Abstract
Monocyte/macrophage lineage cells are highly plastic and can differentiate into various cells under different environmental stimuli. Bone-resorbing osteoclasts are derived from the monocyte/macrophage lineage in response to receptor activator of NF-κB ligand (RANKL). However, the epigenetic signature contributing to the fate commitment of monocyte/macrophage lineage differentiation into human osteoclasts is largely unknown. In this study, we identified RANKL-responsive human osteoclast-specific superenhancers (SEs) and SE-associated enhancer RNAs (SE-eRNAs) by integrating data obtained from ChIP-seq, ATAC-seq, nuclear RNA-seq and PRO-seq analyses. RANKL induced the formation of 200 SEs, which are large clusters of enhancers, while suppressing 148 SEs in macrophages. RANKL-responsive SEs were strongly correlated with genes in the osteoclastogenic program and were selectively increased in human osteoclasts but marginally presented in osteoblasts, CD4+ T cells, and CD34+ cells. In addition to the major transcription factors identified in osteoclasts, we found that BATF binding motifs were highly enriched in RANKL-responsive SEs. The depletion of BATF1/3 inhibited RANKL-induced osteoclast differentiation. Furthermore, we found increased chromatin accessibility in SE regions, where RNA polymerase II was significantly recruited to induce the extragenic transcription of SE-eRNAs, in human osteoclasts. Knocking down SE-eRNAs in the vicinity of the NFATc1 gene diminished the expression of NFATc1, a major regulator of osteoclasts, and osteoclast differentiation. Inhibiting BET proteins suppressed the formation of some RANKL-responsive SEs and NFATc1-associated SEs, and the expression of SE-eRNA:NFATc1. Moreover, SE-eRNA:NFATc1 was highly expressed in the synovial macrophages of rheumatoid arthritis patients exhibiting high-osteoclastogenic potential. Our genome-wide analysis revealed RANKL-inducible SEs and SE-eRNAs as osteoclast-specific signatures, which may contribute to the development of osteoclast-specific therapeutic interventions.
Keywords: Osteoclasts, super-enhancers, enhancer RNAs, Rheumatoid arthritis
Subject terms: Osteoimmunology, Epigenetics in immune cells
Introduction
Osteoclasts are large, multinucleated cells derived from myeloid cells and are the primary cells responsible for bone resorption and bone homeostasis [1–3]. Dysregulated osteoclast differentiation and function lead to bone destruction in pathological conditions, including rheumatoid arthritis (RA) [4–6]. Macrophage-colony stimulating factor (M-CSF) and receptor activator of nuclear factor kappa-Β ligand (RANKL) are key regulators of osteoclast differentiation, survival, and activity. RANKL binds to its receptor, RANK [2] to activate canonical osteoclastogenic signaling pathways and induce osteoclastogenic transcriptional and epigenetic programs during osteoclastogenesis [7–11]. RANKL- or RANK-deficient mice exhibit severe osteopetrosis with impaired osteoclast activity [12–14]. The mechanisms by which RANKL induces osteoclast differentiation have been extensively studied, and the key transcription factors that are involved in initiating and maintaining the osteoclastogenic program have well characterized. Among these transcription factors, nuclear factor of activated T cells, c1 (NFATc1), is a master regulator of osteoclastogenesis [15], and NFATc1 deficiency leads to the acquisition of an osteoporotic phenotype in mice [15–17]. However, NFATc1 and other key factors driving osteoclastogenesis are also expressed and function in other cell types; however, the cell-specific regulatory mechanisms controlling the expression of these factors in osteoclasts remain poorly characterized.
Environmental stimuli trigger immediate and/or chronic changes in gene transcription, leading to the differentiation of cells into various subsets with distinct functions and phenotypes. Epigenetic regulation plays a critical role in cell-type-specific gene expression that establishes physiological phenotypes of cells and contributes to human diseases [11, 18, 19]. Considerable efforts have been made to understand epigenetic regulation in osteoclasts [7–11, 20]. Nishikawa et al. demonstrated that RANKL-induced S-adenosylmethionine (SAM) production mediated DNA methylation via de novo DNA methyltransferase 3a (Dnmt3a) during osteoclastogenesis and that inhibiting Dnmt3a modulated osteoclastogenesis and ovariectomy-induced bone loss [20]. Recent studies, including our research, have shown that targeting bromodomain and extraterminal (BET) proteins, chromatin readers, using small-molecule inhibitors suppressed osteoclastogenesis and attenuated bone destruction in inflammatory arthritis [21, 22]. However, targeting epigenetic regulators broadly affects many cell types since most cells utilize similar chromatin regulators to regulate epigenetic mechanisms. Therefore, unraveling the osteoclast-specific regulatory mechanism is important for understanding the epigenetic network mediating the control of osteoclast formation and activity and for identifying a safe therapeutic target.
Enhancers are cis-regulatory elements that carry transcription factor-binding motifs [23]. Enhancers control cell-type specific transcriptional activation or suppression independent of the orientation or distance to their target promoters [23]. Superenhancers (SEs) are clusters of enhancers that are critical to cell identity and lineage commitment by driving high-level expression of genes encoding key regulators of cell identity and cell fate [24–28]. SEs are hubs that recruit large protein complexes, including the transcriptional apparatus, transcription factors, coactivators and chromatin regulators, and thus promote the expression of cell fate-related genes [29]. Histone 3 acetylation at lysine 27 (H3K27ac), an active enhancer mark, is highly enriched in SEs [30]. In addition, SEs are sensitive to the dynamic changes in pioneer master regulators during differentiation [31]. A recent study identified SEs in mouse osteoclasts base on ChIP-seq data of PU.1, a myeloid pioneer transcription factor [32]. However, human osteoclast-specific SEs have not yet been identified. Moreover, recent data have revealed that enhancers produce noncoding RNA species called enhancer RNAs (eRNAs) [33–36]. eRNAs are transcribed from active tissue-specific enhancers, including superenhancers, and their biological functions have been discovered in many different cells [37–50]. However, the function and mechanism of superenhancers and SE-associated eRNAs in RANKL-stimulated human osteoclasts have yet to be firmly established.
In our present study, we investigated the SE landscape during human osteoclastogenesis and defined RANKL-responsive SEs and SE-associated eRNAs in human osteoclasts. In addition to key osteoclastogenic transcription factors enriched in RANKL-responsive SEs, we found basic leucine zipper ATF-like transcription factor (BATF)-binding motifs in RANKL-responsive SEs. Moreover, we discovered that BATF1/3 is a positive regulator of osteoclasts. We also identified eRNAs transcribed from superenhancers (named SE-eRNAs) in human osteoclasts. Furthermore, RANKL-responsive SE-eRNAs were found in RA synovial macrophages, which are prone to differentiate into osteoclasts. Accordingly, knocking down SE-eRNAs in the vicinity of the NFATc1 gene suppressed NFATc1 expression and RANKL-induced osteoclastogenesis. Our study offers the first evidence of a potential role for RANKL-responsive SE-eRNAs in the reprogramming of myeloid cells to drive their differentiation into human osteoclasts and provides insights into the development of hyperactive osteoclasts in a pathological setting.
Materials and methods
Human studies
Human synovial fluid (SF) samples were collected from RA patients as previously described [51]. The protocol was approved by the Ethics Committee of Seoul National University Hospital Institutional Review Board (1511-094-721) and by the Hospital for Special Surgery Institutional Review Board (2016-957, 2016-958, and 2016-139). A diagnosis of RA was based on the 1987 revised criteria presented by the American College of Rheumatology [52]. Information about patients’ medications was limited, and therefore, we were unable to correlate our findings with patient therapies.
Cell culture
Peripheral blood mononuclear cells (PBMCs) obtained from blood leukocyte preparations were purchased from the New York Blood Center, or mononuclear cells in the SF of RA patients were isolated by density gradient centrifugation with Ficoll (Invitrogen, Carlsbad, CA). CD14+ cells were obtained via isolation using anti-CD14 magnetic beads, as recommended by the bead manufacturer (Miltenyi Biotec, CA). Human CD14+ cells were cultured in α-MEM medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS, HyClone; SH30070.03) and 1% L-glutamine with 20 ng/ml M-CSF for 24 hours to generate osteoclast precursor cells (OCPs). The purity of the monocytes was >97%, as verified by flow cytometry analysis [21].
For human osteoclastogenesis assays, cells were added to 96-well plates at a seeding density of 5 × 104 cells per well in triplicate. Osteoclast precursors were incubated with 20 ng/ml M-CSF and 40 ng/ml human soluble RANKL (PeproTech, Rocky Hill, NJ) for as many as 5 days in α-MEM supplemented with 10% FBS and 1% L-glutamine. Cytokines were replenished every 3 days. Each day, cells were fixed and stained for tartrate-resistant acid phosphatase (TRAP) using an acid phosphatase leukocyte diagnostic kit (Sigma; 387 A) as recommended by the manufacturer. Multinucleated (more than 3 nuclei) TRAP-positive osteoclasts were counted in triplicate wells. All cell cultures were established via a previously published method with some modifications [21].
Gene expression analysis
For real-time qPCR, DNA-free RNA was obtained using an RNeasy Mini Kit from QIAGEN with DNase treatment, and 0.5 mg of total RNA was reverse transcribed using a First-Strand cDNA Synthesis kit (Fermentas, Hanover, MD). Real-time PCR was performed in triplicate using an iCycler iQ thermal cycler and detection system (Applied Biosystems, Carlsbad, CA) following the manufacturer’s protocols. The expression of the analyzed genes was normalized relative to the levels of TBP. The primer sequences are listed in Supplementary Table 1.
Immunoblot
Whole cell extracts were prepared by lysis in buffer containing 1x Lamin sample buffer (Bio–Rad) and 2-mercaptoethanol (Sigma). Proteins were separated on 7.5% SDS‒PAGE gels, transferred to polyvinylidene difluoride (PVDf) membranes (Millipore; ISEQ00010), and detected by the antibodies listed in the figure legends. NFATc1 (556602, BD Pharmingen) and α-tubulin (T9026, Sigma Aldrich).
TRAP-staining
Cells were fixed and stained for TRAP using am acid phosphatase leukocyte diagnostic kit (Sigma) as recommended by the manufacturer. Nuclei were stained with methyl green. Multinucleated (more than three nuclei), TRAP-positive osteoclasts were counted in triplicate wells.
RNA interference
A total of 0.2 nmol of antisense LNAs (LNATM GapmeR) and siRNAs (BATF1 and BATF3) were designed and synthesized by Qiagen (Exiqon) and Ambion, respectively. To knock down human Se-RNA, NFATc1 or control siRNA was transfected into primary human CD14+ monocytes with the Amaxa Nucleofector Program Y-001 using a Human Monocyte Nucleofector kit (Amaxa), as previously described [53]. The custom antisense-LNA GapmeR sequence for SE-RNA:NFATc1_1 was 5’TGTTTACACGCTAACAGGATGACAGCAGACACTGTGTGAAATCAGTCAGT3’ (Fig. 7); the SE-RNA:NFATc1_2 sequence was 5’ATTCAGCTCCACTTA3’ (Supplementary Fig. 5); the predesigned siRNA sequence for BATF1 (Ambion, siRNA ID# 3851) was 5’GGGAACGGUUAUUUUUCUAtt3’; and the predesigned siRNA BATF3 (Ambion, siRNA ID# 116229) was 5’GCUAGUAGGUUCUGCUGUUtt3’ (Fig. 3).
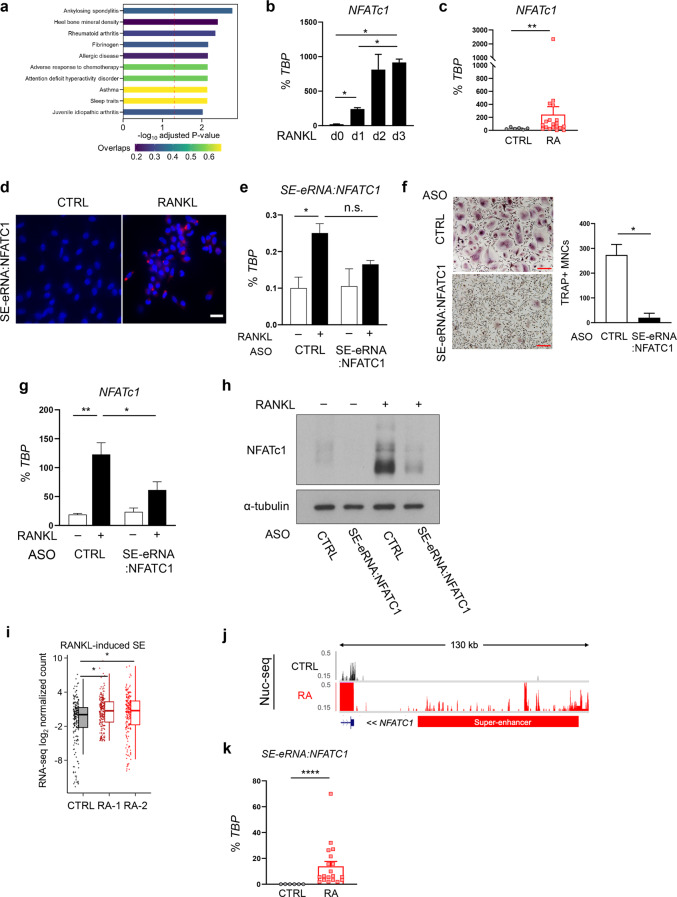
Fig. 7.
RANKL-induced SE-eRNAs are enriched in synovial CD14+ cells from RA patients. a Disease ontology for dREG peak-associated genes with corresponding adjusted p values. RT‒qPCR analysis of NFATc1 expression during osteoclastogenesis (b) and in RA synovial CD14+ cells (c), normalized to the expression of TBP mRNA. d Fluorescence in situ hybridization (FISH) with human osteoclasts. DAPI is shown in blue, and signals for SE-eRNA:NFATc1 are shown in red. Scale bar: 50 μm. e–h The expression of SE-eRNA:NFATc1 was knocked down by electroporation with antisense-LNA GapmeR in human OCPs. e The expression of SE-eRNA:NFATc1 was measured by RT‒qPCR under the indicated conditions. f Osteoclastogenesis assay. Cells were subsequently cultured with M-CSF and RANKL for three days. The left panel shows representative images of TRAP-stained cells. The right panel shows the percentage of TRAP+ multinuclear cells (MNCs: more than three nuclei) per control. Scale bar: 200 µm. g NFATc1 mRNA was measured by RT‒qPCR under the indicated conditions. h Immunoblot analysis of NFATc1 expression in human CD14+ cells transfected with control or SE-eRNA:NFATc1-specific siRNA and treated for 24 h with RANKL. α-Tubulin was the loading control. i Box plots showing PRO-seq signals in RANKL-induced SEs in 2 different RA synovial CD14+ cell types (RA-1, RA-2) and CD14+ cells from healthy controls (CTRL). j Representative tracks showing the nuclear seq data of RA synovial CD14+ cells in the proximity of NFATc1 (RA: RA synovial OCPs, CTRL: disease control). k SE-eRNA:NFATc1 was measured via RT‒qPCR (see Methods). The data are shown as the means ± SEMs. *p < 0.05, **p < 0.01, ****p < 0.0001, n.s., not significant, as determined by one-way ANOVA (b, e, g), Student’s t test (c, f, k), or Kruskal‒Wallis test with Bonferroni correction (i)
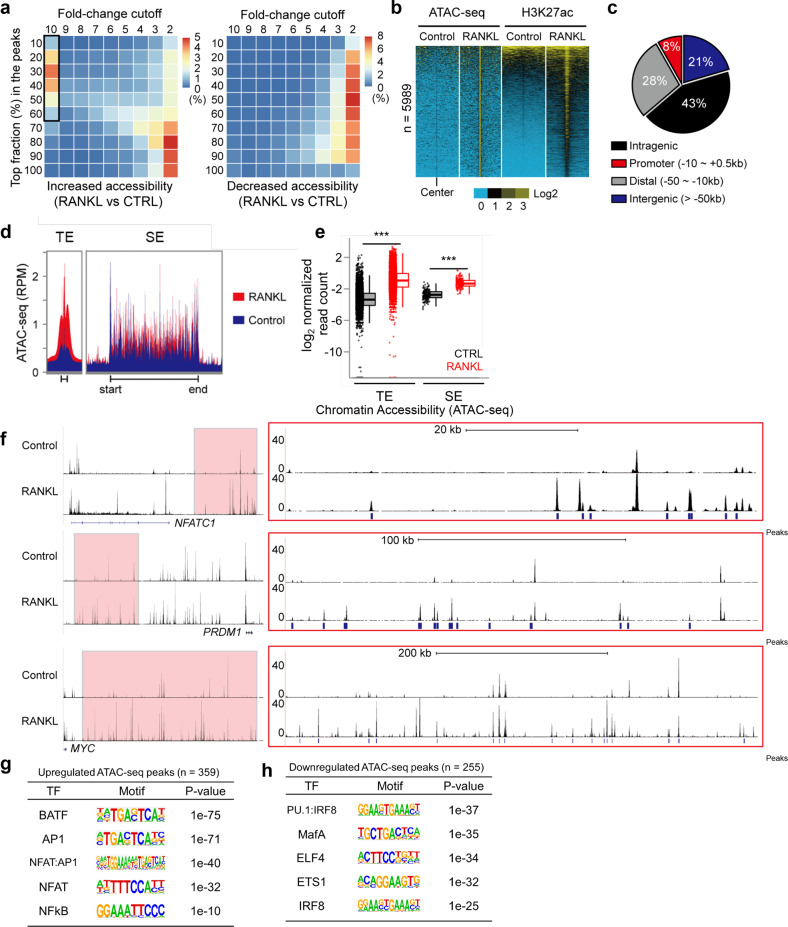
Fig. 3.
Dynamic chromatin accessibility at RANKL-sensitive SEs. a Heatmaps showing differential chromatin accessibility (ATAC-seq) after RANKL (40 ng ml−1) treatment; left panel: increased density of ATAC-seq peaks in RANKL (the black box indicates highly inducible ATAC-seq peaks); right panel: decreased density of ATAC-seq peaks in RANKL. b Heatmaps showing read density determined by ATAC-seq and H3K27ac ChIP-seq in areas ± 2 kb peak centers in RANKL-inducible ATAC-seq peaks (n = 5989, a box from A) in human OCPs treated with or without RANKL (40 ng ml−1). c Pie chart showing the genomic location of RANKL-inducible ATAC-seq peaks. d Read density plot based on ATAC-seq across TE and SE domains under the indicated conditions. e Box plots showing ATAC-seq read densities at TEs and SEs under the indicated conditions. ***p < 0.01 by Kruskal‒Wallis test with Bonferroni correction. f Representative tracks based on ATAC-seq data obtained for the regions in proximity of NFATc1, PRDM1 and MYC loci. Red boxes indicate RANKL-induced SE regions (left). Enlarged ATAC-seq tracks and peaks at SE domains (right). g, h Motif analysis of RANKL-regulated ATAC-seq peaks within SE regions
Fluorescence in situ hybridization
Fluorescence Se-RNA:NFATc1 probes for RNA–FISH were generated according to the protocols presented by Biosearch Technologies. Hybridization was carried out according to a protocol presented Biosearch Technologies. The stained cells were imaged using a Zeiss Axioplan microscope (Zeiss) with an attached Leica DC 200 digital camera (Leica).
ChIP-sequencing
Cell DNA was crosslinked for 5 min at room temperature by the addition of one tenth of the volume of an 11% formaldehyde solution (11% formaldehyde; 50 mM HEPES, pH 7; 100 mM NaCl; 1 mM EDTA, pH 8.0; and 0.5 mM EGTA, pH 8.0) to the growth medium, followed by 5 min of quenching with 100 mM glycine. The cells were pelleted at 4 °C and washed with ice-cold PBS. The DNA-crosslinked cells were lysed with lysis buffer (50 mM HEPES-KOH, pH 7.5; 140 mM NaCl; 1 mM EDTA; 10% glycerol; 0.5% NP-40; and 0.25% Triton X-100) with protease inhibitors on ice for 10 min and were washed with washing buffer (10 mM Tris-HCl, pH 8.0; 200 mM NaCl; 1 mM EDTA; and 0.5 mM EGTA) for 10 min. The lysis samples were resuspended and sonicated in sonication buffer (10 mM Tris-HCl, pH 8.0; 100 mM NaCl; 1 mM EDTA; 0.5 mM EGTA;, 0.1% sodium deoxycholate; and 0.5% N-lauroyl sarcosine) using a Bioruptor (Diagenode) with 30 s on and 30 s off with high-power output for 12 cycles. After sonication, the samples were centrifuged at 12,000 r.p.m. for 10 min at 4 °C, and 1% sonicated-cell extracts were saved as input. Precleared extracts were then incubated with 5 mg of an anti-H3K27ac antibody (ab4729) and RNA polymerase II (pol II, MMS-126R; Abcam) overnight at 4 °C. After overnight incubation, antibody-bound agarose beads were washed twice with sonication buffer, once with sonication buffer with 500 mM NaCl, once with LiCl wash buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA; 250 mM LiCl; and 1% NP-40) and once with TE with 50 mM NaCl. After washing, the DNA was eluted in freshly prepared elution buffer (1% SDS and 0.1 M NaHCO3). Cross-linking was reversed by overnight incubation at 65 °C. RNA and protein were digested using RNase A and proteinase K, respectively, and DNA was purified with a ChIP DNA Clean & Concentrator kit (Zymo Research). Ten nanograms of purified immunoprecipitated DNA per sample was ligated with adaptors, and 100–300-bp DNA fragments were purified to prepare DNA libraries with an Illumina TruSeq ChIP Library Prep Kit following the manufacturer’s instructions. The ChIP libraries were sequenced (50-bp single end reads) using the Illumina HiSeq 2500 Sequencer at the Weill Cornell Medicine Epigenomic Core Facility per the manufacturer’s recommended protocol.
ATAC-sequencing
ATAC-seq was performed as previously described. To prepare nuclei, we spun 50,000 cells at 500 × g for 5 min, washed the cells with 50 mL of cold 1× PBS, and then centrifuged them at 500 × g for 5 min. Cells were lysed with cold lysis buffer (10 mM Tris-HCl, pH 7.4; 10 mM NaCl; 3 mM MgCl2; and 0.1% IGEPAL CA-630). Immediately after lysis, nuclei were spun at 500 × g for 10 min in a refrigerated centrifuge. Immediately following nuclei preparation, the pellet was resuspended in transposase reaction mix (Illumina). The transposition reaction was carried out for 30 min at 37 °C. Directly following transposition, each sample was purified using a Qiagen MinElute kit. Then, we amplified library fragments using 1× NEB Next PCR Master Mix and custom Nextera PCR primers as previously described under the following PCR conditions: 72 °C for 5 min; 98 °C for 30 s; and thermocycling at 98 °C for 10 s, 63 °C for 30 s, and 72 °C for 1 min. The libraries were purified using a Qiagen PCR cleanup kit to yield a final library concentration of ~30 nM in 20 ml. Libraries were amplified for a total of 10–13 cycles and subjected to high-throughput sequencing on an Illumina HiSeq 2500 Sequencer (single end).
ChIP-seq and ATAC-seq analysis
The quality of reads was assessed with FastQC (v0.11.9). Low-quality reads were filtered with skewer (v.0.2.2). The reads were then mapped to the reference genome GRCh37 (hg19) with bowtie2 (v2.3.5.1) set to the “-very-sensitive” parameter. Ambiguous alignments were removed with SAMtools (v1.10) with “view -q 10”, and then, the alignments were sorted according to the coordinates via the SAMtools sorting function. The identification of superenhancers or typical enhancers was performed with HOMER set to “findPeaks -style super -typical”. For the visualization of the average coverage of the peaks, HOMER annotatePeaks.pl was run with the “-hist -size given -d” options. To build genomic coverage tracks, the alignment files were processed into bedGraph files with BEDTools (v2.27.1) genomecov, converted to bigWig format with BedSort and bedGraphToBigWig (UCSC tools), and then uploaded to the UCSC genome browser. HOMER annotatePeaks.pl was used to assign each interval to the nearest gene, and the list is presented as a Chow-Ruskey plot prepared with the R vennerable package. The ChIP-seq read count at enhancer regions was determined with BEDTools multicov and then normalized to the RPKM on the basis of sequencing depth and interval length. Enhancer sensitivity to IBET was determined with HOMER getDifferentialPeaks set to the -F 1.5 option.
RNA-sequencing
Nuclear RNA was first extracted using an RNeasy Mini Kit (Qiagen). True-seq sample preparation kits (Illumina) were then used to generate libraries with multiplexed barcode adaptors. All samples passed a quality control analysis performed with a Bioanalyzer 2100 (Agilent). Paired-end reads were obtained on the Illumina HiSeq 2500/1500 at the Weill Cornell Medical College Genomics Resources Core Facility and Macrogen Korea. The reads were aligned to hg19 using STAR aligner (v2.7.3a) with default parameters. We used BEDTools multicov to count reads that were mapped within intervals, and then, the counts were normalized to FPKM considering both sequencing depth and interval length. The read count matrix was further analyzed with DESeq2 to identify cases of differential transcription.
PRO-sequencing
Cells were incubated under a nuclear run-on reaction condition (5 mM Tris-HCl pH 8.0; 2.5 mM MgCl2; 0.5 mM DTT; 150 mM KCl; 0.5% Sarkosyl; and 0.4 units/μl of RNase inhibitor) with obtained biotin-NTPs and rNTPs (18.75 μM rATP, 18.75 μM rGTP, 1.875 μM biotin-11-CTP, and 1.875 μM biotin-11-UTP for uPRO; 18.75 μM rATP, 18.75 μM rGTP, 18.75 μM rUTP, 0.75 μM CTP, and 7.5 μM biotin-11-CTP for pChRO) for 5 min at 37 °C. Run-On RNA was extracted using TRIzol and fragmented with 0.2 N NaOH for 15 min on ice. The RNA fragments were neutralized, and the buffer was exchanged by passing it through P-30 columns (Bio–Rad). The 3′ RNA adaptor (/5Phos/NNNNNNNNGAUCGUCGGACUGUAGAACUCUGAAC/3InvdT/) at a 5 μM concentration was ligated for 1 hour at room temperature using T4 RNA ligase (NEB), and then, 2 consecutive streptavidin bead binding and extraction cycles were performed. The extracted RNA was converted to cDNA via template switch reverse transcription with 1 μM RP1-short RT primer (GTTCAGAGTTCTACAGTCCGA), 3.75 μM RTP-Template Switch Oligo (GCCTTGGCACCCGAGAATTCCArGrGrG), and 1x Template Switch Enzyme and Buffer (NEB) at 42 °C for 30 min. After SPRI bead clean-up, the cDNA was amplified via PCR for as many as 20 cycles using primers compatible for use in Illumina small RNA sequencing. Low-quality reads and Illumina small RNA 3’ adaptors were removed. PRO-seq reads were aligned to the hg19 genome with STAR aligner set to the default options. The 5’-end positions of each alignment strand were processed into bedGraph tracks with BEDTools genomecov −5 -strand+ (or −) and then converted to bigWig formatted files. The bigWig files and the pretrained support vector matrix (SVM) that was provided with the program were run as inputs in the dREG program. Among the resulting dREG peaks, peaks with scores greater than 0.7 were selected for further analysis. The remaining dREG peaks were then run in dREG-HD. “Relaxed” dREG-HD peaks from all the samples were merged via cat command, BedSort and BEDTools merge -d 100. Read counts of each merged dREG-HD site that were extended by 250 bases from the center were assessed with BEDTools multicov. A differential expression analysis was performed with DESeq2, and differential dREG peaks were identified as those with an adjusted p value less than 0.01.
Data accessions
H3K27ac ChIP-seq data for osteoblasts and CD4+ T cells are available at the ENCODE project web site (www.encodeproject.org) with entry codes ENCFF000CVS and ENCFF000CVU for osteoblasts and ENCFF017MGJ and ENCFF416ZFL for CD4+ T cells. The data obtained in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) database with GSE number GSE203587. The next-generation sequencing experiments performed in this study are presented in Supplementary Fig. 1a. All the data are based on two (ChIP-seq and Nuc-seq) or three (ATAC-seq and PRO-seq) replicate experiments.
Statistical analyses
All statistical analyses were performed with GraphPad Prism 8 or R (v4.1.0) software. Detailed information about the statistical analysis, including the tests and values used, is provided in the figure legends. Shapiro‒Wilk normality tests were performed; for data in a Gaussian distribution, we performed appropriate parametric statistical tests, and for those that were not in an equal variance-Gaussian distribution, we performed nonparametric statistical tests. We performed nonparametric Kruskal‒Wallis tests with Bonferroni correction for comparisons of read density in each region. A p value less than 0.05 was considered to be statistically significant.
Results
RANKL regulates SEs in the vicinity of master osteoclast regulators in human osteoclasts
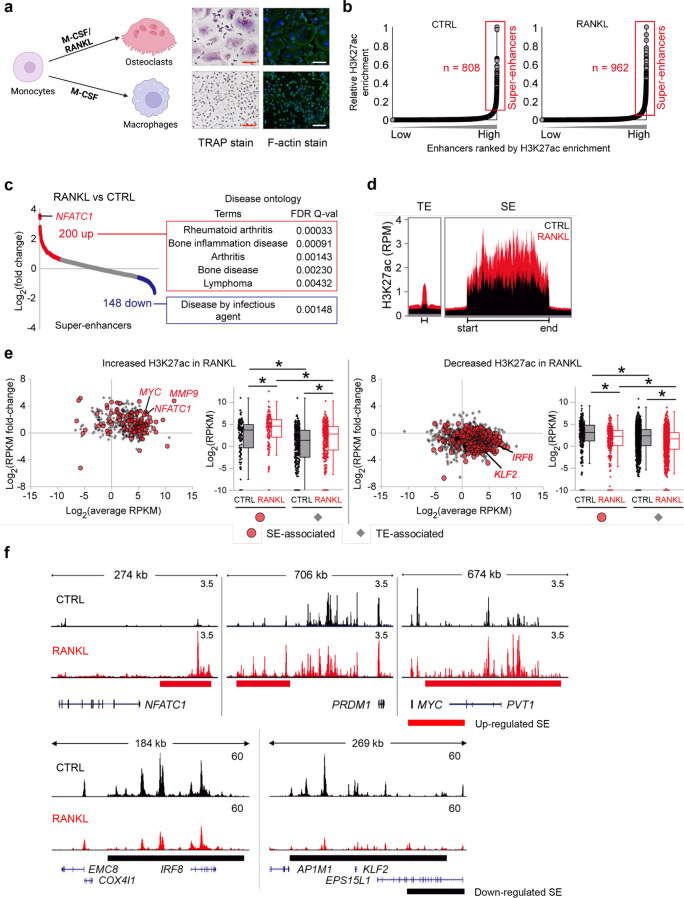
RANKL is a key driver that induces human osteoclast differentiation [21]. Human CD14+ monocytes differentiate into tartrate-resistant acid phosphatase (TRAP)-positive multinuclear osteoclasts with characteristic actin ring formation after M-CSF and RANKL stimulation (Fig. 1a). We sought to identify cell-specific programs in human osteoclasts. Superenhancers (SE) reprogramming via microenvironmental stimuli contributes to the acquisition of a distinct cell-specific phenotype in health and disease [25]. Therefore, we hypothesized that osteoclast-specific SEs are established early in the differentiation program induced by RANKL stimulation and contribute to osteoclast-specific gene expression. SEs exhibit higher enrichment of histone 3 lysine 27 acetylation (H3K27ac) and DNase I hypersensitive regions than typical enhancers [24]. To catalog SEs, we performed H3K27ac ChIP-seq after CD14+ cells were differentiated with or without RANKL stimulation (Supplementary Fig. 1a). A bioinformatic analysis using the rank ordering of superenhancer (ROSE) algorithm [24] defined 808 SEs in CD14+ monocytes (CTRL group) and 962 SEs in RANKL-treated OCPs (the RANKL group) (Fig. 1b). Among SEs in osteoclasts, 200 SEs were induced by RANKL relative to control condition and were closely correlated with gene sets related to bone disease presenting pathological bone resorption, such as RA and bone inflammatory diseases (Fig. 1c). Notably, 148 SEs that were suppressed by RANKL were associated with diseases caused by infectious agents and the regulation of immune system processes (Fig. 1c, Supplementary Fig. 1b). As expected, the 200 aforementioned upregulated SEs exhibited greater enrichment of H3K27ac reads than typical enhancers (TEs) (Fig. 1d). Among all SEs and TEs, basal abundance of H3K27ac peaks was higher in SEs than in TEs, and RANKL treatment increased or decreased H3K27ac enrichment in both SEs and TEs (Fig. 1e). SEs have been previously shown to be closely associated with the expression of master transcription factors (TFs) that control cell identity and fate [24, 26]. Notably, SEs formed after RANKL treatment were located in the vicinity of NFATC1, a master regulator of osteoclastogenesis [15], and key regulators of osteoclast differentiation, such as MYC [54], PR/SET domain 1 (PRDM1) [55], and matrix metalloproteinase-9 (MMP9) [56] (Fig. 1e, f). In contrast, the H3K27ac read density near negative regulators, such as interferon regulatory factor 8 (IRF8) [57], Kruppel-like factor 2 (KLF2) [58], and TNF receptor superfamily member 1B (TNFRSF1B) [59], was decreased in response to RANKL (Fig. 1e, f). These results suggest that RANKL-regulated SEs are closely associated with both positive and negative TFs and promote osteoclast differentiation.
Fig. 1.
Identification of RANKL-sensitive super enhancers (SEs) in human osteoclasts. Human CD14+ monocytes were cultured overnight with M-CSF (20 ng ml−1) and then treated with M-CSF (CTRL) or M-CSF with RANKL (40 ng ml−1, RANKL) for three days (a) or one day (b–f). a Left panel: Schematic showing the differentiation of osteoclasts. Middle panel: osteoclasts were identified as TRAP-positive multinuclear cells (>3 nuclei). Right panel: Phalloidin staining showing actin ring formation in osteoclasts. b–f ChIP-seq of H3K27ac was performed. b Distribution of H3K27ac ChIP-seq enrichment scores under the indicated conditions. Enhancer regions are plotted in increasing order based on their input-normalized H3K27ac ChIP-seq signal intensity. SEs are defined as the population of enhancers above the inflection point of the curve. c A RANKL-sensitive SE was characterized by a >1.5-fold change in H3K27ac read density induced by RANKL (left panel). Disease ontology for RANKL-sensitive SE-associated genes with corresponding q-values (right panel). d Read density plot of H3K27ac ChIP-seq across typical enhancer (TE) and SE domains. e Graphs showing the differential H3K27ac ChIP-seq signals among enhancer domains between RANKL and CTRL groups. Increased (left) or decreased (right) H3K27ac ChIP-seq signals after RANKL treatment are shown. Red dots indicate the signals from SE regions, and gray dots indicate the signals from TE regions. Boxplots showing the changes in H3K27ac ChIP-seq signal intensity under the indicated conditions. *p < 0.01 by Kruskal‒Wallis test with Bonferroni correction. Representative SEs are highlighted with their associated genes. f Representative tracks of H3K27ac ChIP-seq at NFATc1, PRDM1, MYC, IRF8, and KLF2 loci under the indicated conditions. Red and black bars indicate upregulated and downregulated SEs, respectively
RANKL-induced SEs are osteoclast specific and mark osteoclast identity genes
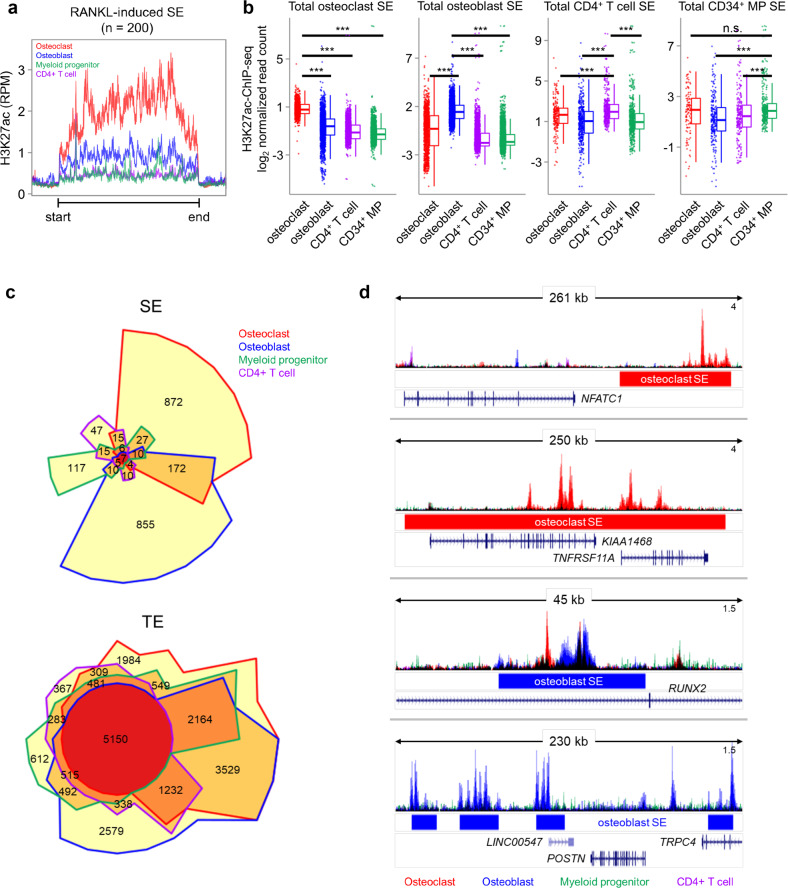
To further examine whether the SEs identified in osteoclasts show cell-type specificity, we extended the study of RANKL-regulated SEs to other cell types related to bone inflammation and RA. To this end, we retrieved datasets with H3K27ac ChIP-seq on osteoblasts, CD34+ progenitors and CD4+ T cells from the ENCODE project and identified SEs in each cell type. In the 200 identified RANKL-induced SEs, the H3K27ac read density was enriched exclusively in human osteoclasts, but in the other cell types (Fig. 2a). A set of SEs in osteoblasts, CD34+ progenitors and CD4+ T cells showed that H3K27ac was highly enriched in cell-type specific SEs (Fig. 2b). Chow–Ruskey diagrams of TE-associated and SE-associated genes in osteoclasts, osteoblasts, CD34+ progenitor cells, and T cells revealed that TEs shared genes that were activated in all four cell types (Fig. 2c, the red circle in the middle), while SEs overlapped in a few cell types and spanned domains that were almost all cell-type specific (Fig. 2c). For example, osteoclast-specific SEs, but not SEs in other cell types, were associated with the genes encoding NFATC1 and TNFRSF11A, which are well- characterized osteoclast genes (Fig. 2d). In contrast, osteoblast-specific SEs were associated with the genes encoding runt-related transcription factor 2 (RUNX2) and periostin (POSTN), which are required for osteoblast differentiation (Fig. 2d). A Gene Ontology analysis on the set of genes associated with SEs in osteoblasts and CD4+ T cells showed that significant biological process terms were highly enriched in cell-type specific function (Supplementary Fig. 2). This result suggests that the genes associated with RANKL-induced SE formation are highly cell-type specific compared to TE-associated genes.
Fig. 2.
RANKL-sensitive SEs exhibit osteoclast specificity. a H3K27ac ChIP-seq distribution for osteoclasts, osteoblasts, CD34+ progenitors and CD4+ T cells within RANKL-induced osteoclast SEs. Each color indicates a different cell type. b Box plots showing H3K27ac ChIP-seq signal intensities among SE domains in the indicated cell type. ***p < 0.001, n.s., not significant, as determined by Kruskal‒Wallis test with Bonferroni correction. c Chow–Ruskey diagram showing SE- and TE-associated genes in osteoclasts (red border), osteoblasts (blue border), CD34+ progenitors (green border) and CD4+ T cells (purple border). The color of the borders around each intersection corresponds to the cell types with overlapping genes. The red circle in the middle represents the overlap of all four cell types. Lighter shades of red, orange, and yellow represent the overlap of fewer cell types. The area of each intersection is proportional to the number of genes within the intersection. d Representative tracks of H3K27ac ChIP-seq data in the vicinity of SEs in the indicated cell types: osteoclasts (red bar), osteoblasts (blue bar), CD34+ progenitors (green bar) or CD4+ T cells (purple bar). Track colors represent the cell types
Chromatin accessibility is dynamically regulated in osteoclast SEs
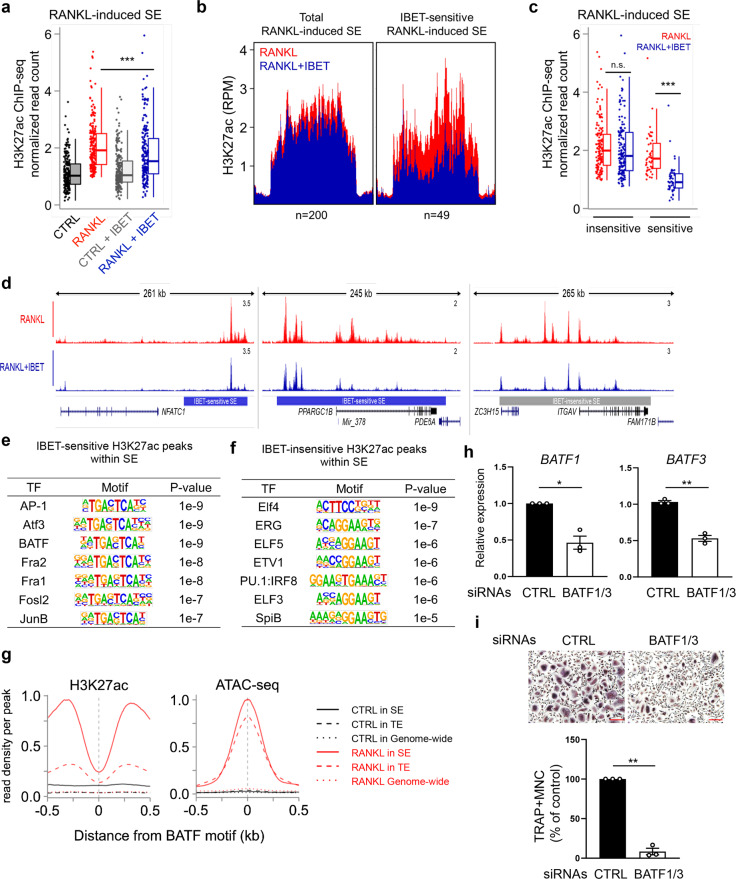
Active enhancers are cis-regulatory elements that modulate the quality and quantity of gene expression [23]. These regions are characterized primarily by specific combinations of posttranslational modifications of histone proteins (H3K27ac and H3K4me1) and “open” chromatin, on which major TFs can bind [30]. Thus, we performed assay of transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) to investigate chromatin changes during osteoclast differentiation. We first binned the ATAC-Seq peaks on the basis of their respective fold changes in response to RANKL (Fig. 3a). Increased chromatin accessibility was associated with both a higher peak fold change (Fig. 3a, black box in the left panel) and at least a twofold change in response to RANKL, but fewer ATAC-seq peaks were uniformly distributed within two-fold change (Fig. 3a, right panel). Highly open chromatin regions (the black box in Fig. 3a, n = 5899) were correlated with increased densities of H3K27ac reads, suggesting that the regulation of enhancers during osteoclast differentiation is closely related to local chromatin accessibility (Fig. 3b). Moreover, the most regulated peaks were located in potential enhancer regions, including distal, intragenic, or intergenic regions, not in promoters (Fig. 3c). Chromatin accessibility was increased in both TEs and SEs in response to RANKL but was significantly increased in TEs with a relatively narrow width (>500 bp) (Fig. 3d, e). These results suggest that chromatin accessibility is regulated in several specific regions in SEs with a wide width (>12.5 kb) marked by H3K27ac (Fig. 1c). Accordingly, relatively narrow ATAC-seq peaks, in contrast to H3K27ac, was upregulated in RANKL-sensitive SEs near the genes NFATC1, PRDM1, and MYC (Fig. 3f). To gain insight into the mechanisms involved in SE regulation, we investigated TF-binding motifs in open chromatin in RANKL-regulated SEs. Open chromatin within RANKL-induced SEs was significantly enriched with TF motifs bound by BATF, activator protein-1 (AP-1), NFAT:AP-1, NFAT, and NF-kB, which have been shown to be important TFs for osteoclast differentiation [2] (Fig. 3g). Sequence motifs bound by TFs regulating osteoclast differentiation, such as PU.1:IRF8, MAF BZIP transcription factor A (MAFA), and IRF8, were enriched in down-regulated ATAC-seq peaks in SEs (Fig. 3h). These results suggest that RANKL-sensitive SEs may be formed as a results of RANKL-dependent signaling TF binding to specific open chromatin regions.
BET protein inhibition differentially regulates SE-eRNA expression
BET protein inhibition effectively suppresses osteoclastogenesis and arthritic bone erosion [21]. Given that bromodomain-containing protein 4 (BRD4) occupancy at SEs in certain cell types is highly sensitive to BET inhibition [25], we reasoned that I-BET151-mediated suppression of osteoclastogenesis results from disrupted SE formation, which drives target gene expression in human osteoclasts. We performed H3K27ac ChIP-seq in the presence of I-BET151 and examined the intensity of the H3K27ac signals in predefined osteoclast-specific SEs. As expected, I-BET151 treatment significantly decreased H3K27ac signals intensity in RANKL-induced SEs (Fig. 4a). Notably, we found that I-BET151 treatment did not cause a decrease in H3K27ac peaks in all RANKL-induced SEs, whereas significant changes in 49 of the 200 identified SEs contributed to an overall decrease in H3K27ac read densities in RANKL-induced SEs (Fig. 4b, c). For instance, SEs in the vicinity of the NFATC1 and peroxisome proliferator-activated receptor gamma coactivator 1-beta (PPARGC1B) genes were highly sensitive to I-BET151 treatment, while SEs near integrin subunit alpha V (ITGAV) were relatively resistant to inhibition (Fig. 4d). Concomitantly, the expression of NFATc1 and PPARGC1B was suppressed by I-BET151 treatment, while I-BET151 exerted a minimal effect on ITGAV expression (Supplementary Fig. 3a). A motif analysis further showed that IBET-sensitive H3K27ac peaks in RANKL-induced SEs were enriched with TF motifs bound by AP-1, activating transcription factor 3 (ATF3), and BATF (Fig. 4e), is similar to the results of the motif enrichment analysis performed with accessible chromatin (Fig. 3g). IBET-insensitive H3K27ac peaks were also enriched in PU.1 family motifs (Fig. 4e). The identified I-BET-sensitive SEs were associated with AP-1 family transcription factors that play key roles in osteoclastogenesis [60]. BATF was consistently found in SE regions (Figs. 3g, 4e). However, the role of BATF in osteoclasts remains unclear. We thus examined the chromatin landscape near BATF-, Fos- and NF-kB-binding motif sites. Similar to those of the key osteoclastogenic transcription factors Fos and NF-kB (Fig. 3g and Supplementary Fig. 3b), the BATF-binding motif site in RANKL-induced SEs was surrounded by robust H3K27ac and ATAC-seq signals compared to those in TEs, indicating that the BATF-binding site is in active SEs (Fig. 4g). BATFs belong to the AP-1 family and comprise three members: BATF, BATF2, and BATF3 [61, 62]. BATF1 expression was initially diminished and later increased, while BATF3 expression was induced by RANKL during osteoclastogenesis (Supplementary Fig. 3c). However, BATF2 expression was not detected (data not shown). BATF1 and BATF3 have been shown to compensate for each other [63]. To assess the role of BATF in osteoclastogenesis, we knocked down both BATF1 and BATF3 in CD14+ cells using small interfering RNAs (siRNAs). KD cells were cultured with M-CSF and RANKL to drive their differentiation into osteoclasts. Intriguingly, decreased BATF1/3 expression suppressed osteoclastogenesis (Fig. 4h, i), suggesting that BATF1/3 are positive regulators of osteoclast differentiation. Taken together, these results suggest that BET proteins modulate osteoclast differentiation by selectively regulating RANKL-induced SE formation.
Fig. 4.
Sensitivity to iBET leads to differential regulation of SE-eRNA expression. a Box plot displaying read densities of H3K27ac ChIP-seq in RANKL-sensitive SE domains in the presence or absence of I-BET151 (500 nM). b Distribution of H3K27ac ChIP-seq signals in all RANKL-induced SEs (left, n = 200) and I-BET151-sensitive SEs with 1.5-fold H3K27ac abundance changes at 200 RANKL-sensitive SEs (right, n = 49). c Box plots showing H3K27ac ChIP-seq signals among SE domains under the indicated conditions. d Representative tracks of IBET-sensitive or -insensitive SEs under the indicated conditions. The normalized read counts of H3K27ac obtained by ChIP-seq are shown. e, f Motif enrichment analysis of I-BET151-sensitive or I-BET151-insensitive SEs. g Overall sequencing coverage of H3K27ac and ATAC-seq around BATF-binding motifs. The dotted line, dashed line, and solid lines represent coverage around motifs in the genome, motifs in TEs, and motifs in SEs, respectively. The black line represents the control group (CTRL), and the red line represents the RANKL-treated group. h, i Osteoclastogenesis assay with human OCPs transfected with control or BATF1-/3-specific siRNA and treated with RANKL (40 ng ml−1) (n = 3). Scale bar: 200 µm. h BATF1/3 knockdown efficiency in human OCPs was measured by RT‒qPCR. i The left panel shows representative images of TRAP+ osteoclasts. The right panel shows the percentage of TRAP+ multinuclear cells (MNCs: more than three nuclei) per control, normalized relative to the number of osteoclasts differentiated under control siRNA conditions. Scale bar: 100 µm. ***p < 0.001, **p < 0.01, or *p < 0.05 by Kruskal‒Wallis test with Bonferroni correction (c) or Student’s t test (h, i)
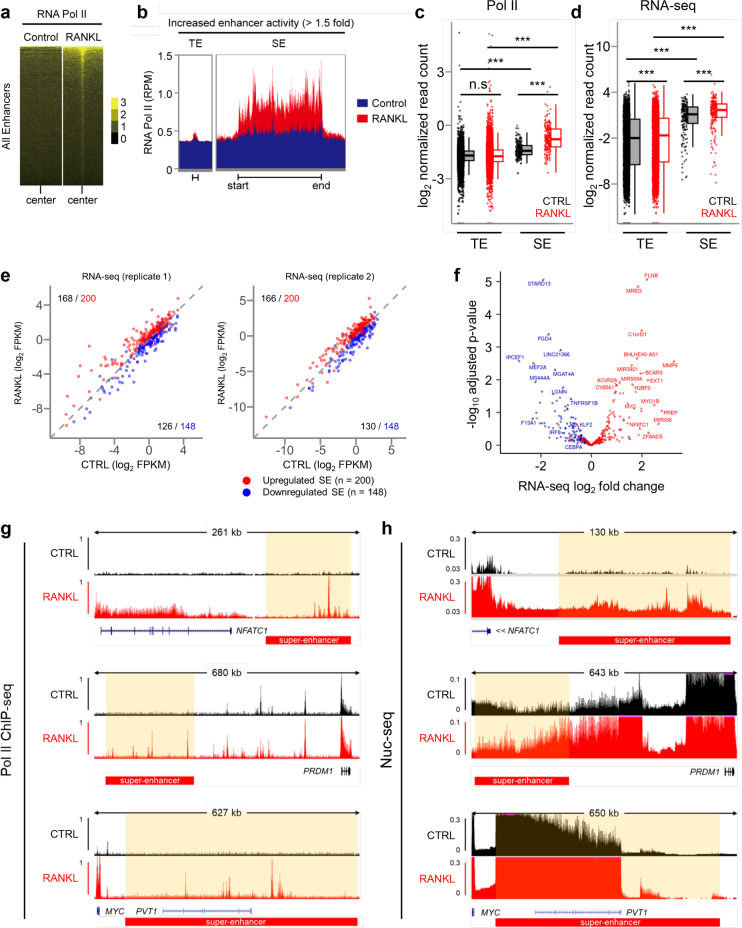
RNA Polymerase II is recruited to osteoclast SE regions to promote enhancer RNA transcription
Several studies have shown that enhancers are transcribed by RNA polymerase II (Pol II) and that enhancer RNAs are associated with superenhancers [64, 65]. Therefore, we performed ChIP-seq and RNA-seq to investigate how Pol II recruitment and eRNA expression are associated with SEs and TEs across the human osteoclast genome. As indicated by ChIP-seq reads, Pol II was enriched in both types of enhancers in human osteoclasts (Fig. 5a). However, many RANKL-induced SEs were occupied by a large proportion of Pol II compared to the amount of Pol II on TEs (Fig. 5b, c). The evidence that this high level of Pol II occupying RANKL-induced SEs suggests that these large cis-element domains may promote the transcription of eRNA, which drives the high-level expression of SE-associated genes. Therefore, to measure eRNA expression in osteoclasts, we isolated nuclear RNA and examined nascent transcripts by nuclear RNA (Nuc)-seq, as previously described [66]. We found that the presence of higher levels of eRNAs transcribed from SEs than from TEs and that RANKL treatment further increased the expression of eRNA encoded by SEs (Fig. 5d). The expression of eRNA was increased in 83–84% RANKL-induced SEs, while 85–88% of SEs with downregulated expression was related to decreased eRNA expression (Fig. 5e). The correlated expression of SEs and eRNAs included a set of genes encoding major TFs important to osteoclasts, such as NFATC1 and MYC (Fig. 5f). We also found that both Pol II recruitment and eRNA expression were enriched in SE regions in proximity to key osteoclast regulators, such as NFATC1, PRDM1, and MYC (Fig. 5g, h), suggesting that osteoclast-specific SE activity may be inferred from eRNA expression.
Fig. 5.
Active RNA Pol II recruitment and transcription in RANKL-sensitive superenhancers in human osteoclasts. a Heatmaps showing the enrichment of RNA Pol II ChIP-seq signals in RANKL-sensitive enhancer regions. b Distribution of RNA Pol II ChIP-seq signals across RANKL-sensitive TE and SE domains under the indicated conditions. c Box plots depict the quantitated normalized tag counts of RNA pol II ChIP-seq signals in TE and SE domains under the indicated conditions. d Box plots depicting the quantitated nascent transcripts of nuclear RNA-seq signals in TE and SE domains under the indicated conditions. e Scatter plot showing the transcript levels of genes associated with RANKL-sensitive SEs, depending on whether they are upregulated or downregulated. Red dots indicate the genes related to RANKL-induced SEs, and blue dots indicate the genes related to RANKL-suppressed SEs. f Volcano plot showing the RNA-seq analysis of differentially expressed SE-associated genes. Red dots show genes associated with upregulated SEs, and blue dots show genes associated with downregulated SEs. g, h Representative tracks of RNA Pol II ChIP-seq (g) and RNA-seq (h) in the proximity of NFATc1, PRDM1, and MYC loci under the indicated conditions. Red boxes depict the RANKL-sensitive SE region. ***p < 0.001, n.s., not significant as determined by Kruskal‒Wallis test with Bonferroni correction (c, d)
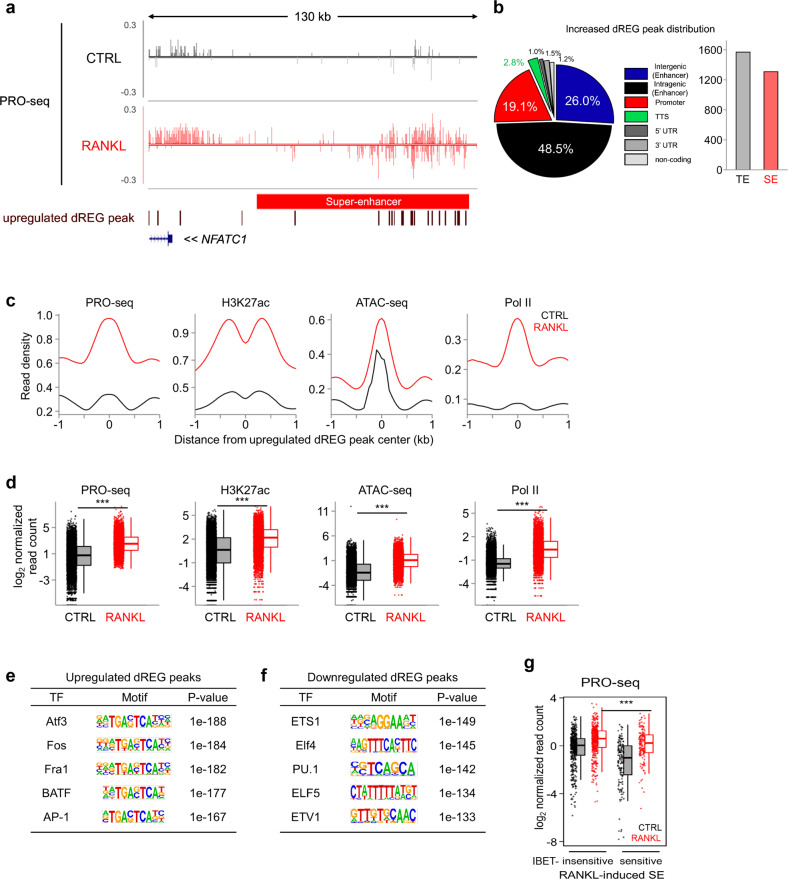
Global identification and characteristics of SE-eRNA in human osteoclasts
To obtain a better resolution map of eRNA at RANKL-sensitive SEs in osteoclasts, we performed a precision nuclear run-on and sequencing (PRO-seq) assay [67], which offers single nucleotide resolution of nascent RNA 3’ ends. Similar to those obtained with Nuc-seq data (Fig. 5h), nascent transcripts were highly enriched at NFATC1-associated SEs as well as PRDM1- and MYC-associated SEs (Fig. 6a, Supplementary Fig. 4). Using discriminative regulatory-element detection from gro-seq (dREG) analysis [68], a sensitive machine learning method that predicts active transcriptional regulatory elements and eRNA loci, we found that most dREG peaks were distributed in potential enhancer regions (intergenic: 26%, intragenic 48.5%), while promoter regions included 19.1% dREG peaks (Fig. 6a, b). A total of 1,308 dREG peaks were found in the SE region, and 1,569 dREG peaks were located in the TE region, suggesting that a significant amount of eRNAs was transcribed in the SE region (Fig. 6b). The predicted eRNA loci derived from dREG analysis were highly occupied by several markers of active enhancers, including H3K27ac, open chromatin (as determined by ATAC-seq), and Pol II (Fig. 6c, d). RANKL stimulation markedly increased the signaling intensities of H3K27ac, Pol II ChIP-seq, and ATAC-seq, indicating that the dREG analysis successfully predicted eRNA loci in both SE and TE (Fig. 6c, d). We also found that dREG within RANKL-induced enhancers was significantly enriched with TF motifs bound by AP-1 family members, such as ATF3, Fos, Fra1, and BATF (Fig. 6e). In contrast, down-regulated dREG regions were enriched with TF motifs bound by PU.1 family TFs such as ETS1, Elf4, and PU.1 (Fig. 6f). RANKL significantly increased the expression of eRNAs in both I-BET-sensitive and I-BET-insensitive SEs. However, the abundance of RANKL-induced eRNAs in I-BET-sensitive SEs was higher than that in I-BET-insensitive SEs (Fig. 6g). These results suggest that BET proteins may associate with SE-eRNA transcription by interacting with SEs during osteoclastogenesis. Taken together, these data suggest that RANKL-regulated signaling input is associated with SE-eRNA expression.
Fig. 6.
Identification of dREG peaks in human osteoclasts. a Representative tracks of PRO-seq at the NFATc1-associated SE domain under the indicated conditions (CTRL: control, and RANKL: RANKL treatment). An increase in the number of dREG peaks in PRO-seq (brown). b Pie chart showing the genomic location of RANKL-inducible dREG peaks obtained from the PRO-seq data. A bar graph shows the number of dREG peaks in TE and SE domains. c Aggregate plots showing the mean Pro-seq, H3K27ac, Pol II ChIP-seq, and ATAC-seq signals centered on an increased number of dREG peaks under the indicated conditions. d Box plots showing PRO-seq, H3K27ac, Pol II ChIP-seq, and ATAC-seq signals centered on an increased number of dREG peaks under the indicated conditions. ***p < 0.001 by Kruskal‒Wallis test with Bonferroni correction. Motif analysis of RANKL-induced (e) and RANKL-suppressed (f) dREG peaks. g Box plots showing PRO-seq signals among RANKL-induced SE domains under the indicated conditions
SE-eRNA regulates the expression of SE-associated TF in human osteoclasts
Consistent with the H3K27ac ChIP-seq data (Fig. 1c), a Gene Ontology analysis revealed that dREG peaks were closely associated with osteoclast- and bone-related diseases, including rheumatoid arthritis (RA) (Fig. 7a). NFATc1 is a master regulator of osteoclasts [15]. During osteoclastogenesis, NFATc1 mRNA expression is induced by RANKL in a time-dependent manner (Fig. 7b). Freshly isolated synovial CD14+ cells from RA patients show higher osteoclastogenic potential and express NFATc1 protein without RANKL stimulation [69]. Consistently, a quantitative RT‒PCR analysis showed increased expression of NFATc1 in synovial CD14+ cells from RA compared to that in disease-control CD14+ cells from healthy donors (Fig. 7c). To evaluate the functional role of SE-eRNAs in human osteoclasts, we first examined NFATC1-associated SE-eRNAs (SE-eRNA:NFATC1) via fluorescence in situ hybridization (FISH) with antisense probes. SE-eRNA:NFATC1 was found only in RANKL-treated cells (Fig. 7d). Subsequently, we found that knocking down SE-eRNA:NFATC1 with antisense oligonucleotides (ASO-LNA) suppressed the differentiation of human OCPs into multinucleated TRAP-positive cells (Fig. 7e, f, Supplementary Fig. 5a, b, see Methods for details). Moreover, knocking down SE-eRNA:NFATC1 abrogated the RANKL-induced expression of NFATc1 mRNA (Fig. 7g, Supplementary Fig. 5c) and protein (Fig. 7h, Supplementary Fig. 5d) in human OCPs. We reasoned that RANKL-responsive SE-eRNAs are associated with the high osteoclastogenic potential of synovial CD14+ cells in RA patients. To this end, we investigated the expression of SE-eRNAs in RA synovial CD14+ cells. Nuc-seq data derived from the synovial CD14+ cells of RA patients or disease-control donors revealed that the expression of some RANKL-sensitive SE-eRNAs was highly upregulated in the synovial CD14+ cells of RA patients (Fig. 7i). Among these SE-eRNAs, nascent RNA expression within NFATC1-associated SEs (SE-eRNA:NFATC1) was increased in the synovial CD14+ cells from RA patients (Fig. 7j). Moreover, a quantitative RT‒PCR analysis showed increased expression of eRNAs in NFATC1-associated SEs in the RA synovial CD14+ cells compared to that in the disease-control CD14+ cells from healthy donors (Fig. 7k). Our previous study showed that administration of I-BET151 suppressed arthritic bone erosion and NFATc1 expression [21]. In the present study, I-BET151 treatment also suppressed the expression of SE-eRNA:NFATC1 (Supplementary Fig. 5e), suggesting that I-BET-mediated inhibition of NFATc1 expression might be mediated, in part, by a decrease in the expression of SE-eRNA:NFATC1. We also found that the levels of SE-eRNA:PRDM1 and SE-eRNA:MYC were significantly increased in RA synovial OCPs (Supplementary Fig. 5f). Taken together, these data suggest that eRNAs transcribed from NFATC1-associated SEs are indispensable for the transcription of NFATC1 and osteoclast differentiation and that the presence of RANKL-responsive SE-eRNAs in RA synovial CD14+ cells may contribute to the high osteoclastogenic potential of RA synovial macrophages.
Discussion
Dysregulation of osteoclast differentiation and activity has been linked to pathological bone diseases and arthritic bone destruction [2]. The major transcription factors and key molecules driving osteoclastogenesis have been extensively studied, and global or conditional knockout mice exhibited defects in bone homeostasis and pathological bone destruction [2]. However, globally targeting these key factors is often accompanied by nonspecific effects, and conditional deletion approaches may not be options for the treatment of human patients. Identifying cell-specific signatures in human osteoclasts is fundamental to developing osteoclast-specific therapeutic interventions for pathological bone resorption in humans. In the present study, we identified 348 RANKL-responsive SEs and SE-associated eRNAs in early human osteocla (osteoclasts) as osteoclast-specific epigenetic programs. RANKL-responsive SEs were enriched by BATF-binding motifs and BET proteins, which are important regulators of osteoclastogenesis. Targeting SE-eRNAs in the proximity of the NFATc1 gene suppressed NFATc1 expression and osteoclastogenesis. Moreover, we found RANKL-responsive SE-eRNAs in synovial macrophages from RA patients. Our study provides a better understanding of the cell-specific programs driving osteoclastogenesis and insights into the control of pathological bone resorption, and suggests that manipulating SE-eRNAs may be a novel, osteoclast-specific therapeutic strategy for treating osteoclast-mediated bone diseases.
Epigenetic mechanisms are increasingly appreciated for their critical roles in transcriptional regulation in human health and disease. Among them, superenhancers play a crucial role in regulating the expression of genes associated with cell identity and cell fate [24–28]. In this study, we identified RANKL-responsive SEs formation in human osteoclasts. Interestingly, we found that the majority of these SEs were induced in an osteoclast specific manner, as they were not detected in osteoblasts, CD4+ T cells, or CD34+ progenitor cells. In our analysis, we found SE specificity in CD34+ progenitors to be ambiguous. CD34+ progenitor cells differentiate into different cell types, including monocytes, osteoclasts, T cells, and even osteoprogenitor cells [70–72]. Therefore, it is possible that CD34+ progenitor cells may share common features in the histone code with other cells, which may be retained in subsequently differentiated cells. However, the importance of shared features between progenitors and differentiated cells remains unknown. Human and mouse osteoclasts share key transcription factors that drive osteoclastogenesis, including NFATc1 [2]. RANKL transduces the same signaling pathways, including the NF-kB and MAPK pathways, in both human and mouse osteoclasts. Although genes proximal to SEs are important for tissue or cell identity, SEs among different species do not exhibit sequence conservation [73], and the overall sequence similarity in noncoding areas between different species varies extensively [74, 75]. Our study suggests the possibility that SEs of osteoclast identity genes may vary among species. For example, although we identified superenhancers in the vicinity of the NFATc1 gene, specifically at the intergenic region of the NFATc1 gene C-terminus in human osteoclasts, Carey et al. showed NFATc1 superenhancer loci in mouse osteoclasts by specifically focusing on two intronic enhancers of NFATc1 [32]. However, whether SE-associated genes are modulated by the same regulatory regions in human and mouse cells is unclear and remains an area for future research.
SEs carry large numbers of transcription factor-binding sites, and master transcription factors and BET proteins contribute to SE formation [25]. In our analysis, NFATc1-, AP-1, BATF- and NF-kB-binding motifs were top-ranked motifs in RANKL-responsive SEs. Among these TFs, the crucial contribution of NFATc1, AP-1, and NF-kB in osteoclasts has been established. Carey et al. showed that PU.1 and MITF coordinate the expression of essential transcription factors in mouse bone marrow-derived osteoclasts and that the PU.1-Eomes-MITF complex is enriched in two intronic NFATc1 enhancers in mouse osteoclasts [10, 32]. In our study, the PU.1 motif was not among the top-ranked TF motifs in RANKL-responsive SEs. However, we found that PU.1-binding motifs preexisted in both RANKL-induced and RANKL-repressed SEs under control conditions (data not shown), which was consistent with a previous report showing that PU.1 reshaped the chromatin landscape by occupying all open regulatory regions in multipotent myeloid precursor cells, including closed sites in these regions [76–78]. Moreover, PU.1 can activate or repress transcription via cooperative action with other TFs. Izawa et al. showed that osteoclast-specific PU.1-binding sites overlapped with NFATc1-binding sites [79]. In addition to the known regulators of osteoclastogenesis, we identified that the BATF-binding motif was highly enriched in RANKL-responsive SEs, suggesting that BATF may be required for SE formation in human osteoclasts. However, the role of BATF proteins in osteoclasts has not yet been characterized. We found that BATF deficiency attenuated the osteoclast differentiation of human cells and identified that BATF is a previously unknown regulator of osteoclastogenesis. Defining the mechanistic link between BATF and SE formation requires more investigation, but consistent with our observations, the BATF3 module has been found in SEs in anaplastic large cell lymphoma [80]. The role of key TFs in osteoclasts has been extensively studied with a specific focus on their function in activating a promoter. Our data suggest that, in addition to their well-defined functions, TFs might regulate osteoclastogenesis by controlling SE formation. BET proteins are also highly enriched in SEs [24]. Indeed, in a previous study, targeting BRD4, a BET protein, with small-molecule inhibitors, including JQ1 or I-BET, suppressed osteoclast differentiation and attenuated joint inflammation and bone erosion in murine models of inflammatory arthritis [21, 81]. Our study showed that RANKL-responsive SEs are preferentially lost in the presence of a BET inhibitor. Given our previous findings showing a positive role for BET proteins in NFATc1 expression and osteoclasts, our present data suggest that BET proteins might regulate gene expression and osteoclastogenesis, in part, by controlling SE formation.
Noncoding RNAs significantly contribute to cellular functions and differentiation [82, 83]. The expression and role of noncoding RNAs such as microRNAs, long intergenic noncoding (linc) RNAs, and intronic eRNAs in osteoclasts and in osteoporosis have been documented [84–87]. Although several noncoding RNA species have been identified in mouse and human osteoclasts, SE-eRNAs and their functions in osteoclastogenesis and bone resorption in human cells have remained largely unexplored. We found that eRNAs were transcribed from SEs in human osteoclasts. Sakaguchi et al. identified 87 putative eRNAs in mouse osteoclasts using cap-analysis of gene expression (CAGE), and deleting the region carrying intronic eRNAs in Nfatc1 genes suppressed Nfatc1 gene expression [86]. These findings suggest that the location of eRNAs of the same gene differs between mouse osteoclasts and human osteoclasts, supporting the importance of studies with human cells. Advances in gene editing technology, such as CRISPR‒Cas9, have enabled direct manipulation of SEs in human cells [88, 89]. However, directly targeting superenhancer regions in humans may disrupt the chromatin landscape and thus remains questionable as an approach. eRNAs are nascent RNA transcripts that are easily targeted. We showed that knocking down SE-eRNA in the vicinity of the NFATc1 gene suppressed NFATC1 mRNA expression and osteoclastogenesis. Thus, osteoclast-specific SE-eRNAs may be potential targets for safe and effective therapeutics in bone diseases. Further comprehensive functional analysis of other SE-eRNAs and characterization of the mechanism of action of RANKL-responsive SEs are needed. Taken together, our study provides the first evidence showing a link between SE-eRNAs and human osteoclast differentiation.
Rheumatoid arthritis (RA) is a chronic autoimmune disease that primarily affects the joints, as well as various bodily systems [90]. Bone erosion is a key clinical feature of RA [6], leading to joint destruction and impaired mobility [91–94]. Osteoclasts are the major cell type responsible for the bone erosion in RA and are found at sites of structural bone damage in inflammatory arthritis [95]. An increase in the number and activity of osteoclasts plays a crucial role in rapid bone erosion in RA [96, 97]. Recent advances in genome-wide sequencing and high-dimensional molecular profiling have provided insights into RA pathogenesis and the contribution of cell types and signaling pathways to its pathogenesis. In addition, tissues from RA patients have been thoroughly analyzed by various techniques, including genome-wide sequencing, histological analysis, and flow cytometry, which have provided data on patient-specific pathogenesis of RA. Notably, RA synovial CD14+ cells show high osteoclastogenic potential and express high levels of NFATc1 [69]. We showed that nascent eRNAs in RA synovial CD14+ cells overlapped with RANKL-responsive SE-eRNAs, suggesting that high levels of osteoclast SE-eRNAs may be associated with the higher osteoclastogenic potential of RA synovial macrophages. In summary, our study is the first to identify SEs and SE-eRNAs in human osteoclasts and provides a better understanding of human osteoclast biology, thereby opening new therapeutic avenues for human pathological bone destruction.
Supplementary information
Acknowledgements
We thank Dr. Lionel Ivashkiv for providing human patient samples. This work was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MSIP; No. 2020R1A2C1006101 and No. 2020M3A9B603885111 to SP) and by the Tow Foundation (to K-HP-M). Figure 1a was generated by BioRender.
Author contributions
SB and KK designed and performed experiments and bioinformatics analysis, analyzed the data, and wrote the manuscript. KK performed the bioinformatics analysis and contributed his expertise. HK, ML, BO and KK performed experiments and analyzed data. HK, SM and JHC contributed their expertise. EYL, SHP and K-HP-M designed and supervised the study, and wrote the manuscript. K-HP-M conceptualized the study. All authors reviewed and provided input on the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Seyeon Bae, Kibyeong Kim.
Contributor Information
Eun Young Lee, Email: elee@snu.ac.kr.
Sung Ho Park, Email: parksu@unist.ac.kr.
Kyung-Hyun Park-Min, Email: ParkminK@hss.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00959-x.
References
- 1.Tsukasaki M, Takayanagi H. Osteoimmunology: evolving concepts in bone-immune interactions in health and disease. Nat Rev Immunol. 2019;19:626–42. doi: 10.1038/s41577-019-0178-8. [DOI] [PubMed] [Google Scholar]
- 2.Park-Min KH. Mechanisms involved in normal and pathological osteoclastogenesis. Cell Mol life Sci: CMLS. 2018;75:2519–28. doi: 10.1007/s00018-018-2817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annu Rev Pathol. 2008;3:457–84. doi: 10.1146/annurev.pathmechdis.3.121806.151431. [DOI] [PubMed] [Google Scholar]
- 4.Goldring SR. Pathogenesis of bone and cartilage destruction in rheumatoid arthritis. Rheumatol (Oxf) 2003;42(Suppl 2):ii11–16. doi: 10.1093/rheumatology/keg327. [DOI] [PubMed] [Google Scholar]
- 5.Sato K, Takayanagi H. Osteoclasts, rheumatoid arthritis, and osteoimmunology. Curr Opin Rheumatol. 2006;18:419–26. doi: 10.1097/01.bor.0000231912.24740.a5. [DOI] [PubMed] [Google Scholar]
- 6.Schett G, Gravallese E. Bone erosion in rheumatoid arthritis: mechanisms, diagnosis and treatment. Nat Rev Rheumatol. 2012;8:656–64. doi: 10.1038/nrrheum.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasui T, Hirose J, Tsutsumi S, Nakamura K, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J Bone Min Res. 2011;26:2665–71. doi: 10.1002/jbmr.464. [DOI] [PubMed] [Google Scholar]
- 8.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202:1261–9. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasui T, Hirose J, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation. Ann N. Y Acad Sci. 2011;1240:7–13. doi: 10.1111/j.1749-6632.2011.06245.x. [DOI] [PubMed] [Google Scholar]
- 10.Carey HA, Hildreth BE, Samuvel DJ, Thies KA, Rosol TJ, Toribio RE, et al. Eomes partners with PU.1 and MITF to Regulate Transcription Factors Critical for osteoclast differentiation. iScience. 2019;11:238–45. doi: 10.1016/j.isci.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park-Min KH. Epigenetic regulation of bone cells. Connect Tissue Res. 2017;58:76–89. doi: 10.1080/03008207.2016.1177037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Sarosi I, Yan XQ, Morony S, Capparelli C, Tan HL, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc Natl Acad Sci USA. 2000;97:1566–71. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–23. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 14.Dougall WC, Glaccum M, Charrier K, Rohrbach K, Brasel K, De Smedt T, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13:2412–24. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Developmental cell. 2002;3:889–901. doi: 10.1016/S1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 16.Winslow MM, Pan M, Starbuck M, Gallo EM, Deng L, Karsenty G, et al. Calcineurin/NFAT signaling in osteoblasts regulates bone mass. Developmental cell. 2006;10:771–82. doi: 10.1016/j.devcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, et al. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118:3775–89. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Disco. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 19.Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet. 2019;20:109–27. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa K, Iwamoto Y, Kobayashi Y, Katsuoka F, Kawaguchi S, Tsujita T, et al. DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nat Med. 2015;21:281–7. doi: 10.1038/nm.3774. [DOI] [PubMed] [Google Scholar]
- 21.Park-Min KH, Lim E, Lee MJ, Park SH, Giannopoulou E, Yarilina A, et al. Inhibition of osteoclastogenesis and inflammatory bone resorption by targeting BET proteins and epigenetic regulation. Nat Commun. 2014;5:5418. doi: 10.1038/ncomms6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng S, Zhang L, Tang Y, Tu Q, Zheng L, Yu L, et al. BET Inhibitor JQ1 Blocks Inflammation and Bone Destruction. J Dent Res. 2014;93:657–62. doi: 10.1177/0022034514534261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williamson I, Hill RE, Bickmore WA. Enhancers: from developmental genetics to the genetics of common human disease. Dev Cell. 2011;21:17–19. doi: 10.1016/j.devcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stower H. Gene expression: Super enhancers. Nat Rev Genet. 2013;14:367. doi: 10.1038/nrg3496. [DOI] [PubMed] [Google Scholar]
- 28.Shin HY. Targeting Super-Enhancers for Disease Treatment and Diagnosis. Mol Cells. 2018;41:506–14. doi: 10.14348/molcells.2018.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold PR, Wells AD, Li XC. Diversity and Emerging Roles of Enhancer RNA in Regulation of Gene Expression and Cell Fate. Front Cell Dev Biol. 2019;7:377. doi: 10.3389/fcell.2019.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–66. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 31.Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–70. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carey HA, Hildreth BE, Geisler JA, Nickel MC, Cabrera J, Ghosh S, et al. Enhancer variants reveal a conserved transcription factor network governed by PU.1 during osteoclast differentiation. Bone Res. 2018;6:8. doi: 10.1038/s41413-018-0011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko JY, Oh S, Yoo KH. Functional Enhancers As Master Regulators of Tissue-Specific Gene Regulation and Cancer Development. Mol Cells. 2017;40:169–77. doi: 10.14348/molcells.2017.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–89. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang HC, Huang HC, Juan HF, Hsu CL. Investigating the role of super-enhancer RNAs underlying embryonic stem cell differentiation. BMC Genomics. 2019;20:896. doi: 10.1186/s12864-019-6293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J, Zhou H, Gerdt C, Tan M, Colson T, Kaye KM, et al. Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc Natl Acad Sci USA. 2016;113:14121–6. doi: 10.1073/pnas.1616697113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez-Dominguez JR, Knoll M, Gromatzky AA, Lodish HF. The Super-Enhancer-Derived alncRNA-EC7/Bloodlinc Potentiates Red Blood Cell Development in trans. Cell Rep. 2017;19:2503–14. doi: 10.1016/j.celrep.2017.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, YW, Lee, S, Yun, J & Kim, A Chromatin looping and eRNA transcription precede the transcriptional activation of gene in the beta-globin locus. Biosci Rep. 2015;35. 10.1042/BSR20140126. [DOI] [PMC free article] [PubMed]
- 40.Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc Natl Acad Sci USA. 2015;112:12776–81. doi: 10.1073/pnas.1512995112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–25. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–48. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drabløs F, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–4. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imamura K, et al. Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J. 2018;37. 10.15252/embj.201797723. [DOI] [PMC free article] [PubMed]
- 46.Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, et al. Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Mol Cell. 2017;66:568–76 e564. doi: 10.1016/j.molcel.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng JH, Pan DZ, Tsai ZT, Tsai HK. Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Sci Rep. 2015;5:12648. doi: 10.1038/srep12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren C, Liu F, Ouyang Z, An G, Zhao C, Shuai J, et al. Functional annotation of structural ncRNAs within enhancer RNAs in the human genome: implications for human disease. Sci Rep. 2017;7:15518. doi: 10.1038/s41598-017-15822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–20. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon RA, Grigoriev G, Lee A, Kalliolias GD, Ivashkiv LB. The interferon signature and STAT1 expression in rheumatoid arthritis synovial fluid macrophages are induced by tumor necrosis factor alpha and counter-regulated by the synovial fluid microenvironment. Arthritis rheumatism. 2012;64:3119–28. doi: 10.1002/art.34544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis rheumatism. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 53.Park-Min KH, Serbina NV, Yang W, Ma X, Krystal G, Neel BG, et al. FcgammaRIII-dependent inhibition of interferon-gamma responses mediates suppressive effects of intravenous immune globulin. Immunity. 2007;26:67–78. doi: 10.1016/j.immuni.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 54.Bae S, Lee MJ, Mun SH, Giannopoulou EG, Yong-Gonzalez V, Cross JR, et al. MYC-dependent oxidative metabolism regulates osteoclastogenesis via nuclear receptor ERRalpha. J Clin Invest. 2017;127:2555–68. doi: 10.1172/JCI89935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyauchi Y, Ninomiya K, Miyamoto H, Sakamoto A, Iwasaki R, Hoshi H, et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med. 2010;207:751–62. doi: 10.1084/jem.20091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu L, et al. Osteoclast-mediated bone resorption is controlled by a compensatory network of secreted and membrane-tethered metalloproteinases. Sci Transl Med. 2020;12. 10.1126/scitranslmed.aaw6143. [DOI] [PMC free article] [PubMed]
- 57.Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X, et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med. 2009;15:1066–71. doi: 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim I, Kim JH, Kim K, Seong S, Kim N. The IRF2BP2-KLF2 axis regulates osteoclast and osteoblast differentiation. BMB Rep. 2019;52:469–74. doi: 10.5483/BMBRep.2019.52.7.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun L, Lian JX, Meng S. MiR-125a-5p promotes osteoclastogenesis by targeting TNFRSF1B. Cell Mol Biol Lett. 2019;24:23. doi: 10.1186/s11658-019-0146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–40. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 61.Dorsey MJ, Tae HJ, Sollenberger KG, Mascarenhas NT, Johansen LM, Taparowsky EJ. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene. 1995;11:2255–65. [PubMed] [Google Scholar]
- 62.Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–63. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- 63.Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, KC W, Albring JC, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–7. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiao S, Huang Q, Ren H, Yang M. The mechanism and function of super enhancer RNA. Genesis. 2021;59:e23422. doi: 10.1002/dvg.23422. [DOI] [PubMed] [Google Scholar]
- 65.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhaliwal NK, Mitchell JA. Nuclear RNA Isolation and Sequencing. Methods Mol Biol. 2016;1402:63–71. doi: 10.1007/978-1-4939-3378-5_7. [DOI] [PubMed] [Google Scholar]
- 67.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–3. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Chu T, Choate LA, Danko CG. Identification of regulatory elements from nascent transcription using dREG. Genome Res. 2019;29:293–303. doi: 10.1101/gr.238279.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yarilina A, Xu K, Chen J, Ivashkiv LB. TNF activates calcium-nuclear factor of activated T cells (NFAT)c1 signaling pathways in human macrophages. Proc Natl Acad Sci USA. 2011;108:1573–8. doi: 10.1073/pnas.1010030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abdallah BM, Al-Shammary A, Skagen P, Abu Dawud R, Adjaye J, Aldahmash A, et al. CD34 defines an osteoprogenitor cell population in mouse bone marrow stromal cells. Stem Cell Res. 2015;15:449–58. doi: 10.1016/j.scr.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 71.Tjonnfjord GE, Veiby OP, Steen R, Egeland T. T lymphocyte differentiation in vitro from adult human prethymic CD34+ bone marrow cells. J Exp Med. 1993;177:1531–9. doi: 10.1084/jem.177.6.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matayoshi A, Brown C, DiPersio JF, Haug J, Abu-Amer Y, Liapis H, et al. Human blood-mobilized hematopoietic precursors differentiate into osteoclasts in the absence of stromal cells. Proc Natl Acad Sci USA. 1996;93:10785–90. doi: 10.1073/pnas.93.20.10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng Y, Kang H, Luo J, Zhang Y. A Comparative Analysis of Super-Enhancers and Broad H3K4me3 Domains in Pig, Human, and Mouse Tissues. Front Genet. 2021;12:701049. doi: 10.3389/fgene.2021.701049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, et al. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- 75.Hardison RC, Oeltjen J, Miller W. Long human-mouse sequence alignments reveal novel regulatory elements: a reason to sequence the mouse genome. Genome Res. 1997;7:959–66. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- 76.Ungerback J, Hosokawa H, Wang X, Strid T, Williams BA, Sigvardsson M, et al. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1) Genome Res. 2018;28:1508–19. doi: 10.1101/gr.231423.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Natoli G, Ghisletti S, Barozzi I. The genomic landscapes of inflammation. Genes Dev. 2011;25:101–6. doi: 10.1101/gad.2018811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Izawa N, Kurotaki D, Nomura S, Fujita T, Omata Y, Yasui T, et al. Cooperation of PU.1 With IRF8 and NFATc1 Defines Chromatin Landscapes During RANKL-Induced Osteoclastogenesis. J Bone Min Res. 2019;34:1143–54. doi: 10.1002/jbmr.3689. [DOI] [PubMed] [Google Scholar]
- 80.Liang HC, Costanza M, Prutsch N, Zimmerman MW, Gurnhofer E, Montes-Mojarro IA, et al. Super-enhancer-based identification of a BATF3/IL-2R-module reveals vulnerabilities in anaplastic large cell lymphoma. Nat Commun. 2021;12:5577. doi: 10.1038/s41467-021-25379-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang QG, Qian J, Zhu YC. Targeting bromodomain-containing protein 4 (BRD4) benefits rheumatoid arthritis. Immunol Lett. 2015;166:103–8. doi: 10.1016/j.imlet.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 82.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu W, Li Z, Cai Z, Xie Z, Li J, Li M, et al. LncRNA-mRNA expression profiles and functional networks in osteoclast differentiation. J Cell Mol Med. 2020;24:9786–9797. doi: 10.1111/jcmm.15560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Silva AM, Moura SR, Teixeira JH, Barbosa MA, Santos SG, Almeida MI. Long noncoding RNAs: a missing link in osteoporosis. Bone Res. 2019;7:10. doi: 10.1038/s41413-019-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakaguchi Y, Nishikawa K, Seno S, Matsuda H, Takayanagi H, Ishii M. Roles of Enhancer RNAs in RANKL-induced Osteoclast Differentiation Identified by Genome-wide Cap-analysis of Gene Expression using CRISPR/Cas9. Sci Rep. 2018;8:7504. doi: 10.1038/s41598-018-25748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim JH, Kim N. Regulation of NFATc1 in Osteoclast Differentiation. J Bone Metab. 2014;21:233–41. doi: 10.11005/jbm.2014.21.4.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu Y, Zeng J, Roscoe BP, Liu P, Yao Q, Lazzarotto CR, et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med. 2019;25:776–83. doi: 10.1038/s41591-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS, et al. Rheumatoid arthritis. Nat Rev Dis Prim. 2018;4:18001. doi: 10.1038/nrdp.2018.1. [DOI] [PubMed] [Google Scholar]
- 91.van der Heijde DM. Joint erosions and patients with early rheumatoid arthritis. Br J Rheumatol. 1995;34(Suppl 2):74–78. doi: 10.1093/rheumatology/XXXIV.suppl_2.74. [DOI] [PubMed] [Google Scholar]
- 92.Machold KP, Stamm TA, Nell VP, Pflugbeil S, Aletaha D, Steiner G, et al. Very recent onset rheumatoid arthritis: clinical and serological patient characteristics associated with radiographic progression over the first years of disease. Rheumatology. 2007;46:342–9. doi: 10.1093/rheumatology/kel237. [DOI] [PubMed] [Google Scholar]
- 93.Ejbjerg B, Narvestad E, Rostrup E, Szkudlarek M, Jacobsen S, Thomsen HS, et al. Magnetic resonance imaging of wrist and finger joints in healthy subjects occasionally shows changes resembling erosions and synovitis as seen in rheumatoid arthritis. Arthritis rheumatism. 2004;50:1097–106. doi: 10.1002/art.20135. [DOI] [PubMed] [Google Scholar]
- 94.Alippe Y, Mbalaviele G. Omnipresence of inflammasome activities in inflammatory bone diseases. Semin Immunopathol. 2019;41:607–18. doi: 10.1007/s00281-019-00753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gravallese EM, Harada Y, Wang JT, Gorn AH, Thornhill TS, Goldring SR. Identification of cell types responsible for bone resorption in rheumatoid arthritis and juvenile rheumatoid arthritis. Am J Pathol. 1998;152:943–51. [PMC free article] [PubMed] [Google Scholar]
- 96.Pettit AR, Ji H, von Stechow D, Muller R, Goldring SR, Choi Y, et al. TRANCE/RANKL knockout mice are protected from bone erosion in a serum transfer model of arthritis. Am J Pathol. 2001;159:1689–99. doi: 10.1016/S0002-9440(10)63016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Redlich K, Hayer S, Ricci R, David JP, Tohidast-Akrad M, Kollias G, et al. Osteoclasts are essential for TNF-alpha-mediated joint destruction. J Clin Investig. 2002;110:1419–27. doi: 10.1172/JCI15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.