Abstract
Purpose:
To develop techniques and establish a workflow using hyperpolarized carbon-13 (13C) MRI and the pyruvate-to-lactate conversion rate (kPL) biomarker to guide MR-transrectal ultrasound fusion prostate biopsies.
Methods:
The integrated multiparametric MRI (mpMRI) exam consisted of a 1-min hyperpolarized 13C-pyruvate EPI acquisition added to a conventional prostate mpMRI exam. Maps of kPL values were calculated, uploaded to a picture archiving and communication system and targeting platform, and displayed as color overlays on T2-weighted anatomic images. Abdominal radiologists identified 13C research biopsy targets based on the general recommendation of focal lesions with kPL >0.02(s−1), and created a targeting report for each study. Urologists conducted transrectal ultrasound-guided MR fusion biopsies, including the standard 1H–mpMRI targets as well as 12–14 core systematic biopsies informed by the research 13C-kPL targets. All biopsy results were included in the final pathology report and calculated toward clinical risk.
Results:
This study demonstrated the safety and technical feasibility of integrating hyperpolarized 13C metabolic targeting into routine 1H–mpMRI and transrectal ultrasound fusion biopsy workflows, evaluated via 5 men (median age 71 years, prostate-specific antigen 8.4 ng/mL, Cancer of the Prostate Risk Assessment score 2) on active surveillance undergoing integrated scan and subsequent biopsies. No adverse event was reported. Median turnaround time was less than 3 days from scan to 13C-kPL targets, and scan-to-biopsy time was 2 weeks. Median number of 13C targets was 1 (range: 1–2) per patient, measuring 1.0 cm (range: 0.6–1.9) in diameter, with a median kPL of 0.0319 s−1 (range: 0.0198–0.0410).
Conclusions:
This proof-of-concept work demonstrated the safety and feasibility of integrating hyperpolarized 13C MR biomarkers to the standard mpMRI workflow to guide MR-transrectal ultrasound fusion biopsies.
Keywords: hyperpolarized 13C MRI, prostate cancer, MR-guided TRUS fusion biopsy
1 |. INTRODUCTION
Multiparametric prostate MRI (mpMRI) is a standard-of-care imaging tool in the clinical workup of men with either known or suspected prostate cancer. Many practice guidelines such as National Comprehensive Cancer Network, American Urological Association, and European Association of Urology1 have incorporated mpMRI into the workflow of prostate cancer diagnosis, although the indications for which mpMRI should be ordered, and at what frequency, still vary from guideline to guideline. This stems from divergent views regarding mpMRI’s overall role and importance in the prostate cancer risk assessment.
Active surveillance is a preferred management strategy for men with low-risk, and selected intermediate-risk, localized prostate cancer to minimize treatment-associated morbidity without compromising oncologic outcomes. This is evidenced by the 10-year cancer-specific survival rate of nearly 99% for men undergoing active surveillance.2 Whereas conventional mpMRI is widely used to guide prostate biopsies, its role in the surveillance setting has long been a subject of controversy.3,4 Although mpMRI-guided confirmatory biopsy led to decreased active surveillance failures,5 a few randomized and retrospective studies failed to confirm the clinical utility of mpMRI in active surveillance.6–8 These contradicting results not only lead to discrepant endorsement among guidelines regarding the use of surveillance mpMRI but highlighted the unmet need to improve both the diagnostic accuracy and yield of mpMRI in this setting.
Hyperpolarized (HP) carbon-13 (13C)-pyruvate MRI is a new rapid molecular imaging technique that can detect increased pyruvate-to-lactate conversion rates (kPL) in clinically significant, aggressive prostate cancer as compared to more indolent tumors.9,10 The emerging technology uses dynamic nuclear polarization to increase the SNR of 13C-enriched compounds by more than 50,000-fold through means of temporarily rearranging the spins to increase their nuclear polarization.11,12 This enables interrogation of previously inaccessible in vivo metabolic pathways with unprecedented signal quality and quantitative accuracy. Several studies have explored the technical aspects of HP 13C MRI in men with localized and metastatic prostate cancer,13–15 as well as correlating 13C markers to histological and molecular signatures of this malignancy.16,17
MR-guided transrectal ultrasound (TRUS) fusion biopsy was designed to utilize MRI’s superior tissue contrast compared to ultrasound to detect prostatic lesions and improve prostate cancer diagnosis and grading. The fusion technology combines targeting information from a prior diagnostic MR exam with real-time, intraprocedural ultrasound guidance for accurate prostate tissue sampling.18 MR-guided TRUS fusion biopsies, together with systematic biopsies, are usually conducted in urologists’ offices as a simple outpatient procedure without requiring an operating room or general anesthesia.
Thus far, HP 13C MRI of localized prostate cancer has been studied primarily in the context of a high-risk cohort, looking at pathological correlation with the prostatectomy specimen.16,17 Prior to this study, it had not been applied to prospectively guide confirmatory or surveillance biopsies. This technical development project aimed to integrate the metabolically defined research targets based on HP 13C-pyruvate MRI into mpMRI workflow and guide MR-TRUS fusion biopsies to improve the identification of clinically significant disease.
2 |. METHODS
2.1 |. Hyperpolarized 13C MRI and standard-of-care multiparametric MRI protocols
The MRI exams were conducted on a clinical 3 Tesla MRI scanner (MR750, GE Healthcare, Chicago IL) equipped with multinuclear spectroscopy capability. The MR coils and imaging setup have been previously described.19 Briefly, HP 13C imaging was performed using a clamshell Helmholtz transmitter and an endorectal coil for receive. For proton imaging, the body coil was used as a transmitter, and signal reception was accomplished using a 4-channel torso array combined with the endorectal probe. The endorectal receiver was a specialized proton (1H)/13C dual-element design.19
The proton mpMRI exam consisted of T1-weighted fast spin echo (FSE), T2-weighted FSE (in-plane resolution = 0.35 mm, 3 mm slices, TR/TE = 6000/102 ms), 3D T2-FSE, and small FOV (Field-of-view optimized and constrained undistorted single-shot, FOCUS) DWI (TR/TE = 4000/78, pixel bandwidth = 1305, b = 0 and 600 s/mm2, and 3 mm slice thickness) sequences. Dynamic contrast-enhanced imaging using a 3D fast spoiled gradient-recalled echo (SPGR) sequence (TR/TE = 3.5/0.9 ms, flip angle = 5°, and 3 mm slice thickness) with gadobutrol (Bayer Pharmaceuticals, Leverkusen, Germany) was acquired as the final series of the exam after the conclusion of the 13C-pyruvate acquisition and other proton imaging. The multiparametric portion of the exam was consistent with the standard mpMRI20 indicated for prostate cancer care at the University of California, San Francisco (UCSF).
For the hyperpolarized 13C pyruvate study, pharmacy kits containing a mixture of 1.432 g of GMP grade [1-13C] pyruvic acid (MilliporeSigma, Miamisburg OH) and 28 mg electron paramagnetic agent (AH111501; GE Healthcare, Oslo, Norway) were prepared and polarized in a 5 Tesla clinical-research polarizer (SPINLab, GE Healthcare, Chicago IL) at 0.77 K for 2.5–3 hours. Subsequently, the pyruvic acid was rapidly dissoluted and neutralized with tris-EDTA buffer solution, yielding sterile doses meeting release criteria of pyruvate concentration (median: 248 mM, 238–271 mM), polarization (median: 38.4%, 34.6%–40.6%), pH (median: 7.7, 7.4–8.2), temperature (<37 °C), and electron paramagnetic agent concentration (median: 1.5 μM, 0.7–1.9 μM). Following terminal sterilization, pharmaceutical release was issued after confirming quality control parameters and filter bubble point test as previously published.21 The dosage of pyruvate solution delivered to the patient was 0.43 mL/kg at an injection rate of 5 mL/s (up to 40 mL), immediately followed by a 20 mL saline flush at the same rate.
A symmetric EPI sequence was prescribed for the HP 13C study.22 Key sequence parameters included TR/TE = 1 s/25.2 ms; resolution = 6.5–8 mm in-plane, 8 mm through-plane; FOV = 10.4 × 10.4–12.8 × 12.8 cm in-plane, and 11.2 cm through-plane. Independent flip angles were applied to pyruvate (15°) and lactate (30°) resonances. The acquisition started 10 s after the completion of the pyruvate injection and saline flush. A rapid, low-resolution T2-FSE axial series (in-plane resolution = 0.35 × 0.8 mm, 3 mm slices, ~1 min length) was acquired immediately after the 13C scan to measure possible motion shift during the exam, which could result from patient movements or bladder filling, and the resulting misalignment in between series. 13C center frequency and power were calibrated using a built-in 8 M urea phantom inside the endorectal coil. Transmit power was increased to ~133% of the nominally calibrated power to compensate for the inductive coupling losses between clamshell transmitter and the 13C element of the endorectal coil.
2.2 |. Image processing
The 13C-pyruvate EPI images were reconstructed using the GE Orchestra Toolbox (GE Healthcare, Chicago IL). Nyquist ghost correction was conducted using the 1H reference scan method, as previously described.22 Global phases of the pyruvate and lactate images were independently calculated and accounted for. The HP 13C metabolic biomarker, kPL, was quantified using an inputless 2-site exchange model that makes no assumptions about the dynamic or bolus profile of pyruvate but rather derives kPL values solely based on measured pyruvate and lactate signals for improved robustness.23
Any misalignment was manually calculated between the high-quality T2-weighted series, acquired near the beginning of the exam, and the quick, post-13C-injection T2-FSE series, and the 13C data were shifted accordingly in 3D to compensate for motion offset. This improved the alignment precision between the kPL maps and the high-quality T2 series on the targeting software Dynacad (Philips Invivo Corp., Gainsville, FL), and thereby allowed more accurate lesion identification and outlining.
2.3 |. Transfer of 13C-pyruvate MRI data to PACS and targeting software
The kPL maps were interpolated to the T2-FSE resolution, matching the exact same registration and matrix size as the T2 series for easy visualization and overlays. A predefined series number was assigned to the kPL maps. For example, if the high-quality T2 were series 5, the kPL map would have been series 502.
The kPL map series was labeled as nondiagnostic HP-13C kPL (diffusion) to easily distinguish it from clinical 1H mpMRI sequences. The color kPL map was overlaid on T2-weighted images. Using the keyword “(Diffusion)” masqueraded the kPL maps as a diffusion-weighted series. This nomenclature allowed us to repurpose the built-in color overlay function on Dynacad (Philips Invivo Corp.; referred to as “fusion” overlays on the UI) that was intended for overlay of DWI as a pseudo color map over grayscale T2-weighted images.
The kPL maps were first uploaded to the picture archiving and communication system (PACS). Following a quality control check on one of the PACS workstations to ensure correct registration, the kPL maps were transferred to the Dynacad (Philips Invivo Corp.), a commercial targeting software that radiologists routinely use to identify and outline suspicious prostate MRI lesions as a part of biopsy planning. All the processing and data transfer occurred between our internal radiology network and PACS system and were therefore compliant with the Health Insurance Portability and Accountability Act.
2.4 |. Prostate lesion targeting and fusion biopsy
Figure 1 illustrates the workflow of HP 13C MR research targeting of the prostate. The conventional 1H mpMRI exam was interpreted by board-certified radiologists according to the standard departmental workflow, and any lesions were categorized using Prostate Imaging Reporting and Data System (PIRADS) v2.1.24 Any suspicious lesions were outlined in Dynacad (Philips Invivo Corp.) in preparation for targeted biopsy. One of 3 board-certified abdominal radiologists, each with more than 10 years of experience reading prostate MRI, additionally outlined the 13C research targets on the kPL-T2w color overlay displayed on Dynacad (Philips Invivo Corp.) (Figure 2A). The 13C research targets were identified and delineated following the general recommendation of delineating focal lesions on the kPL map with kPL value >0.02(s−1), a biomarker of suspected cancer.17 The recommended kPL threshold of 0.02(s−1) for this initial technical development study was selected based on a prior high-risk cohort of patients (Supporting Information Figure S1), who after 13C-pyruvate MRI scans underwent radical prostatectomy with step-section histopathology (gold standard), representing the kPL differences of high- versus low-grade tumors (corrected for different MR sequence TEs25). Radiologists’ experience and judgment play an important role in target selection (similar to selection of 1H MRI targets). Their discretion to include/exclude a target is allowed based on lesion focality, shape, and visual features, as well as likelihood of tumor presence in a given anatomical zone. The 1H mpMRI-defined clinical targets and 13C-defined research targets were independently labeled for urologist review.
FIGURE 1.
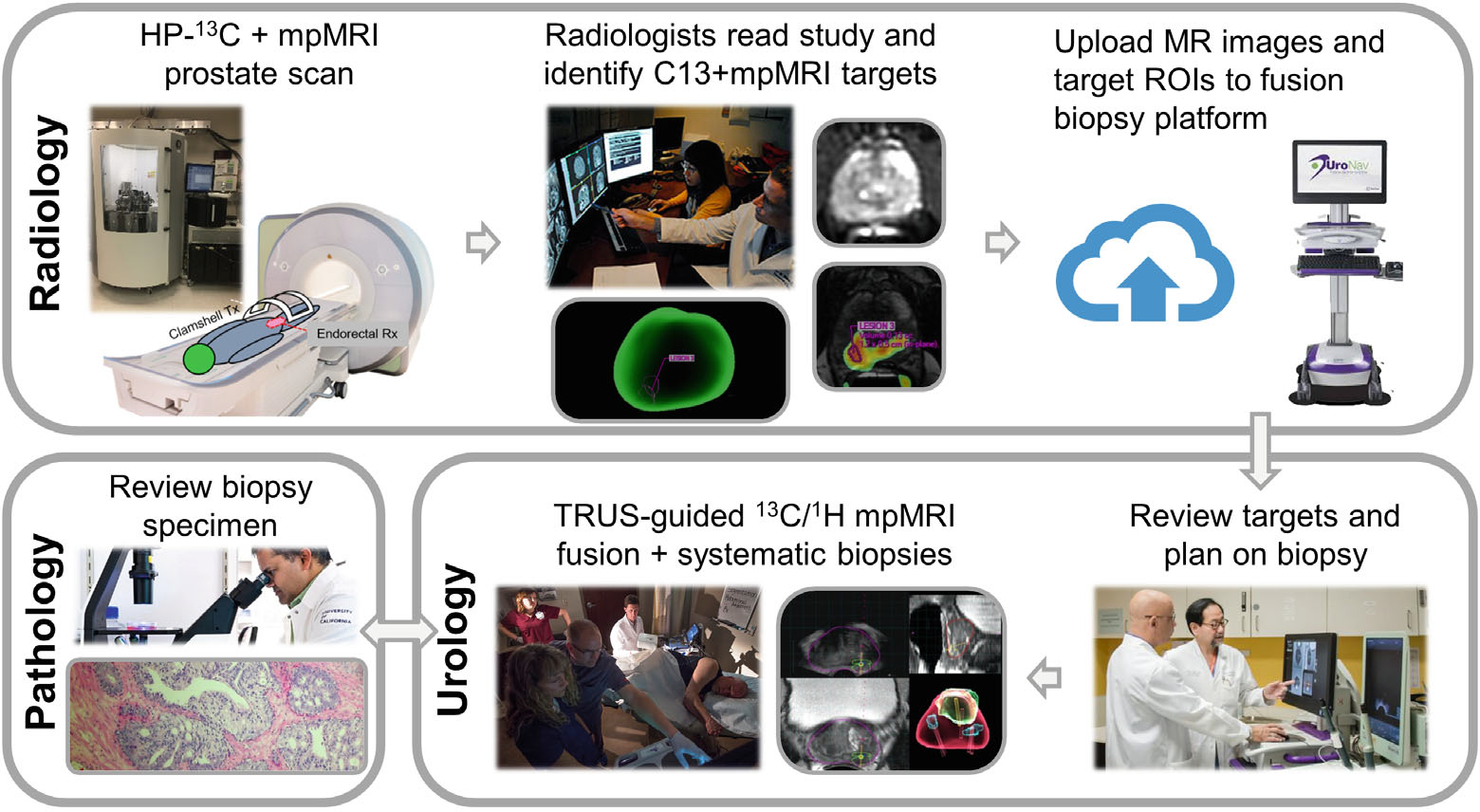
The workflow developed in this project for HP 13C MR research targeting of prostate biopsies based on abnormally high pyruvate-to-lactate conversion kPL values. The HP 13C MR exam and research targeting were integrated into the SOC MRI fusion and systematic biopsy procedures at our institution. First, the patient undergoes an integrated mpMRI exam of the prostate, including a 1-min acquisition following the HP 13C-pyruvate injection. The kPL map is calculated and uploaded to PACS and a software targeting platform (Dynacad, Philips Invivo Corp., Gainsville FL). A radiologist reads the study and outlines the research targets based on 13C kPL findings, in addition to those from the PIRADS lesions based on the 1H mpMRI. The targets and a report are uploaded to the fusion biopsy system (UroNav, Philips Invivo Corp., Gainsville FL) in the urologist’s offices, where they review the targeting and plan for the procedure. After US/MRI fusion-guided biopsies are performed, the tissue specimens are submitted to Pathology for processing and diagnosis. Thus, the HP 13C research biopsy integration takes advantage of the existing infrastructure and minimizes the additional workload for the researchers and clinicians involved. 13C, carbon-13; HP, hyperpolarized; kPL, pyruvate-to-lactate conversion rate; mp, multiparametric; PACS, picture archiving and communication system; PIRADS, Prostate Imaging Reporting and Data System; SOC, standard- of- care; US, ultrasound.
FIGURE 2.
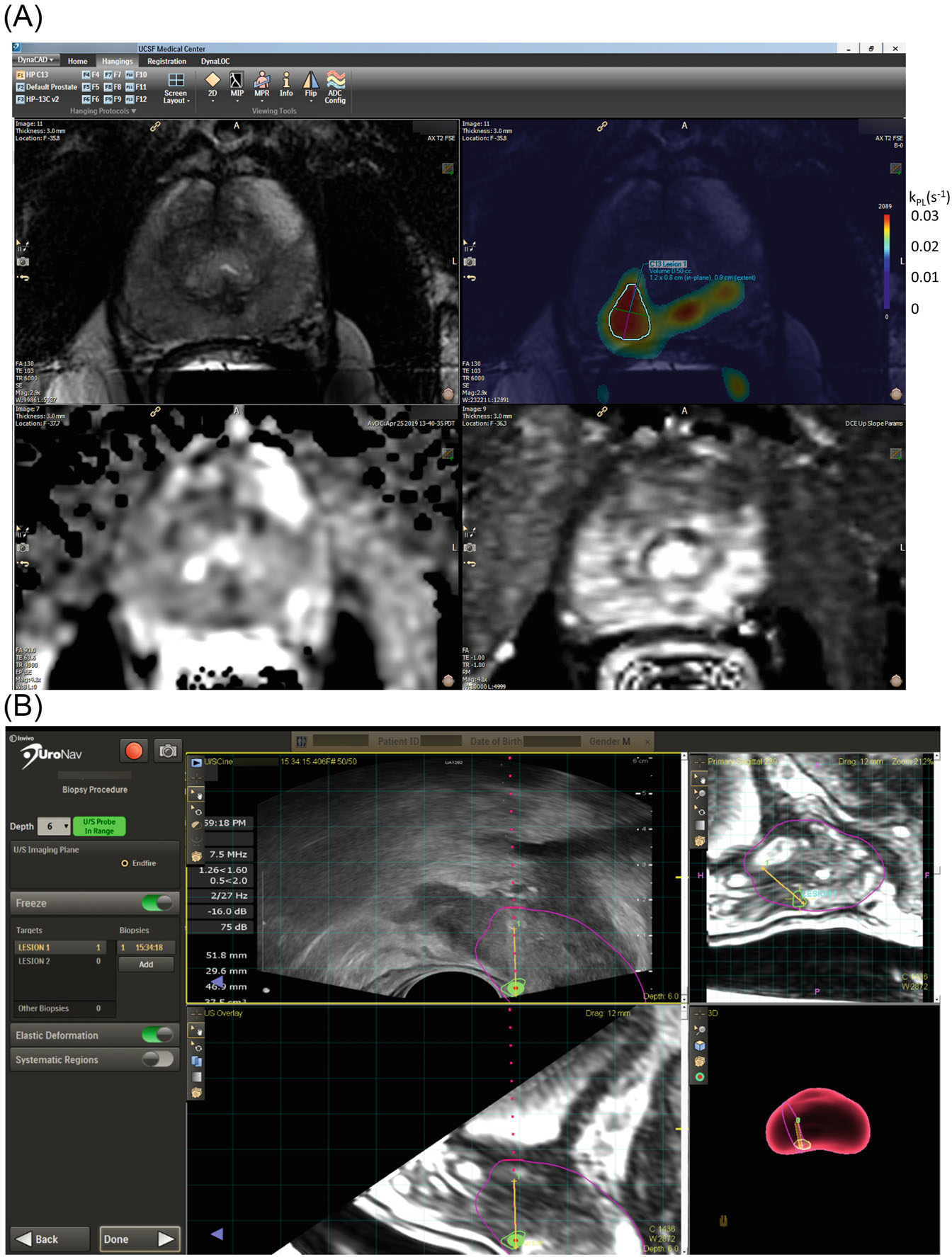
(A) A representative targeting protocol using a commercial prostate biopsy targeting platform (Dynacad, Philips Invivo Corps.). This protocol can also be found in Supporting Information Video S1. The 3D kPL image series was named with the keyword “diffusion” to allow a fusion overlay, displaying kPL as a pseudocolor over T2-weighted series. The overlay was displayed side by side with T2, ADC, and DCE maps, enabling the radiologist to correlate between series and outline 3D ROI for both 1H PIRADS and 13C research biopsy targets. Whereas the recommended kPL threshold for identifying potentially high-grade 13C lesions was set to 0.02(s−1), the lowest value of the heatmap was set to 0.01 for display purposes. This is designed to provide radiologists context on the shape/size of the lesion. The corresponding kPL scales is shown next to the original color bar. (B) Both clinical and research targets are transferred to a commercial TRUS-fusion biopsy platform (UroNav, Philips Invivo Corps.), where the research biopsy targets were counted as systematic biopsies. The urologist sampled these targets under TRUS fusion guidance during a biopsy session, assisted by TRUS-MRI fusion (left panel: US axial, top right: MR sagittal) and 3D-rendered segmentation (bottom right panel) of the prostate. The biopsied tissues were submitted for histopathology analyses. 1H, proton; ROI, region of interest; TRUS, transrectal ultrasound.
Both 13C research and standard 1H mpMRI targeted biopsies were conducted by 1 of the 3 board-certified urological oncologists, each with more than 10 years of experience, who used a commercial fusion-biopsy platform (Uronav, Philips Invivo Corp., Gainsville, FL) following the standard mpMRI-targeted biopsy procedure at our institution (Figure 2B). The transrectal biopsy approach was used by our urologists per institutional practice. The MRI-generated overlays delineating 13C/1H targets were fused with real-time TRUS images in the UroNav system (Philips Invivo Corp.) to guide the biopsy sampling. Systematic TRUS biopsies (12–14 cores) were also conducted in the same session, and the biopsy cores of any 13C research target replaced the systematic biopsy in the same sextant. As such, the 13C research biopsy did not increase the total number of cores.
A sample overlay and targeting procedure is illustrated in Supporting Information Video S1.
2.5 |. Patient characteristics
Five patients with biopsy-confirmed prostate cancer were enrolled (NCT03933670). Eligible patients were 18 years or older, had a biopsy-confirmed diagnosis of prostate cancer, ECOG score 0 or 1, and were either candidates for or currently on an active surveillance protocol as defined by the UCSF urologic oncology practices at the time of enrollment.26 The key exclusion criteria entailed prior treatments for prostate cancer, biopsy within 14 days prior to 13C MRI, poorly controlled hypertension, or contraindication for endorectal coil placement. The patient studies were conducted with informed consent in compliance with Food and Drug Administration- and Institutional Review Board-approved protocols (NCT03933670).
2.6 |. Pathology assessment
The specimens from the 13C research biopsies were submitted to pathology along with the other samples from the same session. All formalin-fixed–paraffin-embedded cores were read by experienced urologic pathologists in a standardized fashion27 and included in the final pathology report. UCSF–Cancer of the Prostate Risk Assessment (CAPRA) score28 was recalculated based on the updated biopsy findings and the most recent prostate-specific antigen values.
3 |. RESULTS
3.1 |. Safety and technical feasibility
Integrated MRI exams and the ensuing biopsies for all 5 patients were safe, successful, and without adverse events. The kPL (HP 13C metabolic biomarker) map was calculated and uploaded to PACS/Dynacad (Philips Invivo Corp.) along with the mpMRI exam. Image postprocessing and upload of kPL maps were done the same day, typically within 1–2 h after the end of the exam. The average turnaround time for the MRI report and targeting was less than 3 days after each exam. A 13C research biopsy targeting report (a representative instance given in Figure 3) was generated and showed the target locations in the 3D segmented prostate, as well as the 13C-kPL/T2 overlay, diffusions, and T1-weighted images arranged side by side to assist the urologists planning the biopsies.
FIGURE 3.
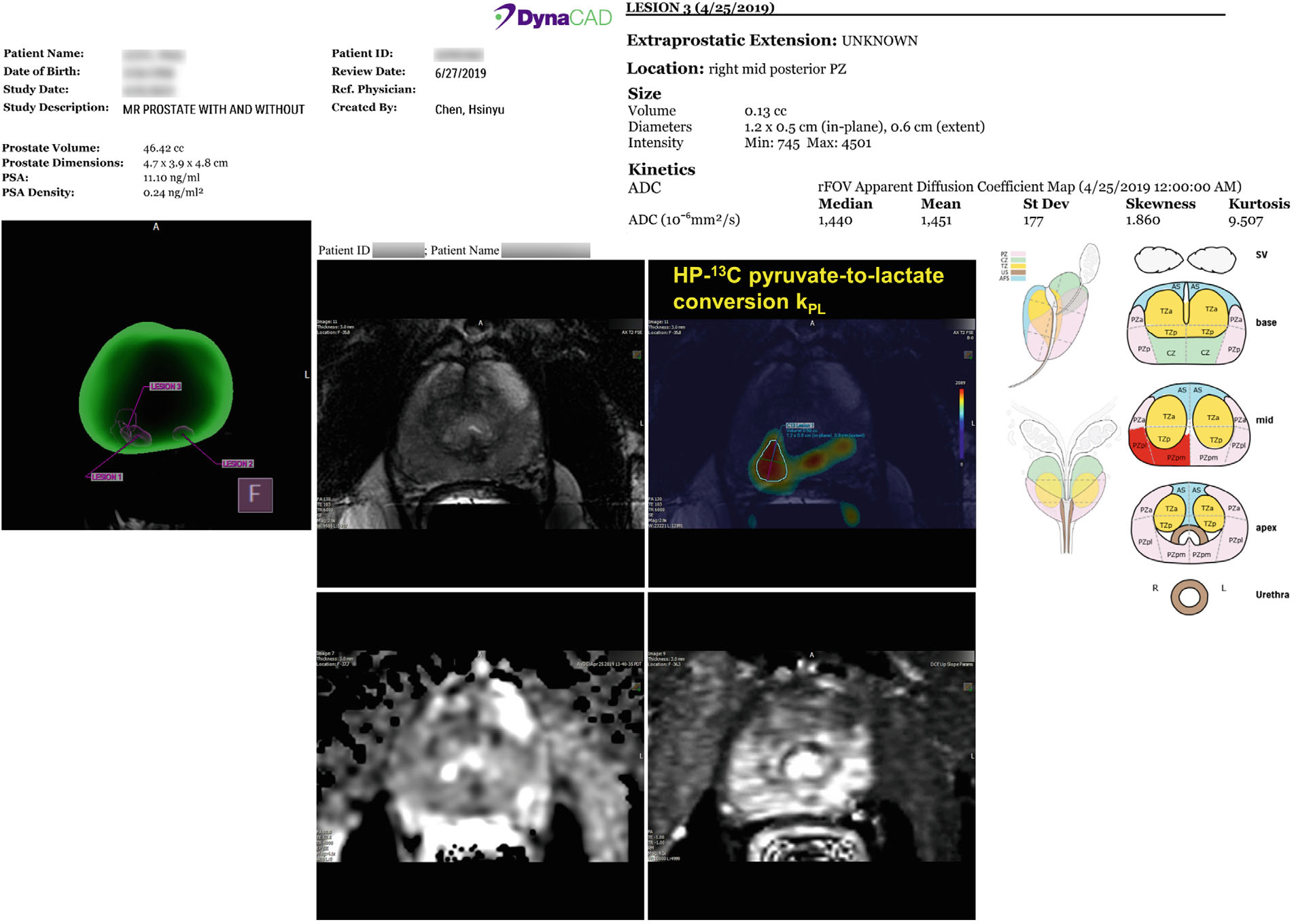
Shows a representative biopsy targeting report a radiologist created using DynaCAD (Philips Invivo Corp.). The report and targets were then sent to the UroNav system (Philips Invivo Corp.) to assist the urological oncologist to identify the 1H mpMRI (PIRADs) and 13C research targets and plan for the biopsy procedure. The report, shown as montage here, illustrates the target locations on the 3D segmented prostate for visual reference (left panel). In the center panel, the 13C-kPL/T2 overlay, DWI, and T1-weighted images are arranged side by side. A 13C-kPL lesion was identified and outlined at the right mid-PZ. The right panel reports the automatically calculated volumes and mean ADC values over the outlined 13C target. PZ, peripheral zone.
3.2 |. HP 13C MRI targeting and MR-guided TRUS fusion biopsy
The patient demographics (N = 5) and clinical characteristics are summarized in Figure 4. The patients enrolled in this study had low- to intermediate-risk disease with median age 71 (range: 62–79), prostate-specific antigen 8.4 ng/mL (range: 1.3–17), and CAPRA score 2 (range: 1–3). The median number of 13C research targets was 1 (range: 1–2), and that of proton mpMRI was 1 (range: 0–2). The 13C targets measured 1 cm (range: 0.6–1.9) in diameter; the median kPL was 0.0319 s−1 (range: 0.0198–0.0410); and the intralesion distribution is given in Table 1. The mean kPL in the segmented prostate excluding 13C targets was 0.0110±0.0022 s−1. The index lesions on proton mpMRI had a median PIRADS score 4 (range: 2–5). The 1H targets had a median 1.1 cm (range: 0.9–2) diameter. All 5 patients underwent TRUS/MRI fusion and systematic biopsies after the integrated mpMRI exam, with 2–3 cores sampled per target.
FIGURE 4.
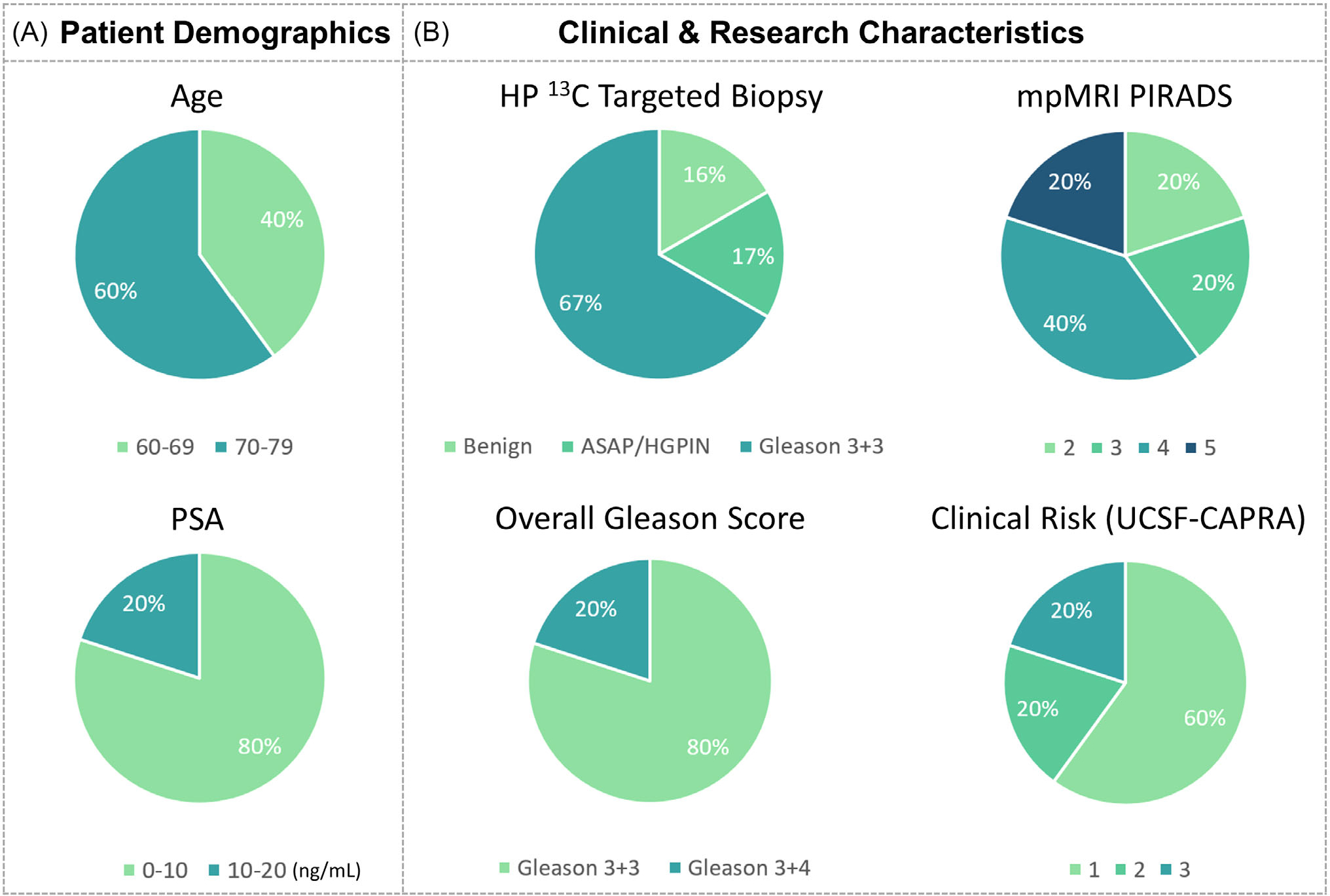
(A) Pie chart summarizing the serum PSA and age of this initial cohort. (B) Pie chart summarizing the pathologic characteristics of the HP 13C research biopsies, PIRADS scores of 1H mpMRI biopsies, overall Gleason score, and clinical risk (CAPRA score). The Gleason 3+4 findings in patient 3 in the left midgland (contralateral to the 13C target) was small-volume (1 out of 4 cores in the sextant, <5% involvement per core) and only detected by systematic biopsy. The kPL value per lesion was calculated from the maximum voxel. CAPRA, Cancer of the Prostate Risk Assessment; PSA, prostate-specific antigen.
TABLE 1.
Summary of the clinical characteristics and biopsy findings from 3 representative patients in this study. (The remaining 2 patients can be found in Supporting Information Table S1). All integrated 13C-1H mpMRI studies and the associated biopsies were safe and successful without adverse events. The median number of 13C targets was 1 (range: 1–2) per patient, measuring 1 cm (range:0.6–1.9) in diameter, with a median kPL of 0.0319 s−1 (range: 0.0198–0.0410). Low-grade cancer involvement was found in the cores corresponding to the 13C targets in 3 out of 5 patients. These findings were consistent with clinically low- to intermediate-risk disease
| Patient No. | Patient 1 | Patient 2 | Patient 3 |
|---|---|---|---|
| PSA | 9.6 | 8.4 | 4.2 |
| UCSF CAPRA score | 1 | 1 | 3 |
| Risk group | Low | Low | Intermediate |
| No. of 13C targets | 1 | 1 | 1 |
| kPL at 13C target(s−1) | 0.0378 (σ = 0.0050) | 0.0198 (σ = 0.0018) | 0.0325 (σ = NA) |
| No. of (+) cores/total cores in targeted + systematic biopsy | 4/17 | 5/18 | 10/18 |
| Overall grade | 3 + 3 | 3 + 3 | 3 + 4 |
| No. of (+) 13C cores/total 13C cores | 1/2 | 1/2 | 0/3 |
| 13C core grade | 3 + 3 | 3 + 3 | Benign |
1H, proton; 13C, carbon-13; kPL, pyruvate-to-lactate conversion rate; mp MRI, multiparametric MRI; NA, nonapplicable; PSA, prostate-specific antigen; UCSF CAPRA score, University of California, San Francisco–Cancer of the Prostate Risk Assessment score.
3.3 |. Correlation between kPL and histopathologic findings from biopsy
Overall, 1 patient (patient 3) had Gleason 3+4, and 4 patients (patient 1,2,4,5) had 3+3 disease (number of biopsy cores positive for cancer per patient, median: 4/17, range: 3/19–10/18), combining pathological findings from standard and 13C research-targeted biopsies (Table 1) (Figure 5) (Supporting Information Table S1). On a per patient basis, the maximum involvement of any core was 16%–52% (median: 16%). The cores sampled from 13C research targets were Gleason 3+3 in 4 patients (patient 1,2,4,5), with median 16% involvement (range: 1%–16%). One patient (patient 5) had atypical small acinar proliferation and high-grade prostatic intraepithelial neoplasia among the 13C cores in 1 target, and the 13C target in another patient (patient 3) contained benign prostate tissue.
FIGURE 5.
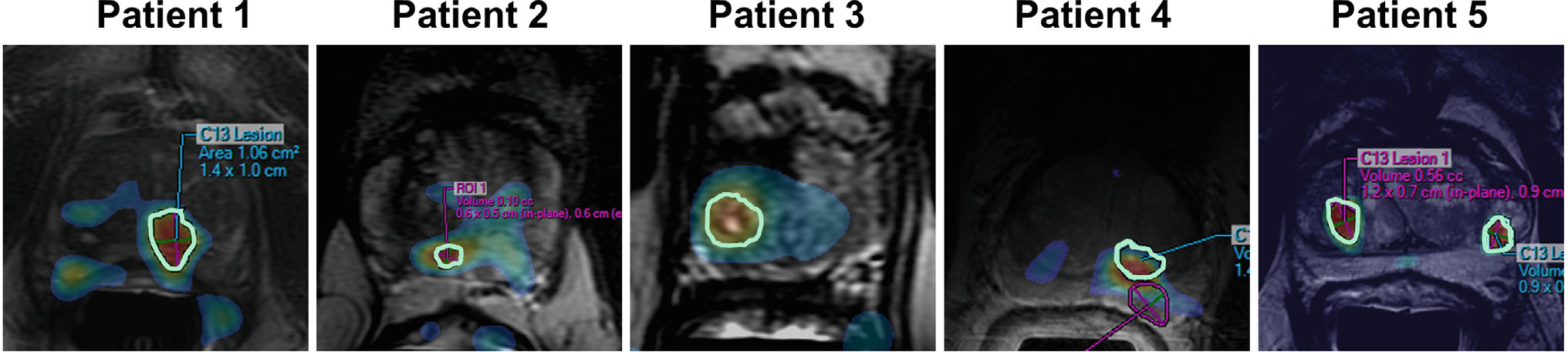
HP 13C-kPL targets from the 5 cases summarized in Table 1 and Supporting Information Table S1
Figure 6 illustrates a representative case (patient 1) who underwent fusion plus systematic biopsies with a 13C research target (kPL = 0.0378 s−1) at the left mid-apex peripheral zone and a 1H mpMRI target (PIRADS 4) at the right mid-base transition zone. Pathological diagnosis of the tissue sample from the 13C target was Gleason 3+3 cancer (16% involvement, 1/2 cores), whereas that from the 1H-MRI target was described in the histology report as “rare atypical glands.” Systematic biopsy found 3/12 cores with low volume 3+3 disease. Altogether, patient 1 had CAPRA score of 1, consistent with the clinically low-risk diagnosis.
FIGURE 6.
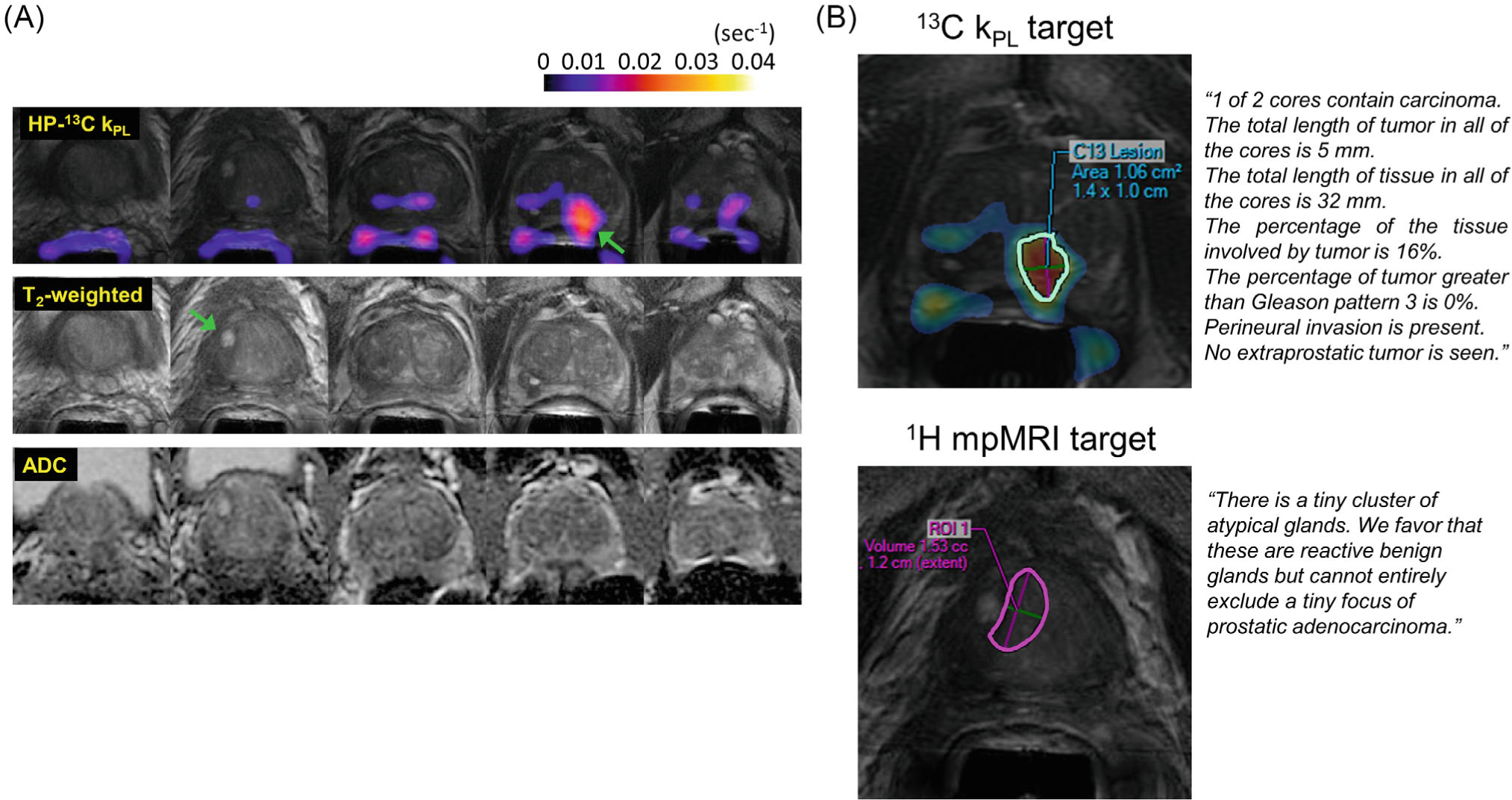
(A) Shows an example of an integrated HP-13C research and standard 1H mpMRI study (patient 1) including key multiparametric T2-weighted, diffusion, and kPL images identifying the biopsy target. One 1H target (PIRADS 4) was identified at right mid-base transition zone and one 13C research target (kPL = 0.0378 s−1) at left mid-apex peripheral zone, as indicated by the arrows. (B) 13C and 1H mpMRI biopsy targets as drawn by an experienced abdominal radiologist. Pathological diagnosis of the tissue sample from the 13C target was Gleason 3+3 cancer (16% involvement, 1/2 cores), whereas that from the 1H-MRI target was described in the histology report as “rare atypical glands”
Taken together with other clinical biomarkers, the biopsy findings in these 5 patients were consistent with clinically low- to intermediate-risk disease (summarized in Table 1), indicating that these patients are appropriate candidates for active surveillance. Four patients continued active surveillance after the study, whereas 1 patient later elected to undergo definitive treatment.
4 |. DISCUSSION
We investigated for the first time the safety and technical feasibility of integrating a rapid, 1-min hyperpolarized 13C-pyruvate MRI acquisition into standard-of-care 1H mpMRI exams for guiding TRUS fusion prostate biopsies. No HP 13C MRI- or biopsy-related adverse events were reported in this initial cohort, which is in agreement with the excellent safety record of 600+ hyperpolarized 13C MRI studies conducted worldwide thus far on patients and volunteers, as well as that of the active surveillance biopsy procedure at our institution and many other centers globally.29,30 These results support future investigations focused on determining whether HP 13C-1H mpMRI could benefit men who are either candidates for or undergoing active surveillance of prostate cancer.
Using the existing infrastructure, we incorporated the rapid HP 13C-pyruvate scan and the associated metabolic biomarker kPL into the routine prostate MRI workflow, for which the major components includes image postprocessing, radiology read with lesion targeting, MR-guided TRUS fusion biopsies, and finally pathology evaluation. The integrable nature of this approach proved effective to improve efficiency and minimize additional effort.
The new 13C targeting feature took advantage of the existing overlay and lesion outlining functions on a commercial, out-of-the-box prostate mpMRI processing, and targeting platform (Dynacad, Philips Invivo Corp.). This not only enabled easy deployment of the 13C targeting capabilities on any PACS workstation within our radiology network, without the burden of additional software installations or modifications, but also allowed directly exporting the 13C targets from the PACS/Dynacad (Philips Invivo Corp., Gainsville FL) to the fusion biopsy platform (UroNav, Philips Invivo Corp.) in the urological oncologists’ offices, along with the 1H mpMRI targets. We envision the same rationale and workflow are generalizable to other commercial targeting and fusion biopsy platforms31,32 in a vendor-independent fashion. Our approach is presumably suitable for either transrectal or transperineal biopsy techniques because most commercial platforms support both by default.33
The 13C research biopsy results were readily incorporated in the pathology report, and the pathological information of the 13C biopsy cores, including the Gleason scores, total percentage of tumor involvement, percentage of Gleason ≥4 pattern, and presence of adverse pathological features, were directly factored into the patients’ clinical risk calculations, such as the UCSF-CAPRA score utilized at our institution. This paves the way for easy incorporation into routine clinical practice in the future.
Five patients were enrolled to test this new approach in a pilot, technical feasibility study. Interestingly, 4 out of 6 13C-kPL targets did not correlate to a PIRADS lesion on 1H mpMRI on a per-lesion basis. The discrepancy is consistent with the knowledge that HP 13C MRI offers unique, complementary information based on prostate tumor metabolism in addition to the anatomical and functional features provided by 1H mpMRI. It also highlights the need to investigate, in a larger cohort, whether a HP 13C-1H mpMRI protocol may overcome the known limitation in sensitivity with conventional prostate MRI for detection of occult but clinically significant tumors in the surveillance setting.34
Our study design substituted systematic biopsy cores with 13C-targeted biopsy cores in the same sextant. The rationale was to reduce oversampling bias as a confounder when comparing the diagnostic accuracy between men who received fusion biopsies, including the 13C targets, versus those who only received standard-of-care targeted and systematic biopsies. Our approach was consistent with that taken by the Active Surveillance Magnetic Resonance Imaging (ASIST) trial,35 also designed to evaluate the utility of mpMRI-guided biopsy. An additional benefit was to fulfill the technique development aim for 13C-guided fusion biopsy without introducing additional biopsy-related morbidity to the men who participated in this technical feasibility study.
Distinct from standard mpMRI series, the 13C-kPL series was labeled nondiagnostic on PACS. On Dynacad (Philips Invivo Corp.), the kPL target lesions were labeled as C13 lesion #, setting them apart from the 1H mpMRI targets, which were named lesion 1, lesion 2, etc. Using separate labels reduced equivocal nomenclature and improved communications between imaging researchers and the patients’ multidisciplinary care team by making the distinction between 13C research imaging results and associated target lesions from those of 1H mpMRI.
Whereas this study successfully demonstrated the safety and technical feasibility of guiding fusion prostate biopsies using the HP 13C MRI-derived kPL biomarker, a few limitations and challenges need to be acknowledged. First, a consensus in the field of prostate mpMRI is trending away from the use of endorectal coils. Similar to current conventional 1H MRI coils, new flexible 13C array coils are becoming commercially available to provide wider spatial coverage and higher SNR. Combining with the recent developments in 13C parallel imaging36 and denoising algorithms,37,38 these advancements will likely obviate the need of endorectal coils for future 13C-pyruvate prostate studies.
Second, the kPL cutoff value (0.02 s−1), representing the dichotomy between high-grade prostate adenocarcinoma and low-grade tumor/benign tissue, was derived from the histopathology of a high-risk cohort who underwent radical prostatectomy.17 This high-risk cohort likely possesses quite distinct underlying biology from the lower-risk, active surveillance population who may benefit the most from HP 13C MRI. Whether the same kPL cutoff value is appropriate for detecting occult high-grade disease in the surveillance population thus needs to be further explored and validated. Additionally, developing and testing a PIRADS-like grading schema for HP 13C would benefit future clinical research but will require a broader evaluation in larger patient populations, as well as input from research radiologists on the methods developed in this project.
Although replacing the systematic cores with 13C-kPL cores reduces oversampling bias, a potential downside would be the missed opportunity to determine whether the standard 12–14 core systematic TRUS biopsy would have also found the 3+3 disease as the 13C-guided biopsy in the same sextant on an individual-patient basis.
The primary factor affecting biopsy accuracy is the imperfect registration of MR-TRUS software fusion and mechanical deflection of the biopsy needles, which can similarly affect 1H mpMRI targeted biopsies.39 Therefore, biopsies are not considered a gold standard like postprostatectomy step-section histopathology is.
Finally, this technical feasibility study was neither designed nor powered to calculate the sensitivity/specificity of HP 13C MRI-guided biopsy, and it would not be appropriate to report the clinical utility or biological significance results given the limited data and lack of a gold standard. The clinical question of whether 13C MRI improves diagnostic accuracy of prostate cancer needs to be tested in a future larger-scale clinical trial. We believe such a trial is feasible given multiple NCI-designated cancer centers are now equipped with HP 13C MRI capabilities and are either already performing or planning to conduct 13C prostate cancer research.40,41 Assuming HP 13C MRI is proven to increase detection of clinically significant prostate cancer, whether this improvement ultimately translates into better disease-specific outcome will require long-term follow-up studies.
5 |. CONCLUSION
This technical development study demonstrated the feasibility of adding HP 13C-pyruvate MRI to guide TRUS fusion prostate biopsies. HP 13C MRI biomarkers were integrated into the diagnostic mpMRI workflow, complete with identification of 13C research targets and sampling of these targets in fusion biopsies. These initial results support future studies in a larger cohort of patients to evaluate the role of HP 13C MRI–guided targeted biopsy for improving prostate cancer risk stratification.
Supplementary Material
FIGURE S1. (A) An example case showing the comparison of kPL in the 13C targeted lesion versus segmented prostate outside of the lesion. Pathological diagnosis of the biopsy tissue was Gleason 3 + 3 tumor with 16% involvement. (B) kPL dichotomy between pathologist-defined low-grade prostate cancer (PCa) (Gleason ≤3 + 4), and high-grade prostate cancer (Gleason score≥4 + 3). *p = 0.034; **p = 0.0003. The recommended kPL threshold = 0.02(s−1) used in our study was corrected for the different MR sequence echo times between the EPSI acquisition in the cited reference versus the EPI in our study.24 Figure reproduced with permission17
TABLE S1. Summary of the clinical characteristics and biopsy findings from Patient 4 and 5
VIDEO S1. This video illustrates the protocol to create 13C-kPL/T2 fusion overlays. The new 13C biopsy targeting feature takes advantage of the existing overlay and lesion outlining functions on a commercial, out-of-the-box prostate mpMRI processing and targeting platform (Dynacad). This not only enabled easy deployment of the 13C biopsy targeting capabilities on any PACS workstation within our radiology network - without the burden of additional software installations or modifications, but also allows directly exporting the 13C targets from the PACS/Dynacad to the fusion biopsy platform (UroNav)
ACKNOWLEDGMENT
We would like to thank Priscilla Chan, Francesca De Las Alas, Romelyn Delos Santos, Evelyn Escobar, Mary Frost, Jasmine Hu, Yaewon Kim, Philip Lee, and Kimberly Okamoto for their assistance with the patient studies.
This work was supported by the National Institutes of Health (NIH) grants U01CA232320, U01EB026412, R01CA238379, and P41EB013598; and the American Cancer Society (ACS) grant 131715-RSG-18-005-01-CCE.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
REFERENCES
- 1.Mottet N, van den Bergh RCN, Briers E, et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate Cancer-2020 update. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2021;79:243–262. [DOI] [PubMed] [Google Scholar]
- 2.Hamdy FC, Donovan JL, Lane JA, et al. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med. 2016;375:1415–1424. [DOI] [PubMed] [Google Scholar]
- 3.Ploussard G, Renard-Penna R. MRI-guided active surveillance in prostate cancer: not yet ready for practice. Nat Rev Urol. 2021;18:77–78. [DOI] [PubMed] [Google Scholar]
- 4.Stavrinides V, Giganti F, Emberton M, Moore CM. MRI in active surveillance: a critical review. Prostate Cancer Prostatic Dis. 2019;22:5–15. [DOI] [PubMed] [Google Scholar]
- 5.Klotz L, Pond G, Loblaw A, et al. Randomized study of systematic biopsy versus magnetic resonance imaging and targeted and systematic biopsy in men on active surveillance (ASIST): 2-year postbiopsy follow-up. Eur Urol. 2020;77:311–317. [DOI] [PubMed] [Google Scholar]
- 6.Chesnut GT, Vertosick EA, Benfante N, et al. Role of changes in magnetic resonance imaging or clinical stage in evaluation of disease progression for men with prostate cancer on active surveillance. Eur Urol. 2020;77:501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu CE, Lonergan PE, Washington SL, et al. Multiparametric magnetic resonance imaging alone is insufficient to detect grade reclassification in active surveillance for prostate cancer. Eur Urol. 2020;78:515–517. [DOI] [PubMed] [Google Scholar]
- 8.Ma TM, Tosoian JJ, Schaeffer EM, et al. The role of multiparametric magnetic resonance imaging/ultrasound fusion biopsy in active surveillance. Eur Urol. 2017;71:174–180. [DOI] [PubMed] [Google Scholar]
- 9.Albers MJ, Bok R, Chen AP, et al. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008;68:8607–8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sriram R, Van Criekinge M, DeLos SJ, et al. Elevated tumor lactate and efflux in high-grade prostate cancer demonstrated by hyperpolarized (13)C magnetic resonance spectroscopy of prostate tissue slice cultures. Cancers (Basel). 2020;12:537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ardenkjaer-Larsen JH, Fridlund B, Gram A, et al. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100:10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golman K, in ‘t Zandt R, Thaning M. Real-time metabolic imaging. Proc Natl Acad Sci U S A. 2006;103:11270–11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen HY, Larson PEZ, Gordon JW, et al. Technique development of 3D dynamic CS-EPSI for hyperpolarized (13) C pyruvate MR molecular imaging of human prostate cancer. Magn Reson Med. 2018;80:2062–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon JW, Chen HY, Autry A, et al. Translation of Carbon-13 EPI for hyperpolarized MR molecular imaging of prostate and brain cancer patients. Magn Reson Med. 2019;81:2702–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen HY, Aggarwal R, Bok RA, et al. Hyperpolarized (13)C-pyruvate MRI detects real-time metabolic flux in prostate cancer metastases to bone and liver: a clinical feasibility study. Prostate Cancer Prostatic Dis. 2020;23:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granlund KL, Tee SS, Vargas HA, et al. Hyperpolarized MRI of human prostate cancer reveals increased lactate with tumor grade driven by monocarboxylate transporter 1. Cell Metab. 2020;31:105–114.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korn N, Larson PEZ, Chen H-Y, et al. The rate of hyperpolarized [1–13C] pyruvate to [1–13C] lactate conversion distinguishes high-grade prostate cancer from low-grade prostate cancer and normal peripheral zone tissue in patients. In Proceedings of the 26th Annual Meeting of ISMRM, Paris, France, 2018:0280. [Google Scholar]
- 18.Logan JK, Rais-Bahrami S, Turkbey B, et al. Current status of MRI and ultrasound fusion software platforms for guidance of prostate biopsies. BJU Int. 2014;114:641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson SJ, Kurhanewicz J, Vigneron DB, et al. Metabolic imaging of patients with prostate cancer using hyperpolarized [1-C-13]pyruvate. Sci Transl Med. 2013;5:198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran GN, Leapman MS, Nguyen HG, et al. Magnetic resonance imaging-ultrasound fusion biopsy during prostate cancer active surveillance. Eur Urol. 2017;72:275–281. [DOI] [PubMed] [Google Scholar]
- 21.Autry AW, Gordon JW, Chen HY, et al. Characterization of serial hyperpolarized (13)C metabolic imaging in patients with glioma. Neuroimage Clin. 2020;27:102323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon JW, Vigneron DB, Larson PE. Development of a symmetric echo planar imaging framework for clinical translation of rapid dynamic hyperpolarized (13) C imaging. Magn Reson Med. 2017;77:826–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson PEZ, Chen HY, Gordon JW, et al. Investigation of analysis methods for hyperpolarized 13C-pyruvate metabolic MRI in prostate cancer patients. NMR Biomed. 2018;31:e3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turkbey B, Rosenkrantz AB, Haider MA, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol. 2019;76:340–351. [DOI] [PubMed] [Google Scholar]
- 25.Chen HY, Gordon JW, Bok RA, et al. Pulse sequence considerations for quantification of pyruvate-to-lactate conversion kPL in hyperpolarized 13C imaging. NMR Biomed. 2019;32:e4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welty CJ, Cowan JE, Nguyen H, et al. Extended followup and risk factors for disease reclassification in a large active surveillance cohort for localized prostate cancer. J Urol. 2015;193:807–811. [DOI] [PubMed] [Google Scholar]
- 27.Klein EA, Cooperberg MR, Magi-Galluzzi C, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66:550–560. [DOI] [PubMed] [Google Scholar]
- 28.Cooperberg MR, Pasta DJ, Elkin EP, et al. The University of California, San Francisco cancer of the prostate risk assessment score: a straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol. 2005;173:1938–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooperberg MR, Zheng Y, Faino AV, et al. Tailoring intensity of active surveillance for low-risk prostate cancer based on individualized prediction of risk stability. JAMA Oncol. 2020;6:e203187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeb S, van den Heuvel S, Zhu X, Bangma CH, Schroder FH, Roobol MJ. Infectious complications and hospital admissions after prostate biopsy in a European randomized trial. Eur Urol. 2012;61:1110–1114. [DOI] [PubMed] [Google Scholar]
- 31.Baco E, Rud E, Eri LM, et al. A randomized controlled trial to assess and compare the outcomes of two-core prostate biopsy guided by fused magnetic resonance and Transrectal ultrasound images and traditional 12-core systematic biopsy. Eur Urol. 2016;69:149–156. [DOI] [PubMed] [Google Scholar]
- 32.Filson CP, Natarajan S, Margolis DJ, et al. Prostate cancer detection with magnetic resonance-ultrasound fusion biopsy: the role of systematic and targeted biopsies. Cancer. 2016;122:884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang J, Yan H, Li J, Wang X, Chen H, Zheng X. Transperineal versus transrectal prostate biopsy in the diagnosis of prostate cancer: a systematic review and meta-analysis. World J Surg Oncol. 2019;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson DC, Raman SS, Mirak SA, et al. Detection of individual prostate cancer foci via multiparametric magnetic resonance imaging. Eur Urol. 2019;75:712–720. [DOI] [PubMed] [Google Scholar]
- 35.Klotz L, Loblaw A, Sugar L, et al. Active surveillance magnetic resonance imaging study (ASIST): results of a randomized multicenter prospective trial. Eur Urol. 2019;75:300–309. [DOI] [PubMed] [Google Scholar]
- 36.Feng Y, Gordon JW, Shin PJ, et al. Development and testing of hyperpolarized (13)C MR calibrationless parallel imaging. J Magn Reson. 2016;262:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brender JR, Kishimoto S, Merkle H, et al. Dynamic imaging of glucose and lactate metabolism by 13 C-MRS without hyperpolarization. Sci Rep. 2019;9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen HY, Autry AW, Brender JR, et al. Tensor image enhancement and optimal multichannel receiver combination analyses for human hyperpolarized (13) C MRSI. Magn Reson Med. 2020;84:3351–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cool DW, Zhang X, Romagnoli C, Izawa JI, Romano WM, Fenster A. Evaluation of MRI-TRUS fusion versus cognitive registration accuracy for MRI-targeted, TRUS-guided prostate biopsy. AJR Am J Roentgenol. 2015;204:83–91. [DOI] [PubMed] [Google Scholar]
- 40.Kurhanewicz J, Vigneron DB, Ardenkjaer-Larsen JH, et al. Hyperpolarized (13)C MRI: path to clinical translation in oncology. Neoplasia. 2019;21:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brindle KM, Keshari KR. Editorial commentary for the special issue: technological developments in hyperpolarized (13)C imaging-toward a deeper understanding of tumor metabolism in vivo. MAGMA. 2021;34:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. (A) An example case showing the comparison of kPL in the 13C targeted lesion versus segmented prostate outside of the lesion. Pathological diagnosis of the biopsy tissue was Gleason 3 + 3 tumor with 16% involvement. (B) kPL dichotomy between pathologist-defined low-grade prostate cancer (PCa) (Gleason ≤3 + 4), and high-grade prostate cancer (Gleason score≥4 + 3). *p = 0.034; **p = 0.0003. The recommended kPL threshold = 0.02(s−1) used in our study was corrected for the different MR sequence echo times between the EPSI acquisition in the cited reference versus the EPI in our study.24 Figure reproduced with permission17
TABLE S1. Summary of the clinical characteristics and biopsy findings from Patient 4 and 5
VIDEO S1. This video illustrates the protocol to create 13C-kPL/T2 fusion overlays. The new 13C biopsy targeting feature takes advantage of the existing overlay and lesion outlining functions on a commercial, out-of-the-box prostate mpMRI processing and targeting platform (Dynacad). This not only enabled easy deployment of the 13C biopsy targeting capabilities on any PACS workstation within our radiology network - without the burden of additional software installations or modifications, but also allows directly exporting the 13C targets from the PACS/Dynacad to the fusion biopsy platform (UroNav)


