Abstract
Survival of Salmonella enterica serovar Typhimurium within host phagocytic cells is a critical step in establishing systemic infection in mice. Genes within Salmonella pathogenicity island 2 (SPI-2) encode a type III secretion system that is required for establishment of systemic infection. Several proteins encoded by SPI-2 have homology to type III secreted proteins from enteropathogenic Escherichia coli and Yersinia and, based on that homology, are predicted to be secreted through the SPI-2 type III secretion system. We have investigated the roles of two of these proteins, SseC and SseD. We demonstrate here that the SseD protein is required for systemic Salmonella infection of the mouse, and we confirmed the virulence requirement for the SseC protein. Experiments were performed, using cellular fractionation and immunoblotting, to identify the subcellular location of the SseC and SseD proteins. Both proteins were found to localize predominantly to the bacterial cell membrane. In addition, our work revealed that SseC and SseD are exposed to the extracellular environment and are loosely associated with the bacterial membrane. Furthermore, localization of SseC and SseD to the bacterial membrane was found to require a functional SPI-2 type III secretion system. Collectively, these results indicate that the SseC and SseD proteins are secreted by the SPI-2 type III secretion system to the bacterial membrane in order to perform their virulence functions.
Salmonella enterica serovar Typhimurium is a causative agent of gastroenteritis in humans (20). This pathogen also causes systemic infection in mice, which serves as an experimental model for the study of human typhoid fever. Many of the steps leading to establishment of systemic Salmonella infection have been identified. Upon colonization of the small intestine by Salmonella, the bacteria invade M cells of Peyer's patches, penetrate into the environment of the lymphoid follicle, and are subsequently taken up by macrophages (12). The ability of Salmonella to survive within this host phagocytic cell is believed to be a critical element in establishment of systemic infection (2). Following entry into the lymphatic system, Salmonella disseminates to the liver and spleen, where unchecked growth causes death of the host (12).
Many genes required for the various stages of Salmonella pathogenesis have been identified. A number of these virulence genes are found clustered in the chromosome in regions called pathogenicity islands. Two of these pathogenicity islands encode type III secretion systems (TTSS), and their function in Salmonella virulence is being extensively studied. Salmonella pathogenicity island 1 (SPI-1) contains genes required for bacterial invasion of M cells and epithelial cells (see reference 4 for a review). A second island, Salmonella pathogenicity island 2 (SPI-2), is essential for systemic virulence and is believed to enable the bacteria to survive within macrophages.
Genes carried within SPI-2 were originally identified in a signature-tagged transposon mutagenesis screen designed to locate genes required for survival of the pathogen within mice (8). Analysis of several mutations within SPI-2 has indicated that the genes on the pathogenicity island are required for bacterial survival within the macrophage. Strains carrying mutations in SPI-2 genes are attenuated for virulence when inoculated by the oral, intraperitoneal, or intravenous route, indicating that the genes are required for stages of infection after invasion of the intestinal mucosa (3, 7, 9, 10, 18, 22). SPI-2 mutants have a reduced rate of replication in the murine liver and spleen (21) and replication defects in primary murine macrophages and in macrophage cell lines have also been reported (3, 10, 18). Experiments looking at the expression of SPI-2 genes indicate that they are induced several hours after uptake by macrophages (3) and are induced in vitro by starvation conditions that presumably mimic the environment of the phagosomal compartment of a macrophage (1, 5).
Based on homology to type III secretion proteins in other organisms, genes within SPI-2 are predicted to encode secreted, structural, regulatory, or chaperone proteins. However, the function of only a handful of genes in SPI-2 has been established by experimental approaches. Two proteins, SpiC and SseB, are secreted outside of the bacterium by the TTSS (1, 24). SpiC is translocated across the phagosomal membrane and into the cytosol of Salmonella-infected J774 cells. Within the host cell, SpiC appears to function by interfering with membrane trafficking. The SseB protein is secreted to the bacterial cell surface, where it is loosely associated with the membrane. It was suggested that SseB may be part of a structure through which proteins are translocated, much like its homolog in enteropathogenic Escherichia coli (EPEC), EspA, which is a component of a filamentous secretion organelle. In addition, two proteins encoded by genes located outside of SPI-2, SspH1 and SspH2, are secreted through this TTSS (16). These two proteins contribute to the pathogenesis of salmonellosis in calves.
SseC and SseD are two additional SPI-2 proteins that are predicted to be secreted by Salmonella based on homology to secreted proteins in EPEC and Yersinia. SseC is 24% identical and 48% similar to EspD of EPEC (10). EspD is secreted by the TTSS of EPEC into the culture supernatant and is required for the attaching and effacing phenotype of EPEC (15). Other evidence suggests that EspD is part of the recently described secretion filament (14). The SseD protein is 27% identical and 57% similar to EspB of EPEC (10). EspB is exported by the TTSS and is translocated into the cytoplasm of host cells (23, 26). Both SseC and SseD also have homology to YopB of Yersinia pseudotuberculosis, a protein that is required for delivery of Yop proteins into the host cell (6).
The TTSS encoded by SPI-2 is an important component of the virulence strategy of Salmonella and contributes to systemic infection and replication within macrophages. However, the mechanism by which it performs this function is largely unknown. To elucidate this mechanism, it is essential to define the Salmonella proteins that are secreted and translocated into the host cell. Therefore, we have investigated the role of SseC and SseD in virulence and determined their localization in the bacterial cell.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. Liquid cultures of bacteria were grown in Lennox broth (LB) (Difco) or minimal medium with limiting magnesium (pH 5.0) (MgM) (1) as indicated. Antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 25 μg ml−1; chloramphenicol, 20 μg ml−1.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH12S | mcrA Δ(mrr-hsdRMS-mcrBC) F′ lacIqlacZΔM15 | Gibco-BRL |
| ER2508 | ion::Tn10Δ(lac)U169 hsdS20 ara 14 fhuA galK2 rpsL20 xyl5 mtl1 supE44 leuB6 | New England Biolabs |
| Serovar Typhimurium | ||
| SL1344 | Mouse virulent | 27 |
| JK14 | SL1344 ssaT::mTn5, Kanr | This work |
| JK20 | SL1344 ssaV::cam,Cmr | This work |
| JK21 | SL1344 sseC::aphT, Kanr | This work |
| JK22 | SL1344 sseD::aphT, Kanr | This work |
| HH110 | 12023s ssaV::cam,Cmr | 21 |
| P9B7 | 12023s ssaT::mTn5, Kanr | 9 |
| Plasmids | ||
| pBDJ129 | Mini-F ori, P1 replicative ori, variable copy vector, Cmr | 13 |
| pBDJ200 | ColE1 ori, lacIq, Ampr | 13 |
| pJK51 | sseD in pGEM-T Easy, Ampr | This work |
| pJK53 | sseC in pGEM-T Easy, Ampr | This work |
| pJK76 | sseC in pACYC184, Cmr | This work |
| pJK77 | sseD in pACYC184, Cmr | This work |
Ampr, ampicillin resistant; Kanr, kanamycin resistant; Cmr, chloramphenicol resistant; Strr streptomycin resistant.
Construction of mutants.
The sseC mutant was constructed by PCR amplification of sseC from the SL1344 chromosome using primers sseC2 (5′-GTGCTACGTTACTCGCTTCC) and sscA1 (5′-CAGTATGATAGCGCAGAAAACG). The 2-kb PCR product was cloned into the vector pGEM-T Easy (Promega, Madison, Wis.), resulting in plasmid pJK53. A kanamycin resistance gene (aphT) lacking a transcriptional terminator was excised from pUC4KD (13) with BamHI, filled in, and cloned into the NheI site within sseC. The aphT gene cassette was inserted into sseC so that transcription of sseC and aphT was oriented in the same direction. Such gene insertions have been found by our laboratory to have minimal transcriptional effects on downstream genes (13). The sseC::aphT fragment was removed from pGEM-T Easy by digestion with EagI and cloned into the EagI site of pBDJ129. This plasmid was introduced into SL1344 carrying pBDJ200. Selection for bacteria containing double crossovers of the mutation into the chromosome was done as described previously (13), and the presence of the mutation in the Salmonella chromosome was confirmed by PCR. The strain with the defined mutation in sseC was designated JK21. This mutation does not affect the expression of sseD, the gene immediately downstream of sseC, as determined by Western blot analysis of the strain for production of SseD (data not shown).
The sseD mutant was constructed by PCR amplifying sseD from the SL1344 chromosome using primers sseD1 (5′-GAAACGGCAATGATGTGCGG) and sseD2 (5′CTGCCATGAGGCGTAACCAC). The 2-kb product was cloned into pGEM-T Easy (resulting in plasmid pJK51), and a filled-in BamHI fragment containing the aphT cassette was cloned into the 42-bp deletion within sseD created by digestion with EcoRV. The sseD::aphT fragment was cloned into pBDJ129 as above, followed by selection for allelic exchange into the SL1344 chromosome. The strain with the defined mutation in sseD was named JK22.
The ssaV::cam mutation of strain HH110 and the ssaT::mTn5 mutation of strain P9B7 were moved into S. enterica serovar Typhimurium SL1344 by P22-mediated transduction and designated JK20 and JK14, respectively.
Construction of plasmids.
Plasmids expressing SseC and SseD were used to complement JK21 and JK22. Plasmids expressing SseC and SseD from the lac promoter in pGEM-T Easy were unstable in Salmonella (data not shown); therefore, the sseC and sseD genes were cloned into the low-copy vector pACYC184 and expressed from the uninducible promoter of the tetracycline resistance gene. The sseC sequence was removed from pJK53 by digestion with SalI and SphI and cloned into the same sites within the Tetr gene of pACYC184. The sseD sequence was cloned from pJK51 into pACYC184 using the same procedure. Expression of SseC and SseD from these plasmids was confirmed by Western blotting (data not shown).
Mouse virulence.
BALB/c mice, 6 to 8 weeks old (Harlan Sprague), were used for all animal infection experiments. The mice were housed and maintained by the University of Iowa Department of Laboratory Animal Medicine. Bacteria were prepared for the experiment by being grown in LB overnight at 37°C and then diluted 1:100 into fresh medium and grown to an optical density at 600 nm (OD600) of 0.4 to 0.5. Cultures were diluted in phosphate-buffered saline and injected intraperitoneally into mice in a volume of 100 μl. Appropriate dilutions of the cultures were plated on LB plates to quantitate the inoculum dose. Survival of infected mice was monitored for 21 days.
Purification of protein and generation of antibodies.
Recombinant SseC and SseD proteins were generated by constructing protein fusions to the maltose binding protein (MBP) using the pMAL-c2 vector (New England Biolabs, Beverly, Mass.). To clone into the fusion vector, sseC and sseD were PCR amplified with primers sseC3 (5′-GATCATAATATTATGAATCGAATTCACAGTAATA) plus sseC4 (5′-GACTTGTCTAGATTAAGCGCGATAGCCAG) and sseD3 (5′-GATCATGAACTGATTCTATGGAAGCGAGTAACGTAGC) plus sseD4 (5′-GTCAGTAAGCTTTACCTCGTTAATGCCCGGAG) and cloned into pGEM-T Easy. sseC and sseD were subsequently cloned into the Xmnl site of pMAL-c2 so that an in-frame fusion with malE was generated. Evidence that the desired fusions were constructed was obtained by sequencing each protein fusion plasmid. Recombinant protein was expressed from the protease-deficient E. coli strain ER2508 and affinity purified by binding to an amylose resin column as specified by the manufacturer. Purified proteins were submitted to Elmira Biologicals (Iowa City, Iowa) for production of polyclonal sheep antibodies. Antibodies were purified from sheep serum collected after the second booster injection of MBP-SseC or MBP-SseD. To purify the antibody, recombinant protein was applied to a polyvinylidene difluoride membrane and incubated with serum overnight at 4°C. The filters were washed, and the antibody was dissociated by treatment with 0.2 M glycine (pH 3.0). The antibody was neutralized with sodium hydroxide and concentrated in a Microcon-10 concentrator (Millipore, Bedford, Mass.).
Fractionation of cells.
Bacterial cultures were grown overnight in the indicated media with aeration, and cells were harvested by centrifugation. Supernatant proteins were isolated as described previously (19). Cell pellets were resuspended in a buffer consisting of 20 mM Tris-HCl, 200 mM NaCl, and 1 mM EDTA and frozen at −80°C. Cells were thawed on ice and lysed by sonication until the suspension became viscous. Cellular debris was removed by centrifugation at 9,000 × g for 15 min. The supernatant was centrifuged at 100,000 × g for 60 min at 4°C to separate the membrane proteins (pellet) from the soluble cytoplasmic and periplasmic proteins as described by Mobley et al. (17). Membrane proteins were resuspended in polyacrylamide gel electrophoresis (PAGE) loading buffer, and soluble proteins were concentrated using Centriplus-10 and Microcon-10 concentrators. Where indicated, cell pellets were treated with xylene or proteinase K prior to the fractionation protocol as described previously (1).
As a separation purity control, a culture of SL1344 was fractionated and the activity of the membrane protein NADH dehydrogenase in the membrane and soluble fractions was determined. Samples were prepared in the same way as above, except that a 20 mM sodium phosphate (pH 6.8) solution was used to resuspend the cells and membrane proteins. The protein concentration from each fraction was determined using protein assay solution (Bio-Rad, Hercules, Calif.). Protein from each fraction was incubated with 2 mM NADH for 15 min at room temperature, and the OD340 was determined. The NADH dehydrogenase activity was expressed as micromoles of NADH degraded per minute per milligram of protein.
Western blotting.
Proteins were separated by sodium dodecyl sulfate-PAGE (12.5% polyacrylamide). The equivalent of 10 ml of an OD600 = 0.4 culture of each fraction was loaded into a well. Proteins were transferred to nitrocellulose by electroblotting and incubated overnight with a 1:10,000 dilution of SseC or SseD primary antibody. Alkaline phosphatase-conjugated rabbit anti-sheep immunoglobulin G (Jackson ImmunoResearch, West Grove, Pa.) was used as a secondary antibody. Detection was carried out using the Immun-Star chemiluminescent protein detection system (Bio-Rad) followed by imaging with Kodak BioMAX film or the FujiFilm LAS-1000 luminescent imager and accompanying software. The relative intensity of specific bands was quantitated with FujiFilm ImageGauge 3.12 software. As estimated by running the Benchmark prestained protein ladder (Gibco-BRL), SseC ran at 62 kDa and SseD ran at 19 kDa. Purified SseC or SseD protein was run on all gels as an internal size standard.
RESULTS
SseD is required for virulence.
The virulence requirement of several SPI-2 genes predicted to encode secreted effector proteins has been determined (10); however, the virulence requirement of the gene encoding SseD is unknown. To determine the role of SseD in virulence, we constructed a chromosomal mutation in sseD (called JK22) that has minimal polar effects on downstream genes and measured its virulence in mice. We also constructed a chromosomal mutation in sseC (called JK21) and tested its virulence in mice to confirm its phenotype. We infected groups of five mice intraperitoneally with at least 105 CFU of JK21, JK21 complemented with plasmid pJK76, JK22, or JK22 complemented with plasmid pJK77 and monitored survival of the mice for 21 days. Five mice were infected with 100 CFU (about four times the 50% lethal dose [LD50]) (19) of the wild-type strain SL1344 as a control for the virulence of the strains. The control mice infected with SL1344 died within 5 days. The mice infected with JK21 carrying pJK76 died within 3 days, and those infected with JK22 carrying pJK77 all died within 5 days (Table 2). In contrast, none of the mice infected with JK21 or JK22 died, indicating that the LD50 of the sseC mutant was increased 7,800-fold to >2 × 105 and that the LD50 of the sseD mutant was increased more than 10,000-fold to >2.5 × 105 CFU. In addition, the experiment indicated that each mutant became significantly more virulent after the introduction of a plasmid carrying the appropriate single intact gene, suggesting that the chromosomal mutations were nonpolar.
TABLE 2.
Mouse virulence of sseC and sseD mutants
| Strain (genotype) | Plasmid (genotype) | Inoculum (CFU) | Survival (no. surviving/ total no.)a |
|---|---|---|---|
| SL1344 (wild type) | 1.0 × 102 | 0/5 | |
| JK21 (sseC::aphT) | 2.0 × 105 | 5/5 | |
| JK21 (sseC::aphT) | pJK76 (sseC+) | 1.1 × 105 | 0/5 |
| JK22 (sseD::aphT) | 2.5 × 105 | 5/5 | |
| JK22 (sseD::aphT) | pJK77 (sseD+) | 1.3 × 105 | 0/5 |
Mice were inoculated intraperitoneally with the indicated number of bacteria, and survival was determined after 21 days.
SseD is induced by growth in minimal medium.
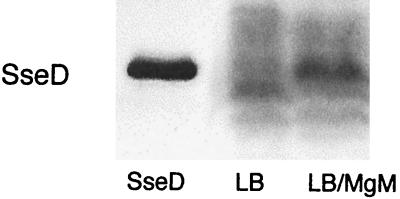
The growth conditions for induction of SPI-2 gene expression and protein localization have been previously established. In vivo, SPI-2 genes are induced by growth inside macrophages for several hours (3). In vitro, these genes are induced by growth in minimal medium with limiting divalent cations (1, 5). Furthermore, localization of SseB to the cell surface occurs as a result of growth under acidic conditions (1). We performed experiments to determine whether we could induce protein expression by growth under the same conditions. A culture of the wild-type Salmonella strain SL1344 was grown overnight in LB, washed in phosphate-buffered saline, and induced for 6 h in MgM. Samples were collected after overnight growth in LB and after 6 h of induction in MgM. Western blotting of these samples with SseD antiserum indicated that SseD was present after the 6-h induction in MgM but not after growth in LB (Fig. 1). Since the pH 5.0 MgM induced the expression of SseD, cells were grown in this medium in future experiments. SseC was not included in the original experiments, but we subsequently found that it is also expressed in MgM, as described in the following section.
FIG. 1.
Expression of SseD. Samples from SL1344 culture grown in LB (lane LB) or grown in LB followed by a 6-h induction in MgM (pH 5) (lane LB/MgM) were electrophoresed, transferred to a membrane, and probed with SseD antiserum. Purified recombinant SseD was run as a size standard (lane SseD).
SseC and SseD are localized to the bacterial membrane.
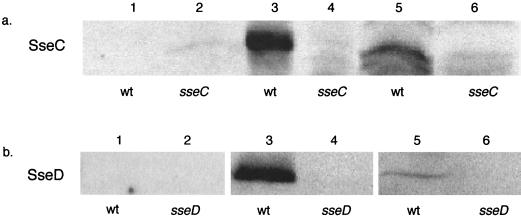
We next sought to determine where SseC and SseD are localized in the bacterium and whether they are secreted into the culture medium under in vitro growth conditions. The wild-type strain SL 1344 was grown in MgM (pH 5.0), and proteins from the culture supernatant, bacterial membranes, and cytoplasm/periplasm were collected as described in Materials and Methods. Coomassie blue staining of these fractions, following sodium dodecyl sulfate-PAGE, revealed the presence of many proteins in each fraction except for the supernatant fraction (data not shown). Western blotting of these fractions with SseC (Fig. 2a) or SseD (Fig. 2b) antiserum indicated that neither protein was secreted into the culture supernatant under these growth conditions (lane 1). The majority of SseC and SseD was found in the membrane fraction (lane 3), although some protein (more for SseC than SseD) could be detected in the soluble fraction (lane 5). As a control for specificity of the antiserum, immunoblots of protein fractions isolated from sseC (JK21) and sseD (JK22) mutants did not detect protein at the size of SseC or SseD (lanes 2, 4, and 6). As a control for the purity of the fractions, the activity of the membrane protein NADH dehydrogenase was measured. We found that 100% of its activity was in the membrane fraction, demonstrating that no detectable levels of membrane proteins are contaminating the soluble fraction.
FIG. 2.
Localization of SseC and SseD. (a) Culture supernatant (lanes 1 and 2), cell membrane (lanes 3 and 4), or soluble protein (lanes 5 and 6) fractions from SL1344 (wt) or JK21 (sseC) grown in MgM were probed with SseC antiserum. (b) Culture supernatant (lanes 1 and 2), cell membrane (lanes 3 and 4), or soluble protein (lanes 5 and 6) fractions from SL 1344 (wt) or JK22 (sseD) grown in MgM were probed with SseD antiserum.
SseC and SseD are not localized properly in secretion apparatus mutants.
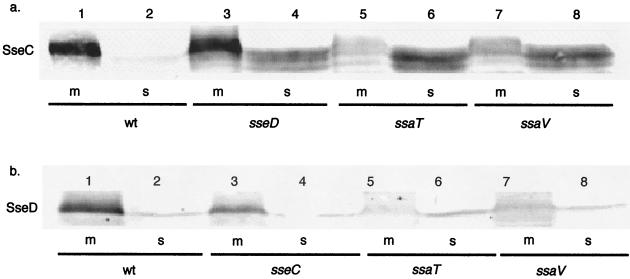
Next, we examined whether the localization of SseC and SseD is affected by mutations in other SPI-2 genes. SsaT and SsaV are predicted to form part of the type III secretion apparatus. A transposon insertion in ssaT results in the loss of mouse virulence (9), indicating that ssaT and/or the downstream gene, ssaU, is important in SPI-2 function. A nonpolar mutation in ssaV also reduces Salmonella virulence, indicating that ssaV is also important in SPI-2 function (21). We analyzed membrane and soluble protein fractions collected from wild-type (SL1344) cells and the sseD (JK22), ssaT (JK14), and ssaV (JK21) mutant strains for the presence of SseC by immunoblotting with SseC antiserum and determining the relative concentrations of each band as described in Materials and Methods (Fig. 3a). We consistently observed that the SseC protein from soluble fractions migrated more quickly in PAGE than did the SseC protein from membrane fractions. The reason for this finding is unclear, but one possible explanation is that when SseC is removed from its usual membrane association, its mobility is altered. Significant amounts of SseC were found in the membrane fraction of the wild type (113.4 relative units) and the sseD mutant (95.4 relative units) (lanes 1 and 3). The supernatant fraction of the parent strain had 0.0 relative unit of SseC, but the sseD mutant had 18.0 relative units of SseC in the soluble fraction. Interestingly, the majority of the SseC protein was found in the soluble fraction of the ssaT and ssaV mutants (87.0 and 78.5 relative units, respectively) (lanes 6 and 8). No SseC protein above background could be detected in the membrane of the ssaT mutant (0.0 relative unit), and we detected 8.6 relative units in the membrane of the ssaV mutant.
FIG. 3.
Localization of SseC and SseD in SPI-2 mutants. (a) Cell membrane (m) and soluble (s) protein fractions from SL1344 (wt), JK22 (sseD), JK14 (ssaT), or JK20 (ssaV) were probed with SseC antiserum. (b) Fractions from SL1344 (wt), JK21 (sseC), JK14 (ssaT), or JK20 (ssaV) were probed with SseD antiserum. Relative units of SseC protein in panel a, as measured by Fujifilm ImageGauge, were as follows: lane 1, 113.4; lane 2, 0.0; lane 3, 95.4; lane 4, 18.0; lane 5, 0.0; lane 6, 87.0; lane 7, 8.6; lane 8, 78.5. Relative units of SseD protein in panel b, as measured by Fujifilm ImageGauge, were as follows: lane 1, 107.4; lane 2, 1.0; lane 3, 69.1; lane 4, 1.0; lane 5, 7.0; lane 6, 1.0; lane 7, 9.0; lane 8, 1.0.
A similar set of experiments was performed to examine SseD localization using SseD antiserum (Fig. 3b). SseD was found in the membrane fraction of wild-type (107.4 relative units) and sseC mutant (69.1 relative units) cells (lanes 1 and 3), while little protein above background was detected in the membrane fractions of the apparatus mutants ssaT and ssaV (7.0 and 9.0 relative units, respectively) (lanes 5 and 7). There was not a comparable increase in the level of SseD protein above background in the soluble fraction, which may be due to degradation of SseD when it is not properly localized. These data indicate that a functional TTSS is required for proper localization of SseC and SseD to the bacterial membrane.
SseC and SseD are secreted to the bacterial cell surface.
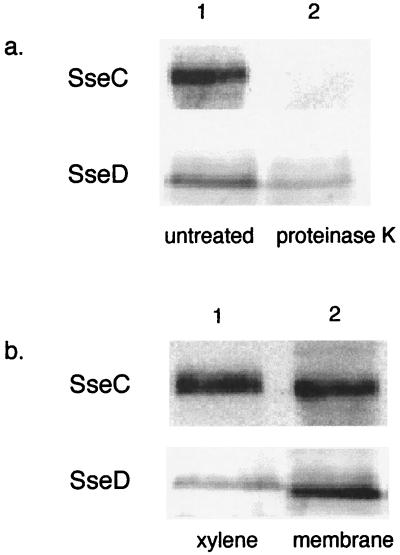
Our cell fractionation studies indicate that SseC and SseD are localized to the bacterial membrane. As one approach to determine whether SseC and SseD are exposed to the extracellular medium, we treated whole bacteria with proteinase K as described in Materials and Methods. We found that SseC and SseD were degraded by proteinase K treatment, but to different extents. SseC was completely degraded by proteinase K treatment, while a small percentage of SseD was still detectable in the membrane after proteinase K treatment (Fig. 4a). Other workers have reported that SseB can be found on the bacterial cell surface under certain growth conditions, in a location where it can be stripped from the cell by treatment with xylene (1). This finding was interpreted to mean that the protein is loosely associated with the bacterial membrane. We used this same experimental technique to determine whether SseC and SseD have a similar association with the membrane. Our experiments revealed that about 50% of the SseC protein in the membrane could be extracted from the cell by xylene (Fig. 4b). SseD could also be extracted from the membrane by xylene, but to a lesser extent (Fig. 4b). These experimental results indicate that SseC and SseD are exposed to the extracellular environment, where they are loosely associated with the bacterial membrane.
FIG. 4.
SseC and SseD are exposed to the extracellular medium. (a) SL1344 cell cultures grown in MgM (pH 5) were either incubated with proteinase K or incubated under identical conditions without the addition of proteinase K. Membrane fractions from untreated cells (lane 1) and proteinase K-treated cells (lane 2) were probed with SseC and SseD antisera. (b) SL1344 cell cultures grown in MgM (pH 5) were treated with xylene. Proteins stripped away from the cell by xylene (lane 1) and residual membrane proteins isolated from the treated cell pellet (lane 2) were detected using SseC and SseD antisera.
Experiments were performed to determine whether SseC or SseD was secreted in vivo after infection of J774 cells. We were unable to detect SseD in J774 cells after 6 h of infection in Western blot experiments (data not shown). Likewise, secretion of SseC into J774 cells was not detected by immunofluorescence microscopy (data not shown).
DISCUSSION
In the work presented here, we demonstrate that sseD is required for the mouse virulence phenotype of Salmonella. An infectious dose of 7,800 or 10,000 times the LD50 of the wild-type strain was not sufficient to cause lethal infection by sseC and sseD mutants, respectively. This adds to the list of SPI-2 genes that are known to be essential for virulence. The virulence requirement of several predicted effector proteins has been determined in other studies. Hensel et al. demonstrated that mice infected with sseC, sseB, or sseA mutants survived a dose of up to 104 CFU, while mutants with mutations in sseF and sseG were less attenuated but still showed a defect in virulence (10). In addition, the LD50 of a spiC (ssaB) mutant was reported to be greater than 3.6 × 106 CFU (24).
Because of their role in virulence and their homology to type III secreted effector proteins in other bacteria, we have examined aspects of SseC and SseD localization in greater detail. We have found that SseC and SseD are induced by growth in minimal medium with limiting magnesium, as has been reported for other SPI-2 proteins. However, even after growth in MgM, both proteins are still expressed at low levels. A large amount of cell culture (10 ml) was required to detect SseC and SseD expression by Western blotting. This requirement may indicate that the in vitro growth conditions being used are still not optimal for induction of these proteins or, alternatively, that the bacteria require only low levels of expression of these proteins for their function.
A focus of our work was to determine the cellular localization of SseC and SseD. We found that both proteins are located in the bacterial membrane after growth in MgM. SseC was associated rather loosely with the cell and could be completely digested with proteinase K and extracted by treatment with xylene. SseD was also susceptible to these treatments, but to a lesser extent. These observations indicate that SseC and SseD are secreted to the bacterial cell surface during growth in MgM and are similar to findings made about the localization of SseB (1). In addition, localization of SseC and SseD to the bacterial membrane required a functional TTSS.
We think it is important to emphasize that SseC and SseD are type III secreted proteins; they are secreted to the bacterial membrane, and this secretion relies on a functional type III secretion apparatus. This differs slightly from what is often observed with proteins secreted from other TTSSs, because secretion into the growth medium can often be detected. For example, the TTSSs of several organisms, including Yersinia, EPEC, Shigella, and Salmonella (SPI-1), secrete proteins into the culture supernatant (11). Secretion of proteins into the culture supernatant by the SPI-2 TTSS has not been reported, perhaps suggesting that the in vitro growth conditions that induce expression are still not optimized for protein secretion. The SPI-2 genes differ from many of the other TTSS genes that have been studied in that they are induced and exert their effects within a macrophage. These properties seem to suggest that the SPI-2 TTSS of internalized Salmonella mediates secretion of proteins into the phagosomal compartment and/or translocation of proteins across the phagosomal membrane into the host cystosol. Some aspect of this intracellular environment is not being re-created by growth in liquid culture to signal protein secretion into the growth medium, and perhaps this signal is provided only by a macrophage after phagocytosis.
Proteins that are secreted by TTSSs generally function in one of two ways: as effector proteins that are translocated into the host cell and alter host cell processes or as translocators that are necessary for injecting effector proteins into the host cell. With the finding that SseC and SseD are secreted, these proteins become candidates for either effectors or translocators. Techniques used to detect translocation of effector proteins into host cells have included fractionation and immunoblotting of infected host cells, immunofluorescence microscopy, and measurement of intracellular enzyme activity of cyaA fusions to secreted proteins (16, 24). We used the first two techniques mentioned above to search for the presence of SseC and SseD within cells, but those experiments were unsuccessful in detecting the proteins. Techniques with greater sensitivity may be able to detect SseC or SseD within host cells. Alternatively, it may be that the proteins are not translocated into host cells but instead act as translocators similar to their homolog, YopB, from Yersinia (6).
We now know of four proteins encoded by SPI-2 that are secreted by Salmonella: SseB, SseC, SseD, and SpiC. SpiC is an effector protein that is translocated into Salmonella-infected macrophages and interferes with normal membrane trafficking in the host cell, including phagosome-lysosome fusion (24). SseB expression is induced by growth in minimal media like several other SPI-2 proteins, and it is secreted to the surface of Salmonella after acid induction (1). In a separate study, it was found that SseB is necessary for Salmonella to prevent NADPH oxidase localization and oxyradical formation at the phagosomal membrane of murine macrophages (25). However, at this point it is unclear whether this effect is due to the direct activity of SseB within the macrophage or whether SseB is required for translocating other effector proteins into the macrophage. It has also been observed that our sseD mutant is defective in NADPH oxidase assembly (A. Gallois, personal communication). These new findings serve as further evidence that Salmonella survives within macrophages due, in part, to its ability to interfere with the generation of reactive oxygen intermediates. Further experiments to determine whether SseD is directly responsible for the defect in NADPH oxidase assembly or whether a more general defect in type III protein translocation is the cause will be performed in the future.
In this study, we have demonstrated that SseC and SseD are required for systemic infection of mice by Salmonella and that both proteins are secreted to the bacterial surface in a type III-dependent fashion. Continued work to investigate these and other SPI-2 secreted proteins will be critical in elucidating the mechanism by which SPI-2 mediates Salmonella survival within macrophages.
ACKNOWLEDGMENTS
We thank D. Holden for providing strain HH110 and S. Falkow for sending us strain P9B7.
J.R.K. is supported by NIH predoctoral training grant GM08629. This work was supported by NIH grant AI38268 and a grant from the Roy J. Carver Charitable Trust (to B.D.J.)
REFERENCES
- 1.Beuzon C R, Banks G, Deiwick J, Hensel M, Holden D W. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999;33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 2.Buchmeier N A, Heffron F. Intracellular survival of wild-type Salmonella typhimurium and macrophage-sensitive mutants in diverse populations of macrophages. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 4.Darwin K H, Miller V L. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev. 1999;12:405–428. doi: 10.1128/cmr.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 6.Hakansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 7.Hensel M, Nikolaus T, Egelseer C. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol Microbiol. 1999;31:489–498. doi: 10.1046/j.1365-2958.1999.01190.x. [DOI] [PubMed] [Google Scholar]
- 8.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 9.Hensel M, Shea J E, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden D W. Functional analysis of ssaJ and the ssaK/IU operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 10.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang F C, Holden D W. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 11.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones B D, Falkow S. Salmonellosis: host immune responses and bacterial virulence determinants. Annu Rev Immunol. 1996;14:533–561. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 13.Klein J R, Fahlen T F, Jones B D. Transcriptional organization and function of invasion genes within Salmonella pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect Immun. 2000;68:3368–3376. doi: 10.1128/iai.68.6.3368-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai L C, Wainwright L A, Stone K D, Donnenberg M S. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao E A, Scherer C A, Tsolis R M, Kingsley R A, Adams L G, Baumler A J, Miller S I. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 17.Mobley H L T, Jones B D, Jerse A E. Cloning of urease gene sequences from Providencia stuartii. Infect Immun. 1986;54:161–169. doi: 10.1128/iai.54.1.161-169.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 20.Rubin R H, Weinstein L. Salmonellosis: microbiologic, pathogenic, and clincal features. New York, N.Y: Stratton Intercontinental Medical Book Corp.; 1977. [Google Scholar]
- 21.Shea J E, Beuzon C R, Gleeson C, Mundy R, Holden D W. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor K A, O'Connell C B, Luther P W, Donnenberg M S. The EspB protein of enteropathogenic Escherichia coli is targeted to the cytoplasm of infected HeLa cells. Infect Immun. 1998;66:5501–5507. doi: 10.1128/iai.66.11.5501-5507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchiya K, Barbieri M, Funato K, Shah A, Stahl P, Groisman E. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 1999;18:3924–3933. doi: 10.1093/emboj/18.14.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez T A, Xu Y, Jones C J, Holden D W, Lucia S M, Dinauer M C, Mastroeni P, Fang F C. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science. 2000;287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 26.Wolff C, Nisan I, Hanski E, Frankel G, Rosenshine I. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1998;28:143–155. doi: 10.1046/j.1365-2958.1998.00782.x. [DOI] [PubMed] [Google Scholar]
- 27.Wray C, Sojka W J. Experimental Salmonella typhimurium in calves. Res Vet Sci. 1978;25:139–143. [PubMed] [Google Scholar]