Abstract
One challenge in the engineering of biological systems is to be able to recognise the cellular stress states of bacterial hosts, as these stress states can lead to suboptimal growth and lower yields of target products. To enable the design of genetic circuits for reporting or mitigating the stress states, it is important to identify a relatively reduced set of gene biomarkers that can reliably indicate relevant cellular growth states in bacteria. Recent advances in high-throughput omics technologies have enhanced the identification of molecular biomarkers specific states in bacteria, motivating computational methods that can identify robust biomarkers for experimental characterisation and verification. Focused on identifying gene expression biomarkers to sense various stress states in Bacillus subtilis, this study aimed to design a knowledge integration strategy for the selection of a robust biomarker panel that generalises on external datasets and experiments. We developed a recommendation system that ranks the candidate biomarker panels based on complementary information from machine learning model, gene regulatory network and co-expression network. We identified a recommended biomarker panel showing high stress sensing power for a variety of conditions both in the dataset used for biomarker identification (mean f1-score achieved at 0.99), as well as in a range of independent datasets (mean f1-score achieved at 0.98). We discovered a significant correlation between stress sensing power and evaluation metrics such as the number of associated regulators in a B. subtilis gene regulatory network (GRN) and the number of associated modules in a B. subtilis co-expression network (CEN). GRNs and CENs provide information relevant to the diversity of biological processes encoded by biomarker genes. We demonstrate that quantitatively relating meaningful evaluation metrics with stress sensing power has the potential for recognising biomarkers that show better sensitivity and robustness to an extended set of stress conditions and enable a more reliable biomarker panel selection.
Keywords: Transcriptomics analysis, System biology, Biomarker discovery, Machine learning
1. Introduction
Bioengineering applications often use bacteria as the host organisms to create high-value products such as pharmaceuticals, biofuels, fine chemicals, etc. [1]. While grown under optimal user-defined conditions, bacteria still undergo periods of cellular stresses that may lead to suboptimal growth and lower yields of target products [[2], [3], [4]]. One of the goals of the biotechnology industry is to recognise these detrimental cellular states so that strategies for mitigating the damage can be engineered [5,6]. Detrimental states can be characterised and recognised using a variety of techniques, from morphological and biochemical assays [7,8] to omics methods [9]. In the context of omics methods, panels of genes whose expression are indicative of certain cellular states can be used as transcriptional biomarkers.
By measuring the expression of a few key biomarker genes using amplicon panels [10,11] or qPCR [12,13], instead of characterising the global transcription using genome-scale RNA-seq or Microarray, the cellular states can be assayed at reduced costs. Moreover, while these sequencing measures are not easily carried out in real-time, having cellular state biomarkers is particularly attractive for single-cell measurement technologies that seek to develop ‘live cell’ biosensors using flow cytometry [14,15] or microfluidics systems [[16], [17], [18]]. These ‘live cell’ biosensors require the biomarker panels to be small, consisting of only a few key genes. Small biomarker panels are essential because live monitoring of cellular state currently relies on the use of reporter genes, for which a limited number of distinct systems are available.
Transcriptomic methods such as microarrays and RNA-seq are used to reveal gene expression patterns under different environmental conditions. These experiments can provide the genome-scale data necessary to identify transcriptional biomarkers for distinguishing physiological and biochemical states in a bacterial cell. Statistical tests such as differential expression analysis have discovered the biomarker genes indicative of certain stress states for model bacteria used in the biotechnology industry, e.g., Escherichia coli [19,20] and Bacillus subtilis [21,22]. Other biomarker studies in biomedical domains [[23], [24], [25]] have exploited the massive transcriptomics data by applying machine learning methods such as classification and feature selection models [26]. We recently proposed a pipeline of machine learning models to extract diverse cellular states from condition-dependent transcriptomics data and identify gene biomarkers that can classify different stress states for B. subtilis [27].
Transcriptomics data are high dimensional, with the number of features vastly exceeding the number of samples. Hence it is likely to discover multiple sets of gene features that can potentially serve as biomarkers to discriminate samples in groups of interest. To assess these candidate biomarker panels, most studies on biomarker discovery have applied a single criterion, e.g., fold change in differential expression analysis [28] or cross-validation performance in machine learning model [29]. Our previous study [27] highlighted a minimal biomarker panel that achieved the highest performance in discriminating different cellular states for B. subtilis. However, individual scoring criteria are inherently biased as they capture limited information. A good biomarker panel for an in vivo validation should present multiple properties, for example, good discriminative capacity between cellular states and high relevance in the gene regulatory control of the organism. Therefore, there is a need for better panel selection methods that can rank candidate biomarker panels by integrating a variety of data-driven and domain-based criteria, to select a robust and reproducible biomarker panel.
One source of information that can be used to assess biomarkers, but has often been neglected, are models of gene regulation such as gene regulatory networks (GRN). A gene regulatory network is a collection of molecular species and their interactions, which together govern gene expression levels of mRNA and control gene-product abundance. GRNs play an important role in all kinds of cellular processes, including cell cycle, metabolism, and signal transduction. A well-curated GRN has already been described for B. subtilis [30]. This GRN has been rigorously validated experimentally and includes thousands of genes and hundreds of regulatory factors. GRNs such as this can reveal the regulatory circuits that are modulated by the identified biomarker genes, or which can, in-turn, regulate biomarker gene expression. By mapping biomarkers in a GRN it is possible to assess the diversity of cellular processes that the biomarkers are involved in.
As our current knowledge of regulatory interactions within the cell is still limited, co-expression networks (CEN), which are inferred from the correlations of expression patterns across different conditions [31,32] can also provide additional information about transcriptional relationships between genes and their products. By grouping genes that are highly interconnected we can identify modules of genes with similar functions and relate the genes of unknown function to well-studied genes [33]. Therefore, CEN can be used to study the functional diversity of biomarkers as a supplementary method to complement GRNs.
We hypothesised that to identify a robust biomarker panel, a multimodal approach combining prior known information about gene interaction and co-expression with computational data-driven methods is crucial. We designed a two-step approach to rank and select a robust biomarker panel comprising key genes. Firstly, we applied the Bacillus subtilis biomarker identification model (BIM), which we developed previously, to obtain a pool of candidate biomarker panels with satisfactorily high performance in predicting the stress state of a sample. Secondly, for each biomarker panel we integrated complementary information from BIM, GRN and CEN, developing a recommendation system to identify the biomarker panel with a few key genes to sense stress conditions. We successfully validated the robustness of the recommended panel on nine external datasets covering 10 stress conditions. These findings suggest that our in silico biomarker recommendation system, integrating multi-source knowledge and data-driven techniques, can facilitate in vitro experiments to sense cellular states for monitoring stress conditions in B. subtilis.
2. Material and methods
2.1. Datasets and data processing
For discovering biomarker panels, we used a tiling array dataset [34] that assays the transcriptomes of B. subtilis strain BSB1 measured under diverse conditions, including alternative nutrient shifts, lifestyle changes and adaptation to various stimuli. The wide range of conditions can lead to distinct transcriptional states in bacteria. Some of these conditions can cause bacterial stress, enabling the identification of biomarker genes specific to different stress states. Although, more recently, RNA-seq technology has also generated many transcriptomics datasets containing different conditions respectively, the integration of these small datasets was challenging since they are experimentally more diverse and would introduce between-experiment noise when included. The unified tiling array dataset from Nicolas and co-workers (referred to as the Nicolas dataset) was preferable for exploring the transcriptional profiles across conditions as between-experiment noises were minimised by conducting all experiments following standard operating procedures and estimating the gene expression quantities using the same signal processing protocols. The experimental procedures and signal processing protocols are described in Supplementary Material SOM1-2 from Ref. [34].
Nicolas and co-workers computed an aggregate expression index for each of 5875 transcribed regions as the median log 2 expression signal intensity of probes lying entirely within the corresponding region. These expression values were pre-processed with quantile normalisation that makes the data distribution across samples identical for reducing the between-sample variations. The raw data in Gene Expression Omnibus (GSE27219) and pre-processed data was made available at:
http://genome.jouy.inra.fr/basysbio/bsubtranscriptome.
To enable the discovery of relevant cellularlieing states using unsupervised machine learning, we further processed data using the processing steps as elaborately explained in our previous work [27]. Briefly, these steps included: a) filtering genes that were invariant across conditions; b) removing genes and samples related to late-stage sporulation conditions, as sporulation produces a very strong transcriptional response across a large number of genes that would mask many other cellular states we are interested in capturing; c) normalising the expression quantities by subtracting the corresponding reference conditions within each experiment; to produce a processed condition-dependent gene expression data (Fig. 1a) for downstream analysis. The data processed using the above steps, containing 2536 genes and 180 samples, can be downloaded from:
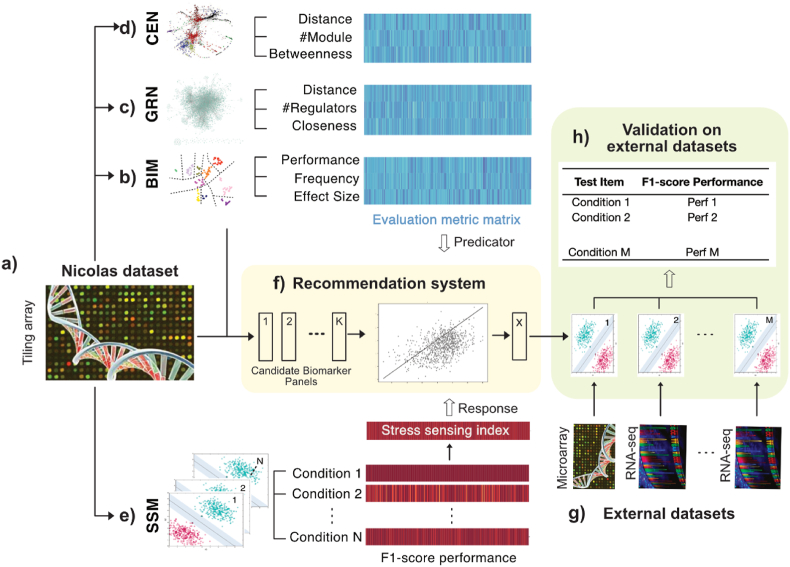
Fig. 1.
Overall approach. a) Nicolas dataset, which measures condition-dependent gene expression profiles using Tiling array technology, is used for biomarker identification and optimisation. b) A Biomarker Identification Model (BIM) is used to identify a pool of candidate biomarker panels and extract biomarker evaluation metrics: Performance, Frequency, Effect Size. c) A Gene Regulatory Network (GRN) is used to extract biomarker evaluation metrics: Distance, number of Regulator, Closeness. d) A Co-Expression Network (CEN) is used to extract biomarker evaluation metrics: Distance, number of Module, Betweenness. e) A Stress Sensing model (SSM) produces a stress sensing index for each candidate biomarker panel. The stress sensing index is calculated as the average f1-score performance on predicting N stress conditions in the Condition-dependent gene expression data. f) The recommendation system takes a set of candidate biomarker panels as input and recommends an optimal biomarker panel. This recommendation system is trained to relate evaluation metrics with the stress sensing index. g) A set of external datasets, collected in different centres with RNA-seq and Microarray technologies, are used for validation. h) Validation on external datasets is performed by assessing the performance of the recommended biomarker panel predicting M conditions included in the condition-specific gene expression datasets.
https://github.com/neverbehym/biomaker-recommendation-system.
We also used nine condition-specific gene expression datasets (Fig. 1g) for the validation of the biomarker identification model. These external datasets, independent from Nicolas dataset used for model training, were generated over past years using RNA-seq data or Microarray data in multi-centres including our own laboratory. Each external datasets consisted of mRNA samples collected under a test condition treated with a specific environmental stress perturbation as opposed to a control condition without the treatment. The details of experimental conditions and Gene Expression Omnibus (GEO) session IDs for all datasets used in this study can be found in Supplementary Table 1. While most tests conditions in the external datasets were seen in Nicolas dataset, some conditions were new and not studied in Nicolas dataset. The selection of these external datasets allowed us to validate the generalisability of the identified biomarker panel on data collected by different technologies, which include independent samples grown in conditions present in the training data and in additional conditions not present in the training data.
2.2. Overall approach
We proposed a in silico pipeline (Fig. 1) to select a small and robust biomarker panel to sense a variety of stress states for B. subtilis. We used the Nicolas dataset (Fig. 1a) for training a recommendation system and a set of external datasets (Fig. 1g) for validation of the recommended biomarker panel. Nine evaluation metrics were derived from a Biomarker Identification Model (Fig. 1b), a Gene Regulatory Network (Fig. 1c) and a Co-Expression Network (Fig. 1d), and a stress sensing index was calculated by applying a Stress Sensing Model (Fig. 1e). The recommendation system (Fig. 1f) can select a biomarker panel from a pool of candidate panels by ranking the panels based on the evaluation metrics and coupling these evaluation metrics with the stress sensing index. We reported the performance of the selected biomarker panel to predict an extended list of conditions from external datasets for validation (Fig. 1h).
2.3. Biomarker identification model
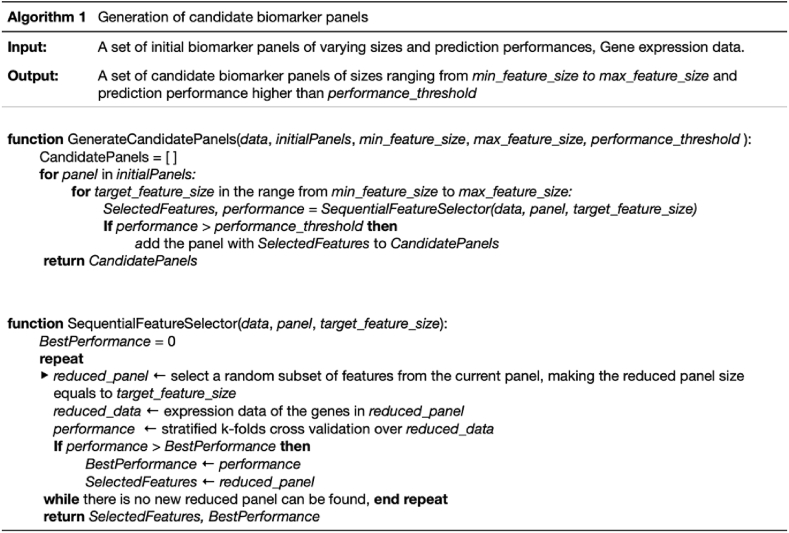
In our previous study [27], we proposed a Biomarker Identification Model (BIM), which was applied to the Nicolas dataset to discover biomarker panels indicative of the cellular states for B. subtilis. This BIM first discovers different cellular states introduced by a wide range of conditions using UMAP dimension reduction [35] and Leiden clustering [36] methods. The BIM then identifies panels of biomarkers indicative of these cellular states using our RGIFE model [37], a recursive feature elimination-style feature selection algorithm. Here we improved the BIM to enable the identification of a sufficient number of biomarker panels of small size and high predictive performance by performing an additional feature elimination and evaluation process (Algorithm 1). On each of the 500 biomarker panels of different sizes and prediction performances we identified from our previous work [27], we iteratively removed individual genes while optimising cross-validation performance until reaching the desired panel size ranging from min_feature_size to max_feature_size. Here the cross-validation performance was computed as the micro f1-score in 10-folds stratified cross-validation test. We set min_feature_size as 5, which was the minimal size of the initial 500 biomarker panels returned by RGIFE. We set max_feature_size as 10 as this was the smallest number of genes required to achieve a f1-score of 1. We retained the panels with cross-validation performance more than performance_threshold of 0.9 to ensure a high prediction capacity for all candidate biomarker panels. This enabled as many as 949 candidate panels for the evaluation of our biomarker recommendation methods.
We extracted three metrics from this refined BIM to evaluate candidate biomarker panels (Fig. 1b): a) Performance is used to measure the prediction performance of a given biomarker panel on distinct cellular states in the Biomarker Identification Model. It is calculated as 10-folds cross-validation f1-score for predicting ten cellular states with Random Forest classifier. b) Frequency reflects the consistency of a biomarker panel being selected across multiple repetitions of running BIM starting from different random states. As a high-throughput dataset tends to give false positive results, a biomarker panel with a higher Frequency value is expected to have less chance of being spurious signals. We first computed, for each gene, the frequency of appearing in the pool of candidate biomarker panels and then calculated Frequency as the average frequency values across all genes in a panel. c) Effect Size evaluates the overall strength of differential expressions in the biomarkers between distinct cellular states. A biomarker panel with a higher Effect Size value will likely make a reporter system with stronger signals. It is calculated as the average value of for all genes in each biomarker panel. As defined in Equation (1), is used to compute Cohen's d score, which measures the standardised difference in biomarker expression between two cellular states i and j.
| (1) |
2.4. Gene regulatory network
To assess the diversity of cellular processes in which the biomarkers are involved with prior knowledge, we curated a gene regulatory network for B. subtilis by incorporating the network reconstructed by Faria and co-workers [30] with the latest Subtiwiki regulon database (http://subtiwiki.uni-goettingen.de/v3/regulation/export). This network consists of 6704 edges, indicating the interactions between 294 regulators and 2816 targeted genes under various mechanisms such as transcriptional factors, RNA switches, riboswitches, and small regulatory RNAs (Supplementary Table 2). The network comprises 35 components, with the largest one containing 2798 nodes.
We extracted three evaluation metrics from GRN (Fig. 1c): a) Number of regulators is the number of transcriptional regulators in GRN associated with the genes from a given biomarker panel. b) Distance indicates how distant any two genes from a given biomarker panel are in GRN. The distance between connected genes is calculated as the number of least hops in the network. The distance between unconnected genes is set as m+1, where m is the largest distance between any two connected genes in the network. c) Closeness is calculated as the average node closeness centrality across genes from a given biomarker panel in GRN.
2.5. Co-expression network
To study the functional diversity of biomarkers diagnostic of the transcriptional relationships discovered in GRNs, we constructed the co-expression network (CEN) that captures the expression similarity patterns across conditions. We applied weighted gene co-expression network analysis using the R package WGCNA [32]. The resulting network is a fully connected and weighted network, with nodes being studied genes and edges reflecting the similarity of gene expression profiles across various conditions. First, we computed the adjacency value as in Equation (2), where is similarity strength based on Pearson correlation and β is soft-thresholding power index. We tuned the parameter β = 4 to maximise the scale-free topology criterion [38]. Second, we identified modules, i.e., clusters of highly connected genes, with the Dynamic Tree Cut method [39]. We tuned the parameters detectCutHeight, mergeCutHeight, minModuleSize in blockwiseModules function to maximise the average Overlap score across all modules in a network. The Overlap score for a given module is calculated in Equation (3), where measures the overlap level between and a most concordant regulon in the GRN. We identified 55 modules with similar co-expression patterns as indicated by different colours, leaving 245 genes unassigned (grey). We summarised the profiles of these modules by studying the central genes with high intramodular connectivity, the highly overlapped regulons, and the overrepresented biological process in Gene Ontology. Please find the details in Supplementary Table 3.
| (2) |
| (3) |
We extracted three evaluation metrics from the CEN (Fig. 1d): The number of module (#Module) is the number of different modules in the CEN that are associated with the genes from a given biomarker panel. Distance measures the average distance between pairs of biomarker genes in the CEN. The cost of each edge is calculated as the inverse of co-expression strength, and the distance between two genes is set as the least sum of costs for the path connecting them. Betweenness is the average node betweenness centrality of biomarker genes in the CEN.
2.6. Stress sensing model
The Stress Sensing Model (SSM) is used to measure the overall performance of a biomarker panel in predicting stress states induced by a variety of test conditions (Fig. 1e). While BIM identifies biomarkers indicative of different cellular states revealed by applying unsupervised machine learning methods on condition-dependent transcriptomes, SSM assesses the biomarkers using the condition labels provided. We first applied the Linear Support Vector Machine (SVM) model to classify gene expression patterns of a biomarker under each test condition from their gene expression patterns under the corresponding control condition. We computed the cross-validation f1-score performance to reflect the single-stress sensing power of this biomarker panel. We then computed the stress sensing index that reflects the multi-stress sensing power as the average f1-score performance across all test conditions. We applied SMM to measure the multi-stress sensing power of a pool of candidate biomarker panels identified by BIM, assessing the overall prediction performance in 13 test conditions included in Nicolas dataset (Table 1a).
Table 1.
a) Conditions studied in the Nicolas datasets during the training process. b) Conditions studied in the external datasets during the validation process, in which some are similar to the conditions covered in the training dataset while some are different.
| a) | Test Conditions | Sample Size |
b) | Test Conditions | Covered in Nicolas dataset | Sample Size |
||
|---|---|---|---|---|---|---|---|---|
| Test | Control | Test | Control | |||||
| Nicolas dataset | Anaerobic | 6 | 3 | External datasets | Antibiotic (amphotericin, SynAnt49, YydF) | N | 9 | 9 |
| Antibiotic (Mitomycin) | 6 | 6 | ||||||
| Biofilm | 4 | 5 | Cold | Y | 5 | 4 | ||
| Cold | 6 | 6 | Deep Starvation | N | 3 | 4 | ||
| Germination | 8 | 6 | Glycine betaine | N | 6 | 6 | ||
| Heat | 6 | 6 | Hydroxyurea | N | 6 | 6 | ||
| Low motility | 8 | 6 | Heat | Y | 12 | 9 | ||
| Oxidative (Diamide, H2O2, Paraquat) | 15 | 6 | Oxidative (H2O2) | Y | 5 | 5 | ||
| Potassium | N | 4 | 4 | |||||
| Salinity | 6 | 6 | Pressure | N | 11 | 5 | ||
| Shift from glucose | 25 | 9 | Salinity | Y | 6 | 6 | ||
| Shift to glucose | 24 | 5 | ||||||
| Starvation | 9 | 8 | ||||||
| Stationary | 9 | 9 | ||||||
To mitigate the over-optimistic estimation of prediction performance due to limited dataset sizes, we performed 100 repetitions of the leave one out cross-validation and applied Synthetic Minority Over-sampling Technique (SMOTE) [40] to oversample the minority class to the same size as the majority class in the training process.
2.7. Recommendation system
We built a recommendation system (Fig. 1f) that takes a pool of candidate biomarker panels as input and produces a recommended panel as the output. There are four modules in the recommendation system, i.e., Biomarker Identification Model (BIM), Gene Regulatory Network (GRN), Co-Expression Network (CEN) and Stress Sensing Model (SSM) as described in sections 2.3, 2.4, 2.5, 2.6. We trained a regression model with Elastic Net regularisation to predict the stress sensing index of a biomarker panel with 9 evaluation metrics generated from BIM, GRN, CEN. Except for the GRN, all other modules used the Nicolas dataset as their source. The list of candidate biomarker panels, the evaluation metrics and stress sensing indices for all panels are provided in https://github.com/neverbehym/biomaker-recommendation-system/data. The Elastic Net Regression Model is illustrated in Equation (4), where α is the mixing parameter between L1 regularisation and L2 regularisation, is coefficient shrinkage parameter. We tuned the model parameters = 0.2, = 1e-4 to optimise the 10-folds cross-validation performance. A recommendation score was calculated as the predicted stress sensing index value (Equation (5)) for each candidate biomarker panel. The recommendation system then selects the panel with the highest recommendation scores as the optimal biomarker panel.
| (4) |
| (5) |
To understand the respective impact of each evaluation metric on the recommendation system, we calculated the SHapley Additive exPlanations (SHAP) values [41], reflecting the overall importance of each feature in a linear regression model. While model coefficients can also describe how the response values will change on the value of an input feature, the coefficients depend on the scale of the input features. Taking the distribution of feature values in regard, SHAP values are computed as the difference between the expected model output and the partial dependence plot at the feature's value, and thus can better measure feature impacts on the model.
2.8. External validation
We ran external validation (Fig. 1h) to test the robustness of the optimal biomarker panel selected by the recommendation system (see in 2.7) on several B. subtilis gene expression datasets independent of Nicolas dataset used for model training. We estimated the stress sensing power of the recommended biomarker panel specific to the conditions covered in these external datasets (Table 1b) by applying the stress sensing model (see in 2.6).
To assess the ability of the recommendation system to prioritise an optimal biomarker panel over other candidate biomarkers and random genes, we computed the stress sensing index for the remaining candidate biomarker panels and a set of random gene panels in comparison with the recommended biomarker panel. This set of random gene panels was generated by repeatedly random sampling an equal number of genes as in each candidate biomarker panel.
3. Results
3.1. Assessment of the evaluate metrics for candidate biomarker panels
We discovered 949 candidate biomarker panels that can discriminate 10 cellular states in B. subtilis by rigorously running the Biomarker identification model (see 2.3). These candidate biomarker panels have all achieved classification performance of more than 0.9 (f1-score) in cross-validation tests. The sizes of these panels, i.e., the number of genes each biomarker panel consists of, ranged from 6 to 10.
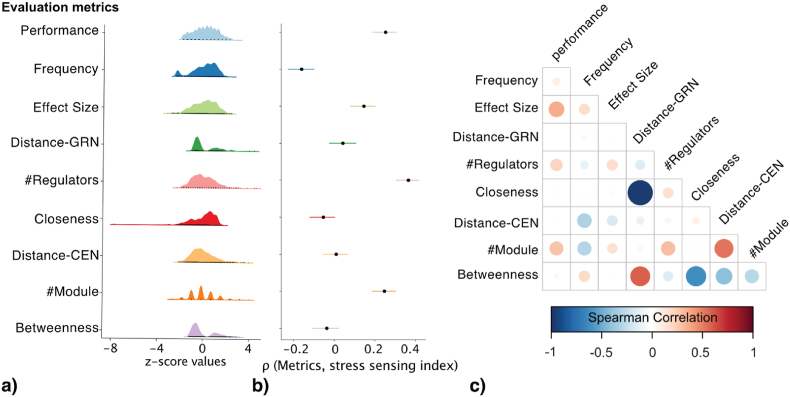
We derived a set of metrics that evaluated different properties of the candidate biomarker panels: Performance for classification performance to distinguish different cellular states; Frequency for the stability of genes being selected as a biomarker; Effects Size for the strength of distinguishability; Number of regulators (#Regulators) and Number of modules (#modules) for the diverse biological modalities in GRN and CEN; Distance-GRN and Distance-CEN for the coverage diameter in GRN and CEN; Closeness and Betweenness for the significance by centrality measurements in GRN and CEN. These evaluation metrics varied across candidate biomarker panels, with large dynamic ranges seen in the distribution (Fig. 2a). The correlation analysis between these metrics and stress sensing index (Fig. 2b) and the correlation analysis within these metrics (Fig. 2c) indicated they are complementary measurements and that individually they are not sufficiently predictive of stress sensing power. The initial assessment of this set of selected metrics showed the potential in predicting the stress sensing power of the biomarkers by incorporating them in a computational model.
Fig. 2.
Evaluation metrics for biomarker panels. a) The distribution plots for different evaluation metrics. b) The Spearman correlation between each evaluation metric and stress sensing index is shown as the dot with the line indicates a 95% confidence interval. c) The correlation map between 9 evaluation metrics.
3.2. Recommendation system to select a robust biomarker panel
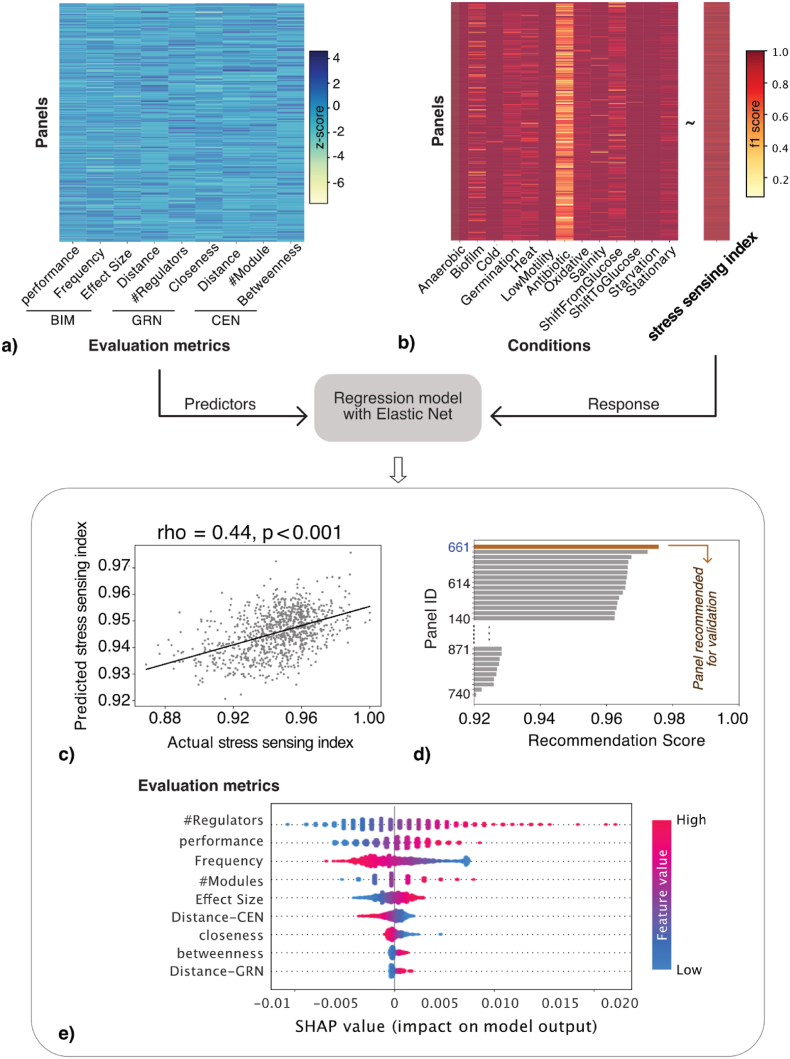
To build a recommendation system capable of selecting the optimal biomarker panel based on evaluation metrics and stress sensing index, we trained a regression model with Elastic Net regularisation to couple the evaluation metrics with the stress sensing index (Fig. 3a–b). We observed a significant correlation (Spearman rho = 0.44, p-value<0.001) between the actual stress sensing index and predicted stress sensing index produced by the trained regression model (Fig. 3c). The trained model assigned a recommendation score as the predicted stress sensing index (Equation (5)) to each candidate biomarker panel. Panel 661, which achieved the highest score, was selected as the recommended panel (Fig. 3d). This recommended biomarker panel showed great prediction performance in classifying 13 stress conditions covered in Nicolas dataset, with mean f1-score achieved at 0.99 (i.e. actual stress sensing index).
Fig. 3.
Recommendation of an optimal biomarker panel. a) The heatmap of 9 evaluation metrics for 949 panels. These metrics, derived from the Biomarker Identification model (BIM), Gene Regulatory Network (GRN) and Co-Expression Network (GEN), are Z-score standardised. b) The heatmap of classification performances (f1-score) in distinguishing 13 stress conditions respectively from control conditions, based on which stress sensing index is computed. The feature metrics of the biomarker panels are fitted in a linear regression model with Elastic Net regularisation to predict the stress sensing index based on 13 conditions. c) The scatter plot of predicted stress sensing index against actual stress sensing index. The Spearman correlation is 0.44 with significant effect (p-value <0.001). d) The bar plot of RecommendScore in 949 candidate biomarker panels, sorted in descending order. The system selects Panel 661 with highest Recommend Score as the recommended biomarker panel. e) The SHAP summary plot of feature importance given by the trained regression model. All the instances are displayed as dots, with the colour indicating the original feature value and the x-axis SHAP value indicating the impact of this feature metric on the model.
We analysed the respective impact of each evaluation metric on the outcome of the recommendation system, i.e., recommendation score (Fig. 3e). Number of regulators, which reflects how diverse the transcriptional regulatory parts are associated with the biomarker genes, showed the most significant impact on the model. The recommendation system gives a higher score for a biomarker panel consisting of genes that are involved in more regulatory parts. Number of Modules, which indicates the coverage of different modules in the co-expression network, similarly presented a positive correlation with the recommendation scores. Performance, Frequency, Effect Size, as the evaluation metrics extracted from the Biomarker identification model, respectively indicates the prediction power for classifying distinct cellular states, the uniqueness in candidate solutions, the strength of differential expression, were the remaining features among the top 5 contributors for ranking the candidate biomarker panels.
3.3. Validation of the recommended biomarker panel on external datasets
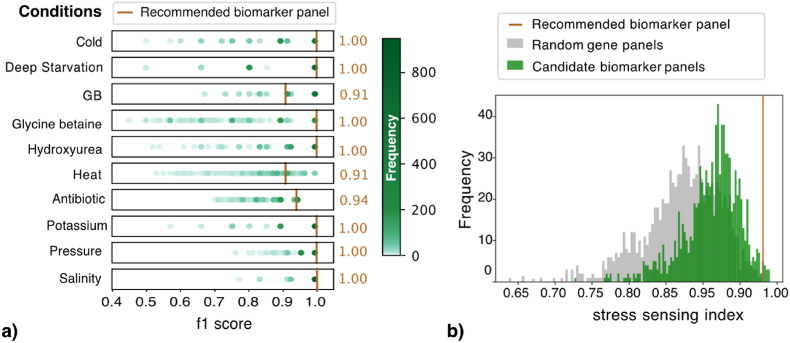
We validated the performance of the recommended biomarker panel on external datasets to classify samples grown under 10 test conditions from the samples grown under the corresponding control conditions (Table 1b). Despite variations observed in distributions of classification performance across candidate biomarker panels for all conditions, the recommended biomarker panel achieved 100% prediction accuracy for 7 conditions and still good prediction performance for the remaining 3 conditions (Fig. 4a). Note that half of the conditions (e.g., Pressure, Hydroxyurea, Potassium, Glycine betaine) we tested here were not covered in Nicolas dataset used for the model training. This shows that the proposed knowledge-based recommendation system holds the potential of prioritising biomarker genes responsive to an extended set of treatment conditions that were not seen in the biomarker identification process.
Fig. 4.
Validation performance of recommended biomarker panel on external datasets. a) A group of scatter histograms with each shows the distribution of validation performance (f1-score calculated on an external dataset) for a specific condition across 949 candidate biomarker panels. The hue of a dot reflects the frequency of occurrence for the corresponding value range. The validation performance of the recommended biomarker panel is marked as brown vertical lines with the value listed at the right for each condition. b) The grey histogram shows the distribution of stress sensing index across random gene panels, computed using external datasets. The green histogram shows the distribution of stress sensing index across candidate biomarker panels. A brown line marks the stress sensing index for the recommended biomarker panel.
To summarise the overall stress sensing power of a given biomarker panel, we averaged the prediction performance over all test conditions as the stress sensing index. We found that the stress sensing index for the recommended panel (0.98) is higher than 98% of the candidate biomarker panels and 99.5% of the random gene panels (Fig. 4b).
3.4. Biological characterisation of the recommended biomarker panel
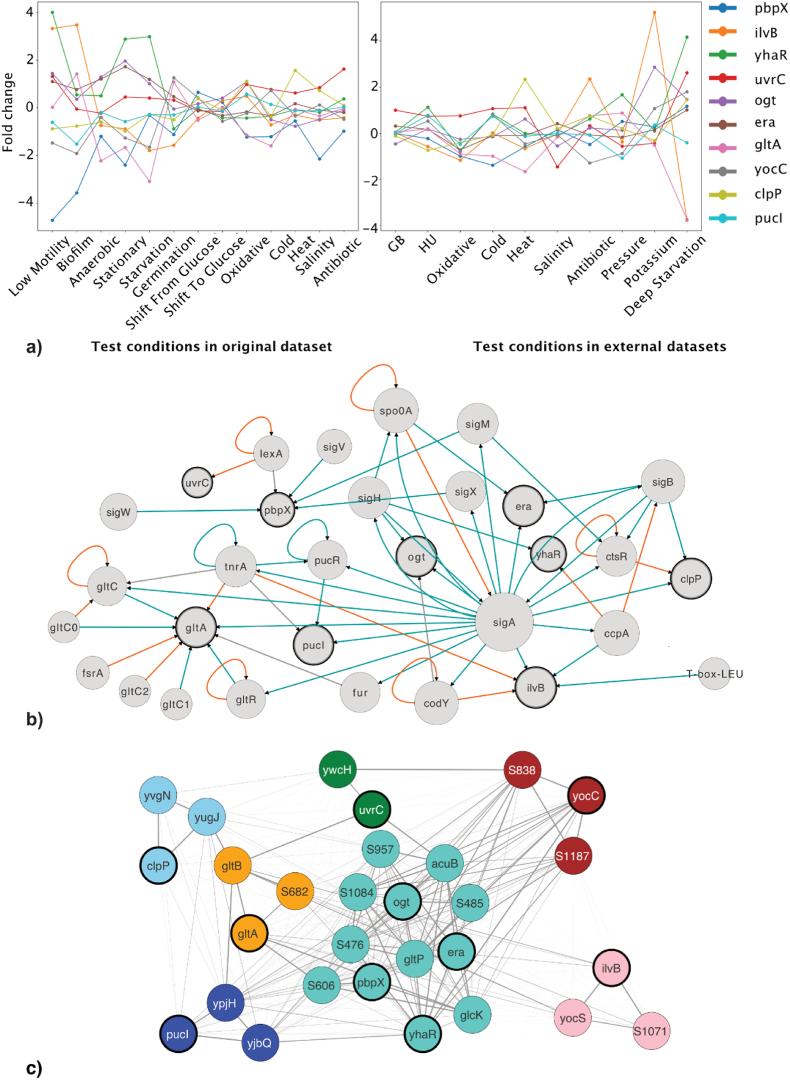
By training the recommendation system to predicate the stress sensing power of biomarker panels with evaluation metrics, we selected the biomarker panel with the highest predicted stress sensing index for in-vitro validation. As shown in Table 2, this recommended biomarker panel consists of 10 genes with various functions corresponding to DNA repair, endopeptidase activity, the synthesis of essential proteins, etc. These recommended biomarker genes presented varying alterations in transcription in response to different stress conditions, which combined can be used to distinguish a specific stress condition (Fig. 5a).
Table 2.
Gene functions and products of the recommended biomarker panel.
| Gene name | Gene function | Gene product |
|---|---|---|
| yhaR | putative dehydratase | |
| ogt | DNA repair | O6-methylguanine DNA alkyltransferase |
| pbpX | endopeptidase | penicillin-binding protein X |
| gltA | glutamate biosynthesis | glutamate synthase (large subunit) |
| yocC | conserved hypothetical protein | |
| era | ribosome assembly | GTP-binding protein |
| ilvB | biosynthesis of branched-chain amino acids | acetolactate synthase (large subunit) |
| uvrC | DNA repair after UV damage | excinuclease ABC (subunit C) |
| clpP | protein degradation | ATP-dependent Clp protease proteolytic subunit |
| pucI | purine utilization | allantoin permease |
Fig. 5.
The functional profiles of recommended biomarker genes. a) The line charts show the fold changes of 10 biomarker gene expressions in test conditions against the control conditions. We tested 13 treatment conditions in the original training dataset and 10 in external datasets. b) The smallest component of Gene Regulatory Network that connects the recommended biomarker genes (highlighted in bold borderline). The node size is proportional to node degree centrality in this network. An edge indicates the regulation from a source node to a target node, with the orange line denoting suppression and green denoting activation. c) A subnet of Co-expression Network composed of recommended biomarker genes highlighted in bold borderline and their closest neighbour genes. The edge line width represents the adjacent value, thicker edge meaning higher similarity in co-expression patterns and longer distance in the network. The node colour indicates the specific modules to which it belongs.
The evaluation metrics #Regulators and #Modules, which have shown a significant impact on the stress sensing power of the biomarkers in the recommendation system, are relevant to the diversity of biological processes that biomarker genes are entailed. Therefore, studying involved regulatory parts and co-expression modalities can reveal the functional profiles of the biomarkers. We extracted the smallest component that connects the biomarkers from the Gene Regulatory Network, in which 20 sigma factors or transcriptional regulators were found associated with the regulations of these biomarker genes (Fig. 5b). This includes sigA for household regulation, sigB for the general stress response, sigH for transiting to stationary growth phase, sigM/sigV/sigX for extracytoplasmic functions, ccpA/codY/tnrA for carbon, nitrogen or amino acid regulations, lexA/ctsR for DNA repair or heat shock response, etc. In complementary analysis, we also studied the Co-expression Network's subnet consisting of biomarkers and their closest neighbours (Fig. 5c). We found that 6 biomarker genes belong to unique modules while 4 are from the largest module (turquoise, 440 genes). As seen in the Supplementary Table 3B, these modules are associated with diverse biological processes, ranging from alternative carbon metabolism, various amino acid biosynthesis to protein repair and regulation of cell morphogenesis, etc.
4. Discussion
The objective of this study was to propose a computational method for selecting a robust biomarker panel that can be reproducibly applied in future studies to predict a wider range of stress conditions for B. subtilis. From the Nicolas dataset which includes the B. subtilis transcriptomes collected in many conditions and a biomarker identification model which selects gene signatures to discriminate distinct cellular states, we obtained the candidate biomarker panels for multi-stress conditions. By quantitatively relating a collection of evaluation metrics that capture complementary information of the biomarker with the stress sensing power of the biomarkers in a regression model, we built a biomarker recommendation system which ranks the candidate biomarker panels. The recommended biomarker panel, selected by computational models that were trained with Nicolas dataset, showed great performance to predict 10 stress conditions in external datasets. While in this work we have focused on a specific application domain, multi-stress sensing panel for B. subtilis, our pipeline is general-purpose and could be applied to comparable datasets of different species or cellular states.
Although the accumulation of large-scale omics data recently has enabled the discovery of molecular biomarkers with high prediction performance, the high-dimensionality and high-noise characteristics of omics data have also posed challenges in stability and reproducibility of the machine learning methods applied. It is not uncommon to find little overlap between existing biomarker solutions produced by different datasets or different methods. The biomarker recommendation system we presented in this paper adopts a knowledge-based strategy that assesses the existing solutions using prior knowledge and multi-source criteria. The knowledge–based strategy and recommendation system proposed here can be readily applied to any biomarker discovery study where multiple biomarker solutions achieving comparable performance by traditional data-driven criteria, e.g., prediction accuracy, may be produced.
The key to successfully identifying biomarker solutions with improved robustness and confidence, however, lies in selecting of a set of effective evaluation metrics to assess biomarkers. In the case of identifying biomarkers indicative of various stress states in bacteria, it is important to utilise knowledge that can reveal the functional diversity of biomarker genes. Therefore, we incorporated the metrics extracted Gene Regulatory Network and Co-expression Network that are potentially relevant to diverse biological processes the biomarker genes may be involved in, which is likely the reason why the recommended biomarker panel showed great prediction accuracy in extended stress conditions including even conditions unseen in training data. We also want to highlight that this recommendation system can be adapted to incorporate any new evaluation metrics that are complementary and relevant.
While the recommended biomarker panel showed great prediction accuracy in diverse stress conditions from several independent datasets, the effectiveness of our results is constrained by the validation datasets used. Although we have maximally explored the relevant B. subtilis gene expression profiles that are publicly available for external validation, these datasets are all small, consisting of few samples grown under a single stress condition versus the corresponding control samples. They also vary on assay platforms (including RNA-seq and Microarray) and bacteria strains (ranging from wide type to BSB1 and JH642). To verify the efficacy of the putative biomarkers it is essential to perform in-vitro validation in addition to in-silico validation. Our future work includes performing RT-qPCR assays to test the predictive power of the identified biomarker panel to discriminate a number of stress conditions of interests.
To summarise, we showed that we could identify a robust biomarker that achieved improved prediction accuracy in external datasets by quantitatively evaluating the candidate biomarkers based on multi-source criteria incorporating various data-driven metrics and prior biological knowledge. Therefore, the in silico methods we proposed in this paper can be applied to recommend an optimal biomarker panel indicative of various stress states in Bacillus subtilis for in vitro validation and implementation with increased confidence.
Credit author statement
Yiming Huang: Conceptualization; Methodology; Software; Validation; Formal analysis; Investigation; Data curation; Writing – original draft, Writing – review & editing; Visualization; Project administration. Nishant Sinha: Methodology; Validation; Writing – review & editing. Anil Wipat: Validation; Funding acquisition; Resources; Supervision; Writing – review & editing. Jaume Barcadit: Conceptualization; Funding acquisition; Resources; Supervision; Writing – review & editing.
Code availability
Code and Jupyter notebook are available at: https://github.com/neverbehym/biomaker-recommendation-system.
Declaration of competing interest
The authors declare no conflict interests.
Acknowledgements
This work was funded by the Engineering and Physical Sciences Research Council (EPSRC) ‘Synthetic Portabolomics: Leading the way at the crossroads of the Digital and the Bio Economies (EP/N031962/1)’. The authors would like to thank Wendy Smith, David Markham and other ICOS members for valuable feedback and discussions.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2022.12.001.
Contributor Information
Yiming Huang, Email: Y.Huang61@newcastle.ac.uk.
Jaume Bacardit, Email: Jaume.Bacardit@newcastle.ac.uk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Katz L., Chen Y.Y., Gonzalez R., Peterson T.C., Zhao H., Baltz R.H. Synthetic biology advances and applications in the biotechnology industry: a perspective. J Ind Microbiol Biotechnol. 2018;45(7):449–461. doi: 10.1007/s10295-018-2056-y. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt F.R. Optimization and scale up of industrial fermentation processes. Appl Microbiol Biotechnol. 2005;68(4):425–435. doi: 10.1007/s00253-005-0003-0. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay A. Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol. 2015;23(8):498–508. doi: 10.1016/j.tim.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Wehrs M., Tanjore D., Eng T., Lievense J., Pray T.R., Mukhopadhyay A. Engineering robust production microbes for large-scale cultivation. Trends Microbiol. 2019;27(6):524–537. doi: 10.1016/j.tim.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 5.Borkowski O., Ceroni F., Stan G.B., Ellis T. Overloaded and stressed: whole-cell considerations for bacterial synthetic biology. Curr Opin Microbiol. 2016;33:123–130. doi: 10.1016/j.mib.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Bonilla C.Y. Generally stressed out bacteria: environmental stress response mechanisms in gram-positive bacteria. Integr Comp Biol. 2020;60(1):126–133. doi: 10.1093/icb/icaa002. [DOI] [PubMed] [Google Scholar]
- 7.Pepperkok R., Ellenberg J. Microscopy for Systems Biology. 2006;7(September):690–697. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 8.Otto O., et al. Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat Methods. 2015;12(3):199–202. doi: 10.1038/nmeth.3281. [DOI] [PubMed] [Google Scholar]
- 9.Yurkovich J.T., Palsson B.O. Quantitative -omic data empowers bottom-up systems biology. Curr Opin Biotechnol. 2018;51:130–136. doi: 10.1016/j.copbio.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Gohl D.M., et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. 2016;34(9):942–949. doi: 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- 11.Gołębiewski M., Tretyn A. Generating amplicon reads for microbial community assessment with next-generation sequencing. J Appl Microbiol. 2020;128(2):330–354. doi: 10.1111/jam.14380. [DOI] [PubMed] [Google Scholar]
- 12.Raynor M.P., Stephenson S.A., Pittman K.B., Walsh D.C.A., Henderson M.A., Dobrovic A. Identification of circulating tumour cells in early stage breast cancer patients using multi marker immunobead RT-PCR. J Hematol Oncol. 2009;2:24. doi: 10.1186/1756-8722-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klett H., et al. Identification and validation of a diagnostic and prognostic multi-gene biomarker panel for pancreatic ductal adenocarcinoma. Front Genet. 2018;9(APR):1–14. doi: 10.3389/fgene.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Díaz M., Herrero M., García L.A., Quirós C. Application of flow cytometry to industrial microbial bioprocesses. Biochem Eng J. 2010;48(3):385–407. doi: 10.1016/j.bej.2009.07.013. [DOI] [Google Scholar]
- 15.Heins A.L., Hoang M.D., Weuster-Botz D. Advances in automated real-time flow cytometry for monitoring of bioreactor processes. Eng Life Sci. 2021;(October):1–19. doi: 10.1002/elsc.202100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaiser M., et al. Monitoring single-cell gene regulation under dynamically controllable conditions with integrated microfluidics and software. Nat Commun. 2018;9(1) doi: 10.1038/s41467-017-02505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dusny C., Grünberger A. Microfluidic single-cell analysis in biotechnology: from monitoring towards understanding. Curr Opin Biotechnol. 2020;63:26–33. doi: 10.1016/j.copbio.2019.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Sampaio N.M.V., Blassick C.M., Andreani V., Lugagne J.B., Dunlop M.J. Dynamic gene expression and growth underlie cell-to-cell heterogeneity in Escherichia coli stress response. Proc Natl Acad Sci U S A. 2022;119(14) doi: 10.1073/pnas.2115032119. 2020.09.14.297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S., Nam D., Jung J.Y., Oh M.K., Sang B.I., Mitchell R.J. Identification of Escherichia coli biomarkers responsive to various lignin-hydrolysate compounds. Bioresour Technol. 2012;114:450–456. doi: 10.1016/j.biortech.2012.02.085. [DOI] [PubMed] [Google Scholar]
- 20.Rau M.H., Bojanovič K., Nielsen A.T., Long K.S. Differential expression of small RNAs under chemical stress and fed-batch fermentation in E. coli. BMC Genom. 2015;16(1):1–16. doi: 10.1186/s12864-015-2231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagler K., et al. Identification of differentially expressed genes during Bacillus subtilis spore outgrowth in high-salinity environments using RNA sequencing. Front Microbiol. 2016;7(OCT) doi: 10.3389/fmicb.2016.01564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostertz J., Scharf C., Hecker M., Homuth G. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology (Road Town, V I (Br)) 2004;150(2):497–512. doi: 10.1099/mic.0.26665-0. [DOI] [PubMed] [Google Scholar]
- 23.Tabl A.A., Alkhateeb A., ElMaraghy W., Rueda L., Ngom A. A machine learning approach for identifying gene biomarkers guiding the treatment of breast cancer. Front Genet. 2019;10(MAR) doi: 10.3389/fgene.2019.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith B.P., et al. Identification of early liver toxicity gene biomarkers using comparative supervised machine learning. Sci Rep. 2020;10(1):1–27. doi: 10.1038/s41598-020-76129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcos-Zambrano L.J., et al. Applications of machine learning in human microbiome studies: a review on feature selection, biomarker identification, disease prediction and treatment. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.634511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang J., Alelyani S., Liu H. Data classification: algorithms and applications. CRC Press; New York: 2014. Feature selection for classification: a review; pp. 37–64. [Chapter 2], no. [DOI] [Google Scholar]
- 27.Huang Y., Smith W., Harwood C., Wipat A., Bacardit J. 2021. Computational strategies for the identification of a transcriptional biomarker panel to sense cellular growth states in Bacillus subtilis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalman M.R., Deeter A., Nimishakavi G., Duan Z.H. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinf. 2012;13(Suppl 2) doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ou F.S., Michiels S., Shyr Y., Adjei A.A., Oberg A.L. Biomarker discovery and validation: statistical considerations. J Thorac Oncol. Apr. 2021;16(4):537–545. doi: 10.1016/j.jtho.2021.01.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faria J.P., et al. Reconstruction of the regulatory network for Bacillus subtilis and reconciliation with gene expression data. Front Microbiol. Mar. 2016;7(MAR) doi: 10.3389/fmicb.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart J.M., Segal E., Koller D., Kim S.K. A gene-coexpression network for global discovery of conserved genetic modules. Science. 1979;302(5643):249–255. doi: 10.1126/science.1087447. 2003. [DOI] [PubMed] [Google Scholar]
- 32.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9 doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P., Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierre Nicolas A.L., Mäder Ulrike, Dervyn Etienne, Rochat Tatiana. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 1979:2012. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- 35.McInnes L., Healy J., Melville J. 2018. UMAP: uniform manifold approximation and projection for dimension reduction. [Google Scholar]
- 36.Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lazzarini N., Bacardit J. RGIFE: a ranked guided iterative feature elimination heuristic for the identification of biomarkers. BMC Bioinf. 2017;18(1):1–22. doi: 10.1186/s12859-017-1729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B., Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1) doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 39.Langfelder P., Zhang B., Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics. 2008;24(5):719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 40.Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: synthetic minority over-sampling technique nitesh. J Artif Intell Res. 2002;16(Sept. 28):321–357. [Google Scholar]
- 41.Lundberg S.M., Lee S.-I. A unified approach to interpreting model predictions. NeurIPS Proceedings. 2017;10 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.