Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly transmissible and potentially fatal virus. So far, most comprehensive analyses encompassing clinical and transcriptional manifestation have concentrated on the lungs. Here, we confirmed evident signs of viral infection in the lungs and spleen of SARS-CoV-2-infected K18-hACE2 mice, which replicate the phenotype and infection symptoms in hospitalized humans. Seven days post viral detection in organs, infected mice showed decreased vital signs, leading to death. Bronchopneumonia due to infiltration of leukocytes in the lungs and reduction in the spleen lymphocyte region were observed. Transcriptome profiling implicated the meticulous regulation of distress and recovery from cytokine-mediated immunity by distinct immune cell types in a time-dependent manner. In lungs, the chemokine-driven response to viral invasion was highly elevated at 2 days post infection (dpi). In late infection, diseased lungs, post the innate immune process, showed recovery signs. The spleen established an even more immediate line of defense than the lungs, and the cytokine expression profile dropped at 7 dpi. At 5 dpi, spleen samples diverged into two distinct groups with different transcriptome profile and pathophysiology. Inhibition of consecutive host cell viral entry and massive immunoglobulin production and proteolysis inhibition seemed that one group endeavored to survive, while the other group struggled with developmental regeneration against consistent viral intrusion through the replication cycle. Our results may contribute to improved understanding of the longitudinal response to viral infection and development of potential therapeutics for hospitalized patients affected by SARS-CoV-2.
Keywords: immune-mediated response, SARS-CoV-2, transcriptome profiling
INTRODUCTION
Over the past 20 years, three outbreaks of the severe acute respiratory syndrome coronavirus (SARS-CoV) infection caused by three closely related coronaviruses—SARS-CoV-1, MERS-CoV (Middle East respiratory syndrome coronavirus), and SARS-CoV-2 have threatened humanity (Abdelrahman et al., 2020; Blanco-Melo et al., 2020; de Wit et al., 2016; Masters, 2006). Among these, interest in SARS-CoV-2 has intensified due to the ongoing coronavirus disease 2019 (COVID-19) pandemic (Kirtipal et al., 2020; Wu et al., 2020; Zhu et al., 2020). Infection with SARS-CoV-2 is characterized by a range of symptoms including fever, cough, and general malaise in majority of the cases (Blanco-Melo et al., 2020; Chen et al., 2020; Mohamadian et al., 2021). More severe cases of COVID-19 develop acute respiratory distress syndrome and acute lung injury, leading to morbidity and mortality resulting from damage of the alveolar lumen, which leads to inflammation and pneumonia (Chen et al., 2020; Peiris et al., 2003; Wölfel et al., 2020; Xu et al., 2020).
Coronaviruses are single-stranded and positive-sense enveloped RNA viruses with an extensively sized genome of around 30 kilobases (Masters, 2006). The nucleocapsid protein, which ranges from 43 to 50 kDa, is the protein component of the helical nucleocapsid that binds genomic RNA in a beads-on-a-string fashion (Masters, 2006). The coronavirus virion is composed of three distinctive protein components, i.e., envelope protein, membrane protein, and spike (S) protein (Masters, 2006). SARS-CoV-2 invades cells either by cell-surface membrane fusion or receptor-mediated endocytosis (Qinfen et al., 2004). Both mechanisms require the binding of its S protein to the angiotensin converting enzyme 2 (ACE2), mediated by the receptor-binding domain (Cui et al., 2019; Walls et al., 2020). Since human ACE2 (hACE2) supports SARS-CoV-2 binding, it cannot infect conventional laboratory strains of mice efficiently (Cleary et al., 2020; Wan et al., 2020). Therefore, K18-hACE2-transgenic mice in which hACE2 expression is controlled by epithelial cell cytokeratin-18 (K18) promoter were developed for the study of SARS-CoV pathogenesis (Bao et al., 2020; McCray et al., 2007; Moreau et al., 2020; Winkler et al., 2020; Yinda et al., 2021). The lung tissues from infected hACE2 mice around 3 dpi displayed gross lesions and signs of interstitial pneumonia, characterized by thickened alveolar septa accompanied by infiltration of inflammatory cells such as neutrophils and macrophages (Bao et al., 2020; Winkler et al., 2020; Yinda et al., 2021; Zheng et al., 2022). Although the lungs were previously shown to have morbid signatures in this mouse model (Bao et al., 2020; Oladunni et al., 2020; Winkler et al., 2020; Yinda et al., 2021), detailed immune-mediated response to virus infection is not yet completely understood. Furthermore, the few preceded transcriptome analyses of SARS-CoV-2 infected mouse were either limited to specific time point of HFH4 (hepatocyte nuclear factor-3/forkhead homologue 4)-hACE2 transgenic mice showing variances in susceptibility (Jiang et al., 2020; Zheng et al., 2022) or focused on major organ, lungs (Winkler et al., 2020). In this study, we evaluated the overall clinical pathogenesis and transcriptome profiling of the lungs and spleen of SARS-CoV-2-infected K18-hACE2 transgenic mice. Extensive analysis of these organs will enable better understanding of the organ-specific response against SARS-CoV-2 infiltration.
MATERIALS AND METHODS
Mice and in vivo experiment
K18-hACE2 mice were purchased from Jackson Laboratories. All animal experiments were conducted in a BSL3 (biosafety level 3) facility in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The protocol was approved by the Institutional Animal Care and Use Committee of the Department of Laboratory Animal Resources of Yonsei University College of Medicine accredited by AAALAC (Association for Assessment and Accreditation of Laboratory Animal Care) International (No. 001071). To establish SARS-CoV-2 (NCCP 43326) infection in the K18-hACE2 mice, 12-week-old male mice were anaesthetized using a zoletil-rompun mixture (4:1) and intranasally inoculated with 1 × 106 plaque-forming units (PFU) of SARS-CoV-2. The body temperature and weight were measured daily until 7 days post infection (dpi) using an electronic scale and an implantable programmable temperature transponder (BMDS, USA). For animal welfare, mice with body temperature under 25°C were considered dead.
Plaque assay
Vero cells were purchased from the Korean Cell Line Bank and cultured in Dulbecco’s minimum essential medium (DMEM) supplemented with 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% fetal bovine serum (FBS) at 37°C in a 5% CO2 incubator. For plaque assay, Vero cells were seeded in a 6-well plate 1 day before the assay. Following this, cells were infected with SARS-CoV-2 serially diluted in serum-free medium for 1 h with gentle agitation every 15 min. The cells were then overlaid with DMEM containing 1% SeaPlaqueTM agarose (Lonza, Switzerland), 2% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. After 3 days of incubation until observation of clear plaques, 4% paraformaldehyde was used for fixation, and a 0.5% crystal violet-20% methanol solution was used for staining the cells. The number of plaques observed was multiplied by the dilution factor to calculate the virus titer.
Histopathological analysis
For histopathological analysis, SARS-CoV-2-infected K18-hACE2 mice were sacrificed at 1, 2, 5, and 7 dpi. Weight of the tissue was measured immediately after sacrifice using an electronic scale and recorded as a percentage of the total body weight. Collected tissue samples were fixed in 10% formalin with gentle agitation over 24 h and embedded in paraffin. Paraffin slides with 4 μm width were deparaffinized by immersion in xylene and then rehydrated in ascending grades of ethanol. H&E staining was performed by dipping the slides in 0.1% Mayer’s hematoxylin for 10 min followed by 0.5% eosin. After staining, the following sequential steps were performed: washing in distilled water until the eosin stopped streaking and immersion in 50% ethanol 10 times, in 70% ethanol 10 times, in 95% ethanol for 30 s, and in 100% ethanol for 1 min. An experienced mouse pathologist histopathologically analyzed the H&E slides based on the extent and severity of lesion.
RNA sequencing and bioinformatic analyses
CLC Genomics Workbench 9.5.3 software (Qiagen, Germany) was used to map the reads to the mouse genome (mm10, build name GRCm38) and the SARS-CoV-2 viral genome (GenBank No. MN985325.1) and generate gene expression values in the normalized form of transcripts per kilobase million (TPM) (Supplementary Fig. S1). All differentially expressed genes (DEGs) were chosen based on P < 0.001, and absolute 2-fold change based on statistical analysis (t-test and ANOVA), except for analysis of the spleen at 5 dpi. For selection of DEGs in analysis of the spleen at 5 dpi, P < 0.005, absolute 2-fold change was used based on statistical analysis (t-test and ANOVA). R Studio v3.6.3 was used to analyze RNA sequencing (RNA-seq) data, including hierarchical clustering and principal components and correlation analyses. Gene Ontology (GO) enrichment analysis was performed using R package clusterProfiler (Yu et al., 2012). The P < 0.05 (Benjamini-Hochberg) was considered statistically significant. Immune cell deconvolution was conducted using R package immunedeconv (Sturm et al., 2020).
RESULTS
Lungs and spleen of K18-hACE2 mice are highly susceptible to SARS-CoV-2 infection
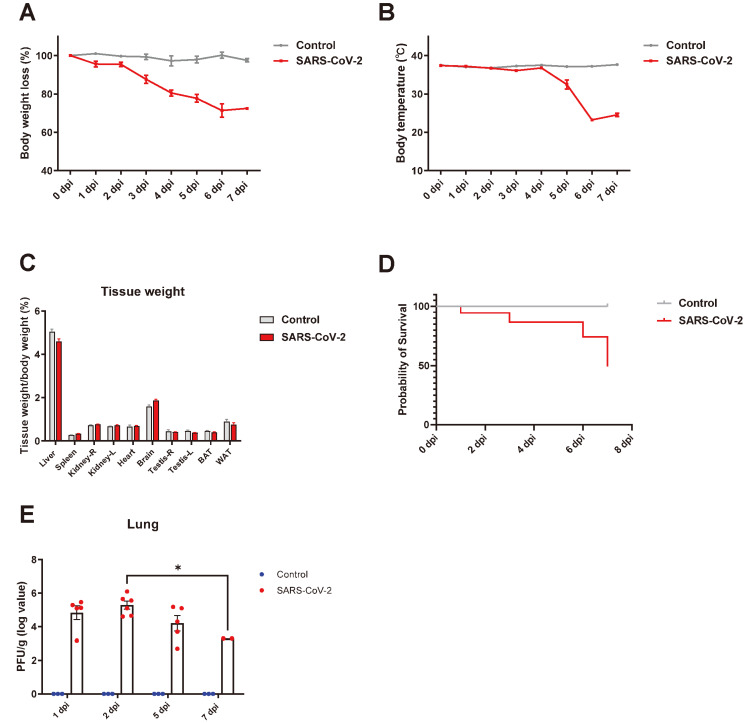
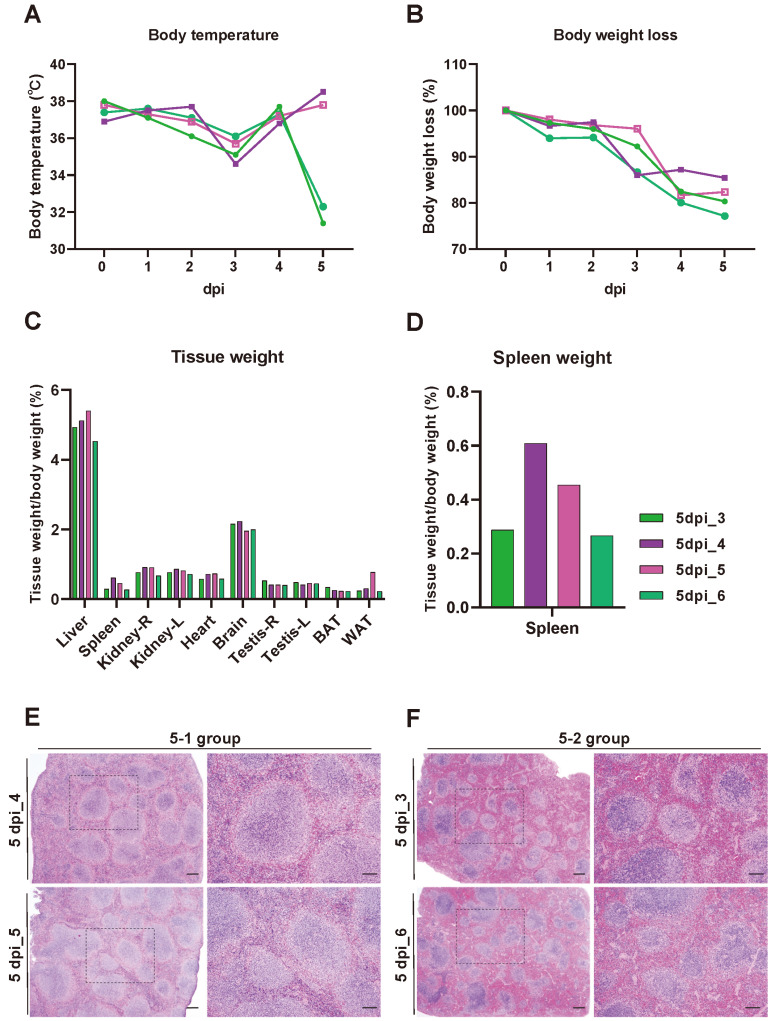
To define the clinical signs after SARS-CoV-2 infection in K18-hACE2 mice, their body weight and temperature were measured daily until 7 dpi. Consistent with previous studies, the body weight of SARS-CoV-2-infected K18-hACE2 mice gradually decreased during infection, and a loss of over 20% of total body weight was observed at 7 dpi compared with that at 0 dpi (Fig. 1A). The body temperature of SARS-CoV-2-infected mice dropped below 30°C after 4 dpi (Fig. 1B), and half of them died within 7 dpi (Fig. 1D). Unlike the loss of body weight, tissue weight normalized with total body weight did not change significantly in SARS-CoV-2-infected mice compared with non-infected control mice (Fig. 1C). Interestingly, SARS-CoV-2 was detected in the lung specimens at 1 dpi when the clinical signs showed no apparent change, and the value of PFU significantly increased at 2 dpi before gradual reduction (Fig. 1E). As per design, non-infected control mice lacked SARS-CoV-2.
Fig. 1. SARS-CoV-2 infected in K18-hACE2 transgenic mice.
(A-D) K18-hACE2 C57BL/6 transgenic mice were mock-infected with PBS (n = 8) or infected with 1.0 × 106 PFU of SARS-CoV-2 (n = 24) by intranasal. Body weight loss (A), body temperature (B), and survival rate (D) were measured at 0 to 7 dpi. Relative tissue weight with body weight in different organs (liver, spleen, kidney, heart, brain, testis, brown-fat adipose tissue [BAT], and white-fat adipose tissue [WAT]) was measured during autopsy (C). (E) Viral titer in lung was determined by plaque assay, PFU/g, at 1, 2, 5, 7 dpi. *P < 0.05.
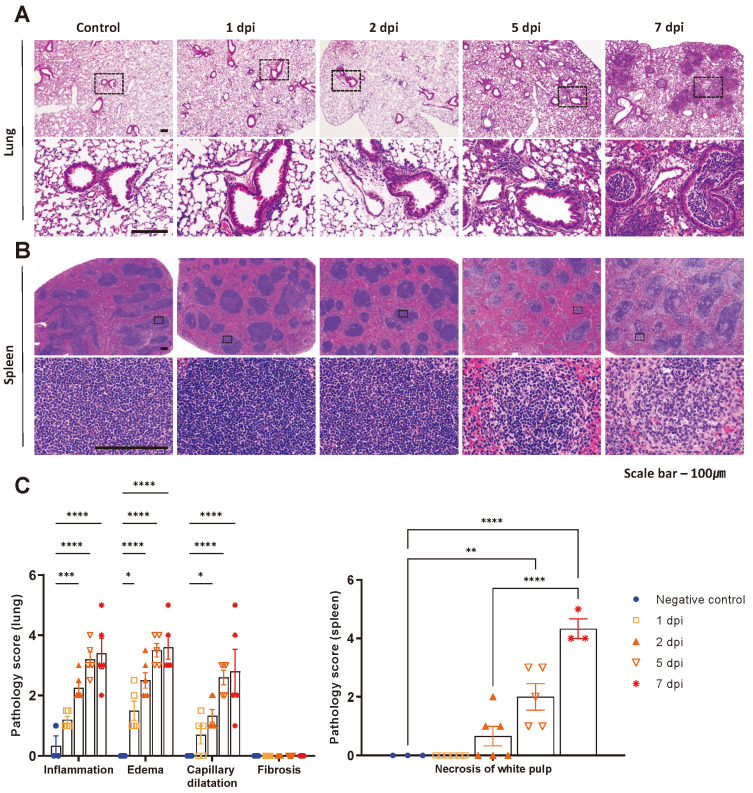
To further evaluate the pathogenesis of SARS-CoV-2 infection in K18-hACE2 mice, the mice were sacrificed at 1, 2, 5, and 7 dpi. We then assessed the pathological score of the lungs, which are highly permissive to SARS-CoV-2, and the spleen, a representative immune organ. As shown in histopathological images, disease progression was distinct in the two organs (Figs. 2A and 2B). In the lungs, several lesions, including alveolar pneumonia, vascular edema, and capillary dilatation developed after SARS-CoV-2 infection (Figs. 2A and 2C). Especially at 5 and 7 dpi, infiltration of mononuclear cells and neutrophils along with thickening of the alveolar septa were prominent. A few of the K18-hACE2 mice that survived the infection until 7 dpi showed severe bronchopneumonia, with the lumen of the bronchioles filled with immune cells (Fig. 2A). In the spleen, white pulp atrophy characterized by decrease in B- and T-cell region was observed during infection. Indeed, numerous lymphocytes exhibited necrotic features characterized by nuclear pyknosis/karyorrhexis at 7 dpi (Figs. 2B and 2C).
Fig. 2. Pathological analysis of lung and spleen in SARS-CoV-2 infected K18-hACE2 mice.
(A and B) H&E stained lungs and spleen sections from mock-infected or 1.0 × 106 PFU SARS-CoV-2 infected mice. Scale bars = 100 μm. (C) To diagnose lung pathology score, inflammation, edema, capillary dilatation, and fibrosis were measured in lung. In spleen, necrosis of white pulp was diagnosed for SARS-CoV-2 derived lesion at 1, 2, 5, and 7 dpi. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Transcriptional profiles associated with early and late responses to SARS-CoV-2 infection in the lungs and spleen
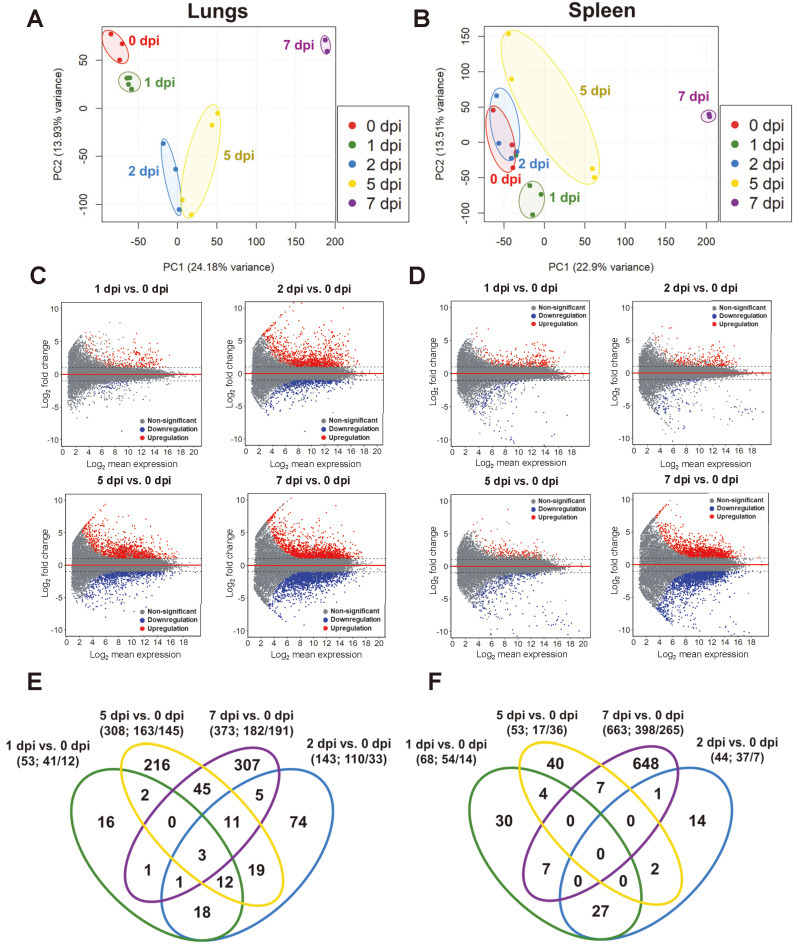
A previous study on K18-hACE2 transgenic mice infected with SARS-CoV-2 reported that the lungs were the most heavily infected organ with numerous consistent and severe histopathological changes (Winkler et al., 2020). RNA-seq analysis of the lung homogenates of K18-hACE2 transgenic mice showed distinct immune signatures associated with the early (2 and 4 dpi) and late (7 dpi) period of the SARS-CoV-2 infection (Winkler et al., 2020). Upregulated genes during the late period of infection demonstrated pronounced enrichment in cytokine-mediated signaling, neutrophil activation, cellular response to type II interferon, and Toll-like receptor signaling (Winkler et al., 2020). Although the spleen is not the most severely infected tissue, moderate levels of viral RNA were observed after the infection (Winkler et al., 2020). In this regard, to assess differences in the transcriptome profile of the lungs and spleen in response to SARS-CoV-2, we performed RNA-seq of lung and spleen homogenates of K18-hACE2 transgenic mice at 0 (mock), 1, 2, 5, and 7 dpi (Figs. 3A and 3B).
Fig. 3. Distinct transcriptional signatures in the lungs and spleen of SARS-CoV-2-infected K18-hACE2 transgenic mice.
(A and B) Principal component analysis plot for RNA-seq data of the lungs and spleen isolated at 0, 1, 2, 5, and 7 dpi; 730 and 780 differentially expressed genes (DEGs) were selected [P < 0.001 (ANOVA) and |fold change| > 2], and normalized TPM (transcripts per million) were used for PCA (principal component analysis). (C and D) MA plots comparing the total number of genes from samples taken on days 1, 2, 5, and 7 versus negative control. Blue indicates genes passing a 5% false discovery rate (FDR) threshold. Red horizontal line of axes marks log2|fold change| = 0. Each dot in the MA plot represents a single gene. (E and F) Venn diagram of overlapping genes identified in differential expression analysis at 0 dpi compared with that at 1, 2, 5, and 7 dpi.
For quality assessment, we evaluated the correlation of our RNA-seq data and previously published data of SARS-CoV-2-infected samples prior to analysis (Supplementary Fig. S2). GSE154104 was retrieved from the GEO (Gene Expression Omnibus) database. Briefly, GSE154104 extracted RNA from lung tissues of transgenic K18-hACE2 mice at 0, 2, 4, and 7 dpi for RNA-seq analysis (Winkler et al., 2020). Our data and GSE154104, which utilized lung homogenates isolated from the same transgenic mouse model, showed a moderate positive correlation (Supplementary Fig. S2). However, neither internal lung data nor GSE154104 was positively correlated with internal spleen data. This discrepancy may have resulted from an organ-specific response to viral infection. We also confirmed the reproducibility of the expression profile in identical gene sets for enriched cytokine-mediated signaling, neutrophil activation, cellular response to type II interferon, and Toll-like receptor signaling utilizing our lung and spleen sample (Supplementary Fig. S3). Expression of gene sets initiated in the early period of infection in lungs and spleen (at 1 and 2 dpi), but expression levels at 7 dpi were quite variable in contrast to a previous study in same transgenic mouse model (Winkler et al., 2020).
Numerous genes were differentially expressed at all time points compared with that in mock-infected animals for both organs, which were most prominent at 7 dpi. DEGs gradually increased in the lung from 2 dpi, though a large number of genes were expressed abruptly at 7 dpi in the spleen (Figs. 3C and 3D). This expression pattern was also observed in principal component analysis, showing that 1, 2, and 5 dpi samples were clustered together in the spleen, whereas clusters in the lungs apparently separated following the passage of time after infection (Figs. 3A and 3B). Direct two-group comparison analysis demonstrated that the expression of 730 DEGs in lungs steadily increased over time as follows: 53 (1 dpi vs 0 dpi), 143 (2 dpi vs 0 dpi), 308 (5 dpi vs 0 dpi), and 373 (7 dpi vs 0 dpi). Significant differences in the number of upregulated and downregulated genes were not observed in the lungs except at 2 dpi (Fig. 3E). At 2 dpi, 110 genes were upregulated, while 33 genes were downregulated. Clinical data showed the highest viral concentration at 2 dpi in the lungs (Fig. 1E). We assumed that prominent differences in the number of upregulated and downregulated genes at 2 dpi might have been associated with immediate host response to incessant viral intrusion in lungs. However, the numbers of genes similarly expressed at 1, 2, and 5 dpi in the spleen and spiked at 7 dpi were 68 (1 dpi vs 0 dpi), 44 (2 dpi vs 0 dpi), 53 (5 dpi vs 0 dpi), and 663 (7 dpi vs 0 dpi) (Fig. 3F). Especially, at 7 dpi, 398 genes were highly expressed, whereas the expression of 265 genes declined. We estimated that the expression pattern at 7 dpi will be a crucial factor to understand the transcriptional profile in the spleen.
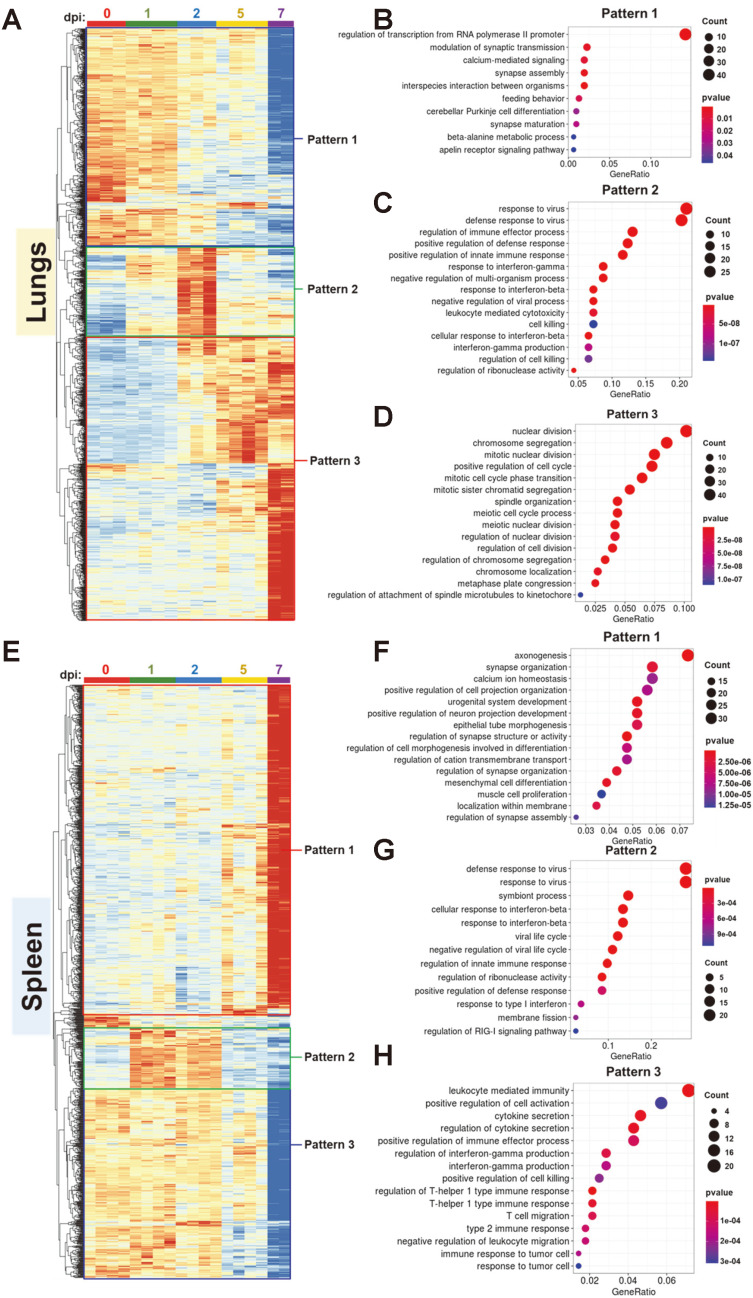
Multicomparison analysis identified 1,294 and 1,167 differentially expressed genes in the lungs and spleen, respectively. Hierarchical clustering revealed distinct expression patterns in the lungs and spleen. In the lungs, three patterns were identified: 1) decrease over time (pattern 1); 2) initial increase during the early period and then decrease when approaching the late period (pattern 2); and 3) gradual increase over time (pattern 3) (Fig. 4A). GO analysis of DEGs showed considerable enrichment of genes associated with regulation of transcription from RNA polymerase II following pattern 1 (Fig. 4B), defense response to virus following pattern 2 (Fig. 4C), and nuclear division following pattern 3 (Fig. 4D). Most downregulated genes involved in transcription regulation (pattern 1) were in the zinc finger protein (ZFP) family. The ZFP family may have inhibited SARS-CoV-2 dissemination by directly neutralizing SARS-CoV-2 effector proteins or assisting host cellular defense through activating type I interferon signaling and several antiviral effector genes (Esposito et al., 2022). Furthermore, a recent study reported that lung epithelial cells showed a significantly decreased ZFP activity in patients with mild and severe SARS-CoV-2 infection when compared to healthy controls (Esposito et al., 2022). Downregulated expression of ZFPs in pattern 1 possibly reflected the severity of infection in the K18-hACE2 transgenic mouse model in late infection. Active and immediate innate immune responses were heightened at 2 dpi followed by gradual alleviation in the lungs, consistent with PFU values of the virus in clinical data (Figs. 1E and 4C). The transcriptional virus concentration was confirmed by mapping reads to the SARS-CoV-2 genome (Supplementary Fig. S4). Furthermore, terms associated with interferon beta and interferon gamma showed significant enrichment, which coincides with the type I and II interferon-stimulated gene signature in the mouse model of SARS-CoV-2 (Israelow et al., 2020; Winkler et al., 2020). Cell proliferation (pattern 3) may be a consequence of the host recovery process after viral perturbation (Fig. 4D). In the spleen, DEGs were also classified into three patterns: 1) significant upregulation at 7 dpi (pattern 1); 2) initial increase during the early period followed by decrease during the late period (pattern 2); and significant downregulation at 7 dpi (pattern 3) (Fig. 4E). Genes involved in axonogenesis, calcium ion homeostasis, regulation of transmembrane cation transport activity, in pattern 1 (Fig. 4F). Although genes that engage in neuron organization represented the highest enrichment, a few genes in this term overlapped with other developmental terms such as urogenital system development or epithelial tube morphogenesis. Thus, we assumed that genes assigned to the term axonogenesis or synapse structural organization should not be interpreted merely as neuronal process in the spleen but as part of the activated developmental process for recovery in the late period of infection. Additionally, the increased expression of genes associated with calcium ion homeostasis or transmembrane cation transport at 7 dpi may be a result of mediation of viral egress and further replication by intracellular calcium/cation channels located in host endolysosomal compartment membranes (Heister and Poston, 2020). Pattern 2 in the spleen included genes related to defense response to virus, cellular response to interferon beta, and regulation of innate immune response (Fig. 4G). The expression pattern and genes were similar to those observed in pattern 2 of the lungs (Figs. 4A-4C); however, upregulation of genes was initiated earlier (1 dpi) and decreased more than that in the lungs in the last period of infection (7 dpi). In other words, in both organs, gene expression associated with immune response increased during the early period (1 and 2 dpi) and decreased during the late period (5 and 7 dpi). In the spleen, genes that sharply decreased at 7 dpi (pattern 3) were also related to a cytokine-driven immune response against the virus. However, terms relevant to interferon beta were observed in pattern 2 while interferon gamma and T-helper type I or II related immunologic terms were only enriched in pattern 3. It seemed that interferon-beta mediated response was heightened in early period and decreased to an extent similar to that in negative control. However, some of the genes related to interferon-gamma response maintained expression until 5 dpi, and others slightly increased at 2 dpi though eventually both dropped sharply at 7 dpi (Figs. 4E and 4H). Substantial downregulation of interferon gamma production at 7 dpi may have contributed to the degree of morbidity in the spleen (Fig. 2C).
Fig. 4. Enriched gene ontology associated with transcriptional response in the lungs and spleen of SARS-CoV-2-infected K18-hACE2 transgenic mice.
(A) Heatmaps from cluster analysis (Euclidean, complete linkage) of 1,294 DEGs in the lungs. DEGs were classified into three expression patterns. Gene expression levels in the heatmaps are z-score normalized values determined from TPM values. (B-D) Gene Ontology (GO) enrichment analysis of genes belonging to each expression pattern in the lungs. GO category representing biological process was analyzed, and the top 15 GO terms were listed after eliminating redundant terms. Dot color and size represent P value and gene ratio (gene counts in specific term/total genes), respectively. (E) Heatmaps from cluster analysis of 1,167 DEGs in the spleen. Three clusters of specific patterns were identified. (F-H) GO enrichment analysis of DEGs in the spleen.
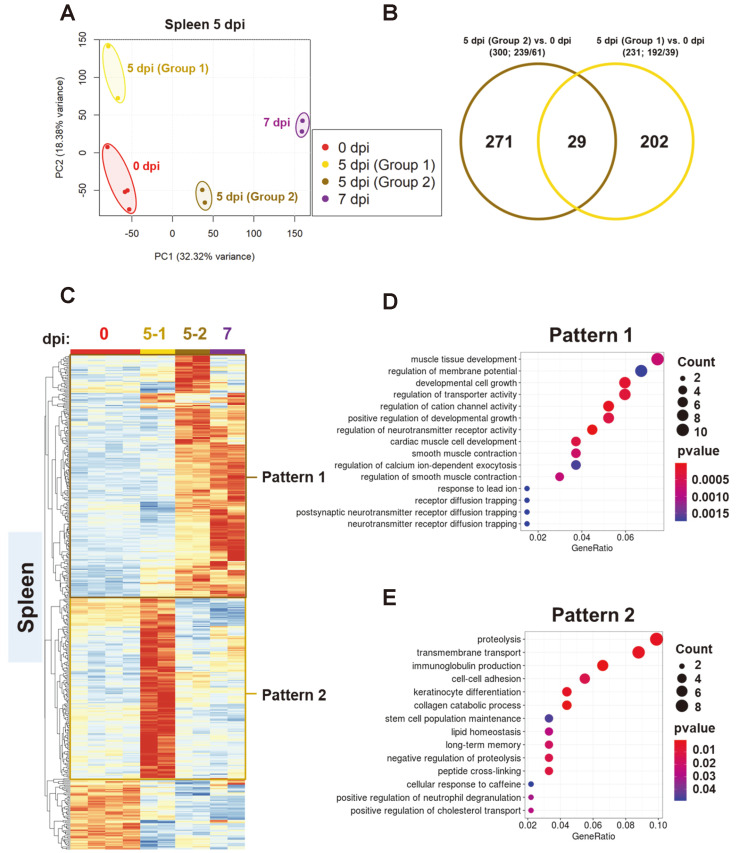
Diverged transcriptional profiles of spleen at 5 dpi provided insights into putative mechanisms contributing to lethality
Infected K18-hACE2 transgenic mice at 5 dpi sacrificed for isolation of spleen were distinguishable as two dissimilar sub-clusters on two dimensional principle component analysis plot. One group of 5 dpi spleen samples (#4, #5) was projected in the vicinity of the negative control, and the other group (#3, #6) was close to the 7 dpi spleen sample (Fig. 5A). In combination with the clinical overview of infected mice, we decided to designate those groups as 5-1 and 5-2 group (Fig. 5A). Statistical comparison of each 5 dpi spleen group and negative control revealed 231 and 300 differentially expressed genes. In both groups, most DEGs were more likely to be upregulated than downregulated; 192 (5-1 vs 0 dpi) and 239 (5-2 vs 0 dpi) (Fig. 5B). We identified group-specific patterns in corresponding groups through hierarchical clustering analysis utilizing 449 DEGs from multicomparison analysis (Fig. 5C). Subsequent GO analysis revealed that DEGs showing elevated expression in the 5-2 group (pattern 1) are related to developmental cell growth and the regulation of transporter activity (Fig. 5D), while highly expressed genes in the 5-1 group (pattern 2) are associated with immunoglobulin production in addition to proteolysis (Fig. 5E). Recurrent emergence of transporter activity and development-related terms may have been triggered by an identical mechanism showing that the 5-2 group is clustered in vicinity with the 7 dpi group in the spleen, as observed in the increased pattern in the late period of spleen infection (pattern 1) (Figs. 4F and 5D). Interestingly, sub-analysis of the negative control and two groups at 5 dpi indicated that proteolysis is upregulated in the 5-1 group, though expression level decreased at 7 dpi (pattern 2) (Fig. 5E). Given that the efficient host cell entry of SARS-CoV-2 is mediated by proteolytic activity of proteases such as TMPRSS2 (Harrison et al., 2020), upregulation of these terms may result from sustained infection and viral life cycle. We assumed that substantial upregulation of proteolysis or transport activity in the late period of the 5-1 group may be an evident consequence of consecutive viral entry or replication assisting further dissemination of virus. Definite attempts to struggle against incessant virus intrusion were also detected in 5-1 group. Upregulated immunoglobulin production, presumably implying activated somatic hypermutation, occurs for affinity maturation, and such humoral immunity may have reinforced the defense of the 5-1 group (pattern 2) (Fig. 5E). Indeed, negative regulation of proteolysis was also enriched in GO term analysis in pattern 2 (Fig. 5E). Later, we thoroughly compared transcriptome profile and clinical manifestation of these groups. The body temperature of all samples was gradually dropping to 3 dpi, and samples #3 and #6 increased slightly after 3 dpi, and then showed a sudden drop in body temperature at 5 dpi. However, samples #4 and #5 returned to normal body temperature (Fig. 6A). Body weight loss was similarly reduced in all samples (Fig. 6B). Although no significant difference was observed in each relative organ weight, the relative spleen weights of samples #4 and #5 were heavier than samples #3 and #6. (Figs. 6C and 6D). In perspective of histopathology, aggravate lesion resulted from disease progression was detected as extreme atrophy and increased necrosis of white pulp in sample #3 and #6 (Figs. 6E and 6F). Combining all of these results, we supposed that 5-1 group might be non-lethal group and could have endured infection and survived longer. Yet, 5-2 group might be more lethal than 5-1 group and could have succumbed to infection and dead eventually.
Fig. 5. Dinstinct the transcriptional profiles of the two spleen groups 5 days post SARS-CoV-2 infection in K18-hACE2 transgenic mice.
(A and B) Principal component analysis (PCA) plot for RNA-seq data of total number of genes in the spleen isolated at 0, 5-1 (non-fatal group), 5-2 (fatal group), and 7 dpi; 231 and 300 differentially expressed genes (DEGs) were selected [P < 0.005 (ANOVA) and |fold change| > 2] and normalized TPM (transcripts per million) were used for PCA. (C) Heatmaps from cluster analysis (Euclidean, complete linkage) of 449 DEGs in samples from the two spleen groups 5 dpi compared with that at 0 dpi. DEGs were classified into two expression patterns. Gene expression levels in the heatmaps are z-score normalized values determined from normalized TPM values. (D and E) Gene ontology (GO) enrichment analysis of genes belonging to each expression pattern in the spleen. GO category representing a biological process was analyzed, and the top 15 GO terms were listed after eliminating redundant terms. Dot color and size represent P value and gene ratio (gene counts in specific term/total genes), respectively.
Fig. 6. Differences between 5-1 group (#4 and #5) and 5-2 group (#3 and #6) that identified by clinical characteristics and spleen pathology.
(A-D) Analysis of clinical characteristics in SARS-CoV-2 infected K18-hACE2 mice that sacrificed at 5 dpi. Note the body temperature change (A) and body weight loss (B) at 0-7 dpi. Body temperature of 5-1 group was recovered as normal status after 3 dpi. Note the relative tissue weight with body weight (C), and relative spleen weight with body weight (D). BAT, brown-fat adipose tissue; WAT, white-fat adipose tissue. (E and F) Spleen histopathology in SARS-CoV-2 infected K18-hACE2 mice that sacrificed at 5 dpi. Compared with 5-1 group (E), the spleen weight of #3 and #6 was relatively reduced. Severe atrophy and necrosis of white pulp were increased in 5-2 group (F). Scale bars = 200 μm and 100 μm (inset).
Changes in immune cells in lungs and spleen induced by SARS-CoV-2 infection
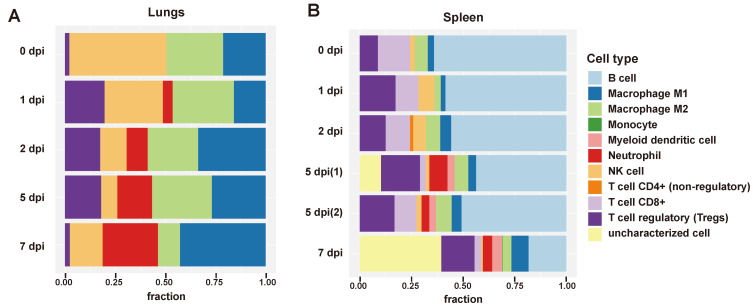
To investigate relative abundance and composition of specific immune cell types in the lungs and spleen of SARS-CoV-2-infected K18-hACE2 transgenic mice, we employed an integrated in silico approach that combines seven state-of-the-art computational methods for deconvolution of cell type fractions from bulk RNA-seq (Sturm et al., 2020). In the lungs, an increase in the number of neutrophils in the infected mice indicated progression of viral infection. While the fraction of neutrophils increased at 7 dpi, proportion of regulatory T cells decreased. A previous review suggested that defective function of regulatory T cells can be associated with exaggerated neutrophil activity (Okeke and Uzonna, 2019). Pro-inflammatory M1 macrophages fractions were prominent at the height of viral infection (2 dpi) in the lungs, while pro-healing M2 macrophages decreased close to death (7 dpi), suggesting a quantitative balance of polarized macrophages in regulated assimilation with the infection (Fig. 7A). In the spleen, the largest secondary lymphoid organ that is involved in hematopoiesis and red blood cell clearance, B cell and CD8+ T cell population is notably higher than that in the lungs. Similar to that in the lungs, inflammatory activity is a characteristic function of NK cells, neutrophils, and macrophages in the spleen. Corresponding to experimental data, uncharacterized elevation of cell portion in the spleen as exacerbation of pathogenesis is accompanied by significant necrosis of lymphocytes during the late period of infection (Figs. 2B and 2C, Fig. 7B). In the two spleen sample groups at 5 dpi, activated immune response was shown as increased proportion of neutrophils in 5-1 group. Furthermore, a decreased portion of CD8+ T cells in the 5-1 group could have been driven by persistent stimulation of T cells following extended duration of infection (Rha and Shin, 2021). Except for those two cell types, no pronounced difference in immune cell composition and fraction was observed between the two distinct groups of 5 dpi spleen tissue samples (Fig. 7B).
Fig. 7. Comprehensive evaluation of transcriptome-based immune cell type quantification in SARS-CoV-2-infected lungs and spleen of K18-hACE2 transgenic mice.
(A) Immune deconvolution results utilizing gene signatures from bulk RNA-seq data known to be present in a given tissue to identify immune cell composition in the lungs. Bars are arranged in a longitudinal fashion, each indicating 0, 1, 2, 5, and 7 days post viral infection. Length of the colored bar represents the relative fraction of specific immune cell type. Normalized TPM was used for immune cell deconvolution analysis. (B) Immune cell deconvolution results of the spleen samples. Five days post infection of the spleen is split into 5-1 and 5-2 groups.
DISCUSSION
In this study, we performed a comprehensive comparison between the lungs and spleen of SARS-CoV-2-infected K18-hACE2 mice that simulate the severity of COVID-19 in human patients.
We identified that SARS-CoV-2 infection in K18-hACE2- transgenic mice causes severe pulmonary disease and lesions in the spleen. Following intranasal SARS-CoV-2 inoculation, infected transgenic mice rapidly lost weight after 4 dpi and finally succumbed to the disease at 7 dpi. Body temperature dropped abruptly after 4 dpi, confirming definite signs of reaching a moribund stage. Substantial amount of viral RNA was detected at 1 dpi, which peaked at 2 dpi followed by gradual decline until 7 dpi, using the plaque assay. Histopathology of the lungs corroborated severe infection with evident inflammation and vasodilation caused by leukocyte infiltration. The spleen showed lesions in the white pulp characterized by a decrease in the cellularity and reduction in the area of lymphocytes.
Transcriptome analyses of the lung and spleen homogenates revealed obvious evidence of active innate immune response against the virus. Upon viral intrusion, immune response along with active transcriptional regulation of chemokines was most intense at 2 dpi in the lungs and gradually abated, reaching a moderate level in the late period of infection. Defense mechanism of the spleen initiated from 1 dpi, much earlier than that of the lungs. Expression of cytokines and inflammatory features of the spleen were restored to basal level or even lower at 7 dpi. Genes involved in the operation of the antiviral defense system in both organs include interferon-stimulated genes such as Oas1a, Oas1b, Oas1g, Oas2, Oas3, Oasl1, Mx1, Mx2, Ifit1, Ifit1bl1, Ifit1bl2, Ifit3b, Cxcl10, Isg15, Rsad2, Ddx60, and Zbp1. Recent studies also emphasized the selective modulation of these genes and their contribution to the severity of infection (McClain et al., 2021; Pairo-Castineira et al., 2021; Park, 2021). In addition to these reproducible results, the novelty of transcriptome analysis in this study lies in the longitudinal transcriptome signature of the spleen. Especially, the enriched pathways relevant to interferon beta and gamma present a distinctive expression pattern in the spleen. A substantial decrease in interferon gamma production could have exacerbated pathological phenotypes of viral infection at 7 dpi. Nevertheless, we cannot ignore that prolonged expression of interferon gamma could have also affected activation and maintenance of the subsequent immune response. For instance, responsiveness to interferon gamma is important for the differentiation and development of helper T cell function (Tau et al., 2000), and terms associated with helper T cell were also observed in enrichment analysis (pattern 3). This result implies that the spleen performs a much more elaborate modulation of interferon over time, which requires further research. In the spleen, cation and transporter-associated terms were especially increased in the late period of infection compared to that in the lungs. This warrants further investigation on whether this phenomenon is a consequence of viral dissemination in the host cellular environment or lymphocytes stimulation (Badou et al., 2005).
In addition to the immune response, both organs showed organ-specific enrichment response and immune cell type profile. In a time-dependent manner, innate immune cells such as neutrophils invigorated the inflammatory response in the lungs, which triggered cell proliferation for recovery. This observation is noteworthy considering the fact that a previous study suggested that neutrophils also contribute to the symptoms observed in COVID-19 patients, as demonstrated by CXCL2 and CXCL8 induction (Blanco-Melo et al., 2020). The spleen, on the contrary, seems to be different from the lungs due to the intervention of adaptive immune response by lymphocytes together with the innate immune response. The ongoing antiviral activity may have forced a proportion of B cells to become ultimately exhausted owing to the sustained infection. Cooperation of distinctive immune cells may have been a critical factor to determine severity of illness in both organs. However, this result should be interpreted with caution because deconvolution results only offer a relative snapshot of composition, and in silico results can be inaccurate as the deconvolution tool was initially designed for human tumor samples rather than those from mice.
Especially at 5 days post viral infection, the spleen showed noticeable differences between the 5-1 and 5-2 groups. We hypothesized that persistent developmental process could have occurred throughout entire period in the 5-2 group, which implicates body struggled to restore severely devastated tissue. Most of identified genes are known to be engaged in activating mitogenic responses and regulating proliferation in cardiac muscle or neuron; Adra1b, Auts2, Cacng7, Cpne9, Mapt, and Slit1. But the others were also known to be associated with vascular contraction or dilation; Prkg1, and Edn1, which have possibly been stimulated by chemokines released in immune process. However, there are some pitfalls in complete interpretation since we sacrificed mice precisely at 5 dpi, thus we could not trace and confirm changes in expression levels of genes assigned to enriched pathway in the later period. Furthermore, only few genes were enriched. In an effort to eradicate fundamental culprit of sustained infection, 5-1 group tried to impede viral entry itself. Genes included in the negative regulation of proteolysis of 5-1 group, namely Stfa1, Stfa2, and Stfa3 are known to encode stefins. Previous research reported that stefins act as fast and tight binding inhibitors of endopeptidases cathepsin L and papain (Mihelic et al., 2006). Peptidases such as cathepsin L are already known to have increased levels in SARS-CoV-2-infected patients and are thus discussed as a promising target for new anti-COVID-19 drug development (Zhao et al., 2021). The 5-1 group was also characterized by humoral immune response resulting from clonal expansion of lymphocytes. Previous reports showed that COVID-19 patients at different stages of recovery demonstrated various preferences for B cell receptor genes (Wen et al., 2020). Genes such as Igkv4-92, Igkv1-35, Igkv8-18, Igkv8-26, Igkv18-36, and Igkv4-54 were involved in this pathway, and these isotypes can be utilized as potential pharmaceutical targets.
Even though this study offered insights into physiological and pathological response against viral infection in an environment resembling that for hospitalized humans, further investigation is required. Some significant genes might have been overlooked in this study because they lack expression or statistical significance to pass the threshold for DEGs. In addition, since reliable quantification of ACE2 expression is impossible due to the non-native K18 promoter, a similar study scheme might have to be conducted in mouse model expressing hACE2 under a different type of promoter (Winkler et al., 2020).
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This study was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIT) (2020M3A9D5A01082439 and 2018R1A5A2025079, to H.Y.G., 2016M3A9D5A01952416 to K.T.N., and Bio & Medical Technology Development Program 2021M3H9A1038083 to K.T.N.).
Footnotes
AUTHOR CONTRIBUTIONS
J.-Y.S., K.T.N., and J.K.S. conceived and designed experiments and supervised the research. J.A.K. and H.Y.G. analyzed RNA sequencing data. S.-H.K., J.S.S., H.N., H.J., J.K., D.J., J.J.K., D.O., S.Y., S.G.L., Y.W.L., H.J.J., I.H.P., J.O., S.-H.S., Y.J.L., S.-M.H., S-H.A., J.-Y.B., J.C., S.Y.K., Y.B.K., J.-Y.H., H.-J.L., H.B.K., D.G.J., D.S., M.S., M.-S.P., K.-S.C., J.W.P., J.-W.Y., J.-S.S., and H.-Y.L. performed experiments and analyzed the data. J.A.K., S.-H.K., and H.Y.G. wrote the original draft of the manuscript. J.-Y.S., K.T.N., H.Y.G., and J.K.S. reviewed and edited the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza A respiratory viruses. Front. Immunol. 2020;11:552909. doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badou A., Basavappa S., Desai R., Peng Y.Q., Matza D., Mehal W.Z., Kaczmarek L.K., Boulpaep E.L., Flavell R.A. Requirement of voltage-gated calcium channel beta4 subunit for T lymphocyte functions. Science. 2005;307:117–121. doi: 10.1126/science.1100582. [DOI] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary S.J., Pitchford S.C., Amison R.T., Carrington R., Robaina Cabrera C.L., Magnen M., Looney M.R., Gray E., Page C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020;177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., D'Abrosca G., Antolak A., Pedone P.V., Isernia C., Malgieri G. Host and viral zinc-finger proteins in COVID-19. Int. J. Mol. Sci. 2022;23:3711. doi: 10.3390/ijms23073711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heister P.M., Poston R.N. Pharmacological hypothesis: TPC2 antagonist tetrandrine as a potential therapeutic agent for COVID-19. Pharmacol. Res. Perspect. 2020;8:e00653. doi: 10.1002/prp2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Song E., Mao T., Lu P., Meir A., Liu F., Alfajaro M.M., Wei J., Dong H., Homer R.J., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217:e20201241. doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirtipal N., Bharadwaj S., Kang S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. 2020;85:104502. doi: 10.1016/j.meegid.2020.104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain M.T., Constantine F.J., Henao R., Liu Y., Tsalik E.L., Burke T.W., Steinbrink J.M., Petzold E., Nicholson B.P., Rolfe R., et al. Dysregulated transcriptional responses to SARS-CoV-2 in the periphery. Nat. Commun. 2021;12:1079. doi: 10.1038/s41467-021-21289-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Jr., Wohlford-Lenane C., Jr., Hickey M., Jr., Manzel L., Jr., Shi L., Jr., Netland J., Jr., Jia H.P., Jr., Halabi C., Jr., Sigmund C.D., Jr., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihelic M., Teuscher C., Turk V., Turk D. Mouse stefins A1 and A2 (Stfa1 and Stfa2) differentiate between papain-like endo- and exopeptidases. FEBS Lett. 2006;580:4195–4199. doi: 10.1016/j.febslet.2006.06.076. [DOI] [PubMed] [Google Scholar]
- Mohamadian M., Chiti H., Shoghli A., Biglari S., Parsamanesh N., Esmaeilzadeh A. COVID-19: virology, biology and novel laboratory diagnosis. J. Gene Med. 2021;23:e3303. doi: 10.1002/jgm.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau G.B., Burgess S.L., Sturek J.M., Donlan A.N., Petri W.A., Mann B.J. Evaluation of K18-hACE2 mice as a model of SARS-CoV-2 infection. Am. J. Trop. Med. Hyg. 2020;103:1215–1219. doi: 10.4269/ajtmh.20-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeke E.B., Uzonna J.E. The pivotal role of regulatory T cells in the regulation of innate immune cells. Front. Immunol. 2019;10:680. doi: 10.3389/fimmu.2019.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladunni F.S., Park J.G., Pino P.A., Gonzalez O., Akhter A., Allué-Guardia A., Olmo-Fontánez A., Gautam S., Garcia-Vilanova A., Ye C., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591:92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- Park S.H. An impaired inflammatory and innate immune response in COVID-19. Mol. Cells. 2021;44:384–391. doi: 10.14348/molcells.2021.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Yuen K.Y., Osterhaus A.D., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Qinfen Z., Jinming C., Xiaojun H., Huanying Z., Jicheng H., Ling F., Kunpeng L., Jingqiang Z. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73:332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rha M.S., Shin E.C. Activation or exhaustion of CD8(+) T cells in patients with COVID-19. Cell. Mol. Immunol. 2021;18:2325–2333. doi: 10.1038/s41423-021-00750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm G., Finotello F., List M. Immunedeconv: an R package for unified access to computational methods for estimating immune cell fractions from bulk RNA-sequencing data. Methods Mol. Biol. 2020;2120:223–232. doi: 10.1007/978-1-0716-0327-7_16. [DOI] [PubMed] [Google Scholar]
- Tau G.Z., von der Weid T., Lu B., Cowan S., Kvatyuk M., Pernis A., Cattoretti G., Braunstein N.S., Coffman R.L., Rothman P.B. Interferon gamma signaling alters the function of T helper type 1 cells. J. Exp. Med. 2000;192:977–986. doi: 10.1084/jem.192.7.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.W., Wu X.X., Jiang X.G., Xu K.J., Ying L.J., Ma C.L., Li S.B., Wang H.Y., Zhang S., Gao H.N., et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Port J.R., Bushmaker T., Offei Owusu I., Purushotham J.N., Avanzato V.A., Fischer R.J., Schulz J.E., Holbrook M.G., Hebner M.J., et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021;17:e1009195. doi: 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M.M., Yang W.L., Yang F.Y., Zhang L., Huang W.J., Hou W., Fan C.F., Jin R.H., Feng Y.M., Wang Y.C., et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target. Ther. 2021;6:134. doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.S., Wang Q., Min J., Shen X.R., Li Q., Zhao Q.C., Wang X., Jiang R.D., Geng R., Chen Y., et al. Single-cell landscape of lungs reveals key role of neutrophil-mediated immunopathology during lethal SARS-CoV-2 infection. J. Virol. 2022;96:e0003822. doi: 10.1128/jvi.00038-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.