan N, Becker JB, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. 2016;137:1398–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Yan X, Yang M, Liu J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111. [PMC free article] [PubMed] [Google Scholar]
- [30].Liu H, Tao L, Zhang J, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer. 2018;142:769–778. [DOI] [PubMed] [Google Scholar]
- [31].Lee SH, Sung JY, Yong D, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 2016;102:89–95. [DOI] [PubMed] [Google Scholar]
- [32].Han MK, Zhou Y, Murray S, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. 2014;2:548–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Molyneaux PL, Cox MJ, Wells AU, et al. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir Res. 2017;18:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Molyneaux PL, Maher TM. The role of infection in the pathogenesis of idiopathic pulmonary fibrosis. Eur Respir Rev. 2013;22:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Molyneaux PL, Cox MJ, Willis-Owen SAG, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Muhlebach MS, Zorn BT, Esther CR, et al. Initial acquisition and succession of the cystic fibrosis lung microbiome is associated with disease progression in infants and preschool children. PLoS Pathog. 2018;14:e1006798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Byun MK, Chang J, Kim HJ, et al. Differences of lung microbiome in patients with clinically stable and exacerbated bronchiectasis. PLoS One. 2017;12:e0183553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20:1279–1290. [DOI] [PubMed] [Google Scholar]
- [39].Huang YJ, Nariya S, Harris JM, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. 2015;136:874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sze MA, Dimitriu PA, Suzuki M, et al. Host response to the lung microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2015;192:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Weinberg F, Dickson RP, Nagrath D, et al. The lung microbiome: a central mediator of host inflammation and Metabolism in lung cancer patients? Cancers (Basel). 2021;13:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liang H, Li X, Yu X, et al. Facts and fiction of the relationship between preexisting tuberculosis and lung cancer risk: a systematic review. Int J Cancer. 2009;125:2936–2944. [DOI] [PubMed] [Google Scholar]
- [43].Zhang R, Chen L, Cao L, et al. Effects of smoking on the lower respiratory tract microbiome in mice. Respir Res. 2018;19:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Panzer AR, Lynch SV, Langelier C, et al. Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically ill trauma patients. Am J Respir Crit Care Med. 2018;197:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gregory AC, Sullivan MB, Segal LN, et al. Smoking is associated with quantifiable differences in the human lung DNA virome and metabolome. Respir Res. 2018;19:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Steed E, Balda MS, Matter K. Dynamics and functions of tight junctions. Trends Cell Biol. 2010;20:142–149. [DOI] [PubMed] [Google Scholar]
- [47].Invernizzi R, Lloyd CM, Molyneaux PL. Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology. 2020;160:171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hall MW, Joshi I, Leal L, et al. Immune modulation in COVID-19: strategic considerations for personalized therapeutic intervention. Clin Infect Dis. 2020;74(1):144-148. doi: 10.1093/cid/ciaa904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol. 2015;16:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tay MZ, Poh CM, Rénia L, et al. The trinity of COVID-19: immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cantuti-Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Santos LD, Antunes KH, Muraro SP, et al. TNF-Mediated alveolar macrophage necroptosis drives disease pathogenesis during respiratory syncytial virus infection. Eur Respir J. 2021;57:1–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Antunes KH, Fachi JL, de Paula R, et al. Microbiota-Derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat Commun. 2019;10:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Martin TR, Frevert CW. Innate immunity in the lungs. Proc Am Thorac Soc. 2005;2:403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhou Z, Ren L, Zhang LI, et al. Overly exuberant innate immune response to SARS-CoV-2 infection. 2020. 10.2139/ssrn.3551623 [DOI] [Google Scholar]
- [57].Zhou Z, Ren L, Zhang L, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liao M, Liu Y, Yuan J, et al. Single-Cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. [DOI] [PubMed] [Google Scholar]
- [59].Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jamilloux Y, Henry T, Belot A, et al. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun Rev. 2020;19:102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Seif F, Khoshmirsafa M, Aazami H, et al. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Azkur AK, Akdis M, Azkur D, et al. Immune response to SARS‐CoV‐2 and mechanisms of immunopathological changes in COVID‐19. Allergy. 2020;75:1564–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pavel AB, Glickman JW, Michels JR, et al. Th2/th1 cytokine imbalance is associated with higher COVID-19 risk mortality. Front Genet. 2021;12:706902. doi: 10.3389/fgene.2021.706902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mazzoni A, Maggi L, Capone M, et al. Cell‐mediated and humoral adaptive immune responses to SARS‐CoV‐2 are lower in asymptomatic than symptomatic COVID‐19 patients. Eur J Immunol. 2020;50:2013–2024. [DOI] [PubMed] [Google Scholar]
- [66].Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kalfaoglu B, Almeida-Santos J, Tye CA, et al. T-Cell hyperactivation and paralysis in severe COVID-19 infection revealed by single-cell analysis. Front Immunol. 2020;11:2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tian Y, Sun K, Meng T, et al. Gut microbiota may not be fully restored in recovered COVID-19 patients after 3-month recovery. Front Nutr. 2021;8:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ferreira C, Viana SD, Reis F. Is gut microbiota dysbiosis a predictor of increased susceptibility to poor outcome of COVID-19 patients? an update. Microorganisms. 2021;9:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Chen J, Vitetta L. Modulation of gut microbiota for the prevention and treatment of COVID-19. J Clin Med. 2021;10:2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yeoh YK, Zuo T, Lui G-Y, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zuo T, Liu Q, Zhang F, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Khan M, Mathew BJ, Gupta P, et al. Gut dysbiosis and IL-21 response in patients with severe COVID-19. Microorganisms. 2021;9:1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Moreira-Rosário A, Marques C, Pinheiro H, et al. Gut microbiota diversity and C-Reactive protein are predictors of disease severity in COVID-19 patients. bioRxiv. 2021;12:705020. doi: 10.3389/fmicb.2021.705020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Luu M, Weigand K, Wedi F, et al. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci Rep. 2018;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhou Y, Shi X, Fu W, et al. Gut microbiota dysbiosis correlates with abnormal immune response in moderate COVID-19 patients with fever. J Inflamm Res. 2021;14:2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Tao W, Zhang G, Wang X, et al. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med Microecol. 2020;5:100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Haiminen N, Utro F, Seabolt E, et al. Functional profiling of COVID-19 respiratory tract microbiomes. Sci Rep. 2021;11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Törnquist K, Asghar MY, Srinivasan V, et al. Sphingolipids as modulators of SARS-CoV-2 infection. Front Cell Dev Biol. 2021;9:1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Santos AF, Póvoa P, Paixão P, et al. Changes in glycolytic pathway in SARS-COV 2 infection and their importance in understanding the severity of COVID-19. Front Chem. 2021;9:685196. doi: 10.3389/fchem.2021.685196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Codo AC, Davanzo GG, de Brito Monteiro L, et al. Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent axis. Cell Metab. 2020;32:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Viciani E, Gaibani P, Castagnetti A, et al. Critically ill patients with COVID-19 show lung fungal dysbiosis with reduced microbial diversity in patients colonized with Candida spp. Int J Infect Dis. 2022;117:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Calderaro A, Buttrini M, Montecchini S, et al. Detection of SARS-CoV-2 and other infectious agents in lower respiratory tract samples belonging to patients admitted to intensive care units of a tertiary-care hospital, located in an epidemic area, during the Italian lockdown. Microorganisms. 2021;9:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhong H, Wang Y, Shi Z, et al. Characterization of respiratory microbial dysbiosis in hospitalized COVID-19 patients. Cell Discov. 2021;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hoque MN, Sarkar M, Hasan M, et al. SARS-CoV-2 infection reduces human nasopharyngeal commensal microbiome with inclusion of pathobionts. Sci Rep. 2021;11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hoque MN, Rahman MS, Ahmed R, et al. Diversity and genomic determinants of the microbiomes associated with COVID-19 and non-COVID respiratory diseases. Gene Rep. 2021;23:101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Hoque MN, Akter S, Mishu ID, et al. Microbial co-infections in COVID-19: associated microbiota and underlying mechanisms of pathogenesis. Microb Pathog. 2021;156:104941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Lin X, Gong Z, Xiao Z, et al. Novel coronavirus pneumonia outbreak in 2019: computed tomographic findings in two cases. Korean J Radiol. 2020;21:365–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Man WH, de Steenhuijsen Piters WAA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Mostafa HH, Fissel JA, Fanelli B, et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. Mbio. 2020;11: 1- 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Zhang H, Ai J-W, Yang W, et al. Metatranscriptomic characterization of Coronavirus disease 2019 identified a host transcriptional classifier associated with immune signaling. Clin Infect Dis. 2021;73:376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Han Y, Jia Z, Shi J, et al. The active lung microbiota landscape of COVID-19 patients. medRxiv. 2020;2008.2020.20144014. doi: 10.1101/2020.08.20.20144014. [DOI] [Google Scholar]
- [95].Lloréns-Rico V, Gregory AC, Van Weyenbergh J, et al. Clinical practices underlie COVID-19 patient respiratory microbiome composition and its interactions with the host. Nat Commun. 2021;12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Tsitsiklis A, Zha B, Byrne A, et al. Impaired immune signaling and changes in the lung microbiome precede secondary bacterial pneumonia in COVID-19. Res Sq 2021. doi: 10.21203/rs.3.rs-380803/v1. [DOI] [Google Scholar]
- [97].Maes M, Higginson E, Pereira-Dias J, et al. Ventilator-Associated pneumonia in critically ill patients with COVID-19. Crit Care. 2021;25:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kolhe R, Sahajpal NS, Vyavahare S, et al. Alteration in nasopharyngeal microbiota profile in aged patients with COVID-19. Diagnostics. 2021;11:1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Liu J, Liu S, Zhang Z, et al. Association between the nasopharyngeal microbiome and metabolome in patients with COVID-19. Synth Syst Biotechnol. 2021;6:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhang L, Fan Y, Su H, et al. Chlorogenic acid methyl ester exerts strong anti-inflammatory effects via inhibiting the COX-2/NLRP3/NF-κB pathway. Food Funct. 2018;9:6155–6164. [DOI] [PubMed] [Google Scholar]
- [101].Gaibani P, Viciani E, Bartoletti M, et al. The lower respiratory tract microbiome of critically ill patients with COVID-19. Sci Rep. 2021;11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Merenstein C, Liang G, Whiteside SA, et al. Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. mBio. 2021;12:e01777-21. 10.1128/mBio.01777-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Iebba V, Zanotta N, Campisciano G, et al. Profiling of oral microbiota and cytokines in COVID-19 patients. Front Microbiol. 2021; 12:671813. doi: 10.3389/fmicb.2021.671813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Soffritti I, D’-Accolti M, Fabbri C, et al. Oral microbiome dysbiosis is associated with symptoms severity and local immune/inflammatory response in COVID-19 patients: a cross-sectional study. Front Microbiol. 2021;12:1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lansbury L, Lim B, Baskaran V, et al. Co-Infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zhu X, Ge Y, Wu T, et al. Co-Infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Conte L, Toraldo DM. Targeting the gut–lung microbiota axis by means of a high-fibre diet and probiotics may have anti-inflammatory effects in COVID-19 infection. Ther Adv Respir Dis. 2020;14:1753466620937170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Li J, Zhao J, Wang X, et al. Novel angiotensin-converting enzyme-inhibitory peptides from fermented bovine milk started by Lactobacillus helveticus KLDS. 31 and Lactobacillus casei KLDS. 105: purification, identification, and interaction mechanisms. Front Microbiol. 2019;10:2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Minato T, Nirasawa S, Sato T, et al. B38-CAP is a bacteria-derived ACE2-like enzyme that suppresses hypertension and cardiac dysfunction. Nat Commun. 2020;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ceccarelli G, Borrazzo C, Pinacchio C, et al. Oral bacteriotherapy in patients with COVID-19: a retrospective cohort study. Front Nutr. 2021;7:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kumova OK, Fike AJ, Thayer JL, et al. Lung transcriptional unresponsiveness and loss of early influenza virus control in infected neonates is prevented by intranasal Lactobacillus rhamnosus GG. PLoS Pathog. 2019;15:e1008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Eguchi K, Fujitani N, Nakagawa H, et al. Prevention of respiratory syncytial virus infection with probiotic lactic acid bacterium Lactobacillus gasseri SBT2055. Sci Rep. 2019;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Liu F, Ye S, Zhu X, et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J Med Case Rep. 2021;15:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Biliński J, Winter K, Jasiński M, et al. Rapid resolution of COVID-19 after faecal microbiota transplantation. Gut. 2022;71:230–232. [DOI] [PubMed] [Google Scholar]
- [116].Munshi R, Hussein MH, Toraih EA, et al. Vitamin D insufficiency as a potential culprit in critical COVID‐19 patients. J Med Virol. 2021;93:733–740. [DOI] [PubMed] [Google Scholar]


