Abstract
While the link between serum proteins and cancer has been studied in an effort to enable early-stage cancer detection, factors that might perturb this link has been poorly understood. To ask this question, we performed serum protein profiling on a prospective cohort of 601 individuals with or without lung, pancreatic, or colorectal cancers and identified ten distinct serum protein signatures with distinct link to the patient metadata. Importantly, we discovered that a positive history of alcohol consumption is a major factor that diminishes the sensitivity of serum protein-mediated liquid biopsy in early-stage malignancies, resulting in a 44% decline in the sensitivity of detecting American Joint Committee on Cancer (AJCC) stage I malignancies. Our data provide evidence that patient lifestyle can affect the sensitivity of liquid biopsy and suggest the potential need for abstinence from alcohol before measurement during serum protein-based cancer screening.
Keywords: Cancer, Lung cancer, Pancreatic cancer, Colorectal cancer, Alcohol, Biomarker, Serum protein
Cancers; Lung cancer; Pancreatic cancer; Colorectal cancer; Alcohol; Biomarkers; Seroproteins.
1. Introduction
The detection of cancer in its early stage is considered one of the holy grails of patient care in the fight against the disorder. One of the reasons behind this popular notion is because earlier stages of malignancies generally arise from a few driver mutations and are comprised of a less heterogeneous population of cancer cells, which leads to a better prognosis of patients after treatment [1]. Previous studies have thus investigated changes that specifically occur at the earliest stages of neoplastic progression and identified inflammation [2, 3, 4] and angiogenesis [5, 6] as notable examples. Inflammation is considered to contribute to neoplastic progression via creating a tumor-promoting microenvironment [4, 6], which often entails a constitutively active angiogenic switch in the lesion. This change typically commences during the premalignant stage of cancer [5], and is often probed to study early-stage cancer detection.
At the intersection between angiogenesis and inflammation lies serum proteins comprised of diverse growth factors, cytokines, and soluble forms of surface receptors. Hence, serum proteins have long been studied as potential diagnostic tools for cancer [7, 8, 9, 10], and increasing number of serum protein-based liquid biopsies have entered large-scale clinical trials [11, 12, 13]. Despite an extensive effort to establish a link between serum proteins and the likelihood of neoplastic progression in individuals, factors that can affect or even disrupt this link have been poorly understood.
Herein, we performed an extensive serum protein profiling on a prospective cohort that involved 601 individuals with or without lung, pancreatic, or colorectal cancers and identified positive history of alcohol consumption as a key factor diminishing the sensitivity of serum protein-based blood tests in early-stage cancers. The perturbation was driven by a positive correlation between serum protein concentrations of cancer patients who have consumed alcohol in the past and those of healthy individuals without cancer, which led to the reversing of the alterations induced by cancer occurrence alone. In contrast, the consideration of the individual's history of alcohol consumption enhanced the sensitivity of these blood tests, providing the evidence that patient lifestyle can strongly affect cancer detection by liquid biopsy. These data suggest the potential need for abstinence from alcohol in serum protein-based cancer screening.
2. Results
2.1. Identification of serum protein signatures in cancer patients
Our study involved 601 individuals with or without three of the most lethal types of cancers that account for 39% of all cancer mortalities: lung, pancreas, and colorectum [14]. Fifteen out of 420 cancer patients were later determined as having a non-primary tumor, of which protein concentration profiles we excluded for further investigations (Table S1). Serum protein markers previously described to be associated with at least one of the three cancer types studied were identified based on an exhaustive literature search (Table S2). Concentrations of these forty-eight protein biomarkers were determined by a multiplex immunoassay [15], which resulted in a total of 28,848 measurements. Clinical metadata of participants, which included sex, age, body mass index (BMI), history of tobacco smoking, history of alcohol consumption, prior history of malignancy, family history of malignancy, diabetes, and hypertension, were prospectively collected for analysis. Raw data are presented in Table S3.
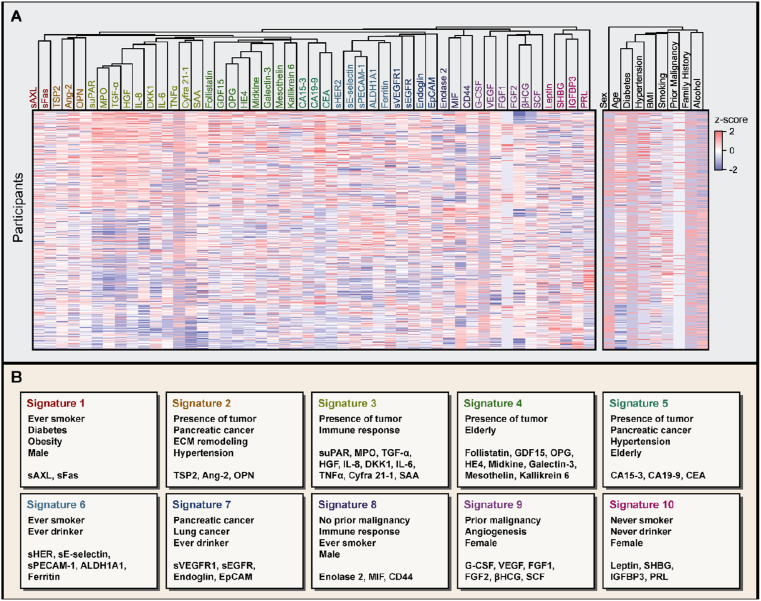
To establish serum protein signatures in cancer patients, we performed cohort-wide hierarchical clustering of serum proteins (Materials and methods). Euclidean distance-based hierarchical clustering revealed ten prominent serum protein signatures (signatures 1–10; Figure 1A). Each signature had varying number of member proteins, which ranged from the minimum of two to the maximum of ten. Complete list of member proteins for each signature and its signature-specific attributes are described in Figure 1B.
Figure 1.
Multicancer cohort reveals ten prominent serum protein signatures. (A) Heatmap of serum protein concentrations and individual metadata in the cohort. Each column has been hierarchically clustered using the Euclidean distance. Ten co-regulated signatures of serum proteins are color-coded. Each row represents 586 participants who met the inclusion criteria. (B) Signatures and their signature-specific attributes. Members of each signature are presented in panels. SeroHet signatures encompassed expansive aspects of cancer biology, including stage, cancer type, immune response, angiogenesis, and extracellular matrix remodeling.
Signatures covered expansive aspects of cancer biology. Signatures 3 and 8 were strongly associated with immune response, with cytokines being the predominant member of these signatures. IL-6, IL-8, and TNFα, which are well-known proinflammatory cytokines that are often co-regulated [16], all belonged to signature 3. Analogously, both MIF and its co-receptor CD44, which are often overexpressed under inflammatory stimuli [17], belonged to signature 8. Signature 9 was highly enriched with growth factors associated with angiogenesis, including VEGF and FGFs [18], while signature 2 was enriched with serum proteins associated with extracellular matrix (ECM) remodeling, including angiopoietin-2 (Ang-2), osteopontin (OPN), and thrombospondin 2 (TSP2) [19, 20, 21]. Overall, these links to diverse cancer-related processes corroborate the biological relevance of classifying co-regulated serum biomarkers into a set of serum protein signatures.
2.2. Patient metadata strongly regulates serum protein signatures
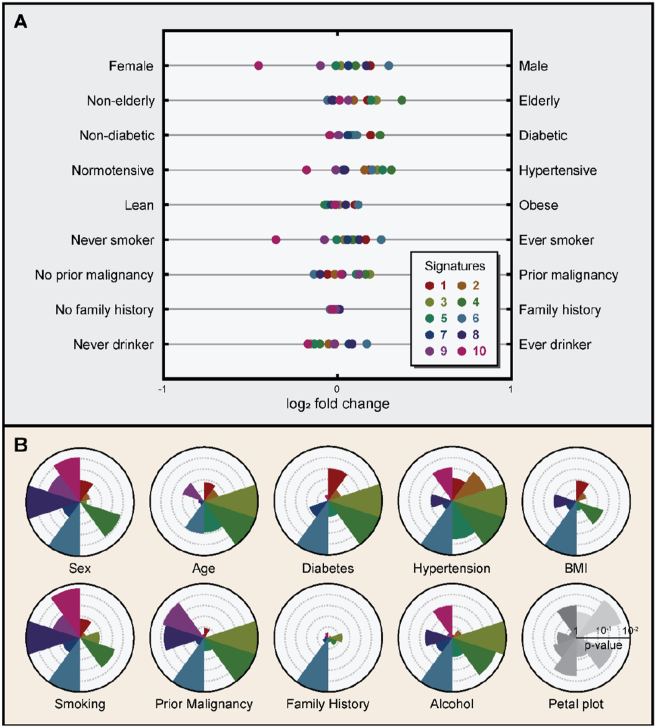
To further understand the biological relevance of these signatures, we performed metadata analysis on nine clinical parameters prospectively collected (Materials and methods). For each parameter, binarization was performed to enable statistical comparison between two distinct groups of individuals with different clinical characteristics: female versus male (sex), non-elderly versus elderly (age), non-diabetic versus diabetic (diabetes), normotensive versus hypertensive (hypertension), lean versus obese (BMI), never smoker versus ever smoker (tobacco smoking), no prior malignancy versus prior malignancy (prior history of malignancy), no family history versus family history (family history of malignancy), and never drinker versus ever drinker (alcohol consumption). Criteria for the binarization are further detailed in Materials and methods. In accordance with previous literatures [22, 23, 24, 25, 26, 27, 28], sex, tobacco smoking, and hypertension significantly contributed to interpatient serum protein heterogeneity in participants, as shown by the signature-wise fold changes (Figure 2A) and their corresponding p-values (Figure 2B). By contrast, family history had the least contribution to interpatient serum protein heterogeneity (Figure S1), with signature 6 being the only signature that was significantly associated (p-value = 0.0108). Signature 6 was associated with all metadata studied except for the age of participants and was strongly overrepresented in ever-smokers (p-value < 0.001), providing a potential explanation for the observed negative correlation between signature 6 expression and family history of malignancy (Figure 2A). Individuals without the family history of malignancy may require stronger association with potent carcinogenic factors, such as tobacco smoking, to confer carcinogenesis. For individuals without the family history of malignancy, tobacco smoking was a stronger predictor of a potential neoplastic progression compared to age.
Figure 2.
Association of signatures to patient metadata. (A) Forest plot showing the log2 fold changes for each signature. Signatures are color-coded and represented as a single dot. Each row represents different types of patient metadata studied. (B) Petal plot showing the −log10 p-values for each signature. The scale of the axis is indicated in the bottom right. Each signature is presented in a clockwise order, beginning from the top. Signatures are color-coded. The outermost circle represents the threshold for the statistical significance (p-value < 0.01).
Many previously reported associations between serum proteins and patient metadata have been reproduced, substantiating the validity of our observations. For instance, serum protein markers previously known to correlate with tobacco smoking, including aldehyde dehydrogenase 1 family member A1 (ALDH1A1) [26, 28] and ferritin [27], were rediscovered by the signature analysis in this cohort. Furthermore, lung cancer patients exhibited 50% increased likelihood of positive history of tobacco smoking compared to pancreatic or colorectal cancer patients (p-value < 0.001, Fisher's exact test). In general, never-smokers overexpressed signature 10, while ever-smokers overexpressed signatures 6 and 8. As previously described, sex also had a strong influence on serum protein concentrations, with female participants being enriched with signatures 9 and 10, while male participants were enriched with signatures 1, 6, and 8. Prolactin (PRL) and sex hormone binding globulin (SHBG), which are two well-known sex hormone-associated serum proteins typically overexpressed in female individuals [22, 25], could also be rediscovered in our cohort. Alcohol consumption also had a substantial influence on the serum protein signatures of individuals, with signature 6 being overrepresented in ever-drinkers and signatures 3, 4, and 10 being overrepresented in never-drinkers. The observed positive correlation between alcohol consumption and serum ferritin level corroborates previous reports [29, 30]. ALDH1A1, an aldehyde dehydrogenase known to function in ethanol detoxification [31, 32], also positively correlated with chronic alcohol consumption, corroborating our analysis.
2.3. Serum protein signatures 2–5 are diagnostic of early-stage cancers
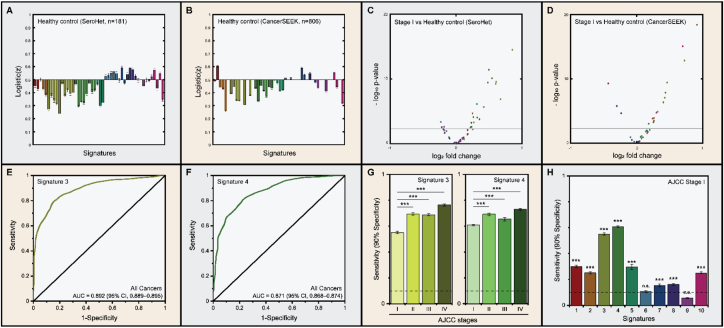
To understand the role of each signature in serum protein-based cancer detection, we compared signatures of the American Joint Committee on Cancer (AJCC) stage I cancer patients to those of healthy individuals without cancer. In order to confirm that the observed serum protein signatures are independent of cohorts, we also analyzed signatures in the CancerSEEK cohort [11], which also included patients with lung, pancreatic, and colorectal cancers. Strong positive correlation was observed between serum protein signatures from individuals with lung, pancreatic, or colorectal cancers in the two cohorts (Spearman's ρ = 0.716, p-value < 0.001; Figures S2 and 3), indicating that the observed differences in serum protein signatures are reproducible. In both cohorts, signatures 2–5 were overexpressed in cancer patients compared to healthy individuals without cancer (Figure 3A, B). Considering that signature 2 is associated with extracellular matrix remodeling and that signature 3 is associated with inflammatory immune response, the observed serum protein difference between cancer patients and healthy individuals are highly predicted. Volcano plot between healthy individuals and clinical stage I cancer patients in both cohorts revealed more serum proteins upregulated by the presence of tumor compared to vice versa (Figure 3C, D), suggesting the potential role of these serum protein signatures in early-stage cancer detection. Indeed, all serum protein included in the CancerSEEK detection algorithm, except for PRL, belonged to signatures 2–5 [11]. Strong modulation of serum level of PRL by sex and tobacco smoking [22, 23, 24] prevented its membership to these signatures, despite its consistent overexpression in cancer patients from both cohorts. Since PRL has the least similarity to other members of signature 10, as shown by the Euclidean distance between each serum protein (Figure 1A), further expansion of serum biomarker panels may reveal novel cancer-associated serum protein signature containing PRL.
Figure 3.
Early-stage cancer detection by signatures. (A) Logistic(z) values of healthy individuals compared to cancer patients in the current study. (B) Logistic(z) values of healthy individuals compared to cancer patients in CancerSEEK cohort. Signatures 2–5 were underrepresented in healthy individuals compared to cancer patients in both cohorts. (C) Volcano plot between clinical stage I cancer patients and healthy individuals without cancer in SeroHet cohort. The horizontal axis indicates the log2 fold change in the normalized protein concentration of AJCC stage I cancers compared to healthy individuals. (D) Volcano plot between clinical stage I cancer patients and healthy individuals without cancer in CancerSEEK cohort. The horizontal axis indicates the log2 fold change in the normalized protein concentration of AJCC stage I cancers compared to healthy individuals. Horizontal line represents the Bonferroni-corrected threshold for p-value at a target family-wise error rate of 0.05. Upregulation of signatures 3 and 4 in clinical stage I cancer patients are evident in both cohorts. (E) ROC curves on the detection of cancer using SeroHet signature 3. AUC value of the curve was 0.892 (95% CI, 0.889–0.895). (F) ROC curves on the detection of cancer using signature 4. AUC value of the curve was 0.871 (95% CI, 0.868–0.874). (G) Average sensitivity at 90% specificity of detecting cancer with each AJCC stage using signatures 3 and 4. The sensitivity of the classifier positively correlated with cancer progression in both cases. (H) Average sensitivity at 90% specificity of detecting AJCC stage I cancer using each signature. SeroHet signatures 3 and 4 were highly diagnostic of cancer at AJCC stage I. Error bar, 95% CI.
Assessment of the potential of signatures in early-stage cancer detection was done by a logistic regression (Materials and methods), as has been previously described [11, 12]. To ensure the reproducibility of our observations, we performed 1,000 iterations of 10-fold cross-validation for each signature (Figure S4). Notably, signatures 3 and 4, which had the greatest number of member proteins, were the most sensitive in detecting cancer, regardless of the cancer type (Figure S5). The average area under the receiver operating characteristic (ROC) curve, hereafter abbreviated as AUC, was 0.892 (95% confidence interval (CI), 0.889–0.895; Figure 3E) for signature 3 and 0.871 (95% CI, 0.868–0.874; Figure 3F) for signature 4. The detection performance increased with cancer progression in both serum protein signatures (Figure 3G), which is a phenomenon that has been consistently reported in serum protein-based liquid biopsies [11, 12, 13, 33, 34]. The average sensitivity of clinical stage I cancer detection at 90% specificity was 55% (95% CI, 54–56%) for signature 3 and 61% (95% CI, 60–61%) for signature 4, which were highest among all serum protein signatures studied (Figure 3H). Metadata previously known to positively correlate with cancer occurrence, which included the history of prior malignancy, aging [14], hypertension [35, 36, 37], and diabetes [38], were also enriched with signatures 3 and 4. By contrast, signatures 6 and 9 exhibited poor performance in detecting cancer, suggesting that angiogenic growth factors and soluble forms of cell adhesion molecules are relatively poor diagnostic markers for cancer. The observed poor association between signature 6 and the presence of tumor was despite signature 6 being the most upregulated signature in ever-smokers (Figure 2A).
2.4. Chronic alcohol consumption perturbs the performance of liquid biopsy
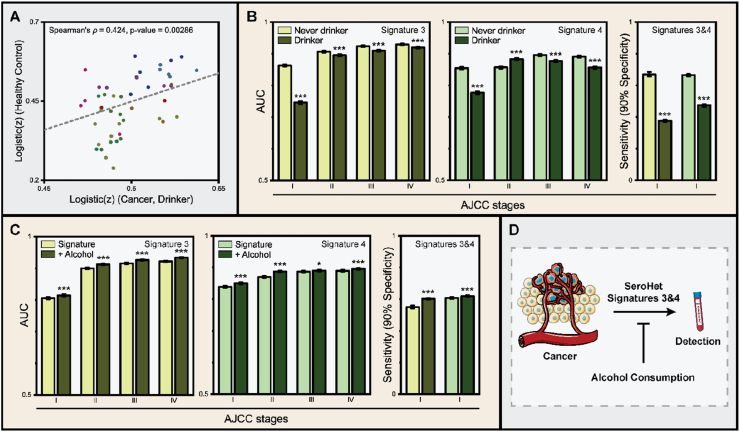
Strikingly, participants with the history of alcohol consumption showed extensive downregulation of signatures 3 and 4 (Figure 2A), which may have detrimental effects on liquid biopsy-based cancer detection. To assess whether alcohol consumption perturbs cancer detection, we first performed correlation analysis of serum protein concentrations of cancer patients with the history of chronic alcohol consumption and healthy individuals without cancer. Indeed, we observed a positive correlation between the two groups of participants (Spearman's ρ = 0.424, p-value = 0.00286), indicating that cancer patients who consistently consumed alcohol phenocopy healthy individuals, at least in these cancer-associated serum biomarkers studied. Intrigued by this result, we re-analyzed ROC curves for signatures 3 and 4 and dissected them based on the patient's history of alcohol consumption. To our surprise, consistent underperformance of cancer detection was observed in cancer patients with the history of alcohol consumption (Figure 4B). Alcohol consumption had the strongest detrimental effect in early-stage cancer detection, as the AUC for signatures 3 and 4 were decreased by 14% and 9% in clinical stage I cancer patients, respectively, upon chronic alcohol consumption. In terms of the average sensitivity at 90% specificity, signature 3 exhibited a striking 44% decline (p-value < 0.001), while signature 4 exhibited 29% decline upon chronic alcohol uptake (p-value < 0.001).
Figure 4.
Alcohol consumption perturbs cancer detection by liquid biopsy. (A) Correlation between serum protein profiles of healthy individuals without cancer and cancer patients with the history of chronic alcohol intake. Spearman's ρ was 0.424 (p-value = 0.00286), indicating that ever-drinker with cancer phenocopy healthy individuals without cancer. (B) Performance of cancer detection using signatures 3 and 4 by the history of alcohol consumption. Individuals with positive history of chronic alcohol intake underperformed in these blood tests. (C) Performance of cancer detection with and without the binary parameter on the history of chronic alcohol consumption. The inclusion of the parameter enhanced the performance of blood tests in all stages. (D) Schematic diagram illustrating the effect of chronic alcohol consumption in liquid biopsy-based cancer detection. Error bar, 95% CI.
Based on this observation, we wondered whether the inclusion of the history of alcohol consumption as a novel binary parameter could enhance the performance of cancer detection by liquid biopsy. To test this hypothesis, we performed 1,000 iterations of 10-fold cross-validation in the presence of the parameter and compared its performances to those without (Materials and methods). The performance of liquid biopsies was significantly enhanced with the inclusion of the new parameter (Figure 4C), regardless of the clinical stage of the malignancy. The average sensitivity at 90% specificity was enhanced by 10% in signature 3 (p-value < 0.001) and by 2% in signature 4 (p-value < 0.001). Overall, our data consistently indicate that chronic alcohol consumption can extensively perturb liquid biopsy-based early-stage cancer detection by forcing the serum protein concentration profiles of ever-drinker cancer patients to mimic those of healthy individuals without cancer (Figure 4D). To the best of our knowledge, this is the first direct evidence that patient lifestyle can have an extensive influence on the performance of cancer detection by liquid biopsy [39, 40] and necessitates further analyses of serum protein signatures in larger cohorts.
3. Discussion
Herein, we presented a comprehensive analysis of serum protein signatures in a prospective cohort that involved 601 participants with or without three of the most lethal cancers (lung, pancreas, and colorectum). We identified ten distinct serum protein signatures in cancer patients, which covered expansive aspects of cancer biology, including sex, age, obesity, tobacco smoking, alcohol consumption, family history, immune response, angiogenesis, and extracellular matrix remodeling (Figure 1B and Figure 2A, B).
The biological relevance of these serum protein signatures was studied by a comprehensive metadata analysis (Figure 2A, B). Sex and tobacco smoking had the most influence on serum protein signatures, while family history had the least. Sex- and smoking-dependent variations in these signatures were in consistent with previous reports [22, 25, 26, 27, 28]. Although our serum protein signatures only pertained to 48 serum proteins studied in this cohort, investigators can also assess the membership of their novel serum protein of interest in each signature using the MATLAB-based SeroHet software (https://github.com/hanjunlee21/SeroHet). As signatures 2–5 were highly enriched in cancer patients with clinical stage as early as stage I (Figure 3A–D), further expansion of these signatures will be valuable in achieving liquid biopsy-based cancer screening in clinics.
Our comprehensive analysis on cancer-associated serum protein signatures also provides investigators with resource to efficiently design high-performance liquid biopsy panels. Due to the scarcity of information on how serum proteins are regulated in cancer patients, rational design of serum protein panels has long been a formidable task for cancer biologists [9, 41]. Since we have identified multiple distinct signatures that are highly diagnostic of early-stage cancers, investigators may efficiently improve the performances of pre-existing blood tests by adding serum proteins that belong to signatures that are underrepresented in the previous liquid biopsy panel. In this sense, our study lays a practical foundation for signature-based rational design of high-performance liquid biopsies.
Importantly, based on an extensive metadata analysis, we discovered that the detection of cancer in patients with the history of chronic alcohol consumption is much harder compared to patients without the history of chronic alcohol consumption, as the serum protein concentration profiles of the former strongly phenocopy those of healthy individuals without cancer. The observed decrease in the expression of signatures 3 and 4 upon chronic alcohol intake (Figure 2A) was specific to cancer patients, as alcohol consumption exhibited less pronounced effects on the expression of these signatures in healthy individuals without cancer (Figures S6, S7). Downregulation of proinflammatory cytokines, such as IL-8 and TNFα, upon chronic alcohol consumption has been previously demonstrated [42]. By contrast, signature 6 showed an identical trend in both healthy individuals and cancer patients, with chronic alcohol consumption being positively correlated with the overexpression of signature 6 in both groups of participants (Figure 2A, Figure S6). This is in agreement with previous reports, which demonstrated upregulation of ferritin and ALDH1A1 upon chronic alcohol consumption [26, 27, 28]. Overall, our data indicate that there is both universal and cancer-specific regulation of serum protein signatures by chronic alcohol uptake, of which combined effects result in high resemblance of serum protein signatures of cancer patients with the history of alcohol consumption to those of healthy individuals without cancer. Examination of the history of chronic alcohol uptake enhanced the sensitivity of liquid biopsy in all stages, providing the direct evidence that patient lifestyle can affect the performance of blood tests. However, the inclusion of the new binary parameter on the history of alcohol consumption did not completely abolish the inequality in the performance of liquid biopsies, suggesting the need to develop better tools to identify cancers in ever-drinkers.
One potential approach is to implement abstinence from alcohol before each blood test. Acute alcohol intake is known to impair the release of proinflammatory cytokines, such as IL-6 and TNFα, in response to inflammatory stimuli [43], of which effect is known to persist days after administration. Since many liquid biopsy panels rely on the inflammatory response to the tumor, acute alcohol intake can have detrimental effects on the detection of cancer by these blood tests. However, it remains to be studied whether acute alcohol intake can indeed impair the performance of serum protein-based liquid biopsies, as our study instead focused on chronic alcohol consumption in cancer patients. Since prebiopsy lifestyle intervention is a commonly utilized strategy to manage cancer in clinics [44], short-term alcohol intervention, such as a preanalytical week-long cessation [43], is a viable option to enhance the performance of liquid biopsies. Another approach is to identify novel serum protein signatures that are differentially regulated in ever-drinker cancer patients compared to healthy individuals. Inclusion of the members of these signatures may significantly enhance the performance of serum protein-based liquid biopsies in detecting early-stage cancers. However, whether application of these approaches can indeed boost the performance of serum protein-based liquid biopsies await further clinical trials.
3.1. Limitations of the study
One major downfall of the study is the relatively small size of the cohort that we have utilized to uncover serum protein signatures in cancer patients. Rather, our study focused on increasing the number of serum proteins and types of metadata to ensure comprehensive analysis on serum protein signatures. Overall, our study involved 28,848 independent measurements on forty-eight different serum protein markers, which to date is one of the widest serum protein panels utilized in multicancer cohorts [11]. Another major downfall is the inherent bias in the selection of healthy individuals. Other implications other than cancer, such as viral infection, can modulate serum protein signatures and potentially affect the performance of liquid biopsies, but its validation requires further investigations. Finally, there were correlation of clinical parameters that were inherent to the population. For instance, female individuals in our cohort were an order of magnitude less likely to have a history of tobacco smoking compared to male individuals (p-value < 0.001, Fisher's exact test), which was a trend that is inherent to the population that we studied. Nevertheless, this study provides a comprehensive view on the regulation of serum proteins in cancer patients and provides the first evidence that patient lifestyle can strongly perturb the practice of cancer screening. We envision these observations to be invaluable in deciphering the role of serum proteins in cancer, a second leading cause of death for humankind.
4. Materials and methods
4.1. Study design
The current study included 601 East Asian individuals with or without three of the most lethal types of cancer worldwide: lung, pancreas, and colorectum. To ensure the representability of the cohort, every patient was re-evaluated of the primary origin of the tumor. 15 out of 480 cancer patients were later determined as having a non-primary tumor, of which protein concentration profiles we excluded for further investigations. After the participants provided written informed consent, peripheral blood samples as well as patient demographic and clinical data were prospectively collected. The participants did not consume anything but water 8–10 h before the fasting blood test. Peripheral blood samples were sent to the technicians as cryopreserved vials containing 500 μL of serum. The biospecimen and data used in the study was provided by the Seoul National University Bundang Hospital Human Bioresource Center. Participants were diagnosed with hypertension if the systolic blood pressure was greater than or equal to 140 mmHg or the diastolic blood pressure was greater than or equal to 90 mmHg, following repeated examination (n ≥ 2). Participants were diagnosed with diabetes if either the fasting plasma glucose level was greater than or equal to 126 mg/dL, the casual glucose level was greater than or equal to 200 mg/dL, or the hemoglobin A1c level was greater than or equal to 6.5%. Obesity was diagnosed if the body mass index was greater than or equal to 25 kg/m3, per Asians of the Regional Office for the Western Pacific Region of the World Health Organization criteria [45]. Cancer staging was based on an 8th edition of the AJCC manual. Detailed information regarding the cohort is shown in Table S1. This study was approved by the Institutional Review Board of Seoul National University Bundang Hospital (X-2001-586-906) and compiled with the Health Insurance Portability and Accountability Act.
4.2. Quantification of serum proteins
The Luminex 200 system with xPONENT 4.3 (Luminex, Austin TX) was utilized to determine the concentration of each protein marker within the blood samples of individuals. Luminex bead-based multiplex immunoassays (Millipore, Bilerica NY) were split into a total of six panels: human angiogenesis/growth factor panel 1 (Millipore, Bilerica NY; catalog number, HANG2MAG-12K; angiopoietin-2, G-CSF, endoglin, FGF1, follistatin), human angiogenesis panel 2 (Millipore, Bilerica NY; catalog number, HAGP1MAG-12K; sAXL, sHER2, sE-selectin, TSP2, sEGFR, suPAR, sVEGFR1, sPECAM-1, OPN), human cancer/metastasis biomarker panel 1 (Millipore, Bilerica NY; catalog number, HCMBMAG-22K; GDF15, DKK1, enolase 2, OPG), human circulating cancer biomarker panel 1 (Millipore, Bilerica NY; catalog number, HCCBP1MAG-58K; CA15-3, CA19-9, MIF, leptin, IL-6, CEA, IL-8, HGF, sFas, TNFα, PRL, SCF, Cyfra21-1, FGF2, β-HCG, HE4, TGF-α, VEGF), human circulating cancer biomarker panel 3 (Millipore, Bilerica NY; catalog number, HCCBP3MAG-58K; galectin-3, myeloperoxidase, SHBG, IGFBP3, ferritin), and human circulating cancer biomarker panel 4 (Millipore, Bilerica NY; catalog number, HCCB4MAG-58K; mesothelin, midkine, kallikrein 6, ALDH1A1, EpCAM, CD44). Less than 25 μl of serum was used per panel for the bead-based multiplex immunoassay. Multiplex immunoassay procedures were performed according to manufacturer's instructions. Briefly, each well was prewetted by filling with 100 μl of washing buffer for 10 min. After removal of washing buffer, 25 μL of each standard, quality control, and assay buffer were added to appropriate wells, followed by the addition of 25ul of matrix solution. 25 μL of mixed beads were added to each well and incubated on a plate shaker overnight at 4 °C. The plate was washed with 200 μL of washing buffer 2 times and then 25 μL of detection antibodies were added and incubated on plate shaker for 1 h at room temperature. 25 μL of streptavidin-phycoerythrin were added and incubated on plate shaker for 30 min at room temperature and then washed with 200 μL of washing buffer 2 times. 100 μL sheath fluid were added to all wells, and the beads were resuspended on a plate shaker for 5 min at room temperature. The level of human serum amyloid A1 was determined by a solid phase sandwich enzyme-linked immunosorbent assay (ELISA) using a human serum amyloid A1 DuoSet ELISA kit (R&D Systems, Minneapolis MN; catalog number, DY3019-05). Concentration of each marker was determined after performing 5-parameter curve fits on logarithmized data using SoftMax Pro (version 5.4). All measurements were made within eight batches of immunoassay sessions, of which batch identifiers were documented in Table S3.
4.3. Hierarchical clustering
Hierarchical clustering was performed on OriginPro (version 2020b). Group average-clustering based on Euclidean distances was performed. Each parameter was mean-centered normalized before analysis, and the z-scores for each parameter were color-coded in the final heatmap. Individual metadata have been binarized and mean-centered normalized before hierarchical clustering.
4.4. Statistics
All statistical analyses were performed on OriginPro, if not explicitly stated otherwise. A two-tailed Welch's t-test was used when comparing the difference between two groups, as each group exhibited unequal variance. To assess the statistical significance of a given sensitivity at 90% specificity, a two-tailed one-sample Student's t-test was used against the baseline sensitivity of 10%. Spearman's correlation coefficient was used when assessing the correlation between two profiles of serum protein concentrations. ROC curves were drawn on OriginPro, with score and state values being generated from the SeroHet software, which performs 1,000 iterations of 10-fold cross-validation. Logistic regression was performed using the fitglm function of MATLAB assuming the logistic binomial model of probability. Binary parameter for chronic alcohol consumption was set as one if the individual has a positive history of chronic alcohol intake and as zero if the individual has a negative history of chronic alcohol intake. Bland-Altman test was used to derive p-values from 95% confidence intervals and was performed as previously described [46]. Signature-wise Bonferroni correction was utilized to assess the statistical significance of fold changes for each serum protein marker. ∗p-value < 0.05; ∗∗p-value < 0.01; and ∗∗∗p-value < 0.001 were considered statistically significant.
Declarations
Author contribution statement
Hanjun Lee: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Seo Yihl Kim; Dong wook Kim: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Young Soo Park: Contributed reagents, materials, analysis tools or data.
Jin-Hyeok Hwang; Sukki Cho: Conceived and designed the experiments; Analyzed and interpreted the data.
Je-Yoel Cho: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
Professor Je-Yoel Cho and Jin-Hyeok Hwang were supported by the Ministry of Science and ICT, South Korea [NRF-2021R1A5A1033157 & NRF-2017R1A2B2012191].
Data availability statement
All original codes are available at https://github.com/hanjunlee21/SeroHet and archived at https://doi.org/10.5281/zenodo.4898421. All figures introduced in this study can be reproduced using the SeroHet software.
Any additional information regarding the study is available from the lead contact.
Declaration of interest's statement
The authors declare no competing interests.
Additional information
Supplementary content related to this article has been published online at https://doi.org/10.1016/j.heliyon.2022.e12359.
Acknowledgements
We thank S. Yum for assistance in the management of clinical data prospectively collected from lung cancer patients. We are grateful to J.-M. An, J.Y. Kim, and E.-H. Yeon for their technical assistance. We also thank M.S. Lawrence for his critical review of the manuscript.
Contributor Information
Jin-Hyeok Hwang, Email: woltoong@snu.ac.kr.
Sukki Cho, Email: tubincho@snu.ac.kr.
Je-Yoel Cho, Email: jeycho@snu.ac.kr.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Haber A., Daniel, Gray S., Nathanael, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 2.Qian B.-Z., Pollard J.W. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Visser K.E., Eichten A., Coussens L.M. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D., Weinberg A., Robert Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Menakuru S.R., Brown N.J., Staton C.A., Reed M.W.R. Angiogenesis in pre-malignant conditions. Br. J. Cancer. 2008;99:1961–1966. doi: 10.1038/sj.bjc.6604733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D., Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/s0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 7.Yurkovetsky Z., Skates S., Lomakin A., Nolen B., Pulsipher T., Modugno F., Marks J., Godwin A., Gorelik E., Jacobs I., et al. Development of a multimarker assay for early detection of ovarian cancer. J. Clin. Oncol. 2010;28:2159–2166. doi: 10.1200/JCO.2008.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand R.E., Nolen B.M., Zeh H.J., Allen P.J., Eloubeidi M.A., Goldberg M., Elton E., Arnoletti J.P., Christein J.D., Vickers S.M., et al. Serum biomarker panels for the detection of pancreatic cancer. Clin. Cancer Res. 2011;17:805–816. doi: 10.1158/1078-0432.CCR-10-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kälin M., Cima I., Schiess R., Fankhauser N., Powles T., Wild P., Templeton A., Cerny T., Aebersold R., Krek W. Novel prognostic markers in the serum of patients with castration-resistant prostate cancer derived from quantitative analysis of the pten conditional knockout mouse proteome. Eur. Urol. 2011;60:1235–1243. doi: 10.1016/j.eururo.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 10.Wingren C., Sandstrom A., Segersvard R., Carlsson A., Andersson R., Lohr M., Borrebaeck C.A.K. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012;72:2481–2490. doi: 10.1158/0008-5472.CAN-11-2883. [DOI] [PubMed] [Google Scholar]
- 11.Cohen J.D., Li L., Wang Y.X., Thoburn C., Afsari B., Danilova L., Douville C., Javed A.A., Wong F., Mattox A., et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen J.D., Javed A.A., Thoburn C., Wong F., Tie J., Gibbs P., Schmidt C.M., Yip-Schneider M.T., Allen P.J., Schattner M., et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. U.S.A. 2017;114:10202–10207. doi: 10.1073/pnas.1704961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang T., Ren S.X., Zhou C.C. Multi-cancer blood testing combined with PET-CT: road for hope to screen for cancer and guide intervention. Signal Transduct. Targeted Ther. 2020;5:2. doi: 10.1038/s41392-020-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 15.Graham H., Chandler D.J., Dunbar S.A. The genesis and evolution of bead-based multiplexing. Methods. 2019;158:2–11. doi: 10.1016/j.ymeth.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Ma Y.F., Ren Y., Dai Z.J., Wu C.J., Ji Y.H., Xu J.R. IL-6, IL-8 and TNF-alpha levels correlate with disease stage in breast cancer patients. Adv. Clin. Exp. Med. 2017;26:421–426. doi: 10.17219/acem/62120. [DOI] [PubMed] [Google Scholar]
- 17.Yoo S.A., Leng L., Kim B.J., Du X., Tilstam P.V., Kim K.H., Kong J.S., Yoon H.J., Liu A.H., Wang T., et al. MIF allele-dependent regulation of the MIF coreceptor CD44 and role in rheumatoid arthritis. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E7917–E7926. doi: 10.1073/pnas.1612717113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cross M.J., Claesson-Welsh L. FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol. Sci. 2001;22:201–207. doi: 10.1016/s0165-6147(00)01676-x. [DOI] [PubMed] [Google Scholar]
- 19.Bornstein P., Armstrong L.C., Hankenson K.D., Kyriakides T.R., Yang Z.T. Thrombospondin 2, a matricellular protein with diverse functions. Matrix Biol. 2000;19:557–568. doi: 10.1016/s0945-053x(00)00104-9. [DOI] [PubMed] [Google Scholar]
- 20.Hu B., Guo P., Fang Q., Tao H.Q., Wang D.G., Nagane M., Huang H.J.S., Gunji Y., Nishikawa R., Alitalo K., et al. Angiopoietin-2 induces human glioma invasion through the activation of matrix metalloprotease-2. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8904–8909. doi: 10.1073/pnas.1533394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He B., Mirza M., Weber G.F. An osteopontin splice variant induces anchorage independence in human breast cancer cells. Oncogene. 2006;25:2192–2202. doi: 10.1038/sj.onc.1209248. [DOI] [PubMed] [Google Scholar]
- 22.Guyda H.J., Friesen H.G. Serum prolactin levels in humans from birth to adult life. Pediatr. Res. 1973;7:534–540. doi: 10.1203/00006450-197305000-00007. [DOI] [PubMed] [Google Scholar]
- 23.Terkel J., Blake C.A., Hoover V., Sawyer C.H. Pup survival and prolactin levels in nicotine-treated lactating rats. Proc. Soc. Exp. Biol. Med. 1973;143:1131–1135. doi: 10.3181/00379727-143-37485. [DOI] [PubMed] [Google Scholar]
- 24.Andersson K., Eneroth P., Fuxe K., Mascagni F., Agnati L.F. Effects of chronic exposure to cigarette-smoke on amine levels and turnover in various hypothalamic catecholamine nerve-terminal systems and on the secretion of pituitary-hormones in the male-rat. Neuroendocrinology. 1985;41:462–466. doi: 10.1159/000124220. [DOI] [PubMed] [Google Scholar]
- 25.Hakkinen K., Pakarinen A. Muscle strength and serum testosterone, cortisol and SHBG concentrations in middle-aged elderly men and women. Acta Physiol. Scand. 1993;148:199–207. doi: 10.1111/j.1748-1716.1993.tb09549.x. [DOI] [PubMed] [Google Scholar]
- 26.Patel M., Lu L., Zander D.S., Sreerama L., Coco D., Moreb J.S. ALDH1A1 and ALDH3A1 expression in lung cancers: correlation with histologic type and potential precursors. Lung Cancer. 2008;59:340–349. doi: 10.1016/j.lungcan.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 27.Lee S.H., Kim E.Y., Lee S.K., Hong J.Y., Kim Y.S. The relationship between serum ferritin levels, smoking, and lung function in Korean: analysis of the Fourth Korea National Health and Nutrition Examination Survey (KNHANES IV) Chest. 2011;140:568A. [Google Scholar]
- 28.Rossi A., Voigtlaender M., Klose H., Schluter H., Schon G., Loges S., Paolini M., Bokemeyer C., Reck M., Tarro G., Binder M. High aldehyde dehydrogenase levels are detectable in the serum of patients with lung cancer and may be exploited as screening biomarkers. J. Oncol. 2019;11 doi: 10.1155/2019/8970645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kristenson H., Fex G., Trell E. Serum ferritin, gammaglutamyl-transferase and alcohol-consumption in healthy middle-aged men. Drug Alcohol Depend. 1981;8:43–50. doi: 10.1016/0376-8716(81)90085-5. [DOI] [PubMed] [Google Scholar]
- 30.Milman N., Kirchhoff M. Relationship between serum ferritin, alcohol intake, and social status in 2235 Danish men and women. Ann. Hematol. 1996;72:145–151. doi: 10.1007/s002770050153. [DOI] [PubMed] [Google Scholar]
- 31.Chan A.W.K. Racial-differences in alcohol sensitivity. Alcohol Alcohol. 1986;21:93–104. [PubMed] [Google Scholar]
- 32.Yoshida A., Dave V., Ward R.J., Peters T.J. Cytosolic aldehyde dehydrogenase (ALDH1) variants found in alcohol flushers. Ann. Hum. Genet. 1989;53:1–7. doi: 10.1111/j.1469-1809.1989.tb01116.x. [DOI] [PubMed] [Google Scholar]
- 33.Tredan O., Manuel M., Clapisson G., Bachelot T., Chabaud S., Bardin-dit-Courageot C., Rigal C., Biota C., Bajard A., Pasqual N., et al. Patients with metastatic breast cancer leading to CD4(+) T cell lymphopaenia have poor outcome. Eur. J. Cancer. 2013;49:1673–1682. doi: 10.1016/j.ejca.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann J.N., Landgren O., Landy R., Kemp T.J., Santo L., McShane C.M., Shearer J.J., Lan Q., Rothman N., Pinto L.A., et al. A prospective study of circulating chemokines and angiogenesis markers and risk of multiple myeloma and its precursor. JNCI Cancer Spectr. 2020;4:9. doi: 10.1093/jncics/pkz104. pkz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow W.H., Gridley G., Fraumeni J.F., Jarvholm B. Obesity, hypertension, and the risk of kidney cancer in men. N. Engl. J. Med. 2000;343:1305–1311. doi: 10.1056/NEJM200011023431804. [DOI] [PubMed] [Google Scholar]
- 36.Lee S.Y., Kim M.T., Jee S.H., Im J.S. Does hypertension increase mortality risk from lung cancer? A prospective cohort study on smoking, hypertension and lung cancer risk among Korean men. J. Hypertens. 2002;20:617–622. doi: 10.1097/00004872-200204000-00017. [DOI] [PubMed] [Google Scholar]
- 37.Lindgren A., Pukkala E., Nissinen A., Tuomilehto J. Blood pressure, smoking, and the incidence of lung cancer in hypertensive men in North Karelia, Finland. Am. J. Epidemiol. 2003;158:442–447. doi: 10.1093/aje/kwg179. [DOI] [PubMed] [Google Scholar]
- 38.Giovannucci E., Harlan D.M., Archer M.C., Bergenstal R.M., Gapstur S.M., Habel L.A., Pollak M., Regensteiner J.G., Yee D. Diabetes and Cancer A consensus report. Diabetes Care. 2010;33:1674–1685. doi: 10.2337/dc10-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belpomme D., Irigaray P., Sasco A.J., Newby J.A., Howard V., Clapp R., Hardell L. The growing incidence of cancer: role of lifestyle and screening detection (Review) Int. J. Oncol. 2007;30:1037–1049. doi: 10.3892/ijo.30.5.1037. [DOI] [PubMed] [Google Scholar]
- 40.Balavarca Y., Weigl K., Thomsen H., Brenner H. Performance of individual and joint risk stratification by an environmental risk score and a genetic risk score in a colorectal cancer screening setting. Int. J. Cancer. 2020;146:627–634. doi: 10.1002/ijc.32272. [DOI] [PubMed] [Google Scholar]
- 41.Cima I., Schiess R., Wild P., Kaelin M., Schuffler P., Lange V., Picotti P., Ossola R., Templeton A., Schubert O., et al. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3342–3347. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arbabi S., Garcia I., Bauer G.J., Maier R.V. Alcohol (ethanol) inhibits IL-8 and TNF: role of the p38 pathway. J. Immunol. 1999;162:7441–7445. [PubMed] [Google Scholar]
- 43.El-Guindy N.B.D., de Villiers W.J., Doherty D.E. Acute alcohol intake impairs lung inflammation by changing pro- and anti-inflammatory mediator balance. Alcohol. 2007;41:335–345. doi: 10.1016/j.alcohol.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostroff J.S., Burkhalter J.E., Cinciripini P.M., Li Y.L., Shiyko M.P., Lam C.Y., Hay J.L., Dhingra L.K., Lord-Bessen J., Holland S.M., Manna R. Randomized trial of a presurgical scheduled reduced smoking intervention for patients newly diagnosed with cancer. Health Psychol. 2014;33:737–747. doi: 10.1037/a0033186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Organization W.H. 2000. The Asia-Pacific Perspective: Redefining Obesity and its Treatment. [Google Scholar]
- 46.Altman D.G., Bland J.M. How to obtain the P value from a confidence interval. BMJ. 2011;343 doi: 10.1136/bmj.d2304. d2304–d2304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All original codes are available at https://github.com/hanjunlee21/SeroHet and archived at https://doi.org/10.5281/zenodo.4898421. All figures introduced in this study can be reproduced using the SeroHet software.
Any additional information regarding the study is available from the lead contact.