Abstract
Genome-wide studies related to neurological disorders and neurodegenerative diseases have pointed to the role of epigenetic changes such as DNA methylation, histone modification, and noncoding RNAs. DNA methylation machinery controls the dynamic regulation of methylation patterns in discrete brain regions.
Objective
This review aims to describe the role of DNA methylation in inhibiting and progressing neurological and neurodegenerative disorders and therapeutic approaches.
Methods
A Systematic search of PubMed, Web of Science, and Cochrane Library was conducted for all qualified studies from 2000 to 2022.
Results
For the current need of time, we have focused on the DNA methylation role in neurological and neurodegenerative diseases and the expression of genes involved in neurodegeneration such as Alzheimer's, Depression, and Rett Syndrome. Finally, it appears that the various epigenetic changes do not occur separately and that DNA methylation and histone modification changes occur side by side and affect each other. We focused on the role of modification of DNA methylation in several genes associated with depression (NR3C1, NR3C2, CRHR1, SLC6A4, BDNF, and FKBP5), Rett syndrome (MECP2), Alzheimer's, depression (APP, BACE1, BIN1 or ANK1) and Parkinson's disease (SNCA), as well as the co-occurring modifications to histones and expression of non-coding RNAs. Understanding these epigenetic changes and their interactions will lead to better treatment strategies.
Conclusion
This review captures the state of understanding of the epigenetics of neurological and neurodegenerative diseases. With new epigenetic mechanisms and targets undoubtedly on the horizon, pharmacological modulation and regulation of epigenetic processes in the brain holds great promise for therapy.
Keywords: DNA methylation, Neurological disorders, Alzheimer’s, Depression, ADHD, Rett syndrome
1. Introduction
The neurological and neurodegenerative disorders, including many sporadic and hereditary disorders, are characterized by the progressive loss of neurons’ structure and function, often associated with neuronal death. The causes of neurological and neurodegenerative diseases are complicated and related to many factors, such as age, heredity, lifestyle, and environmental factors (Gilmore et al., 2008, Re et al., 2012).
Among epigenetic components, DNA methylation could be a significant epigenetic marker that has been most broadly examined (Lu et al., 2013a). DNA methylation is a post-replication alteration that frequently happens in cytosines of the CpG dinucleotide sequence, leading to the exchange of a methyl group from S-adenyl methionine to a cytosine (Jin and Liu, 2018). When DNA is symmetrically methylated, the methyl groups alter DNA structure. The main consequence of methyl alteration is that a variety of transcription factors cannot recognize the DNA and hence induce repression of transcription (Prokhortchouk and Defossez, 2008). DNA methylation in the mammalian nervous system regulates neural stem cell fate, brain development native function, neurodevelopmental disorders, and neurodegenerative diseases (Hirabayashi and Gotoh, 2010, Urdinguio et al., 2009, Xu and Li, 2012). DNA methylation is also associated with memoryand ischemia-induced damage (Xu and Li, 2012). Recent studies reported that DNA methylation changes are associated with cancer and neurological disorders (Lu et al., 2013a, Xu and Li, 2012). These epigenetic modifications regulate the networks of essential genes that mediate physiological processes and represent a simple and rational method to prevent or even treat these disorders (Landgrave-Gómez et al., 2015). New evidence suggests that altering metabolism through exercise or a variety of diets such as ketogenic diets, low-carbohydrate diets, and intermittent fasting can change the concentrations of various metabolites, some of which may modulate the activity of proteins that causes epigenetic modifications (Shimazu et al., 2013, Shyh-Chang et al., 2013).
In this brief overview, we accompanied the emergence of a new understanding of DNA methylation mechanisms and their implications for CNS function and dysfunction. Research in the previous two decades discovered an emerging outline of the relationship between numerous epigenetic pathways and neurological or neurodegenerative disorders. We will begin by outlining the epigenetics and DNA methylation, then focus intensively on recent progress made in the study of DNA methylation in major neurological disorders such as schizophrenia, depression, attention deficit hyperactivity disorder (ADHD), Alzheimer’s disease (AD), and Rett syndrome, as well as the role of DNA methylation in therapeutic approaches for the treatment of these disorders.
2. Epigenetics and DNA methylation
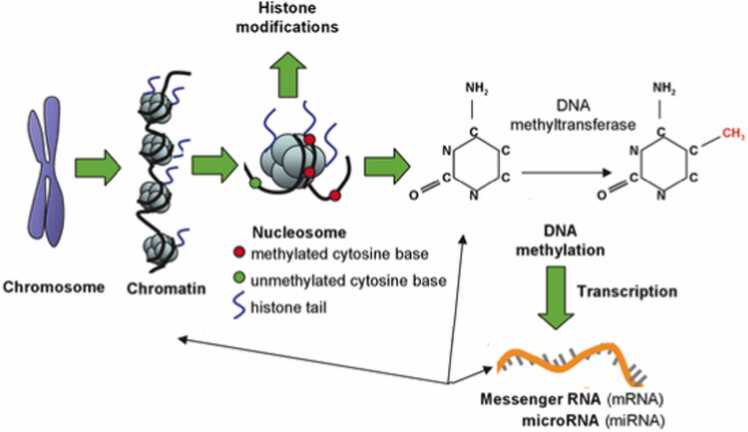
Epigenetics refers to mechanisms that regulate gene expression without altering the primary DNA sequence. Epigenetic changes affect gene activation in response to environmental cues, which is essential for the primary cell and tissue differentiation (Iridoy Zulet et al., 2017, Abdi et al., 2018). According to epigenetic theory, the genome and the environment can work together to influence regulatory mechanisms that control gene expression by modifying epigenetic DNA marks that can persist for a lifetime (Weinhold, 2006, Kanherkar et al., 2014). Furthermore, the stochastic accumulation of epigenetic changes is linked to aging (Huidobro et al., 2013) as well as sporadic neurological disorders (Wang et al., 2008), for which aging is currently recognized as a significant risk factor (Nussbaum and Ellis, 2003). Human cells undergo epigenetic changes throughout their lives, as previously stated. In identical twins with the same hereditary load, diverse epigenetic patterns are accumulated depending on the environmental factors they are exposed to, for example, diet, tobacco, or exercise. This causes discernible differences in the phenotypes of both twins, indicating different susceptibilities to disease or disease outcomes (Fraga et al., 2005). DNA methylation, histone modification, and noncoding RNA action are all critical epigenetic mechanisms (Fig. 1). DNA methylation is the most studied epigenetic mark, and its relationship to disease development has been extensively researched (Iridoy Zulet et al., 2017). The DNA methylation process is a reversible mechanism wherein methyl groups (–CH3) are delivered to cytosines positioned in CpG (5′-Cytosine-phosphate-guanosine-3′) nucleotides turning these cytosines into 5-methylcytosines (5mC) (Martínez-Iglesias et al., 2020, Alipour et al., 2020). DNA methylation is catalyzed by specific enzymes known as de novo DNA methyltransferases (DNMTs), and it occurs at the expense of ATP and S-adenosylmethionine as methyl donors (Moore et al., 2013).
Fig. 1.
Schematic diagram of Epigenetics Modification.
DNA methylation is an essential part of the epigenetic system, which organizes changes in numerous genes and helps control the expression of genes in all vertebrates (Altuna et al., 2019a). Most cytosine methylation occurs in cytosine phosphate guanine (CpG) islands, which are found in both eukaryotes and prokaryotes. Currently, five states of the cytosine base are known: 5-carboxylcytosine (5cC), 5-formylcytosine (5fC), 5-Hydroxymethylcytosine (5hmC), 5-methylcytosine (5mC), and unmodified cytosine (C). After the unaltered form, the most prevalent state of cytosine in the brains is 5mC, which is mainly found in the CpG dinucleotides. CpG islands are found in more than 60% of mammalian gene promoters (Prasad and Jho, 2019). 5mC was assumed to be related to the suppression of gene expression, while 5hmC, which causes DNA demethylation, was associated with enhanced gene expression. As more study is done, the role of methylation in gene expression depends on the CpG region in the genome. Most of the time, methylation at the gene’s promoter is negatively associated with gene expression (Ogino et al., 2009, Aziz et al., 2020).
DNMTs are tissue- and cell-specifically expressed during neural development as well as in active neurogenesis (Feng et al., 2007) and adult stem cell niches (Kang, 2011), where they have been implicated in neural plasticity and survival (Ooi et al., 2007). Once methylation is established, proteins from the methyl-CpG-binding domain (MBD) family are recruited in methylated loci to stimulate the recruitment of histone modulatory variables (Jones and Takai, 2001, Klose and Bird, 2004), indicating a synergistic modulation of numerous epigenetic marks (Mehler, 2008).
The MBD proteins are also recruited in brain development functions in adults (Chahrour et al., 2008). The most common consequence of DNA methylation is the silencing of genes and noncoding genomic regions, mainly when gene promoters are influenced (Chahrour et al., 2008, Yasui et al., 2007).
This epigenetic process is widespread in brain cells. The results of previous studies indicated that 5hmC is distinctly different from 5mC in its chromatin dependence during neural stem cell (NSC) development. 5-hydroxymethylcytosine (5hmC) has been proposed that it is both an intermediate state in the demethylation process and a significant epigenetic impact on neurological disorders (Chen et al., 2014, Cheng et al., 2015). But in general, the process of DNA demethylation and the enzymes that catalyze this reaction, although DNA demethylases such as cytidine deaminase caused by activation, are only partially understood after the past decade (Bhutani et al., 2010) or the DNA demethylating activity of TET1 (a member of TETs) (Tahiliani et al., 2009) have been identified.
Evidence demonstrates the critical role of DNA methylation in common diseases. Researchers have attempted to use DNA methylation as a biomarker to distinguish epigenetic changes related to disease status, including neurological disorders (Jin and Liu, 2018). In neurons and the nervous system, the overall balance between DNA methylation, demethylation and hydroxymethylation creates different neural patterns in processes such as learning or memory, and their dysregulation may be associated with neurological disorders (Wang et al., 2008).
2.1. DNA methylation in neurological disorders
Epigenetic modifications form long-lasting cellular memories in the brain, which are used to translate the mechanisms and responses to environmental stimuli (Akyürek et al., 2021, Kim and Kaang, 2017). Microglia, a type of immune cells, populate the central nervous system (CNS) during early fetal development and can self-renew locally. They remain in the CNS microenvironment throughout life, accounting for 10–15% of all CNS cells (Li and Barres, 2018). There are generally accepted classifications of these cells in the brain in response to infection or tissue injury, pro-inflammatory phenotype (M1-phenotype), and in response to neurodegenerative diseases, the anti-inflammatory phenotype (M2-phenotype) (Nimmerjahn et al., 2005, Xu et al., 2015). Neuron-microglia crosstalk in the CNS affects homeostasis and neuronal function in the healthy brain, so the differentiation of specific phenotypes and activation states of these phenotypes play an essential role in CNS health and disease (Esteller, 2008, Petralla et al., 2021). As a result of neuronal stimulation, several transcription factors cause epigenetic revolutions in microglia, followed by chromatin remodeling and the formation of a distinct microglial phenotype, which may have implications for neuronal activity, maturation, and synaptic networks. Inflammatory status and tissue-specific Transcription Factors such as PU.1, CEBP, IRF8, SMAD2/3, and SALL1 (Holtman et al., 2017) are examples that promote the expression of IL-6 and tumor necrosis factor (TNF), NF-κB, NF-AT, and STAT1/3 and cause the histone H3 K4monomethylated (H3K4me1) and histone H3 K9lysine acetylation (H3K9ac). So this procedure is responsible for gene expression enhancers and chromatin being modified (Holtman et al., 2017, Veremeyko et al., 2019).
Abnormal DNA methylation patterns are associated with a wide range of human neurological diseases, including several neuropsychiatric illnesses (schizophrenia, depression, ADHD), neurodevelopmental disorder (Rett syndrome), neurodegenerative disease (Alzheimer's disease), and cognitive impairment (Qureshi and Mehler, 2013, Weng et al., 2013).
Recent research has revealed a link between DNA methyltransferases and pain processing. Following a peripheral nerve injury, the level of DNA methyltransferases, DNMT3a and DNMT1, increased in the dorsal root ganglia (DRG). By stimulating DRG, these enzymes can promote the gene Kcna2, which influences the voltage-gated potassium channel, resulting in spinal cord sensitization and neuropathic pain symptoms (Sun et al., 2019, Zhao et al., 2017). A decrease in DNA methyltransferases has been detected in various CNS disorders, affecting the BDNF DNA methylation status in the hippocampus, which modulates learning and memory (Guo et al., 2011, Nguyen et al., 2007). Post-mitotic neurons and glial cells make up a large portion of the brain’s cells, both of which have a limited ability to divide. Mature neurons have DNMT1 and DNMT3A expressions. In both development and disease, DNMT3B is required for the dynamic programming of epigenetic regulation (Gao et al., 2020, Ng et al., 1999).
Mutations in DNMT1 have been identified in hereditary sensory and autonomic neuropathy type 1 (HSAN1) syndrome, with other neuropathies and autosomal dominant cerebellar ataxia, deafness, and narcolepsy (ADCA-DN) (Sun et al., 2014, Kernohan et al., 2016). DNMT1 activity is critical for maintaining DNA methylation, chromatin stability, and gene regulation. Thus, the mutation of DNMT1 impairs DNA methyltransferase activity and decreases heterochromatin binding in the G2 cell cycle stage, resulting in extensive hypomethylation and local hypermethylation (Jin and Robertson, 2013). This may explain its complex pathogenesis in the nervous system. In addition, the decline in the DNMT3a2 expression in the hippocampus has been associated with intellectual and cognitive disorders indicating the essential role of DNMT3a2 in memory formation and mental function (Klein et al., 2011). The results of one study determined that the process of amyotrophic lateral sclerosis- mesenchymal stromal cells (ALS-MSCs) can be modulated by inhibiting overexpressed DNMTs. This approach may provide better efficacy in stem cell therapy (Oh et al., 2016). Another study also showed that methyltransferase levels increased in the demyelinated hippocampus of multiple sclerosis patients, while demethylation enzymes decreased (Chomyk et al., 2017).
DNA methylation can be recognized by a variety of methyl-CpG-binding domain (MBD) proteins “epigenome readers” such as methyl-CpG binding protein 2 (MECP2) and methyl-CpG-binding domain proteins 1–4 (MBD1–4). MECP2 is an X-linked gene that codes for a nuclear protein that binds to methylated DNA and acts as a broad suppressor interacting with histone-modifying complexes (Goffin et al., 2011). Mutations in the MECP2 gene and subsequently irregular expression of MECP2 are the leading cause of the Rett syndrome, a neurological X-linked disorder mainly affecting females. MECP2 mutations have also been associated with a wide range of other neurodevelopmental diseases, including X-linked mental retardation, and autism, representing that mutation in MECP2 has extensive consequences and leads to various neurodevelopmental disorders (Bostick et al., 2007, Amir et al., 1999, Gonzales and LaSalle, 2010).
Abnormal DNA methylation patterns have also been reported in several neurodegenerative disorders. Alzheimer’s disease (AD) is a common neurodegenerative illness marked by progressive dementia that may be due to abnormal DNA methylation. In some AD brain research, elevated levels of S-adenosylhomocysteine have been detected, which inhibits DNMT s activity (Kennedy et al., 2004). Furthermore, DNA hypomethylation has been identified, in promoter CpGs of AD-related genes such as presenilin 1(PS1), APP, and β-site APP-cleaving enzyme1 (BACE1), resulting in abnormal upregulation of these genes, leading to accumulation of Aβ (Kennedy et al., 2004). Moreover, hypermethylation is also observed at a specific position in the promoter of specific genes, such as methylenetetrahydrofolate and apolipoprotein E reductase, in the brain of AD patients. As a result, it can be said that the change of methylation level is one of the causes of Alzheimer's disease related to genes (Wang et al., 2008).
Previous studies also demonstrate abnormal DNA methylation in various psychiatric diseases (Grayson and Guidotti, 2013). Postmortem methylome profiling of brains from patients with schizophrenia has demonstrated intense alterations in the DNA methylation profile, including genes that are related to pathogenesis (Mill et al., 2008). Predominantly, hypermethylation of glutamic acid decarboxylase 67 (GAD67) and RELN promoter regions was linked to decreased expression of these genes in schizophrenia patients (Guidotti et al., 2000). Table 1.
Table 1.
The relationship between neurological diseases with genes involved and their methylation.
| Neurologic disorders | Methylation status | Specific genetic loci | Probable effect | symptoms |
|---|---|---|---|---|
| Rett syndrome | Unknown | MECP2 gene | Loss of the activity of the MECP2 gene Reduced BDNF (Na and Monteggia, 2011) | Seizures, cerebral palsy, Repetitive, stereotyped hand movements |
| Alzheimer’s disease | Hypomethylation Hypermethylation |
PS1, BACE1, APP Neprilysin (NEP) |
Upregulation of PS1, BACE1, APP Aβ accumulation (Qazi et al., 2018) |
Progressive dementia |
| Stroke | Hypermethylation | Global methylation (Krupinski et al., 2018) | Sudden onset focal dysfunction | |
| Multiple sclerosis | Hypomethylation | PAD2 | PAD2 Upregulation (Calabrese et al., 2012) |
several neurologic symptoms: muscular weakness, visual symptoms, tremors, intestinal and urinary disorders, cognitive abnormalities |
| Epilepsy | Hypermethylation | reelin | Decreased reelin expression (Henshall and Kobow, 2015) | seizure |
| Parkinson’s disease | Hypomethylation | SNCA intron1 | Increased expression of SNCA (Jowaed et al., 2010) | Rigidity, tremors, shaking, difficulty in walking |
| Immunodeficiency, Centromeric Instability, and Facial Anomalies Syndrome Type1 (ICF1) syndrome | Hypomorphic mutation of DNMT3B | DNMT3B | Reduced the activity of DNMT3B (De Greef et al., 2011) | Mental retardation syndrome |
| ICF2 | Hypomethylation | ZBTB24 | hypomethylation of minor satellite DNA and Centromeric instability (Hardikar et al., 2020) | Mental retardation syndrome |
| ADCA-DN | Hypomethylation | DNMT1 | reduced DNMT1 activity (Kernohan et al., 2016) | Autosomal dominant abnormality with Cerebellar ataxia |
| Amyotrophic lateral sclerosis (ALS) | Hypomethylation | VEGF, SOD1 | No transcriptional silencing (Lu et al., 2013b) | Weakness and atrophy of the muscles |
| schizophrenia | Hypermethylation | GAD67 RELN |
Decreased expression of GAD67and RELN (Grayson and Guidotti, 2013) | Hallucinations, Disorganized thinking |
| Fragile X syndrome | Hypermethylation | FMR1 | FMR1 inactivation (Kumari et al., 2020) | Intellectual disability |
| HSAN1 | Hypomethylation | DNMT1 | Reduced DNMT1 activity (Sun et al., 2014) | Multiple neuropathies |
MECP2: methyl CpG binding protein 2, BDNF: brain-derived neurotrophic factor, PS1: presenilin 1, BACE1: beta-secretase 1, APP: amyloid precursor protein, NEP: neprilysin or neutral endopeptidase, PAD2: peptidyl arginine deiminase, SNCA: Alpha-synuclein, DNMT: DNA methyltransferase, ZBTB24: zinc finger and BTB domain-containing protein 24, VEGF: vascular endothelial growth factor, SOD1: superoxide dismutase type 1, GAD67: glutamate decarboxylase-67, FMR: fragile X mental retardation, Immunodeficiency, Centromeric Instability, and Facial Anomalies Syndrome Type1; ICF1, Immunodeficiency, Centromeric Instability, and Facial Anomalies Syndrome Type 2; ICF2, Amyotrophic lateral sclerosis; ALS. Amyotrophic lateral sclerosis; ALS.
2.2. DNA methylation and depression
Depression is one of the most common psychiatric disorders in the world (Reszka et al., 2021). Depressive symptoms in adolescence have long-term consequences for brain development and can cause severe social and educational problems. Furthermore, depression is linked to cerebrovascular diseases such as stroke (Guo et al., 2021, Xiang et al., 2021). Major depressive disorder (MDD) is a complex, debilitating psychiatric condition with a high prevalence of 3.63%. Symptoms include anxiety, sadness, hopelessness, and emptiness, feelings of guilt or loss and worthlessness, irritability or frustration, loss of interest or pleasure in most routine activities, sleep disturbances, reduced or increased appetite, low energy, difficulty thinking and concentrating, impaired cognition, physical pity, and physical pity. Genetic and environmental risk factors influence depression (Li et al., 2021, Sales et al., 2021, Borçoi et al., 2021). Epidemiologic studies show that genetic factors increase the risk of depression. On the other hand, studies have shown a strong relationship between specific genes and environmental factors in the development of depressive disorder (Sun et al., 2021). Evidence suggests that the onset of depression is increased by around 60% by exposure to stressful events (Sales et al., 2021). Exposure to stress can modify DNA methylation patterns and affects brain plasticity and emotion (Reszka et al., 2021). One of the main neurobiological mechanisms of depression is dysregulated and dysfunctional stress response system (such as hypothalamic-pituitary-adrenal (HPA) axis activity and glucocorticoid receptor (GR) sensitivity) to show the adaptive change (Guo et al., 2021). Moreover, prenatal depressive symptoms might influence fetal epigenetic programming (Kallak et al., 2021). Prenatal depression is associated with differential methylation in GNAS, CTNNA2, OSBPL10, and 5-HTTLPR (Sales et al., 2021, Drzymalla et al., 2021). Mothers with persistent perinatal depression have hypermethylation of OXTR in the saliva. However, this is the only marker associated with perinatal depression in mothers, but no causal effect has been proven (Sales et al., 2021). Maternal depression leads to increased neonatal DNA methylation in the glucocorticoid receptor gene (NR3C1) and BDNF IV promoter (Braithwaite et al., 2015). Preclinical studies have reported stress-induced hypermethylation and reduced gene expression, indicating that exposure to stress conditions in early life leads to persistent epigenetic changes and influences neural and behavioral patterns in adulthood (Sales et al., 2021, Drzymalla et al., 2021). Studies show that epigenetic processes such as DNA methylation are heritable. This could explain the heritability of depression (Juruena et al., 2021). Exposure to stress results in the modification of DNA methylation in several genes associated with depression, including the glucocorticoid receptor (NR3C1 or GR), mineralocorticoid receptor (NR3C2 or MR), corticotrophin-releasing hormone receptor 1 (CRHR1), serotonin transporter (SLC6A4 or 5-HTT), brain-derived neurotrophic factor (BDNF), and FK506-binding protein 5 (FKBP5) (Borçoi et al., 2021, Drzymalla et al., 2021, Ding and Dai, 2019). Different studies have reported the significant relationship between alteration in DNA methylation of FKBP5 with depressive symptoms (Guo et al., 2021, Han et al., 2017, Höhne et al., 2015). These alterations in gene expression may lead to significant modifications in neural and behavioral functions (Sales et al., 2021). Homer1a expression in the hippocampus and cingulate gyrus of patients with major psychiatric disorders including major depression (Leber et al., 2017). Also, Stratum lacunosum glial cells displayed reduced Homer1a expression in bipolar disorder when compared to major depression (Leber et al., 2017). Deletion of synaptic plasticity protein Homer1a results in depression-like behavior and various antidepressant treatments induce its expression (Sun et al., 2021). Gestational stress increases the expression of DNMTs and DNA methylation of BDNF, thereby inducing depressive-like and anxiety-like phenotypes by downregulation of BDNF expression in the hippocampus of the offspring (Zheng et al., 2016).
Also, maternal neglecting or separation stress resulted in hypermethylation of DNA in the hippocampus and the protein phosphatase one catalytic subunit (PP1C) and adenosine A2a receptor (A2AR) promoter in the nucleus accumbens. Upregulation of A2AR is associated with synaptic dysfunction in depression (Carvalho et al., 2019). On the contrary, some studies indicate an association between DNA hypomethylation and stress. For instance, maternal separation stress increased DNA methylation in NR3C1 and Syn I genes, followed by increased NR3C1 mRNA in the hypothalamus, Syn I mRNA, and protein levels in the amygdala, and decreased in the nucleus accumbens (Holmes et al., 2019). Thus, it can be concluded that the alteration followed by DNA methylation depends on various factors, including the type of stressor, age, sex, brain structure, gene, and region (Sales et al., 2021, Drzymalla et al., 2021). Epigenetic modifications of 5-mC and 5-hmC are abundantly found in the brain and are directly associated with depression (Reszka et al., 2021). Transposable elements of Alu and LINE-1, and 5-mC and 5-hmC, have been considered potential biomarkers in mental disorders such as MDD. Interestingly, 5-mC is associated with the downregulation of genes, while 5-hmC is correlated with demethylation and increased transcription (Misiak et al., 2019).
Studies showed a low level of 5-hmC in patients with MDD and a high level of 5-mC in BD type I patients. While in another study on patients with BD and MDD, there was a reduced level of 5-mC and a significant reduction of 5-mC and 5-hmC in major depression (Reszka et al., 2021). In a study by Liu et al., hypomethylation of LINE-1 was observed in the blood of MDD patients (Liu et al., 2016). The mechanism of action of some drugs used as mood stabilizers and antidepressants is based on modifying DNA methylation at specific CpG sites (Goud Alladi et al., 2018).
A review study showed a significant association between DNA methylation and depression risk. Hypermethylation of BDNF, CRMP2, NR3C1, and SLC6A4 is associated with Depression and MDD (Xiang et al., 2021, Li et al., 2021, Borçoi et al., 2021, Li et al., 2019, Schiele et al., 2021, Sanwald et al., 2021, de Assis Pinheiro et al., 2021). Several studies have investigated DNA methylation of some critical genes modulating depressive symptoms, including PTPRN2 (correlated with mood state disturbances), HES5 (associated with MDD and suicide), GATA2 (related to depressive behavior in rats), DGKA (differed significantly between MDDs and controls), NIPA2 (increased risk of MDD), PRDM7 (important in aging and Alzheimer), KCNIP1 (regulate neuronal membrane excitability), GRIK2 (related to mood disorders and depressive symptoms) (Wang et al., 2021). Moreover, HELZ2 and ZNF624 gene expressions differed differentially between MDDs and health controls (Wang et al., 2021). Most patients resist treatment with conventional anti-depressant drugs; according to the results, epigenetic markers can be used in drug responses for psychiatric disorders (Zhou et al., 2021). Characterizing specific DNA methylation patterns identifies novel biomarkers for subtyping psychiatric disorders and the decision of optimal drug choice (Yamagata et al., 2021). For instance, DNA methylation of FKBP5 is a potential marker for the treatment response to mindfulness-based stress reduction in post-traumatic stress disorder (Bishop et al., 2018). Some drugs, such as Clozapine and Sulpiride, activate DNA demethylation in brain tissue. Zhou et al.,2021 found that antidepressant drugs increased DNA methylation in BDNF promoters in patients with MDD and BD (Zhou et al., 2021).
DNA methylation of multiple immune-related loci in patients with depression shows the association between inflammation and depression (Crawford et al., 2018). The study by Sun and colleagues conducted the correlation between promoter methylation of Homer1a and depression-like behaviors. Some antidepressant drugs act through the induction of Homer1a. Moreover, DNA methylation of CpG sites around the binding sites for CRE in Homer1 promoter results in major depressive disorder (Sun et al., 2021).
2.3. DNA methylation and ADHD
ADHD is a heterogeneous disorder with a complex and multifactorial background. Numerous genetic and environmental factors and their interactions play a critical role in the pathophysiology of this disease (Barkley, 1998). Recently, genetic risk factors for ADHD have been identified, which include genes involved in neurotransmitter transport, neurodevelopment, growth processes, cell adhesion, and ion transport (Demontis et al., 2019, Rovira et al., 2020). In addition to genetic risk factors, the onset and persistence of ADHD are also associated with environmental factors (Jin and Liu, 2018), such as low birth weight (Faraone et al., 2005, Karimi-Nazarabad et al., 2015), maternal stress during pregnancy (Humphreys et al., 2019, Palladino et al., 2019), and toxin exposure (Williams and Ross, 2007, Murgatroyd et al., 2009).
Notably, the environment can interact with the genome through epigenetic changes, such as DNA methylation (Murgatroyd et al., 2009, Plazas-Mayorca and Vrana, 2011), which is highly sensitive in early life (Bauer et al., 2016). The role of altered DNA methylation in ADHD has been evaluated primarily through candidate gene studies (Hamza et al., 2019). In addition, the first extensive epigenome communication studies (EWAS) to diagnose ADHD and population symptoms have been performed primarily on relatively small groups of children (Mooney et al., 2020) and adolescents (Meijer et al., 2020).
Epigenetic studies focusing on adult ADHD are rare. Quantitative studies have targeted candidate genes for ADHD, such as norepinephrine transporters (Sigurdardottir et al., 2021), dopamine transporters (Keitel et al., 2018), and serotonin receptors (Perroud et al., 2016). A single EWAS has been performed for ADHD symptoms in the general adult population (Toikumo et al., 2019) and ADHD status (Rovira et al., 2020). Chang et al.’s study of twins showed that the genetic contribution to ADHD varies from childhood to adulthood (Chang et al., 2013), and Meijer et al. Showed that epigenetic differences could distinguish between persistent ADHD and transient ADHD (Meijer et al., 2020). Meijer and colleagues performed targeted bisulfite sequencing for 37 candidate genes to investigate differential DNA methylation between adults with ADHD and healthy individuals. They found that, unlike EWAS, this approach provides information on the methylation level of all CpG sites in target areas (Weiß et al., 2021). More studies are needed to understand better whether people are prone to hyperactivity without a genetic background and simply by the DNA methylation status of neurons.
The results of the studies showed that the further analysis of DNA methylation in ADHD can help identify the biomarkers of the disease and potentially the mechanisms of the disease, with the results of some studies pointing to the relationship between the methylation level of promoter of GART and SON, SLC7A8, MARK2, ERC2 and CREB5 genes (Mooney et al., 2020, Neumann et al., 2020) and DRD4 and 5-HTT regions (van Mil et al., 2014) and the disease. Eventually multi-positional algorithms will be essential for discovery of clinically valuable biomarkers.
2.4. DNA methylation and Alzheimer’s disease
AD is the leading cause of dementia and also one of the most pressing public health issues in our life. By 2050, it is anticipated to have reached a global prevalence of over 91 million AD cases. Although the pathogenesis of AD is yet unknown, the most commonly recognized theory is the amyloid pathway, in which the accumulation of tangles and plaques is claimed to play a crucial role in the disease’s course and development (Ozaki and Niida, 2019).
However, other characteristics, including phospholipid metabolism, cholesterol, and abnormal calcium, frequently appear before the accumulation of tangles and plaques appear early in disorder. The analysis of genome set and disorder cascade analysis obtained from the findings of “epigenome-wide association studies (EWAS)” proposed biological functions involving the amyloid-β protein precursor (APP) degradation, tau adhesion molecules, lipid-related mechanisms, and brain immune functions in the pathogenesis of Alzheimer’s disorder (Wei et al., 2020). Moreover, AD is seen as a multifaceted illness that results from the combination of genetic and environmental variables, which are influenced by epigenetic processes (Altuna et al., 2019b).
There is increasing evidence that epigenetic variation plays a significant role in the development of Alzheimer’s disease, although gene mutations account for just 5% of all cases. Furthermore, recent methodological developments can employ EWAS in complex disorders phenotypes, such as AD. Epigenetics reversibly regulates gene expression and may be inherited through cell division (Prasad and Jho, 2019). DNA methylation is a vital epigenetic pattern that manages changes in specific genes and helps regulate gene expression in vertebrates, which is the best-studied example of epigenetics modifications in AD (Wei et al., 2020).
The link between AD and DNA methylation has been studied extensively. In the peripheral blood and brain, distinct methylations of genes were discovered in control groups and AD patients. The APP gene was the only one that was consistently hypermethylated in both the blood and the brain, suggesting that it might be the most effective diagnostic biomarker of blood for AD. Furthermore, there was an increase in the APP gene expression in AD patients (Wei et al., 2020, Iwata et al., 2014). In addition, Coppieters et al (Coppieters et al., 2014). found a positive correlation between global levels of 5mC and amyloid-beta in the brain of patients with AD.
An in vitro investigation has shown that APP hypermethylation is related to higher expression in AD brains. Although other studies reported no change in relative hypomethylation in various areas of the APP gene in individuals with AD, the methylation-detecting techniques utilized in this research were not sufficiently sensitive, affecting the credibility of the findings. This finding will need further investigation to be confirmed (Wei et al., 2020).
Altuna et al (Altuna et al., 2019b). proposed that altered methylation of DNA in the AD hippocampus happens at particular regulating areas that can be critical for neuronal differentiation, supporting the idea that adult hippocampus neurogenesis may have a role in the development of AD.
Several genes have been discovered to be differentially methylated in AD brain autopsy samples using “Illumina Infinium Human Methylation450K arrays”, including those genes previously identified as carrying genetic variations for AD, such as BIN1 (amphiphysin II) or ANK1 (ankyrin-1) (Altuna et al., 2019b, Lunnon et al., 2014).
Interestingly, some of these DNA methylation patterns are available in the early AD stages, indicating that such modifications may play a role in the disease’s development. Overall, these studies add to our knowledge of the pathophysiology of AD (Altuna et al., 2019b).
2.5. DNA methylation and Rett syndrome
Rett syndrome (RTT) is a common mental disability that occurs once per 10,000–22,000 girls. It is marked by a stage of average growth and development until approximately one year, followed by a fast regression that includes stereotypic hand wringing, irregular breathing, ataxia, autism, slowed head growth or microcephaly, lack of acquired motor and verbal abilities, seizures. Despite these symptoms, patients live until maturity (Kriaucionis and Bird, 2003).
According to a recent study, mutations of MeCP2 cause RTT syndrome (MIM 312750), a juvenile neurological illness that is amongst the most prevalent due to mental impairment in women. Extensive RTT patient screening indicated that 80% of patients with RTT syndrome are related to detectable mutations in MeCP2 gene, including insertions, deletions, nonsense, and missense (Goffin et al., 2011, Kriaucionis and Bird, 2003). Moreover, MeCP2 is overexpressed in the postnatal brain, suggesting that methylation-dependent gene regulation can play an essential role in the development of the mammalian central nervous system (Chen et al., 2001). Furthermore, several genes become silenced when the promoters of these genes are methylated. Hence, using transient transfection research, scientists assumed that MeCP2 gene was a transcriptional suppressor and could suppress the transcription in both cells and in-vitro (Esteller, 2008, Petralla et al., 2021). To investigate MeCP2’s suppression properties, scientists monitored reporter gene expression with a fusion of the GAL4 DNA- binding domain to the various parts of MeCP2 gene (Petralla et al., 2021). A domain with 100 amino acids was observed in the middle of the protein, which is in charge of transcriptional suppression (TRD).
Additionally, it revealed that the binding of MeCP2 was capable of suppressing the transcription (from up to 2000 bp of the transcription start site (TSS)) (Kriaucionis and Bird, 2003). The DNA electrophoretic mobility shift assay (EMSA) was utilized by W. Gabel et al (Gabel et al., 2015). to evaluate the MeCP2 binding to different forms of methylated DNA. Consistent with similar studies, MeCP2 showed high affinity to mCG DNA and not hmCG, which proves that MeCP2 might not preferentially bind to hmCG in neurons. In contrast, MeCP2 binds to mCA, hmCA, and mCG with high affinity compared to binding to mCC and mCT with low affinity (the same affinity to unmethylated DNA), respectively. This tight binding between MeCP2 to mCG, mCA, and hmCA indicates the potential role of MeCP2 in regulating long gene expression in the brain through binding to the referred sites. Also, a thin-layer chromatography, and Tet-assisted bisulfite sequencing (TAB-seq) analysis, showed that the methylation form of hmCA is rare in the brain (Gabel et al., 2015).
The frequency of hCG and mCH in the neuronal genome and the level of MeCP2 protein were significantly increased over the postnatal period. As such, this suggests that MeCP2 could play a role in the maturation of neurons by binding to hmCG and/or mCH methylated DNA (Szulwach et al., 2011, Kriaucionis and Heintz, 2009).
2.6. The role of DNA methylation in Therapeutic approaches to neurological diseases
Epigenetically targeted drugs, in general, and DNA methylation-targeted drugs, in particular, may have distinct pharmacological and toxicological properties. DNA methylation is the best-studied epigenetic mechanism in eukaryotic cells. Mutations in genes can cause epigenetic dysfunction leading to certain neurodevelopmental disorders (Stirzaker and Armstrong, 2021). Some altered epigenetic patterns are directly associated with the presence of a mutation in an epigenetic gene involved in a neurodevelopmental disorder. It has been reported that DNA methyltransferase activity is high in neurons, and its activity may contribute to induced ischemic brain damage in mice (Dolen et al., 2019). DNA-demethylating drugs are currently considered as a treatment option (Nuzziello and Liguori, 2021). These drugs may be suitable for various neurodegenerative and neurodevelopmental diseases, such as fragile X syndrome. Histone deacetylase (HDAC) inhibitors are the recent focus for researchers (Kumari et al., 2020). Epigenetic mechanisms are a central process in determining cell fate (Sivalingam and Samikkannu, 2020). However, there are no new epigenetic regulators of development nor known mechanisms to be used for development. This is a rich area for additional research, especially regarding noncoding RNAs and their role in CNS development (Sivalingam and Samikkannu, 2020).
All current approaches to modifying DNA methylation levels target the endogenous enzymatic machinery responsible for adding and removing mCs from DNA in some way. The use of constitutive and conditional gene knockout mouse models and viral-mediated RNA knockdown or overexpression techniques has revealed much about the importance of active DNA methylation during neurodevelopment and in the functioning adult CNS (Kaur et al., 2022). The contribution of the epigenome in protection against neurodegenerative diseases such as AD or PD have been demonstrated (Suchy et al., 2010). For example, supplementation of S-Adenosyl methionine (SAM) in a transgenic mouse model (SOD1-G93A) of amyotrophic lateral sclerosis (ALS) delayed the onset of motor neuron pathology. HDACi also facilitated disease progression in ALS animal models (Suchy et al., 2010). Sodium phenylbutyrate significantly extended survival in G93A transgenic ALS mice (Ryu et al., 2005).
Research into Huntington’s disease (HD), a neurodegenerative disorder caused by a trinucleotide repeat expansion in the gene (HTT) encoding the huntingtin protein, found that mutant huntingtin interacts directly with HAT proteins, resulting in altered histone acetylation (Jiang et al., 2006). Numerous studies have shown that treatment with HDACi halts progressive neuronal degeneration in both fly and mouse HD models. Several selective HDACi and other compounds are investigated (Chopra et al., 2012, Duan, 2013).
Researchers now know that, while DNMT3a/b are frequently responsible for de novo methylation and DNMT1 for its maintenance, these roles are not mutually exclusive, and knocking out both DNMT1 and DNMT3a in adult forebrain neurons is required to elicit dysfunction in long-term plasticity and deficits in learning and memory (Mattei et al., 2022). A Tet1 knockout mouse model and RNA knockdown experiments recently demonstrated that Tet1-mediated mC oxidation is required for memory and the regulation of activity-related genes in the dorsal hippocampus, including Fos and Arc (Zhan et al., 2022). Previous studies have shown that targeting the epigenome, especially with small drug molecules, can cross the blood-brain barrier and delays the onset and progression of symptoms in animal models of neurodegenerative disease (Fischer, 2014). As suggested in many reports, the multicentric and multicellular effects exerted by most drugs, the possibility of unexpected side effects, and the anatomic and metabolic differences between humans and rodents are reasons for concern (Fischer, 2014). This suggests that further studies are needed to clarify the most appropriate therapeutic approaches, including the use of selective inhibitors, timing, dosing regimen, a better understanding of the interplay between histone tail modifications and other mechanisms regulating gene expression, and evaluation of potential side effects (Fischer, 2014). In addition, the route of drug administration varied across studies, are not expressed in the same way in brain regions affected by AD, PD, or other neurodegenerative diseases. Although HDAC2 and HDAC6 may represent promising drug targets in AD, it remains unknown which drug or dosing regimen is most effective, and similar conclusions can be drawn for other neurodegenerative diseases (Harrison and Dexter, 2013).
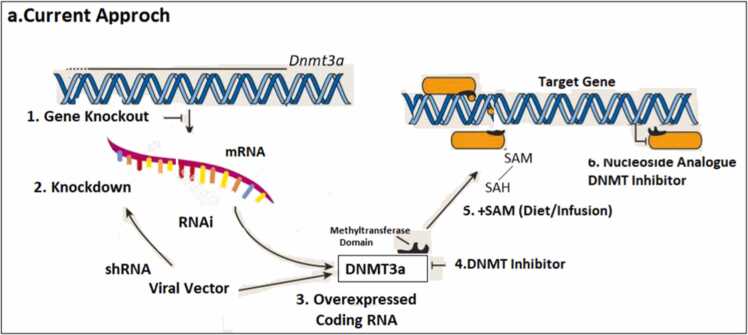
Indeed, while recent tools such as fluorescence-activated cell sorting and next-generation sequencing have greatly improved the ability to measure epigenetic changes with cellular and genetic precision (see sidebar, Measuring the Epigenome), our approaches (Fig. 2) to manipulating DNA methylation are far less sophisticated. In contrast, as demonstrated for various DNMT proteins and Tet1, genetic processes such as traditional gene knockout animal lines, small hairpin RNA knockdown, and virally mediated gene overexpression are capable of exhibiting complete isoform selectivity (Sagarkar et al., 2022).
Fig. 2.
Current Approaches to manipulating DNA methylation.
These approaches still have limitations because they do not provide precise temporal control over methylation status. Furthermore, because they presumably affect methylation status on a genome-wide scale, these approaches still lack target specificity. Another limitation to our knowledge is that the drugs have been tested in animal models carrying mutations in human genes such as SOD1, which account for only about 1–2% of human cases. In addition, histone code writers or deletion proteins could be potential drug development targets, but studies are limited. Natural compounds found in the diet, including folate, vitamins, polyphenols, and flavonoids, alter the availability of methyl groups and affect the activity of DNMTs, thus representing potential “epigenetic” preventive factors for neurodegeneration.
3. Conclusion
In this brief overview, we reviewed the emergence of a new understanding of epigenetic molecular mechanisms and their implications for CNS function and dysfunction. Studies have shown how various environmental factors, such as stress, smoking, radiation, diets, and medications throughout life can affect epigenetics and thereby act as strong determinants of human health in the coming decades. Research in the previous two decades has discovered an emerging outline of the relationship between numerous epigenetic pathways and neurological disorders. However, changes in chromatin structure are probable to happen in many loci of the genome, and it is imperative to conduct epigenetic studies at the genome level to examine this issue in animal models like the Alzheimer’s model.
Perhaps the results of this study and the studies mentioned can lead to advances in treatment approaches and tools needed by person-centered medicine in neurological disorders. Therapeutic approaches aimed at creating of global epigenomic maps in neurological disorders in histone modification patterns, DNA methylation, and RNA expression in primary tissues and cell types of all major lineages in the human cell body would be valuable. Finally, it appears that the various epigenetic changes do not occur separately and that DNA methylation and histone modification changes occur side by side and affect each other. A complete understanding of these epigenetic changes and their interactions will lead to better treatment strategies for neurological disorders such as hyperactivity and mental health problems in patients with ADHD, Alzheimer’s disease, stress, and depression. By studying the mechanisms and targets of epigenetics, especially DNA methylation, drug modulation and regulation of epigenetic processes, there will be many promises for the treatment of neurological diseases.
References
- Abdi A., Khabazi A., Sakhinia E., Alipour S., Talei M., Babaloo Z. Evaluation of SOCS1 methylation in patients with Behcet’s disease. Immunol. Lett. 2018;203:15–20. doi: 10.1016/j.imlet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Akyürek E., Uysal B., Gülden G., Taştan C. The effect of molecular genetic mechanisms on drug addiction and related new generation CRISPR gene engineering applications. Gene. 2021;2:50–60. [Google Scholar]
- Alipour S., Sakhinia E., Khabbazi A., Samadi N., Babaloo Z., Azad M., et al. Methylation status of interleukin-6 gene promoter in patients with Behçet's disease. Reumatol. Clin. 2020;16(3):229–234. doi: 10.1016/j.reuma.2018.06.006. [DOI] [PubMed] [Google Scholar]
- Altuna M., Urdánoz-Casado A., Sánchez-Ruiz de Gordoa J., Zelaya M.V., Labarga A., Lepesant J.M., et al. DNA methylation signature of human hippocampus in Alzheimer’s disease is linked to neurogenesis. Clin. Epigenet. 2019;11(1):1–16. doi: 10.1186/s13148-019-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuna M., Urdánoz-Casado A., Sánchez-Ruiz de Gordoa J., Zelaya M.V., Labarga A., Lepesant J.M.J., et al. DNA methylation signature of human hippocampus in Alzheimer’s disease is linked to neurogenesis. Clin. Epigenet. 2019;11(1):91. doi: 10.1186/s13148-019-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- de Assis Pinheiro J., Freitas F.V., Borçoi A.R., Mendes S.O., Conti C.L., Arpini J.K., et al. Alcohol consumption, depression, overweight and cortisol levels as determining factors for NR3C1 gene methylation. Sci. Rep. 2021;11(1):1–9. doi: 10.1038/s41598-021-86189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz S.G.-G., Aziz S.G.-G., Khabbazi A., Alipour S. The methylation status of TNF-α and SOCS3 promoters and the regulation of these gene expressions in patients with Behçet’s disease. Biomarkers. 2020;25(5):384–390. doi: 10.1080/1354750X.2020.1754912. [DOI] [PubMed] [Google Scholar]
- Barkley R.A. Attention-deficit hyperactivity disorder. A Handb. Diagn. Treat. 1998 doi: 10.1038/scientificamerican0998-66. [DOI] [PubMed] [Google Scholar]
- Bauer T., Trump S., Ishaque N., Thürmann L., Gu L., Bauer M., et al. Environment‐induced epigenetic reprogramming in genomic regulatory elements in smoking mothers and their children. Mol. Syst. Biol. 2016;12(3):861. doi: 10.15252/msb.20156520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N., Brady J.J., Damian M., Sacco A., Corbel S.Y., Blau H.M. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J.R., Lee A.M., Mills L.J., Thuras P.D., Eum S., Clancy D., et al. Methylation of FKBP5 and SLC6A4 in relation to treatment response to mindfulness based stress reduction for posttraumatic stress disorder. Front. Psychiatry. 2018;9:418. doi: 10.3389/fpsyt.2018.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borçoi A.R., Mendes S.O., Moreno I.A.A., Gasparini dos Santos J., Freitas F.V., Pinheiro J.A., et al. Food and nutritional insecurity is associated with depressive symptoms mediated by NR3C1 gene promoter 1F methylation. Stress. 2021:1–8. doi: 10.1080/10253890.2021.1923692. [DOI] [PubMed] [Google Scholar]
- Bostick M., Kim J.K., Estève P.-O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317(5845):1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Braithwaite E., Kundakovic M., Ramchandani P., Murphy S., Champagne F. Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation. Epigenetics. 2015;10(5):408–417. doi: 10.1080/15592294.2015.1039221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese R., Zampieri M., Mechelli R., Annibali V., Guastafierro T., Ciccarone F., et al. Methylation-dependent PAD2 upregulation in multiple sclerosis peripheral blood. Multiple Sclerosis. Journal. 2012;18(3):299–304. doi: 10.1177/1352458511421055. [DOI] [PubMed] [Google Scholar]
- Carvalho K., Faivre E., Pietrowski M.J., Marques X., Gomez-Murcia V., Deleau A., et al. Exacerbation of C1q dysregulation, synaptic loss and memory deficits in tau pathology linked to neuronal adenosine A2A receptor. Brain. 2019;142(11):3636–3654. doi: 10.1093/brain/awz288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M., Jung S.Y., Shaw C., Zhou X., Wong S.T., Qin J., et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z., Lichtenstein P., Asherson P.J., Larsson H. Developmental twin study of attention problems: high heritabilities throughout development. JAMA Psychiatry. 2013;70(3):311–318. doi: 10.1001/jamapsychiatry.2013.287. [DOI] [PubMed] [Google Scholar]
- Cheng Y., Bernstein A., Chen D., Jin P. 5-Hydroxymethylcytosine: a new player in brain disorders? Exp. Neurol. 2015;268:3–9. doi: 10.1016/j.expneurol.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.Z., Akbarian S., Tudor M., Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27(3):327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chen Y., Damayanti N.P., Irudayaraj J., Dunn K., Zhou F.C. Diversity of two forms of DNA methylation in the brain. Front Genet. 2014;5:46. doi: 10.3389/fgene.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomyk A.M., Volsko C., Tripathi A., Deckard S.A., Trapp B.D., Fox R.J., et al. DNA methylation in demyelinated multiple sclerosis hippocampus. Sci. Rep. 2017;7(1):1–10. doi: 10.1038/s41598-017-08623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra V., Quinti L., Kim J., Vollor L., Narayanan K.L., Edgerly C., et al. The sirtuin 2 inhibitor AK-7 is neuroprotective in Huntington’s disease mouse models. Cell Rep. 2012;2(6):1492–1497. doi: 10.1016/j.celrep.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters N., Dieriks B.V., Lill C., Faull R.L., Curtis M.A., Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer's disease human brain. Neurobiol. Aging. 2014;35(6):1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- Crawford B., Craig Z., Mansell G., White I., Smith A., Spaull S., et al. DNA methylation and inflammation marker profiles associated with a history of depression. Hum. Mol. Genet. 2018;27(16):2840–2850. doi: 10.1093/hmg/ddy199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 2019;51(1):63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Dai J. Advance in stress for depressive disorder. Depressive disorders: mechanisms. Meas. Manag. 2019:147–178. doi: 10.1007/978-981-32-9271-0_8. [DOI] [PubMed] [Google Scholar]
- Dolen E.K., McGinnis J.H., Tavory R.N., Weiss J.A., Switzer R.L. Disease-associated mutations G589A and V590F relieve replication focus targeting sequence-mediated autoinhibition of DNA methyltransferase 1. Biochemistry. 2019;58(51):5151–5159. doi: 10.1021/acs.biochem.9b00749. [DOI] [PubMed] [Google Scholar]
- Drzymalla E., Gladish N., Koen N., Epstein M., Kobor M., Zar H., et al. Association between maternal depression during pregnancy and newborn DNA methylation. medRxiv. 2021 doi: 10.1038/s41398-021-01697-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W. Targeting sirtuin-1 in Huntington’s disease: rationale and current status. CNS Drugs. 2013;27(5):345–352. doi: 10.1007/s40263-013-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Faraone S.V., Perlis R.H., Doyle A.E., Smoller J.W., Goralnick J.J., Holmgren M.A., et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Feng J., Fouse S., Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr. Res. 2007;61(7):58–63. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Fischer A. Targeting histone-modifications in Alzheimer's disease. What is the evidence that this is a promising therapeutic avenue? Neuropharmacology. 2014;80:95–102. doi: 10.1016/j.neuropharm.2014.01.038. [DOI] [PubMed] [Google Scholar]
- Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L., et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. USA. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel H.W., Kinde B., Stroud H., Gilbert C.S., Harmin D.A., Kastan N.R., et al. Disruption of DNA-methylation-dependent long gene repression in Rett syndrome. Nature. 2015;522(7554):89–93. doi: 10.1038/nature14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Emperle M., Guo Y., Grimm S.A., Ren W., Adam S., et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 2020;11(1):1–14. doi: 10.1038/s41467-020-17109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore J.L., Yi X., Quan L., Kabanov A.V. Novel nanomaterials for clinical neuroscience. J. NeuroImmune Pharmacol. 2008;3(2):83–94. doi: 10.1007/s11481-007-9099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin D., Allen M., Zhang L., Amorim M., Wang I.T., Reyes A.R., et al. Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat. Neurosci. 2011;15(2):274–283. doi: 10.1038/nn.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M.L., LaSalle J.M. The role of MeCP2 in brain development and neurodevelopmental disorders. Curr. Psychiatry Rep. 2010;12(2):127–134. doi: 10.1007/s11920-010-0097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goud Alladi C., Etain B., Bellivier F., Marie-Claire C. DNA methylation as a biomarker of treatment response variability in serious mental illnesses: a systematic review focused on bipolar disorder, schizophrenia, and major depressive disorder. Int. J. Mol. Sci. 2018;19(10):3026. doi: 10.3390/ijms19103026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D.R., Guidotti A. The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology. 2013;38(1):138–166. doi: 10.1038/npp.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greef J.C., Wang J., Balog J., Den Dunnen J.T., Frants R.R., Straasheijm K.R., et al. Mutations in ZBTB24 are associated with immunodeficiency, centromeric instability, and facial anomalies syndrome type 2. Am. J. Hum. Genet. 2011;88(6):796–804. doi: 10.1016/j.ajhg.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A., Auta J., Davis J.M., Gerevini V.D., Dwivedi Y., Grayson D.R., et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch. Gen. Psychiatry. 2000;57(11):1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- Guo J.U., Ma D.K., Mo H., Ball M.P., Jang M.-H., Bonaguidi M.A., et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011;14(10):1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wang W., Guo Y., Du X., Shi G., Lu C. Associations of FKBP5 polymorphisms and methylation and parenting style with depressive symptoms among Chinese adolescents. BMC Psychiatry. 2021;21(1):1–11. doi: 10.1186/s12888-021-03576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza M., Halayem S., Bourgou S., Daoud M., Charfi F., Belhadj A. Epigenetics and ADHD: toward an integrative approach of the disorder pathogenesis. J. Atten. Disord. 2019;23(7):655–664. doi: 10.1177/1087054717696769. [DOI] [PubMed] [Google Scholar]
- Han K.-M., Won E., Sim Y., Kang J., Han C., Kim Y.-K., et al. Influence of FKBP5 polymorphism and DNA methylation on structural changes of the brain in major depressive disorder. Sci. Rep. 2017;7(1):1–12. doi: 10.1038/srep42621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardikar S., Ying Z., Zeng Y., Zhao H., Liu B., Veland N., et al. The ZBTB24-CDCA7 axis regulates HELLS enrichment at centromeric satellite repeats to facilitate DNA methylation. Protein Cell. 2020;11(3):214–218. doi: 10.1007/s13238-019-00682-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison I.F., Dexter D.T. Epigenetic targeting of histone deacetylase: therapeutic potential in Parkinson's disease? Pharmacol. Ther. 2013;140(1):34–52. doi: 10.1016/j.pharmthera.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Henshall D.C., Kobow K. Epigenetics and epilepsy. Cold Spring Harb. Perspect. Med. 2015;5(12) doi: 10.1101/cshperspect.a022731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y., Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 2010;11(6):377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- Höhne N., Poidinger M., Merz F., Pfister H., Brückl T., Zimmermann P., et al. FKBP5 genotype-dependent DNA methylation and mRNA regulation after psychosocial stress in remitted depression and healthy controls. Int. J. Neuropsychopharmacol. 2015;18(4):pyu087. doi: 10.1093/ijnp/pyu087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes L., Jr, Shutman E., Chinaka C., Deepika K., Pelaez L., Dabney K.W. Aberrant epigenomic modulation of glucocorticoid receptor gene (NR3C1) in early life stress and major depressive disorder correlation: systematic review and quantitative evidence synthesis. Int. J. Environ. Res. Public Health. 2019;16(21):4280. doi: 10.3390/ijerph16214280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtman I.R., Skola D., Glass C.K. Transcriptional control of microglia phenotypes in health and disease. J. Clin. Investig. 2017;127(9):3220–3229. doi: 10.1172/JCI90604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huidobro C., Fernandez A.F., Fraga M.F. Aging epigenetics: causes and consequences. Mol. Asp. Med. 2013;34(4):765–781. doi: 10.1016/j.mam.2012.06.006. [DOI] [PubMed] [Google Scholar]
- Humphreys K.L., King L.S., Sacchet M.D., Camacho M.C., Colich N.L., Ordaz S.J., et al. Evidence for a sensitive period in the effects of early life stress on hippocampal volume. Dev. Sci. 2019;22(3) doi: 10.1111/desc.12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iridoy Zulet M., Pulido Fontes L., Ayuso Blanco T., Lacruz Bescos F., Mendioroz, Iriarte M. Epigenetic changes in neurology: DNA methylation in multiple sclerosis. Neurologia. 2017;32(7):463–468. doi: 10.1016/j.nrl.2015.03.011. [DOI] [PubMed] [Google Scholar]
- Iwata A., Nagata K., Hatsuta H., Takuma H., Bundo M., Iwamoto K., et al. Altered CpG methylation in sporadic Alzheimer's disease is associated with APP and MAPT dysregulation. Hum. Mol. Genet. 2014;23(3):648–656. doi: 10.1093/hmg/ddt451. [DOI] [PubMed] [Google Scholar]
- Jiang H., Poirier M.A., Liang Y., Pei Z., Weiskittel C.E., Smith W.W., et al. Depletion of CBP is directly linked with cellular toxicity caused by mutant huntingtin. Neurobiol. Dis. 2006;23(3):543–551. doi: 10.1016/j.nbd.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Jin Z., Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5(1):1–8. doi: 10.1016/j.gendis.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B., Robertson K.D. DNA methyltransferases, DNA damage repair, and cancer. Epigenetic Alter. Oncog. 2013:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Jowaed A., Schmitt I., Kaut O., Wüllner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients' brains. J. Neurosci. 2010;30(18):6355–6359. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juruena M.F., Gadelrab R., Cleare A.J., Young A.H. Epigenetics: a missing link between early life stress and depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2021;109 doi: 10.1016/j.pnpbp.2020.110231. [DOI] [PubMed] [Google Scholar]
- Kallak T.K., Bränn E., Fransson E., Johansson Å., Lager S., Comasco E., et al. DNA methylation in cord blood in association with prenatal depressive symptoms. Clin. epigenetics. 2021;13(1):1–14. doi: 10.1186/s13148-021-01054-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K.-S. Epigenetic regulations in adult stem cells: the role of DNA methyltransferase in stem cell aging. Epigenomics. 2011;3(6):671–673. doi: 10.2217/epi.11.92. [DOI] [PubMed] [Google Scholar]
- Kanherkar R.R., Bhatia-Dey N., Csoka A.B. Epigenetics across the human lifespan. Front. Cell Dev. Biol. 2014;2:49. doi: 10.3389/fcell.2014.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi-Nazarabad M., Goharshadi E.K., Entezari M.H., Nancarrow P. Rheological properties of the nanofluids of tungsten oxide nanoparticles in ethylene glycol and glycerol. Microfluid. Nanofluidics. 2015;19(5):1191–1202. [Google Scholar]
- Kaur G., Rathod S.S.S., Ghoneim M.M., Alshehri S., Ahmad J., Mishra A., et al. DNA methylation: a promising approach in management of alzheimer’s disease and other neurodegenerative disorders. Biology. 2022;11(1):90. doi: 10.3390/biology11010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keitel C., Benwell C.S., Thut G., Gross J. No changes in parieto‐occipital alpha during neural phase locking to visual quasi‐periodic theta‐, alpha‐, and beta‐band stimulation. Eur. J. Neurosci. 2018;48(7):2551–2565. doi: 10.1111/ejn.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B., Bottiglieri T., Arning E., Ziegler M., Hansen L., Masliah E. Elevated S-adenosylhomocysteine in Alzheimer brain: influence on methyltransferases and cognitive function. J. Neural Transm. 2004;111(4):547–567. doi: 10.1007/s00702-003-0096-5. [DOI] [PubMed] [Google Scholar]
- Kernohan K.D., Schenkel L.C., Huang L., Smith A., Pare G., Ainsworth P., et al. Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clin. Epigenet. 2016;8(1):1–6. doi: 10.1186/s13148-016-0254-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kaang B.-K. Epigenetic regulation and chromatin remodeling in learning and memory. Exp. Mol. Med. 2017;49(1) doi: 10.1038/emm.2016.140. e281-e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C.J., Botuyan M.-V., Wu Y., Ward C.J., Nicholson G.A., Hammans S., et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet. 2011;43(6):595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Bird A.P. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J. Biol. Chem. 2004;279(45):46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S., Bird A. DNA methylation and Rett syndrome. Hum. Mol. Genet. 2003;12 doi: 10.1093/hmg/ddg286. Spec No 2:R221-7. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupinski J., Carrera C., Muiño E., Torres N., Al-Baradie R., Cullell N., et al. DNA methylation in stroke. Update of latest advances. Comput. Struct. Biotechnol. J. 2018;16:1–5. doi: 10.1016/j.csbj.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari D., Sciascia N., Usdin K. Small molecules targeting H3K9 methylation prevent silencing of reactivated FMR1 alleles in fragile X syndrome patient derived cells. Genes. 2020;11(4):356. doi: 10.3390/genes11040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgrave-Gómez J., Mercado-Gómez O., Guevara-Guzmán R. Epigenetic mechanisms in neurological and neurodegenerative diseases. Front. Cell. Neurosci. 2015;9(58) doi: 10.3389/fncel.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber S.L., Llenos I.C., Miller C.L., Dulay J.R., Haybaeck J., Weis S. Homer1a protein expression in schizophrenia, bipolar disorder, and major depression. J. Neural Transm. 2017;124(10):1261–1273. doi: 10.1007/s00702-017-1776-x. [DOI] [PubMed] [Google Scholar]
- Liu S., Du T., Liu Z., Shen Y., Xiu J., Xu Q. Inverse changes in L1 retrotransposons between blood and brain in major depressive disorder. Sci. Rep. 2016;6(1):1–10. doi: 10.1038/srep37530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Barres B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018;18(4):225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- Li M., D’Arcy C., Li X., Zhang T., Joober R., Meng X. What do DNA methylation studies tell us about depression? A systematic review. Transl. Psychiatry. 2019;9(1):1–14. doi: 10.1038/s41398-019-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang T., Chen S., Yue Y., Xu Z., Yuan Y. DNA methylations of brain-derived neurotrophic factor exon VI are associated with major depressive disorder and antidepressant-induced remission in females. J. Affect. Disord. 2021;295:101–107. doi: 10.1016/j.jad.2021.08.016. [DOI] [PubMed] [Google Scholar]
- Lunnon K., Smith R., Hannon E., De Jager P.L., Srivastava G., Volta M., et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer's disease. Nat. Neurosci. 2014;17(9):1164–1170. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Liu X., Deng Y., Qing H. DNA methylation, a hand behind neurodegenerative diseases. Front Aging Neurosci. 2013;5:85. doi: 10.3389/fnagi.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Liu X., Deng Y., Qing H. DNA methylation, a hand behind neurodegenerative diseases. Front. Aging Neurosci. 2013;5:85. doi: 10.3389/fnagi.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Iglesias O., Carrera I., Carril J.C., Fernández-Novoa L., Cacabelos N., Cacabelos R. DNA Methylation in Neurodegenerative and Cerebrovascular Disorders. Int J. Mol. Sci. 2020;21(6) doi: 10.3390/ijms21062220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattei A.L., Bailly N., Meissner A. DNA methylation: a historical perspective. Trends Genet. 2022 doi: 10.1016/j.tig.2022.03.010. [DOI] [PubMed] [Google Scholar]
- Mehler M.F. Epigenetics and the nervous system. Ann. Neurol.: Off. J. Am. Neurol. Assoc. Child Neurol. Soc. 2008;64(6):602–617. doi: 10.1002/ana.21595. [DOI] [PubMed] [Google Scholar]
- Meijer M., Klein M., Hannon E., Van der Meer D., Hartman C., Oosterlaan J., et al. Genome-wide DNA methylation patterns in persistent attention-deficit/hyperactivity disorder and in association with impulsive and callous traits. Front. Genet. 2020;11:16. doi: 10.3389/fgene.2020.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J., Tang T., Kaminsky Z., Khare T., Yazdanpanah S., Bouchard L., et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82(3):696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mil N.H., Steegers-Theunissen R.P., Bouwland-Both M.I., Verbiest M.M., Rijlaarsdam J., Hofman A., et al. DNA methylation profiles at birth and child ADHD symptoms. J. Psychiatr. Res. 2014;49:51–59. doi: 10.1016/j.jpsychires.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Misiak B., Ricceri L., Sąsiadek M.M. Transposable elements and their epigenetic regulation in mental disorders: current evidence in the field. Front. Genet. 2019;10:580. doi: 10.3389/fgene.2019.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney M.A., Ryabinin P., Wilmot B., Bhatt P., Mill J., Nigg J.T. Large epigenome-wide association study of childhood ADHD identifies peripheral DNA methylation associated with disease and polygenic risk burden. Transl. Psychiatry. 2020;10(1):1–12. doi: 10.1038/s41398-020-0710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L.D., Le T., Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C., Patchev A.V., Wu Y., Micale V., Bockmühl Y., Fischer D., et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Na E.S., Monteggia L.M. The role of MeCP2 in CNS development and function. Horm. Behav. 2011;59(3):364–368. doi: 10.1016/j.yhbeh.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann A., Walton E., Alemany S., Cecil C., González J.R., Jima D.D., et al. Association between DNA methylation and ADHD symptoms from birth to school age: a prospective meta-analysis. Transl. Psychiatry. 2020;10(1):1–11. doi: 10.1038/s41398-020-01058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen S., Meletis K., Fu D., Jhaveri S., Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn.: Off. Publ. Am. Assoc. Anat. 2007;236(6):1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- Ng H.-H., Zhang Y., Hendrich B., Johnson C.A., Turner B.M., Erdjument-Bromage H., et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999;23(1):58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308(5726):1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Nussbaum R.L., Ellis C.E. Alzheimer's disease and Parkinson's disease. N. Engl. J. Med. 2003;348(14):1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- Nuzziello N., Liguori M. Medical Epigenetics. Elsevier,; 2021. Pharmacoepigenomics in neurodegenerative diseases; pp. 559–581. [Google Scholar]
- Ogino S., Nosho K., Shima K., Baba Y., Irahara N., Kirkner G.J., et al. p21 expression in colon cancer and modifying effects of patient age and body mass index on prognosis. Cancer Epidemiol. Biomark. Prev. 2009;18(9):2513–2521. doi: 10.1158/1055-9965.EPI-09-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y.S., Kim S.H., Cho G.-W. Functional restoration of amyotrophic lateral sclerosis patient-derived mesenchymal stromal cells through inhibition of DNA methyltransferase. Cell. Mol. Neurobiol. 2016;36(4):613–620. doi: 10.1007/s10571-015-0242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K., Niida S. Genetic Background for Alzheimer's Disease: Knowledge Accumulated from AD GWAS. Brain Nerve. 2019;71(10):1039–1051. doi: 10.11477/mf.1416201403. [DOI] [PubMed] [Google Scholar]
- Palladino V.S., McNeill R., Reif A., Kittel-Schneider S. Genetic risk factors and gene–environment interactions in adult and childhood attention-deficit/hyperactivity disorder. Psychiatr. Genet. 2019;29(3):63–78. doi: 10.1097/YPG.0000000000000220. [DOI] [PubMed] [Google Scholar]
- Perroud N., Zewdie S., Stenz L., Adouan W., Bavamian S., Prada P., et al. Methylation of serotonin receptor 3A in ADHD, borderline personality, and bipolar disorders: link with severity of the disorders and childhood maltreatment. Depress Anxiety. 2016;33(1):45–55. doi: 10.1002/da.22406. [DOI] [PubMed] [Google Scholar]
- Petralla S., De Chirico F., Miti A., Tartagni O., Massenzio F., Poeta E., et al. Epigenetics and communication mechanisms in microglia activation with a view on technological approaches. Biomolecules. 2021;11(2):306. doi: 10.3390/biom11020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plazas-Mayorca M.D., Vrana K.E. Proteomic investigation of epigenetics in neuropsychiatric disorders: a missing link between genetics and behavior? J. Proteome Res. 2011;10(1):58–65. doi: 10.1021/pr100463y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Jho E.H. A concise review of human brain methylome during aging and neurodegenerative diseases. BMB Rep. 2019;52(10):577–588. doi: 10.5483/BMBRep.2019.52.10.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk E., Defossez P.-A. The cell biology of DNA methylation in mammals. Biochim. Et. Biophys. Acta (BBA)-Mol. Cell Res. 2008;1783(11):2167–2173. doi: 10.1016/j.bbamcr.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Qazi T.J., Quan Z., Mir A., Qing H. Epigenetics in Alzheimer’s disease: perspective of DNA methylation. Mol. Neurobiol. 2018;55(2):1026–1044. doi: 10.1007/s12035-016-0357-6. [DOI] [PubMed] [Google Scholar]
- Qureshi I.A., Mehler M.F. Epigenetic mechanisms governing the process of neurodegeneration. Mol. Asp. Med. 2013;34(4):875–882. doi: 10.1016/j.mam.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reszka E., Jabłońska E., Lesicka M., Wieczorek E., Kapelski P., Szczepankiewicz A., et al. An altered global DNA methylation status in women with depression. J. Psychiatr. Res. 2021;137:283–289. doi: 10.1016/j.jpsychires.2021.03.003. [DOI] [PubMed] [Google Scholar]
- Re F., Gregori M., Masserini M. Nanotechnology for neurodegenerative disorders. Maturitas. 2012;73(1):45–51. doi: 10.1016/j.maturitas.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Rovira P., Demontis D., Sánchez-Mora C., Zayats T., Klein M., Mota N.R., et al. Shared genetic background between children and adults with attention deficit/hyperactivity disorder. Neuropsychopharmacology. 2020;45(10):1617–1626. doi: 10.1038/s41386-020-0664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Smith K., Camelo S.I., Carreras I., Lee J., Iglesias A.H., et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti‐apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J. Neurochem. 2005;93(5):1087–1098. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- Sagarkar S., Bhat N., Sapre M., Dudhabhate B., Kokare D.M., Subhedar N.K., et al. TET1-induced DNA demethylation in dentate gyrus is important for reward conditioning and reinforcement. Mol. Neurobiol. 2022:1–17. doi: 10.1007/s12035-022-02917-0. [DOI] [PubMed] [Google Scholar]
- Sales A.J., Guimarães F.S., Joca S.R. DNA methylation in stress and depression: from biomarker to therapeutics. Acta Neuropsychiatr. 2021;33(5):217–241. doi: 10.1017/neu.2021.18. [DOI] [PubMed] [Google Scholar]
- Sanwald S., Widenhorn-Müller K., Schönfeldt-Lecuona C., Montag C., Kiefer M. Factors related to age at depression onset: the role of SLC6A4 methylation, sex, exposure to stressful life events and personality in a sample of inpatients suffering from major depression. BMC Psychiatry. 2021;21(1):1–14. doi: 10.1186/s12888-021-03166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiele M.A., Zwanzger P., Schwarte K., Arolt V., Baune B.T., Domschke K. Serotonin transporter gene promoter hypomethylation as a predictor of antidepressant treatment response in major depression: a replication study. Int. J. Neuropsychopharmacol. 2021;24(3):191–199. doi: 10.1093/ijnp/pyaa081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T., Hirschey M.D., Newman J., He W., Shirakawa K., Le Moan N., et al. Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339(6116):211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Locasale J.W., Lyssiotis C.A., Zheng Y., Teo R.Y., Ratanasirintrawoot S., et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdardottir H., Kranz G., Rami-Mark C., James G., Vanicek T., Gryglewski G., et al. Association of norepinephrine transporter methylation with in vivo NET expression and hyperactivity–impulsivity symptoms in ADHD measured with PET. Mol. Psychiatry. 2021;26(3):1009–1018. doi: 10.1038/s41380-019-0461-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalingam K., Samikkannu T. Neuroprotective effect of piracetam against cocaine-induced neuro epigenetic modification of DNA methylation in astrocytes. Brain Sci. 2020;10(9):611. doi: 10.3390/brainsci10090611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker C., Armstrong N.J. Twin and Family Studies of Epigenetics. Elsevier,; 2021. Evaluation and measurement of epigenetic modifications in population-based studies; pp. 17–39. [Google Scholar]
- Suchy J., Lee S., Ahmed A., Shea T.B. Dietary supplementation with S-adenosyl methionine delays the onset of motor neuron pathology in a murine model of amyotrophic lateral sclerosis. Neuromolecular Med. 2010;12(1):86–97. doi: 10.1007/s12017-009-8089-7. [DOI] [PubMed] [Google Scholar]
- Sun L., Gu X., Pan Z., Guo X., Liu J., Atianjoh F.E., et al. Contribution of DNMT1 to neuropathic pain genesis partially through epigenetically repressing Kcna2 in primary afferent neurons. J. Neurosci. 2019;39(33):6595–6607. doi: 10.1523/JNEUROSCI.0695-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Verkaik-Schakel R.-N., Biber K., Plösch T., Serchov T. Antidepressant treatment is associated with epigenetic alterations of Homer1 promoter in a mouse model of chronic depression. J. Affect. Disord. 2021;279:501–509. doi: 10.1016/j.jad.2020.10.040. [DOI] [PubMed] [Google Scholar]
- Sun Z., Wu Y., Ordog T., Baheti S., Nie J., Duan X., et al. Aberrant signature methylome by DNMT1 hot spot mutation in hereditary sensory and autonomic neuropathy 1E. Epigenetics. 2014;9(8):1184–1193. doi: 10.4161/epi.29676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach K.E., Li X., Li Y., Song C.-X., Wu H., Dai Q., et al. 5-hmC–mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toikumo S., Malan-Muller S., Swart P., Womersley J., van den Heuvel L., Tromp G., et al., editors. Epigenome-Wide Association Study on Posttraumatic Stress Disorder Diagnosis and Metabolic Syndrome in a South African Population. European Neuropsychopharmacology; 2019: Elsevier RADARWEG 29, 1043 NX AMSTERDAM, NETHERLANDS.
- Urdinguio R.G., Sanchez-Mut J.V., Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8(11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Veremeyko T., Yung A.W.Y., Dukhinova M., Strekalova T., Ponomarev E.D. The role of neuronal factors in the epigenetic reprogramming of microglia in the normal and diseased central nervous system. Front. Cell. Neurosci. 2019:13. doi: 10.3389/fncel.2019.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li W., Wu Y., Tian X., Duan H., Li S., et al. Genome-wide DNA methylation and gene expression analyses in monozygotic twins identify potential biomarkers of depression. Transl. Psychiatry. 2021;11(1):1–10. doi: 10.1038/s41398-021-01536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-C., Oelze B., Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PloS One. 2008;3(7) doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold B. National Institute of Environmental Health Sciences; 2006. Epigenetics: the science of change. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Zhang L., Zeng Y. DNA methylation in Alzheimer's disease: in brain and peripheral blood. Mech. Ageing Dev. 2020;191 doi: 10.1016/j.mad.2020.111319. [DOI] [PubMed] [Google Scholar]
- Weiß A.L., Meijer M., Budeus B., Pauper M., Hakobjan M., Groothuismink J., et al. DNA methylation associated with persistent ADHD suggests TARBP1 as novel candidate. Neuropharmacology. 2021;184 doi: 10.1016/j.neuropharm.2020.108370. [DOI] [PubMed] [Google Scholar]
- Weng Y.-L., An R., Shin J., Song H. Ming G-l. DNA Modif. Neurol. Disord. Neurother. 2013;10(4):556–567. doi: 10.1007/s13311-013-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.H., Ross L. Consequences of prenatal toxin exposure for mental health in children and adolescents. Eur. Child Adolesc. Psychiatry. 2007;16(4):243–253. doi: 10.1007/s00787-006-0596-6. [DOI] [PubMed] [Google Scholar]
- Xiang D., Sun S., Wang G., Liu Z. Effects of CRMP2 DNA methylation in the hippocampus on depressive-like behaviors and cytoskeletal proteins in rats. Front. Cell. Neurosci. 2021;15:77. doi: 10.3389/fncel.2021.644663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Li X. DNA methylation in neurodegenerative disorders. Curr. Geriatr. Rep. 2012;1(4):199–205. [Google Scholar]
- Xu Y., Xu Y., Wang Y., Wang Y., He L., Jiang Z., et al. Telmisartan prevention of LPS-induced microglia activation involves M2 microglia polarization via CaMKKβ-dependent AMPK activation. Brain, Behav., Immun. 2015;50:298–313. doi: 10.1016/j.bbi.2015.07.015. [DOI] [PubMed] [Google Scholar]
- Yamagata H., Ogihara H., Matsuo K., Uchida S., Kobayashi A., Seki T., et al. Distinct epigenetic signatures between adult-onset and late-onset depression. Sci. Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-81758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui D.H., Peddada S., Bieda M.C., Vallero R.O., Hogart A., Nagarajan R.P., et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc. Natl. Acad. Sci. 2007;104(49):19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan W., Li Y., Yuan J., Zhi N., Huang Y., Liu Y., et al. New Insights into TETs in psychiatric disorders. Int. J. Mol. Sci. 2022;23(9):4909. doi: 10.3390/ijms23094909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.-Y., Liang L., Gu X., Li Z., Wu S., Sun L., et al. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat. Commun. 2017;8(1):1–15. doi: 10.1038/ncomms14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Fan W., Zhang X., Dong E. Gestational stress induces depressive-like and anxiety-like phenotypes through epigenetic regulation of BDNF expression in offspring hippocampus. Epigenetics. 2016;11(2):150–162. doi: 10.1080/15592294.2016.1146850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li M., Wang X., He Y., Xia Y., Sweeney J.A., et al. Drug response-related DNA methylation changes in schizophrenia, bipolar disorder, and major depressive disorder. Front. Neurosci. 2021:15. doi: 10.3389/fnins.2021.674273. [DOI] [PMC free article] [PubMed] [Google Scholar]