Abstract
Brachyspira (Serpulina) hyodysenteriae induces a mucohemorrhagic diarrheal disease in pigs. The production of a beta-hemolysin has been considered a major virulence attribute of this organism. Previous reports have failed to correlate a specific cloned gene sequence with a purified beta-hemolytic protein sequence. Thus, questions still remain concerning the structural gene sequence of the hemolysin. To answer this question unequivocally, the beta-hemolytic toxin was purified from extracts of log-phase spirochetes, and the N-terminal amino acid sequence was determined (K-D-V-V-A-N-Q-L-N-I-S-D-K) and compared with the translated sequences of previously cloned genes, tlyA to tlyC. The lack of homology between tlyA to tlyC translated sequences and the purified beta-hemolytic toxin sequence resulted in the study that is reported here. A degenerate probe was designed based on the N-terminal amino acid sequence of the purified beta-hemolysin and used to screen a B. hyodysenteriae genomic library. Three overlapping clones were identified, and one was sequenced to reveal an open reading frame coding for a putative 8.93-kDa polypeptide containing the N-terminal sequence of the purified beta-hemolysin. To distinguish this gene from the tlyA to tlyC genes, it has been designated hlyA. A hemolysis-negative Escherichia coli strains containing hlyA was beta-hemolytic on blood agar media. Also, the hemolytic activity of the recombinant protein had identical protease and lipase sensitivities and electrophoretic mobility to those of native B. hyodysenteriae beta-hemolysin. Based on sequence analysis, the translated protein had a pI of 4.3, an α-helical structure, and a phosphopantetheine binding motif. Hybridization analysis of genomic DNA indicated that the hlyA gene was present in B. hyodysenteriae and B. intermedia but was not detected in B. innocens, B. pilosicoli, or B. murdochii under high-stringency conditions. The location of hlyA on the chromosomal map was distinct from the locations of tlyA, tlyB, and tlyC.
Brachyspira (Serpulina) hyodysenteriae, the etiologic agent of swine dysentery, induces a mucohemorrhagic diarrheal disease in susceptible pigs and a severe cecitis in mice (7, 11). Virulence attributes associated with B. hyodysenteriae include the production of a beta-hemolysin, the presence of a biologically active lipooligosaccharide, motility, and the presence of an NADH oxidase (5, 12, 19, 20, 22, 23). Beta-hemolytic activity has been one of the major phenotypic characteristics used to distinguish B. hyodysenteriae from avirulent species of Brachyspira. Numerous methods have been reported for hemolysin production and purification, most involving traditional protein purification techniques (13–15, 23). These studies have been at variance in their reports of the apparent molecular mass of the isolated hemolytic factor, ranging from 19 to 74 kDa. The beta-hemolysin has been shown to be physicochemically similar to streptolysin S; it is oxygen stable, heat labile, and active over a wide pH range, requires an appropriate carrier for its isolation (usually RNA-core), and is sensitive to both protease and lipase digestion (14, 23).
The biologic activity of the beta-hemolysin recovered from B. hyodysenteriae has been studied primarily in vitro. Early work indicated that the binding of the hemolysin to erythrocytes was temperature independent while lytic activity required physiologic temperatures and that erythrocyte swelling occurred prior to lysis (22). While proteolytic or phospholipase-like activity has not been associated with the beta-hemolysin of B. hyodysenteriae, hemoglobin release was detected prior to cell swelling, indicating a cytolytic mechanism involving membrane perturbation and leakage (22, 23). Studies with osmoprotectants indicated that a pore with a diameter of 1.0 to 1.1 nm was formed by the beta-hemolysin (10). Additional studies from this laboratory have indicated that purified beta-hemolysin affects the integrity of epithelial cell monolayers and induces colonic epithelial cell damage similar to that caused by infection with B. hyodysenteriae (9, 26).
Previously, three separate genes from B. hyodysenteriae have been identified and associated with beta-hemolysis based on their ability to induce a hemolytic phenotype in Escherichia coli (7, 11). These three genes, referred to as tlyA, tlyB, and tlyC, encode gene products with apparent molecular masses of 26.9, 93.3, and 30.8 kDa, respectively. Furthermore, it was demonstrated that tlyA was detected in the pathogenic B. hyodysenteriae but not the nonpathogenic B. innocens. The tlyB gene product was reported to have homology to Clp proteins, a class of intracellular proteins that display proteolytic activities as well as being regulators of proteolysis. None of these three genes, however, have been linked directly by protein sequencing to a B. hyodysenteriae-specific gene product that displays hemolytic activity or to the native beta-hemolysin. Likewise, none of the recombinant products synthesized from the cloned genes have been shown to possess the biochemical properties attributed to the B. hyodysenteriae beta-hemolysin (13, 14, 22, 23), with the possible exception that E. coli harboring tly containing plasmids have hemolytic activity (18, 25). Recent studies identifying the cryptic E. coli hemolysin SheA (3) raise the possibility that tlyA, tlyB, and tlyC code for proteins that induce SheA or other cryptic E. coli hemolysins. In fact, in an earlier study, a cloned Salmonella gene, slyA, was originally thought to be a hemolysin gene (16) but was later identified as a transcriptional regulator that activated the expression of sheA in E. coli K-12 (3, 17, 21).
To correlate the beta-hemolysin protein sequence with one of the tly genes, the native protein was purified and the N-terminal amino acid sequence was obtained. Unexpectedly, this sequence was not present in any of the three translated tly genes identified previously. Further studies resulted in the cloning of the gene, hlyA, that encoded the purified beta-hemolysin (i.e., HlyA), and these studies revealed a DNA sequence with no homology to tlyA, tlyB, or tlyC. The coding sequence of hlyA is much smaller than those of the previously reported tly genes, and this is directly correlated with the biochemical analyses of the native beta-hemolysin.
MATERIALS AND METHODS
Bacteria.
B. hyodysenteriae strain B204, serotype 2, was grown as previously described (5). Spirochetes were grown in 3-liter volumes to a concentration of approximately 109 organisms per ml as determined with a Petroff-Hauser bacterial counting chamber. Active motility of greater than 75% of the organisms was desired and interpreted as an indication of a culture within the log phase of growth. E. coli strains were routinely grown in Luria-Bertani (LB) broth or on LB agar medium. Phage plates for LE392 (6) consisted of standard LB agar base supplemented with 2.5 mM CaCl2 with LB soft agar overlays. E. coli SOLR [e14-(mcrA) Δ(mcrCB-hsdSMR-mrr)171 sbcC recB recJ uvrC umuC::Tn5(Kanr) lac gyrA96 relA1 thi-1 endA1 λR [F′ proAB laclq ZΔM15] Su−] was the Lambda ZAP II recipient for plasmid excision (Stratagene, La Jolla, Calif.). E. coli BL21(λDE3) is a λ DE3 lysogen which carries T7 RNA polymerase under lac promoter control.
Hemolysin preparation and purification.
Crude hemolysin was prepared by a modification of a method described previously (13). A 3-liter volume of B. hyodysenteriae culture was centrifuged at 10,000 × g at 4°C for 20 min. Bacteria were suspended in beta-hemolysin extraction buffer (phosphate-buffered saline [140 mM NaCl, 8 mM Na2HPO4, 1.5 mM KH2PO4, 2.7 mM KCl; pH 7.2], 0.05% RNA-core [Sigma Chemical Co., St. Louis, Mo.], 1 mM glucose, 1 mM MgSO4) at 1/20 of the original culture volume and held at 37°C for 30 min with sufficient agitation to maintain suspension. Cells were pelleted by centrifugation, the supernatant was retained and frozen, and the extraction was repeated three to five times depending on the extent of cell vigor as assessed by observing motility. Supernatants were pooled, vacuum filtered through a 0.22-μm-pore-size filter (Gelman Sciences, Ann Arbor, Mich.), and stored at −70°C until further purification. Throughout all manipulations, beta-hemolysin-containing solutions were maintained at 4°C or less and assayed periodically for hemolytic activity. The beta-hemolysin-containing supernatant was reduced in volume and desalted by repeated concentration and reconstitution with sterile deionized water, utilizing a forced-flow ultrafiltration device with an approximate molecular mass cutoff filter of 3 kDa (Filtron Technologies, Northborough, Mass.). This concentrated, crude hemolysin was loaded onto preparative, nondenaturing 12% polyacrylamide slab gels and electrophoresed until the dye front reached the bottom of the gel. The portion of the gel that contained the hemolytic activity was identified by placing a 2-mm-wide slice (top-to-bottom) of the gel onto blood agar and incubating it at 37°C for 20 min. The polyacrylamide gel was then sliced (side-to-side, 3 to 5 mm wide) to excise the portion of the gel containing the hemolytic activity. The hemolysin was electroeluted from the acrylamide slices into Tris-glycine buffer (37.6 mM Tris-HCl, 50 mM glycine [pH 7.5]). The supernatant was then reduced in volume and desalted by ultrafiltration, as described above. The process was repeated, so that beta-hemolysin preparations were electrophoresed and electroeluted twice. Preparations were analyzed by size exclusion capillary electrophoresis, and the N-terminal amino acid sequence analysis was determined by the Edman reaction. The amino acid sequence was then used to construct a degenerate oligonucleotide probe for subsequent hybridization and gene identification. The protease and lipase sensitivity of hemolytic preparations was assessed as previously described (23).
Library construction and screening.
The B. hyodysenteriae genomic library was constructed in Lambda ZAP II using 5- to 10-kb randomly sheared DNA fragments whose ends were polished by DNA polymerase. EcoRI linkers were added using DNA ligase, and the fragments were cloned into Lambda ZAP II EcoRI-digested, dephosphorylated DNA. Following packaging, the library was amplified using standard techniques (24).
To screen the library, the phage were grown at a density of 500 plaques per 85-mm-diameter petri dish. Nitrocellulose disks were used to lift the plaques, and the DNA was denatured with 0.1 M NaOH and baked at 80°C. The lifts were hybridized under standard conditions (24) overnight at 47°C. Positive plaques were purified by standard techniques and rescreened by the above method.
DNA manipulations.
Plasmids containing cloned B. hyodysenteriae chromosomal DNA fragments were excised in vitro from the corresponding recombinant Lambda ZAP II plaques with helper phage R408 and introduced into E. coli SOLR as specified by the manufacturer (Stratagene). Restriction maps of each cloned fragment were constructed by standard techniques.
DNA sequencing and analysis.
Prior to DNA sequence analysis, the region of the three recombinant clones reactive with the degenerate oligonucleotide probe was first determined. This was accomplished by DNA-DNA hybridization using plasmid DNAs digested with EcoRI-EcoRV-HindIII or with ClaI-HindIII. Following separation of the fragments by agarose gel electrophoresis, blotting to a nylon membrane, and hybridization with the degenerate oligonucleotide probe, the region containing the amino-terminal portion of the hemolysin was identified and the sequence of this region was determined in plasmid pISM1236 (see Fig. 2). The reverse primer (5′-CAGGAAACAGCTATGACC-3′) and two custom primers designated TV2 (5′-CAAAATAATAGTCGCCCTCAAC-3′) and TV7 (5′-TAGCTGTTGACGGCGGAATG-3′) were used to obtain complete overlapping sequence. The DNA sequence data were assembled and analyzed for open reading frames and sequence homologies using MacVector software, version 5.0.2. The sequence was also analyzed for sequence motifs using PROSITE software (http://www.genome.ad.jp/SIT/MOTIF.html) (1).
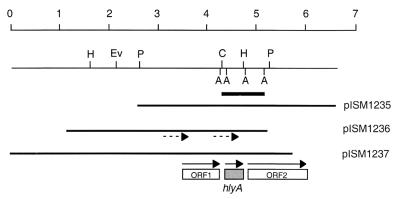
FIG. 2.
Physical maps of hlyA recombinant clones from the Lambda ZAP II genomic library. The plasmid designations are also given. The scale at the top is in kilobases. The sequences used for probing chromosomal DNAs are indicated by the heavy bar. The boxes represent open reading frames; the gray box is hlyA. The solid arrows indicate the direction of transcription. The dotted arrows indicate positions of the sequencing primers TV2 (left) and TV7 (right). Only AseI sites relevant to the hybridization probe are shown. ORF, open reading frame; A, AseI; C, ClaI; Ev, EcoRV; H, HindIII; P, PstI. Not shown is the EcoRI site in the vector multiple-cloning site on the right side of all clones.
Chromosomal mapping.
Restriction endonuclease digestion, pulsed-field gel electrophoresis, and Southern blot analysis of DNA prepared in agarose beads were done as described previously (27). The blots were hybridized and washed at 60°C.
DNA hybridization.
Hybridization was performed to determine if other Brachyspira species contained homologs of the beta-hemolysin gene identified in this study in B. hyodysenteriae. Genomic DNAs of six different serpulinal species, B. innocens strain B256, an unnamed pathogenic chicken isolate, strain C-1 (proposed name, B. alvinipulli), B. intermedia strain PWS/A (an isolate from a pig with nondysenteric colitis), B. murdochii strain 155-20 (a nonpathogenic isolate from a pig), B. pilosicoli strain P43/678 (an end-on-attaching spirochete isolated from a pig with colitis), and B. hyodysenteriae strain B204, were obtained from Neil Jensen (National Animal Disease Center, Ames, Iowa). All of the strains analyzed, except for the strongly beta-hemolytic B. hyodysenteriae B204, are moderately to weakly beta-hemolytic. DNA from each of these spirochetal species was digested individually with four different restriction enzymes: MboI, HindIII, AseI, and SspI. Portions (2 μg) of DNA from each reaction mixture were electrophoresed through 1% agarose gels and blotted to nylon membranes (Hybond-N; Amersham, Arlington Heights, Ill.). DNA was fixed to nylon membranes by UV cross-linking using a Stratagene 1800 UV cross-linker. A ClaI-EcoRI 0.95-kb fragment from plasmid pISM1236 (see Fig. 1) containing the entire open reading frame of the beta-hemolysin gene was used in these hybridization studies. This fragment was isolated by agarose gel electrophoresis following plasmid digestion and was purified using a Qiagen (Chatsworth, Calif.) column. The probe was labeled with 32P by random-primer labeling using the Klenow fragment of DNA polymerase (Amersham). The membrane blots were hybridized overnight and washed twice at 65°C. Autoradiographs were exposed for 9 h at −80°C.
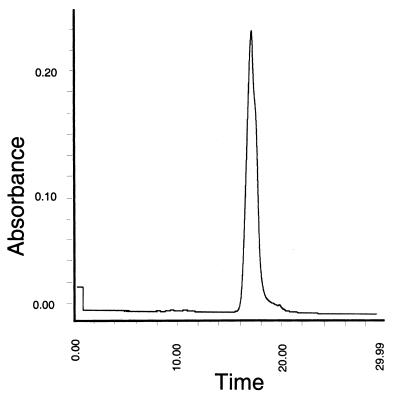
FIG. 1.
Chromatogram of purified beta-hemolysin. A sample (approximately 10 nl) of the purified beta-hemolysin (10 HU/μl) was subjected to size exclusion capillary electrophoresis. A single protein peak was detected by UV spectral analysis at 214 nm.
Nucleotide sequence accession number.
The nucleotide sequence of the B. hyodysenteriae hlyA gene has been assigned GenBank accession number U94886.
RESULTS AND DISCUSSION
Hemolysin purification and amino acid sequencing.
For the purposes of biological and biochemical characterization, the crude preparations of beta-hemolysin obtained from B. hyodysenteriae B204 cells (13) were purified by ultrafiltration concentration followed by two successive separations on native polyacrylamide gels and electroelution. Hemolytic activity of the purified fractions was maintained throughout the purification process (Table 1). Analysis of the final hemolytically active fraction by size exclusion capillary electrophoresis revealed a single polypeptide peak at 214 nm with an apparent molecular mass between 19 and 21 kDa (Fig. 1). This was consistent with the results of Kent et al. (13), suggesting that this gene product was smaller than the tlyA to tlyC gene products. The purified hemolysin was then analyzed for its N-terminal amino acid sequence. The results indicated that a single polypeptide had been obtained with an N-terminal sequence of K-D-V-V-A-N-Q-L-N-I-S-D-K. No similar sequences are present in the predicted products of tlyA to tlyC (25). Thus, a different gene product was responsible for beta-hemolytic activity of B. hyodysenteriae.
TABLE 1.
Purification of the B. hyodysenteriae beta-hemolysin
| Fraction | Hemolytic activitya
|
% Protein | HU per mg of protein | |
|---|---|---|---|---|
| (HU/μl) | HU per mg (dry wt) | |||
| Unconcentrated crude | <1 | 20 | 1 | 2,000 |
| Concentrated crude | 11 | 2,200 | 28.4 | 11,957 |
| CEb | 17 | 3,400 | 11 | 30,910 |
HU, hemolytic units.
CE, capillary electrophoresis fraction (purified).
Cloning of the B. hyodysenteriae hemolysin gene.
To identify the specific gene encoding the beta-hemolysin, a degenerate oligonucleotide probe [5′-AAAGATGT(A/T)GT(A/T)GC(A/T)AATCA-3′] was designed from the N-terminal sequence of the purified beta-hemolysin for screening the genomic library. When tested by DNA hybridization, the probe recognized a single HindIII chromosomal fragment (data not shown). Screening of the B. hyodysenteriae genomic library resulted in eight positive plaques representing three independent clones. Following excision of the DNA into pBluescript plasmids, the three cloned fragments were restriction mapped (Fig. 2). The three plasmids were designated pISM1235 to pISM1237. Each of the cloned fragments contained a ClaI-HindIII fragment of approximately 0.45 kb, which reacted positively with the degenerate probe (data not shown). Colonies of the E. coli host strain SOLR were converted from a nonhemolytic to a hemolytic phenotype when carrying these plasmids (Fig. 3). Control strains containing either pBluescript SK(−) (Stratagene) or a pBluescript SK(−) derivative with an unrelated 4-kb insert of B. hyodysenteriae chromosomal DNA were hemolytically negative on blood agar. They were also negative by DNA hybridization when probed with the degenerate oligonucleotide (data not shown).
FIG. 3.
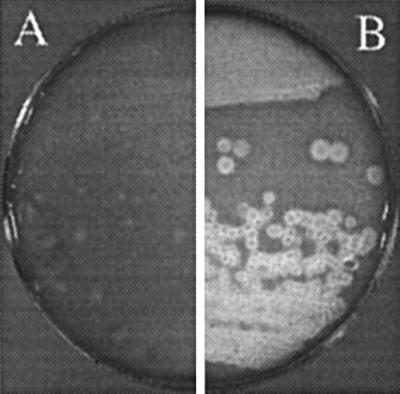
Hemolytic activity of recombinant E. coli SOLR(pISM1236). The strains were streaked to obtain single colonies onto bovine blood agar plates and grown for 48 h at 37°C. (A) SOLR (negative control); (B) SOLR(pISM1236).
Nucleotide sequence analysis of plasmid pISM1236 yielded an ORF with the sequence shown in Fig. 4. The amino acid sequence derived from the purified B. hyodysenteriae hemolysin was found 7 amino acids downstream (Fig. 4). The protein sequence data were unambiguous; there was no suggestion that proteolytic degradation had occurred in the purified protein sample used for N-terminal sequencing or that there was more than a single protein in the sample. Therefore, either the protein sequence MALIDEI functions as an unusual signal sequence in B. hyodysenteriae and the protein undergoes a cleavage event between I7 and K8 or translation of this gene begins 21 bp downstream of an AUG codon. To differentiate between this gene and the genes identified by ter Huurne et al. (25), designated tlyA to tlyC, this gene has been designated hlyA in keeping with conventional nomenclature.
FIG. 4.
Nucleotide sequence and deduced amino acid sequence of the B. hyodysenteriae hlyA gene. The structural gene begins at nucleotide 78 at the underlined ATG codon and ends at nucleotide 312 at the bold TAA codon. The numbers refer to the DNA sequence. A potential ribosomal binding site (RBS) and Pribnow box are indicated. The singly underlined amino sequence represents the phosphopantetheine binding motif. The doubly underlined amino acid sequence is the N-terminal sequence derived from purified beta-hemolysin.
Sequence analysis of hlyA.
The hlyA predicted gene product had a molecular mass of 8.93 kDa, and it appeared to contain an unusual 7-amino-acid signal sequence that was cleaved from the mature secreted protein as determined by protein sequencing (Fig. 4). The translated product had a pI of 4.3 and a predicted rod-shaped or linear conformation arising from an α-helical structure (data not shown). The linear nature of the hlyA gene product may explain the disagreement between the predicted molecular mass and that estimated by size exclusion capillary electrophoresis in this study (19 to 21 kDa); i.e., linear proteins tend to appear larger than globular proteins of similar size by size exclusion chromatography. Discrepancies in the previously reported molecular masses of the beta-hemolysin of B. hyodysenteriae may also be attributable to the predicted rod shape of this molecule (13), or, depending on the ionic strength of the buffer and the protein concentration, multimeric forms of the protein could also explain these differences. The translation product contained a conserved phosphopantetheine binding site (amino acid residues 32 through 47), suggesting that there is posttranslational modification of the beta-hemolysin (Fig. 4). Posttranslational modification (e.g., fatty acid binding) would also be consistent with the loss of hemolytic activity following lipase treatment (data not shown). It is not known if hlyA is part of an operon or is expressed independently of nearby genes.
DNA hybridization analysis.
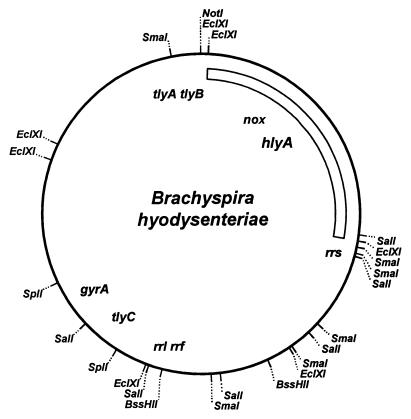
hlyA was localized on the 3.2-Mbp physical and genetic map of B. hyodysenteriae B78T by Southern blot hybridization. This gene is located on the EclA, SalA, and SmaB fragments (27), thus placing it in a region poorly defined by mapped restriction sites. Figure 5 shows the region of the chromosome represented by these fragments. The nox gene, which encodes NADH oxidase, is found in the same region of the genome. hlyA is not adjacent to tlyA, tlyB, or tlyC, three other genes reported to encode hemolytic activity in recombinant E. coli (25).
FIG. 5.
Localization of the hlyA gene on the combined physical and genetic map of B. hyodysenteriae B78T. The physical map of B. hyodysenteriae B78T is shown with landmark restriction sites. The locations of several previously mapped genes are shown. The bar indicates the region containing the hlyA gene.
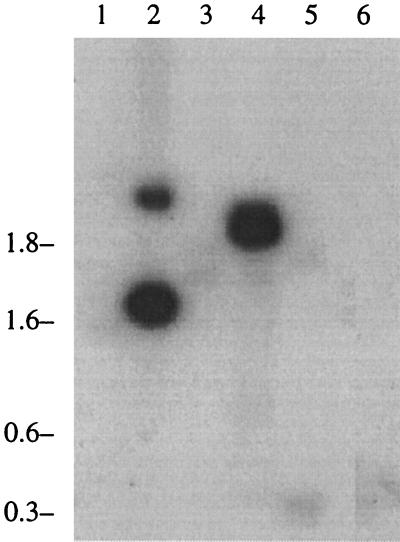
Genomic DNA from six different species of Brachyspira was digested with one of four restriction enzymes, electrophoresed, and hybridized with a hlyA-specific probe derived from the gene sequence. Results from all four enzymes were similar. A Southern blot experiment performed with one of those enzymes, AseI, is shown in Fig. 6. B. intermedia strain PWS/A (lane 4), which has an intermediate beta-hemolytic pattern, was the only species other than B. hyodysenteriae (lane 2) to show significant binding of the probe. This organism is representative of what was formerly known as the “intermediate types” of B. hyodysenteriae. The pathogenicity of these isolates, now collectively categorized as B. intermedia, has been equivocal in the hands of some researchers; other reports have indicated that these strains are clearly pathogenic but, compared to B. hyodysenteriae, induce a different and milder clinical disease called porcine spirochetal colitis in experimentally infected swine (2, 4, 8). In the present studies, there was a positive association between the presence of the hlyA beta-hemolysin gene and pathogenicity in porcine Brachyspira isolates.
FIG. 6.
Hybridization analysis of chromosomal DNA from different Brachyspira species. Chromosomal DNA from different Brachyspira species was digested with AseI, and the fragments were separated by agarose electrophoresis and transferred to a nylon membrane. Hybridization was performed using a 0.95-kb ClaI-EcoRI fragment from plasmid pISM1236 as described in Materials and Methods and shown in Fig. 2. Lanes: 1, B. pilosicoli P43/678; 2, B. hyodysenteriae B204; 3, B. murdochii 155-20; 4, B. intermedia PWS/A; 5, strain C-1; 6, B. innocens B256. Molecular masses (in kilobases) are given on the left.
Analysis of recombinant HlyA protein.
E. coli strains containing plasmids pISM1235 to pISM1237 were hemolytic on blood agar (Fig. 3). HlyA does not appear to be readily released from recombinant E. coli, however, because the appearance of hemolytic colonies occurred only after 48 h of incubation, presumably as cells died. In comparison to the release of the native hemolysin from B. hyodysenteriae, it was necessary to treat recombinant E. coli with lysozyme, EDTA, and sonication to obtain sufficient quantities of the hemolysin for analysis (data not shown). The inability of recombinant E. coli strains to efficiently secrete the beta-hemolysin may relate to the unusual N-terminal sequence of HlyA, which is not present in the mature protein but is present in the coding sequence. It is likely that this 7-amino-acid sequence at the N-terminal end of HlyA serves as a signal sequence in B. hyodysenteriae but does not facilitate secretion by E. coli. By contrast, the native beta-hemolysin was recovered in supernatant fluid following incubation of log-phase B. hyodysenteriae in extraction buffer and removal of the bacterial cells by centrifugation. There was no enzymatic or mechanical disruption of the spirochetes required to obtain the hemolysin, suggesting that the activity was secreted or released from viable cells. The native and recombinant hemolysin shared additional physicochemical characteristics in that both were resistant to inactivation by trypsin digestion but were inactivated by nonspecific protease or lipase treatment as previously described (23). In combination, these data and observations suggest that the hemolytic activity of the recombinant E. coli was the direct result of the hlyA gene expression and not the induction of a cryptic hemolysin from the host E. coli strain.
Relationship of hlyA to tlyA, tlyB, and tlyC.
Previous investigators have reported the cloning and sequencing of three putative hemolysin genes from B. hyodysenteriae (18, 25). Identification of these genes depended solely on induction of a hemolytic phenotype in E. coli. Given the recent report of a cryptic hemolysin in E. coli K-12 (i.e., the sheA product) controlled by DNA binding proteins (3), questions should be raised regarding the nature of the tly Brachyspira gene products. The tlyA to tlyC gene products could represent regulatory proteins that upregulate sheA or other unknown cryptic hemolysins in E. coli. For instance, tlyB has homology to ClpB, a class of regulatory proteins and proteases. In addition, there has not been a report of a direct link between the purified recombinant hemolytic activity from tly-containing clones and a specific protein in B. hyodysenteriae. In contrast, our studies correlate N-terminal amino acid sequence from purified, active beta-hemolysin with the cloned gene DNA sequence. Additionally, hlyA does not map near the tly genes (Fig. 5), nor does it reside within the lysogenic phage of B. hyodysenteriae (T. B. Stanton, personal communication). Whether the previously described tly genes (18, 25) code for other hemolysins or serve as regulatory proteins in B. hyodysenteriae can be answered only by more defined biochemical and genetic studies. The biologic activity of purified HlyA, however, has been shown to induce murine colonic lesions similar to those caused by B. hyodysenteriae and to disrupt the integrity of epithelial cell monolayers (9, 26).
In summary, a beta-hemolysin was purified from B. hyodysenteriae and an unambiguous N-terminal amino acid sequence was obtained. Construction of an oligonucleotide probe from that amino acid sequence facilitated identification, cloning, and sequencing of the gene, hlyA, encoding the protein. Analysis of that sequence provided information about the physicochemical properties of the hlyA gene product. There was no relationship between hlyA and three previously identified B. hyodysenteriae genes (i.e., tlyA to tlyC), suggesting that hlyA was unique. Based on sequence analysis, the hlyA gene and gene product did have homology to plant and bacterial acyl carrier proteins. DNA-DNA hybridization performed on six different species of Brachyspira indicated that at least one other species, B. intermedia, possessed the hlyA gene. This study provides the first direct linkage between a purified hemolytic protein from B. hyodysenteriae and its cognate gene sequence.
ACKNOWLEDGMENTS
We acknowledge Thad Stanton, National Animal Disease Center, USDA, Ames, Iowa, for supplying the Lambda ZAP II library and oligonucleotide and for useful discussions on cloning of the hemolysin gene. We also thank Mary Jo Schmerr, National Animal Disease Center, for helpful discussions on protein purification and for assistance in capillary electrophoresis. We also thank Tina VanDyk and Jill Randolph for technical assistance.
D.L.H. was supported by USDA National Needs Graduate Fellowship 88-38420-3832. This work was supported by funds from USDA CSRS, section 1433.
REFERENCES
- 1.Bairoch A. PROSITE—a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20:2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binek M, Szynkiewicz Z M. Physiological properties and classification of strains of Treponema sp. isolated from pigs in Poland. Comp Immunol Microbiol Infect Dis. 1984;7:141–148. doi: 10.1016/0147-9571(84)90019-5. [DOI] [PubMed] [Google Scholar]
- 3.del Castillo F J, Leal S C, Moreno F, del Castillo I. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted haemolysin. Mol Microbiol. 1997;25:107–115. doi: 10.1046/j.1365-2958.1997.4391813.x. [DOI] [PubMed] [Google Scholar]
- 4.Fellstrom C, Gunnarsson A. Phenotypic characterisation of intestinal spirochaetes isolated from pigs. Res Vet Sci. 1995;59:1–4. doi: 10.1016/0034-5288(95)90021-7. [DOI] [PubMed] [Google Scholar]
- 5.Greer J M, Wannemuehler M J. Comparison of the biological responses induced by lipopolysaccharide and endotoxin of Treponema hyodysenteriae and Treponema innocens. Infect Immun. 1989;57:717–723. doi: 10.1128/iai.57.3.717-723.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 7.Harris D L, Glock R D, Christensen C R, Kinyon J M. Innoculation of pigs with Treponema hyodysenteriae (new species) and reproduction of the disease. Vet Med Small Anim Clin. 1972;67:61–64. [PubMed] [Google Scholar]
- 8.Hudson M J, Alexander T J, Lysons R J. Diagnosis of swine dysentery: spirochaetes which may be confused with Treponema hyodysenteriae. Vet Rec. 1976;99:498–500. doi: 10.1136/vr.99.25-26.498. [DOI] [PubMed] [Google Scholar]
- 9.Hutto D L, Wannemuehler M J. A morphologic comparison of the effects of Serpulina hyodysenteriae or its native hemolysin on murine cecal mucosa. Vet Pathol. 1999;36:412–422. doi: 10.1354/vp.36-5-412. [DOI] [PubMed] [Google Scholar]
- 10.Hyatt D R, Muir S, Joens L A. Conference for Research Workers in Animal Diseases. 1992. Biophysical characteristics of the Serpulina hyodysenteriae native hemolysin; p. 15. [Google Scholar]
- 11.Joens L A, Glock R D. Experimental infection in mice with Treponema hyodysenteriae. Infect Immun. 1979;25:757–760. doi: 10.1128/iai.25.2.757-760.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy M J, Rosnick D K, Ulrich R G, Yancey R J. Association of Treponema hyodysenteriae with porcine intestinal mucosa. J Gen Microbiol. 1988;134:1565–1576. doi: 10.1099/00221287-134-6-1565. [DOI] [PubMed] [Google Scholar]
- 13.Kent K A, Lemcke R M, Lysons R J. Production, purification and molecular weight determinations of the haemolysin of Treponema hyodysenteriae. J Med Microbiol. 1988;27:215–224. doi: 10.1099/00222615-27-3-215. [DOI] [PubMed] [Google Scholar]
- 14.Knoop F C. Investigation of a hemolysin produced by enteropathogenic Treponema hyodysenteriae. Infect Immun. 1981;31:193–198. doi: 10.1128/iai.31.1.193-198.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemcke R M, Burrows M R. Studies on a haemolysin produced by Treponema hyodysenteriae. J Med Microbiol. 1982;15:205–214. doi: 10.1099/00222615-15-2-205. [DOI] [PubMed] [Google Scholar]
- 16.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig A, Tengel C, Bauer S, Bubert A, Benz R, Mollenkopf H J, Goebel W. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol Gen Genet. 1995;249:474–486. doi: 10.1007/BF00290573. [DOI] [PubMed] [Google Scholar]
- 18.Muir S, Koopman M B H, Libby S J, Joens L A, Heffron F, Kusters J G. Cloning and expression of a Serpula (Treponema) hyodysenteriae hemolysin gene. Infect Immun. 1992;60:529–535. doi: 10.1128/iai.60.2.529-535.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuessen M E, Birmingham J R, Joens L A. Biological activity of a lipopolysaccharide in the pathogenicity of Treponema hyodysenteriae. Infect Immun. 1982;37:138–142. doi: 10.1128/iai.37.1.138-142.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nuessen M E, Joens L A, Glock R D. Involvement of lipopolysaccharide in the pathogenicity of Treponema hyodysenteriae. J Immunol. 1983;131:997–999. [PubMed] [Google Scholar]
- 21.Oscarsson J, Mizunoe Y, Uhlin B E, Haydon D J. Induction of haemolytic activity in Escherichia coli. Mol Microbiol. 1996;20:191–199. doi: 10.1111/j.1365-2958.1996.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 22.Saheb S A, Lafleur L. Characteristics of the interaction of a treponemal hemolysin with rabbit erythrocytes. Biochimie. 1980;62:787–793. doi: 10.1016/s0300-9084(80)80134-9. [DOI] [PubMed] [Google Scholar]
- 23.Saheb S A, Massicotte L, Picard B. Purification and characterization of Treponema hyodysenteriae hemolysin. Biochimie. 1980;62:779–785. doi: 10.1016/s0300-9084(80)80133-7. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.ter Huurne A A H M, Muir S, Vanhouten M, Vanderzeijst B A M, Gaastra W, Kusters J G. Characterization of three putative Serpulina hyodysenteriae hemolysins. Microb Pathog. 1994;16:269–282. doi: 10.1006/mpat.1994.1028. [DOI] [PubMed] [Google Scholar]
- 26.Wannemuehler M J, Akili D, Hutto D, Harkins K, Crump M, Schmerr M J. The pathgenic potential of a β-hemolysin from Serpulina hyodysenteriae. J Cell Biochem Suppl. 1994;18A:63. [Google Scholar]
- 27.Zuerner R L, Stanton T B. Physical and genetic map of the Serpulina hyodysenteriae B78(T) chromosome. J Bacteriol. 1994;176:1087–1092. doi: 10.1128/jb.176.4.1087-1092.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]