Graphical abstract
Keywords: In vitro-in vivo extrapolation IVIVE, Benzo[a]pyrene, DNA adduct formation
Highlights
-
•
Link between DNA adducts and BaP exposure.
-
•
Strategy for quantitative in vitro-in vivo extrapolation for carcinogen BaP.
-
•
Discussion of problems in quantitative in vitro-in vivo extrapolation.
Abstract
To reduce the need for animal tests, in vitro assays are often used as alternative methods. To derive toxic doses for higher tier organisms from in vitro assay results, quantitative in vitro-in vivo extrapolation (qIVIVE) based on physiological-based toxicokinetic (PBTK) models is typically the preferred approach. Such PBTK models require many input parameters to address the route from dose to target site concentration. However, respective data is very often not available. Hence, our aim is to call attention to an alternative way to build a link between animal (in vivo) and cell-derived (in vitro) toxicity data. To this end, we selected the carcinogenic chemical benzo[a]pyrene (BaP) for our study. Our approach relates both in vitro assay and in vivo data to a main intermediate marker structure for carcinogenicity on the subcellular level – the BaP-DNA adduct BaP-7,8-dihydrodiol-9,10-epoxide-deoxyguanosine. Thus, BaP dose is directly linked to a measure of the toxicity-initiating event. We used Syrian hamster embryo (SHE) and Balb/c 3T3 cell transformation assay as in vitro data and compared these data to outcomes of in vivo carcinogenicity tests in rodents. In vitro and in vivo DNA adduct levels range within three orders of magnitude. Especially metabolic saturation at higher doses and interspecies variabilities are identified and critically discussed as possible sources of errors in our simplified approach. Finally, our study points out possible routes to overcome limitations of the envisaged approach in order to allow for a reliable qIVIVE in the future.
Introduction
Most toxicity data for risk assessment of chemicals are still derived from animal tests. The need to replace this practice has not only financial and ethical reasons. It is by now often prescribed by law, whenever possible, like in the European chemicals registration legislation (REACH). Although referencing to animal tests for human risk assessment does not seem ideal (Andersen et al., 2019, Balls, 2020), it is mostly the only source of accepted data for risk and hazard assessment of chemicals. In vitro cell-based assays are promising alternatives but typically provide (internal) assay-specific effect concentrations as a measure of toxicity. For regulatory purposes, however, information about a critical external dose is required instead (Grech et al., 2017) in order to derive thresholds below which chemicaĺs risk and hazard are sufficiently low.
To convert in vitro effect concentrations into external doses, quantitative in vitro-in vivo-extrapolation (qIVIVE) is the current strategy to take absorption, distribution, metabolism, and excretion (ADME) into account. The key component of such approaches is mostly a physiology-based toxicokinetic (PBTK) model. By use of many parameters, including rate constants for absorption, membrane permeability, biotransformation (i.e., metabolic clearance), and others, as well as partitioning data and physiological parameters (e.g., for the blood flow), PBTK models aim to estimate an external dose from a point-of-departure concentration obtained from in vitro testing. Some working groups developed tailor-made PBTK models for chemicals of interest (Heredia-Ortiz et al., 2011, Li et al., 2017, Louisse et al., 2010, Turley et al., 2019), but also generic models (i.e., irrespective of certain substances) like SimCyp (Jamei et al., 2009) or the US EPA published httk R package (Pearce et al., 2017) are commercially available.
The readout of an in vitro assay is usually related to one of the key events along the so-called adverse outcome pathway (AOP) (Ankley et al., 2010). The AOP concept often serves as a mechanism-informed guideline for the development of non-animal alternatives (OECD., 2014, Villeneuve et al., 2014), and links the molecular (toxicity) initiating event (MIE) through a cascade of key events with the observed adverse effect. At least to our understanding, in vitro data used as input for PBTK modeling reflects the concentration of a chemical’s active species as close to the location of the MIE as possible - for instance, an internal cell concentration. A PBTK-based qIVIVE can only be validated by comparing extrapolated external doses or tissue concentrations with respective animal test-derived or human-based data. However, if quantitative in vitro and in vivo data at the level of the MIE are accessible, one could investigate if in vitro and in vivo toxicity tests are based on comparable extents of the MIE.
In order to investigate this further, we searched for an example of a chemical that is well characterized in terms of mode of action (MoA) and quantitative toxicity data (in vitro and in vivo). The polycyclic aromatic hydrocarbon benzo[a]pyrene (BaP) is well characterized regarding the effect pathway, metabolization, and adverse effects. It is ubiquitously present in the environment since its results from incomplete combustion of organic matter such as wood or charcoal. Humans are further exposed via tobacco smoke and smoked or grilled food, among others. BaP is classified as a confirmed human carcinogen (Group 1) by the International Agency for Research on Cancer (IARC - International Agency for Research on Cancer, 2010, Straif et al., 2005). Its genotoxicity is mainly based on the cytochrome P450 (CYP)-catalyzed oxidation to its active form 7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydro-benzo[a]pyrene (BPDE), which can bind covalently to DNA purine bases (Sayer et al., 1991). Without sufficient repair, chemical alteration of DNA can lead to mutations, which, if they occur in critical genes, enable the formation of tumor cells. Mechanistic information on cancer development by BaP is given in more detail in the Supporting Information (S1).
In this example, the MIE of cancer genesis is the covalent binding of BPDE to the DNA. The so-formed DNA adducts can be measured in test animals as well as in cells from in vitro tests, providing a quantitative measure for the turn-over rate of the MIE. DNA adduct formation yields (describing the formation of DNA adducts depending on the BaP concentration) are reported in the literature for both in vivo and in vitro studies (Bjelogrlic et al., 1994, Daniel et al., 1983, Ginsberg and Atherholt, 1990, Godschalk et al., 2000, Kulkarni et al., 1986, Marie-Desvergne et al., 2010, Marie et al., 2008, Moore et al., 1987, Motwani et al., 2020, Shiizaki et al., 2013, Topinka et al., 2008). Thereby the number of DNA adducts at a given BaP concentration or dose can be calculated. By reference to this DNA adduct level in the in vitro assay and the in vivo study, a direct link appears possible. For this study, we used only the DNA adduct of BPDE to the N2-nitrogen of deoxy-guanosine (N2-dGuo) for calculation because it is formed to the highest extent and consistently and reliably quantified in the literature (Marie et al., 2008). Many data are available for both in vitro and in vivo genotoxicity due to the use of BaP as a positive control in genotoxicity tests, for example. In vivo studies were conducted excessively when animal testing was less restricted than it is nowadays. Hence, different in vivo carcinogenicity studies are available.
In this study, we used carcinogenicity tests in rodents as in vivo and rodent cell transformation assays (CTA) in Syrian hamster embryo cells (SHE) and the Balb/c 3T3 cell line as in vitro data sources. In in vivo rodent carcinogenicity tests, the animals are typically exposed to the test chemical and dissected and examined for tumors after a defined incubation duration. CTAs are considered as (currently the best) in vitro alternative to predict carcinogenicity in vivo using just one assay instead of a test battery. In CTAs, in general, cells are seeded on dishes and incubated with test agents. Morphologic transformation of cells or cell colonies refers to alterations in the genetic material. It is a multistage process that closely models the various stages of in vivo carcinogenesis. SHE and Balb/c 3T3 have shown a high predictive performance for carcinogenic substances (Combes et al., 1999). The SHE assay was accepted for regulatory purposes in weight of evidence approaches (ECVAM, 2005). A respective OECD guidance document is also available (OECD, 2015). The Balb/c 3T3 assay was recommended for regulatory acceptance by the EURL ECVAM (2004). By linking BaP-DNA adduct levels to exposure concentrations in vitro and in vivo, we want to call attention to a possible alternative route for qIVIVE, including current limitations of this approach.
Methods
Selection of studies and data for the comparison of in vivo and in vitro data
In vitro data from CTAs with SHE (LeBoeuf et al., 1996, Maire et al., 2012b, Pant et al., 2012) and Balb/c 3T3 (Atchison et al., 1982, Dunkel et al., 1981, Sakai and Sato, 1989, Tanaka et al., 2012) cells were collected from the literature. The SHE assay is conducted with non-immortalized cells extracted from Syrian hamster embryos. Morphologic transformation in the growing colonies is expressed in an extensive, random-oriented, multilayered cell growth with crisscrossing at the colony center and on the perimeter (OECD, 2015). In the SHE assay, BaP is used as positive control in the OECD guidance document. Good accordance between in vivo genotoxicity test results of chemicals and cell transformation in the in vitro assay was shown (Pienta et al., 1977). There are mainly-two different protocols for the SHE assay, 24 h and 7 days exposure time, respectively, to distinguish between so-called inducer and promotor substances. BaP is active in both. Here, only 7-day-assay data were used. The investigated BaP concentrations in the SHE assays (LeBoeuf et al., 1996, Maire et al., 2012b, Pant et al., 2012) ranged from 0.01 to 45 µg/mL.
In the Balb/c 3T3 assay, an immortalized embryonic mouse cell line is used. Here, a change of the cells’ phenotypic features undergoing the first steps of the conversion from normal cells to neoplastic-like cell foci with oncogenic properties can be observed. The average number of foci per culture dish serves as quantitative endpoint (ECVAM, 2004). In this in vitro assay, BaP is not officially suggested as a positive control but also gives a positive response. The investigated BaP concentrations (Atchison et al., 1982, Dunkel et al., 1981, Sakai and Sato, 1989, Tanaka et al., 2012) ranged from 0.0005 to 15 µg/mL. In both CTAs, no additional metabolizing material was used, and BaP is converted into the carcinogen BPDE by cellular metabolism only.
As in vivo data sources, animal test studies were selected from the literature. We included only tests that were conducted in rodents (rat or mouse) in this study. Moreover, we used only studies where BaP was applied as a single dose due to the comparability to the DNA adduct assays, where also single doses were used. Additionally, experiments with only one dosing level were excluded as we needed the different data points of a dose–response curve for comparison. Nine studies were finally included in our dataset (Cavalieri et al., 1991, Cavalieri et al., 1988, Deutsch-Wenzel et al., 1983, Grimmer et al., 1987, Pott, 1973, Topping et al., 1981, Wenzel-Hartung et al., 1990). Original data from literature and calculated DNA adduct levels for in vitro and in vivo experiments are summarized in the Supporting Information (Table S2).
Conversion of nominal concentrations into freely dissolved concentrations in in vitro assays
From the cell transformation assays in the literature, only nominal concentrations were reported. Exposure control was not conducted (or reported). Following the widely accepted free-drug theory, only the freely dissolved concentration cfree of a chemical is available to cause an effect (Trainor, 2007). The freely dissolved concentration can be reduced by loss due to vaporization, sorption to well-plates or dishes, cells, and medium components. Volatilization was found to be negligible by checking the medium-air-partition coefficient to be greater than 10,000 L/L, as recommended by Escher et al. (Escher et al., 2019). Loss due to sorption to culture dishes was estimated according to Fischer et al. (2018) and was found to be smaller than 1%, estimated with highest concentration (100 mmol/L), lowest percentage of FBS (10%), and longest assay duration (168 h) used in this study. Further, the estimation of cell mass in the in vitro assays was not possible because it changes during colony formation. In culture medium, the main targets for sorption are typically plasma proteins and lipids provided by the components of fetal bovine serum (FBS). To estimate cfree in the in vitro assay, the used medium volume needs to be known but was not given in the used in vitro adduct formation study (Kulkarni et al., 1986). Only the fraction of FBS in the medium was reported. Therefore, it was possible to calculate correction factors (CF) between each CTA and DNA adduct assay, taking only the FBS components as the dominating sorptive sink into account. According to Eqs. (1), (2) these correction factors were calculated with fu as fraction unbound, VFSA as the volume fraction of serum albumin, and VFPL as the volume fraction of phospholipid in the assay, KSA/w as serum albumin-water and KPL/w as phospholipid-water partition coefficient (in L/L). Eq. (1) is valid with the assumption that the water volume is nearly equal to the total volume. For the Balb/c 3T3 assay, conditions were comparable to those in the in vitro DNA adduct assay (10% FBS in medium). In this case, there was no need to use any correction factor.
| (1) |
| (2) |
Equilibrium partitioning within the reported assay composition was assumed. Note that in this approach, cfree in the medium equals the internal cell (water) concentration, and active transport is neglected. Partition coefficients of BaP to serum albumin and phospholipid were estimated by linear solvation energy relationships obtained from the UFZ LSER database (Ulrich et al., 2017). The following average FBS composition was assumed for the partitioning calculation: 52.76 mL/L serum albumin and 1.57 mL/L phospholipid (adapted from Fischer et al. (Fischer et al., 2017)). We assumed that the remaining component in the medium is only water. Correction factors are given in the Supporting Information (Table S2).
Calculation of the number of DNA adducts in the in vitro assays and in vivo tests
No information on DNA adduct levels was given in the selected in vitro and in vivo effect studies. Thus, nominal BaP concentrations in vitro and BaP doses in vivo needed to be converted into DNA adduct levels using different studies, where respective adducts were determined. In vitro and in vivo DNA adduct formation yields were obtained from the literature (Bjelogrlic et al., 1994, Kulkarni et al., 1986, Marie-Desvergne et al., 2010), where endometrium tissue slices of Syrian hamster, A/HeJ mice, C57BL/6 mice, and Sprague-Dawley rats were exposed to BaP, respectively, and subsequently, DNA adducts were quantified. To this end, only the mainly formed adduct of BPDE to the N2-nitrogen of deoxy-guanosine was considered because this specific DNA adduct was consistently and reliably quantified in the respective studies (Bjelogrlic et al., 1994, Kulkarni et al., 1986, Marie-Desvergne et al., 2010). Hence, we refer only to this specific adduct as DNA adduct in the following.
In vitro and in vivo DNA adduct formation was assumed to be directly proportional to used concentrations or administered dose per test animal. From each in vitro assay and each in vivo test series, all BaP concentrations were converted into DNA adduct levels, according to Eq. (3).
| (3) |
The most common unit for the DNA adduct level is pmol adduct per mg DNA (Bjelogrlic et al., 1994, Daniel et al., 1983, Ginsberg and Atherholt, 1990, Godschalk et al., 2000, Moore et al., 1987, Motwani et al., 2020, Shiizaki et al., 2013, Tapiainen et al., 1996). In Marie-Desvergne et al. (2010), the number of DNA adducts was reported in adducts per 108 normal nucleosides. This unit was converted into pmol adduct/mg DNA according to Eqs. (4), (5) using the Avogadro constant NA and an average molecular mass of 650 g/mol per nucleoside (bitesizebio.com, 2014).
| (4) |
Or:
| (5) |
Other DNA adduct formation studies were also found in the literature (Daniel et al., 1983, Ginsberg and Atherholt, 1990, Godschalk et al., 2000, Marie et al., 2008, Moore et al., 1987, Motwani et al., 2020, Shiizaki et al., 2013, Tapiainen et al., 1996, Topinka et al., 2008). We listed the outcomes and metadata of these experiments in the Supporting Information (Table S2).
Calculation of EC50-like DNA adduct values
Concentration-response relationships were determined for each data set individually. In the case of in vitro assays, the individual data were normalized to the observed maximum response over all studies to harmonize the upper limit of the concentration–response model. Additionally, a decrease in response after reaching the individual maximum in each study was considered to be caused by acute cytotoxicity. Therefore respective data was excluded from the analysis. EC50 values were calculated by applying a two-parameter log-logistic model (see Eq. (6)) using RStudio (version 1.4.1717, basic R version 4.1.0) and the drc package (version 3.1–0). In order to improve model fits for studies with only a few data points, negative controls were included in the analysis for all cases with regard to consistency.
| (6) |
Outliers for the box-whisker-plot were removed after Rosner's test (package EnvStats, version 2.4.0).
Results
DNA adduct levels were calculated according to Eq. (3) based on the following three simplifications. First, for both in vitro and in vivo, a linear relationship between applied BaP concentration or dose and DNA adduct level is assumed. Second, DNA adduct levels in endometrial tissue slices resulting from a defined BaP exposure are expected to be comparable to respective adduct levels in vitro and in vivo. Finally, we assume that the genotoxic responses in vitro and in vivo are predominantly caused by the BPDE-dGuo DNA adduct. These simplifying assumptions lead to some uncertainties in our approach, which are discussed in detail below (see Discussion).
For in vitro assays, the nominal concentration was first corrected according to Eqs. (1), (2) to take sorption processes into account. Fig. 1 shows the scheme applied for calculations. The nominal concentration of 1 µM BaP in mouse and hamster endometrium in vitro assays yielded 0.096 and 0.0732 pmol DNA adducts/mg DNA, respectively (Kulkarni et al., 1986). In vivo, 1.6 pmol DNA adducts/mg DNA were measured after 24 h in C57BL/6 mice after dermal application of 62.5 µg/mouse. For an intravenously administered dose of 40 µmol/kg body weight (BW) in Sprague-Dawley rats, 13.1 DNA adducts per 108 normal nucleosides were determined, corresponding to 0.202 pmol DNA adducts/mg DNA (Marie-Desvergne et al., 2010) at 24 h.
Fig. 1.
Calculation scheme for the determination of DNA adducts in vitro and in vivo.
In the manner of a dose–response curve, in vitro morphological transformation frequency (MTF) or the average number of foci per dish, respectively, were correlated to the estimated number of DNA adducts. The obtained data are shown in Fig. 2.
Fig. 2.
In vitro SHE (A) (LeBoeuf et al., 1996, Maire et al., 2012b, Pant et al., 2012) and Balb/c 3T3 (B) (Atchison et al., 1982, Dunkel et al., 1981, Sakai and Sato, 1989, Tanaka et al., 2012) assay effects (% MTF and average foci per dish) at the calculated adduct levels. Dotted grey lines delineate inconclusive results, caused by acute cytotoxicity. Note that the adduct levels were plotted on a logarithmic scale.
The calculated DNA adduct levels for the SHE assays ranged from 2.5*10-3 to 8.6 pmol/mg DNA. In each SHE assay, vehicle control is carried along, which is usually between 0 and 0.5% MTF. By definition, positive MTF results must be significantly higher than the MTF of the control and higher than 0.6% (Maire et al., 2012a) and we inserted 0.6% MTF as acceptance criteria in Fig. 2 (dashed line). The series of LeBoeuf et al., 1996, Maire et al., 2012b (Bioreliance + BASF), and Pant et al. (2012) (Harlan CCR) show quite a small margin of concentration and no significant increase in the MTF. Hence, these data were excluded from our studies. The other curves show an increase over the entire range of DNA adduct levels, whereas every curve contains data points differing from the overall trend. Regarding the three datasets, which include low BaP concentrations (Maire et al., 2012b (Metz 1–3)), the SHE assay cannot detect lower MTFs than the ones caused by 10-3 pmol DNA adducts/mg DNA.
The Balb/c 3T3 assay showed significant response at DNA adducts levels of 10-4 to 4 pmol/mg DNA, which is basically in the same range as in the SHE assays. All datasets show an increase in the number of foci. Very low (Atchison 1982 and Sakai 1989 (Atchison et al., 1982, Sakai and Sato, 1989)) as well as higher increases (3 Tanaka 2012 sets (Tanaka et al., 2012)) in the number of foci can be seen in the different studies. In the Tanaka (2012) set, the number of foci conspicuously decreases again at very high DNA adduct levels. This can also be seen in some of the SHE-CTA data sets and is probably caused by acute cytotoxicity (Hoffmann et al., 2012). These concentrations are out of the applicability range of the respective in vitro test and are not further considered in this work.
SHE and Balb/c 3T3 assay data vary substantially in their responses, and assay curves cover a wide range of DNA adducts (about five orders of magnitude).
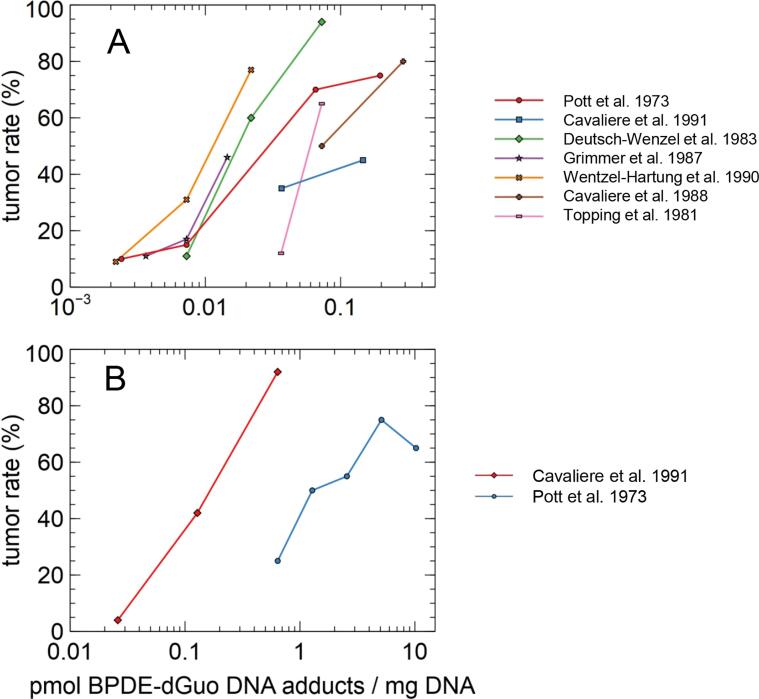
In vivo test results (tumor rate in animals) are depicted in Fig. 3 for the respective calculated DNA adduct levels. Studies with rats and mice are evaluated separately. The studies revealed low to very high tumor incidences between 4 and 94%. The calculated DNA adduct levels ranged from 0.002 to 10.2 pmol/mg DNA.
Fig. 3.
In vivo tumor rates (Cavalieri et al., 1991, Cavalieri et al., 1988, Deutsch-Wenzel et al., 1983, Grimmer et al., 1987, Pott, 1973, Topping et al., 1981, Wenzel-Hartung et al., 1990) for rats (A) and mice (B) vs calculated DNA adduct levels.
DNA adduct levels are widely differing over more than three orders of magnitude. While values in the rat studies seem to be more congruent than the values in the mouse studies, both species show a difference of three orders of magnitude in the DNA adduct levels. Only two mouse studies were included, which does not allow for a conclusion on the variability itself.
Our results show that both in vitro and in vivo test systems were independently designed with BaP doses yielding between 0.0001 and 10 pmol adducts/mg DNA. Fig. 4 shows the responses of each assay after normalization to the overall maximum response. The highest average increase of the normalized response occurs between 0.001 and 0.01 pmol DNA adducts/mg DNA for SHE and Balb assays and for the rat studies. Only for the mouse studies, the range of highest increase is higher between 0.01 and 0.1 pmol DNA adducts/mg DNA. Still, as shown by the colored areas, the variations in the response between the individual studies are considerably high, ranging mostly over far more than 30% normalized response in the respective DNA adduct level range.
Fig. 4.
Comparison of DNA adduct levels for the different in vitro and in vivo tests. The data points show the average response value in the respective DNA adduct range, normalized to the overall maximum response value. The colored areas depict the respective minimum and maximum in each range. Ranges were set to 0.0001–0.001, 0.001–0.1, 0.1–1, and 1–10 pmol DNA adduct/mg DNA. See separated graphs in Supporting Information S1.
We included further analysis of the datasets using a concentration–response fit model for each assay or animal study. A log-logistic curve was fitted to calculate an EC50- (or ED50)-like DNA adduct value that causes 50% of the maximum response. Three studies (LeBoeuf et al., 1996, Maire et al., 2012b, Pant et al., 2012), which did not show an overall increase in the response, could not be fitted. The results, ordered by test setup, are shown in a box-whisker-plot in Fig. 5. Compared to the wide range of DNA adducts calculated from the raw data, in vitro and in vivo data match quite well. All median EC50-like values are within one order of magnitude (rats: 0.025; SHE assays: 0.26). Variations were present already in the original test datasets and were somewhat diminished in the case of the SHE assay through the cfree correction.
Fig. 5.
Box-whisker-plot of EC50-like DNA adduct levels that cause 50 % of the maximum response. Outliers were removed after applying a Rosner’s outlier test.
Discussion
With this work, we aimed to link in vitro and in vivo results for the genotoxic effect of BaP. The toxicity-initiating event, the formation of DNA adduct BPDE-dGuo, was the central connection point between in vitro and in vivo experiments. Due to the simplifications mentioned above, this approach still has some uncertainties, which are discussed in the following paragraphs.
Linearity between dose and DNA adduct formation
Our approach assumes that DNA adduct formation (in vitro and in vivo) is directly proportional to the applied concentration or administered dose. Moreover, our calculations were based on only one DNA adduct formation yield for each DNA adduct calculation, assuming further linearity between dose and adduct formation. These simplifications may overlook species-specific toxicokinetics as well as the potential impact of metabolic saturation and DNA repair mechanisms on the DNA adduct level. However, the in vivo data of Tapiainen et al. (Tapiainen et al., 1996), where 50, 100, 300, 500, or 750 µg BaP/mouse were dermally applied, could be interpreted to be linear over the whole range. However, it might also be the case that metabolic saturation occurs for higher doses. The data of Bjelogrlic et al. (1994) confirm the metabolic saturation at higher doses (500 µg/animal, S2). Since the dose of 40 µmol/kg b.w. in Marie-Desvergne et al. (2010) is relatively high, it remains unclear if the measured DNA amounts are affected by metabolic saturation. However, no data with lower doses or different dose levels were available in literature for rats, and the amounts of DNA adducts levels calculated in this work might be underestimated.
Linearity between applied BaP concentration and DNA adduct level has been shown in vitro for lower concentrations. In Shiizaki et al. (Shiizaki et al., 2013), BPDE-dGuo adducts were measured in HepG2 cells with 0.5,1, 2.5, 5, or 10 µM BaP, showing exact linearity. But, Marie et al. (Marie et al., 2008) published two DNA adduct formation yields at 10 and 50 µM BaP in HepG2 cells, indicating that DNA adduct formation increases less than expected at higher concentrations. However, at high concentrations, cell assay data get increasingly inconclusive, probably due to acute cytotoxic effects, so this limitation of the assumption seems to be acceptable.
Time dependency of DNA adduct level
As outlined above, binding of the reactive BaP metabolite, BPDE, to the DNA is the genotoxicity initiating step. However, DNA adducts are often recognized and repaired by cellular repair mechanisms. Both DNA adduct formation and DNA repair result in a time-dependent DNA adduct level. In some in vivo adduct studies (Bjelogrlic et al., 1994, Marie-Desvergne et al., 2010), the respective time dependency of DNA adduct levels was investigated. Therein, the highest adduct levels were observed after 18–24 h, followed by a slight decrease afterward. We selected the DNA adduct level at 24 h for our work. In addition, distribution patterns of BaP and its metabolites in the organism are time-dependent. Further, repair mechanisms may have a substantial impact on the number of DNA adducts at different time points. To implement repair mechanisms into a quantitative extrapolation, additional data from metabolomics approaches, e.g., are needed for the test systems (Madureira et al., 2014). By selecting only one time point for calculating the DNA adducts our approach may appear too simple. However, Marie-Desvergne et al. (2010) demonstrated that the DNA adducts in rats are relatively stable between 24 and 72 h, thus showing that adduct formation and decomposition through repair mechanisms proceed to equal extents. However, the duration of the carcinogenicity experiments varied between 20 weeks and 28 months (see Supporting Information Table S2), where DNA adducts are certainly phased out.
In vitro, the peak adduct level is reached much earlier than in vivo because no distribution process is necessary to transport the chemical to the different tissues and organs. Marie et al. (2008) observed the highest adduct level in HepG2 cells already at the first sampling point, 4 h after a 24 h treatment period, and it decreased rapidly afterward. It is plausible that in cells and tissues with different metabolic capacities, the time course of DNA adducts differs substantially. However, from the in vitro adduct formation study included in our calculations (Kulkarni et al., 1986), we do not know the time-dependency of the formation of DNA adducts, so we used the only available 18 h-value.
For the selection of literature data, we decided to use only single dosing in vivo tests to be comparable to the in vitro assays, where BaP is dosed only once at the beginning of the test. Generally, for carcinogenesis, the area under the time-concentration curve of genotoxic chemicals is supposed to be decisive for the carcinogenic property (Westberg et al., 2015).
Variability within each test setup
Even though the SHE assay is a standardized CTA, differences are observed between the different studies (Fig. 2A). Also, the Balb/c 3T3 assay data vary (Fig. 2B). CTAs are generally not intended to provide quantitative output but rather state a positive or negative expected outcome with respect to carcinogenicity. However, one should be aware of the variability of the in vitro assays if a quantitative assessment is done. The SHE cell transformation assay exists in two different variants of exposure time (24 h and 7 d). Further, cfree needs to be determined reliable as well as the pH for ionizable chemicals. For both, an implemented exposure control appears helpful. It should be noted that two different protocols were established for the SHE cell transformation assay according to the culture medium pH (pH 6.7, LeBoeuf et al. 1996 or pH 7.0, Maire et al. 2012b).
In the in vivo studies, the different administration routes may explain the variations (Fig. 3) to some extent. Tumors were only partially counted in the tissue corresponding to the administration route. Depending on the absorption and distribution of BaP for a given administration route, differences in the adduct formation may arise. These differences were not regarded in our analysis and may lead to uncertainties in our extrapolation approach.
Variations between the test setups of different cell types and species
In vitro tests and the DNA adduct formation studies were conducted with cells from different tissues. CTAs used primary stem cells or an embryonic cell line, whereas DNA adduct formation yields were measured in endometrium tissue slices (Kulkarni et al., 1986). The latter showed differences in the determined adducts up to factor 30 for the endometrium cells of different species (Sprague-Dawley rat 0.01, A/HeJ mouse 0.07, Syrian golden hamster 0.10, human 0.28 pmol BPDE adducts/mg DNA; S2), keeping in mind that the absolute DNA adduct levels are quite small. Daniel et al. (1983) observed differences up to factor 8 for bladder and trachee-bronchus cells of different species (rat, hamster, dog, monkey, human). The variability is even confirmed for different fish cell lines using rainbow trout, bluegill fry, and brown bullhead fish cells (factor of 10 at 120 h) (Smolarek et al., 1987). Balb/c 3T3 is an immortalized cell line that tends to show different metabolic behavior in relation to primary cells and, thus, to animals used in in vivo studies (Lilienblum et al., 2008). In particular, differences in their capabilities have been demonstrated for BaP metabolization (Genies et al., 2013, Shah et al., 2016). Further, tissue differences were reported in vivo. Besides the DNA adduct formation yield in rat lung after intravenous injection used in this study, Marie-Desvergne et al. (2010) also provided values in rat liver and blood, suggesting a difference of factor four between liver and lung.
Carcinogenesis by BaP other than via BPDE-DNA adducts
A further assumption we made in our approach is that both the transformation response in the CTAs and the tumor incidence correlate with BPDE-dGuo DNA formation. It is shown in several studies that the N2-dGuo adduct is mainly formed (Marie et al., 2008, Peltonen and Dipple, 1995, Piberger et al., 2018) and Shukla et al. showed that most of the mutations arise from this adduct (Shukla et al., 1997). Further, Slaga et al. (1979) demonstrated that trans(+)BPDE-dGuo adducts caused up to 70% of the tumor-initiating activity in mouse skin (Slaga et al., 1979). However, in vivo tumor formation is far more complex than the morphological transformation of a cell colony (Smets, 1980). Although the BPDE-dGuo DNA adduct is the major driver of carcinogenesis (Peltonen & Dipple, 1995), tumor formation caused by BaP is not necessarily based only on this marker structure. Less abundant DNA adducts and formation of reactive oxygen species may also contribute to BaP genotoxicity (see detailed MoA in Supporting Information, S1). Further, BaP is a prototypic ligand of the aryl hydrocarbon receptor (AhR) that is expressed by many different cell types and conveys distinct molecular effects (Stockinger et al., 2021). AhR activation enhances, among others, CYP enzyme expression. These enzymes, in turn, catalyze both toxification of BaP to the genotoxic BPDE and detoxification to the non-genotoxic 3-OH-BaP. Furthermore, the AhR activation may affect cells of the immune system, leading to immune deregulation that may suppress tumor identification and elimination (Leclerc et al., 2021). This represents a non-genotoxic carcinogenic effect of BaP. However, until now, no suitable in vitro tests specifically addressing non-genotoxic carcinogenicity are available (Jacobs et al., 2020).
Further validation of the DNA adduct level approach
To assess the critical factors discussed above and to reduce the uncertainty of the DNA adduct level approach, further validation is, of course, needed. Typically, alternative approaches are validated through comparison with traditional methods, which could, in this case, be a PBTK-based qIVIVE. However, formation of the BPDE-DNA adduct from BaP is at least a four-step process (when neglecting side processes), covering the CYP-mediated formation of an initial BaP-epoxide, its hydrolysis, which is followed by BPDE formation through further epoxidation, and its final reaction with the DNA. A respective four-step PBTK model would require numerous rate constants and partition coefficients as input parameters, keeping in mind that for a PBTK model for the one-step formation of 3-hydroxy-BaP from BaP 46 parameters were considered (Heredia-Ortiz et al., 2011). Reliable input data for such complex PBTK models is, unfortunately, almost completely lacking. Furthermore, the result of the one-step PBTK model of Heredia-Ortiz et al. (2011) deviates up to one order of magnitude from the respective experimental data (Heredia-Ortiz et al., 2011). Thus, for a four-step model, uncertainty in the range of multiple orders of magnitude has to be expected, disqualifying it further as a validation tool for the DNA adduct level approach.
One promising way to further validate our approach could be the experimental determination of DNA adduct levels in different cell lines or tissues after exposure to different BaP concentrations. This would inform about intra- and inter-species variabilities and a possibly cell-specific relationship between BaP exposure and DNA adduct level (linear vs non-linear through metabolic saturation).
Conclusions
In conclusion, DNA adduct levels appear as a promising link for in vivo and in vitro carcinogenicity. They serve as a measure of the MIE and, as such, should cover all pre-MIE processes, which otherwise need to be modeled by multi-parametric PBTK approaches. However, metabolic saturation and repair mechanisms are not addressed by this DNA adduct level approach, while it well shows intra- and interspecies variabilities. All these points and toxicokinetics need to be considered additionally when performing a qIVIVE approach, and should be investigated in future. Currently, there are still many data gaps, even for the long-studied BaP, which hamper a suitable quantitative extrapolation.
CRediT authorship contribution statement
Martin Gerhards: Writing – original draft, Data curation, Formal analysis, Visualization. Alexander Böhme: Writing – review & editing, Conceptualization. Kristin Schubert: Writing – review & editing. Bernhard Kodritsch: Formal analysis, Visualization. Nadin Ulrich: Conceptualization, Data curation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We want to acknowledge the statistical service of the Helmholtz center for environmental research UFZ for valuable input on the statistical evaluation. The presented work contributes to the PhD College InCeTo and to the topic area Chemicals In The Environment (CITE) funded by the Helmholtz Research Program.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crtox.2022.100097.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
All data generated or analyzed during the present study are included in this published article. Data, associated metadata, and calculation tools are also available from the corresponding author.
References
- Andersen M.E., McMullen P.D., Phillips M.B., Yoon M., Pendse S.N., Clewell H.J., Hartman J.K., Moreau M., Becker R.A., Clewell R.A. Developing context appropriate toxicity testing approaches using new alternative methods (NAMs) ALTEX. 2019;36(4):523–534. doi: 10.14573/altex.1906261. [DOI] [PubMed] [Google Scholar]
- Ankley G.T., Bennett R.S., Erickson R.J., Hoff D.J., Hornung M.W., Johnson R.D., Mount D.R., Nichols J.W., Russom C.L., Schmieder P.K., Serrrano J.A., Tietge J.E., Villeneuve D.L. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 2010;29(3):730–741. doi: 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- Atchison M., Chu C.-S., Kakunaqa T., Van Duuren B.L. Chemical cocarcinogenesis with the use of a subclone derived from Balb/3T3 Cells with catechol as cocarcinogen. JNCI - J. Natl. Cancer I. 1982;69(2):503–508. doi: 10.1093/jnci/69.2.503. [DOI] [PubMed] [Google Scholar]
- Balls M. It's time to reconsider the principles of humane experimental technique. Altern. Lab. Anim. 2020;48(1):40–46. doi: 10.1177/0261192920911339. [DOI] [PubMed] [Google Scholar]
- bitesizebio.com. (2014) [Internet]. How To Calculate The Number Of Molecules In Any Piece of DNA, accessed on May 05, 2022. https://bitesizebio.com/20669/how-to-calculate-the-number-of-molecules-in-any-piece-of-dna/.
- Bjelogrlic N.M., Mäkinen M., Stenbäck F., Vähäkangas K. Benzo[a]pyrene-7,8-diol-9,10-epoxide-DNA adducts and increased p53 protein in mouse skin. Carcinogenesis. 1994;15(4):771–774. doi: 10.1093/carcin/15.4.771. [DOI] [PubMed] [Google Scholar]
- Cavalieri E., Rogan E., Sinha D. Carcinogenicity of aromatic hydrocarbons directly applied to rat mammary gland. J. Cancer Res. Clin. Oncol. 1988;114(1):3–9. doi: 10.1007/BF00390478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri E., Higginbotham S., RamaKrishna N., Devanesan P., Todorovic R., Rogan E., Salmasi S. Comparative dose—response tumorigenicity studies of dibenzo[a, l]pyrene versus 7, 12-dimethylbenz[a]anthracene, benzo[a]pyrene and two dibenzo[a, l]pyrene dihydrodiols in mouse skin and rat mammary gland. Carcinogenesis. 1991;12(10):1939–1944. doi: 10.1093/carcin/12.10.1939. [DOI] [PubMed] [Google Scholar]
- Combes R., Balls M., Curren R., Fischbach M., Fusenig N., Kirkland D., Lasne C., Landolph J., LeBoeuf R., Marquardt H., McCormick J., Müller L., Rivedal E., Sabbioni E., Tanaka N., Vasseur P., Yamasaki H. Cell Transformation assays as predictors of human carcinogenicity: the report and recommendations of ECVAM Workshop 39. ATLA - Altern. Lab. Anim. 1999;27(5):745–767. doi: 10.1177/026119299902700505. [DOI] [PubMed] [Google Scholar]
- Daniel F.B., Schut H.A.J., Sandwisch D.W., Schenck K.M., Hoffmann C.O., Patrick J.R., Stoner G.D. Interspecies comparisons of benzo(a)pyrene metabolism and DNA-adduct formation in cultured human and animal bladder and tracheobronchial tissues. Cancer Res. 1983;43(10):4723–4729. [PubMed] [Google Scholar]
- Deutsch-Wenzel R.P., Brune H., Grimmer G., Dettbarn G., Misfeld J. Experimental studies in rat lungs on the carcinogenicity and dose-response relationships of eight frequently occurring environmental polycyclic aromatic hydrocarbons. JNCI - J. Natl. Cancer I. 1983;71(3):539–544. [PubMed] [Google Scholar]
- Dunkel V.C., Pienta R.J., Sivak A., Traul K.A. Comparative neoplastic transformation responses of Balb/3T3 cells, Syrian hamster embryo cells, and rauscher murine leukemia virus-infected Fischer 344 Rat embryo cells to chemical carcinogens. JNCI - J. Natl. Cancer I. 1981;67(6):1303–1315. doi: 10.1093/jnci/67.6.1303. [DOI] [PubMed] [Google Scholar]
- EURL ECVAM – European Union (2004). Tracking System for Alternative methods towards Regulatory acceptance: In vitro BALB/c 3T3 Cell Transformation Assay. EU Reference Laboratory for Alternatives to Animal Testing (EURL ECVAM), Protocol 137, accessed on May 03, 2022. https://tsar.jrc.ec.europa.eu/test-method/tm2004-07.
- Escher B.I., Glauch L., König M., Mayer P., Schlichting R. Baseline toxicity and volatility cutoff in reporter gene assays used for high-throughput screening. Chem. Res. Toxicol. 2019;32(8):1646–1655. doi: 10.1021/acs.chemrestox.9b00182. [DOI] [PubMed] [Google Scholar]
- EURL ECVAM – European Union (2005). Tracking System for Alternative methods towards Regulatory acceptance: Syrian hamster embryo cell transformation assay at pH 6.7. EU Reference Laboratory for Alternatives to Animal Testing (EURL ECVAM), Protocol 136, accessed on May 03, 2022. https://tsar.jrc.ec.europa.eu/test-method/tm2005-02.
- Fischer F.C., Henneberger L., König M., Bittermann K., Linden L., Goss K.-U., Escher B.I. Modeling exposure in the Tox21 in vitro bioassays. Chem. Res. Toxicol. 2017;30(5):1197–1208. doi: 10.1021/acs.chemrestox.7b00023. [DOI] [PubMed] [Google Scholar]
- Fischer F.C., Cirpka O.A., Goss K.-U., Henneberger L., Escher B.I. Application of experimental polystyrene partition constants and diffusion coefficients to predict the sorption of neutral organic chemicals to multiwell plates in in vivo and in vitro bioassays. Environ. Sci. Technol. 2018;52(22):13511–13522. doi: 10.1021/acs.est.8b04246. [DOI] [PubMed] [Google Scholar]
- Genies C., Maître A., Lefèbvre E., Jullien A., Chopard-Lallier M., Douki T. The extreme variety of genotoxic response to benzo[a]pyrene in three different human cell lines from three different organs. PLoS ONE. 2013;8(11):e78356. doi: 10.1371/journal.pone.0078356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg G.L., Atherholt T.B. DNA adduct formation in mouse tissues in relation to serum levels of benzo(a)pyrene-diol-epoxide after injection of benzo(a)pyrene or the diol-epoxide. Cancer Res. 1990;50(4):1189–1194. [PubMed] [Google Scholar]
- Godschalk R.W., Moonen E.J., Schilderman P.A., Broekmans W.M., Kleinjans J.C., Van Schooten F.J. Exposure-route-dependent DNA adduct formation by polycyclic aromatic hydrocarbons. Carcinogenesis. 2000;21(1):87–92. [PubMed] [Google Scholar]
- Grech A., Brochot C., Dorne J.-L., Quignot N., Bois F.Y., Beaudouin R. Toxicokinetic models and related tools in environmental risk assessment of chemicals. Sc. Total Environ. 2017;578:1–15. doi: 10.1016/j.scitotenv.2016.10.146. [DOI] [PubMed] [Google Scholar]
- Grimmer, G., Naujack, K., & Dettbarn, G. (1987). Contribution to the study into the causes of exogenical carcinoma of the urinary bladder-profile analysis of aromatic amines at the place of work. Beitrag zur Ursachenforschung exogen bedingter Blasencarcinome-Profilanalyse aromatischer Amine am Arbeitsplatz. Schriftenreihe der Bundesanstalt fuer Arbeitsschutz. Forschung. no. Fb 511. Wirtschaftsverlag, Bremerhaven, Germany.
- Heredia-Ortiz R., Bouchard M., Marie-Desvergne C., Viau C., Maître A. Modeling of the internal kinetics of benzo(a)pyrene and 3-hydroxybenzo(a)pyrene biomarker from rat data. Toxicol. Sci. 2011;122(2):275–287. doi: 10.1093/toxsci/kfr135. [DOI] [PubMed] [Google Scholar]
- Hoffmann S., Hothorn L.A., Edler L., Kleensang A., Suzuki M., Phrakonkham P., Gerhard D. Two new approaches to improve the analysis of BALB/c 3T3 cell transformation assay data. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2012;744(1):36–41. doi: 10.1016/j.mrgentox.2011.12.002. [DOI] [PubMed] [Google Scholar]
- IARC - International Agency for Research on Cancer (2010). Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, 92, 1-853, Lyon, France. https://publications.iarc.fr/110. [PMC free article] [PubMed]
- Jacobs M.N., Colacci A., Corvi R., Vaccari M., Aguila M.C., Corvaro M., Delrue N., Desaulniers D., Ertych N., Jacobs A., Luijten M., Madia F., Nishikawa A., Ogawa K., Ohmori K., Paparella M., Sharma A.K., Vasseur P. Chemical carcinogen safety testing: OECD expert group international consensus on the development of an integrated approach for the testing and assessment of chemical non-genotoxic carcinogens. Arch. Toxicol. 2020;94(8):2899–2923. doi: 10.1007/s00204-020-02784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamei M., Marciniak S., Feng K., Barnett A., Tucker G., Rostami-Hodjegan A. The Simcyp® Population-based ADME Simulator. Expert Opin. Drug Metab. Toxicol. 2009;5(2):211–223. doi: 10.1517/17425250802691074. [DOI] [PubMed] [Google Scholar]
- Kulkarni M.S., Calloway K., Irigaray M.F., Kaufman D.G. Species Differences in the Formation of Benzo(a)pyrene-DNA Adducts in Rodent and Human Endometrium. Cancer Res. 1986;46(6):2888–2891. [PubMed] [Google Scholar]
- LeBoeuf R.A., Kerckaert G.A., Aardema M.J., Gibson D.P., Brauninger R., Isfort R.J. The pH 6.7 Syrian hamster embryo cell transformation assay for assessing the carcinogenic potential of chemicals. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 1996;356(1):85–127. doi: 10.1016/0027-5107(95)00199-9. [DOI] [PubMed] [Google Scholar]
- Leclerc D., Staats Pires A.C., Guillemin G.J., Gilot D. Detrimental activation of AhR pathway in cancer: an overview of therapeutic strategies. Curr. Opin. Immunol. 2021;70:15–26. doi: 10.1016/j.coi.2020.12.003. [DOI] [PubMed] [Google Scholar]
- Li H., Zhang M., Vervoort J., Rietjens I.M.C.M., van Ravenzwaay B., Louisse J. Use of physiologically based kinetic modeling-facilitated reverse dosimetry of in vitro toxicity data for prediction of in vivo developmental toxicity of tebuconazole in rats. Toxicol. Lett. 2017;266:85–93. doi: 10.1016/j.toxlet.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Lilienblum W., Dekant W., Foth H., Gebel T., Hengstler J.G., Kahl R., Kramer P.J., Schweinfurth H., Wollin K.M. Alternative methods to safety studies in experimental animals: role in the risk assessment of chemicals under the new European Chemicals Legislation (REACH) Arch. Toxicol. 2008;82(4):211–236. doi: 10.1007/s00204-008-0279-9. [DOI] [PubMed] [Google Scholar]
- Louisse J., de Jong E., van de Sandt J.J.M., Blaauboer B.J., Woutersen R.A., Piersma A.H., Rietjens I.M.C.M., Verwei M. The use of in vitro toxicity data and physiologically based kinetic modeling to predict dose-response curves for in vivo developmental toxicity of glycol ethers in rat and man. Toxicol. Sci. 2010;118(2):470–484. doi: 10.1093/toxsci/kfq270. [DOI] [PubMed] [Google Scholar]
- Madureira D.J., Weiss F.T., Van Midwoud P., Helbling D.E., Sturla S.J., Schirmer K. Systems toxicology approach to understand the kinetics of Benzo(a)pyrene uptake, biotransformation, and DNA adduct formation in a liver cell model. Chem. Res. Toxicol. 2014;27(3):443–453. doi: 10.1021/tx400446q. [DOI] [PubMed] [Google Scholar]
- Maire M.-A., Pant K., Phrakonkham P., Poth A., Schwind K.-R., Rast C., Bruce S.W., Sly J.E., Bohnenberger S., Kunkelmann T., Schulz M., Vasseur P. Recommended protocol for the Syrian hamster embryo (SHE) cell transformation assay. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2012;744(1):76–81. doi: 10.1016/j.mrgentox.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Maire M.-A., Pant K., Poth A., Schwind K.-R., Rast C., Bruce S.W., Sly J.E., Kunz-Bohnenberger S., Kunkelmann T., Engelhardt G., Schulz M., Vasseur P. Prevalidation study of the Syrian hamster embryo (SHE) cell transformation assay at pH 7.0 for assessment of carcinogenic potential of chemicals. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2012;744(1):64–75. doi: 10.1016/j.mrgentox.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Marie C., Maître A., Douki T., Gateau M., Tarantini A., Guiraud P., Favier A., Ravanat J.-L. Influence of the metabolic properties of human cells on the kinetic of formation of the major benzo[a]pyrene DNA adducts. J. Appl. Toxicol. 2008;28(5):579–590. doi: 10.1002/jat.1306. [DOI] [PubMed] [Google Scholar]
- Marie-Desvergne C., Maître A., Bouchard M., Ravanat J.-L., Viau C. Evaluation of DNA adducts, DNA and RNA oxidative lesions, and 3-hydroxybenzo(a)pyrene as biomarkers of DNA damage in lung following intravenous injection of the parent compound in rats. Chem. Res. Toxicol. 2010;23(7):1207–1214. doi: 10.1021/tx100081p. [DOI] [PubMed] [Google Scholar]
- Moore C.J., Pruess-Schwartz D., Mauthe R.J., Gould M.N., Baird W.M. Interspecies differences in the major DNA adducts formed from benzo(a)pyrene but not 7,12-dimethylbenz(a)anthracene in rat and human mammary cell cultures. Cancer Res. 1987;47(16):4402–4406. [PubMed] [Google Scholar]
- Motwani H.V., Westberg E., Lindh C., Abramsson-Zetterberg L., Törnqvist M. Serum albumin adducts, DNA adducts and micronuclei frequency measured in benzo[a]pyrene-exposed mice for estimation of genotoxic potency. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2020;849 doi: 10.1016/j.mrgentox.2019.503127. [DOI] [PubMed] [Google Scholar]
- OECD. (2015). Guidance document on the in vitro syrian hamster embryo (SHE) cell transformation assay. OECD Series on Testing and Assessment, No. 214, OECD Publishing, Paris.
- OECD. (2014). The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins. OECD Series on Testing and Assessment, No. 168, OECD Publishing, Paris. 10.1787/9789264221444-en. [DOI]
- Pant K., Bruce S.W., Sly J.E., Kunkelmann T., Kunz-Bohnenberger S., Poth A., Engelhardt G., Schulz M., Schwind K.-R. Prevalidation study of the Syrian hamster embryo (SHE) cell transformation assay at pH 6.7 for assessment of carcinogenic potential of chemicals. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2012;744(1):54–63. doi: 10.1016/j.mrgentox.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Pearce R.G., Setzer R.W., Strope C.L., Wambaugh J.F., Sipes N.S. httk: R Package for High-Throughput Toxicokinetics. J. Stat. Softw. 2017;79(4):1–26. doi: 10.18637/jss.v079.i04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen K., Dipple A. Polycyclic aromatic hydrocarbons: chemistry of DNA adduct formation. J. Occup. Environ. Med. 1995;37(1):52–58. doi: 10.1097/00043764-199501000-00008. [DOI] [PubMed] [Google Scholar]
- Piberger A.L., Krüger C.T., Strauch B.M., Schneider B., Hartwig A. BPDE-induced genotoxicity: relationship between DNA adducts, mutagenicity in the in vitro PIG-A assay, and the transcriptional response to DNA damage in TK6 cells. Arch. Toxicol. 2018;92(1):541–551. doi: 10.1007/s00204-017-2003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienta R.J., Poiley J.A., Lebherz W.B., 3rd. Morphological transformation of early passage golden Syrian hamster embryo cells derived from cryopreserved primary cultures as a reliable in vitro bioassay for identifying diverse carcinogens. Int. J. Cancer. 1977;19(5):642–655. doi: 10.1002/ijc.2910190508. [DOI] [PubMed] [Google Scholar]
- Pott, F. (1973). Untersuchungen zur Kanzerogenität von polyzyklischen aromatischen Kohlenwasserstoffen im Tierexperiment. (Investigations on the carcinogenicity of polycyclic aromatic hydrocarbons in animal studies). Zentralbl. Bakteriol. Hyg. I Abt. Orig. B, 157, 34–43. [PubMed]
- Sakai A., Sato M. Improvement of carcinogen identification in BALB/3T3 cell transformation by application of a 2-stage method. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 1989;214(2):285–296. doi: 10.1016/0027-5107(89)90172-3. [DOI] [PubMed] [Google Scholar]
- Sayer J.M., Chadha A., Agarwal S.K., Yeh H.J., Yagi H., Jerina D.M. Covalent nucleoside adducts of benzo [a] pyrene-7,8-diol-9,10-epoxides: structural reinvestigation and characterization of a novel adenosine adduct on the ribose moiety. J. Org. Chem. 1991;56(1):20–29. [Google Scholar]
- Shah U.-K., Seager A.L., Fowler P., Doak S.H., Johnson G.E., Scott S.J., Scott A.D., Jenkins G.J.S. A comparison of the genotoxicity of benzo[a]pyrene in four cell lines with differing metabolic capacity. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2016;808:8–19. doi: 10.1016/j.mrgentox.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Shiizaki K., Kawanishi M., Yagi T. Dioxin suppresses benzo [a] pyrene-induced mutations and DNA adduct formation through cytochrome P450 1A1 induction and (±)-anti-benzo[a]pyrene-7,8-diol-9,10-epoxide inactivation in human hepatoma cells. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2013;750(1–2):77–85. doi: 10.1016/j.mrgentox.2012.09.008. [DOI] [PubMed] [Google Scholar]
- Shukla R., Liu T., Geacintov N.E., Loechler E.L. The major, N2-dG adduct of (+)-anti-B[a]PDE shows a dramatically different mutagenic specificity (predominantly, G –> A) in a 5'-CGT-3' sequence context. Biochemistry. 1997;36(33):10256–10261. doi: 10.1021/bi970541. [DOI] [PubMed] [Google Scholar]
- Slaga T.J., Bracken W.J., Gleason G., Levin W., Yagi H., Jerina D.M., Conney A.H. Marked differences in the skin tumor-initiating activities of the optical enantiomers of the diastereomeric benzo(a)pyrene-7,8-diol-9,10-epoxides. Cancer Res. 1979;39(1):67–71. [PubMed] [Google Scholar]
- Smets L.A. Cell transformation as a model for tumor induction and neoplastic growth. Biochimica et Biophysica Acta (BBA) - Rev Cancer. 1980;605(1):93–111. doi: 10.1016/0304-419X(80)90022-0. [DOI] [PubMed] [Google Scholar]
- Smolarek T.A., Morgan S.L., Moynihan C.G., Lee H., Harvey R.G., Baird W.M. Metabolism and DNA adduct formation of benzo[a]pyrene and 7,12-dimethylbenz[a]anthracene in fish cell lines in culture. Carcinogenesis. 1987;8(10):1501–1509. doi: 10.1093/carcin/8.10.1501. [DOI] [PubMed] [Google Scholar]
- Stockinger B., Shah K., Wincent E. AHR in the intestinal microenvironment: safeguarding barrier function. Nat. Rev. Gastroenterol. Hepatol. 2021;18:559–570. doi: 10.1038/s41575-021-00430-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straif K., Baan R., Grosse Y., Secretan B., El Ghissassi F., Cogliano V., WHO International Agency for Research on Cancer Monograph Working Group Carcinogenicity of polycyclic aromatic hydrocarbons. Lancet Oncol. 2005;6(12):931–932. doi: 10.1016/s1470-2045(05)70458-7. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Bohnenberger S., Kunkelmann T., Munaro B., Ponti J., Poth A., Sabbioni E., Sakai A., Salovaara S., Sasaki K., Thomas B.C., Umeda M. Prevalidation study of the BALB/c 3T3 cell transformation assay for assessment of carcinogenic potential of chemicals. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2012;744(1):20–29. doi: 10.1016/j.mrgentox.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Tapiainen T., Järvinen K., Pääkkö P., Bjelogrlic N., Vähäkangas K. TPA decreases the p53 response to benzo[a]pyrene-DNA adducts in vivo in mouse skin. Carcinogenesis. 1996;17(6):1377–1380. doi: 10.1093/carcin/17.6.1377. [DOI] [PubMed] [Google Scholar]
- Topinka J., Marvanová S., Vondráček J., Sevastyanova O., Nováková Z., Krčmář P., Pěnčíková K., Machala M. DNA adducts formation and induction of apoptosis in rat liver epithelial ‘stem-like’cells exposed to carcinogenic polycyclic aromatic hydrocarbons. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 2008;638(1–2):122–132. doi: 10.1016/j.mrfmmm.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Topping D.C., Martin D.H., Nettesheim P. Determination of cocarcinogenic activity of benzo [e] pyrene for respiratory tract mucosa. Cancer Lett. 1981;11(4):315–321. doi: 10.1016/0304-3835(81)90097-5. [DOI] [PubMed] [Google Scholar]
- Trainor G.L. The importance of plasma protein binding in drug discovery. Expert Opin. Drug Discov. 2007;2(1):51–64. doi: 10.1517/17460441.2.1.51. [DOI] [PubMed] [Google Scholar]
- Turley A.E., Isaacs K.K., Wetmore B.A., Karmaus A.L., Embry M.R., Krishan M. Incorporating new approach methodologies in toxicity testing and exposure assessment for tiered risk assessment using the RISK21 approach: Case studies on food contact chemicals. Food Chem. Toxicol. 2019;134 doi: 10.1016/j.fct.2019.110819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, N., Endo, S., Brown, T. N., Watanabe, N., Bronner, G., Abraham, M. H., & Goss, K. U. (2017). UFZ-LSER database v 3.2 [Internet]. http://www.ufz.de/lserd.
- Villeneuve D.L., Crump D., Garcia-Reyero N., Hecker M., Hutchinson T.H., LaLone C.A., Landesmann B., Lettieri T., Munn S., Nepelska M., Ottinger M.A., Vergauwen L., Whelan M. Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol. Sci. 2014;142(2):312–320. doi: 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel-Hartung R., Brune H., Grimmer G., Germanise P., Timm J., Wosniok W. Evaluation of the carcinogenic potency of 4 environmental polycyclic aromatic compounds following intrapulmonary application in rats. Exp. Pathol. 1990;40(4):221–227. doi: 10.1016/s0232-1513(11)80302-6. [DOI] [PubMed] [Google Scholar]
- Westberg E.A.C., Singh R., Hedebrant U., Koukouves G., Souliotis V.L., Farmer P.B., Segerbäck D., Kyrtopoulos S., Törnqvist M.Å. Adduct levels from benzo[a]pyrenediol epoxide: Relative formation to histidine in serum albumin and to deoxyguanosine in DNA in vitro and in vivo in mice measured by LC/MS–MS methods. Toxicol. Lett. 2015;232(1):28–36. doi: 10.1016/j.toxlet.2014.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this published article. Data, associated metadata, and calculation tools are also available from the corresponding author.