Rationale:
Pneumonic-type lung cancer (PTLC) is a special type of lung cancer with cough and expectoration as the main clinical symptoms and inflammatory signals as the main imaging manifestations. PTLC can be easily misdiagnosed as pneumonia, and the diagnosis and treatment are always delayed. Metagenomic next-generation sequencing (mNGS), as an emerging and effective method to identify occult pathogens, has been gradually adopted by clinicians.
Patient concerns:
A 58-year-old woman with recurrent cough and expectoration was admitted to hospital on January 12th, 2022. She reported that she was diagnosed with pneumonia half a month ago, after treatment with expectorant and antibiotics for 5 days, the symptoms were relieved. However, the symptoms worsened again 10 days after stopping the drugs. On the current presentation, she denied exposure to patients with infection of COVID-19, smoking history, night sweats, weight loss, rash, joint pain, fever, and shortness of breath.
Diagnoses:
The patient was diagnosed with non-mucinous pneumonic-type lung adenocarcinoma according to the clinical symptoms, changes of CT scans after treatment and cytopathology examinations.
Interventions and outcomes:
The patient was initially diagnosed with pulmonary infection according to computerized tomography (CT) scan. Expectorant and antibiotics used. However, the symptoms worsened again 10 days after stopping the drugs. On her return visit, the CT scan did not showed obvious consolidation absorption and was similar to the previous imaging findings. mNGS was performed to detect the occult pathogens. None pathogen was detected, however, 39 copy number variations were found in Human Chromosomal Instability Analysis of mNGS indicating the presence of tumor cells. The cytopathology findings confirmed the presence of lung adenocarcinoma (non-mucinous adenocarcinoma). She was treated with targeted antitumor drugs, and the CT scan after 20 days of targeted antitumor therapy showed obvious absorption of the lesions.
Lessons:
mNGS may have potential value to screen tumor cells in bronchoalveolar lavage fluid of patients with PTLC, especially in the patients whose samples in bronchioli cannot be collected using existing sampling tools.
Keywords: bronchoalveolar lavage fluid, human chromosomal instability, metagenomic next-generation sequencing, pneumonic-type lung cancer, screening
1. Introduction
Pneumonic-type lung cancer (PTLC) is a special type of lung cancer characterized by ground glass shadows or consolidation in imaging; mucinous adenocarcinoma is the most common pathological type of PTLC.[1,2] However, not all PTLC are mucinous adenocarcinomas, 2.9% of the patients with invasive non-mucinous adenocarcinoma also present with pneumonia in imaging.[1] Patients with PTLC can be easily misdiagnosed as common pneumonia by clinicians,[2] especially the radiologists without a great deal of experience in the township hospitals.
Metagenomic next-generation sequencing (mNGS) is rapidly transitioning from scientific research clinical practice,[3,4] and it has become an emerging and effective approach to identifying occult pathogens from a variety of sample types.[5,6] Most mNGS related studies have focused on the clinical values of mNGS for diagnosis of occult pathogens.[7,8] We present here an unconventional use of mNGS: warning of cancer via human chromosomal instability analysis of bronchoalveolar lavage fluid in a patient with PTLC. To the best of our knowledge, this is the first report of warning of cancer in a patient with PTLC using mNGS.
2. Case report
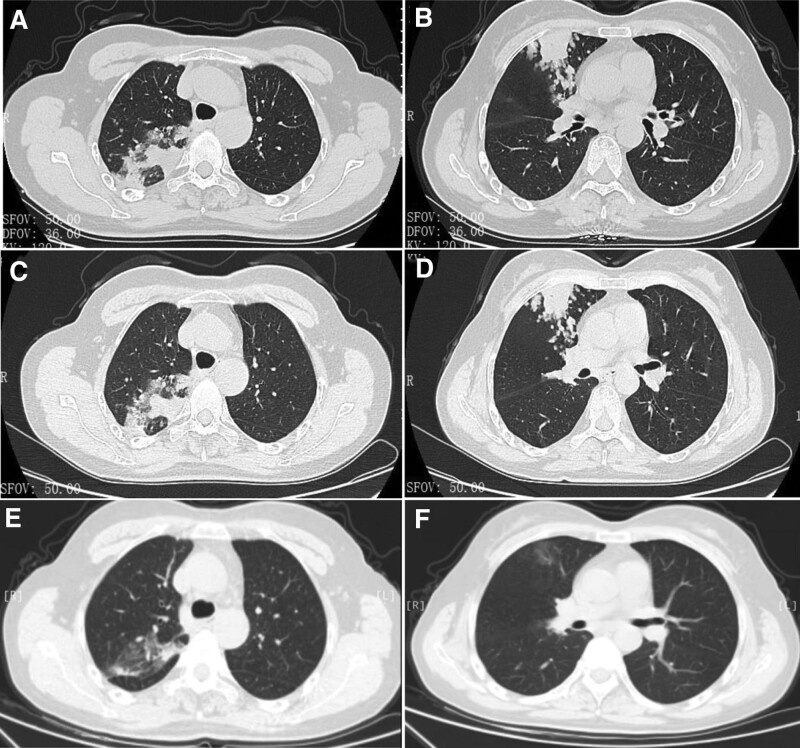
On December 25th, 2021, a 58-year-old woman with cough and expectoration for more than 10 days visited respiratory outpatient department. Chest computerized tomography (CT) scan showed scattered nodular high-density shadows in both lungs, flaky high-density shadows in right upper and lower lobes, a ground glass-like high-density shadow (15 mm) in right upper lobe (Fig. 1A and B). The patient was diagnosed with pneumonia and admitted to hospital that day. On physical examination, the patient presented cough, expectoration, afebrile, normal weight. Vital signs were within the normal range, including a pulse rate of 83 beats/min and a respiratory rate of 20 times/min. The chest examination indicated coarse breath sounds on both lungs without obvious moist rales and rhonchi. The cardiac examination showed normal heart sounds without murmurs and expansion of cardiac boundary. Neurologic examination showed no abnormalities. She denied exposure to patients with infection of COVID-19, smoking history, night sweats, weight loss, rash, joint pain, fever, and shortness of breath. In general, the patient presented a good state of spirit.
Figure 1.
The chest CT scans of the patient. (A and B) The chest CT scan at initial diagnosis. (C and D) The chest CT scan after antibiotic therapy. (E and F) The chest CT scan after 20 days of targeted antitumor therapy. CT = computerized tomography.
The blood tests showed that carcinoma embryonic antigen, carbohydrate antigen 199, carbohydrate antigen 125, cytokeratin 19 fragment, neuron-specific enolase, and squamous cell carcinoma associated antigen were all within the normal ranges. Sputum acid-fast bacillus smear test showed negative. Treatment with expectorant and antibiotics relieved her symptoms and she was discharged 5 days later. She was requested to make a return visit in 1 month.
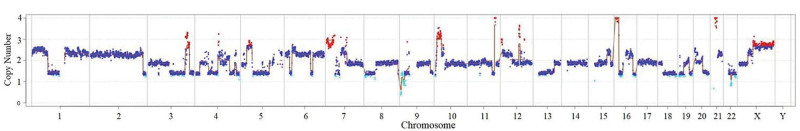
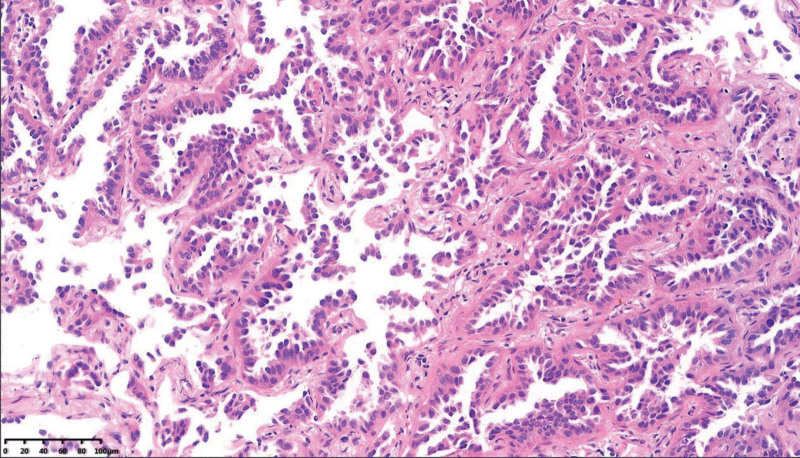
On the return visit (January 12th, 2022), the CT scan did not showed obvious consolidation absorption and was similar to the previous imaging findings (Fig. 1C and D). The patient was diagnosed with pulmonary infection (tuberculosis or other disease?) and admitted to hospital again. Endobronchial ultrasonography (EBUS) with guide–sheath was performed, and bronchoalveolar lavage fluid was detected to look for pathogens using mNGS. The mNGS results showed that none pathogen was detected, however, 39 copy number variations were found in the human chromosomal instability analysis of mNGS indicating the presence of tumor cells (Fig. 2 and Table 1). Four days later T4N0M0 lung adenocarcinoma (non-mucinous adenocarcinoma) was confirmed according to the pathologic findings and previous CT reports (Fig. 3).
Figure 2.
The result of human chromosomal instability analysis of bronchoalveolar lavage fluid using mNGS. Copy number > 2: duplication of chromosome; Copy number < 2: deletion of chromosome. mNGS = metagenomic next-generation sequencing.
Table 1.
List of copy number variations of tumor cells of the patient.
| Reads counts | G+C bases content | Copy number variations | Position and magnification of variations |
|---|---|---|---|
| 17705748 | 0.397 | 7p22.3-p14.2(dup_36.9Mb) | chr7: 0-36900000_3.03_dup |
| 10p14-p12.1(dup_20.9Mb) | chr10: 6700000-27600000_3.10_dup | ||
| 21q11.1-q21.3(dup_13.5Mb) | chr21: 14300000-27800000_3.99_dup | ||
| 1p36.33-p31.1(dup[mos]_70.1Mb) | chr1: 700000-70800000_2.45_dup[mos] | ||
| 1p31.1-p11.2(del[mos]_50.6Mb) | chr1: 70900000-121500000_1.42_del[mos] | ||
| 1q21.1-q44(dup[mos]_103.85Mb) | chr1: 145400000-249250621_2.35_dup[mos] | ||
| 3q11.1-q25.33(del[mos]_66.9Mb) | chr3: 93500000-160400000_1.40_del[mos] | ||
| 3q26.33-q29(del[mos]_17.5Mb) | chr3: 180400000-197900000_1.42_del[mos] | ||
| 4p16.3-p15.32(del[mos]_17.1Mb) | chr4: 0-17100000_1.38_del[mos] | ||
| 4q13.3-q22.3(del[mos]_23.9Mb) | chr4: 72600000-96500000_1.41_del[mos] | ||
| 4q31.22-q32.3(del[mos]_21.8Mb) | chr4: 148100000-169900000_1.43_del[mos] | ||
| 4q34.2-q35.2(del[mos]_13.9Mb) | chr4: 177200000-191100000_1.37_del[mos] | ||
| 5p14.3-p12(dup[mos]_23.7Mb) | chr5: 21600000-45300000_2.65_dup[mos] | ||
| 5q11.1-q23.1(del[mos]_71.5Mb) | chr5: 49400000-120900000_1.46_del[mos] | ||
| 6p25.3-p22.3(dup[mos]_19.0Mb) | chr6: 100000-19100000_2.27_dup[mos] | ||
| 6p22.3-p22.1(del[mos]_10.1Mb) | chr6: 19100000-29200000_1.42_del[mos] | ||
| 6p22.1-p11.1(dup[mos]_29.6Mb) | chr6: 29200000-58800000_2.39_dup[mos] | ||
| 6q11.1-q21(dup[mos]_49.5Mb) | chr6: 61800000-111300000_2.28_dup[mos] | ||
| 6q22.31-q27(dup[mos]_49.9Mb) | chr6: 121100000-171000000_2.28_dup[mos] | ||
| 7p14.2-p11.1(del[mos]_21.2Mb) | chr7: 36900000-58100000_1.70_del[mos] | ||
| 7q11.21-q11.23(dup[mos]_12.7Mb) | chr7: 63300000-76000000_2.46_dup[mos] | ||
| 8p23.3-p11.1(del[mos]_43.7Mb) | chr8: 100000-43800000_1.37_del[mos] | ||
| 9q33.3-q34.3(del[mos]_11.1Mb) | chr9: 130000000-141100000_1.46_del[mos] | ||
| 11q22.3-q23.2(del[mos]_10.6Mb) | chr11: 103900000-114500000_1.41_del[mos] | ||
| 12p13.32-p11.1(dup[mos]_31.1Mb) | chr12: 3800000-34900000_2.30_dup[mos] | ||
| 13q11-q31.2(del[mos]_68.9Mb) | chr13: 19200000-88100000_1.43_del[mos] | ||
| 15q24.2-q26.1(dup[mos]_15.7Mb) | chr15: 76600000-92300000_2.36_dup[mos] | ||
| 16p13.11-p11.2(del[mos]_17.9Mb) | chr16: 16200000-34100000_1.37_del[mos] | ||
| 16q23.1-q24.3(del[mos]_12.1Mb) | chr16: 78100000-90200000_1.34_del[mos] | ||
| 18q11.1-q23(del[mos]_59.58Mb) | chr18: 18500000-78077248_1.38_del[mos] | ||
| 19p13.3-p12(del[mos]_16.0Mb) | chr19: 5200000-21200000_1.38_del[mos] | ||
| 19q13.32-q13.43(del[mos]_13.4Mb) | chr19: 45700000-59100000_1.38_del[mos] | ||
| 20q11.23-q13.33(del[mos]_27.8Mb) | chr20: 35200000-63000000_1.44_del[mos] | ||
| 21q21.3-q22.3(dup[mos]_20.3Mb) | chr21: 27800000-48100000_2.31_dup[mos] | ||
| 22q11.1-q12.1(del[mos]_11.4Mb) | chr22: 16100000-27500000_1.46_del[mos] | ||
| 22q12.2-q13.33(del[mos]_19.8Mb) | chr22: 31400000-51200000_1.37_del[mos] | ||
| 4q12-q13.2(del[mos]_14.2Mb) | chr4: 55900000-70100000_1.47_del[mos] | ||
| Xp11.4-p11.1(dup[mos]_17.4Mb) | chrX: 41200000-58600000_2.34_dup[mos] | ||
| Xq11.2-q28(dup[mos]_91.7Mb) | chrX: 63300000-155000000_2.69_dup[mos] |
Figure 3.
Lung adenocarcinoma was confirmed by HE staining of lung biopsy tissue, HE = hematoxylin-eeosin.
Due to the EGFR exon 19 deletion-insertion mutation (p.L747_A750delinsP), she was treated with targeted antitumor drugs and without surgery and antibiotics. The CT scan after 20 days of targeted antitumor therapy showed obvious absorption of the lesions (Fig. 1E and F), and confirmed that the inflammatory signals showed in imaging were actually the cancer lesions, not the inflammatory lesions. Finally, non-mucinous pneumonic-type lung adenocarcinoma was diagnosed according to the clinical symptoms, changes of CT scans and pathology examinations.
3. Discussion
With the progress of technology, EBUS combined with rapid on-site evaluation of cytology allow to make a decision rapidly during lung biopsy operation.[9] However, the diagnostic yield in EBUS of patients with cancer is significantly associated with the tumor size.[10] Due to the technical and tracheal structural burdens, the sampling tools cannot get into the smaller bronchioles for sampling, and the smaller lesion involve less bronchioles, the diagnostic yield is affected.[9] Therefore, a more sensitivity and convenient method is needed for this situation.
Human chromosomal instability has been considered as an important cancer marker by many scientific researches. It can be applied to identify cancer in a form of noninvasive detection,[11] however, it has rarely been used in clinical practice.[4] mNGS is a technique method of high throughput sequencing of nucleic acid with fast turn-around time, high sensitivity and specificity, and it is usually used in clinical identification of pathogens in patients with infection diseases. In the present case, pulmonary infection was suspected initially, mNGS was performed to identify the pathogens; then the human chromosomal instability of exfoliated tumor cells in alveolar lavage fluid was sensitively detected by mNGS, indicating the potential value of mNGS for warning of tumor cells in patients with PTLC.
To the best of our knowledge, this is the first report of mNGS as an unconventional approach to warn of cancer via the human chromosomal instability analysis of bronchoalveolar lavage fluid in a patient with non-mucinous pneumonic-type adenocarcinoma. We believe that mNGS an unconventional approach has the potential to further improve diagnosis of lung cancer in patients with PTLC for 2 reasons. First, it is of high sensitivity and high throughput for nucleic-acid sequencing. Second, mNGS could warn of cancer by detecting the chromosomal instability of exfoliated tumor cells of alveolar lavage fluid from patients whose samples in bronchioli cannot be collected using existing sampling tools.
Acknowledgments
The authors appreciate the expertise and contributions of Lixia Wang, Jiadong Yan.
Author contributions
Conceptualization: Huabin Wang.
Data curation: Yuexiang Shui, Huabin Wang.
Funding acquisition: Yuexiang Shui.
Visualization: Huabin Wang.
Writing – original draft: Yuexiang Shui, Huabin Wang.
Abbreviations:
- CT =
- computerized tomography
- EBUS =
- endobronchial ultrasonography
- mNGS =
- metagenomic next-generation sequencing
- PTLC =
- pneumonic-type lung cancer
The present study was approved by the Ethics Committee of Lanxi People’s Hospital, and the patient provided the informed consent.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
The authors have no conflicts of interest to disclose.
This work was supported by Science and Technology Program of Zhejiang Province, China (LGF20H010002). Medical Science and Technology Project of Zhejiang Province (2020KY346). Both of the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
How to cite this article: Shui Y, Wang H. Metagenomic next-generation sequencing as an unconventional approach to warn of tumor cells in a patients with non-mucinous pneumonic-type lung adenocarcinoma: Case report. Medicine 2022;101:51(e32448).
References
- [1].Cha YJ, Kim HR, Lee HJ, et al. Clinical course of stage IV invasive mucinous adenocarcinoma of the lung. Lung Cancer. 2016;102:82–8. [DOI] [PubMed] [Google Scholar]
- [2].Liu Y, Li J, Wang S, et al. Advanced pneumonic-type lung carcinoma: a retrospective study of clinical-radiological-pathological characteristics with survival analysis in a single Chinese hospital. Zhongguo Fei Ai Za Zhi. 2019;22:329–335. Chinese. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wilson MR, Sample HA, Zorn KC, et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380:2327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Guo Y, Li H, Chen H, et al. Metagenomic next-generation sequencing to identify pathogens and cancer in lung biopsy tissue. EBioMedicine. 2021;73:103639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wei P, Gao Y, Zhang J, et al. Diagnosis of lung squamous cell carcinoma based on metagenomic next-generation sequencing. BMC Pulm Med. 2022;22:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4:663–74. [DOI] [PubMed] [Google Scholar]
- [7].Ramachandran PS, Wilson MR. Metagenomics for neurological infections - expanding our imagination. Nat Rev Neurol. 2020;16:547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gu W, Deng X, Lee M, et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. 2021;27:115–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xu CH, Wang JW, Wang W, et al. The diagnosis value of endobronchial ultrasound transbronchial lung biopsy combined with rapid on-site evaluation in peripheral lung cancer. Clin Respir J. 2020;14:447–52. [DOI] [PubMed] [Google Scholar]
- [10].Dooms CA, Verbeken EK, Becker HD, et al. Endobronchial ultrasonography in bronchoscopic occult pulmonary lesions. J Thorac Oncol. 2007;2:121–4. [DOI] [PubMed] [Google Scholar]
- [11].Watkins TBK, Lim EL, Petkovic M, et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature. 2020;587:126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]