Abstract
Extracellular vesicles (EV) as drug delivery nanocarriers are under intense investigation. Although clinical-grade EVs have been produced on a large-scale, low yield and high production costs of natural EVs (nEV) limit the relevant industrial translation. Recent studies show that mechanical extrusion of cells can generate nEV-like cell-derived nanovesicles (CNV) which can also be used as drug nanocarriers. Moreover, in comparison with nEVs, CNVs have similar physicochemical properties. Nevertheless, a comprehensive comparison of cargo between nEVs and CNVs has not been investigated yet. Therefore, the aim of this study is to profile and compare CNVs to nEVs. Our results show that no significant difference was found in size, morphology, and classical markers between nEVs and CNVs derived from MDA-MB-231 cells. Protein sequencing data reveals the similarity of membrane proteins between the two groups was ~71%, while it was ~21% when pertaining to total protein cargo. Notably, a high similarity of membrane proteins was also found between nEVs and CNVs derived from eight additional cancer cell lines. Moreover, analysis of the top 1000 small RNAs with RNA sequencing showed a ~65% similarity between the two groups. Altogether, we infer from the high similarity of membrane proteins and small RNA cargo that CNVs can be a good substitute for nEVs. In brief, our findings support previous studies with a notion that CNVs yield comparable performance with nEVs and could pave the way for clinical implementation of CNV-based therapeutics in the future.
Keywords: Cell engineered vesicles, Extracellular vesicles, Next-generation sequencing, Mass spectrometry, Drug delivery
1. Introduction
Extracellular vesicles (EVs) are cell-derived lipid-bilayer enclosed particles.1–4 Based on size, density, and biogenesis, EVs are generally divided into exosomes, microvesicles, and apoptotic bodies.5 The first two vesicles that hold promise for pathophysiologic and translational discoveries are under intense investigation.6,7 These cells derived natural EVs (nEV) have been exploited for drug delivery.8,9 Compared with micelles, liposomes, and polymeric nanoparticles, autologous nEVs act as a natural delivery system that can evade phagocytosis and exhibit optimal biocompatibility without potential safety issues.10–13 Although endocytosis is the major way of nEV uptake,14–17 nEVs can fuse with the cell membrane and directly deliver drugs into the cytoplasm. By evading lysosomal engulfment, nEVs remarkably enhance the delivery efficiency of vulnerable molecules.18–20 In addition, the small size of nEVs facilitates their extravasation, translocation through physical barriers, and passage through the extracellular matrix.21 Although nEV-based drug delivery is promising, the low yield of nEV posts a major challenge which significantly impedes the relevant industrial and clinical translation.22 Typically, a single cell secretes only ~50 nEVs/min.23,24 To harvest a sufficient amount of EVs, abundant cells and a long culture period are required, which makes applications unrealistic. To address or at least alleviate this technical issue, peers developed cell-derived nanovesicles (CNV) as drug delivery nanocarriers, which are derived from either autologous or allogenic cells. In previous studies, CNVs prepared by extruding cells demonstrate similar characteristics of the nEVs.25 Through mechanical extrusion, ultrasonication, freeze-thawing, and other treatments, donor cells release a large number of CNVs in a short period of time.18,26–28 Briefly, CNVs are formed as a result of the disruption of the cell membrane using shear or frictional forces and reorganization of lipid bilayer-forming vesicles in seconds. It was reported that the generation efficiency of CNVs is enhanced over ~50 to 100 times than that of nEVs.18,25,29 Moreover, recent studies demonstrated that CNVs derived from stem cells have similar biological functions in comparison with that of nEVs.30,31 Accordingly, CNVs derived from stem cells have been tested in regenerative medicine, such as skin rejuvenation and bone repair.27,32 Although the roles of CNVs in drug delivery and regenerative medicine have been demonstrated in numerous studies, a fundamental question as to the similarity between nEVs and CNVs has not been clarified so far. Previous studies only performed a simple comparison between the two, and no significant difference was found in their physicochemical properties.27 Both are nanoscale lipid-bilayer-enclosed vesicles and harbor classical EV protein markers. Nevertheless, a comprehensive comparison that can reveal the differences in membrane proteins, total proteins, and small RNA (smRNA) has not been performed yet. Therefore, the aim of this study is to compare the similarity between nEVs and CNVs based on the high-throughput sequencing data. Our study reveals ~21% of nEV proteins can be identified in CNVs derived from MDA-MB-231 cells. Moreover, all 181 membrane proteins targeted by the proximity barcoding assay (PBA) can be detected from nEVs and CNVs derived from MDA-MB-231 cells and another 8 cancer cell lines. The quantitative analysis revealed the average similarity of 181 membrane proteins between nEVs and CNVs was 70.7%. Furthermore, the top 1000 smRNAs derived from nEVs and CNVs, respectively, demonstrate ~65% similarity. The high similarity of membrane proteins between nEVs and CNVs indicates CNVs are good substitutes for nEVs as drug delivery nanocarriers. Moreover, our findings support that CNVs have similar biological functions in comparison with CNVs. Altogether, the findings we reported here may pave the way for clinical implementation of CNV-based drug delivery or therapeutics in the future.
Theory
Given many studies demonstrated CNVs prepared by mechanical extrusion have biological functions, we hypothesize that the composition of CNVs are similar to that of nEVs. If true, we further speculate that CNVs could be substitutes for nEVs.
2. Materials and methods
Cell culture
Breast cancer cell line MDA-MB-231 cells, pancreatic cancer cell line PANC-1, lung adenocarcinoma cell lines NCI-H441, NCI-H661, NCI-H2228, colon adenocarcinoma cell line HT-29, non-small cell lung carcinoma cell line NCI-H1975, glioblastoma cell line U87, and acute lymphoblastic leukemia cell line CCL-119 were purchased from ATCC. All cells passed the mycoplasma contamination test. MDA-MB-231, PANC-1, NCI-H441, NCI-H1975, and U87 were cultured in DMEM (Corning, USA). HT-29 cells were grown in McCoy’s 5a Modified Medium (Gibco, USA). NCI-H661, NCI-H2228, and CCL-119 cells were maintained in RPMI 1640 medium containing 25 mM HEPES and L-glutamine (GE Healthcare, USA). All mediums were supplemented with 5% (v/v) EV-depleted Fetal Bovine Serum (FBS) (Thermal Fisher, USA), 100 units/ml penicillin, 100 μg/ml streptomycin, and 1% non-essential amino acid. All cell lines were incubated at 37 °C with 5% CO2 and a 95% humidified atmosphere.
Isolation of nEVs
When cell confluency reached ~70%, the cell culture medium was removed and rinsed with PBS thrice. Cells were cultured with non-FBS culture medium for 48 h. Subsequently, the cell culture supernatant (CCS) was centrifuged at 2500 × g at 4 °C for 15 min to remove dead cells followed by a centrifugation step at 16,500 × g for 20 min to discard cellular detritus. Afterwards, the medium was filtered using a 0.22-μm pore filter. The supernatant was ultracentrifuged at 100,000 × g at 4 °C for 4 h. The nEV pellets were resuspended with PBS and stored at −80 °C.
Preparation of CNVs
Cells were cultured with non-FBS culture medium for an additional 48 h. Then, cells in the flasks were treated with trypsin for 3 min to harvest cells. The cell suspension was centrifuged at 500 × g at 4 °C for 5 min. Cell pellets were suspended in hypotonic buffer containing 30 mM Tris-HCL (pH 7.5), 225 mM D-mannitol, 75 mM sucrose, 0.2 mM EGTA, and a proteinase phosphatase inhibitor cocktail. Cells were disrupted using a Dounce homogenizer with a tight-fitting pestle (20 passes). Next, the suspension was centrifuged at 2500 × g at 4 °C for 15 min followed by a centrifugation step at 16,500 × g for 20 min to discard cellular detritus. Afterwards, the medium was filtered using a 0.22-μm pore filter. The supernatant was ultracentrifuged at 100,000 × g at 4 °C for 4 h. The CNV pellets were resuspended with PBS and stored at −80 °C.
Routine characterization of nEVs and CNVs
To characterize the morphology of nEVs and CNVs, 10 μl of samples were loaded on the 400 mesh formvar coated copper grid and allowed to incubate for 3 min at room temperature (RT). Samples were drained out with a filter paper and stained with 1% filtered uranyl acetate solution for 1 min. Prepared samples were imaged with Hitachi transmission electron microscopy (TEM) at an acceleration voltage of 100 kV.33 The size distribution and concentration of nEVs and CNVs were detected by Nanosight NS300. Three 30-s videos were taken. Data was analyzed by the build-in algorithm of a NS300 machine and represented as mean ±SD. Classical EV protein biomarkers, including CD81, TSG101, and GAPDH, in nEVs and CNVs were analyzed with western blot. Briefly, 200 μL of nEVs or CNVs lysates were prepared by adding 50 μL of RIPA lysis buffer on ice. Samples were further mixed with 5× loading buffer and placed at 95 °C for 20 min. Gels were running at 60 V for stacking and 100 V for separating. The proteins were transferred to PVDF membrane using Bio-rad mini blotting system at 25 V for 7 min. The membranes were blocked for 1 h in 5% skimmed milk dissolved in TBS. The proteins were detected by incubation with primary antibody conjugated with HRP (CD81: sc-166029, TSG101: sc-7964, GAPDH: sc-32233, Santa Cruz). The membranes were washed thrice prior to imaging.
Profiling of total proteins with mass spectrometry
Protein concentration of nEVs and CNVs was measured by Pierce BCA Protein Assay (ThermoFisher). Three biological replicates of nEV and CNV protein samples (16 μg/each) were loaded into stain-free SDS-PAGE gels. Gels loaded with nEV and CNV proteins were run at 100 V for 10 min and 60 min, respectively. One gel slice (fraction) for each of the triplicate nEV samples and four gel slices (fractions) for each of the three replicate CNV samples were cut into ~1 mm cubes followed by in-gel digestion and extraction of tryptic peptides.34 The excised gel pieces were washed consecutively with 200 μL of DI water, 100 mM ammonium bicarbonate (Ambic)/acetonitrile (ACN) (1:1), and ACN. The gel pieces were reduced with 70 μL of 10 mM DTT in 100 mM Ambic for 1 h at 56 °C, alkylated with 100 μL of 55 mM Iodoacetamide in 100 mM Ambic at room temperature in the dark for 45 min. Subsequently, the gel slices were dried and rehydrated with 50 μL of trypsin in 50 mM Ambic, 10% ACN (20 ng/μl) at 37 °C for 16 h. The digested peptides were extracted twice with 70 μL of 50% acetonitrile containing 5% formic acid (FA) and once with 70 μL of 90% acetonitrile containing 5% FA. Three extracts from each sample were combined, filtered by a 0.22-μm spinning unit (Corning, USA), and dried down by a Speedvac SC110. The in-gel tryptic digests of nEV proteins were reconstituted in 40 μL of 2% ACN containing 0.5% FA spiked with 50 fmol tryptic digests of yeast enolase as an internal standard. The resulting peptide samples were analyzed by nanoLC-tandem mass spectrometry (MS/MS), which was carried out using an Orbitrap Fusion™ Tribrid™ mass spectrometer equipped with a nanospray Flex Ion Source and coupled with a Dionex UltiMate 3000 RSLCnano system.35,36 The gel extracted peptide samples (9 μL for nEV proteins and 15 μL for each fraction of CNV proteins) were injected onto a PepMap C-18 RP nano trapping column (5 μm, 100 μm i.d × 20 mm) at 20 μL/min flow rate for rapid sample loading and then separated on a PepMap C-18 RP nano column (2 μm, 75 μm × 25 cm) at 35 °C. The tryptic peptides were eluted in a 120 min gradient of 5% to 33% ACN in 0.1% FA at 300 nL/min, followed by a 7 min ramping to 90% ACN-0.1% FA and an 8 min hold at 90% ACN-0.1% FA. The column was re-equilibrated with 0.1% FA for 25 min prior to the next run. The Orbitrap Fusion was operated in positive ion mode with spray voltage set at 1.6 kV and source temperature at 275 °C. External calibration for FT, IT, and quadrupole mass analyzers were performed. In data-dependent acquisition (DDA) analysis, the instrument was operated using FT mass analyzer in MS scan to select precursor ions followed by 3 s “Top Speed” data-dependent CID ion trap MS/MS scans at 1.6 m/z quadrupole isolation for precursor peptides with multiple charged ions above a threshold ion count of 10,000 and normalized collision energy of 30%. MS survey scans at a resolving power of 120,000 (fwhm at m/z 200), for the mass range of m/z 375–1575. Dynamic exclusion parameters were set at 35 s of exclusion duration with ±10 ppm exclusion mass width. All data was acquired under Xcalibur 4.3 operation software.
Analysis of MS data
The DDA raw files for CID MS/MS were subjected to database searches using Proteome Discoverer (PD) 2.5 software with the Sequest HT algorithm. The PD 2.5 processing workflow containing an additional node of Minora Feature Detector for precursor ion-based quantification was used for protein identification and relative quantitation analysis within triplicate samples. The database search was conducted against a Homo sapiens database containing 81,725 sequences downloaded from NCBI, and a human EVs database that has 60,444 sequences downloaded from Vesiclepedia. Two-missed trypsin cleavage sites were allowed. The peptide precursor tolerance was set to 10 ppm and fragment ion tolerance was set to 0.6 Da. Variable modification of methionine oxidation, deamidation of asparagine/glutamine, carboxylation on tryptophan, acetylation on protein N-terminal, and fixed modification of cysteine carbamidomethylation, were set for the database search. Identified peptides were further filtered for a maximum 1% FDR using the Percolator algorithm in PD 2.5 along with additional peptide confidence set to high and peptide mass accuracy ≤5 ppm. Relative quantitation of identified proteins among three replicates in nEVs and CNVs was assessed by the label-free quantitation (LFQ) workflow in PD 2.5. Precursor abundance intensity for each peptide identified by MS/MS was automatically determined and their unique peptides for each protein in each sample were summed and used for calculating the protein abundance by PD 2.5 software with normalization against the spiked yeast enolase protein.
Proximity barcoding assay to profile surface proteins
the surface proteins of nEVs and CNVs derived from nine cancer cell lines were profiled by next-generation sequencing (NGS)-based PBA. Micrometer-sized single-stranded DNA clusters with hundreds of copies of a unique DNA motif were generated via rolling circle amplification (RCA) and used to barcode individual nEVs and CNVs. The protein compositions on the surface of nEVs and CNVs derived from each cell line were converted to DNA sequence information. The NGS was then utilized to decode the DNA sequence and indirectly detect the protein profile. In brief, PBA probes were prepared using antibody-oligonucleotide conjugates, containing 8-nucleotide (nt) proteinTag and 8-nt random unique molecular identifier sequence as moleculeTag. Each antibody (20 μg) was prepared by adding 1 μL of 4 mM Sulfo-SMCC (Thermo Scientific) in dimethyl sulfoxide (DMSO; Sigma–Aldrich) and incubating at RT for 2 h. Subsequently, 3 μL of 100 μM each 5’ thiol-modified oligonucleotide was reduced by adding 12 μL of 100 mM DTT (Sigma–Aldrich) in 1x PBS with 5 mM EDTA and incubating at 37 °C for 1 h. Next, the purification processes for activated antibodies and reduced oligonucleotides were conducted using Zeba Spin Desalting Plates, 7 K MWCO (Thermo Scientific) according to the manufacturer’s recommended procedure. Each purified antibody and oligonucleotide were mixed and directly followed by dialysis in a Slide-A-Lyzer MINI Dialysis Device, 7 K MWCO, 0.1 mL (Thermo Scientific) against 5 L of PBS with constant stirring by a magnetic bar at 4 °C overnight. The conjugates were stored at 1 μM antibody concentration in PBS with 0.1% BSA at 4 °C for the following application.37 Successively, each sample of nEVs and CNVs were mixed with PBA probe in solution followed by washes to remove free antibody conjugates. PBA probes bound to the same exosome were brought in proximity and incorporated the same complexTag from a nearby RCA product by DNA polymerase-mediated extension. Next, the extension products are amplified for preparation of a sequencing library. After sequencing of amplification products, the reads of each sample were sorted by complexTags and proteinTags to identify participating proteins on individual EVs. By counting the total number of different moleculeTags for each of the proteinTags sharing the same complexTag, all detected protein molecules from individual EVs in a sample could be identified and quantified. Quality control steps were used to monitor the quality of sequenced reads in each sample. Individual FASTQ files were processed using FASTX-Toolkit to filter sequences based on quality following the traditional filter standard (Phred Score ≥ Q20)
smRNA sequencing
RNA cargo of nEVs and CNVs derived from MDA-MB-231 cells were extracted using miRNeasy Mini Kit (Qiagen, USA) to prepare five biological replicates in each group. The RNA sequencing was performed at the Cold Spring Harbor Laboratory. Illumina reads were trimmed for quality and adapters with Trimmomatic.38 All the reads were aligned to the human reference genome hg38 with STAR.39 COMPSRA was used to annotate and count smRNA species.40 DESeq2 R package was used to compare the level of similarity among the samples in our dataset, including generating the principal component analysis (PCA) plot, expression heatmap, Euclidian distance plot, and MA plot. All downstream statistical analyses and data visualizations were performed in R (v.3.1.1).
Statistical analyses
Data analyses were carried out using SPSS 23 software program. The statistical significance was determined by Student’s t-test and ANOVA test. All tests were two-sided, and p-value <0.05 were considered statistically significant.
3. Results
Characteristics of nEVs and CNVs derived from MDA-MB-231 cells
The isolated nEVs and CNVs mainly consisted of vesicles within the diameter range of 30–300 nm exhibiting the characteristic saucer-shaped morphology under the TEM (Fig. 1A). The average yield of nEVs and CNVs derived from ~7 × 106 MDA-MB-231 cells was 1.9 × 1011 and 3.9 × 1013, respectively. The yield of CNVs was ~200-fold higher than that of nEVs, which was in line with reported data.25 In addition, classical EV protein markers, including CD81 and TSG101. In addition, nEV exclusion marker, histones (H3), calnexin, and cytochrome C were assessed, which were not detected in nEV. In contrast, a very faint band of cytosolic cytochrome C was detected in CNV. Furthermore, membrane proteins which are involved in immune regulation, including CD59, MHC-I, CD55, CD44 were identified in both in CNVs and nEVs were identified from both nEV and CNV protein lysates (Fig. 1B). The size of nEVs and CNVs was 109.0 ± 0.8 nm and 123.2 ± 1.9 nm, respectively (Fig. 1C). No significant difference in size, morphology, and protein marker was found between nEVs and CNVs.
Fig. 1.
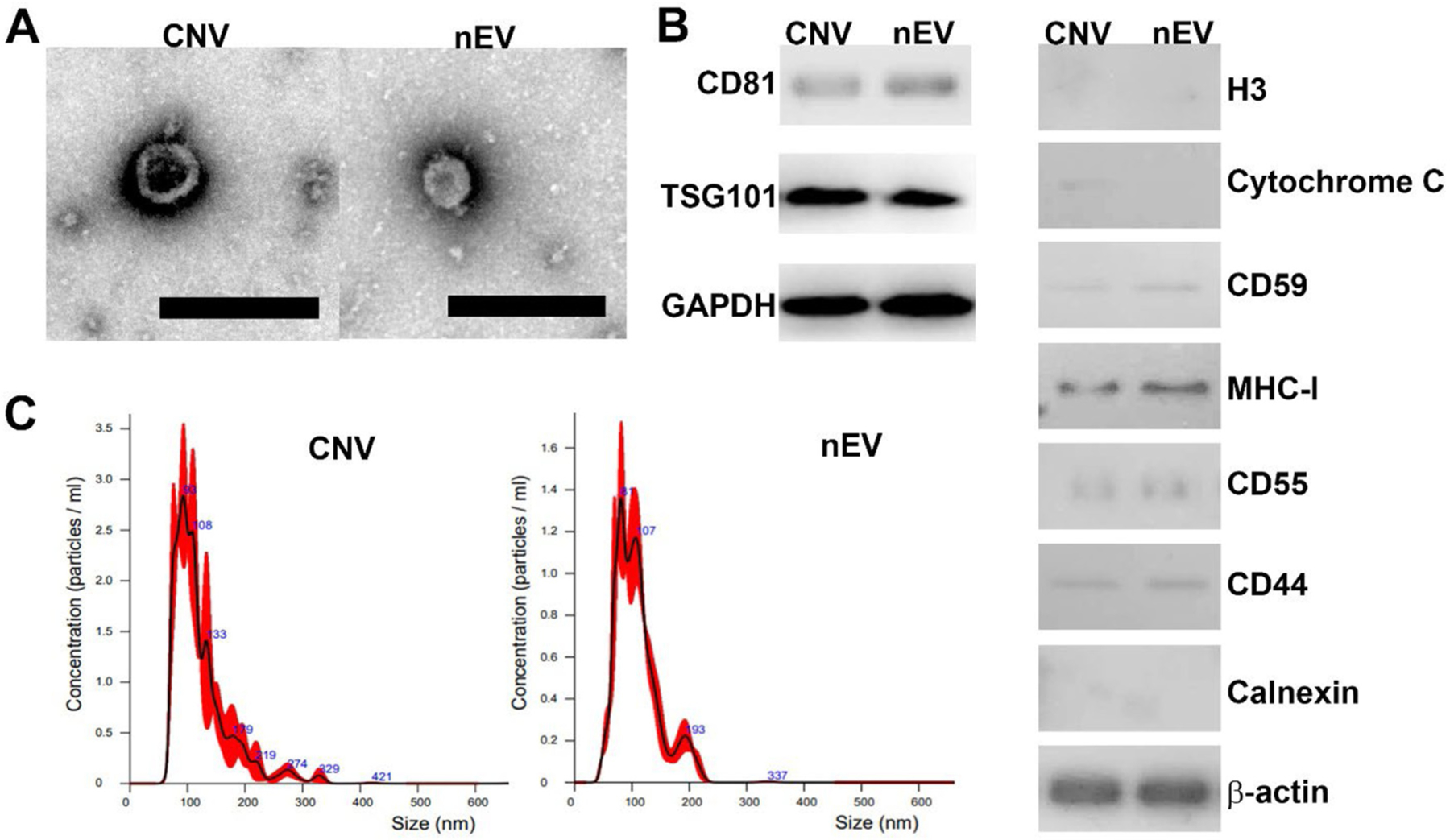
Characterization of nEVs and CNVs. (A) TEM images of CNVs and nEVs, respectively. The scale bar is 200 nm. (B) Western blot analysis of classical EV biomarker proteins, CD81, TSG101, and GAPDH. In addition, nEV exclusion marker, histones (H3) and cytochrome C were assessed. H3, calnexin, and cytochrome C were not detected in nEV, whereas a weak signal of cytosolic cytochrome C was detected in CNV. Furthermore, membrane proteins which are involved in immune regulation, including CD59, MHC-I, CD55, CD44 were identified in both in CNVs and nEVs. (C) Size distribution of CNVs and nEVs derived from MDA-MB-231 cells, respectively.
Similarity of total protein cargo between nEVs and CNVs derived from MDA-MB-231 cells
In nEVs and CNVs, the average protein-to-vesicle ratio was 76.4 μg/1011 and 187.7 μg/1011, respectively. Similarity of total protein cargo, including cytosolic proteins and surface proteins, between nEVs and CNVs, was analyzed with MS. Only proteins with at least two peptides were analyzed, which was considered as a reliable identification. We confidently identified 898 and 2908 proteins from nEVs and CNVs derived from MDA-MB-231 cells (Supplementary table 1). A total of 648 mutual proteins were found between nEVs and CNVs. Therefore, the similarity of total protein cargo between nEVs and CNVs was ~20.5% (Fig. 2A). Next, we compared the top 100 EV proteins in the Vesiclepedia database with proteins only detected in nEVs (nEVs-only), proteins only detected in CNVs (CNVs-only), and mutual proteins detected in both nEVs and CNVs. In comparison, Gallart-Palau et al. identified 86 out of the top 100 EV markers,41 while Sundar et al. detected 28 out of the top 100 EV markers listed in Vesiclepedia.42 The Venn diagram showed that 36 mutual proteins can be found in the top 100 protein list (Fig. 2B). It is noteworthy that the top 100 EV proteins listed in the Vesiclepedia database were derived from various human cell lines and human tissues. Therefore, the top 100 proteins in the Vesiclepedia database may not exactly mirror the profile of proteins derived from MDA-MB-231 nEVs and CNVs in this study. We further performed gene ontology analysis of nEV and CNV proteins and annotated their molecular function and sub-cellular location. The predominant molecular identified in nEVs and CNVs are involved in nucleic acid binding activity, cytoskeletal activity, and signal transduction activity, indicating the molecular functions of nEVs and CNVs are similar to a certain extend (Fig. 2C, Supplementary table 2). Moreover, ~90% of nEV proteins and ~40% of CNV proteins were derived from cellular membranes and cytoskeleton. Approximately 5.2% proteins in nEVs and 5.3% proteins in CNVs were derived from mitochondrion (Supplementary table 3). It is not surprising that significantly a greater number of CNV proteins came from cytosol, which was ~2.1-fold higher than that of nEVs (Fig. 2D). The differences we identified agreed with their respective generation process. In addition, the differences in protein expression between nEVs and CNVs were demonstrated by hierarchical clustering analysis (Fig. 2E). However, no significant difference was found in three intragroup batches (Fig. 2E). The similarity among the three biological replicates of nEVs was 65.5 ± 4.5%. In contrast, the similarity among the three biological replicates of CNVs was 79.1 ± 1.8%. Furthermore, the top 40 mutual proteins were selected out to take the clustering analysis and shown in two heatmaps which also verified the excellent reproducibility of nEV and CNV production (Fig. 2F).
Fig. 2.
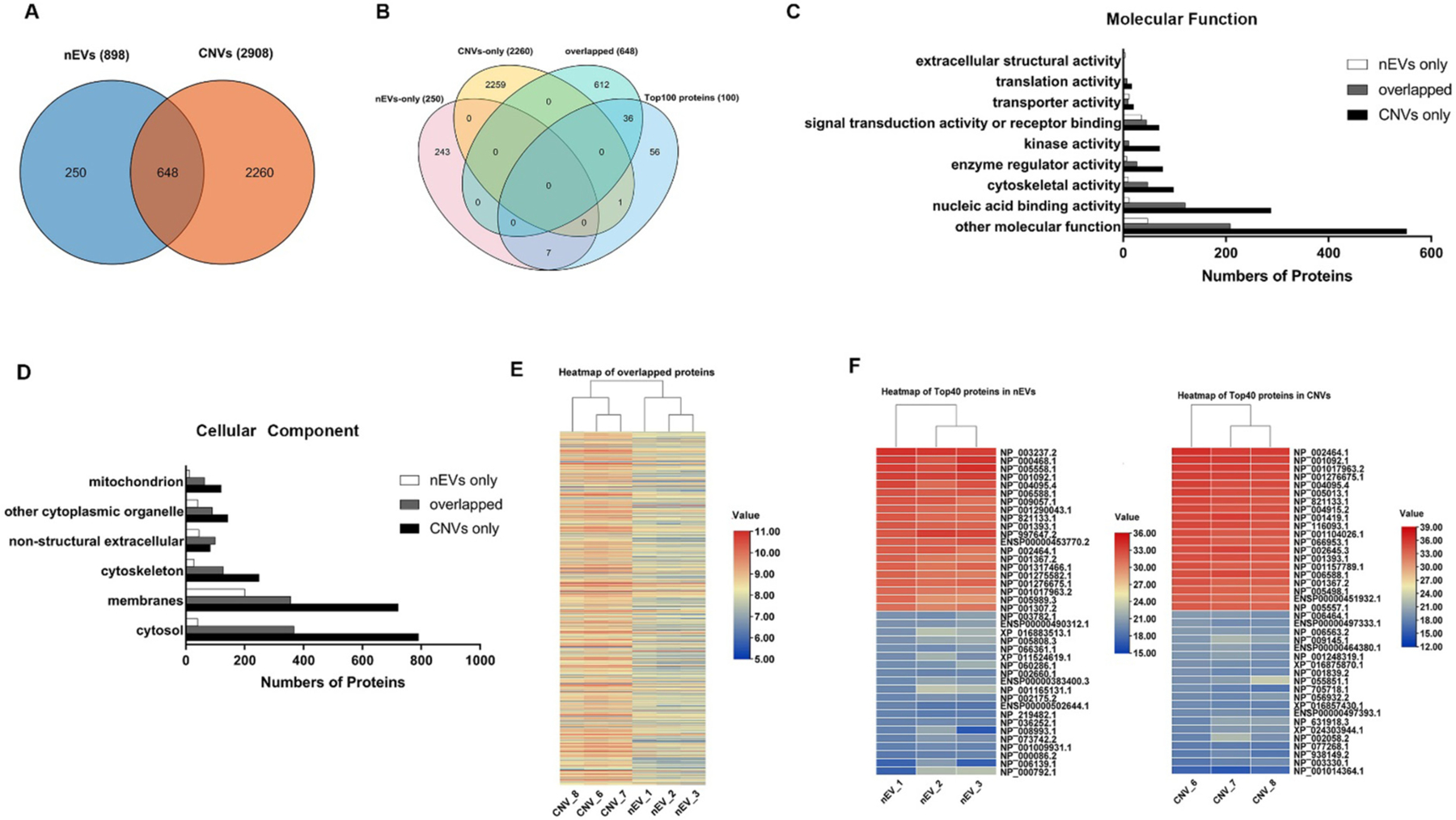
Protein profile of nEVs and CNVs derived from MDA-MB-231 cells detected by MS. (A) Venn diagram depicting the overlap of proteins, which were identified from the 3 biological replicates of nEVs and CNVs. (B) Venn diagram of mutual proteins, nEV-only proteins, CNV-only proteins, and top 100 proteins in Vesiclepedia database. (C) Molecular function of gene ontology (GO) enrichment analysis of proteins detected only in nEVs (white), only in CNVs (black), and in mutual proteins (gray). (D) Cellular component of gene ontology (GO) enrichment analysis of proteins detected only in nEVs (white), only in CNVs (black), and in mutual proteins (gray).(E) Heatmap showing the expression condition of overlapped 648 proteins detected in both nEVs and CNVs. The log10 expression values for overlapped proteins are indicated by colors as shown in the scale, with red indicating a high level of expression and blue, a low level of expression. (F) Heatmap of top 40 proteins within 3 biological replicates of nEVs (left) and CNVs (right). The log2 expression value for the top 40 proteins are indicated by colors as shown in the scale, with red indicating a high level of expression and blue, a low level of expression.
Profiling of surface proteins of nEVs and CNVs derived from nine cancer cell lines
We further investigated 181 surface proteins’ profile of nEVs and CNVs derived from MDA-MB-231 cells and another 8 cancer cell lines. High-quality clean data was obtained following the traditional filter standard of Phred Score ≥ Q20.43 All 181 surface proteins targeted by PBA were detected in nine paired nEV and CNV samples. The hierarchical clustering analysis revealed the membrane protein profiles of 9 cancer cell lines (Fig. 3A). The average similarity in membrane proteins between nEV and CNV derived from respective parental cells was 70.7%, ranging from 59.1% to 84.9% (Fig. 3B, Supplementary table 4).
Fig. 3.
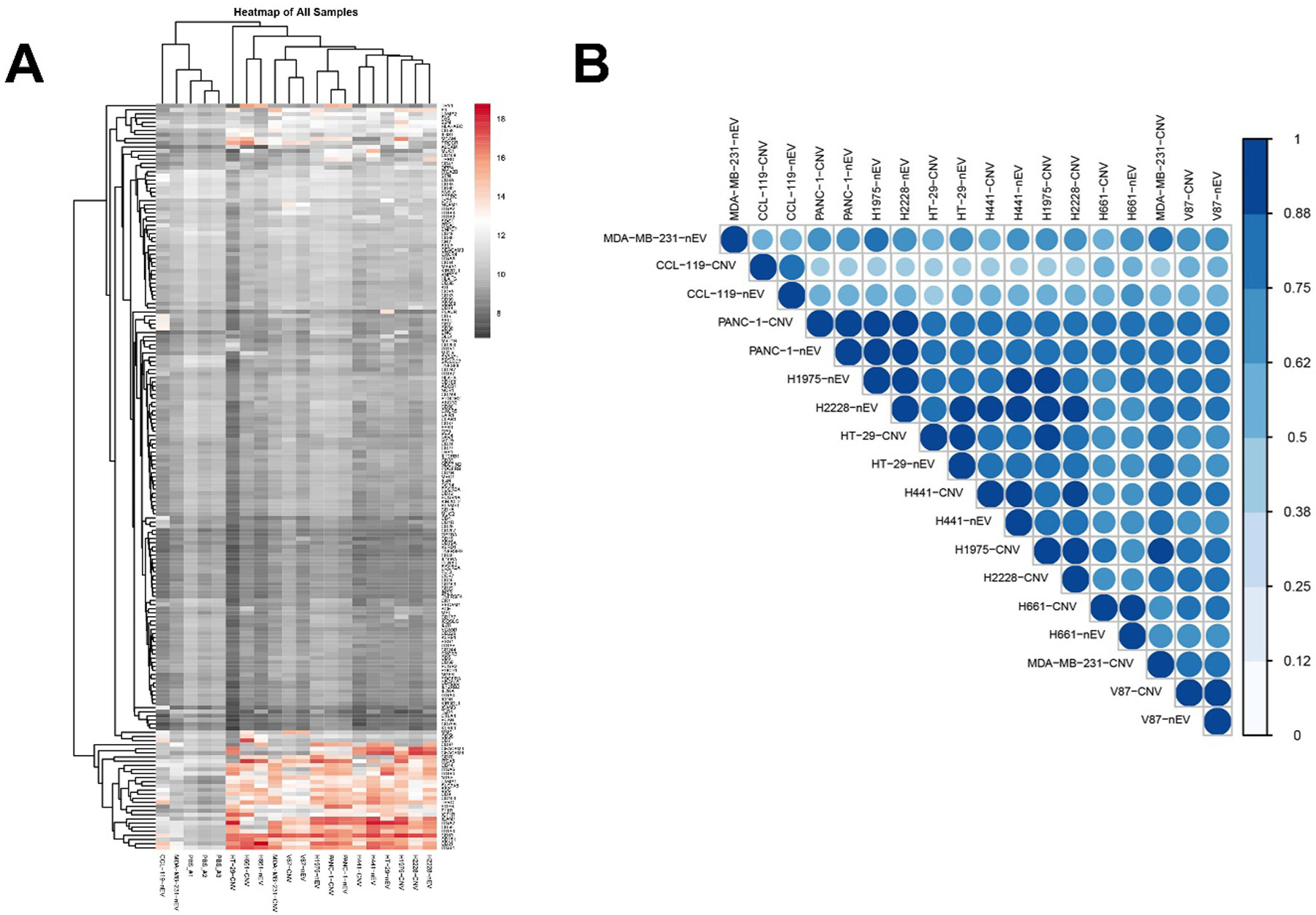
Profiles of surface proteins of nEVs and CNVs derived from 9 cancer cell lines. (A) Heatmap of surface proteins on 9 cancer cell-derived nEVs and CNVs. (B) Correlation analysis of proteins of nEVs and CNVs derived from 9 cancer cell lines. The level of expression was indicated in the scale bar.
smRNA profiling of nEVs and CNVs derived from MDA-MB-231 cells
To compare the similarity of smRNA in nEVs and CNVs derived from MDA-MB-231 cells, RNA-Seq was performed. The average percentage of reads mapped to human total RNA was 77.4% and 81.4% for nEVs and CNVs. We quantified diverse smRNA species from nEVs and CNVs. Differential percentage of piRNA, circRNA, tRNA, miRNA, snoRNA, and snRNA in nEVs and CNVs was identified between two groups (Fig. 4A). Next, the 30 most highly expressed smRNAs were selected out to represent the smRNA expression. Differential expression level was observed between nEVs and CNVs, while the intragroup difference was insignificant (Fig. 4B). We further compared the top 1000 differential gene expressions of smRNAs in nEVs and CNVs. The similarity of smRNA in nEVs and CNVs was 64.7% (Fig. 4C). For visualizing the similarity of all smRNAs extracted from the two groups, the Bland–Altman plot derived MA (log ratio-mean average) plot was employed to show the distribution of measurement results (Fig. 4D). Almost all smRNAs of the two groups were distributed within the 95% consistency limit, demonstrating the two groups of samples have excellent consistency. Similar findings were further identified by the Euclidean distance-based sample clustering plot of smRNAs derived from nEVs and CNVs (Fig. 4E). In brief, >95% similarity in smRNA species was found between nEVs and CNVs, although the percentage of each smRNA species was not exactly the same. Moreover, ~65% similarity in the expression level of top 1000 smRNAs was identified between nEVs and CNVs. The average batch-to-batch variation in five biological replicates of nEVs and CNVs were only 5.4% and 9.3%.
Fig. 4.
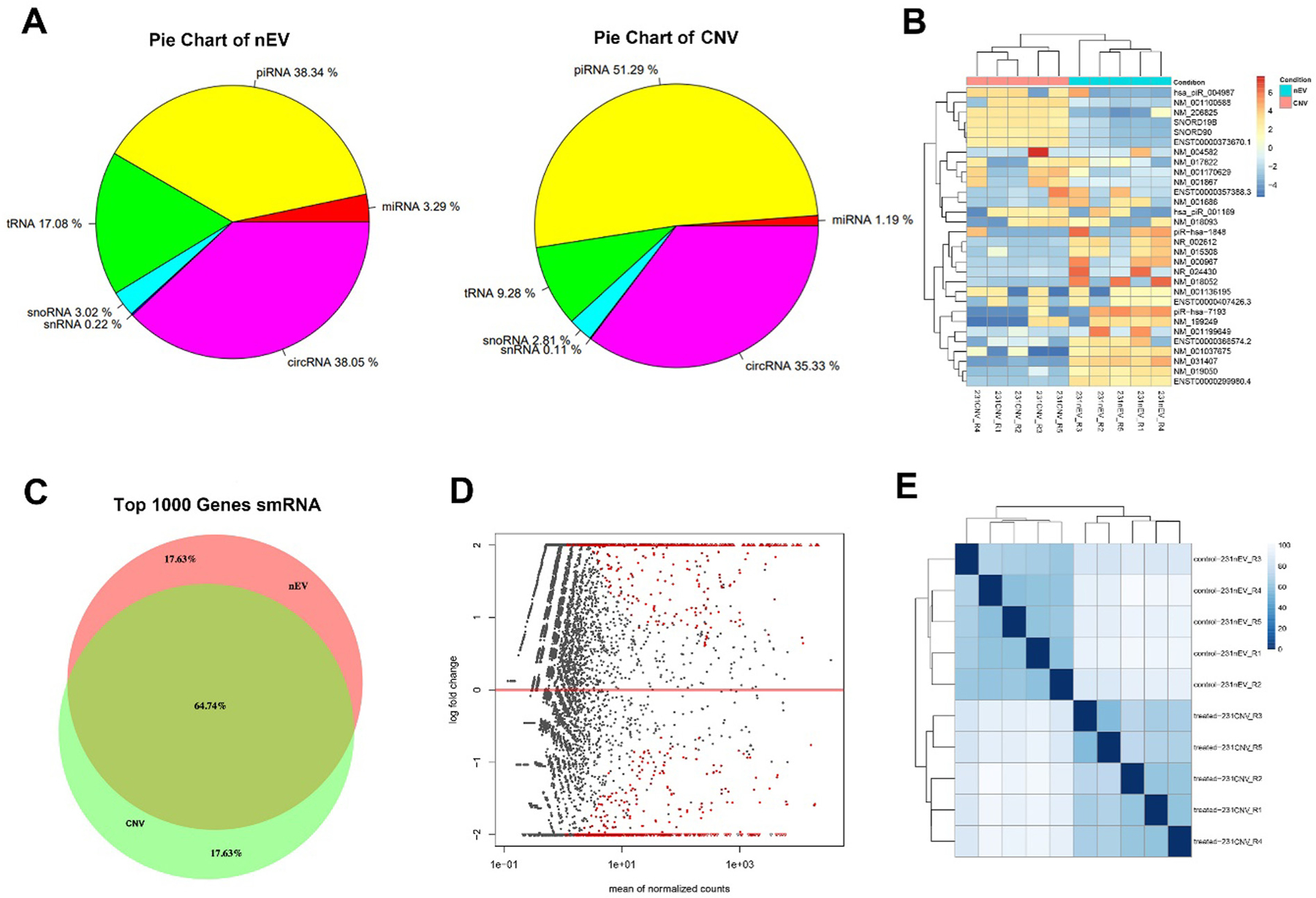
RNAs profiles of MDA-MB-231 derived nEVs and CNVs. (A) Pie chart of smRNA species and their distributions in the nEVs and CNVs. (B) Heatmap showing the expression level of the 30 most highly expressed smRNAs. Left, the CNVs. Right, the nEVs. The expression value for individual genes is indicated by color, as shown in the scale bar, with red indicating a high level of expression and blue a low level of expression. (C) Venn diagram depicting the similarity between top 1000 differential expression smRNAs of nEVs and CNVs. (D) MA plot of smRNAs from nEVs and CNVs. The data was transformed onto M (log ratio) and A (mean average) scale. The red dots represent smRNAs with significant differential expression, while the gray dots represent similar expression. (E) Euclidean distance plot of smRNA from nEVs and CNVs (five replicates). Intensity is shown in the scale bar.
4. Discussion
Clinical grade nEVs put toward the treatment of diseases have been produced on a large-scale, employing good manufacturing practice standards.44 However, the relevant clinical translation of nEVs was hindered by extremely low yield, long turnaround time, and unaffordable costs.45 Recently, CNVs prepared by mechanical extrusion and ultrasonication have been developed as substitutes for nEVs. Meanwhile, a variety of donor cells, such as dendritic cells, mesenchymal stem cells, and autologous tumor cells, have been used as the source for the production of CNVs.26 In addition, the drug delivery efficacy and biological functions of CNVs have been demonstrated by numerous studies.46,47 Briefly, in comparison with nEVs, CNVs show similar biofunctions and properties but also own additional advantages in production, including high yield, short production period, and low cost. Therefore, CNVs may hold great promise in treatment of cancers and other diseases. To validate whether CNVs can be acceptable substitutes for nEVs, the comparison between nEVs and CNVs is a prerequisite. In previous studies, no significant difference was found in their size distribution, morphology, and EV protein markers. Yet, the evidence was insufficient given the protein and RNA cargo have never been comprehensively compared between nEVs and CNVs. To the best of our knowledge, this study is the first attempt to compare nEV and CNV cargo using high-throughput sequencing techniques. Through the analyses of protein and RNA cargo, our findings support the previous reports that CNVs have biofunctions (to a certain extent) and can be used as substitutes for nEVs.
In this study, the mechanical extrusion-based procedure can massively produce CNVs within 5 min, and the yield is ~200-fold higher than that of nEVs. The production efficiency is in line with reported data.25 nEVs and CNVs demonstrated similar physicochemical properties in size, morphology, and EV markers. MS data revealed 21% of similarity in total protein cargo between nEVs and CNVs derived from MDA-MB-231 cells. Biological function annotation of proteins derived from nEVs and CNVs suggests that the majority of mutual proteins are associated with molecular function, nucleic acid binding activities, cytoskeletal activities, and others. Thus, we speculate the biological functions of two entities were similar. Although the value was modest, it was still acceptable given ~40% similarity between protein samples is considered as high similarity.48 Notably, 2908 proteins were identified in CNVs in comparison with 898 nEV proteins. We speculated that sufficient cytosolic proteins were instantaneously encapsulated into CNVs during extrusion. Alternatively, free cytosolic proteins in the supernatant might be co-isolated. The average protein-to-vesicle ratio also indicated protein contaminants in nEV and CNV samples. In contrast, the natural secretion procedure of nEVs can steadily and selectively wrap certain cytosolic proteins.5 The gene ontology analysis supported our findings that the majority of CNV proteins were derived from cytosol. On the contrary, an overwhelming majority of nEV proteins were from cellular membranes. We also speculated that the value might be deteriorated by the inherent detection bias of MS. MS sampling of precursor ions is biased towards more intense ion signals, limiting the consistent detection of low abundant peptides.49 Moreover, we prepared only four gel fractions of CNV sample. Therefore, low abundant proteins in CNV gel fractions might not be effectively detected by MS, which may weaken the comparison power between nEV and CNV protein cargo to a certain degree. In future work, ten gel fractions could be prepared for MS and cargo comparison.33 Later, we further analyzed the intragroup similarity of nEVs and CNVs, respectively. We observed that three biological replicates of CNVs demonstrated significantly higher similarity in comparison with that of nEVs (p < 0.05). It was reported that cells secrete heterogeneous populations of nEVs with different sizes and compositions. The formation of different populations of nEVs may be caused by their complex formation mechanisms and cargo sorting which are not fully understood yet. On the contrary, CNVs were prepared by simple mechanical extrusion of donor cells in 5 min. Theoretically, much fewer variables are involved in CNV production, and thus the intragroup similarity is higher than that of CNVs. Nevertheless, the high similarity of the three biological replicates of nEV revealed the excellent reproducibility of nEVs and CNVs, which can facilitate massive production in industries.
Recent studies demonstrated mechanical methods, such as electro-poration, extrusion, ultrasonication, and freeze/thaw cycles can more efficiently load drugs into EVs in comparison with that of the incubation method.26 The membrane integrity of EVs can be disrupted by mechanical forces, allowing drug loading, and then the membrane is spontaneously restored through hydrophobic–hydrophilic interaction forces between lipid molecules. In our previous study, we observed that cytosolic EV proteins can be depleted. On the contrary, membrane proteins, especially the multi-pass transmembrane proteins, can be preserved on the membranes to a certain extent during drug loading.18 Therefore, in the scenario of EVs as drug delivery nanocarriers, a simple comparison of total protein cargo between nEVs and CNVs cannot truly reflect their performance. Instead, quantitative and qualitative analyses of their membrane proteins could be more informative. On the other hand, certain membrane proteins can assist nEVs or CNVs in evading host immune clearance. For example, CD47 (i.e., “Don’t eat me” signal), CD55, CD59, and other membrane antigens enable nEVs or CNVs to escape phagocytosis by macrophages and block the formation of membrane attachment complexes.50 CD55 and CD59 can bind to C3 and C9, respectively, which can block their insertion into lipid bilayers and significantly inhibit lysis by complements.51 The expression of CD55 and CD59 is considered as a mechanism of better stability in circulation by conferring biological entities with the stability to escape complement mediated destruction.52 Therefore, these antigens can contribute to the stability, low immunogenicity, and long circulatory availability of nEVs and CNVs. We also detected membrane protein MHC-I and CD44 in both nEVs and CNVs. MHC-I is expressed on the cell surface of all nucleated cells and presents peptide fragments derived from intracellular proteins.53 CD44 is highly expressed in many cancers including MDA-MB-231 cells and regulates metastasis.54 In brief, as drug delivery nanocarriers, membrane proteins of nEVs and CNVs are more important than that of cytosolic proteins wrapped within vesicles. The average similarity in membrane proteins between nEVs and CNVs derived from nine cancer cells was 70.7%, indicating drug-loaded CNVs may have very similar performance in comparison with that of drug-loaded nEVs. In addition, Goh et al. used a shearing-based spin cups method to produce cell-derived nanovesicles (CDNs) and investigated their biodistributions versus naturally secreted exosomes through in vitro mouse tumor model. Both biodistribution profiles demonstrated liver as the main accumulation for the engineered CDNs and exosomes, which underline the livers play significant role of the clearance and elimination of both the CDNs and exosomes. Moreover, the CDNs was observed with higher accumulation in tumor site than that of exosomes, indicating CDNs had great targeting and reducing off-targeting effect in tumor model.55 In brief, CNVs produced by extrusion based- or other mechanical method have similar biodistribution behavior and targeting effects as exosomes (or nEVs), demonstrating CNVs could be promising drug delivery nanocarriers. It is noteworthy that we prepared three biological triplicates of nEVs and CNVs in each group. Three samples were pooled together for NGS-based PBA analysis, and thus, we were unable to determine the batch-to-batch variation of membrane proteins in each group.
Lastly, smRNA profiles of nEVs and CNVs derived from MDA-MB-231 cells were investigated. Notably, in this study, we manually extruded MDA-MB-231 cells to generate CNVs at 4 °C for 5 min. Other parameters of mechanical extrusion, such as mechanical force and cycle, were not strictly controlled, which may have led to the relatively high batch-to-batch variation in five biological replicates of CNVs. The commercially automated extruder with optimized parameters is expected to further decrease the variation. As to the ~65% similarity in smRNAs between nEVs and CNVs, we speculated bare smRNAs might influence the comparison. In cell extrusion, smRNAs might escape from the cytosol. These bare smRNAs as contaminants may attach onto CNV membranes, and thus partially distort the sequencing results. Nevertheless, an average of 65% similarity in smRNA between nEVs and CNVs is still high. The value also indicated CNVs may have similar biological functions in regulating protein synthesis and other cellular activities.56 Thus, this finding supports previous studies that reported CNVs derived from stem cells can promote tissue regeneration.27,57,58
5. Conclusions
In summary, the mechanical extrusion method can significantly increase the yield of CNVs with small batch-to-batch variation. It may meet the requirements of massive production and could be translated to clinical use. Meanwhile, our data demonstrated high similarity in membrane proteins and in smRNAs between nEVs and CNVs, which provide first-hand evidence for CNV-based drug delivery and regenerative medicine. In conclusion, CNVs prepared by membrane extrusion are good substitutes for nEVs.
Supplementary Material
Acknowledgments
The work was partially supported by National Cancer Institute 1R01CA230339, 1R37CA255948, and BK20212012. We thank NIH SIG 1S10 OD017992-01 grant support to SZ for the Orbitrap Fusion mass spectrometer. We thank the Swedish Biotechnology company, Vesicode AB, for providing support for the profile and analysis of the membrane proteins of extarcellular vesicles. All authors reviewed the manuscript.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Yi Wen: Designed the research, Conducted, Analyzed data, Wrote the manuscript. Qin Fu: Conducted, Analyzed data, Wrote the manuscript. Ashley Soliwoda: Conducted. Sheng Zhang: Analyzed data, Supervised experiments. Mingfeng Zheng: Analyzed data. Wenjun Mao: Designed the research, Supervised experiments, Wrote the manuscript. Yuan Wan: Designed the research, Supervised experiments, Wrote the manuscript.
Appendix A. Supplementary data
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.vesic.2022.100004. The Supporting Information is available.
RNA sequencing data can be reviewed through the following link https://dataview.ncbi.nlm.nih.gov/object/PRJNA795053?reviewer=2a1apo6sagh8kenmhrngqh5i77
Protein sequencing data was uploaded to ProteomeXchange. Dataset identifier is PXD030826.
References
- 1.Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, Mornet S, Brisson AR. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12:614–627. 10.1111/JTH.12554. [DOI] [PubMed] [Google Scholar]
- 2.Salih M, Zietse R, Hoorn EJ. Urinary extracellular vesicles and the kidney: Biomarkers and beyond. Am J Physiol - Ren Physiol. 2014;306:1251–1259. 10.1152/AJPRENAL.00128.2014/ASSET/IMAGES/LARGE/ZH20111472990003.JPEG. [DOI] [PubMed] [Google Scholar]
- 3.Winck FV, Ribeiro ACP, Domingues RR, Ling LY, Riaño Pachón DM, Rivera C, Brandão TB, Gouvea AF, Santos-Silva AR, Coletta RD, Leme AFP. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. Sci Rep. 2015;51(5):1–13. 10.1038/srep16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu D, Yang F, Xiong F, Gu N. The smart drug delivery system and its clinical potential. Theranostics. 2016;6:1306. 10.7150/THNO.14858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. 10.1124/PR.112.005983. [DOI] [PubMed] [Google Scholar]
- 6.Julich H, Willms A, Lukacs-Kornek V, Kornek M. Extracellular vesicle profiling and their use as potential disease specific biomarker. Front Immunol. 2014;5. 10.3389/FIMMU.2014.00413/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi Liu, Yiqiu Xia, Jillian Smollar, Wenjun Mao, Yuan Wan. The roles of small extracellular vesicles in lung cancer: molecular pathology, mechanisms, diagnostics, and therapeutics. Biochim Biophys Acta Rev Cancer. 2021;1876(1):188539. 10.1016/j.bbcan.2021.188539. [DOI] [PubMed] [Google Scholar]
- 8.Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery ☆. 2016. 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Bunggulawa EJ, Wang W, Yin T, Wang N, Durkan C, Wang Y, Wang G. Recent advancements in the use of exosomes as drug delivery systems 06 biological sciences 0601 biochemistry and cell biology. J Nanobiotechnol. 2018;16. 10.1186/S12951-018-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Che J, Okeke C, Hu Z-B, Xu J. DSPE-PEG: A distinctive component in drug delivery system. Curr Pharm Des. 2015. 10.2174/1381612821666150115144003. [DOI] [PubMed] [Google Scholar]
- 11.Villa F, Quarto R, Tasso R. Extracellular vesicles as natural, safe and efficient drug delivery systems. Pharm. 2019;11:557. 10.3390/PHARMACEUTICS11110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth T, Kullberg M, Malik N, Smith-Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release. 2015;199:145–155. 10.1016/J.JCONREL.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiklander OPB, Nordin JZ, O’Loughlin A, Gustafsson Y, Corso G, Mäger I, Vader P, Lee Y, Sork H, Seow Y, Heldring N, Alvarez-Erviti L, Edvard Smith CI, Le Blanc K, Macchiarini P, Jungebluth P, Wood MJA, El Andaloussi S. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:1–13. 10.3402/JEV.V4.26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287–296. 10.1016/J.APSB.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heusermann W, Hean J, Trojer D, Steib E, von Bueren S, Graff-Meyer A, Genoud C, Martin K, Pizzato N, Voshol J, Morrissey DV, Andaloussi SEL, Wood MJ, Meisner-Kober NC. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J Cell Biol. 2016;213:173–184. 10.1083/JCB.201506084/VIDEO-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joshi BS, de Beer MA, Giepmans BNG, Zuhorn IS. Endocytosis of extra-cellular vesicles and release of their cargo from endosomes. ACS Nano. 2020;14:4444–4455. 10.1021/ACSNANO.9B10033/SUPPL_FILE/NN9B10033_SI_002.AVI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;194(19):213–228. 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 18.Wan Y, Wang L, Zhu C, Zheng Q, Wang G, Tong J, Fang Y, Xia Y, Cheng G, He X, Zheng SY. Aptamer-conjugated extracellular nanovesicles for targeted drug delivery. Cancer Res. 2018;78:798–808. 10.1158/0008-5472.CAN-17-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009. 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193. 10.1007/S00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Den Boorn JG, Schlee M, Coch C, Hartmann G. Sirna delivery with exosome nanoparticles. Nat Biotechnol. 2011;294(29):325–326. 10.1038/nbt.1830. [DOI] [PubMed] [Google Scholar]
- 22.Ingato D, Lee JU, Sim SJ, Kwon YJ. Good things come in small packages: Overcoming challenges to harness extracellular vesicles for therapeutic delivery. J Control Release. 2016;241:174–185. 10.1016/J.JCONREL.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Caradec J, Kharmate G, Hosseini-Beheshti E, Adomat H, Gleave M, Guns E. Reproducibility and efficiency of serum-derived exosome extraction methods. Clin Biochem. 2014;47:1286–1292. 10.1016/J.CLINBIOCHEM.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Wen Y, Chen Y, Wang G, Abhange K, Xue F, Quinn Z, Mao W, Wan Y. Factors influencing the measurement of the secretion rate of extracellular vesicles. Analyst. 2020;145:5870–5877. 10.1039/D0AN01199A. [DOI] [PubMed] [Google Scholar]
- 25.Chul Jang S, Youn Kim O, Min Yoon C, Choi D-S, Roh T-Y, Park J, Nilsson J, Lö tvall J, Kim Y-K, Song Gho Y. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Publ. 2013;7:7698–7710. 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
- 26.Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;386(38):754–763. 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Abhange KK, Wen Y, Chen Y, Xue F, Wang G, Tong J, Zhu C, He X, Wan Y. Preparation of engineered extracellular vesicles derived from human umbilical cord mesenchymal stem cells with ultrasonication for skin rejuvenation. 2019. 10.1021/acsomega.9b03561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Geeurickx E, Tulkens J, Dhondt B, Van Deun J, Lippens L, Vergauwen G, Heyrman E, De Sutter D, Gevaert K, Impens F, Miinalainen I, Van Bockstal PJ, De Beer T, Wauben MHM, Nolte-’t Hoen ENM, Bloch K, Swinnen JV, van der Pol E, Nieuwland R, Braems G, Callewaert N, Mestdagh P, Vandesompele J, Denys H, Eyckerman S, De Wever O, Hendrix A. The generation and use of recombinant extracellular vesicles as biological reference material. Nature Commun. 2019;101(10):1–12. 10.1038/s41467-019-11182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jhan YY, Prasca-Chamorro D, Palou Zuniga G, Moore DM, Arun Kumar S, Gaharwar AK, Bishop CJ. Engineered extracellular vesicles with synthetic lipids via membrane fusion to establish efficient gene delivery. Int J Pharm. 2020;573:118802. 10.1016/J.IJPHARM.2019.118802. [DOI] [PubMed] [Google Scholar]
- 30.Ramasubramanian L, Kumar P, Wang A. Engineering extracellular vesicles as nanotherapeutics for regenerative medicine. Biomol. 2019;10(2020):48. 10.3390/BIOM10010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Q, Hai B, Kelly J, Wu S, Liu F. Extracellular vesicle mimics made from iPS cell-derived mesenchymal stem cells improve the treatment of metastatic prostate cancer. Stem Cell Res Ther. 2021;12:1–13. 10.1186/S13287-020-02097-5/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang CC, Kang M, Lu Y, Shirazi S, Diaz JI, Cooper LF, Gajendrareddy P, Ravindran S. Functionally engineered extracellular vesicles improve bone regeneration. Acta Biomater. 2020;109:182–194. 10.1016/J.ACTBIO.2020.04017.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wan Y, Cheng G, Liu X, Hao S-J, Nisic M, Zhu C-D, Xia Y-Q, Li W-Q, Wang Z-G, Zhang W-L, Rice SJ, Sebastian A, Albert I, Belani CP, Zheng S-Y. Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes. Nat Biomed Eng. 2017;1:58. 10.1038/s41551-017-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Thannhausser TW, Li L, Zhang S. Development of an integrated approach for evaluation of 2-D gel image analysis: Impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis. 2007;28:2080–2094. 10.1002/ELPS.200600524. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Anderson E, Zhang S. Evaluation of six sample preparation procedures for qualitative and quantitative proteomics analysis of milk fat globule membrane. Electrophoresis. 2018;39:2332–2339. 10.1002/ELPS.201800042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinjing Liu, Hsieh Ching-Lin Gelincik Ozkan, et al. Proteomic characterization of outer membrane vesicles from gut mucosa-derived fusobacterium nucleatum. J Proteomics. 2019;20;195: 125–137. 10.1016/j.jprot.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 37.Wu D, Yan J, Shen X, Sun Y, Thulin M, Cai Y, Wik L, Shen Q, Oelrich J, Qian X, Dubois KL, Ronquist KG, Nilsson M, Landegren U, Kamali-Moghaddam M. Profiling surface proteins on individual exosomes using a proximity barcoding assay. Nature Commun. 2019;101(10):1–10. 10.1038/s41467-019-11486-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30:2114–2120. 10.1093/BIOINFORMATICS/BTU170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. 10.1093/BIOINFORMATICS/BTS635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Kho AT, Chase RP, Pantano L, Farnam L, Amr SS, Tantisira KG. COMPSRA: a comprehensive platform for small RNA-seq data analysis. Sci Rep. 2020;101(10):1–7. 10.1038/s41598-020-61495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallart-Palau X, Serra A, Sze SK. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol Neurodegener. 2016;11. 10.1186/S13024-016-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundar IK, Li D, Rahman I. Proteomic analysis of plasma-derived extracellular vesicles in smokers and patients with chronic obstructive pulmonary disease. ACS Omega. 2019;4:10649–10661. 10.1021/ACSOMEGA.9B00966/SUPPL_FILE/AO9B00966_SI_001.ZIP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;126(12):443–451. 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen YS, Lin EY, Chiou TW, Harn HJ. Exosomes in clinical trial and their production in compliance with good manufacturing practice. Tzu-Chi Med J. 2020;32:113. 10.4103/TCMJ.TCMJ_182_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abhange K, Makler A, Wen Y, Ramnauth N, Mao W, Asghar W, Wan Y. Small extracellular vesicles in cancer. Bioact Mater. 2021;6:3705–3743. 10.1016/J.BIOACTMAT.2021.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, Barnes S, Grizzle W, Miller D, Zhang HG. A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18:1606–1614. 10.1038/MT.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian T, Zhang HX, He CP, Fan S, Zhu YL, Qi C, Huang NP, Xiao ZD, Lu ZH, Tannous BA, Gao J. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137–149. 10.1016/J.BIOMATERIALS.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. 10.1101/GR.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehrich BM, Liang Y, Fiandaca MS. Foetal bovine serum influence on in vitro extracellular vesicle analyses. J Extracell Vesicles. 2021;10. 10.1002/JEV2.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Wang L, Zheng M, Zhu C, Wang G, Xia Y, Blumenthal EJ, Mao W, Wan Y. Engineered extracellular vesicles for concurrent anti-PDL1 immunotherapy and chemotherapy. Bioact Mater. 2022;9:251–265. 10.1016/J.BIOACTMAT.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruiz-Argüelles A, Llorente L. The role of complement regulatory proteins (CD55 and CD59) in the pathogenesis of autoimmune hemocytopenias. Autoimmun Rev. 2007;6:155–161. 10.1016/J.AUTREV.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Lu M, Zhao X, Xing H, Xun Z, Yang T, Cai C, Wang D, Ding P. Liposome-chaperoned cell-free synthesis for the design of proteoliposomes: Implications for therapeutic delivery. Acta Biomater. 2018;76:1–20. 10.1016/J.ACTBIO.2018.03.043. [DOI] [PubMed] [Google Scholar]
- 53.Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003;110:163–169. 10.1046/J.1365-2567.2003.01738.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu H, Niu M, Yuan X, Wu K, Liu A. CD44 as a tumor biomarker and therapeutic target. Exp Hematol Oncol. 2020;91(9):1–14. 10.1186/S40164-020-00192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goh WJ, Zou S, Ong WY, Torta F, Alexandra AF, Schiffelers RM, Storm G, Wang JW, Czarny B, Pastorin G. Bioinspired cell-derived nanovesicles versus exosomes as drug delivery systems: a cost-effective alternative. Sci Rep. 2017;71(7):1–10. 10.1038/s41598-017-14725-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang C Novel functions for small RNA molecules. Curr Opin Mol Ther. 2009;11:641./pmc/articles/PMC3593927/ (accessed February 10, 2022). [PMC free article] [PubMed] [Google Scholar]
- 57.Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience. 2015;65:783–797. 10.1093/BIOSCI/BIV084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Corrado C, Barreca MM, Zichittella C, Alessandro R, Conigliaro A. Molecular mediators of RNA loading into extracellular vesicles. Cells. 2021;10:3355. 10.3390/CELLS10123355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


