Abstract
Cryo-focused ion beam (FIB) milling of vitrified specimens is emerging as a powerful method for in situ specimen preparation. It allows for the preservation of native and near-native conditions in cells, and can reveal the molecular structure of protein complexes when combined with cryo-electron tomography (cryo-ET) and sub-tomogram averaging. Cryo-FIB milling is often performed on plunge-frozen specimens of limited thickness. However, this approach may have several disadvantages, including low throughput for cells that are small, or at low concentration, or poorly distributed across accessible areas of the grid, as well as for samples that may adopt a preferred orientation. Here, we present a detailed description of the “Waffle Method” protocol for vitrifying thick specimens followed by a semi-automated milling procedure using the Thermo Fisher Scientific (TFS) Aquilos 2 cryo-FIB/scanning electron microscope (SEM) instrument and AutoTEM Cryo software to produce cryo-lamellae. With this protocol, cryo-lamellae may be generated from specimens, such as microsporidia spores, yeast, bacteria, and mammalian cells, as well as purified proteins and protein complexes. An experienced lab can perform the entire protocol presented here within an 8-hour working day, resulting in two to three cryo-lamellae with target thicknesses of 100–200 nm and dimensions of approximately 12 μm width and 15–20 μm length. For cryo-FIB/SEMs with particularly low-contamination chambers, the protocol can be extended to overnight milling, resulting in up to 16 cryo-lamellae in 24 h.
Graphical abstract:
Keywords: FIB-SEM , HPF , Waffle Method , cryo-ET , Sample preparation , In situ , AutoTEM Cryo
Background
Studying biological systems in close-to-native or in situ states opens wide frontiers towards precise contextual understandings of the mechanistic behaviors of molecular machines. In cryo-electron tomography (cryo-ET) pipelines, the sample is vitrified, which 1) captures and preserves biological processes in their close-to-native state, 2) maintains sample hydration, and 3) protects it from the vacuum of the electron microscope ( Saibil, 2022 ). Vitrification is commonly followed by sample thinning with cryo-focused ion beam (cryo-FIB) milling ( Marko et al., 2007 ) before cryo-ET data acquisition, providing the highest resolution in situ three dimensional (3D) views into the structures of individual protein complexes and cell parts in native contexts ( Wang et al., 2022 ). Cryo-ET also allows for the assessment of the overall morphology of more complex biological systems, including cellular organelles, whole cells, intact organisms, and tissues ( Bauerlein and Baumeister, 2021 ). However, vitrification with common plunge-freezing approaches is limited to samples up to ~5–10 μm thick. For thicker samples, a high-pressure freezer (HPF) must be used, which vitrifies specimens up to at least 60 μm thick ( Harapin et al., 2015 ). HPF vitrification has been successfully applied to multicellular biological samples ( Harapin et al., 2015 ), and its applicability has been extended with the waffle method ( Kelley et al., 2022 ). The waffle method allows for 1) vitrification of samples with thicknesses of tens of microns, 2) elimination of preferred orientation issues, 3) production of large (between 10 μm × 10 μm and 70 μm × 25 μm in x,y) and specimen-dense cryo-lamellae, and 4) an increase in cryo-FIB lamellae production throughput.
Specimens vitrified with an HPF cannot be directly imaged by cryo-ET and need to be thinned down to <300 nm to be transparent enough to the electron beam. During the last decade, cryo-FIB has gained substantial traction for thinning vitrified biological specimens, overtaking the promising but technologically challenging method of cryo-ultramicrotomy ( McDowall et al., 1983 ; Marko et al., 2007 ). Until several years ago, HPF vitrification of large (10+ μm thick) specimens followed by cryo-FIB/SEM and cryo-ET has typically been demonstrated on multicellular C. elegans organisms ( Harapin et al., 2015 ; Mahamid et al., 2015 ; Schaffer et al., 2019 ). Direct cryo-FIB milling of these samples requires a significant amount of continuous milling (>1 day) and has a low success rate. To increase the throughput, Mahamid et al. (2015) and Schaffer et al. (2019) developed a cryo-liftout (cryo-LO) approach, where a section of the specimen is extracted with a micro-manipulator and transferred to a special half-moon grid, for further thinning. Cryo-LO allows for region of interest (ROI) extraction, and decreases milling times per single lamella of ~150 nm thickness and a suitable imaging area of ~8 μm × 7 μm to ~10 h ( Schaffer et al., 2019 ); however, it requires specialized milling equipment and expertise.
To address these issues, we developed the waffle method, as described in Kelley et al. (2022). Here, we present a detailed protocol to support the waffle method, including sample vitrification (Procedure A), and waffle sample milling (Procedure B), that is applicable to a variety of biological samples. Samples prepared with Procedure A are generally tens of microns thick. Milling of these waffle samples does not require specialized instrumentation and can be performed manually or semi-automatically. Procedure B describes semi-automated milling, where milling takes ~2 h per lamellae site, with target thickness of 100–200 nm (as defined by the AutoTEM Cryo software), and approximate dimensions of 12 μm in width × 15–20 μm in length. Procedure B utilizes the MAPS software for defining ROIs and lamellae sites, then TFS AutoTEM Cryo automates the majority of the waffle milling process similarly to freely-available milling software packages ( Buckley et al., 2020 ; Klumpe et al., 2021 ). Waffle lamellae are typically larger in cross-sectional area than lamellae generated with conventional plunge-frozen cryo-FIB-milling, and may be produced at a higher rate (~2,800 μm 2 per 24 h for waffle lamellae versus ~600 μm 2 per 24 h for conventional lamellae; see Notes for calculations).
To increase the quality and efficiency of the protocol, and to ensure that the resulting cryo-ET tomograms contain objects of interest, ROI localization is optionally done by correlating fluorescent images of the sample before or after freezing. Cryo-fluorescent light microscopy (cryo-FLM) is used to illuminate fluorescently-tagged objects in the vitrified sample and may be performed with a stand-alone cryo-FLM, or with an FLM integrated inside the milling instrument (iFLM, Gorelick et al., 2019). Cryo-FLM can be done before (as screening), during (for FIB-milling navigation), and after FIB-milling (for guiding data collection) ( Watanabe et al., 2020 ; Gupta et al., 2021 ). However, it is important to account for autofluorescence for certain samples ( Carter et al., 2018 ). In addition to fluorescence-based approaches, two other methods for localizing or obtaining statistical information of objects of interest in situ exist, but fall outside the scope of this protocol: samples with EM-tags for localization in cryo-ET data ( Silvester et al., 2021 ; Tan et al., 2021 ), and structural mass spectrometry ( Klykov et al., 2022 ).
Materials and Reagents
Figure 1 shows several of the required items listed below together with common laboratory equipment.
Figure 1. Waffle preparation instrumentation.
The items from the Materials and Reagents section are shown with their corresponding numbering. For the waffle assembly, please refer to the original waffle paper by Kelley et al. (2022). Other common cryo-EM laboratory equipment included for completeness (black box on the bottom-right): P10 pipette with corresponding tips, fine-point tweezers, and AutoGrid box opening tool.
-
200-mesh cryo-EM grids (e.g., SPI Supplies, catalog number: 4220C-CF)
Note: Grids need to be coated with an extra ~20 nm thick carbon layer. While most commonly used grid material can be utilized, we typically use 200-mesh copper grids. Larger and smaller mesh sizes will decrease and increase lamellae dimensions, respectively. Grid foil type is not important, as it will be milled through.
Gridboxes (e.g., SubAngstrom, catalog number: GBV01)
Freezer planchettes (hats) type “B” (e.g., Ted Pella, catalog number: 39201)
SEM pin stub (e.g., Ted Pella, catalog number: 16261)
Abrasive sandpaper with grits 1,200, 7,000, 15,000, or similar (e.g., ADVcer, ADV_Sandpaper_Sheets_S01_5_10BUGY)
HPF holder (e.g., Technotrade International, catalog number: 290-1)
POL Metallpflege metal-polish (discontinued, substituted by, e.g., Ted Pella, catalog number: 892)
Filter papers (e.g., Whatman, catalog number: 1004090)
Kimwipe tissues (e.g., Kimtech, catalog number: 34155)
C-clip ring (Thermo Fisher Scientific, catalog number: 1036173)
Cryo-FIB AutoGrid (Thermo Fisher Scientific, catalog number: 1205101)
Yeast, bacteria, mammalian cells, proteins, or protein complexes
-
Concentrating supplies
For cells or organisms, Conical Sterile Polypropylene Centrifuge Tubes (e.g., Thermo Fisher Scientific, catalog number: 339652)
For purified proteins and protein complexes, protein concentrators with MWCO based on the protein size (e.g., Thermo Fisher Scientific, catalog number: 88513)
Respective cell culture or yeast media
Industrial grade dry nitrogen tank
Industrial grade liquid nitrogen tank
1-Hexadecene (e.g., Millipore Sigma, catalog number: 822064)
Equipment
Personal protective equipment for work with liquid nitrogen (e.g., gloves, eye protection, oxygen monitors)
Stereoscope (e.g., Zeiss, catalog number: 455053-0000-000)
Clipping station and clipping tools (e.g., Thermo Fisher Scientific, catalog number: 1000068)
Pipettors and tips (10 μL, 200 μL, 1,000 μL)
Centrifuge (e.g., Thermo Fisher Scientific, catalog number: 75004240)
Tweezers (e.g., Ted Pella, catalog number: 5220)
AutoGrid tweezers (e.g., Ted Pella, catalog number: 47000-600)
Grid transfer dewar (e.g., Thomas Scientific, catalog number: 5028M40)
Liquid Nitrogen dewar with 4 L capacity
-
Liquid Nitrogen dewar or puck system for grid storage (e.g., SubAngstrom, catalog number: GSS02-E)
Note: In our case, storing lamellae separated from other samples reduces contamination.
CCU-010 HV high vacuum coater (e.g., Safematic, catalog number: 100001)
CT-010 carbon fibre evaporation head module (e.g., Safematic, catalog number: 100003)
Plasma Cleaner (e.g., Gatan Inc., Gatan Solarus II Model 955)
Wohlwend Compact 01 HPF or comparable (M. Wohlwend GmbH)
-
Cryo-FIB/SEM dual beam microscope, including relevant grid shuttles (Thermo Fisher Scientific, Aquilos 2)
Note: The standard Aquilos 2 shuttle has a 45° pretilt. We found that the 27° pretilt shuttle supplied with the TFS iFLM module is more convenient for performing precleans (Procedure B, Step 30) because the pretilt allows for higher milling angles without lowering the stage.
-
Anti-contamination cryo-shield/cryo-shutter system (e.g., Delmic CERES Ice Shield, catalog number: DM-2707-999-0003-1)
Note: Such a low-contamination setup is optional. We found it useful in terms of reducing lamellae contamination during automated milling and allowing for automated overnight milling.
Software
-
FIB-milling software:
Microscope operating platform. Instruments from TFS are typically operated with xT. Here, we used xT v. 20.1.1.
Software for defining ROIs and aligning fluorescent signals, when applicable. Here we used MAPS v. 3.16-3.17.
Software for milling automation. Here we used AutoTEM Cryo v. 2.2.
Procedure
Note: Working with liquid nitrogen is potentially dangerous and should be done with caution and proper protection (i.e., cryo-gloves, protective glasses, oxygen monitors, etc.).
-
Sample Vitrification with High-Pressure Freezing (HPF)
Note: For additional instructions and demonstrations of Procedure A, refer to Video 1 .
-
Sample Thinning using cryo-FIB Milling
{Adamowicz, 2021 #1816}Notes:
Steps B1–B6, B8–B12, B14–B23, B25, and B29–B30 are done in xT software, B7, B13, and B23–B24 in MAPS, and B25–B28 and B31–B32 in AutoTEM.
For additional instructions and a demonstration of how to prepare lamellae sites before automated milling, refer to Video 2 . The process of automated milling is demonstrated in Video 3 .
-
Electron beam (EB) settings of 2 kV and 13 pA are kept throughout the whole protocol. Ion beam (IB) starting settings are 30 kV and 10 pA. While the IB voltage is kept the same throughout the protocol, currents are changed according to the text. For imaging, we typically use an ETD detector.
Two-screen recording of xT, MAPS, and AutoTEM windows for the site preparation before automated milling of waffle lamellae ( Procedure B ). After creating a new project in MAPS, a SEM grid overview is recorded (0:08), followed by platinum sputtering and gas injection system (GIS) coating inside the instrument chamber, then defining of preliminary lamellae sites (1:09). Next, the grid is positioned orthogonally to the IB, and precuts are milled for several lamellae sites (1:24). After finishing the precuts, the grid is placed back to the mapping position, and a new grid overview is recorded (3:12) and used to define final lamellae sites (3:33). Then, an AutoTEM project is created, and eucentric positions, milling angle, and preliminary lamellae positions for each site are defined (3:35). Precleans and notch milling are shown starting at 5:25. Final lamellae placement is shown at 8:40.
Before loading the specimen, purge the GIS lines. This step can be done while the FIB/SEM system is cooling down. No sample should be present during GIS purging.
Wake up the system and switch on both beams, then make sure that the “touch alarm” is enabled.
Apply a scan rotation of 180° to the EB and IB.
Select the grid, then go to the predefined “Mapping” position.
Lower the magnification, center on the grid, and focus the EB on the grid.
Increase the magnification until one square is visible, focus the EB, and then link the Z height (press “link Z to FWD”).
-
Create a new project and acquire an overview image of the grid in the EB:
Create and name a new Layer.
Create and name a new Tileset.
-
Define Tileset parameters ( Table 1 ).
Table 1. Parameters for the acquisition of EB grid overviews in MAPS .
Name [Name of your Tileset] Acquisition Type Electron Tiles X,Y 5,7 Tile HFW 600 µm Total HFW 2.76 mm Resolution 1,536 × 1,024 Pixel Size 390.625 nm Dwell 1 µs Frames Check, 1 Reduced Area Uncheck
-
Click “Prepare for sputtering” and run conductive platinum deposition with parameters from Table 2 .
Table 2. Platinum sputter coating parameters .
Current 30.0 mA Pressure 0.10 mbar Voltage Not changeable; values are 1 kV (while EB is 2 kV) Run Time 15 s Click “Recover from Sputtering” when finished to shut down the sputtering functionality (the “Run” button should be active again).
Go to the predefined “Deposition” position.
Add a layer of organometallic platinum (also known as GIS platinum): in the cryo GIS Deposition tab, select a grid and define the timing. With our milling settings, specifically the milling angle, 2 min of GIS works well for lamellae being milled to 200 nm thickness, or 2 min 15 s for 150 nm thickness.
Go back to the “Mapping” position.
-
Define the ROIs and preliminarily lamellae sites.
Notes:
For fluorescently labeled targets, ROIs and lamellae sites are defined based on the presence of fluorescence signals. This can be done either with a grid overview acquired in advance with an external cryo-FLM system, or with an internal FLM module. In both cases, MAPS allows for fluorescent image export, which is aligned with an EB grid overview. The general workflow for using fluorescence in FIB-SEM is described elsewhere, e.g., Gorelick et al. (2019).
In most cases, the grid will have distinct features, which can be used for alignment (e.g., broken squares or the grid center mark). We typically perform three-point alignment. First, rough alignment is performed using three points; then, MAPS allows for fine adjustments. Alignments are usually performed after platinum sputtering and GIS coating.
-
From the “Mapping” position, place the grid orthogonal to the IB:
Notes:
Make sure that “touch alarm” is enabled during all steps. Additionally, one can lower the stage, then rotate it, tilt, and move the stage back up to move to the orthogonal position.
Values are indicated for the standard TFS Aquilos 2 shuttle with 45 ° pretilt. For systems equipped with an iFLM, the shuttle pretilt is 27°, which requires tilting the stage to 25° in Step B14a.
Rotate the stage to the actual value of 108.1°, or relative 180° from the Mapping position, then tilt the stage to 7°, so the IB angle is 90° to the sample.
Save the shuttle position and use this saved position for future navigation.
Wake up the system, which automatically shuts down after sputtering.
With the IB at 10 pA current, navigate to one of the preliminary lamellae sites defined in Step B13.
Adjust the contrast so that any features on the surface of the lamella site are clearly visible, then acquire an image, and focus the IB while the image is being acquired. This will minimize charging.
Check the surface quality. Ideally, the surface should be “flat” and without contamination.
Locate the grid bars. If they are not visible, mill small window into the surface at 15 nA where you expect the grid bars to be ( Figure 3 ). If you hit two or more orthogonal grid bars, draw the corresponding lines, extending to all the squares with preliminary lamellae sites ( Kelley et al., 2022 ).
-
Precuts:
-
For 200-mesh grids where the grid bars are aligned during clipping, define rectangular milling patterns in {x,y} as {22 µm, 37 µm} (top) and {20 µm, 17 µm} (bottom), with the distance between the boxes set to 25 µm. Precut patterns for xT are available as Supplementary File 1 .
Note: We recommend using these extensively-tested pattern dimensions while initially implementing the waffle method.
Mill the patterns at 15 nA (milled precuts are shown in Figure 3D ).
-
Change back to 10 pA, zoom out, navigate to the next preliminary lamella site, and repeat Steps B17–B20.
Go back to the “Mapping” position.
Acquire a new Tileset with the same parameters as in Table 1 .
Delete old preliminary lamellae sites and define new lamellae sites on the new grid overview according to the precut positions.
Open AutoTEM and select the project with the same Project Name created in MAPS.
-
Choose the corresponding template for milling, and apply it to all lamellae sites ( Table 3 ).
Table 3. Milling parameters in AutoTEM Cryo.
Lamella thickness (defined as “Pattern Offset”) (µm) Front Width Overlap (µm) Rear Width Overlap (µm) Milling Current (nA) Pattern Type DCM Rescan Interval (s) Rough Milling 1.0 1.5 1.0 1.0 Rectangle 120 Medium Milling 0.8 0.65 0.5 1.0 CCS 90 Fine Milling 0.6 0.35 0.1 0.5 CCS 60 Finer Milling 0.4 0.05 0.05 0.3 CCS 30 Polishing 1 0.15 N/A N/A 0.03 CCS 30 Polishing 2 0 N/A N/A 0.01 CCS 10 The template can be found as Supplementary File 3 . Initially, we recommend setting the Depth Correction to 100% for 10 μm lamellae length (see Notes for an additional discussion).
Run “Preparation” for all lamellae sites in the “Guided” mode.
Follow the prompts for eucentric tilt and milling angle (20°).
Position the lamellae to guide the notch placement in Step B31.
-
Prepare “Precleans” ( Figure 4 ):
Note: Confirm with EB images that material below the lamellae sites are milled away completely ( Supplementary Figure 1 ).
Notes:
Steps B30b, d assume that apertures are aligned well, so there is no shift between milling patterns after changing the current.
If there is a significant shift of milling windows between 10 pA and 7 nA or 3 nA, take a quick snapshot at the milling current in question to accurately position the rectangular milling pattern.
-
Tilt the shuttle to a high milling angle, so the bottom of any material below each lamella is visible.
Note: The standard Aquilos 2 shuttle has a 45 ° pretilt, and the maximum angle typically is around 25–30 °. To reach the bottom of the material below lamellae, a standard shuttle might require moving the stage down in z.
The iFLM shuttle has a 27 ° pretilt, and tilting the stage to achieve a high milling angle does not require moving the stage down. For the iFLM shuttle, a milling angle of 45 ° can be used as a starting point, followed by milling at 30 °, and finally, at the milling angle in Step B30c.
Define the milling rectangular pattern at 10 pA, and then mill at 7 nA until the bottom of the bulk material is gone ( Figure 4A , B ).
Tilt to the final milling angle (20° in this protocol). Define the milling rectangular pattern at 10 pA and position the pattern a few microns away from the edge of the slab. Mill at 3 nA until the excess material is gone ( Figure 4C ).
-
Notch milling
Align the notch patterns in xT with the initial lamella position in AutoTEM ( Figure 5A ) . This step does not have to be accurate since the lamella position will be redefined later.
The total size of the notch pattern is 3.5 µm in x, 8.1 µm in y, and 3 µm in z. The size of the vertical patterns, except for the box on the side, is 3.5 µm in x and 0.2 µm in y, and vice versa for horizontal patterns. The overlap between patterns is 0.1 µm. The side pattern is 0.2 µm in x and 1.3 µm in y. The spacing between top and bottom parts is 1.1 µm ( Figure 5B ). The pattern can be found as Supplementary File 1 .
Mill at 0.3 nA. With the z depth defined in Step B31b, milling should take ~2 min 15 s in total ( Figure 5C ). The time can be adjusted when using different milling angles.
-
Final lamellae positioning:
Rerun “Image Acquisition” and “Lamella placement.”
Place the lamella ~1 µm away from the notch, with the top and bottom mill boxes overlapping with the notch, as shown in Figure 6 .
Typical lamellae dimensions are 12 µm × 15–20 µm × 150–200 nm.
-
Run in step-wise mode based on the AutoTEM template in Table 3 and Supplementary File 3 .
Note: Step-wise mode is used to minimize contamination growth on lamellae after milling.
-
Overtilts or Final Polishing:
Note: This step is optional and performed depending on the final lamellae thickness and lamellae quality.
If the lamella or GIS layer is damaged, do not perform automated post-milling operations.
Otherwise, define a positive 0.5° overtilt (milling angle goes to 20.5°) in the Polishing 2 step, and run automated milling only for that step. Alternatively, manually place a CCS milling pattern on top of the lamella. In both cases, milling for 1–2 min is sufficient.
Video 1. Sample preparation with the waffle method.

HPF sample preparation with the waffle method, including HPF planchette polishing (0:22) followed by application of 1-Hexadecene (2:25), waffle assembly in the HPF tip holder (2:50), and high pressure freezing of the waffle grid (4:10; Procedure A ). For more information, including intermediate states of the planchettes, please refer to Supplementary Figure 1 in the original waffle paper ( Kelley et al., 2022 ).
Video 2. Setting up milling sites.

Video 3. AutoTEM milling.
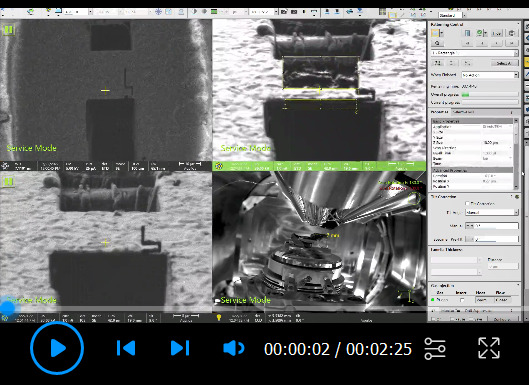
Recording of automated milling in AutoTEM (Step B32d). First, Rough Milling, Medium Milling, Fine Milling, and Finer Milling are performed for each lamella (0:01). Then, Polishing 1 and Polishing 2 are performed for each lamella (0:45). Parameters are listed in Table 3 .
Figure 3. Milling precuts with the corresponding timing as in Video 2 .
Initial milling patterns ( Supplementary File 1 ) are placed at the region of interest (A) . Prior to milling, an additional smaller pattern (yellow pattern) is used to localize the grid bar location (B) . The precut patterns are then adjusted to avoid the grid bars (C). The finished precuts are shown in panel (D) .
Figure 4. Precleans.
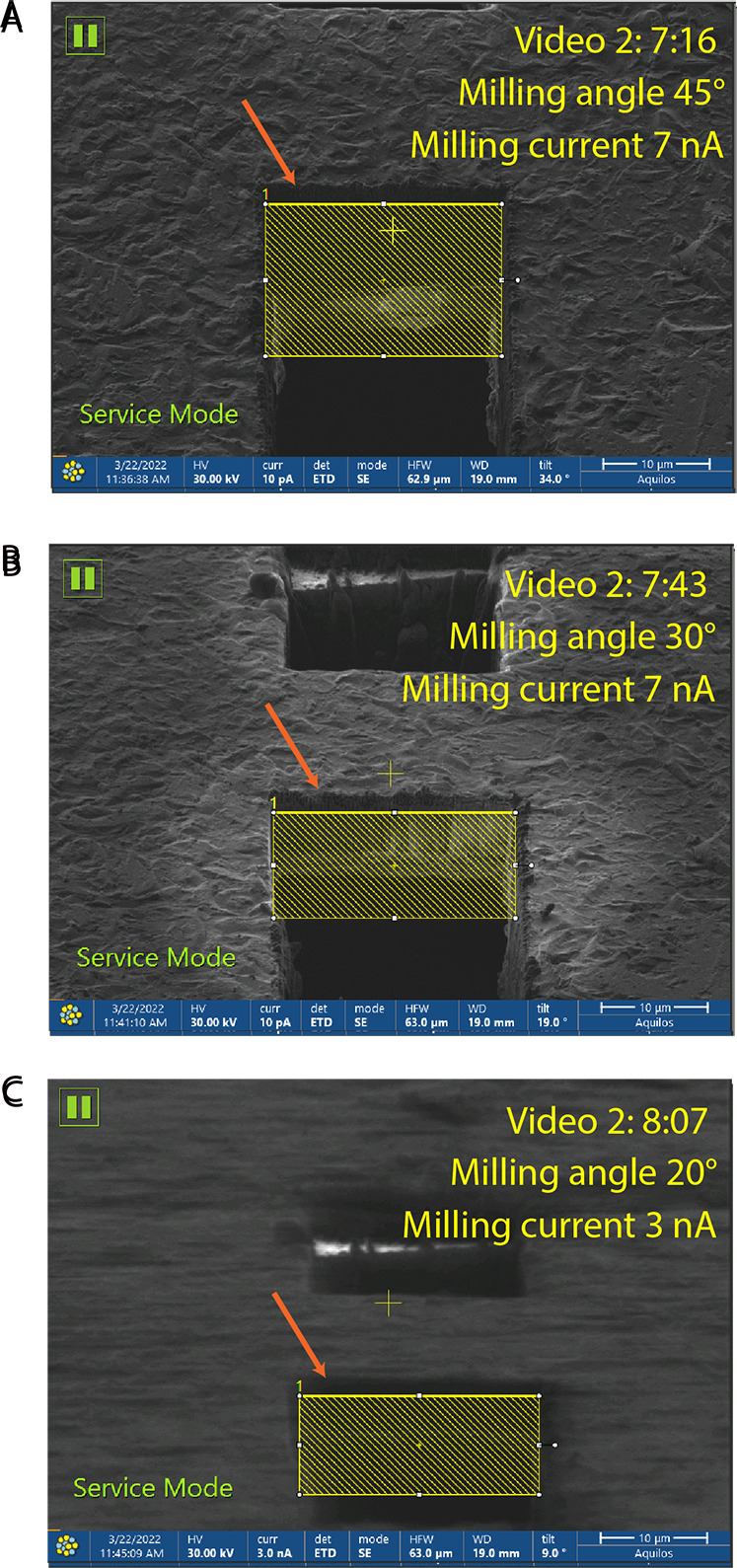
Milling precleans with the corresponding timing, as in Video 2 . (A, B) Milling at 7 nA is performed at high milling angles. (C) The stage is tilted to the milling angle (20° in this protocol), and a further preclean is performed at 3 nA current. In all cases, it is important to place milling patterns slightly below the edge of the slab ( orange arrow ), and to make sure that excess material below the potential lamella is milled away completely.
Figure 5. Milling notch with the corresponding timing as in Video 2 . (A) .

The notch pattern ( Supplementary File 2 ) is placed according to the pre-defined lamella site as in Step B29 and after precleans are done. (B) After digitally zooming on the pattern, the position of the Notch pattern with the defined dimensions is adjusted to be close to the edge of the slab. (C) Example of the milled notch before complete detachment.
Figure 6. Final lamella placement during the Preparation step in AutoTEM with the corresponding timing as in Video 2 .

Typical lamella dimensions are 12 μm in width, 10 µm length and 150 nm thick.
-
Rigidify grids with an extra layer of carbon:
Place grids on a glass slide, grid bar side down. Place the glass slide on the stage of a carbon evaporator, and evaporate 20 nm of carbon onto the film side of the grids.
-
Sand and polish planchettes:
Sand the flat side of Type B planchettes on 1,200, 7,000, and 15,000 grit sandpaper to get rid of the concentric rings present from manufacturing.
Dip the flat side of the planchettes in POL metallpflege metal polish, and rub it by hand in circular motions on a piece of filter paper.
Get rid of any excess polish with a Kimwipe tissue, then rub the planchettes on a piece of filter paper again to ensure all of the polish is gone.
-
Coat planchettes in 1-Hexadecene:
Place all planchettes flat side up on a piece of filter paper.
Apply a droplet of 1-Hexadecene (4–6 μL) to the top of the planchette, then drag the pipette tip so the drop spreads onto the filter paper.
Apply another droplet of 1-Hexadecene on top of the planchette.
Keep the planchettes coated in 1-Hexadecene for at least 15 min before waffle assembly.
-
Prepare a concentrated sample.
Note: The sample should typically be as concentrated as possible while remaining pipette-able. However, the sample does not necessarily need to be viscous.
-
Plasma clean carbon-rigidified grids:
Plasma clean grids with the grid bar side up (extra carbon side down) using a recipe of 80% O 2 gas and 20% H 2 gas for 30 s, with a forward radio frequency (RF) target of 50 W.
-
Waffle assembly:
Blot away 1-Hexadecene from a planchette hat, and place it on the HPF holder tip, flat side up.
Place the grid on the planchette hat, grid bar side up.
Apply 5 µL of the sample to the grid.
Blot away 1-Hexadecene from a second planchette hat, and place it on top of the sample, flat side down.
If necessary, lightly blot away excess sample that has squeezed out of the planchette hats.
Close the HPF tip.
Tighten the assembly with tweezers.
-
HPF:
Note: Typically, with our setup (Wohlwend Compact 01 HPF), the HPF pressure is 2,100 bar.
Transfer the assembled waffle sample to the HPF chamber.
Vitrify the sample.
Transfer the HPF holder into the liquid nitrogen chamber.
-
Loosen the HPF holder tip, and remove the waffle assembly.
Note: Ideally, the planchettes will separate fully from the grid. If the assembly is stuck together, use tweezers to press between the two planchettes; another pair of tweezers may be used to hold the assembly in place. If only one planchette hat separates, grab the grid by the outer rim and carefully lift it off of the other planchette hat.
-
Clipping waffle grids
Note: Waffle clipping is done in the same setup as clipping of grids of conventional samples prior to cryo-FIB/SEM thinning with the addition of a stereoscope.
Mark the notch of the AutoGrid with a marker ( Figure 2A ).This helps with grid bar alignment when clipping. It is also helpful for orienting the lamellae with respect to the TEM tilt axis during loading.
Under a stereoscope and before clipping with a c-clip ring, rotate the grid with tweezers until the grid bars are square to the notch of the AutoGrid ( Figure 2B , C ). It is possible that the grid moves slightly while clipping, which is not detrimental to the method, as the grid bars are aligned primarily to maximize the area within a square where a lamella can be placed.
Figure 2. Waffle grid clipping after HPF.
Properly orientating the grid bars helps to maximize the area within a square in which you can place a lamella. (A) Example of a cryo-FIB AutoGrid with the notch marked. The opposite side of the AutoGrid is shown with a marking that corresponds to the notch, which is visible during the clipping process. (B) Alignment of the grid bars as seen in a stereoscope without sample and (C) with sample in the grid clipping chamber.
Data analysis
Figure 7 shows an example of a final cryo-FIB-prepared waffle method lamella of yeast cells. Generated lamellae are further subjected to imaging with cryo-ET. Examples of the procedure, as well as more examples of milled lamellae, can be found in Kelley et al. (202 2 ).
Figure 7. Example of waffle lamellae of yeast cells with 150 nm target thickness prepared with this protocol.(A) .
Electron beam view with ETD detector, and (B) Ion beam view with ETD detector.
Notes
The protocol is highly reproducible in our hands, with a success rate close to 100% for most of the samples. The most often occurring issue is poor vitrification of the samples. Poor vitrification becomes apparent during TEM data acquisition, not during cryo-FIB/SEM or cryo-FLM. While we are using relatively high milling currents during precleans, the milling windows are placed away from the lamellae sites, and, therefore, we consider that poorly vitrified areas are from insufficient vitrification in the HPF. To reduce poor vitrification issues, we recommend adding cryo-protectant to the sample before the HPF; typically we add 5%–10% glycerol ( Bauerlein et al., 2021 ). If poor vitrification still occurs, we suggest increasing the sample concentration.
Initially, we recommend using the settings as in Supplementary File 3 . However, depending on the sample type, milling time can be reduced. To speed up the milling process, the Depth Correction parameter can be adjusted. Depth Correction should be decreased when material behind the lamella is milled during automated milling (see overmill in Supplementary Figure 1 ).
Other potential issues which may arise are summarized in Supplementary Figure 1 . Avoiding overmilling (Supplementary Figure 1A , B) reduces milling times and gallium implantation of the lamellae. Ensuring that the bulk layer of material is removed during precleans (Supplementary Figure 1A , B) is critical. We notice that lamellae deformations, such as sagging (Supplementary Figure 1B ) and buckling (Supplementary Figure 1C ), often occur due to incomplete notch milling. While we do not currently have a solution for sagging, buckling may be avoided by ensuring that the notch is milled through completely. Double lamellae (Supplementary Figure 1D , E) are often a consequence of displacement of the milling patterns by AutoTEM. While we cannot highlight precise reasons for such a displacement, it is likely caused by errors in cross-correlation during re-navigation to the milling site, and by improper drift correction.
We have found that replacing the TFS pole piece shutter assembly with a Delmic CERES Ice Shield on our TFS Aquilos 2 reduced the lamellae contamination rate by a factor of approximately 3. Prior to this change, we were unable to perform automated milling overnight without accruing considerable contamination on lamellae (presumably from the electron beam pole piece). Our current contamination rate after this upgrade is ~1.5 nm/h, previously ~4.4 nm/h, which allows for up to 16 waffle lamellae to be milled automatically overnight with acceptably low contamination, as opposed to our previous limit of 2–4 waffle lamellae milled during an 8-h workday. As each cryo-FIB/SEM instrument setup is different, we recommend testing contamination during overnight waffle milling. Based on contamination rates, the cryo-FIB/SEM operator may either restrict automated waffle milling to a normal 8-h workday, or find a way to lower the contamination rate in the cryo-FIB/SEM chamber to allow for significantly higher automated overnight waffle milling throughput. Additionally, we did not experience any clogs in the cooling lines during up to 72 h of continuous operation at cryo-temperatures.
Automated cryo-FIB/SEM workflows in the literature (i.e., Tacke et al., 2021) claim approximately 24 lamellae in 24 h, where each lamellae has an imageable area of ~5 μm × 5 μm, resulting in a total imageable area of ~600 μm 2 . The low-contamination waffle protocol we describe here produces 16 lamellae in 24 h, where each lamella has an imageable area of ~11 μm × 16 μm, resulting in a total imageable area of ~2,800 μm 2 .
Acknowledgments
This work was performed at the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NIH NIGMS (GM103310), and NIH (U24GM139171).
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Buckley G. , Gervinskas G. , Taveneau C. , Venugopal H. , Whisstock J. C. and de Marco A. ( 2020 . ). Automated cryo-lamella preparation for high-throughput in-situ structural biology . J Struct Biol 210 ( 2 ): 107488 . [DOI] [PubMed] [Google Scholar]
- 2. Bauerlein F. J. B. , Pastor-Pareja J. C. and Fernández-Busnadiego R. ( 2021 . ). Cryo-Electron Tomography of Native Drosophila Tissues Vitrified by Plunge Freezing . BioRxiv . doi: 10.1101/2021.04.14.437159 [DOI] [Google Scholar]
- 3. Bauerlein F. J. B. and Baumeister W. ( 2021 . ). Towards Visual Proteomics at High Resolution . J Mol Biol 443 ( 20 ): 167 - 187 . [DOI] [PubMed] [Google Scholar]
- 4. Carter S. D. , Mageswaran S. K. , Farino Z. J. , Mamede J. I. , Oikonomou C. M. , Hope T. J. , Freyberg Z. and Jensen G. J. ( 2018 . ). Distinguishing signal from autofluorescence in cryogenic correlated light and electron microscopy of mammalian cells . J Struct Biol 201 ( 1 ): 15 - 25 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gorelick S. , Buckley G. , Gervinskas G. , Johnson T. K. , Handley A. , Caggiano M. P. , Whisstock J. C. , Pocock R. and de Marco A. ( 2019 . ). PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy . Elife 8 : e45919 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gupta T. K. , Klumpe S. , Gries K. , Heinz S. , Wietrzynski W. , Ohnishi N. , Niemeyer J. , Spaniol B. , Schaffer M. , Rast A. , et al. .( 2021 . ). Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity . Cell 184 ( 14 ): 3643 - 3659 e3623 . [DOI] [PubMed] [Google Scholar]
- 7. Harapin J. , Bormel M. , Sapra K. T. , Brunner D. , Kaech A. and Medalia O. ( 2015 . ). Structural analysis of multicellular organisms with cryo-electron tomography . Nat Methods 12 ( 7 ): 634 - 636 . [DOI] [PubMed] [Google Scholar]
- 8. Kelley K. , Raczkowski A. M. , Klykov O. , Jaroenlak P. , Bobe D. , Kopylov M. , Eng E. T. , Bhabha G. , Potter C. S. , Carragher B. and Noble A. J. ( 2022 . ). Waffle Method: A general and flexible approach for improving throughput in FIB-milling . Nat Commun 13 ( 1 ): 1857 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klumpe S. , Fung H. K. , Goetz S. K. , Zagoriy I. , Hampoelz B. , Zhang X. , Erdmann P. S. , Baumbach J. , Muller C. W. , Beck M. , et al. .( 2021 . ). A modular platform for automated cryo-FIB workflows . Elife 10 : e70506 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klykov O. , Kopylov M. , Carragher B. , Heck A. J. R. , Noble A. J. and Scheltema R. A. ( 2022 . ). Label-free visual proteomics: Coupling MS- and EM-based approaches in structural biology . Mol Cell 82 ( 2 ): 285 - 303 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mahamid J. , Schampers R. , Persoon H. , Hyman A. A. , Baumeister W. and Plitzko J. M. ( 2015 . ). A focused ion beam milling and lift-out approach for site-specific preparation of frozen-hydrated lamellas from multicellular organisms . J Struct Biol 192 ( 2 ): 262 - 269 . [DOI] [PubMed] [Google Scholar]
- 12. Marko M. , Hsieh C. , Schalek R. , Frank J. and Mannella C. ( 2007 . ). Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy . Nat Methods 4 ( 3 ): 215 - 217 . [DOI] [PubMed] [Google Scholar]
- 13. McDowall A. W. , Chang J. J. , Freeman R. , Lepault J. , Walter C. A. and Dubochet J. ( 1983 . ). Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples . J Microsc 1 ): 1 - 9 . [DOI] [PubMed] [Google Scholar]
- 14. Saibil H. R. ( 2022 . ). Cryo-EM in molecular and cellular biology . Mol Cell 82 ( 2 ): 274 - 284 . [DOI] [PubMed] [Google Scholar]
- 15. Schaffer M. , Pfeffer S. , Mahamid J. , Kleindiek S. , Laugks T. , Albert S. , Engel B. D. , Rummel A. , Smith A. J. , Baumeister W. ( 2019 . ). A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue . Nat Methods 16 ( 8 ): 757 - 762 . [DOI] [PubMed] [Google Scholar]
- 16. Silvester E. , Vollmer B. , Prazak V. , Vasishtan D. , Machala E. A. , Whittle C. , Black S. , Bath J. , Turberfield A. J. , Grunewald K. , et al. .( 2021 . ). DNA origami signposts for identifying proteins on cell membranes by electron cryotomography . Cell 184 ( 4 ): 1110 - 1121 e1116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tacke S. , Erdmann P. , Wang Z. , Klumpe S. , Grange M. , Plitzko J. and Raunser S. ( 2021 . ). A streamlined workflow for automated cryo focused ion beam milling . J Struct Biol 213 ( 3 ): 107743 . [DOI] [PubMed] [Google Scholar]
- 18. Tan Z. Y. , Cai S. , Noble A. J. , Chen J. K. , Shi J. , and Gan L. ( 2021 . ). Heterogeneous non-canonical nucleosomes predominate in yeast cells in situ . BioRxiv . doi: 10.1101/2021.04.04.438362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Watanabe R. , Buschauer R. , Böhning J. , Audagnotto M. , Lasker K. , Lu T. W. , Boassa D. , Taylor S. , Villa E. ( 2020 . ). The in-situ structure of Parkinson’s disease-linked LRRK 2. Cell 182 ( 6 ): 1508 1518 1518 .e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Z. , Grange M. , Pospich S. , Wagner T. , Kho A.L. , Gautel M. , Raunser S. ( 2022 . ). Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin . Science 6582 ( 375 ): eabn1934 . [DOI] [PubMed] [Google Scholar]