Abstract
Objectives
Our aim was to compare the clinical and virological outcomes in Omicron BA.1- and BA.2-infected patients who received sotrovimab with those in patients who received nirmatrelvir for the prevention of severe COVID-19.
Methods
In this multi-centric, prospective ANRS 0003S CoCoPrev cohort study, patients at a high risk of progression of mild-to-moderate BA.1 or BA.2 COVID-19 who received sotrovimab or nirmatrelvir were included. The proportion of patients with progression to severe COVID-19, time between the start of treatment to negative PCR conversion, SARS-CoV-2 viral decay, and characterization of resistance variants were determined. A multi-variable Cox proportional hazard model was used to determine the time to negative PCR conversion and a mixed-effect model for the dynamics of viral decay.
Results
Amongst 255 included patients, 199 (80%) received ≥3 vaccine doses, 195 (76%) received sotrovimab, and 60 (24%) received nirmatrelvir. On day 28, new COVID-19-related hospitalization occurred in 4 of 193 (2%; 95% CI, 1–5%) sotrovimab-treated patients and 0 of 55 nirmatrelvir-treated patients (p 0.24). One out of the 55 nirmatrelvir-treated patients died (2%; 95% CI, 0–10%). The median time to negative PCR conversion was 11.5 days (95% CI, 10.5–13) in the sotrovimab-treated patients vs. 4 days (95% CI, 4–9) in the nirmatrelvir-treated patients (p < 0.001). Viral decay was faster in the patients who received nirmatrelvir (p < 0.001). In the multi-variable analysis, nirmatrelvir and nasopharyngeal PCR cycle threshold values were independently associated with faster conversion to negative PCR (hazard ratio, 2.35; 95% CI, 1.56–3.56; p < 0.0001 and hazard ratio, 1.05; 95% CI, 1.01–1.08; p 0.01, respectively).
Conclusions
Early administration of nirmatrelvir in high-risk patients compared with that of sotrovimab was associated with faster viral clearance. This may participate to decrease transmission and prevent viral resistance.
Keywords: Anti-viral treatment, COVID-19, Monoclonal antibodies, Nirmatrelvir, Sotrovimab
Introduction
The neutralizing antibody sotrovimab, which targets the spike protein of SARS-CoV-2, and the protease inhibitor nirmatrelvir (associated with ritonavir) have been shown to reduce the risk of COVID-19-related hospitalization and death in unvaccinated out-patients at the risk of progression [1,2]. We recently reported in a prospective, real-life cohort study that after early administration of sotrovimab, the clinical and virological outcomes in high-risk patients with mild-to-moderate COVID-19 due to the Omicron BA.1 and BA.2 sub-lineages were similarly good [3]. Nirmatrelvir, for which the susceptibilities of the BA.1 and BA.2 sub-lineages were comparable with those of the ancestral strain [4], was shown to reduce the risk of severe COVID-19 during the Omicron surge in Israel [5]. Our aim was to compare the clinical and virological outcomes in Omicron BA.1- and BA.2-infected patients with mild-to-moderate COVID-19 who received sotrovimab with those in patients who received nirmatrelvir for the prevention of progression to severe COVID-19.
Methods
Our study was based on the “Agence Nationale de Recherche sur le SIDA et les hépatites virales” (ANRS) “Prévention des complications de la COVID-19” (CoCoPrev) study (NCT04885452 [6]), an ongoing multi-centric, prospective cohort study which includes patients at a high risk of severe COVID-19 and having PCR-proven mild-to-moderate COVID-19 in the first 5 days of symptoms. Treatment initiation was left at the treating physician's discretion. In this study, we included all patients infected with the Omicron BA.1 or BA.2 sub-lineage who received a single infusion of 500 mg of intravenous sotrovimab or 5 days of oral nirmatrelvir between 24 January 2022 and 5 May 2022. The outcome in most patients who received sotrovimab was previously reported [3]. The primary outcome was the proportion of patients with COVID-19-related hospitalization or death on day 28 after treatment administration. The secondary outcomes were the time to negative conversion of nasopharyngeal SARS-CoV-2 PCR (defined as the first negative PCR conversion or with a cycle threshold [Ct] value of >31); predictive factors related to negative PCR conversion; dynamics of viral decay, estimated using the slope of changes over time in the Ct value, which was assessed using PCR; and genotypic characterization of resistant variants (please see supplementary material [3]). The survival time was calculated as the time from the start of treatment until the first negative PCR conversion. A multi-variable Cox proportional hazard model was used to estimate the effect of nirmatrelvir and sotrovimab on the time to negative PCR conversion, and Kaplan Meier curves were drawn. Mixed-effect models were used to estimate the temporal dynamics of the Ct value. The protocol was approved by the “CPP Sud-Est IV” Ethics Committee (Paris, France) and the French Regulatory Authority. Written informed consent was obtained from each patient before enrolment.
Results
The baseline characteristics of 255 consecutive patients are presented in Table S1. Of them, 195 (76%) received sotrovimab and 60 (24%) received nirmatrelvir.
The clinical outcomes 7 and 28 days after treatment administration are presented in Table S2. On day 28, 4 of 193 (2%; 95% CI, 1–5%) sotrovimab-treated patients and 0 of 55 nirmatrelvir-treated patients were hospitalized because of COVID-19 (p 0.24). A single patient from the nirmatrelvir-treated group died of COVID-19 (1/55; 2%; 95% CI, 0–10%). On day 7, a lower proportion of sotrovimab-treated patients experienced improvement of symptoms (127/193 [71%]) compared with that of patients who received nirmatrelvir (49/60 [89%], p 0.02).
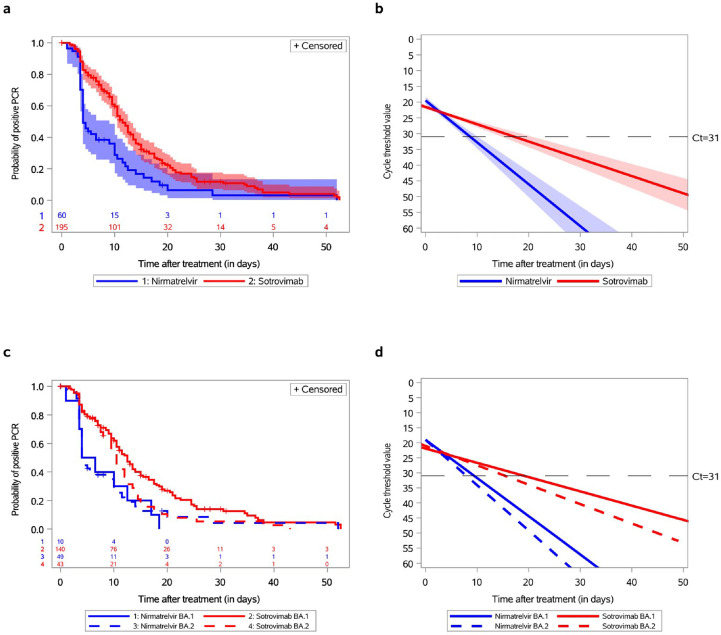
The median time to negative nasopharyngeal PCR conversion was 11.5 days (95% CI, 10.5–13) amongst the patients who received sotrovimab and 4 days (95% CI, 4–9) amongst those who received nirmatrelvir (Fig. 1 (a), p < 0.0001). The slope of Ct values of gene N, adjusted or not for age and immunosuppressive conditions, was consistently steeper in the nirmatrelvir-treated group than in the sotrovimab-treated group (Fig. 1(b), p < 0.001). Finally, the proportion of patients who negatively converted their nasopharyngeal PCR on days 7, 14, and 21 was significantly higher amongst the nirmatrelvir-treated patients (p < 0.0001, p 0.01, and p 0.02, respectively; Table S2). Amongst BA.1-infected patients, the median time to PCR conversion was 12.5 days (95% CI, 10.5–14) in the sotrovimab-treated group and 5 days (95% CI, 1–12.5) in the nirmatrelvir-treated group (Fig. 1(c), p 0.01). Amongst BA.2-infected patients, the median time was 10.5 (95% CI, 8–12.5) and 4 days (95% CI, 4–9), respectively. The slope of the Ct values of gene N was consistently steeper in the patients who received nirmatrelvir than in those who received sotrovimab (Fig. 1(d); p < 0.003 for BA.1-infected patients and p 0.001 for BA.2-infected patients). Nirmatrelvir and the nasopharyngeal PCR Ct values on day 0 were independently associated with faster conversion to negative PCR (hazard ratio, 2.35; 95% CI, 1.56–3.56; p < 0.0001 and hazard ratio, 1.05; 95% CI, 1.01–1.08; p 0.01, respectively) after adjustments for age, immunosuppressive conditions, Omicron sub-lineage, time between the onset of symptoms and initiation of treatment, and Ct value of nasopharyngeal PCR on day 0 (Table 1 ). Although we previously reported the occurrence of mutations in the spike protein in sotrovimab-treated patients [3], no mutation in the protease was observed before conversion to negative PCR amongst 26 nirmatrelvir-treated patients with successful sequence amplification on day 0 and with at least 1 follow-up sample (median time, 7 days). No major side effect was reported by the treating physicians.
Fig. 1.
Time to negative conversion of nasopharyngeal SARS-CoV-2 PCR and changes in cycle threshold (Ct) values of gene N according to treatment as well as Omicron BA.1 and BA.2 sub-lineages. (a) The survival probability plot for the event of negative PCR conversion amongst 60 Omicron-infected patients treated with nirmatrelvir and 195 Omicron-infected patients treated with sotrovimab (not adjusted on variables at baseline). The p for the log-rank test (p < 0.0001) indicates strong evidence of a significant difference between the survival curves. (b) Mixed model for Ct of gene N for 60 Omicron-infected patients treated with nirmatrelvir and 195 Omicron-infected patients treated with sotrovimab (not adjusted on variables at baseline). The p for the slope was <0.0001. After adjustment for age class and immunosuppression status at baseline, the p for the slope remained <0.0001. (c) The survival probability plots for the event of negative PCR conversion for 10 BA.1-infected patients treated with nirmatrelvir vs. 140 BA.1-infected patients treated with sotrovimab (the p for the log-rank test was 0.01) and for 49 BA.2-infected patients treated with nirmatrelvir vs. 43 BA.2-infected patients treated with sotrovimab (the p for the log-rank test was 0.1). The p was 0.06 for the comparison of BA.1- vs. BA.2-infected patients treated with sotrovimab and 0.84 for the comparison of BA.1- vs. BA.2-infected patients treated with nirmatrelvir. (d) Mixed model for Ct of gene N for 10 BA.1-infected and 49 BA.2-infected patients treated with nirmatrelvir and 140 BA.1-infected and 43 BA.2-infected patients treated with sotrovimab. The p for the slope was 0.003 for BA.1-infected patients and 0.001 for BA.2-infected patients. After adjustment for age class and immunosuppression status at baseline, the p for the slope remained at 0.003 and 0.001 respectively. Ct, cycle threshold.
Table 1.
Survival analysis for the effect of nirmatrelvir and sotrovimab on the risk of negative PCR conversion (adjusted for age class, immunosuppression status, Omicron sub-lineage, time between the onset of symptoms and initiation of treatment, and cycle threshold value of nasopharyngeal PCR on day 0)
| Variable | Hazard ratio | 95% CI | p | |
|---|---|---|---|---|
| Treatment | Nirmatrelvir vs. sotrovimab | 2.35 | 1.56–3.56 | <0.0001 |
| Age (y) | ≥80 vs. <80 | 0.71 | 0.45–1.15 | 0.17 |
| Immunosuppressive conditions | Yes vs. no | 0.63 | 0.43–0.91 | 0.01 |
| Omicron sub-lineage | BA.2 vs. BA.1 | 1.51 | 1.12–2.01 | 0.01 |
| Time between symptoms and initiation of treatment | 0.99 | 0.88–1.08 | 0.90 | |
| Ct value of nasopharyngeal PCR on d 0 | 1.05 | 1.01–1.08 | 0.01 | |
Ct, cycle threshold.
Discussion
In this prospective, real-life cohort study of Omicron BA.1- and BA.2-infected patients at a very high risk of progression to severe COVID-19, although the frequency of COVID-19-related complications was equally low, early administration of nirmatrelvir compared with that of sotrovimab was associated with a faster decline in upper SARS-CoV-2 viral load, shorter time to negative nasopharyngeal PCR conversion, and higher proportion of patients with symptom alleviation 7 days after treatment start.
In our study, both the nirmatrelvir- and sotrovimab-treated patients, although at a high risk of severe COVID-19, experienced a low rate of COVID-19 progression, supporting the real-life effectiveness of both the strategies in Omicron BA.1 and BA.2 settings [3,5]. However, nirmatrelvir initiated in the first 5 days after the onset of symptoms accelerates viral clearance and independently increases the probability of negative PCR conversion compared with sotrovimab. Nirmatrelvir also had the greatest anti-viral activity compared with sotrovimab, remdesivir, and molnupiravir in another real-life observational study which was conducted in the same time period in Italy [7]. A shorter time to achieve low viral loads in nirmatrelvir-treated patients compared with that in untreated controls was shown in patients hospitalized for mild-to-moderate COVID-19 due to Omicron [8]. This could have 2 important consequences, in addition to the prevention of COVID-19 progression. First, reduced viral shedding may translate to decreased transmission, with a public health benefit [9]. Second, viral eradication may prevent the emergence of viruses resistant to nirmatrelvir through 3-chymotrypsin-like cysteine protease mutations [10]. Finally, nirmatrelvir used in combined therapies may prevent the emergence of variants escaping anti-spike mono-clonal antibodies in immunocompromised patients [11].
As experienced in another real-life study [7], nirmatrelvir- and sotrovimab-based strategies were not used in the same patients. Nirmatrelvir was prescribed mainly to BA.2-infected patients because the drug was made available under compassionate use authorization in France on 20 January 2022, when the BA.2 sub-lineage was beginning to replace the BA.1 sub-lineage, because lower in vitro neutralizing ability of sotrovimab on the BA.2 sub-lineage was demonstrated [12]. Although sotrovimab was associated with a low incidence of the progression of COVID-19 due to Omicron BA.2 amongst very-high-risk out-patients, with no emergence of mutations [3], the WHO recently made a strong recommendation against its use [13]. The low proportion of transplant recipients amongst nirmatrelvir-treated patients reflects complex ritonavir-mediated drug-drug interactions, representing an important limitation to its use [14]. In patients who could not receive nirmatrelvir, molnupiravir [8], or remdesivir [15], all of which retain activity against the Omicron BA.4 or BA.5 sub-variant dominant since June 2022 [16] could be discussed. The use of bebtelovimab is now hampered by the emergence of the BQ1.1 sub-variant [17].
Although our study is limited by the relatively small number of nirmatrelvir-treated patients, we believe that the high-risk profile of the treated patients and the comparison with sotrovimab are of interest. Although the slopes of the Ct values of gene N of the BA.1 and BA.2-infected patients were consistently steeper amongst the nirmatrelvir-treated patients, the median time to PCR conversion amongst the BA.2-infected patients did not significantly differ between the nirmatrelvir- and sotrovimab-treated patients, presumably because of lack of statistical power. No symptomatic rebound of COVID-19 was observed after nirmatrelvir completion; however, our study was not designed to capture viral load rebound with potential mutations after conversion to negative PCR. Finally, residual confounding biases related to the cohort study's design may have been underestimated.
In conclusion, although both the strategies were associated with a low frequency of progression to severe COVID-19, nirmatrelvir was associated with faster viral clearance compared with sotrovimab.
Author contributions
GM-B (guarantor), A-GM, CS, and CD, LN, AB, FC, and YY (guarantor) were involved in study conception, data extraction, data analysis, interpretation of results, and drafting of the manuscript. SK was involved in data extraction, data analysis, interpretation of results, and drafting of the manuscript. CL-N was involved in data extraction, data analysis, interpretation of results, and drafting of the manuscript. CM, AC, CC, FC, J-PM, GG, AT-D, A-MR, VP, FC, KL, NP-S, PH, AP, GP, AM, VD, MD, JF, and CC were involved in patient inclusion and revising the manuscript. RL was involved in the study conception, interpretation of results, and revising the manuscript.
Transparency declaration
The authors declare that they have no conflicts of interest. The “Agence Nationale de Recherche sur le SIDA et les hépatites virales” (ANRS) "Prévention des complications de la COVID-19" (CoCoPrev) study was conducted with the support of ANRS│MIE and funded by French ministries: Ministère des Solidarités et de la Santé and Ministère de l’Enseignement Supérieur, de la Recherche et de l’Innovation.
Acknowledgements
We thank Pr Yazdan Yazdanpanah and the ANRS-MIE team for their invaluable support and help. This study would have not been possible without the teams involved in the "Prévention des complications de la COVID-19" (CoCoPrev) study and designated as the CoCoPrev Study Group: Magali Garcia, Valentin Giraud, Agathe Metais, France Cazenave-Roblot, and Jean-Philippe Martellosio (CHU de Poitiers); Anne-Marie Ronchetti, Thomas Gabas, Naima Hadjadj, Célia Salanoubat, Amélie Chabrol, Pierre Housset, Agathe Pardon, Anne-Laure Faucon, Valérie Caudwell, and Latifa Hanafi (CHU Sud-Francilien, Corbeil-Essonne); Laurent Alric, Grégory Pugnet, Morgane Mourguet, Eva Bories, Delphine Bonnet, Sandrine Charpentier, Pierre Delobel, Alexa Debard, Colleen Beck, Xavier Boumaza, Stella Rousset, and Aurore Perrot (CHU de Toulouse); Fanny Lanternier, Claire Delage, Elisabete Gomes Pires, Morgane Cheminant, and Nathalie Chavarot (Hôpital Necker, Paris); Anthony Chauvin and Xavier Eyer; Véronique Delcey (Hôpital Lariboisière, Paris); Simon Bessis and Romain Gueneau (Hôpital du Kremlin Bicêtre); Pelagie Thibaut, Marine Nadal, Martin Siguier, Marwa Bachir, and Christia Palacios (Hôpital Tenon, Paris); Valérie Pourcher, Cléa Melenotte, Antoine Faycal, Vincent Berot, Cécile Brin, Siham Djebara, Karen Zafilaza, Stéphane Marot, Sophie Sayon, Valentin Leducq, Isabelle Malet, Elisa Teyssou, and Adélie Gothland (Hôpital de la Pitié Salpétrière, Paris); Karine Lacombe, Yasmine Abi Aad, Thibault Chiarabini, Raynald Feliho, Nadia Valin, Fabien Brigant, Julien Boize, Pierre-Clément Thiébaud, Marie Moreau, and Charlotte Billard (Hôpital St Antoine, Paris); Nathalie De Castro, Geoffroy Liégeon, Blandine Denis, Jean-Michel Molina, and Lucia Etheve (Hôpital Saint Louis, Paris); André Cabié, Sylvie Abel, Ornella Cabras, Karine Guitteaud, and Sandrine Pierre-François (CHU de Martinique); Vincent Dubee, Diama Ndiaye, Jonathan Pehlivan, Michael Phelippeau, and Rafael Mahieu (CHU d’Angers); Alexandre Duvignaud, Thierry Piston, Arnaud Desclaux, Didier Neau, Charles Cazanave (CHU de Bordeaux); Jean-François Faucher, Benjamin Festou, Magali Dupuy-Grasset, Véronique Loustaud-Ratti, and Delphine Chainier (CHU de Limoges); Nathan Peiffer-Smadja, Christophe Choquet, Olivia Da Conceicao, Michael Thy, Lio Collas, Cindy Godard, Donia Bouzid, Vittiaroat Ing, Laurent Pereira, Thomas Pavlowsky, and Camille Ravaut (Hôpital Bichat, Paris); Antoine Asquier-Khati, David Boutoille, Marie Chauveau, Colin Deschanvres, and François Raffi (CHU de Nantes); Audrey Le Bot, Marine Cailleaux, François Benezit, Anne Maillard, Benoit Hue, and Pierre Tattevin (CHU de Rennes); François Coustilleres, Claudia Carvalho-Schneider, Simon Jamard, Laetitia Petit, and Karl Stefic (CHU de Tours); Natacha Mrozek, Clement Theis, Magali Vidal, Leo Sauvat, and Delphine Martineau (CHU de Clermond-Ferrand); Benjamin Lefèvre, Guillaume Baronnet, and Agnès Didier (CHRU de Nancy); Florence Ader, Thomas Perpoint, Anne Conrad, Paul Chabert, and Pierre Chauvelot (CHU de Lyon); Aurélie Martin, Paul Loubet, Julien Mazet, Romaric Larcher, and Didier Laureillard (CHU de Nîmes); Mathilde Devaux (Hôpital de Poissy); Jérôme Frey, Amos Woerlen, Aline Remillon, Laure Absensur-Vuillaume, and Pauline Bouquet (CHU de Metz); Albert Trinh-Duc and Patrick Rispal (Hôpital d’Agen); Philippe Petua and Julien Carillo (Hôpital de Tarbes); Aurore Perrot, Karen Delavigne, Pierre Cougoul, Jérémie Dion, Odile Rauzy (Oncopole, Toulouse), Mathieu Blot, Thibault Sixt, Florian Moretto, Carole Charles, and Lionel Piroth (CHU de Dijon); Sophie Circosta, Lydia Leger, Arulvani Arulananthan, Carine Lascoux, Pascaline Valérie, and Léia Becam (Team Biobanque ANRS-INSERM US19,Villejuif); Yazdan Yazdanpanah, Ventzislava Petrov-Sanchez, Alpha Diallo, Soizic Le Mestre, and Guillaume Le Meut (ANRS-MIE); and Isabelle Goderel, Frédéric Chau, Brahim Soltana, Jessica Chane Tang (IPLESP), Jeremie Guedj (Université de Paris, IAME, INSERM, Paris), and Yvanie Caille (Renaloo).
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2022.12.016.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gupta A., Gonzalez-Rojas Y., Juarez E., Casal M.C., Moya J., Falci D.R., et al. Effect of sotrovimab on hospitalization or death among high-risk patients with mild to moderate COVID-19. JAMA. 2022;327:1236–1246. doi: 10.1001/jama.2022.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond J., Leister-Tebbe H., Gardner A., Abreu P., Bao W., Wisemandle W., et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med. 2022;386:1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin-Blondel G., Marcelin A.G., Soulie C., Kaisaridi S., Lusivika-Nzinga C., Dorival C., et al. Sotrovimab to prevent severe COVID-19 in high-risk patients infected with Omicron BA.2. J Infect. 2022;85:e104–e108. doi: 10.1016/j.jinf.2022.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., et al. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. N Engl J Med. 2022;386:1475–1477. doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arbel R., Sagy Y.W., Hoshen M., Battat E., Lavie G., Sergienko R., et al. Nirmatrelvir use and severe Covid-19 outcomes during the omicron surge. N Engl J Med. 2022;387:790–798. doi: 10.1056/NEJMoa2204919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin-Blondel G., Marcelin A.G., Soulie C., Kaisaridi S., Lusivika-Nzinga C., Dorival C., et al. J Infect. 2022;85:e104–e108. doi: 10.1016/j.jinf.2022.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazzotta V., Cozzi Lepri A., Colavita F., Rosati S., Lalle E., Cimaglia C., et al. Viral load decrease in SARS-CoV-2 BA.1 and BA.2 Omicron sublineages infection after treatment with monoclonal antibodies and direct antiviral agents. J Med Virol. 2022;95 doi: 10.1002/jmv.28186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong C.K., Au I.C., Lau K.T., Lau E.H., Cowling B.J., Leung G.M. Real-world effectiveness of early molnupiravir or nirmatrelvir-ritonavir in hospitalised patients with COVID-19 without supplemental oxygen requirement on admission during Hong Kong’s omicron BA.2 wave: a retrospective cohort study. Lancet Infect Dis. 2022;22:1681–1693. doi: 10.1016/S1473-3099(22)00507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marc A., Kerioui M., Blanquart F., Bertrand J., Mitja O., Corbacho-Monne M., et al. Quantifying the relationship between SARS-CoV-2 viral load and infectiousness. Elife. 2021;10 doi: 10.7554/eLife.69302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iketani S., Mohri H., Culbertson B., Hong S.J., Duan Y., Luck M.I., et al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature. 2022 Nov 9 doi: 10.1038/s41586-022-05514-2. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gliga S., Luebke N., Killer A., Gruell H., Walker A., Dilthey A.T., et al. Clin Infect Dis. 2022. Oct 3 doi: 10.1093/cid/ciac802. Online ahead of print. [DOI] [Google Scholar]
- 12.Bruel T., Hadjadj J., Maes P., Planas D., Seve A., Staropoli I. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med. 2022;28:1297–1302. doi: 10.1038/s41591-022-01792-5. [DOI] [PubMed] [Google Scholar]
- 13.Agarwal A., Rochwerg B., Lamontagne F., Siemieniuk R.A., Agoritsas T., Askie L., et al. A living WHO guideline on drugs for covid-19. BMJ. 2020;370:m3379. doi: 10.1136/bmj.m3379. [DOI] [PubMed] [Google Scholar]
- 14.Lim S., Tignanelli C.J., Hoertel N., Boulware D.R., Usher M.G. Prevalence of medical contraindications to nirmatrelvir/ritonavir in a cohort of hospitalized and nonhospitalized patients with COVID-19. Open Forum Infect Dis. 2022;9:ofac389. doi: 10.1093/ofid/ofac389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottlieb R.L., Vaca C.E., Paredes R., Mera J., Webb B.J., Perez G., et al. Early remdesivir to prevent progression to severe Covid-19 in outpatients. N Engl J Med. 2022;386:305–315. doi: 10.1056/NEJMoa2116846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takashita E., Yamayoshi S., Simon V., Van Bakel H., Sordillo E.M., Pekosz A., et al. Efficacy of antibodies and antiviral drugs against omicron BA.2.12.1, BA.4, and BA.5 subvariants. N Engl J Med. 2022;387:468–470. doi: 10.1056/NEJMc2207519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Planas D., Bruel T., Staropoli I., Guivel-Benhassine F., Porrot F., Maes P., et al. bioRxiv. 2022 Nov 21 doi: 10.1101/2022.11.17.516888. 2022.11.17.516888. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.