Abstract
Liver cancer has a high prevalence, with majority of the cases presenting as hepatocellular carcinoma (HCC). The prognosis of metastatic HCC has hardly improved over the past decade, highlighting the necessity for liver cancer research. Studies have reported the ability of the KiSS1 gene to inhibit the growth or metastasis of liver cancer, but contradictory research results are also emerging. We, therefore, sought to investigate the effects of KiSS1 on growth and migration in human HCC cells. HepG2 human HCC cells were infected with lentivirus particles containing KiSS1. The overexpression of KiSS1 resulted in an increased proliferation rate of HCC cells. Quantitative polymerase chain reaction and immunoblotting revealed increased Akt activity, and downregulation of the G1/S phase cell cycle inhibitors. A significant increase in tumor spheroid formation with upregulation of β-catenin and CD133 was also observed. KiSS1 overexpression promoted the migratory, invasive ability, and metastatic capacity of the hepatocarcinoma cell line, and these effects were associated with changes in the expressions of epithelial mesenchymal transition (EMT)-related genes such as E-cadherin, N-cadherin, and slug. KiSS1 overexpression also resulted in dramatically increased tumor growth in the xenograft mouse model, and upregulation of proliferating cell nuclear antigen (PCNA) and Ki-67 in the HCC tumors. Furthermore, KiSS1 increased the angiogenic capacity by upregulation of the vascular endothelial growth factor A (VEGF-A) and CD31. Based on these observations, we infer that KiSS1 not only induces HCC proliferation, but also increases the metastatic potential by increasing the migratory ability and angiogenic capacity.
Keywords: angiogenesis, epithelial mesenchymal transition, hepatocellular carcinoma, KiSS1, kisspeptin, metastasis
INTRODUCTION
Liver cancer is highly prevalent among the malignancies, the most common being hepatocellular carcinoma (HCC), which accounts for more than 90% of all liver cancer cases (Global Burden of Disease Liver Cancer Collaboration et al., 2017; Llovet et al., 2021). Little improvement in the prognosis of liver cancer patients over the past decade highlights the necessity for liver cancer research (Dhanasekaran et al., 2016). HCC mainly metastasizes to the lungs, lymph nodes, adrenal glands, and bones, and metastatic HCC usually has a poor prognosis (Colecchia et al., 2014; Kummar and Shafi, 2003). In general, HCC metastasis begins with epithelial mesenchymal transition (EMT) (Ogunwobi et al., 2019). Hepatic epithelial cells, which were adherent to the basement membrane, lose their cell adhesion capability and migrate to remote locations (Brabletz et al., 2018; Lamouille et al., 2014). These morphological changes enable the successful migration of HCC cells, promoting dissemination and spread throughout the body (Ogunwobi et al., 2019). Transcription factors that promote EMT, including N-cadherin, Snail, and Slug, are also upregulated during HCC progression (Puisieux et al., 2014; Tiwari et al., 2012). Between hepatic tumor cells and non-tumor stroma, there exists a tumor microenvironment called the extracellular matrix (ECM) (Delire et al., 2018). HCC undergoes intravasation, a process that invades the ECM or vascular endothelial cells, and penetrates blood vessels or lymphatic vessels to spread to other tissues (Kim et al., 2016b; Yang et al., 2011). After extravasation (the process of escaping from within the blood vessels), HCC initiates colonization for tumor growth at a new metastatic site (Rathod et al., 2000). Blood supply to HCC tumors is provided through angiogenesis, which further enhances growth formation and metastasis (Lu et al., 2015). Even after surgical removal at an early stage to prevent HCC metastasis, HCCs have a high recurrence rate due to micrometastasis. Hence, controlling the metastasis of HCC is considered one of the main treatment targets (Imamura et al., 2003).
The KiSS1 gene encodes the kisspeptin protein (also known as metastin), a biologically active peptide (Clarke et al., 2015). Kisspeptin is a peptide ligand of the G-protein coupled receptor (GPR54), which is a carboxy-terminal amidated peptide with 54 amino acid residues (Ohtaki et al., 2001). In studies using melanoma or breast cancer models, this gene was first described as inhibiting metastasis without affecting tumorigenesis (Lee and Welch, 1997a; 1997b; Lee et al., 1996). Since then, various roles of KiSS1 have been elucidated, including regulation of reproductive hormones and kidney function (Clarke et al., 2015; Kauffman, 2009; Oakley et al., 2009; Terasawa et al., 2013).
Although numerous studies reported that KiSS1 inhibits cancer progression and metastasis, it was also shown that this gene may play a dual role as a tumor suppressor or oncogene in the progression of liver cancer. It was further reported that KiSS1 reduces the cell motility and invasiveness and inhibits metastasis by downregulating the expression of MMP-9 (matrix metalloproteinase-9) in HCC (Shengbing et al., 2009), thereby suggesting a tumor suppressor role against HCC. In contrast, emerging studies indicate that KiSS1 may play a role as a promoter in liver cancer. Ikeguchi et al. (2003) first reported about 2-fold upregulation in the mRNA level of KiSS1 in HCC tissues derived from liver cancer patients, as compared to non-cancerous liver. This overexpression of the KiSS1 gene had a statistically significant and positive correlation with the HCC progression and poor prognosis (Ikeguchi et al., 2003). In the same study, it was suggested that KiSS1 expression promotes the acquisition of a more aggressive tumor phenotype of HCC (Ikeguchi et al., 2003). Consistent with these findings, other researchers observed high KiSS1 expression levels in liver cancer patients who had undergone liver transplantation (Schmid et al., 2007). They suggested that these high levels of KiSS1 expression could be represented as an independent prognostic marker of disease-free (Schmid et al., 2007).
Despite these efforts to study the effect of KiSS1 on the development of liver cancer, its role remains unclear. This is because most studies on the role of KiSS1 in liver cancer are still at the initial stage of analyzing the correlation between the gene expression and prognosis, in samples derived from liver cancer patients. It is yet to be ascertained whether KiSS1 induces functional changes by mediating the expression of certain genes in liver cancer. To clarify this point, it is necessary to observe functional traits (such as altered phenotypes) by regulating the expression of KiSS1 in HCC, and to investigate the molecular mechanisms involved. This report focuses on and investigates the effects of KiSS1 overexpression on HCC cell proliferation and liver tumor growth, and its effects on migration and angiogenesis in relation to metastasis.
MATERIALS AND METHODS
Cell lines and culture
HepG2 human liver HCC cell line was purchased from Korean Cell Line Bank (KCLB; Korea). 293T cells line was purchased from American Type Culture Collection (ATCC; USA). These cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat inactivated fetal bovine serum (FBS; Biowest, France), 1% antibiotic-antimycotic solution (Welgene, Korea) in high relative humidity (95%), and controlled CO2 level (5%). Human umbilical vein endothelial cells (HUVECs) were purchased from PromoCell (Germany) and cultured in endothelial cell growth medium (PromoCell). When the cells reached about 70%-80% of cell density, the cell were suspended with 0.1% Trypsin-EDTA (Invitrogen Life Technologies, USA) for 3 min at 37°C, then plated on cell culture dish (SPL Life Sciences, Korea).
Establishment of stable cell line overexpressing KiSS1 gene
Transfection of the KiSS1 gene into 293T cells was conducted according to the manufacturer’s manual (Origene, USA). Briefly, for transformation, 293T cells were stably packaged with lentviral vector pLenti-C-mGFP-P2A-Puro (Origene) containing KiSS1 DNA (Origene). The lentivirus-containing medium was collected from 293T packaging cell line, the medium was filtrated with 0.22 µm low protein binding filter (Millipore, Germany). At the half-confluent of HepG2 cells then, the lentivirus-containing medium transferred to HepG2 cells (50%-60% confluency) with 10 µg/ml hexadimethrine bromide (Sigma-Aldrich, USA). After infection for 18 h, the lentivirus-containing medium was replaced with fresh cell culture medium and further incubated for 48 h. Transfectants were selected with antibiotics free-medium containing 10 µg/ml puromycin (Sigma-Aldrich) for 48 h, and was repeated 3 times. In this study, the KiSS1-overexpressing HepG2 cell line gene was named HepG2-KiSS1 cells. For vector control group, mock transfection was performed without adding only KiSS1 cDNA clone.
Cell counting
The cells were seeded at a cell density of 1 × 105 cells/well in 6-well plates (SPL Life Sciences) and incubated at 37°C in a humidified atmosphere of 5% CO2 containing air. After 96 h, the number of cells was measured using EVETM Automatic Cell Counter (NanoEntek, Korea).
Tumor spheroid formation
The cells were seeded at a cell density of 2 × 103 cells/well with 200 µl of cell culture medium in ultra-low attachment plate (96-well type, round bottom clear; Corning, USA). After 24, 74, and 168 h, the tumor spheroids were photographed using a phase-contrast microscope (Olympus, Japan). The area of tumor spheroids was quantified using cellSens Dimension software (ver. 1.18; Olympus) by drawing the edge of the tumor spheroid and calculating the area.
Transwell migration assay
Transwell migration assay was used as described in a previous studies with slight modifications in order to investigate the migratory ability of the cells (Kim et al., 2017a; 2017b). To avoid changes in the cell number, the cells were treated with 12.5 µg/ml of Mitomycin C (MMC) for 2 h. After incubating with 10 µg/ml of Hoechst 33342 solution for nuclei staining of living cells, the cells were seeded at a density of 2 × 104 cells/well in the upper chamber of a transwell insert (BD Biosciences, USA) in a total of 200 µl of cell culture media. The lower chamber was added with 500 µl of cell culture media. The cells were then incubated in 37°C and humidified CO2 incubator for 72 h. The cells that migrated from the upper chamber to the bottom chamber were photographed using a fluorescence microscope (Olympus). The migrated cell numbers were quantified by cellSens Dimension software.
Transwell invasion assay
Transwell invasion assay was used as described in a previous studies with slight modifications in order to determine the invasive ability of the cells (Kim et al., 2017a; 2017b). After the cells were treated with 12.5 µg/ml MMC, then stained with 10 µg/ml of Hoechst 33342 for 10 min. After 50 µl of 250 µg/ml fibronectin coating of the bottom of a transwell, the cells were seeded at a density of 2 × 104 cells/well in the upper chamber of a transwell insert in a total of 200 µl of cell culture media. The cells that migrated from the upper chamber to the bottom chamber were photographed using a fluorescence microscope. The invaded cell numbers were quantified by cellSens Dimension software.
Collagen invasion assay
In order to investigate the ability of cancer cells to invade the ECM, a collagen invasion assay was conducted with slight modifications based on a previous study (Miyazaki et al., 2019). MMC-treated cells were seeded at a cell density of 5 × 103 cells/well in ultra-low attachment plate and tumor spheroids were formed for 72 h. For preparation of collagen-coated plate, collagen derived from bovine dermis (KOKEN, Japan) was mixed with cell culture media at a ratio of 2:1, and 50 µl of collagen mixture placed in a round bottom 96-well plate (SPL Life Sciences) was solidified in 37°C incubator for 2 h. One spheroid per well was transferred to 96-well plate coated with collagen, and 200 µl of cell culture medium was added. Tumor spheroids, including cells invading the surrounding collagen, were observed under a phase-contrast microscope every 24 h and for up to 96 h. Area of tumor spheroids was quantified by cellSens Dimension software, and calculated the area minus the area of the initial tumor spheroid.
In vitro metastasis assay
To evaluate the metastatic colonization of the cells, we used the in vitro metastasis assay we developed. After coating the outer bottom surface of the 96-well type transwell insert with 12.5 µg/50 µl of fibronectin, the transwell inserts were combined on ultra-low attachment plate. MMC-treated cells were seeded in the upper chamber of the transwell at a density of 2 × 104 cells/well, and culture medium was added to the bottom chamber. To evaluate the ability of cancer cells to form colonies in the bottom chamber after invasion, the spheroids formed in the bottom chamber were observed under a phase-contrast microscope every 24 h and for up to 96 h. The metastasized area was quantified by cellSens Dimension software.
Tube formation assay
In order to investigate the angiogenic capacity of the cells, tube formation assay was used using immortalized HUVECs. Based on our previous study (Kim et al., 2016a; 2017a), primary HUVECs were immortalized by simian virus 40 (SV40) large T antigen in the second passage, and an SV40-HUVEC line was established. MMC-treated HepG2 cells (vector control) or HepG2-KiSS1 cells were seeded in 6-well plate (SPL Life Sciences) at a cell density of 5 × 105 cells/well, and the cell culture medium was replaced with an endothelial cell growth medium after 24 h of cell adhesion. After 24 h of incubation with the medium, the conditioning medium was harvested. After fluorescent staining SV40-HUVECs with 2 µg/ml Calcein-AM (Invitrogen Life Technologies), the cells were seeded at a density of 7.5 × 104 cells/100 µl/well with the cancer cell culture medium in the Matrigel (Corning) coated 24-well plates (Corning). After 3 h, SV40-HUVECs formed tubes, and we observed it under a fluorescence microscope.
Establishment of xenograft mouse model
Five-week-old athymic nude mice were purchased from KOATECH (Korea), and maintained at the Laboratory Animal Research Center of Chungbuk National University under specific pathogen-free conditions. The mice were housed in a temperature- and humidity-controlled environment under a 12-h light-dark cycle, and all animal experiments were performed during the light cycle. All animal experiments were approved by Institutional Animal Care and Use Committee (IACUC) at Chungbuk National University (CBNUA-1565-21-01). After a one-week acclimatization, HepG2 cells (vector control) or HepG2-KiSS1 cells were inoculated subcutaneously in right upper flank of mice (3 × 106 cells/mouse; n = 5 per group). Tumor volume was measured using a digital caliper (CD-15APX; Mitutoyo Korea, Korea) every 2-3 days from 3 days after the inoculation, and calculated by shortest diameter2 × longest diameter × 0.5236 (mm3). Body weight of mice was also measured on the same day. On day 26 post-inoculation, all mice were sacrificed, and tumor were isolated and weighed. Lung, liver, spleen, and kidney of all mice were weighed.
Immunohistochemistry (IHC) and H&E stain
IHC and was performed based on our previous studies, with some modifications (Go et al., 2017; Lee et al., 2021). After the formalin-fixed tumor tissues were embedded in paraffin and cut into 4 mm sections, the tissue sections were deparaffinized in xylene (OCI, Korea) and rehydrated according to the ethanol (OCI) concentration gradient. Antigen retrieval was conducted by rehydrating the slides, followed by incubation with 10 mM sodium citrate buffer (pH 6.0; Sigma-Aldrich) at 121°C and 15 psi (100 kPa) for 5 min. After incubating the slides with 3% hydrogen peroxide (Sigma-Aldrich), blocking was performed with 5% bovine serum albumin (BSA; RMBIO, USA). The tissue sections were incubated overnight with primary antibodies against proliferating cell nuclear antigen (PCNA) (Biolegend, USA), Ki-67 (Biolegend), vascular endothelial growth factor-A (VEGF-A) (Bioss, USA) or cluster of differentiation 31 (CD31; Santa Cruz Biotechnology, USA) at 1:50. The tissue sections were subsequently incubated with biotinylated goat anti-mouse or anti rabbit IgG antibody (H + L) at 1:100 for 30 min at 37°C incubator. The tissue sections were then reacted with Avidin-biotin peroxidase complexes (Vector Labs, USA), followed by incubation with 3,30-diaminobenzidine (DAB) kit (Vector Labs). A counterstain was performed with hematoxylin (Sigma-Aldrich) and observed under a bright microscope (Olympus). DAB positive area was semi-quantified using ImageJ 1.53c software (National Institutes of Health, USA). H&E stain was performed for histological analysis of tumor tissue. The tumor tissue sections that had undergone the above-mentioned rehydrating process were stained with hematoxylin and eosin (Sigma-Aldrich). The slides were observed under a bright microscope.
Total RNA extraction and quantitative polymerase chain reaction (qPCR)
Total RNA of the cells was isolated from cells using TRIzol reagent (Invitrogen Life Technologies), and RNA concentration was measured with a microreader (BioTek Instruments). cDNA synthesis was performed using an PrimeScript RT Master Mix (Takara Bio, Japan), following the manufacturer’s instructions. For quantitative analysis on the mRNA expression, the cDNA was amplified with 10 pmole/µl of each forward and reverse primer using TB Green Premix Ex Taq II (Takara Bio). qPCR was carried out for 40 cycles of 95°C for 15 s, 58°C for 60 s using QuantStudio 3 Real-Time PCR System (Applied Biosystems, USA). Table 1 shows the sequence of primers used in this study. GAPDH was used as an internal control. To visualize the amplified KiSS1 genes, qPCR products were separated in a 1.5% agarose gel and the gel was scanned using Lumino Graph 2 (ATTO Corporation, Japan).
Table 1.
Primer sequences for qPCR
| Gene | Primer sequences (5'→3') |
|---|---|
| GAPDH | Forward: GGTTTCCATAGGACCTGCTG Reverse: TCTTGGGTGTCTCGTCTTCT |
| KiSS1 | Forward: AGCAGCTAGAATCCCTGGG Reverse: GTTCCAGTTGTAGTTCGGCA |
| CCNE1 | Forward: GCACTTTCTTGAGCAACACC Reverse: TCCTCAAGTTTGGCTGCAAT |
| CDKN1A | Forward: CTGAAGTGAGCACAGCCTAG Reverse: AGGGAAAAGGCTCAACACTG |
| CDKN1B | Forward: TCGTCTTTTCGGGGTGTTTT Reverse: TGATCAAATGGACTGGCGAG |
| CDKN2A | Forward: CCCCACTACCGTAAATGTCC Reverse: CAAGAGAAGCCAGTAACCCC |
| CCNB1 | Forward: CATGGTGCACTTTCCTCCTT Reverse: GCTGTGGTAGAGTGCTGATC |
| MKI67 | Forward: TTCGGGCCCCTAAAGTAGAA Reverse: GGTTCGGATGATTTGCCTCT |
| PTEN | Forward: AGATGGCACTTTCCCGTTTT Reverse: TCGGAAACCTCTCTTAGCCA |
| PIK3R1 | Forward: ATGGTTGTTGTTCCCTGTCC Reverse: AGGCAGGAATTTGTGAAGCA |
| Akt2 | Forward: TAAGAAGGACCCCAAGCAGA Reverse: GGACTGGGCGGTAAATTCAT |
| Akt3 | Forward: TAGATGGGTAGGATGGCTGG Reverse: CTCAAATTTGGCCGTGTGAC |
| CD133 | Forward: TGAGACCCAACATCATCCCT Reverse: GCACAGAGGGTCATTGAGAG |
| CD146 | Forward: GGATGGCATTCAAGGAGAGG Reverse: GCATTCAACACCTGTCTCCA |
| CTNNB1 | Forward: TGCAGTTATGGTCCATCAGC Reverse: CCTCACGATGATGGGAAAGG |
| SNAI2 | Forward: GCGATGCCCAGTCTAGAAAA Reverse: CATGCAAATCCAACAGCCAG |
| PRELID1 | Forward: TCGCCAACTACATCGACAAG Reverse: CTCCTCCTGCAATTTCTCCC |
| CDH1 | Forward: TGACAACAAGCCCGAATTCA Reverse: TGACCACACTGATGACTCCT |
| CDH2 | Forward: TGGATGAAGATGGCATGGTG Reverse: TCTGCTGACTCCTTCACTGA |
| CXCR4 | Forward: CCTGCCTGGTATTGTCATCC Reverse: CTCAAACTCACACCCTTGCT |
Automated capillary-based western immunoblot
Total proteins of the cells were isolated by using radio immunoprecipitation assay (RIPA) buffer (ATTO Corporation), and the protein concentration was measured using bicinchoninic acid (Sigma-Aldrich) assay. Immunoblotting was conducted on a JESSTM Simple Western automated nano-immunoassay system (ProteinSimple, USA). A mixture of proteins, fluorescent 5× master mix, 400 mM dithiothreitol (ProteinSimple) and biotinylated molecular weight markers was prepared, and then denatured at 95°C for 5 min. Primary antibodies were diluted with antibody diluent. Table 2 shows the information of the primary antibodies used in this study. The working solutions of HRP-conjugated anti-mouse or rabbit secondary antibodies included in this kit were used. For chemiluminescence detection, luminol-peroxide mix was prepared. Assay protocol was as follows: 25 min for separation time, 20 min for blocking, 45 min for primary antibody reaction, 45 min for secondary antibody reaction. The Compass Simple Western software (ver. 6.0.0; ProteinSimple) was used to obtain the chemiluminescent band images. The band intensities were quantified with CSAnalyzer4 software (ATTO Corporation). GAPDH and its corresponding target proteins were analyzed from the same protein lysate. All protein expression levels were normalized to the GAPDH expression in each band.
Table 2.
List of primary antibodies used in immunoblotting
| Antigen | Type | Dilution | Manufacturer | Catalog No. | MW (kDa) |
|---|---|---|---|---|---|
| GAPDH | MM | 1:100 | Abcam | ab8245 | 35 |
| KiSS1R | RM | 1:20 | Cell Signaling Technology | 13776 | 15 |
| Cyclin E1 | MM | 1:100 | Abcam | ab3927 | 47 |
| PCNA | MM | 1:100 | Biolegend | 307901 | 36 |
| p85α | MM | 1:20 | Santa Cruz Biotechnology | sc-1637 | 85 |
| p-AktSer473 | RM | 1:100 | Cell Signaling Technology | 4060T | 60 |
| Akt 1/2/3 | RM | 1:100 | Abcam | ab179463 | 60 |
| β-catenin | MM | 1:100 | Biolegend | 862602 | 86 |
| CD133 | MM | 1:100 | Biolegend | 372802 | 120 |
| E-cadherin | RP | 1:20 | Abcam | ab15148 | 120 |
| N-cadherin | MM | 1:100 | Biolegend | 844702 | 130 |
| Slug | MM | 1:100 | Santa Cruz Biotechnology | sc-166476 | 30 |
| VEGF-A | RP | 1:100 | Bioss | bs-0279R | 23, 46 |
| CD31 | MM | 1:100 | Santa Cruz Biotechnology | sc-376764 | 130 |
MM, mouse monoclonal; RM, rabbit monoclonal; RP, rabbit polyclonal; MW, molecular weight.
Statistical analysis
All experiments were conducted at least three times, and all data presented as mean ± SEM. Data were statistically analyzed by Student’s t-test or multiple t-test using the Prism 7.0 software (GraphPad Software, USA). Significance was presented at *P < 0.05, **P < 0.01, or ***P < 0.001.
RESULTS
KiSS1 overexpression in HCC cells and characterization
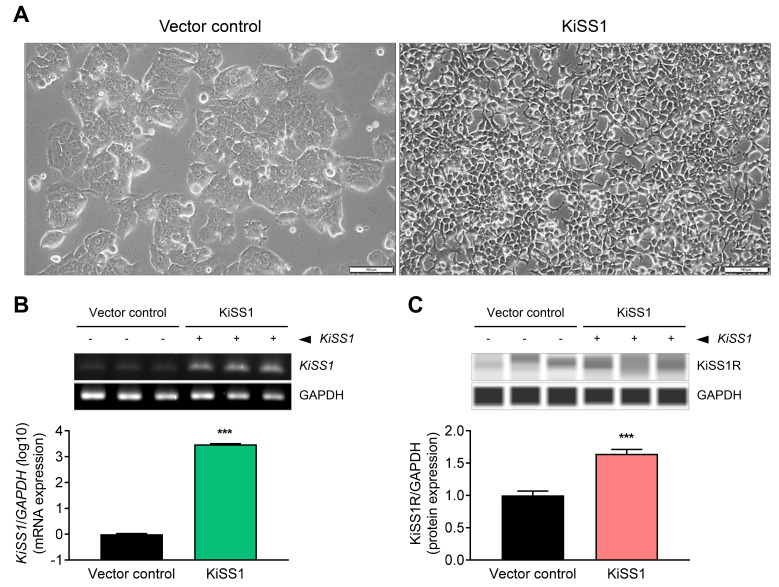
The HepG2-KiSS1 cell line overexpressing KiSS1 was established to investigate the effects of KiSS1 on tumor growth and migration in HCC cells. We observed that KiSS1 overexpression induces an alteration in cells to a mesenchymal morphology (Fig. 1A). Moreover, the KiSS1 gene expression was 2,986 times higher in the HepG2-KiSS1 cells than the control vector-inserted cells (Fig. 1B), and expression of the kisspeptin protein was 1.64 times higher than the control vector-inserted cells (Fig. 1C). Based on these morphological and genetic changes, we investigated the effects of KiSS1 overexpression on functional changes in HCC cells in subsequent experiments.
Fig. 1. KiSS1 overexpression and characterization.
HepG2 (vector control) cells or HepG2-KiSS1 cells overexpressing KiSS1 were prepared using 293T cells transfected with a lentiviral vector designed to express the KiSS1 gene. (A) Comparison of the morphology of both cell lines. Scale bars = 100 µm. (B) Comparison of relative expression levels of the KiSS1 gene in the two cell lines. Gene levels were quantified by applying qPCR, and the genes amplified through qPCR were visualized through electrophoresis. The bar graph is expressed logarithmically (log10). (C) Comparison of relative expression levels of kisspeptin protein in the two cell lines. Protein levels were measured using automated capillary-based Western immunoblot. Data in the bar graph were obtained from at least three repeated experiments. qPCR and immunoblotting data were statistically analyzed by Student’s t-test, and are presented as the mean ± SEM. Significance is presented at ***P < 0.001.
KiSS1 overexpression increases the proliferation of HCC cells
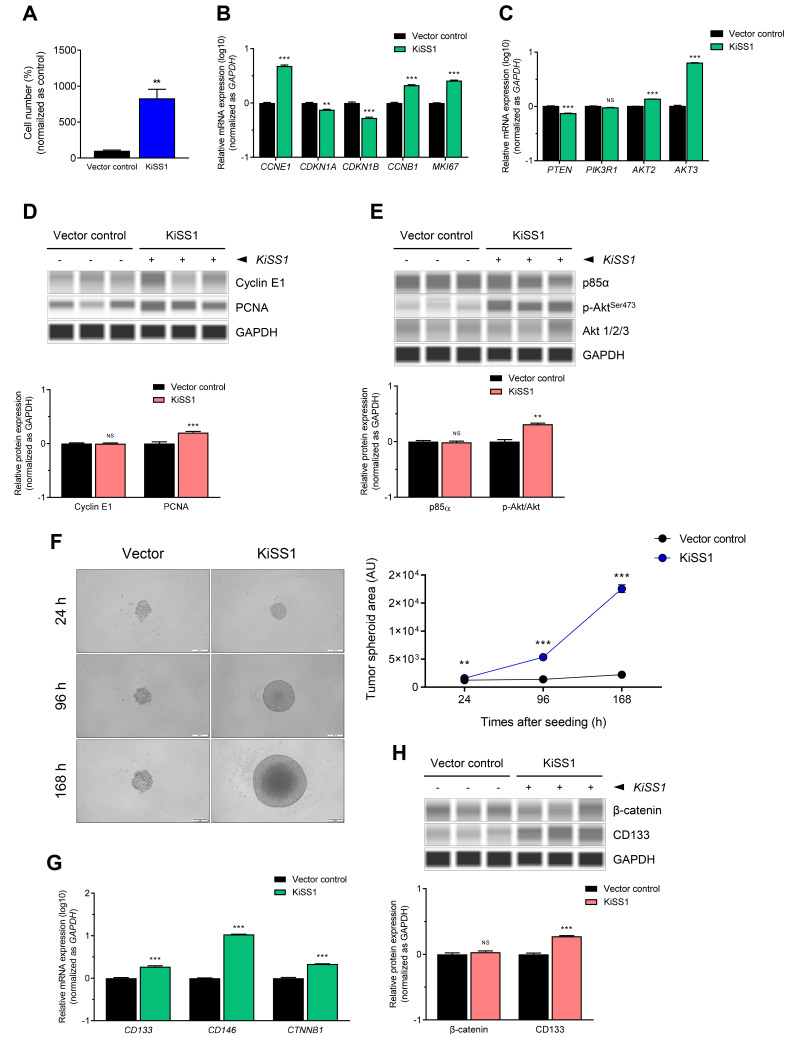
To determine the effect of KiSS1 overexpression on the proliferation of HCC, we first measured for alterations in cell growth rate by achieving cell counting (Fig. 2A). Contrary to our expectations, the cell growth rate of KiSS1-overexpressing cells was about 8.5 times greater than the vector control cells, indicating that KiSS1 dramatically increased the proliferation of HCC cells (Fig. 2A). To investigate the mechanism of cell proliferation, changes in the expressions of cell proliferation-related genes and cell cycle genes were examined. We observed upregulated expressions of CCNE1 and CCNB1 (which promote the G1/S phase), and downregulated expressions of CDKN1A and CDKN1B (G1/S phase inhibitors), resulting in increased accumulation of cells in the S phase (Fig. 2B). Thus, changes in the transcriptional levels of CCNB1 imply the possibility of regulating the G2/M phase by KiSS1 overexpression (Fig. 2B). KiSS1 overexpression also resulted in increased expression of the MKI67 gene, a cell proliferation marker (Fig. 2B). Moreover, KiSS1 overexpression increased the gene expressions of Akt2 and Akt3, which are key modules of PI3K/Akt signaling (Fig. 2C). Similar to these gene expression changes, an increase in PCNA was observed in the KiSS1-overexpressing cells (Fig. 2D). Interestingly, we observed not only increased Akt protein level, but also an increase in serine phosphorylation (Figs. 2C and 2E). However, no significant changes were detected in both the gene and protein levels of the phosphoinositide 3-kinase (PI3K) regulatory subunit p85α, by KiSS1 overexpression (Figs. 2C and 2E). These results indicate that KiSS1 overexpression induces the proliferation of HCC cells through Akt activation, and subsequent modulation of the G1/S phase-related cell cycle genes.
Fig. 2. KiSS1 increases cell proliferation of HCC.
(A) Cell proliferation was determined by counting the cells. Briefly, cells were seeded at a cell density of 1 × 105 cells/well in 6-well plates, and 96 h later, the number of cells was measured. The value of vehicle control was set as 100%. (B, C, and G) mRNA levels were measured by qPCR, and (D, E, and H) protein levels were measured using automated capillary-based Western immunoblot. (B) mRNA expressions of cell cycle genes and proliferation marker genes, and (C) PI3K/Akt signaling genes. (D) Protein expressions of cell cycle genes and PCNA, and (E) PI3K/Akt signaling proteins. (F) Tumor spheroid formation assay. Cells were seeded at a density of 2 × 103 cells/well in ultra-low attachment plate, and cells were allowed to form tumor spheroids. After 24, 72, and 168 h, the tumor spheroids were photographed under phase-contrast microscopy. Scale bars = 200 µm. (G) mRNA expressions and (H) protein expressions of CSC-related proteins. Data in the graphs were obtained from at least three repeated experiments, statistically analyzed by (A) Student’s t-test or (B-H) multiple t-test, and are presented as the mean ± SEM. Significance is presented at **P < 0.01 or ***P < 0.001. NS, not significant; AU, arbitrary unit.
KiSS1 overexpression increases the tumorigenesis of HCC cells
Considering that increased proliferation of HCC cells can result in increased tumorigenesis, we investigated the effect of KiSS1 overexpression on tumorigenesis of HCC cells. We found that KiSS1 overexpression dramatically increases the ability of HCC cells to form tumor spheroids in a time-dependent manner (Fig. 2F). KiSS1-overexpressing cells showed upregulation of the CD133, CD146, and CTNNB1 genes (Fig. 2G), with significant upregulation of the CD133 protein (Fig. 2H). Taken together, these results indicate that KiSS1 increases the tumor spheroid formation of hepatocarcinoma cells by upregulating the expression of CD133, and may thus increase the tumorigenic capacity.
KiSS1 overexpression increases the metastatic capacity by increasing the migratory and invasive ability of HCC cells
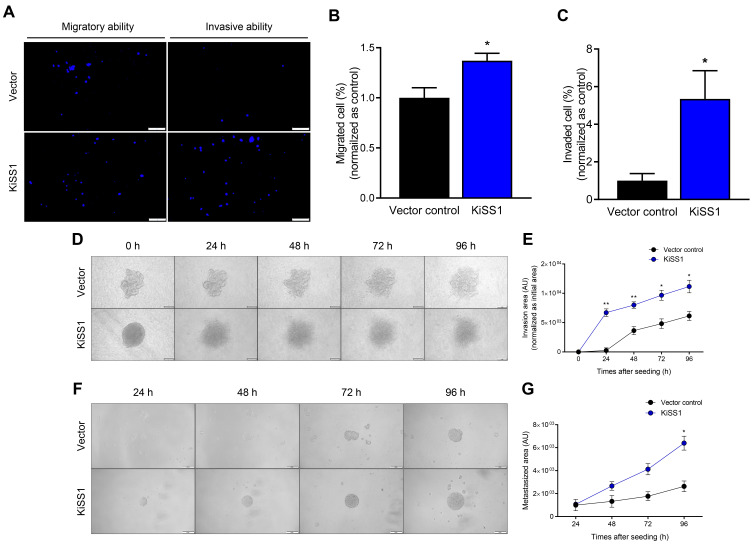
Based on our results that KiSS1 overexpression induces alterations in the mesenchymal morphology of HCC cells (Fig. 1A), we next investigated whether KiSS1 overexpression increases the mobility and invasiveness of HCC cells (Fig. 3). We observed that HepG2-KiSS1 cells migrated faster than vector control cells, and had increased cell invasiveness (Figs. 3A-3C). In the collagen invasion assay, the KiSS1-overexpressing tumor spheroids cultured on collagen penetrated faster and more widely into the collagen, suggesting an increased ability of hepatocarcinoma tumors to invade the ECM (Figs. 3D and 3E). Moreover, we observed that the KiSS1-overexpressing spheroids started to invade more rapidly from 24 h, whereas vector control spheroids started to invade slowly from 48 h (Figs. 3D and 3E). Tumor metastasis generally entails the spread of cancer cells, and results in the formation of metastatic colonies. Thus, the ability to form spheroids at the metastasis site can be evaluated to predict the metastatic ability of cancer cells. This was demonstrated by the observation that after invading from the upper chamber, the KiSS1-overexpressing cells in the bottom chamber (metastatic sites) formed tumor spheroids significantly faster, as compared to the vector control group (Figs. 3F and 3G).
Fig. 3. Effects of KiSS1 overexpression on migration of HCC.
(A) Transwell migration assay and transwell invasion assay. MMC-treated and Hoechst 33342 pre-stained cells were seeded in the upper chamber of a transwell insert with (invasion assay) or without (migration assay) fibronectin coating; 72 h later, the blue fluorescence of cells that migrated to the bottom chamber were photographed using a fluorescence microscope. Scale bars = 200 µm. (B and C) Quantitative graph for (A). (D) Collagen invasion assay and (E) quantitative graph. MMC-treated tumor spheroids were seeded in a 96-well plate coated with collagen, and were photographed using a phase-contrast microscope every 24 h, for up to 96 h. Scale bars = 200 µm. (F) In vitro metastasis assay and (G) quantitative graph. MMC-treated cells were seeded in the upper chamber where a fibronectin-coated transwell insert was placed; an ultra-low attachment plate was then combined with the upper chamber. Spheroids in the bottom chamber were photographed using a phase-contrast microscope every 24 h, for up to 96 h. Data in the graph were obtained from at least three repeated experiments, statistically analyzed by multiple t-test, and are presented as the mean ± SEM. Significance is presented at *P < 0.05 or **P < 0.01. AU, arbitrary unit.
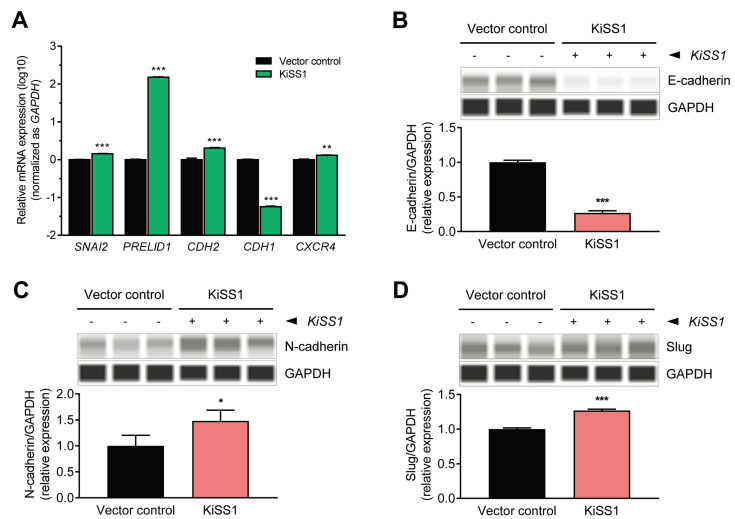
Given that tumor metastasis is initiated after cells undergo a conversion process (EMT) to a mesenchymal phenotype, we sought to investigate whether KiSS1 overexpression regulates the expressions of EMT markers (Fig. 4). We found that the expression of the epithelial marker E-cadherin was downregulated at both the mRNA and protein levels (Figs. 4A and 4B), and the expressions of N-cadherin and slug (mesenchymal markers) were upregulated at both the mRNA and protein levels (Figs. 4A, 4C, and 4D). Increased gene levels of PRELID1 and CXCR4 indicated that KiSS1 overexpression probably promotes EMT in HCC cells. These results indicate that KiSS1 overexpression increases the migratory and invasive ability of liver cancer cells by modulating the expression of EMT genes, and consequently increases the metastatic capacity.
Fig. 4. Altered expressions of EMT-related genes in KiSS1 overexpressing HCC cells.
(A) mRNA expressions of EMT genes (qPCR). Protein expressions of (B) E-cadherin, (C) N-cadherin, and (D) Slug (immunoblotting). Gene levels were quantified through qPCR, and the protein levels were measured using an automated capillary-based Western immunoblot. Data in the graph were obtained from at least three repeated experiments, statistically analyzed by (A) multiple t-test or (B-D) Student’s t-test, and are presented as the mean ± SEM. Significance is presented at *P < 0.05, **P < 0.01, or ***P < 0.001.
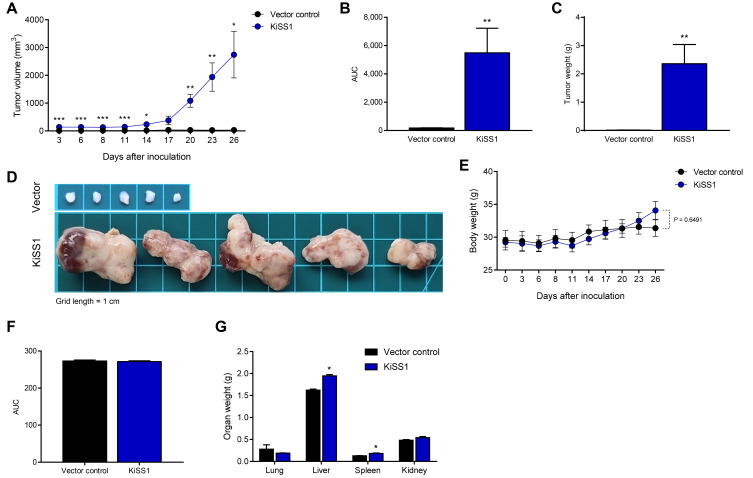
KiSS1 overexpression increases liver tumor growth
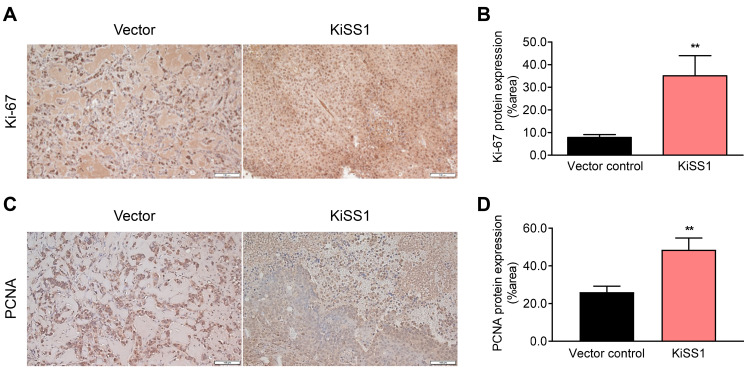
Based on our findings that KiSS1 overexpression increases the proliferation rate of HCC cells and promotes tumorigenicity, we investigated the effect of KiSS1 on in vivo tumor growth using a xenograft mouse model (Fig. 5). As expected, KiSS1 overexpression induced a significant difference in the tumor growth rate (Figs. 5A and 5B); 19 days after inoculation of cancer cells, we observed an 88-fold tumor volume increase, as compared to the vector control group (Figs. 5A and 5B). In addition, KiSS1 overexpression dramatically increased the tumor weight (Fig. 5C), with marked differences as seen macroscopically (Fig. 5D). Although KiSS1 overexpression did not significantly affect mouse body weight during the experimental period (Figs. 5E and 5F), significant increases were obtained in the liver and spleen weights (Fig. 5G), indicating increased side effects associated with increased tumor growth. Although it was observed in Fig. 5D that the absolute size of the vector control tumor was small, this was because the growth of the vector control tumor was low compared to the excessive growth of the KiSS1-overexpressing tumor. This is explained by the relatively low proliferation of the vector control cells shown in Fig. 2A and the relatively small tumor spheroids of the vector control cells shown in Fig. 2F. No tumor metastasis to other organs (including liver and spleen) was found at necropsy of the mice (data not shown). Consistent with the experimental results of increased tumor growth, histological analysis determined the increased expressions of Ki-67 and PCNA in the KiSS1 overexpression group (Fig. 6). These results indicate that KiSS1 overexpression increases the liver tumor growth by upregulating Ki-67 and PCNA.
Fig. 5. Effects of KiSS1 overexpression on tumor growth in the HCC-xenograft mouse model.
(A) Tumor volumes were measured by a digital caliper every 2-3 days per week, from day 3 to 26 after inoculation, and calculated by the formula: shortest diameter2 × longest diameter × 0.5236 (mm3). (B) Area under curve (AUC) of (A). (C) Tumor weights (n = 5). (D) Gross images of tumor. Grid length = 1 cm. (E) Body weights of the mice (n = 5). (F) AUC of (E). (G) Organ weights of the mice (n = 5). Data are presented as the mean ± SEM obtained from five mice per group. Significance is presented at *P < 0.05, **P < 0.01, or ***P < 0.001.
Fig. 6. Effects of KiSS1 overexpression on the expressions of tumor growth-related proteins in the HCC-xenograft mouse model.
IHC was conducted to investigate the protein expressions of Ki-67 and PCNA in tumor tissues. (A) Microscopy of the Ki-67-stained tumor tissue. (B) DAB positive area of Ki-67 expressing cells was semi-quantified using the ImageJ software. (C) Microscopy of PCNA stained tumor tissue. (D) DAB positive area intensity of PCNA expressing cells was semi-quantified using the ImageJ software. Scale bars = 100 µm. Significance is presented at **P < 0.01.
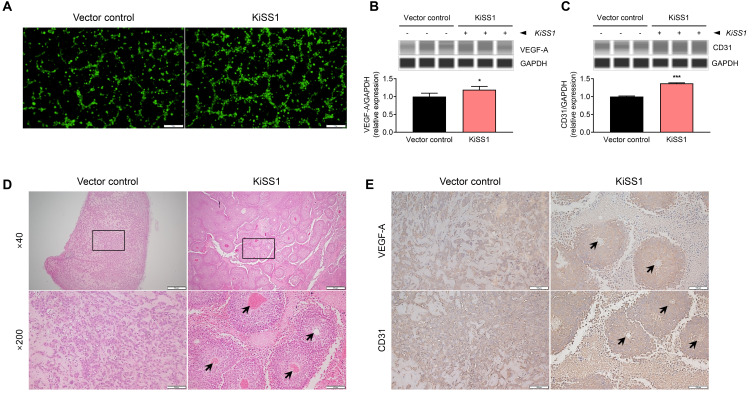
KiSS1 overexpression increases angiogenesis in HCC cells
All the results presented above indicate that KiSS1 overexpression increases not only the growth of hepatocarcinoma cells, but also the metastatic potential. Considering that angiogenesis is essential for cancer cells to grow continuously and secure a metastasis pathway (Kim and Choi, 2021), we sought to investigate whether KiSS1 overexpression affects the angiogenic ability of metastatic HCC cells (Fig. 7). We observed that the culture medium of KiSS1-overexpressing cells increased the tube formation of SV40-HUVECs (Fig. 7A), which is associated with significant upregulation of VEFG-A and CD31 proteins (Figs. 7B and 7C). It is noteworthy that relatively much more blood vessels were found in KiSS1-overexpressing liver tumor sections, in contrast to the absence of evident blood vessels in the tumor sections of the vector control group (Fig. 7D). The significant difference in vascular evidence from macroscopy of the two tumor groups validates these results convincingly (Fig. 7D). Moreover, increased expressions of VEGF-A and CD31 were observed by KiSS1 overexpression in the liver tumor tissue (Fig. 7E). These results indicate that overexpression of KiSS1 in liver tumors increases the angiogenesis by upregulating the expressions of VEGF-A and CD31.
Fig. 7. Effects of KiSS1 overexpression on angiogenesis.
(A) Tube formation assay using SV40-HUVECs. SV40-HUVECs were stained with Calcein-AM, and the cells were seeded in the cancer cell culture medium in Matrigel coated plates. Tube formation in SV40-HUVECs was achieved after 3 h, and were observed under fluorescence microscope. Scale bars = 100 µm. Protein expression levels of (B) VEGF-A and (C) CD31 were determined using automated capillary-based Western immunoblot. Data in the graph were obtained from at least three repeated experiments, statistically analyzed by Student’s t-test, and are presented as the mean ± SEM. Significance is presented at *P < 0.05 or ***P < 0.001. (D) H&E staining of tumor section obtained from xenograft mouse model. HepG2 cells (vector control) or HepG2-KiSS1 cells were inoculated subcutaneously in mice (n = 5 per group). The rectangle in the 40× magnification images indicates the 200× magnification image reproduced below. Histology of blood vessels is denoted by black arrows. Scale bars for 40× = 500 µm; Scale bars for 200× = 100 µm. (E) IHC was conducted to investigate the protein expressions of VEGF-A and CD31 in tumor tissues. Histology of blood vessels is denoted by black arrows. Scale bars = 100 µm.
DISCUSSION
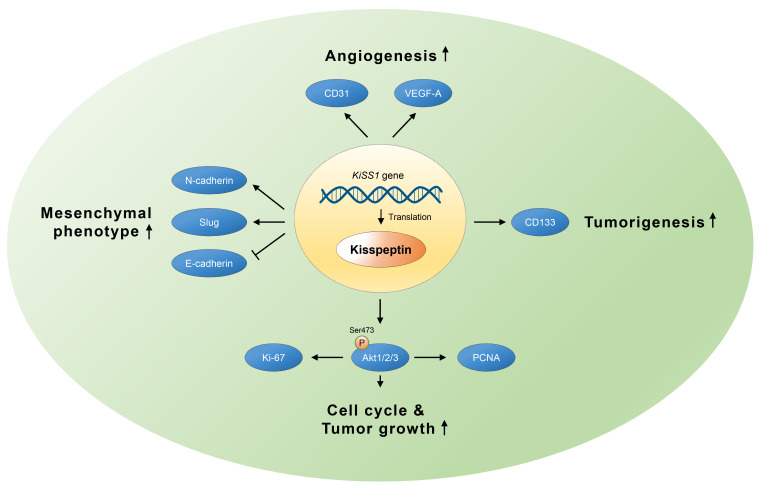
This study investigates the effects of KiSS1 gene on the proliferation and migration of liver tumors. We found that KiSS1 overexpression increased the proliferation rate of HCC cells, which was related to the expression of cell cycle genes involved in the G1/S phase progression, and this cell cycle upregulation was promoted by Akt activity. KiSS1 overexpression increased the liver tumorigenicity in both the cellular and animal models. In addition, KiSS1 overexpression increased the EMT, mobility, invasiveness, and metastatic colonization of HCC cells. The growth and metastasis of these liver tumors may be exacerbated by VEGF- and CD31-mediated angiogenesis. Based on our findings, Fig. 8 summarizes the effects of KiSS1 on HCC progression.
Fig. 8. Schematic diagram presenting the effects of KiSS1 gene on tumor growth and migration of HCC.
Transcription and subsequent translation of KiSS1 into kisspeptin induces cell proliferation and tumorigenesis of HCC, with subsequent promotion of migration and angiogenesis, resulting in metastasis. Increased kisspeptin expression by KiSS1 overexpression increases Akt phosphorylation and upregulates protein levels, which upregulate the expressions of proliferation-inducing proteins such as Ki-67 and PCNA, thereby induce the growth of HCC. Tumorigenesis in liver cancer can be accelerated by kisspeptin-induced CD133. Upregulation of N-cadherin and slug, and downregulation of E-cadherin by kisspeptin, induce EMT and increase cell migration in liver cancer. Kisspeptin may therefore contribute to the angiogenesis of liver cancer by upregulating CD31 and VEGF-A.
As demonstrated in studies conducted in several cancer models, both cancer proliferation and progression are accompanied by cell cycle alterations induced by cell cycle regulators (Leal-Esteban and Fajas, 2020; Rivadeneira et al., 2010). Liver cancer is characterized by alterations in cell cycle regulation through numerous molecular mechanisms (Bisteau et al., 2014). In HCC cells overexpressing KiSS1, we observed altered expressions of cell cycle genes regulating the G1/S phase. Significant changes in the cyclin-dependent kinase (CDK) inhibitor (such as CDKN1A and CDKN1B) were particularly noticeable. It is well known that p21 is transcriptionally regulated by the p53 gene, and it has been reported that the expressions of p21 and p27 are suppressed in liver tumors, preventing apoptosis through cell cycle regulation (Adimoolam et al., 2001; Bhardwaj et al., 1999; Eferl et al., 2003). Moreover, it has been reported that the induction of CDK inhibitors requires the PI3K/Akt pathway, and Akt phosphorylation interferes with the nuclear translocation of p27 and induces G1 arrest (Chang et al., 2003; Liang et al., 2002). The PI3K/Akt pathway is associated with tumorigenesis and is upregulated in several types of liver cancers, and there are considerable accumulated evidence of cell cycle regulation by the PI3K/Akt pathway in liver cancer (Fan et al., 2015; Liu et al., 2015; Wang et al., 2021). Considering these previous studies, we propose that KiSS1 induces the proliferation of liver cancer by inhibiting the expression of cell cycle inhibitors via PI3K/Akt signaling and regulating genes related to the G1/S phase.
Wnt/β-catenin signaling promotes the progression of most malignant tumors including liver cancer, and mutations of genes involved in this pathway are characteristic of hepatobiliary tumors, with the β-catenin mutation found most often in hepatoblastoma (Perugorria et al., 2019; Wen et al., 2020). It was recently identified that hepatic tumorigenesis has similar characteristics to cancer stem cells (CSCs), and that β-catenin mediated cell dedifferentiation into CSCs, which could contribute to liver tumor growth (Pandit et al., 2018). Other studies also reported that this signaling pathway encoded by CTNNB1 plays a major role in HCC and cholangiocarcinoma tumorigenesis (He and Tang, 2020; McGrath et al., 2020). CD133 regulates the self-renewal and proliferation of CSCs more directly than β-catenin, and is one of the best characterized biomarkers (Barzegar Behrooz et al., 2019; Liou, 2019). Since CD133 not only regulates tumorigenesis but is also implicated in cancer metastasis, the KiSS1-induced increase in CD133 in HCC cells observed in this study may provide clues to understanding the mechanism of liver tumor metastasis (Glumac and LeBeau, 2018; Liou, 2019). Thus, we report for the first time, that upregulation of β-catenin and CD133 by KiSS1 overexpression has the potential to induce tumorigenicity and progression of liver cancers.
Tumor metastasis can be initiated from morphological changes in cancer cells, i.e., induction of the EMT program (Mittal, 2018). EMT is one of the highly evolutionarily conserved developmental programs involved in mesenchymal acquisition, which increases tumor mobility (Wu and Zhou, 2008; Yoon et al., 2021). EMT induces loss of epithelial markers such as E-cadherin and ZO-1, and upregulation of mesenchymal markers such as N-cadherin, snail, slug and vimentin, eventually easing the process of migration for cells (Kalluri and Weinberg, 2009; Loh et al., 2019). In addition, cells must undergo EMT in order for tumor cells to penetrate into adjacent cell layers or to intravasate into blood vessels and lymphatic vessels (Yang and Weinberg, 2008). Therefore, based on our observation in the upregulation of E-cadherin, N-cadherin, and slug by KiSS1 overexpression, it can be explained that KiSS1 increases the migratory and invasive ability of HCC by controlling the expressions of EMT regulators, thereby achieving liver tumor metastasis. Moreover, it has recently been reported that HCC metastasis can be promoted by induction of EMT via β-catenin, suggesting that the upregulation of β-catenin by KiSS1 overexpression found in our study may induce metastasis through EMT (Yuan et al., 2020). Interestingly, given that β-catenin is an epithelial marker, the KiSS1-induced tumor metastasis is likely to be induced by collective cell migration (Aman and Piotrowski, 2008; Carvalho et al., 2019; Zhu et al., 2018).
Tumors are supplied with nutrients through the recruitment of new blood vessels via a process called angiogenesis, and can grow and metastasize to other tissues (Bielenberg and Zetter, 2015; Takeda et al., 2002). Of note, all systemic anticancer therapies approved for HCC in the US, EU, and China are molecular targeted therapies with antiangiogenic effects that target VEGF and its receptors (Morse et al., 2019; Zhu et al., 2020). Moreover, according to previous studies, the upregulation of VEGF-A expression induces angiogenesis in HCC (Lu et al., 2015). These data provide a basis for our hypothesis and findings that upregulated VEFG expression in liver tumors overexpressing KiSS1 results in increased angiogenesis in liver tumors. Similar to VEGF, CD31 is highly expressed in liver cancer patients with poor prognosis, and has therefore been used as a diagnostic marker (Qian et al., 2018; Zhang et al., 2021). CD31 is associated with increased microvessel density in HCC, as well as being used as a marker of increased angiogenesis in liver cancer (Bosmuller et al., 2018; Ghanekar et al., 2013). Moreover, it has been reported that CD31 influences the development of CSCs by regulating its expression through CD133 and/or VEGF, and promotes HCC metastasis by inducing EMT (Behrooz and Syahir, 2021; Zhang et al., 2018). Therefore, we report that KiSS1 promotes the formation of new blood vessels in HCC by regulating the expressions of VEGF and CD31, and may serve as a bridge in liver tumor metastasis.
Our study further determined that KiSS1 overexpression resulted in increased cell proliferation and migration ability of HCC. However, in order to understand the exact role of the KiSS1 gene in liver tumor, it was necessary to confirm whether the gene expression is inhibited or knocked out using the RNAi system, which is contrary to our approaches. Moreover, based on the upregulation of metastatic colonization by KiSS1 shown in the in vitro metastasis assay, it is necessary to systematically investigate the effects of the KiSS1 gene on liver cancer metastasis using an orthotropic xenograft model or a metastatic model artificially established by injecting liver cancer cells into the tail vein of mice. Experimental strategies using agonists or antagonists for kisspeptin could be an experimental approach helpful in evaluating the potential of KiSS1 for diagnostic and therapeutic purposes (Stathaki et al., 2019). Taken together, we conclude that in liver cancer, the KiSS1 gene not only increases tumor growth, but also stimulates migration and angiogenesis for metastasis. To date, the role of KiSS1 in liver cancer progression remains unclear. We believe that the findings of this study could contribute to clarifying the mechanism. Continued efforts to elucidate the role of KiSS1 will spur the development of gene therapy or diagnosis using this gene, and development of anticancer drugs that involve the regulation of this gene.
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program (2020R1A2C2006060) and the Global Research and Development Center (GRDC) Program (2017K1A4A3014959) through the National Research Foundation (NRF) of Korea, funded by the Ministry of Science and ICT. This work was also supported by the Sejong Fellowship of National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2021R1C1C2093998).
Footnotes
AUTHOR CONTRIBUTIONS
C.W.K. and K.C.C. conceived experiments. C.W.K., H.K.L., M.W.N., and Y.C. performed experiments. C.W.K. and H.K.L. wrote the manuscript. K.C.C. secured funding. H.K.L. and K.C.C. provided expertise and feedback.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Adimoolam S., Lin C.X., Ford J.M. The p53-regulated cyclin-dependent kinase inhibitor, p21 (cip1, waf1, sdi1), is not required for global genomic and transcription-coupled nucleotide excision repair of UV-induced DNA photoproducts. J. Biol. Chem. 2001;276:25813–25822. doi: 10.1074/jbc.M102240200. [DOI] [PubMed] [Google Scholar]
- Aman A., Piotrowski T. Wnt/beta-catenin and Fgf signaling control collective cell migration by restricting chemokine receptor expression. Dev. Cell. 2008;15:749–761. doi: 10.1016/j.devcel.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Barzegar Behrooz A., Syahir A., Ahmad S. CD133: beyond a cancer stem cell biomarker. J. Drug Target. 2019;27:257–269. doi: 10.1080/1061186X.2018.1479756. [DOI] [PubMed] [Google Scholar]
- Behrooz A.B., Syahir A. Could we address the interplay between CD133, Wnt/beta-catenin, and TERT signaling pathways as a potential target for glioblastoma therapy? Front. Oncol. 2021;11:642719. doi: 10.3389/fonc.2021.642719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj B., Bhardwaj G., Lau J.Y. Expression of p21 and p27 in hepatoma cell lines with different p53 gene profile. J. Hepatol. 1999;31:386. doi: 10.1016/S0168-8278(99)80245-1. [DOI] [PubMed] [Google Scholar]
- Bielenberg D.R., Zetter B.R. The contribution of angiogenesis to the process of metastasis. Cancer J. 2015;21:267–273. doi: 10.1097/PPO.0000000000000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisteau X., Caldez M.J., Kaldis P. The complex relationship between liver cancer and the cell cycle: a story of multiple regulations. Cancers (Basel) 2014;6:79–111. doi: 10.3390/cancers6010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosmuller H., Pfefferle V., Bittar Z., Scheble V., Horger M., Sipos B., Fend F. Microvessel density and angiogenesis in primary hepatic malignancies: Differential expression of CD31 and VEGFR-2 in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Pathol. Res. Pract. 2018;214:1136–1141. doi: 10.1016/j.prp.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Brabletz T., Kalluri R., Nieto M.A., Weinberg R.A. EMT in cancer. Nat. Rev. Cancer. 2018;18:128–134. doi: 10.1038/nrc.2017.118. [DOI] [PubMed] [Google Scholar]
- Carvalho J.R., Fortunato I.C., Fonseca C.G., Pezzarossa A., Barbacena P., Dominguez-Cejudo M.A., Vasconcelos F.F., Santos N.C., Carvalho F.A., Franco C.A. Non-canonical Wnt signaling regulates junctional mechanocoupling during angiogenic collective cell migration. Elife. 2019;8:e45853. doi: 10.7554/eLife.45853.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Lee J.T., Navolanic P.M., Steelman L.S., Shelton J.G., Blalock W.L., Franklin R.A., McCubrey J.A. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- Clarke H., Dhillo W.S., Jayasena C.N. Comprehensive review on kisspeptin and its role in reproductive disorders. Endocrinol. Metab. (Seoul) 2015;30:124–141. doi: 10.3803/EnM.2015.30.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colecchia A., Schiumerini R., Cucchetti A., Cescon M., Taddia M., Marasco G., Festi D. Prognostic factors for hepatocellular carcinoma recurrence. World J. Gastroenterol. 2014;20:5935–5950. doi: 10.3748/wjg.v20.i20.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delire B., Henriet P., Lemoine P., Leclercq I.A., Starkel P. Chronic liver injury promotes hepatocarcinoma cell seeding and growth, associated with infiltration by macrophages. Cancer Sci. 2018;109:2141–2152. doi: 10.1111/cas.13628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanasekaran R., Bandoh S., Roberts L.R. Molecular pathogenesis of hepatocellular carcinoma and impact of therapeutic advances. Res. 2016;5 doi: 10.12688/f1000research.6946.1. F1000 Faculty Rev-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R., Ricci R., Kenner L., Zenz R., David J.P., Rath M., Wagner E.F. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112:181–192. doi: 10.1016/S0092-8674(03)00042-4. [DOI] [PubMed] [Google Scholar]
- Fan B., Yu Y., Zhang Y. PI3K-Akt1 expression and its significance in liver tissues with chronic fluorosis. Int. J. Clin. Exp. Pathol. 2015;8:1226–1236. [PMC free article] [PubMed] [Google Scholar]
- Ghanekar A., Ahmed S., Chen K., Adeyi O. Endothelial cells do not arise from tumor-initiating cells in human hepatocellular carcinoma. BMC Cancer. 2013;13:485. doi: 10.1186/1471-2407-13-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden, of Disease Liver Cancer Collaboration , Akinyemiju T., Abera S., Ahmed M., Alam N., Alemayohu M.A., Allen C., Al-Raddadi R., Alvis-Guzman N., Amoako Y., et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the Global Burden of Disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glumac P.M., LeBeau A.M. The role of CD133 in cancer: a concise review. Clin. Transl. Med. 2018;7:18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go R.E., Kim C.W., Jeon S.Y., Byun Y.S., Jeung E.B., Nam K.H., Choi K.C. Fludioxonil induced the cancer growth and metastasis via altering epithelial-mesenchymal transition via an estrogen receptor-dependent pathway in cellular and xenografted breast cancer models. Environ. Toxicol. 2017;32:1439–1454. doi: 10.1002/tox.22337. [DOI] [PubMed] [Google Scholar]
- He S., Tang S. WNT/beta-catenin signaling in the development of liver cancers. Biomed. Pharmacother. 2020;132:110851. doi: 10.1016/j.biopha.2020.110851. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M., Hirooka Y., Kaibara N. Quantitative reverse transcriptase polymerase chain reaction analysis for KiSS-1 and orphan G-protein-coupled receptor (hOT7T175) gene expression in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2003;129:531–535. doi: 10.1007/s00432-003-0469-z. [DOI] [PubMed] [Google Scholar]
- Imamura H., Matsuyama Y., Tanaka E., Ohkubo T., Hasegawa K., Miyagawa S., Sugawara Y., Minagawa M., Takayama T., Kawasaki S., et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J. Hepatol. 2003;38:200–207. doi: 10.1016/S0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A.S. Sexual differentiation and the Kiss1 system: hormonal and developmental considerations. Peptides. 2009;30:83–93. doi: 10.1016/j.peptides.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.W., Choi K.C. Potential roles of iridoid glycosides and their underlying mechanisms against diverse cancer growth and metastasis: do they have an inhibitory effect on cancer progression? Nutrients. 2021;13:2974. doi: 10.3390/nu13092974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.W., Go R.E., Lee G.A., Kim C.D., Chun Y.J., Choi K.C. Immortalization of human corneal epithelial cells using simian virus 40 large T antigen and cell characterization. J. Pharmacol. Toxicol. Methods. 2016a;78:52–57. doi: 10.1016/j.vascn.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Kim C.W., Hwang K.A., Choi K.C. Anti-metastatic potential of resveratrol and its metabolites by the inhibition of epithelial-mesenchymal transition, migration, and invasion of malignant cancer cells. Phytomedicine. 2016b;23:1787–1796. doi: 10.1016/j.phymed.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Kim C.W., Kim C.D., Choi K.C. Establishment and evaluation of immortalized human epidermal keratinocytes for an alternative skin irritation test. J. Pharmacol. Toxicol. Methods. 2017a;88:130–139. doi: 10.1016/j.vascn.2017.08.005. [DOI] [PubMed] [Google Scholar]
- Kim C.W., Lee H.M., Lee K., Kim B., Lee M.Y., Choi K.C. Effects of cigarette smoke extracts on cell cycle, cell migration and endocrine activity in human placental cells. Reprod. Toxicol. 2017b;73:8–19. doi: 10.1016/j.reprotox.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Kummar S., Shafi N.Q. Metastatic hepatocellular carcinoma. Clin. Oncol. (R. Coll. Radiol.) 2003;15:288–294. doi: 10.1016/S0936-6555(03)00067-0. [DOI] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal-Esteban L.C., Fajas L. Cell cycle regulators in cancer cell metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165715. doi: 10.1016/j.bbadis.2020.165715. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Shin H.J., Koo J., Kim T.H., Kim C.W., Go R.E., Seong Y.H., Park J.E., Choi K.C. Blockade of transforming growth factor beta2 by anti-sense oligonucleotide improves immunotherapeutic potential of IL-2 against melanoma in a humanized mouse model. Cytotherapy. 2021;23:599–607. doi: 10.1016/j.jcyt.2021.01.003. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Miele M.E., Hicks D.J., Phillips K.K., Trent J.M., Weissman B.E., Welch D.R. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J. Natl. Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Welch D.R. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int. J. Cancer. 1997a;71:1035–1044. doi: 10.1002/(SICI)1097-0215(19970611)71:6<1035::AID-IJC20>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Welch D.R. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997b;57:2384–2387. [PubMed] [Google Scholar]
- Liang J., Zubovitz J., Petrocelli T., Kotchetkov R., Connor M.K., Han K., Lee J.H., Ciarallo S., Catzavelos C., Beniston R., et al. PKB/Akt phosphorylates p27, impairs nuclear import of p27 and opposes p27-mediated G1 arrest. Nat. Med. 2002;8:1153–1160. doi: 10.1038/nm761. [DOI] [PubMed] [Google Scholar]
- Liou G.Y. CD133 as a regulator of cancer metastasis through the cancer stem cells. Int. J. Biochem. Cell Biol. 2019;106:1–7. doi: 10.1016/j.biocel.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Liao W., Yuan Q., Ou Y., Huang J. TTK activates Akt and promotes proliferation and migration of hepatocellular carcinoma cells. Oncotarget. 2015;6:34309–34320. doi: 10.18632/oncotarget.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- Loh C.Y., Chai J.Y., Tang T.F., Wong W.F., Sethi G., Shanmugam M.K., Chong P.P., Looi C.Y. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells. 2019;8:1118. doi: 10.3390/cells8101118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Lin N., Chen Z., Xu R. Hypoxia-induced secretion of platelet-derived growth factor-BB by hepatocellular carcinoma cells increases activated hepatic stellate cell proliferation, migration and expression of vascular endothelial growth factor-A. Mol. Med. Rep. 2015;11:691–697. doi: 10.3892/mmr.2014.2689. [DOI] [PubMed] [Google Scholar]
- McGrath N.A., Fu J., Gu S.Z., Xie C. Targeting cancer stem cells in cholangiocarcinoma (Review) Int. J. Oncol. 2020;57:397–408. doi: 10.3892/ijo.2020.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu. Rev. Pathol. 2018;13:395–412. doi: 10.1146/annurev-pathol-020117-043854. [DOI] [PubMed] [Google Scholar]
- Miyazaki K., Oyanagi J., Hoshino D., Togo S., Kumagai H., Miyagi Y. Cancer cell migration on elongate protrusions of fibroblasts in collagen matrix. Sci. Rep. 2019;9:292. doi: 10.1038/s41598-018-36646-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse M.A., Sun W., Kim R., He A.R., Abada P.B., Mynderse M., Finn R.S. The role of angiogenesis in hepatocellular carcinoma. Clin. Cancer Res. 2019;25:912–920. doi: 10.1158/1078-0432.CCR-18-1254. [DOI] [PubMed] [Google Scholar]
- Oakley A.E., Clifton D.K., Steiner R.A. Kisspeptin signaling in the brain. Endocr. Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunwobi O.O., Harricharran T., Huaman J., Galuza A., Odumuwagun O., Tan Y., Ma G.X., Nguyen M.T. Mechanisms of hepatocellular carcinoma progression. World J. Gastroenterol. 2019;25:2279–2293. doi: 10.3748/wjg.v25.i19.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaki T., Shintani Y., Honda S., Matsumoto H., Hori A., Kanehashi K., Terao Y., Kumano S., Takatsu Y., Masuda Y., et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- Pandit H., Li Y., Li X., Zhang W., Li S., Martin R.C.G. Enrichment of cancer stem cells via beta-catenin contributing to the tumorigenesis of hepatocellular carcinoma. BMC Cancer. 2018;18:783. doi: 10.1186/s12885-018-4683-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perugorria M.J., Olaizola P., Labiano I., Esparza-Baquer A., Marzioni M., Marin J.J.G., Bujanda L., Banales J.M. Wnt-beta-catenin signalling in liver development, health and disease. Nat. Rev. Gastroenterol. Hepatol. 2019;16:121–136. doi: 10.1038/s41575-018-0075-9. [DOI] [PubMed] [Google Scholar]
- Puisieux A., Brabletz T., Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- Qian H., Yang L., Zhao W., Chen H., He S. A comparison of CD105 and CD31 expression in tumor vessels of hepatocellular carcinoma by tissue microarray and flow cytometry. Exp. Ther. Med. 2018;16:2881–2888. doi: 10.3892/etm.2018.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod K., Sheth R., Shah P., Rege S. Active contrast extravasation in spontaneous rupture of hepatocellular carcinoma: a rare CT finding. J. Postgrad. Med. 2000;46:35–36. [PubMed] [Google Scholar]
- Rivadeneira D.B., Mayhew C.N., Thangavel C., Sotillo E., Reed C.A., Grana X., Knudsen E.S. Proliferative suppression by CDK4/6 inhibition: complex function of the retinoblastoma pathway in liver tissue and hepatoma cells. Gastroenterology. 2010;138:1920–1930. doi: 10.1053/j.gastro.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K., Wang X., Haitel A., Sieghart W., Peck-Radosavljevic M., Bodingbauer M., Rasoul-Rockenschaub S., Wrba F. KiSS-1 overexpression as an independent prognostic marker in hepatocellular carcinoma: an immunohistochemical study. Virchows Arch. 2007;450:143–149. doi: 10.1007/s00428-006-0352-9. [DOI] [PubMed] [Google Scholar]
- Shengbing Z., Feng L.J., Bin W., Lingyun G., Aimin H. Expression of KiSS-1 gene and its role in invasion and metastasis of human hepatocellular carcinoma. Anat. Rec. (Hoboken) 2009;292:1128–1134. doi: 10.1002/ar.20950. [DOI] [PubMed] [Google Scholar]
- Stathaki M., Stamatiou M.E., Magioris G., Simantiris S., Syrigos N., Dourakis S., Koutsilieris M., Armakolas A. The role of kisspeptin system in cancer biology. Crit. Rev. Oncol. Hematol. 2019;142:130–140. doi: 10.1016/j.critrevonc.2019.07.015. [DOI] [PubMed] [Google Scholar]
- Takeda A., Stoeltzing O., Ahmad S.A., Reinmuth N., Liu W., Parikh A., Fan F., Akagi M., Ellis L.M. Role of angiogenesis in the development and growth of liver metastasis. Ann. Surg. Oncol. 2002;9:610–616. doi: 10.1007/BF02574475. [DOI] [PubMed] [Google Scholar]
- Terasawa E., Guerriero K.A., Plant T.M. Kisspeptin and puberty in mammals. Adv. Exp. Med. Biol. 2013;784:253–273. doi: 10.1007/978-1-4614-6199-9_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari N., Gheldof A., Tatari M., Christofori G. EMT as the ultimate survival mechanism of cancer cells. Semin. Cancer Biol. 2012;22:194–207. doi: 10.1016/j.semcancer.2012.02.013. [DOI] [PubMed] [Google Scholar]
- Wang Z., Cui X., Hao G., He J. Aberrant expression of PI3K/AKT signaling is involved in apoptosis resistance of hepatocellular carcinoma. Open Life Sci. 2021;16:1037–1044. doi: 10.1515/biol-2021-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Wu Y., Awadasseid A., Tanaka Y., Zhang W. New advances in canonical Wnt/beta-catenin signaling in cancer. Cancer Manag. Res. 2020;12:6987–6998. doi: 10.2147/CMAR.S258645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhou B.P. New insights of epithelial-mesenchymal transition in cancer metastasis. Acta Biochim. Biophys. Sin. (Shanghai) 2008;40:643–650. doi: 10.1111/j.1745-7270.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Yang J.D., Nakamura I., Roberts L.R. The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Semin. Cancer Biol. 2011;21:35–43. doi: 10.1016/j.semcancer.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Shin B., Woo H.G. Endoplasmic reticulum stress induces CAP2 expression promoting epithelial-mesenchymal transition in liver cancer cells. Mol. Cells. 2021;44:569–579. doi: 10.14348/molcells.2021.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K., Xie K., Lan T., Xu L., Chen X., Li X., Liao M., Li J., Huang J., Zeng Y., et al. TXNDC12 promotes EMT and metastasis of hepatocellular carcinoma cells via activation of beta-catenin. Cell Death Differ. 2020;27:1355–1368. doi: 10.1038/s41418-019-0421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhang L., Gao Y., Wang Y., Liu Y., Zhang H., Wang Q., Hu F., Li J., Tan J., et al. Role of aneuploid circulating tumor cells and CD31(+) circulating tumor endothelial cells in predicting and monitoring anti-angiogenic therapy efficacy in advanced NSCLC. Mol. Oncol. 2021;15:2891–2909. doi: 10.1002/1878-0261.13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Y., Kong L.Q., Zhu X.D., Cai H., Wang C.H., Shi W.K., Cao M.Q., Li X.L., Li K.S., Zhang S.Z., et al. CD31 regulates metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma via the ITGB1-FAK-Akt signaling pathway. Cancer Lett. 2018;429:29–40. doi: 10.1016/j.canlet.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Zhu G.J., Song P.P., Zhou H., Shen X.H., Wang J.G., Ma X.F., Gu Y.J., Liu D.D., Feng A.N., Qian X.Y., et al. Role of epithelial-mesenchymal transition markers E-cadherin, N-cadherin, beta-catenin and ZEB2 in laryngeal squamous cell carcinoma. Oncol. Lett. 2018;15:3472–3481. doi: 10.3892/ol.2018.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.D., Tang Z.Y., Sun H.C. Targeting angiogenesis for liver cancer: past, present, and future. Genes Dis. 2020;7:328–335. doi: 10.1016/j.gendis.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]