Abstract
In recent years, Alzheimer’s disease (AD) diagnosis using neuroimaging and deep learning has drawn great research attention. However, due to the scarcity of training neuroimaging data, many deep learning models have suffered from severe overfitting. In this study, we propose an ensemble learning framework that combines deep learning and machine learning. The deep learning model was based on a 3D-ResNet to exploit 3D structural features of neuroimaging data. Meanwhile, Extreme Gradient Boosting (XGBoost) machine learning was applied on a voxel-wise basis to draw the most significant voxel groups out of the image. The 3D-ResNet and XGBoost predictions were combined with patient demographics and cognitive test scores (Mini-Mental State Examination (MMSE) and Clinical Dementia Rating (CDR)) to give a final diagnosis prediction. Our proposed method was trained and validated on brain MRI brain images of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset. During the training phase, multiple data augmentation methods were employed to tackle overfitting. Our test set contained only baseline scans, i.e., the first visit scans since we aimed to investigate the ability of our approach in detecting AD during the first visit of AD patients. Our 5-fold cross-validation implementation achieved an average AUC of 100% during training and 96% during testing. Using the same computer, our method was much faster in scoring a prediction, approximately 10 min, than feature extraction-based machine learning methods, which often take many hours to score a prediction. To make the prediction explainable, we visualized the brain MRI image regions that primarily affected the 3D-ResNet model’s prediction via heatmap. Lastly, we observed that proper generation of test sets was critical to avoiding the data leakage issue and ensuring the validity of results.
Keywords: Machine learning, Deep learning, Ensemble learning, Alzheimer’s disease diagnosis
1. Introduction
More than 55 million people were living with Alzheimer’s disease (AD) in 2020 and this number was predicted to exceed 152 million by 2050 (Report, 2018). AD is one of the most common neurodegenerative disorders that causes impairment in cognitive function which can affect not only the patients but also their family, friends, and the society. Pathological changes in the brain of people with AD, including abnormal cell death or synaptic dysfunction, start to develop at least 20 years before symptoms can be observed (Böhle et al., 2019). Although there is currently no cure for AD, accurate AD diagnosis is still needed to inform clinical decisions. Accurate AD diagnosis is also imperative to AD drugs development where subjects’ status must be evaluated before, during, and at the end of clinical trials for treatment effectiveness monitoring.
Several research have been conducted to investigate biomarkers that can be assessed non-invasively for AD diagnosis (Nguyen et al., 2019). Among them, single or multiple neuroimaging modalities based on structural magnetic resonance imaging (MRI), functional MRI (fMRI), and metabolic positron emission tomography (FDG-PET) have yielded the most promising results in research (Mosconi et al., 2007). While MRI provides the biomarkers related to progressive structural damage of the brain such as temporal lobe atrophy or cerebral atrophy caused by AD (Cuingnet et al., 2011), the blood flow observed in fMRI reflects functional disruption of local regions that can lead to AD (Duc et al., 2020, Vemuri et al., 2012); and PET gives the information of cerebral metabolic rates of glucose which associates to neuronal activity (Rodrigues and Silveira, 2014). Depending on the assumption regarding which regions of the brain give complementary information, input handling can be divided into different categories such as voxel-based morphometry (Good et al., 2001, Good et al., 2002, Wang et al., 2015), region of interest (ROI)-based (hippocampal volume) (Colliot et al., 2008, Rusinek et al., 2004, Tapiola et al., 2008), slice-based (Aderghal et al., 2017, Gao et al., 2018, Luo et al., 2017) and patch-based methods (Cheng and Liu, 2017, Li and Liu, 2018, Suk et al., 2014).
In recent years, with the advance in machine learning algorithms and the emergence of deep learning, there have been several works focusing on creating a predictive tool using neuroimaging to assist clinicians in AD diagnosis. In the track of machine learning, support vector machines (SVM) (Boser et al., 1996, Vapnik, 1995) has been the most frequently used method in this domain. Recently, a significant improvement of tree-based algorithms such as random forest (RF) (Breiman, 2001) or extreme gradient boosting (XGBoost) (Chen and Guestrin, 2016) allowed the processing of high-dimensional data efficiently and led to an increasing research interest in employing these algorithms. They can be applied for imagery or non-imagery data (Fulton et al., 2019). In 2014, Moradi et al. (2014) proposed using random forest classifier and biomarkers extracted from MRI and cognitive measures for early detection of AD conversion in MCI patients. The work of (Gray et al., 2012, Moradi et al., 2014) showed that random forest can be applied for manifold learning of pairwise similarities derived from multiple models and it also introduces the concept of feature importance ranking of either region-based or voxel-based features which allows identifying brain regions corresponding to the pathology. Regarding deep learning approaches, many supervised and unsupervised models have been proposed to learn the hidden representation of the image without domain knowledge to diagnose AD. Liu et al. (2018) presented a cascaded convolutional neuron network (CNN) model to first learn the multi-level features from two different modalities (MRI and PET) and then ensemble these features to produce the prediction for patients. In (Hosseini-Asl et al., 2016, Parisot et al., 2018), the authors also demonstrated that CNN could achieve good results in identifying AD. Despite the success of CNN approaches, they require the training of many parameters which easily results in overfitting and needs large training datasets. The works of (Suk et al., 2015, Suk et al., 2014) utilized unsupervised algorithms such as Deep Boltzmann Machine (DBM) and Autoencoders (AE) for transfer learning to generalize the model to unseen instances. Data transformation methods including flipping, cropping and rotation were also employed before the training phase to increase the diversity of the input dataset (Esmaeilzadeh et al., 2018, Farooq et al., 2017, Islam and Zhang, 2018).
To solve the overfitting problem, in this study, we propose a framework combining deep learning and machine learning and employed data augmentation. Our best-performing model achieved an AUC of 96.2% in classifying AD patients vs. cognitively normal (CN) subjects with a much faster scoring time compared to machine learning methods relying on feature extraction. In addition, utilizing an occlusion method, we generated heatmap visualizations to explain where in the MRI images our model looked at to predict patients’ status. This is highly desirable for the application of artificial intelligence systems in medicine.
The rest of this manuscript is organized as follows: Section 2 presents how we collect and preprocess the data along with the proposed methods and evaluation criteria. Results, including classification performance and heatmaps, will be reported in Section 3. Finally, 4, 5 are discussions, conclusions, and future work.
2. Materials and methods
2.1. Data acquisition
The data used for developing the models in our work was retrieved from the ADNI dataset (https://ida.loni.usc.edu). ADNI is a large-scale study focusing on the early detection and progression monitoring of AD. The image data in ADNI has gone through careful quality control, ensuring the reliability for the development and verification of our classification models. We selected patients who have MRI scans data and achieved a pool of 924 T1-weighted images of 462 subjects. A subject may possess more than one MRI scan due to multiple visits. Multiple scans of the same subject i.e., intra-subject scans acquired at various timestamps, introduce insignificant discrepancy compared to inter-subject scans. A target of our research is to detect AD during the first visit of patient. Therefore, only the first visit scans, i.e., baseline scans were included in the testing set. However, for the training set, we included both baseline scans and follow-up scans. This was crucial since the well-known problem of training complicated deep learning models on a modest size dataset is severe overfitting. Including follow-up scans in the training set helped to increase the training set size, thus reducing overfitting. The summary of our dataset is in Table 1.
Table 1.
Demographic details of all participants used in this study.
| Group | Number of subjects | Gender (M/F) | Age (years) | MMSE | CDR |
|---|---|---|---|---|---|
| AD | 231 | 126/105 | 75.52 ± 7.71 | 22.74 ± 3.00 | 0.82 ± 0.36 |
| CN | 231 | 123/108 | 75.25 ± 5.89 | 29.06 ± 1.10 | 0.01 ± 0.13 |
MMSE: Mini-Mental State Examination; CDR: Clinical Dementia Rating.
The selected data was preprocessed using a standard processing pipeline. Firstly, the MRI images were processed with N3 bias field correction in Advanced Normalization Tools (ANTS) (Avants et al., 2008) to eliminate intensity inhomogeneity artifacts. Next, the brain region was extracted using the FSL BET tool (Jenkinson et al., 2005; Smith, 2002). Finally, the brain extracted image was registered into a 1 mm MNI152 standard-space T1-weighted atlas. The results are 3D processed images with the size of (182,218,182).
2.2. Methods
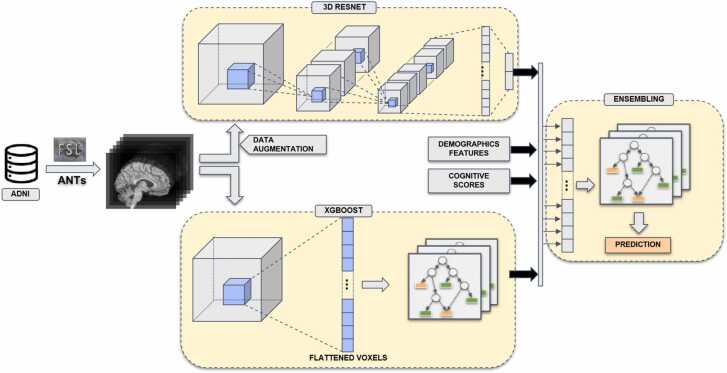
Our framework is a uni-data, multi-model approach, which means we explored features of MRI images by applying different machine learning methods. In the deep learning direction, a well-studied CNN ResNet was applied to exploit the local spatial characteristic of the images. In the second direction, XGBoost, a white-box machine learning algorithm was adopted to analyze the importance per voxel. At the final stage, prediction probabilities of both directions and demographics features and cognitive test scores were combined by XGBoost to produce the final prediction. Fig. 1 depicts an overview of our framework.
Fig. 1.
Overview of our classification framework. The MRI data were taken from ADNI data source and preprocessed by a combination of FSL and ANTs processing tools. The processed data flowed through a deep learning pipeline (top) and a XGBoost pipeline (bottom). Finally, the generated features of the two pipelines were concatenated with demographics features and cognitive scores to construct a combined features for the ensemble model (right). The ensemble model gave the final prediction (AD or CN).
2.2.1. 3D-ResNet
CNNs have been widely used in computer vision and applications whose data contain implicit local relationships. In visual object classification, for example, CNNs perform impressively by analyzing the relationship of groups of neighbor pixels to detect edges, shapes, etc. The MRI images possess the same attribute but in 3D. In our work, we adopted Residual Networks (ResNet) developed by He et al. (2015) and modify its operation to be 3D-compatible. ResNet is built based on residual blocks in which an input can either pass through conventional convolution layers or skip several layers. This prevents the gradient vanishing problem and enables the use of deeper networks. ResNet also frees the user from architecture tuning since it can always shrink itself into a more simplified version by leveraging skip connections during training. In this study, we employed an 18-layer 3D ResNet.
Data augmentation: Although ADNI is one of the most extensive medical image datasets, the amount of data has been still considered scarce for a deep learning approach which typically required thousands of samples to be effectively trained. A limited dataset often leads to overfitting problems, in which the model does not generalize on unseen test data, although it performs well on training data. To overcome this issue, in our work we applied multiple data augmentation methods to virtually expand the data capacity, namely non-baseline augmentation, random flip and rotation, and random cutout. All of them are recognized as semantic-intact methods, which means they do not distort or alter the semantic characteristic of the image data like other augmentation methods such as noise adding, translation, or color augmentation. We consider medical images as very sensitive data, and their characteristics should be preserved as much as possible.
Non-baseline augmentation is a special augmentation method in which the original image data was not manipulated. Instead, we included intra-subject scans from the second visit onwards.
Moreover, the image data were flipped and rotated with a random probability. The rotation angle was a multiplier of 90 degrees. Then, a randomly chosen cube patch (size r × r × r) was cut out from the image. Cutout method (Raj et al., 2022, Singh et al., 2021), an occlusion technique (Chlap et al. 2021), not only helps to enlarge the dataset size but also prevents AI models from only focusing exclusively on some key visual characteristics of the image. For example, a model may only look at the hippocampus region which has been proven to be a key factor in AD diagnosis but ignores other brain regions which may also have signs of disease. The cutout is similar to the drop-out method usually seen in deep neural networks but at the input image level instead of hidden layers. Cutout regularization drives the model to consider more of the image context into consideration when making decisions (Devries and Taylor, 2017). The flip-rotation and cutout probability as well as cutout patch size were chosen based on extensive parameter searching.
2.2.2. Machine learning models
Random Tree Embedding (RTE) (Geurts et al., 2006, Moosmann et al., 2006) is an unsupervised learning method that embeds the input vector non-linearly into lower binary feature space. Each tree of the forest splits at random without using a target variable and its leaves are finally binary encoded based on the category that its value belongs. Combining the encoded vector of all trees in the forest produces the new sparse feature space.
XGBoost is a boosting ensemble learning method in which weak learners are built sequentially to reduce the gradient of the previous learners. Each weak learner in the model is a conventional decision tree. The tree consists of various nodes and at each node, it can be split into branches based on information gain and the last node where the branch stops splitting gives the output. XGBoost is an improved version of Gradient Boosting Machine which introduces a parallel and distributed way of tree learning and adds a regularization term in the loss function.
Using these two methods, RTE and XGBoost, we built three classifiers with a voxel-as-features method: (i) XGB-HC: a 40 × 40 × 40 region of the hippocampus was cropped from the original image, then an XGBoost classifier was built on the flattened extracted region, (ii) XGB: a hippocampus-extended region of size 90 × 90 × 90 was cropped from the original image, then an XGBoost classifier was built on the flattened extracted region, (iii) RTE-XGB: a hippocampus-extended region of size 90×90×90 was cropped from the original image, then an unsupervised random tree embedding was used to embed the flattened image into smaller space and finally, an XGBoost classifier was built on the embedded vector.
2.2.3. Ensemble learning algorithm
The ensemble technique that we adopted in this study is stacked ensemble learning which is also known as super learner. Stacking (Wolpert, 1992) allows for combining different learning algorithms at the base level. Intuitively, if the base models are diversified, the stacked ensemble will be able to learn from different angles, which results in heterogeneous characteristics.
In our study, the predicted probabilities of how likely a person having AD were produced by three machine learning models: XGB-HC, XGB, and RTE-XGB. Then the ENS-1 model was created by ensembling the outputs of the three machine learning models using an XGBoost classifier. Similarly, the ENS-2 model was created by ensembling the outputs of the three machine learning models and the output of the 3D-ResNet model; the ENS-3 model: the outputs of the three machine learning models, the output of the 3D-ResNet model, and demographic information; and finally, the ENS-ALL model: the outputs of the three machine learning models, the output of the 3D-ResNet model, demographic information, and cognitive scores. Table 2 shows the models investigated in this work.
Table 2.
List of abbreviations for models mentioned in this paper.
| Abbreviation | Description | Input | Classification Method |
|---|---|---|---|
| 3D-ResNet | 3D Residual Network | 3D preprocessed image | 3D- ResNet |
| XGB-HC | XGBoostmodelon hippocampus regions | Flattened 3D hippocampus region extracted from 3D preprocessed image | XGBoost |
| XGB | XGBoostmodelon complete image | Flattened 3D hippocampus extended region extracted from 3D preprocessed image | XGBoost |
| RTE-XGB | XGBoost model with input dimension reduction | Flattened 3D hippocampus extended region extracted from 3D preprocessed image | RTE XGBoost |
| ENS-1 | Ensemble model of all XGBoost models |
Predicted probability of XGB-HC, XGB, RTE-XGB |
XGBoost |
| ENS-2 | ENS-1 + 3D-ResNet | Predicted probability of XGB-HC, XGB, RTE-XGB, 3D-ResNet | XGBoost |
| ENS-3 | ENS-1 + 3D-ResNet + demographics features | Predicted probability of XGB-HC, XGB, RTE-XGB, 3D-ResNet, demographic features (age and gender) | XGBoost |
| ENS-ALL | ENS-1 + 3D-ResNet + demographics features + cognitive scores | Predicted probability of XGB-HC, XGB, RTE-XGB, 3D-ResNet, demographic features (age and gender), cognitive test scores (MMSE and CDR) | XGBoost |
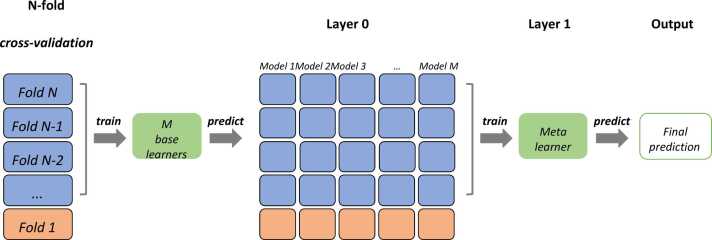
We trained the stacked ensemble model using 5-folds cross-validation. Therefore, for each fold, we combined the prediction of the first level models trained on the rest of the folds as the input to fit a meta learner and tested on the current fold. The detail of how we trained the stacked ensemble learning model with M base learners for fold 1 in 5-folds cross-validation is shown in Fig. 2.
Fig. 2.
Stacked ensemble learning with N-fold cross validation.
2.2.4. Performance evaluation
We suggest that splitting the dataset into training and testing sets should be handled in a per-subject manner instead of a per-scan manner. In this way, we can ensure that all scans of a subject should be located exclusively to one set only and data leakage should not happen. Since the size of the dataset is modest, one well-known technique to evaluate the performance and stability of the model is N-fold cross-validation. In this study, we chose N = 5 and evaluate performance metrics for each fold. Area Under the Curve (AUC) was used as our performance metric since we find it the most stable metric. Other metrics such as accuracy tend to have high variance due to the small test set. To assess the stability of our model, we calculated the standard deviation of AUC across folds.
All models were built by using PyTorch (for deep learning) (Paszke et al., 2019), Scikit-Learn (for machine learning) (Pedregosa et al., 2011) and XGBoost python package (Chen and Guestrin, 2016).
3. Results
3.1. Classification performance
Table 3 summarizes performance of the deep learning models. Naively forward the MRI images through a 2D-ResNet gave a mediocre mean AUC of 0.631. 3D-ResNet boosted the AUC significantly to 0.877 (approx. 39%) by effective 3D convolutional operations. We used the following data augmentation parameters: flip rotate probability = 0.3, cut out probability = 0.8, cut out size = 0.1. With those metrics, we observed a modest boost in performance from mean AUC 0.877 to mean AUC 0.884. However, the performance gap between the training phase and the testing phase was tightened (mean AUC 0.917 in training - data not shown - vs. mean AUC 0.884 in testing, i.e., 0.033 gap) compared to one without data augmentation (mean AUC 0.938 in training - data not shown - vs. mean AUC 0.877 in testing, i.e., 0.061 gap) and the standard deviation of AUC among folds decreased from ± 0.057 to ± 0.039 as well. These results indicated signs of reduced overfitting and improved model stability. In conclusion, 3D-ResNet with data augmentation gave the best results in terms of performance and stability.
Table 3.
Performance summary (AUC) of the deep learning models.
| 2D-ResNet | 3D-ResNet | Augmented 3D-ResNet | |
|---|---|---|---|
| Fold 1 | 0.616 | 0.866 | 0.870 |
| Fold 2 | 0.583 | 0.907 | 0.887 |
| Fold 3 | 0.713 | 0.952 | 0.948 |
| Fold 4 | 0.552 | 0.799 | 0.845 |
| Fold 5 | 0.690 | 0.863 | 0.872 |
| Mean | 0.631 | 0.877 | 0.884 |
| Std | ±0.069 | ±0.057 | ±0.039 |
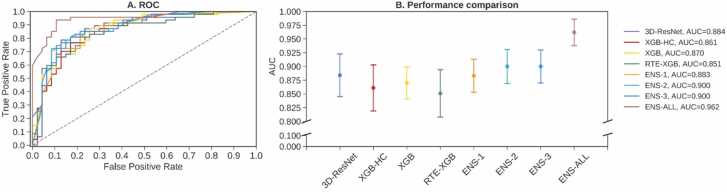
Table 4 summarizes the results of all the models investigated in our work. The machine learning models XGB-HC, XGB, and RTE-XGB with mean AUCs of 0.861, 0.868, and 0.851, respectively, showed good discriminative capability compared to the deep learning model 3D-ResNet which achieved mean AUC of 0.884. Specifically, with only biomarkers from the hippocampus region, the XGB-HC model achieved mean AUC 0.861, which revealed that the hippocampus was a critical biomarker in predicting AD. Taking the hippocampus-extended region as input, the XGB model achieved a mean AUC of 0.868, i.e., only a 0.007 increase compared to the XGB-HC model. The RTE-XGB model, in which the feature space of the hippocampus-extended region was reduced by a random tree embedding before being fed into the XGB classifier, achieved a mean AUC of 0.851, i.e., a 0.017 AUC drop compared to the XGB model. The ensemble of the three machine learning models XGB-HC, XGB, and RTE-XGB, creating ENS-1, resulted in a mean AUC of 0.883, which was comparable to the mean AUC of 0.884 of the 3D-ResNet model. But the ENS-1 model had a lower standard deviation, ± 0.030 for the ENS-1 model vs. ± 0.039 for the 3D-ResNet, thus better stability. Ensembling the 3D-ResNet model and the three machine learning models, creating the ENS-2 model, improved the mean AUC to 0.899 ± 0.031. Adding demographic information (age and gender), creating the ENS-3 model, seemed to yield no effect since there was no significant difference between ENS-2 and ENS-3 models (mean AUC of 0.899 ± 0.031 vs mean AUC of 0.898 ± 0.029, respectively), suggesting that age and gender were not predictive information for AD status. Finally, adding scores from cognitive tests (MMSE and CDR), creating the ENS-ALL model, significantly improved the performance with a mean AUC of 0.962 ± 0.024. The ENS-ALL model was therefore the best-performing model.
Table 4.
Performance summary (AUC) of the proposed models. The ENS-ALL model achieved the highest mean AUC of 0.962.
| 3D-ResNet | XGB-HC | XGB | RTE-XGB | ENS-1 | ENS-2 | ENS-3 | ENS-ALL | |
|---|---|---|---|---|---|---|---|---|
| Fold 1 | 0.870 | 0.857 | 0.867 | 0.850 | 0.866 | 0.878 | 0.878 | 0.951 |
| Fold 2 | 0.887 | 0.866 | 0.865 | 0.819 | 0.891 | 0.898 | 0.903 | 0.981 |
| Fold 3 | 0.948 | 0.927 | 0.916 | 0.923 | 0.931 | 0.953 | 0.947 | 0.985 |
| Fold 4 | 0.845 | 0.837 | 0.840 | 0.818 | 0.870 | 0.889 | 0.888 | 0.970 |
| Fold 5 | 0.872 | 0.816 | 0.851 | 0.844 | 0.858 | 0.880 | 0.876 | 0.926 |
| Mean | 0.884 | 0.861 | 0.868 | 0.851 | 0.883 | 0.899 | 0.898 | 0.962 |
| Std | ±0.039 | ±0.042 | ±0.029 | ±0.043 | ±0.030 | ±0.031 | ±0.029 | ±0.024 |
Fig. 3 illustrates ROC curves (reported for fold 1) and AUCs for the four base learners 3D-ResNet, XGB-HC, XGB, and RTE-XGB and the four ensemble models ENS-1, ENS-2, ENS-3, and ENS-ALL. Similar to the results shown in Table 4, Fig. 3 demonstrated that the performance of the four base learners 3D-ResNet, XGB-HC, XGB, and RTE-XGB were quite comparable. Assembling them and adding demographic information and cognitive test scores, which created ensemble models ENS-1, ENS-2, ENS-3, or ENS-ALL, improved the performance. The ENS-ALL model, which was an ensemble of all four base learners, demographic information, and cognitive test scores, achieved the highest performance of 0.962 ± 0.024 AUC.
Fig. 3.
Classification performance obtained by four base models and four ensemble models. (A) ROC curves (reported for fold 1). (B) mean AUCs and standard deviations of the models.
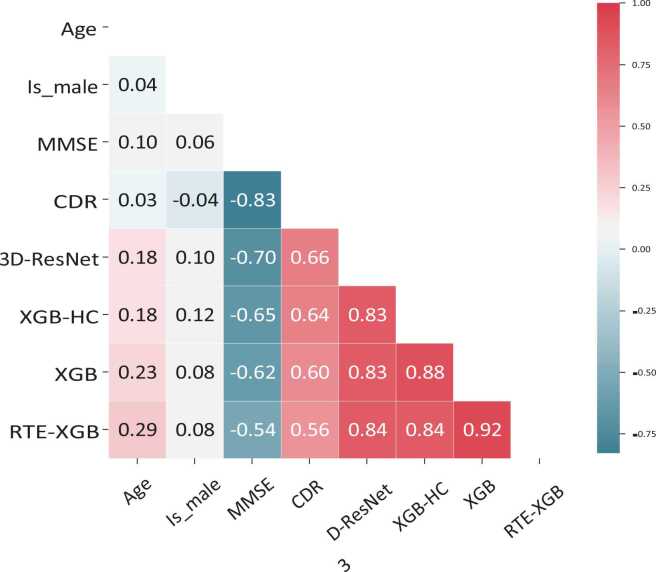
Fig. 4 shows the correlation coefficients between some variables of concern in our study. As expected, the correlation between cognitive test scores, CDR and MMSE, and the predictions was strong (> 0.5). A positive correlation between CDR scores and the models’ predictions indicated a proportional relationship between them. This result was reasonable since the higher the CDR score, the more likely a patient is to be diagnosed with AD. The negative correlation between MMSE and the models’ predictions indicated an inverse proportional relationship between them, which is expected since the higher the MMSE score, the less likely a patient is to be diagnosed with AD. Age had a moderate correlation with the models’ predictions with correlation values from 0.18 to 0.29. Meanwhile, there was a very low correlation between gender and the models’ predictions.
Fig. 4.
Correlation coefficients between age, gender, MMSE score, CDR score and predictions of the base learners 3D-ResNet, XGB-HC, XGB, RTE-XGB (reported for fold 1).
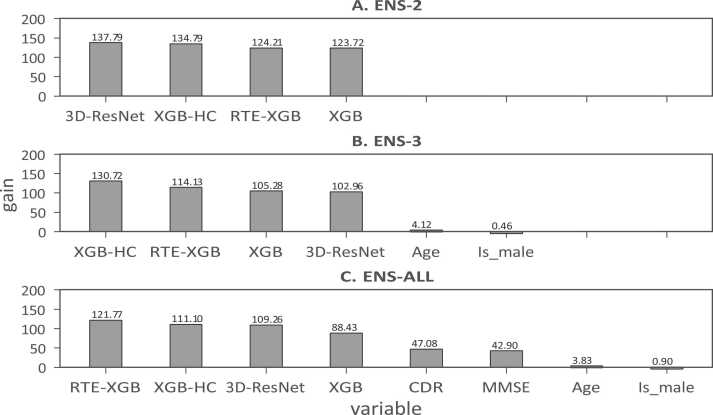
The importance of individual components to the ensemble models ENS-2, ENS-3, and ENS-ALL are shown in Fig. 5. The results suggested that the base learners including 3D-ResNet, XGB-HC, XGB, and RTE-XGB provided equal contributions to the final predictions of the ensemble models ENS-2, ENS-3, and ENS-ALL. We could again observe that cognitive test scores CDR and MMSE played significant roles in the final predictions of the ensemble models. Meanwhile, age and gender just contributed minor information for the final prediction.
Fig. 5.
The importance of the base learners 3D-ResNet, XGB-HC, XGB, RTE-XGB, demographic information (age and gender), and cognitive test scores (CDR and MMSE) to final predictions of the ensemble models ENS-2, ENS-3, and ENS-ALL (reported for fold 1).
3.2. Heatmap visualization
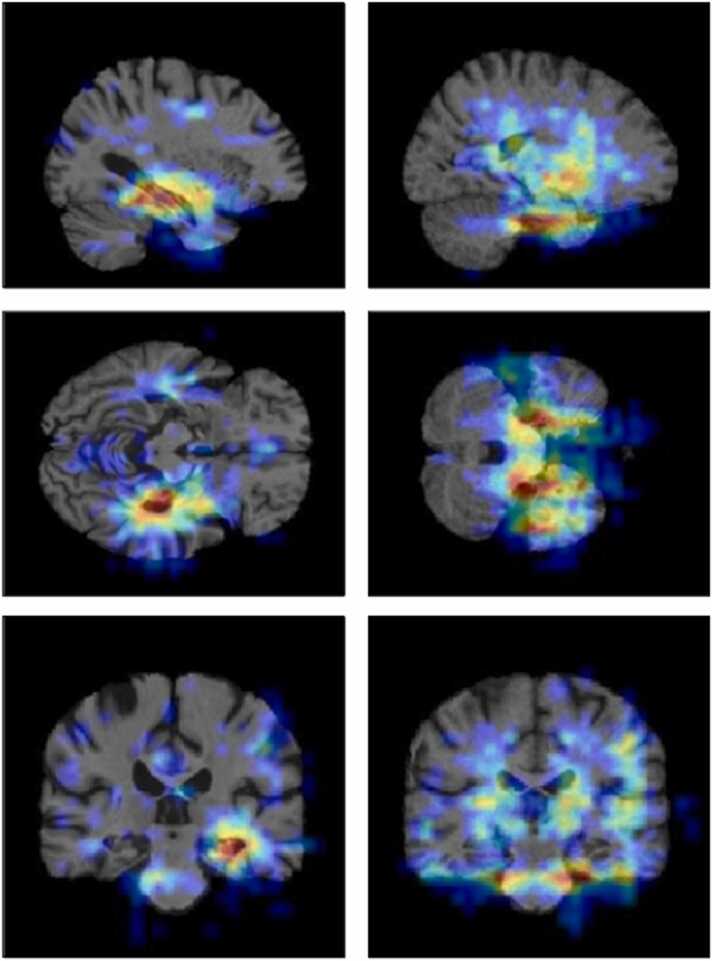
Deep neural network-based approaches have often been considered as “black-boxes” since it is often difficult to understand how deep neural networks arrive at their predictions. These networks could perform very well but what drove their predictions was, in many cases, not obvious. Therefore, the predictions were often not explainable. In medical diagnosis, the explainability of AI’s predictions is critical to gaining doctors’ and patients’ confidence in AI. In this work, we used an occlusion matrix method as the heatmap visualization tool to help highlight the decisive brain regions and make the prediction more transparent. To achieve that, a mask of size m × m × m was slid over the 3D input images. At each sliding position, a corresponding masked input image was passed through the convolution network, and we recorded the level of prediction probability changes compared to unmasked input. Finally, a heatmap of prediction probability deviation was constructed. This visualization method is similar to (Zeiler and Fergus, 2014). The rationale is that the more crucial a brain region is, the more deviated the prediction outcome when that brain region is masked. Fig. 6 depicts heatmap visualization of an AD subject on the left and a CN subject on the right. The heated area (red color) indicates the brain regions which contributed the most to the final prediction. We observed that, for AD patients, the 3D-ResNet model mostly looked at the hippocampus area whereas, for CN subjects, it looked at a broader space without solely relying on any specific region.
Fig. 6.
Heatmap visualization of an AD subject (left column) and a CN subject (right column). The red color regions are the most important regions to the model’s prediction whereas the blue color regions are less important.
4. Discussion
Various methods to identify AD/MCI using either uni-modal or multi-modal neuro-imaging data and artificial intelligence have been proposed. These methods used either state-of-the-art machine learning-based frameworks (Zhang et al., 2011; Kim et al., 2018; Hidalgo-Muñoz et al., 2014; Khazaee et al., 2015; Nguyen et al., 2019) or deep learning-based frameworks (Duc et al., 2020, Etminani et al., 2022, Zhu et al., 2021) to achieve classification accuracies from about 75% to about 95% on the binary classifications. For example, one of the pioneer studies in AD/MCI identification using machine learning framework was proposed by Zhang et al. (2011). The authors introduced multi-kernel SVM classifiers to combine with the biomarkers of three modalities including structural MRI, FDG-PET, and CSF to achieve up to 93.2% accuracy when classifying AD from CN subjects. On the other hand, Etminani et al. (2022) introduced a validated 3D deep learning architecture that predicts the final clinical diagnosis of AD, MCI, and cognitively normal (CN) using fluorine 18-FDG PET and compare the model's performance to that of multiple expert nuclear medicine physicians' readers. The authors have reported that the proposed model was able to obtain an AUC of 96.4% in AD, 71.4% in MCI-AD, and 94.7% in CN in the unseen test dataset, outperforming the physicians’ performance.
The advantage of deep learning-based frameworks is that the extraction of features is not necessary. However, it comes with the cost of high computational expense and sometimes it requires large memories for data and model loading. Another drawback of deep learning frameworks is that it is often not obvious how the models arrived at predictions or what image regions or parameters they considered. This is often known as the “black-box” issue. On the other hand, machine learning models can provide explanations or interpretations to physicians. However, intermediate feature engineering steps that include feature extraction and feature selection are needed for machine learning models to work. The feature extraction task is quite time-consuming and requires domain knowledge.
In our opinion, an ideal AI model for deployment in clinical practice needs to satisfy the three following criteria. First, the AI model needs to have high classification performance. Second, the AI model needs to be able to score a prediction quickly enough while using a reasonable computational cost. And finally, the AI model needs to be explainable, e.g., parameters or image regions that the model considered to score a prediction are displayed to the end-user. Explainability is important to solve the “black-box” issue and increase physicians’ trust in AI models.
In this work, we proposed an ensemble learning method combining deep learning and machine learning. During the training phase, we employed multiple data augmentation methods to tackle the overfitting issue. Among the ensemble model, the ENS-3 model achieved nearly 90% AUC without using any domain-knowledge or cognitive scores (Table 4). Further analysis of the importance of each input revealed that the importance of the base learners was high. The results suggested that our model could provide a reliable prediction of AD based solely on MRI images. Adding cognitive test scores, the ENS-ALL model achieved 96.2% AUC. The results indicate that using the ensemble of multiple classifiers and data is promising for improving diagnostic accuracy. Our model, considered an end-to-end framework, has the advantage of fast prediction time since it could score a prediction within approximately 10 min. Whereas, other machine learning approaches, which require the extraction of sophisticated brain features using toolkits such as FreeSurfer, FSL, etc. before feeding to the machine learning models, usually take many hours to extract features from raw data before scoring a prediction given the same computer. Interestingly, the XGBoost pipeline and standalone voxel-wise features, without any locality relationship among them, achieved acceptable performance. Using an occlusion-based method, we created a heatmap to visualize the MRI image regions that our 3D-ResNet model looked at when scoring a prediction. In summary, our model achieved relatively good performance with a fast scoring time, and the prediction result could be explained by heatmap visualization.
Our study has several limitations. First, although our model is faster in scoring a prediction, our model’s AUC is lower than some of the state-of-the-art machine learning models relying on feature extraction. We believe that with the availability of more data in future, our approach’s AUC could be further improved, and the performance gap will be gradually reduced. Second, this study doesn’t include the detection of mild cognitive impairment (MCI), an earlier stage of AD of which the detection would allow earlier interventions and has drawn great research interest. We plan to include MCI detection in our future studies.
5. Conclusion and future
In this work, we proposed a uni-data, multi-model framework for AD detection. Using the proposed methodology, we have boosted the classification performance and reduced overfitting. Five-fold cross validation implementation achieved on average 100% AUC for training and 96.2% AUC for testing. In detail, an ensemble of machine learning models performed comparably to the 3D-ResNet deep learning model with the test set AUC of 88.3% and 88.4%, respectively. The combination of machine learning models and the 3D-ResNet deep learning model improved the test set AUC to near 90%. Adding demographic information (age and gender) and especially cognitive test scores (MMSE and CDR) further improved the test set AUC to 96.2%. Our end-to-end framework has the advantage of fast scoring time, approximately 10 min to score a prediction, compared to many hour scoring time of feature extraction-based approaches. In future, we plan to extend our model to detect both AD and MCI. We also plan to add brain images acquired from other imaging modalities such as fMRI and PET and other biomarkers to increase the diversity of the individual learners. Transfer learning may also be applied to further reduce overfitting caused by the limited training data.
Conflict of Interest
Declarations of interest: None.
Acknowledgment
This research is funded by International University, VNU-HCM under grant number T2019−05−BME.
Contributor Information
Huong Ha, Email: htthuong@hcmiu.edu.vn.
Nguyen Thanh Duc, Email: thanh.duc.nguyen@mail.mcgill.ca.
Hoan Thanh Ngo, Email: nthoan@hcmiu.edu.vn.
References
- Aderghal, K., Boissenin, M., Benois-Pineau, J., Catheline, G., & Karim, A., 2017, 01, Classification of sMRI for AD Diagnosis with Convolutional Neuronal Networks: A Pilot 2-D+e Study on ADNI. In (p. 690–701).
- Avants B., Tustison N., Song G. Advanced normalization tools (ants) Insight J. 2008:1–35. [Google Scholar]
- Böhle M., Eitel F., Weygandt M., Ritter K. Layer-wise relevance propagation for explaining deep neural network decisions in MRI-based Alzheimer’s Disease classification. Front. Aging Neurosci. 2019:194. doi: 10.3389/fnagi.2019.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boser B., Guyon I., Vapnik V. A training algorithm for optimal margin classifier. Proc. Fifth Annu. ACM Workshop Comput. Learn. Theory. 1996:5. [Google Scholar]
- Breiman, L., 2001, 10, Random forests. Machine Learning.
- Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM,; New York, NY, USA: 2016. XGBoost: A scalable tree boosting system; pp. 785–794. [Google Scholar]
- Cheng, D., & Liu, M., 2017, 09, Classification of Alzheimer’s Disease by cascaded convolutional neural networks using pet images. In (p. 106–113).
- Chlap P., Min H., Vandenberg N., Dowling J., Holloway L., Haworth A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021;65(5):545–563. doi: 10.1111/1754-9485.13261. [DOI] [PubMed] [Google Scholar]
- Colliot O., Ch´etelat G., Chupin M., Desgranges B., Magnin B., Benali H., Leh´ericy S. Discrimination between alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus. Radiology. 2008;248(1):194–201. doi: 10.1148/radiol.2481070876. [DOI] [PubMed] [Google Scholar]
- Cuingnet R., Gerardin E., Tessieras J., Auzias G., Leh´ericy S., Habert M.-O., et al. Automatic classification of patients with Alzheimer’s Disease from structural mri: a comparison of ten methods using the adni database. neuroimage. 2011;56(2):766–781. doi: 10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Devries, T. and Taylor, G.W., 2017, Improved regularization of convolutional neural networks with cutout. CoRR, abs/1708.04552.
- Duc N.T., Ryu S., Qureshi M.N.I., Choi M., Lee K.H., Lee B. 3d-deep learning based automatic diagnosis of Alzheimer’s Disease with joint MMSE prediction using resting-state fMRI. Neuroinformatics. 2020;18(1):71–86. doi: 10.1007/s12021-019-09419-w. [DOI] [PubMed] [Google Scholar]
- Esmaeilzadeh S., Belivanis D.I., Pohl K.M., Adeli E. End-to-end Alzheimer’s Disease diagnosis and biomarker identification. Int. Workshop Mach. Learn. Med. Imaging. 2018:337–345. doi: 10.1007/978-3-030-00919-9_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etminani K., Soliman A., Davidsson A., Chang J.R., Martínez-Sanchis B., Byttner S., Ochoa-Figueroa M. A 3D deep learning model to predict the diagnosis of dementia with Lewy bodies, Alzheimer’s Disease, and mild cognitive impairment using brain 18F-FDG PET. Eur. J. Nucl. Med. Mol. Imaging. 2022;49(2):563–584. doi: 10.1007/s00259-021-05483-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq A., Anwar S., Awais M., Rehman S. A deep cnn based multi-class classification of Alzheimer’s Disease using mri. 2017 IEEE Int. Conf. Imaging Syst. Tech. (Ist. ) 2017:1–6. [Google Scholar]
- Fulton L.V., Dolezel D., Harrop J., Yan Y., Fulton C.P. Classification of Alzheimer’s Disease with and without imagery using gradient boosted machines and resnet-50. Brain Sci. 2019;9(9):212. doi: 10.3390/brainsci9090212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, L., Pan, H., Liu, F., Xie, X., Zhang, Z., & Han, J. (2018, 07). Brain disease diagnosis using deep learning features from longitudinal mr images: Second international joint conference, apweb-waim 2018, Macau, China, July 23–25, 2018, proceedings, part i. In (p. 327–339).
- Geurts P., Ernst D., Wehenkel L. Extremely randomized trees. Mach. Learn. 2006;63:3–42. [Google Scholar]
- Good C.D., Johnsrude I.S., Ashburner J., Henson R.N., Friston K.J., Frackowiak R.S. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage. 2001;14(1):21–36. doi: 10.1006/nimg.2001.0786. [DOI] [PubMed] [Google Scholar]
- Good C.D., Scahill R.I., Fox N.C., Ashburner J., Friston K.J., Chan D., Frackowiak R.S. Automatic differentiation of anatomical patterns in the human brain: validation with studies of degenerative dementias. Neuroimage. 2002;17(1):29–46. doi: 10.1006/nimg.2002.1202. [DOI] [PubMed] [Google Scholar]
- Gray K., Aljabar P., Heckemann R., Hammers A., Rueckert D. Random forest-based similarity measures for multi-modal classification of Alzheimer’s Disease. NeuroImage. 2012:65. doi: 10.1016/j.neuroimage.2012.09.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, K., Zhang, X., Ren, S., & Sun, J. (2015). Deep residual learning for image recognition. CoRR, abs/1512.03385.
- Hidalgo-Muñoz A.R., Ramírez J., Górriz J.M., Padilla P. Regions of interest computed by SVM wrapped method for Alzheimer’s Disease examination from segmented MRI. Front. Aging Neurosci. 2014;6:20. doi: 10.3389/fnagi.2014.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini-Asl, E., Keynton, R., & El-Baz, A. (2016). Alzheimer’s disease diagnostics by adaptation of 3d convolutional network. In 2016 ieee international conference on image processing (icip) (pp. 126–130).
- Islam J., Zhang Y. Brain mri analysis for Alzheimer’s Disease diagnosis using an ensemble system of deep convolutional neural networks. Brain Inform. 2018:5. doi: 10.1186/s40708-018-0080-3. 05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M., Pechaud M., Smith S. BET2: MR-based estimation of brain, skull and scalp surfaces. Elev. Annu. Meet. Organ. Hum. Brain Mapp. 2005:2005. [Google Scholar]
- Khazaee A., Ebrahimzadeh A., Babajani-Feremi A. Identifying patients with Alzheimer’s disease using resting-state fMRI and graph theory. Clin. Neurophysiol. 2015;126(11):2132–2141. doi: 10.1016/j.clinph.2015.02.060. [DOI] [PubMed] [Google Scholar]
- Kim J., et al. Identification of Alzheimer's disease and mild cognitive impairment using multimodal sparse hierarchical extreme learning machine. Hum. Brain Mapp. 2018;39(9):3728–3741. doi: 10.1002/hbm.24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Liu M. Alzheimer’s disease diagnosis based on multiple cluster dense convolutional networks. Comput. Med. Imaging Graph. 2018:70. doi: 10.1016/j.compmedimag.2018.09.009. [DOI] [PubMed] [Google Scholar]
- Liu M., Cheng D., Wang K., Wang Y. Multi-modality cascaded convolutional neural networks for Alzheimer’s Disease diagnosis. Neuroinformatics. 2018:16. doi: 10.1007/s12021-018-9370-4. 03. [DOI] [PubMed] [Google Scholar]
- Luo S., Li X., Li J. Automatic Alzheimer’s Disease recognition from mri data using deep learning method. J. Appl. Math. Phys. 2017;05:1892–1898. 01. [Google Scholar]
- Moosmann F., Triggs B., Jurie F. Fast discriminative visual codebooks using randomized clustering forests. Adv. Neural Inf. Process. Syst. 2006:19. [Google Scholar]
- Moradi E., Pepe A., Gaser C., Huttunen H., Tohka J. Machine learning framework for early mri-based Alzheimer’s conversion prediction in mci subjects. NeuroImage. 2014:104. doi: 10.1016/j.neuroimage.2014.10.002. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosconi L., Brys M., Glodzik-Sobanska L., De Santi S., Rusinek H., De Leon M.J. Early detection of Alzheimer’s Disease using neuroimaging. Exp. Gerontol. 2007;42(1–2):129–138. doi: 10.1016/j.exger.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Nguyen D.T., Ryu S., Qureshi M.N.I., Choi M., Lee K.H., Lee B. Hybrid multivariate pattern analysis combined with extreme learning machine for Alzheimer’s dementia diagnosis using multi-measure rs-fmri spatial patterns. PloS One. 2019;14(2) doi: 10.1371/journal.pone.0212582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisot S., Ktena S.I., Ferrante E., Lee M., Guerrero R., Glocker B., Rueckert D. 06). Disease prediction using graph convolutional networks: Application to autism spectrum disorder and Alzheimer’s Disease. Med. Image Anal. 2018:48. doi: 10.1016/j.media.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Paszke A., Gross S., Massa F., Lerer A., Bradbury J., Chanan G., et al. Pytorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019:32. [Google Scholar]
- Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Duchesnay E. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- Raj R., Mathew J., Kannath S., Rajan J. Crossover based technique for data augmentation. Comput. Methods Prog. Biomed. 2022;218 doi: 10.1016/j.cmpb.2022.106716. [DOI] [PubMed] [Google Scholar]
- Report W.A. World Alzheimer Rep. 2018:2018. [Google Scholar]
- Rodrigues F., Silveira M. Longitudinal FDG-PET features for the classification of Alzheimer’s Disease. 2014 36th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2014:1941–1944. doi: 10.1109/EMBC.2014.6943992. [DOI] [PubMed] [Google Scholar]
- Rusinek H., Endo Y., De Santi S., Frid D., Tsui W.-H., Segal S., De Leon M. Atrophy rate in medial temporal lobe during progression of alzheimer disease. Neurology. 2004;63(12):2354–2359. doi: 10.1212/01.wnl.0000148602.30175.ac. [DOI] [PubMed] [Google Scholar]
- Singh R., Bharti V., Purohit V., Kumar A., Singh A., Singh S. MetaMed: Few-shot medical image classification using gradient-based meta-learning. Pattern Recognit. 2021;120 [Google Scholar]
- Smith S. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk H.-I., Lee S., Shen D. Latent feature representation with stacked auto-encoder for ad/mci diagnosis. Anat. Hefte. 2015;220(2):841–859. doi: 10.1007/s00429-013-0687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk H.-I., Lee S.-W., Shen D. Hierarchical feature representation and multimodal fusion with deep learning for ad/mci diagnosis. NeuroImage. 2014:101. doi: 10.1016/j.neuroimage.2014.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapiola T., Pennanen C., Tapiola M., Tervo S., Kivipelto M., Hänninen T., et al. MRI of hippocampus and entorhinal cortex in mild cognitive impairment: a follow-up study. Neurobiol. Aging. 2008;29(1):31–38. doi: 10.1016/j.neurobiolaging.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Vapnik V.N. Springer-Verlag; Berlin, Heidelberg: 1995. The Nature of Statistical Learning Theory. [Google Scholar]
- Vemuri P., Jones D.T., Jack C.R. Resting state functional MRI in Alzheimer’s Disease. Alzheimer’s Res. Ther. 2012;4(1):1–9. doi: 10.1186/alzrt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-Y., Yu J.-T., Liu Y., Yin R.-H., Wang H.-F., Wang J., Tan L. Voxel-based meta-analysis of grey matter changes in Alzheimer’s Disease. Transl. Neurodegener. 2015;4(1):1–9. doi: 10.1186/s40035-015-0027-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert D. Stacked generalization. Neural Netw. 1992;5:241–259. [Google Scholar]
- Zeiler, M.D. and Fergus, R., 2014, Visualizing and understanding convolutional networks. In European conference on computer vision (pp. 818–833).
- Zhang D., Wang Y., Zhou L., Yuan H., Shen D., Alzheimer's Disease Neuroimaging Initiative Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Sun L., Huang J., Han L., Zhang D. Dual attention multi-instance deep learning for Alzheimer’s disease diagnosis with structural MRI. IEEE Trans. Med. Imaging. 2021;40(9):2354–2366. doi: 10.1109/TMI.2021.3077079. [DOI] [PubMed] [Google Scholar]