Summary
Human pluripotent stem cells (hPSCs) regularly and irreversibly show the erosion of X chromosome inactivation (XCI) by long non-coding RNA (lncRNA) XIST silencing, causing challenges in various applications of female hPSCs. Here, we report reliable methods to reactivate XIST with monoallelic expression in female hPSCs. Surprisingly, we find that the editing of XIST regulatory regions by Cas9-mediated non-homologous end joining is sufficient for the reactivation of XIST by endogenous systems. Proliferated hPSCs with XIST reactivation show XCI from an eroded X chromosome, suggesting that hPSCs with normal dosage compensation might lead to a growth advantage. Furthermore, the use of targeting vectors, including the XIST regulatory region sequences and selection cassette, enables XIST reactivation in hPSCs with high efficiency. XIST-reactivated hPSCs can show the restoration of differentiation potential. Thus, our findings demonstrate that XIST re-expression is a beneficial method to maximize the use of female hPSCs in various applications, such as proper disease modeling.
Keywords: dosage compensation, pluripotent cells, gene editing, DNA methylation, differentiation potential
Graphical abstract
Highlights
-
•
Erosion of XCI by XIST silencing affects neuronal differentiation in female hPSCs
-
•
Editing of XIST promoter regions by Cas9 nuclease causes DNA demethylation
-
•
Editing of XIST promoter regions induces XIST reactivation and de-erosion of XCI
-
•
De-erosion of XCI restores differentiation potential in female hPSCs
Motivation
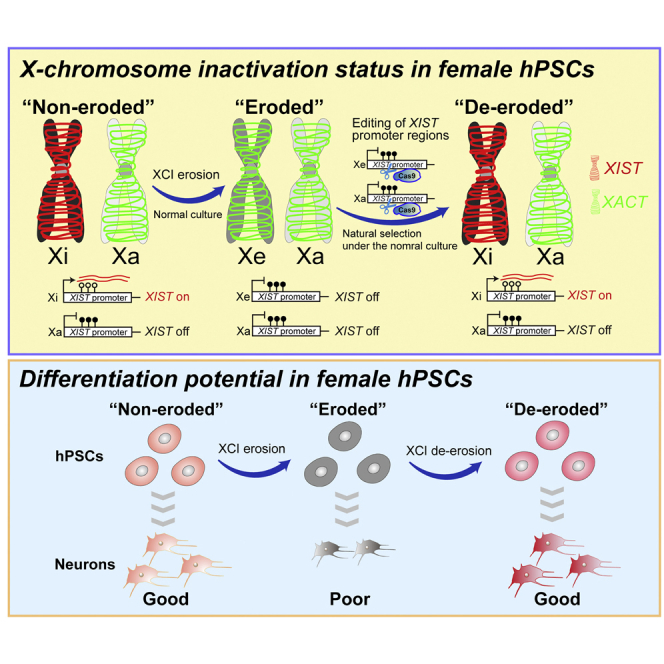
Erosion of X chromosome inactivation (XCI) is caused by XIST silencing. Erosion of XCI is inevitable and irreversible in female hPSCs, causing the limitation of female hPSC utilities. However, there are no reliable and reproducible methods to reactivate XIST, leading to de-erosion in female hPSCs. To resolve the long-standing problem in female hPSCs, we developed methods to restore XIST expression in a monoallelic manner, resulting in the de-erosion of XCI. De-erosion of XCI is beneficial to various applications of female hPSCs including patient-specific iPSCs.
Erosion of X chromosome inactivation (XCI) by XIST silencing is irreversible in female hPSCs. Motosugi et al. demonstrate that editing of XIST promoter regions with hyper DNA methylation by Cas9 nuclease causes demethylation and XIST reactivation. XIST reactivation results in the de-erosion of XCI and restoration of differentiation potential.
Introduction
The generation of human pluripotent stem cells (hPSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), has proved to be important for numerous medical applications and drug discoveries and elucidated basic human biology in vitro (Takahashi et al., 2007; Thomson et al., 1998; Yamanaka, 2020). Although hPSCs are a powerful tool to understand biological phenomena based on genetic background, “erosion” of X chromosome dosage compensation in female hPSCs is an inevitable problem, induced by the irreversible silencing of the long non-coding RNA (lncRNA) XIST; GenBank: NG_016172. XIST is expressed from one of two X chromosomes to archive X chromosome inactivation (XCI), and the expression is maintained in female somatic cells (Augui et al., 2011; Penny et al., 1996). Previous studies have shown that XIST repression in female hPSCs causes the overexpression of X-linked genes and results in a poor differentiation ability (Anguera et al., 2012; Fukuda et al., 2021; Mekhoubad et al., 2012; Patel et al., 2017). Therefore, XIST dysregulation may be one of the largest and most underestimated sources of variability hampering reproducibility and disease modeling in female hPSC research.
While naive conversion from hPSCs has been shown to reactivate XIST, the efficiency depends on the genetic background (Guo et al., 2017; Sahakyan et al., 2017). Furthermore, the selection of the XIST reactivation allele is inconsistent when naive cells revert to the primed hPSC state (An et al., 2020; Sahakyan et al., 2017). Thus, unless effective solutions are identified, XIST silencing can limit the deployment of female hPSC lines in translational efforts. If women are to equally benefit from the potential revolution in human cell therapies, and if hPSCs are to be used to their full potential, it is essential to develop a new method to restore XCI in the eroded X chromosome.
In the present study, we highlighted the importance of XIST expression in female hPSCs upon differentiation. To reduce the undesirable effects of irreversible XIST repression, we have developed a novel and reproducible method to restore XCI by editing XIST regulatory elements in female hPSCs, leading to XIST reactivation by the endogenous expression system. Female hPSCs with XIST reactivation, including X-linked disease iPSCs, undergo restoration of differentiation ability. Thus, the method of reactivating XIST expression is useful for evaluating appropriate phenotypes, including in disease modeling applications, in female hPSCs.
Results
XIST repression in female hPSCs commonly affects the differentiation modules but not the core pluripotency factors
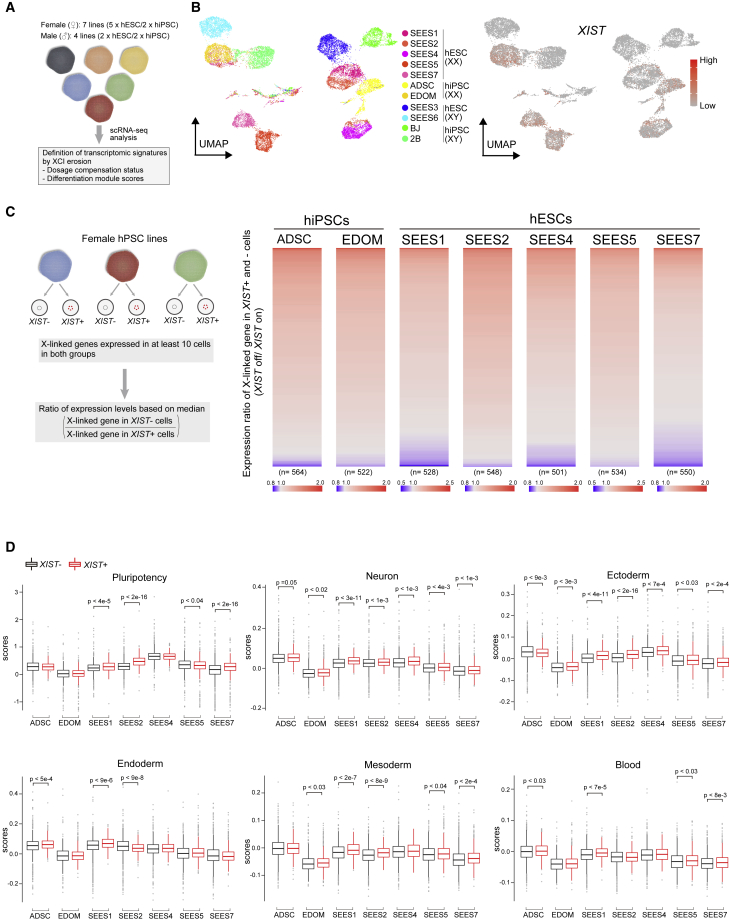
To obtain insights into transcriptomic features based on the XCI status without considering the effect of the genetic background, we performed single-cell RNA sequencing (scRNA-seq) analysis using 11 hPSC lines (seven females and four males), including both ESCs and iPSCs. In female hPSC lines, to compare the transcriptional effects driven by XCI erosion, we used a mixed population composed of XIST+ and XIST– cells with the same passage number (Figure 1A). Dimensionality reduction analysis (Stuart et al., 2019) by uniform manifold approximation and projection (UMAP) revealed that the genetic background was a strong driver for cluster segregation (Figure 1B). The XIST expression status and derivational origins were not major drivers of cluster segregation (Figure 1B). Since UMAP analysis was performed using all transcripts, to examine the effect of XIST expression on the expression dosage of X-linked genes, we calculated the median expression levels in individual X-linked genes between XIST+ and XIST– cells in each female hPSC line. The comparison analysis of the median expression levels showed the tendency of over-dosage of X-linked genes in XIST– cells in all lines examined (Figure 1C). Moreover, the average expression levels of X-linked genes in XIST– hPSCs were significantly upregulated compared with those in XIST+ and male hPSCs (Figure S1A), consistent with the findings from previous reports (Anguera et al., 2012; Mekhoubad et al., 2012).
Figure 1.
XIST repression commonly affects the differentiation modules, but not core pluripotency, in female hPSCs
(A) Analysis scheme of single-cell RNA sequencing (scRNA-seq) analysis in 11 hPSC lines.
(B) UMAP analysis of 11 hPSC lines. Left: UMAP based on genetic background. Right: XIST expression status. The heatmap shows the expression levels.
(C) Expression ratio of X-linked genes in XIST+ and XIST– female hPSCs based on median levels. The X-linked genes expressed in at least ten cells in both XIST+ and XIST– groups were used for the analysis. The heatmap shows the ratio (XIST–/XIST+), indicating that red color is overexpressed in XIST– cells. n indicates the number of X-linked genes analyzed.
(D) Module scores related to differentiation and pluripotency. The pluripotency module was based on the expression of OCT4, SOX2, and NANOG (Wang et al., 2012). The differentiation markers previously identified were used for the module calculation (Bock et al., 2011). The comparisons of each module were based on the XIST expressions status. p values were calculated using Wilcoxon’s rank-sum test.
Next, we investigated if the XIST expression status impacts the expression of the core pluripotency factors OCT4, SOX2, and NANOG (Wang et al., 2012) in hPSCs. Although the extent of expression of each gene differed among the lines, especially that of NANOG, we did not observe statistically significant differences between the factors in XIST+ and XIST– cells (Figure S1B). However, we found that the expression levels of UTF1, linked to the heterogeneity of the hPSC population (Tan et al., 2007), greatly differed among the lines (Figure S1C). The expression levels in XIST+ cells were significantly higher than those in XIST– cells in the SEES2 line (Figure S1C). Interestingly, UMAP analysis in SEES2 showed an XIST+ cluster with UTF1 expression (Figure S1D), suggesting that extensive erosion might be associated with the dysregulation of pluripotency-related gene expression.
Findings from previous studies have suggested that hPSC lines with strong XCI erosion, as evaluated using the DNA methylation status, showed poor differentiation scores, which were calculated using the expression levels in a subset of genes (module) (Patel et al., 2017; Saelens et al., 2018). However, given that the genetic background substantially affects the transcriptomic features, including the erosion status, in hPSCs (Figure 1C), the score comparisons between XIST+ and XIST– cells in the same line would indicate the direct effect of XIST suppression. We calculated the differentiation scores based on findings from previously validated gene sets in hPSCs (Bock et al., 2011) as well as the core pluripotency factors (OCT4/SOX2/NANOG). Interestingly, different from the results in individual gene-expression statuses (Figure S1B), the core pluripotency module showed significant differences between XIST+ and XIST– cells in some lines, but the tendency depended on the lines (Figure 1E). In differentiation modules, the endoderm scores also showed inconsistent tendencies in each line (Figure 1E). However, in other lineages, the differentiation scores of XIST+ cells tended to be higher than those of XIST– cells in most lines (Figure 1E). Especially, the neuronal scores of XIST+ cells were higher than those of XIST– cells in all lines, and the differences in six out of seven lines were statistically significant (Figure 1E). These results indicate that XIST repression does not lead to a consistent tendency in the core pluripotency factors, but it commonly affects the differentiation modules, with a particularly negative effect on neuronal differentiation.
The XIST expression status is linked to the stages of neuronal differentiation
Next, to address whether the molecular characteristics of the neuronal lineages of hPSCs are linked to the phenotypes, we conducted spinal motor neuron differentiation using defined protocols of two-dimensional culture (Klim et al., 2019). We first evaluated the XCI status in differentiating neurons by immunofluorescence with H3K27me3/H2AK119ub, XCI hallmarks that overlap with the XIST RNA (Anguera et al., 2012; Fukuda et al., 2021; Motosugi et al., 2021; Sahakyan et al., 2017), and neuronal cell markers. Unexpectedly, immunofluorescence combined with XIST RNA-fluorescence in situ hybridization (immuno-FISH) analysis revealed that a complete overlap of XIST RNA with XCI hallmarks (H3K27me3/H2AK119ub) was observed only in hPSCs, not in differentiating and primary cells (Figures S1E and S1F). In particular, most motor neurons (MNs) lacked H2AK119ub modification (Figures S1E and S1F). These results indicated that epigenetic modification on Xi depends on cell types even when XIST is statedly expressed. Thus, we conducted an immuno-FISH assay to evaluate if the XIST expression status is linked to the neuronal differentiation stage.
Seven female hPSC lines, composed of a mixture of XIST+ and XIST– cells in each line, were differentiated, and the status was analyzed at two time points (neuronal progenitor/post-mitotic phases) (Figure S2A). To exclude the possibility that the starting number of XIST+ or XIST– cells affects the differentiation status evaluation, we counted the number of marker+/XIST+ to XIST+ cells or marker+/XIST– to XIST– cells in differentiated cells. On day 6 of differentiation, the population of cells with PAX6+/XIST+ was significantly smaller than that with PAX6+/XIST–in most lines (Figures S2B and S2C). However, on day 10 of differentiation, among the seven female hPSC lines, five showed a significantly higher proportion of ISL1+/XIST+ cells than of ISL1+/XIST– cells (Figures S2B and S2C), indicating that XIST repression negatively affected the generation of post-mitotic MNs.
Next, we examined if differentiation into post-mitotic neuronal cells in cortical organoid development is also linked to the XIST expression status (Figure S2D). Immuno-FISH analysis showed that considerably varied phenotypes and organoids from some lines formed a typical morphology, but no marker was detected in male or female lines (Figures S2E–S2G). In the female hPSC lines, at day 90 of differentiation from hPSCs, three lines (ADSC-iPS, SEES1, and SEES7) expressed all markers (SATB2/TBR1/CTIP2) (Bhaduri et al., 2020; Thomsen et al., 2016; Velasco et al., 2019) in at least three organoids (Figures S2F and S2G). Consistent with the results observed in MNs, immuno-FISH analysis in the three female lines revealed that the differentiation potential of XIST+ cells toward cortical neuron markers (SATB2 and TBR1) tended to be greater than that of XIST– cells (Figure S2G).
To evaluate the differentiation status of cortical organoids based on XIST expression in a less biased manner, we analyzed the scRNA-seq data using the HUES66 female line (Velasco et al., 2019). We first examined whether differentiated cells also exhibit the over-dosage of X-linked genes in XIST– cells. Since the differentiating cells show a wide transcriptome diversity (Thomsen et al., 2016), we focused on the cells in the same cluster identified by markers (Bhaduri et al., 2020; Thomsen et al., 2016). We examined the median expression levels of individual X-linked genes in XIST+ and XIST– groups in classified cell types (Figures S2H and S2I). The analysis revealed that the over-dosage of X-linked genes in various cell types derived from HUES66, especially in the radial glial cell population, showed a strong tendency for over-dosage (Figure S2J). These results indicated that XIST repression leads to the overexpression of X-linked genes in differentiating cells, consistent with the findings from previous reports (Patel et al., 2017). Next, we calculated the percentage of XIST+ and XIST– cells in each cell type and compared the ratio. Interestingly, the cells from the initial phase of neuronal differentiation, such as radial glial cells, tended to include a large population in XIST– cells compared with that of XIST+ cells (Figure S2K), whereas the population of XIST+ cells was larger among neuronal cells (Figure S2K), and the XIST gene was identified as a cluster marker of CTIP2 excitatory neurons (Table S1). In the neurons, the number of cells marked by CCNI or NEUROD2 was greater in the XIST– population (Figure S2K). However, trajectory analysis showed that these populations showed a low pseudotime, indicating the initial differentiation phases (Figure S2L). Consistent with the results observed in MNs (Figure S2C), in cortical differentiation from hPSCs, the status of XIST+ cells tended to be more matured compared with that of XIST– cells. Taken together, considering findings from previous reports that showed that XIST repression is irreversible before/after differentiation and negatively affects the differentiation potential of female hPSCs (Anguera et al., 2012; Fukuda et al., 2021; Mekhoubad et al., 2012; Salomonis et al., 2016; Vallot et al., 2015), our findings demonstrated that the high module scores of neuronal lineage in XIST+ female hPSCs are linked to a phenotype.
Experimental results using female hPSCs with autosomal gene mutations differ based on the XIST expression status in hPSCs
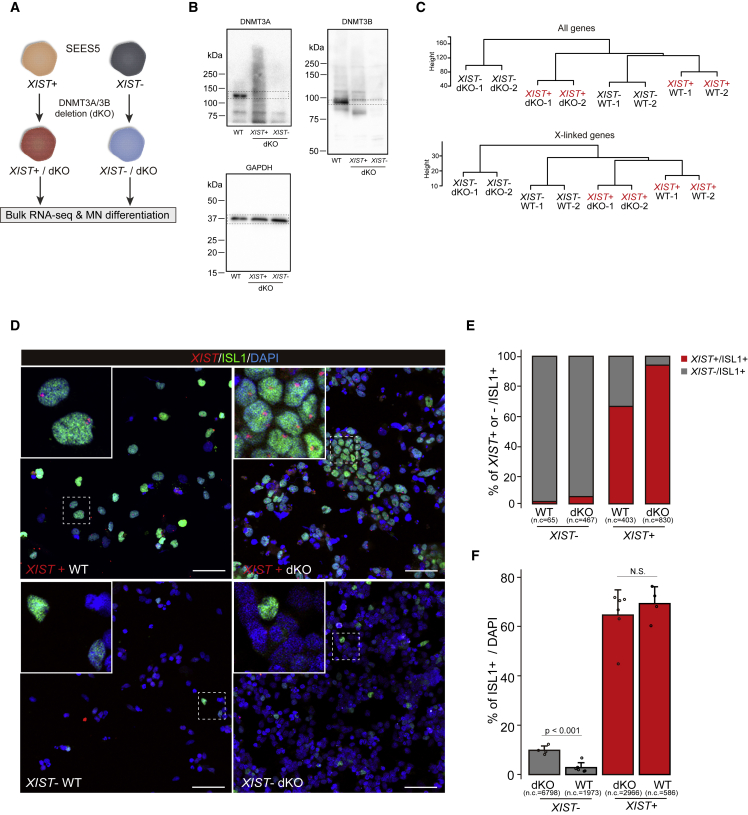
The deficiency of the de novo DNA methyltransferases DNMT3A and 3B impairs MN differentiation in male hPSCs (Ziller et al., 2018). Recently, we found that the deletion of both enzymes prevents XIST silencing in female hPSCs (Fukuda et al., 2021). The findings from these studies led us to question whether the XIST expression status in female hPSCs with autosomal gene defects affects the experimental results. To address this, we generated a DNMT3A/3B double-mutant knockout (dKO) using XIST+ or XIST– EES5 lines (Figures 2A and 2B).
Figure 2.
The X chromosome inactivation (XCI) status affects the results of the autosomal gene functional assay of female hPSCs
(A) Experimental scheme for the generation of a DNMT3A/3B double-mutant knockout (dKO) line. The XIST+ or XIST– SEES5 line was used to examine if the XCI status affects MN differentiation.
(B) Western blot analysis, with GAPDH used as the loading control.
(C) Bulk RNA-seq analysis. Hierarchical clustering analysis using all genes (top) and X-linked genes (bottom).
(D) Immuno-FISH analysis of MNs. ISL1 immunofluorescence combined with XIST RNA-FISH (immuno-FISH) analysis was performed to evaluate the differentiation efficiency. Representative images of the ISL1- and XIST expression statuses in the dKO and wild-type (WT) lines, respectively. Scale bar represents 50 μm.
(E and F) Quantification results of the immuno-FISH analysis. The percentages of XIST+ or XIST– cells with ISL+ are shown (E). The differentiation efficiencies evaluated using ISL1+ cells are shown (F). Each dot and n.c. indicate the percentage in the observed area and the total number of cells analyzed, respectively. Error bars show standard deviations. p values were calculated using Student’s t test.
We first checked the transcriptome status by bulk RNA-sequencing in each dKO line. Hierarchical clustering analysis using all transcripts, including autosomal genes, showed that the dKO samples were close to each other, regardless of the XIST expression status (Figure 2C). However, when clustering analysis was conducted using X-linked genes, the clusters were formed based on the XIST expression status (Figure 2C), indicating that the expression status of X-linked genes depends on the XIST expression status rather than the loss of de novo DNA methyltransferases in the SEES5 line.
The MN differentiation experiments using immuno-FISH revealed that XIST was not reactivated in the majority of MNs upon DNMT3A/3B deletion (Figures 2D and 2E). Meanwhile, MNs derived from XIST+ cells retained XIST expression (Figures 2D and 2E). Surprisingly, on day 14, the number of ISL1+ cells was >60% in both XIST+ lines (Figures 2D and 2F), and the efficiency of the dKO line was comparable to that of wild type (Figures 2D and 2F). However, the efficiency markedly reduced in XIST– lines (<10% in both lines; Figures 2D and 2F). Furthermore, the dKO line in XIST– cells exhibited a significantly higher efficiency compared with the wild-type XIST– line (Figures 2D and 2F). However, given that XIST repression causes XCI erosion, we cannot conclude whether the findings using XIST– lines result from DNMT3A/3B dysfunction and/or XCI erosion. Taken together, these results demonstrated that the XCI status in female hPSCs affects not only X chromosome dosage compensation but also the outcomes of the autosomal gene functional assay.
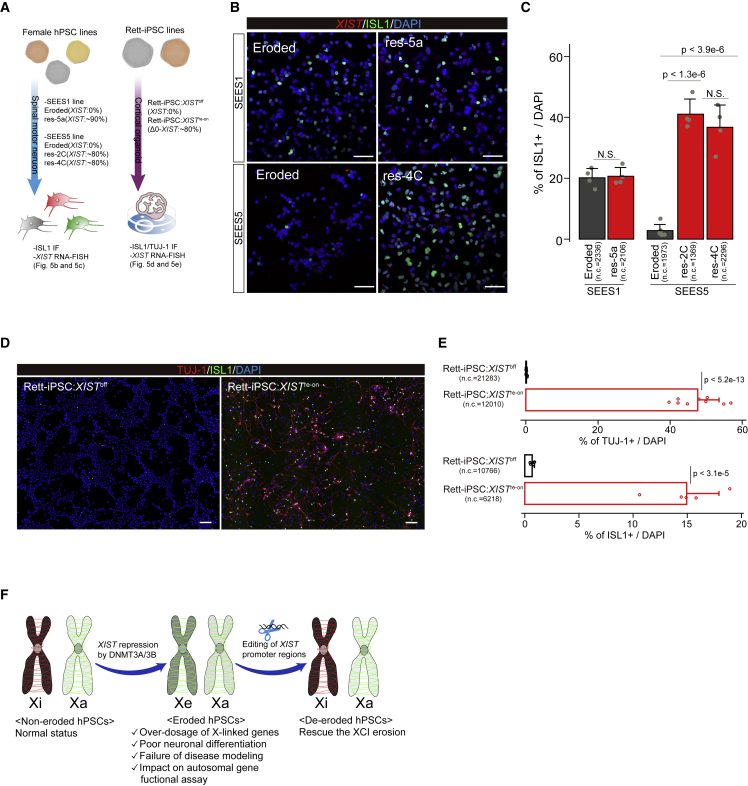
Editing of XIST regulatory elements leads to the de-erosion of female hPSCs
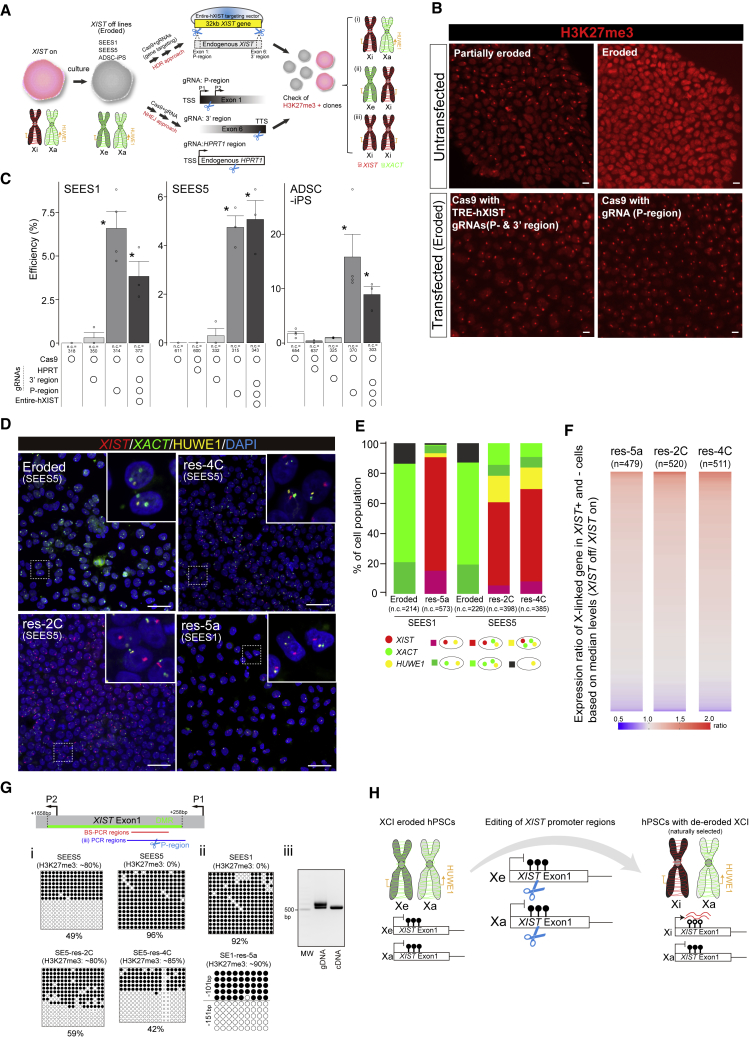
Thus far, our findings have revealed the importance of XIST expression in the cellular phenotype of female hPSCs. However, there is no reliable method to restore XIST expression in the current culture system of hPSCs. Even when naive conversion is applied, the efficiency depends on the genetic background (Guo et al., 2017; Sahakyan et al., 2017). Recently, we found that the endogenous reactivation of XIST by the clustered regularly interspaced short palindromic repeats (CRISPR) activation system can induce XIST transcripts, but the XIST RNA does not act in cis (Fukuda et al., 2021; Motosugi et al., 2021), suggesting that epigenetic modifications affecting XIST transcript behavior might be altered in the XIST gene of eroded female hPSCs. To overcome these challenges, we developed two new approaches to reacquire the XIST function via an endogenous transcription system. The first approach is based on a gene-targeting strategy using CRISPR-Cas9-mediated homology-directed repair (HDR) (Doudna and Charpentier, 2014), for which we introduced a new XIST gene into the original region (Figure 3A). In the second approach, we tested the possibility of non-homologous end joining (NHEJ) (Doudna and Charpentier, 2014)-mediated gene reactivation (Figure 3A). Since the findings of a recent study suggested that Cas9-mediated double-strand breaks (DSBs) induce de novo DNA methylation (DNAme) around guide RNA (gRNA)-targeting sites (Cali et al., 2019), and this indicates that epigenetic status alters before/after DSBs, we inferred that if an opposite event, DNA de-methylation, occurs in XIST– female hPSCs during NHEJ, XIST repressed by DNAme would be reactivated. For the HDR approach, we generated a targeting vector containing the entire 32 kb human XIST gene (Entire-hXIST; Figure 3A). In the NHEJ approach, we designed three sgRNA regions; promoter regions (P regions), 3′ terminal regions (3′ region), and HPRT1 regions (as control) (Figure 3A). Using three female lines without XIST expression (ADSC-iPS, SEES1, and SEES5), we conducted various transfection experiments and assessed the H3K27me3 status.
Figure 3.
Editing of XIST regulatory elements leads to the de-erosion of female hPSCs
(A) Experimental scheme for XIST re-expression in eroded female hPSCs. Gene targeting (top panel: HDR approach) and DSB-mediated XIST reactivation (bottom panel: NHEJ approach) are shown. For the HDR approach, a large construct (Entire-hXIST) containing the full-length human XIST gene was used. The gRNAs were designed at the 5′ promoter regions within exon 1 (P region) and 3′ regions of exon 6 (3′ region). The P region is located between the P1 and P2 promoter regions (Chapman et al., 2014). For the NHEJ approach, the gRNAs of the P region, 3′ region, or HPRT1 region (as negative control) were used, respectively. XIST– female hPSC lines were used, and the H3K27me3 statuses were examined to determine whether XCI was reacquired. The three expected XCI patterns are shown [i-iii]. Xi: inactive X chromosome, Xe: eroded X chromosome, and Xa: active X chromosome.
(B and C) Efficiency test of clones with H3K27me3 foci reacquisition in various combinations. Representative images (B) and quantification results (C) are shown on the left and right, respectively. Clones containing at least ten cells with the H3K27me3 foci were counted as positive clones. Each dot and n.c. indicates an independent experiment and the total number of clones counted, respectively. Error bars show the SD. p values were calculated using Student’s t test. Scale bar represents 10 μm.
(D and E) RNA-FISH analysis. XIST/XACT/HUWE1 were examined. Representative images (D) and quantification results (E) are shown in the left and right, respectively. n.c. refers to the number of cells analyzed. Scale bar represents 50 μm.
(F) Expression ratio of X-linked genes in XIST+ and XIST– female hPSCs based on median levels. The heatmap shows the ratio of X-linked genes (XIST–/XIST+). The X-linked genes expressed in at least ten cells in both the XIST+ and XIST– groups were used for the analysis. n refers to the number of X-linked genes analyzed.
(G) Bisulfite sequencing (BS) analysis around the XIST promoter region. Differentially methylated regions (DMRs) with approximately 50% of the DNA methylated regions were identified in IMR-90 cells (Roadmap Epigenomics project data). BS and PCR regions in (iii) are shown in green, red, and blue, respectively. (i) and (ii) indicate BS analysis in the SEES5 background (i) and SE-res-5a (ii). (iii) PCR analysis using genomic DNA (gDNA) or cDNA from the SE5-res-5a line.
(H) Summary of XCI-reacquired female hPSCs formed via the editing of XIST regulatory regions. After the induction of DSB by Cas9 around the XIST promoter regions with DNAme, the cells with XCI reacquired on the Xe allele were naturally selected in a normal culture.
Seven days after transfection, the HDR approach resulted in the generation of a significantly greater number of H3K27me3+ clones in all three lines compared with that in controls (Figures 3B and 3C). Surprisingly, we found that the NHEJ approach using P region gRNA also resulted in the significant and efficient generation of H3K27me3+ clones (Figures 3B and 3C). Moreover, we did not obtain clones with two H3K27me3 foci (Figure 3B), indicating that XCI with two X chromosomes (Figure 3Aiii) would be harmful to cell viability.
We further determined whether NHEJ-mediated gene reactivation occurs in DNAme-regulated genes other than XIST. We found that GTL2, an imprinted gene regulated by DNAme (Stadtfeld et al., 2010), was repressed in the BJ- and 2B-iPS lines, whereas SEES3 and SEES6 lines expressing GTL2 showed DNA hypomethylated states at the regulatory element known as intergenic differentially methylated regions (IG-DMRs) (Figures S3A and S3B). We designed gRNA targeting the IG-DMR (da Rocha et al., 2008) and performed a transfection experiment (Figure S3C). The qPCR assay demonstrated that the GTL2 expression levels were significantly upregulated in the cells with Cas9/gRNA (IG-DMR) compared with that in control cells in both BJ- and 2B-iPS lines (Figure S3D). The DNAme levels in IG-DMR were reduced in the transfected cells when gRNA targeting IG-DMR was used (Figure S3E). These results indicated that CRISPR-Cas9-mediated NHEJ causes the reactivation of genes repressed by DNAme.
Using one of two approaches, we obtained clones with single H3K27me3 foci in >85% of cells, termed SE1-res-5a (SEES1 line), SE5-res-2C (SEES5 line), and SE5-res-4C (SEES5 line). We used the HDR approach for the generation of SE5-res-2C and SE5-res-4C lines. However, using genotyping PCR, we detected the sequences spanning XIST and exogenous sequences included in Entire-hXIST vector only in the SE5-res-2C line (Figure S4Ai and S4Aii). Sanger sequencing analysis also confirmed the presence of single-nucleotide variants (SNVs) derived from Entire-hXIST only in SE5-res-2C (Figure S4Aiii). We also confirmed the integration of exogenous sequences derived from at 3′ region of XIST in the SE5-res-2C line, but at the 5′ regions of XIST, even though we tested several primers spanning the sequences exogenous and upstream of the XIST transcription start sites, none of them detected the expected amplicon. Meanwhile, the DNA-FISH experiment using the SE5-res-2C line showed two XIST/XACT signals, respectively, indicating no additional XIST integration (Figure S4B). In terms of XIST reactivation in the SE5-res-2C line, recombination occurred in the cells; however, since editing of the P region was sufficient to reactivate XIST via NHEJ, we cannot determine if XIST reactivation resulted from recombination.
We found scars around the P region in the SE5-res-4C line, but the P region was non-scarred in the SE5-res-2C line (Figure S4C), indicating that H3K27me3 foci reacquisition in SE5-res-4C was mediated by NHEJ; however, we did not exclude the possibility that HDR did not occur in the SE5-res-4C line because the available SNV information is limited. In the SE1-res-5a line, we employed the NHEJ approach, and genotyping PCR analysis around the P region revealed the presence of two alleles with different deletions (Figures S4D and S4E).
Next, we examined whether XIST was expressed from an eroded (Figure 3Ai) or active (Figure 3Aii) allele in the three lines. Since HUWE1 is often protected from erosion and is used as an active allele marker (Motosugi et al., 2021; Patel et al., 2017; Sahakyan et al., 2017), we conducted RNA-FISH analysis using XIST, HUWE1, and XACT, a marker of eroded hPSCs (Vallot et al., 2015). In the assay, 68%–98% of XIST+ cells exhibited monoallelic XACT expression, and most of the cells retained HUWE1 expression (Figures 3D and 3E), indicating the occurrence of X-linked gene silencing and expression of XIST from the eroded allele (Figure 3Ai). The scRNA-seq analysis also showed that XIST reactivation results in the downregulation of X-linked genes (Figure 3F). In these clones, we observed reduced DNAme levels and the expression of XIST from an unmethylated allele (Figure 3G), consistent with the findings from a previous report (Fukuda et al., 2021). Taken together, these results demonstrated that XCI erosion can be corrected by XIST reactivation with the endogenous transcriptional system, and cells with normal XCI proliferate naturally under normal cultural conditions without specific selection; presumably, normal dosage compensation leads to a growth advantage (Figure 3H).
An efficient HDR-based screening method for XIST-reactivated hPSCs and maintenance of XIST+ hPSCs
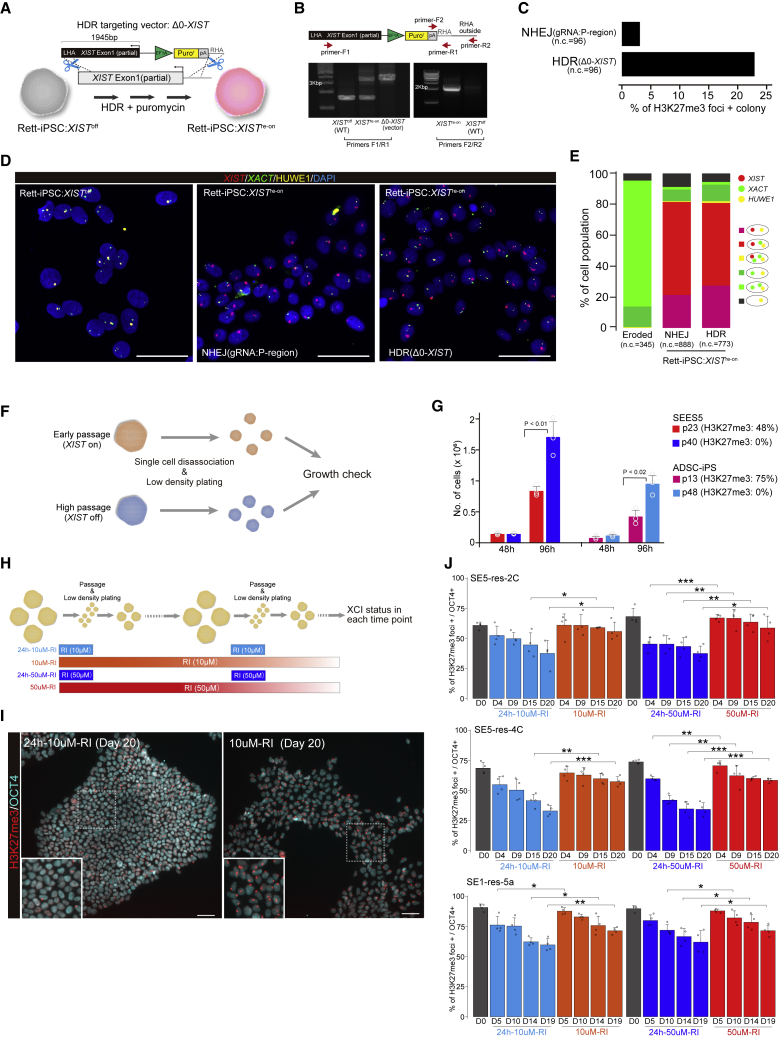
Although the editing of XIST P regions by NHEJ or HDR approaches served as a useful tool for XIST reactivation, the efficiency of obtaining positive clones for H3K27me3 remained low. In particular, the use of Entire-hXIST (large size >30 kb) resulted in poor survivability after transfection (data not shown). To conduct an easy selection with HDR, we generated a small targeting vector containing the sequences at the XIST P regions and a puromycin-resistant gene upstream of the XIST transcription start site (Δ0-XIST; Figure 4A). To test the practicality of X-linked disease modeling of hiPSCs, we used an hiPSC line derived from a patient with Rett syndrome, which is caused by an X-linked mutation in MECP2 (Pohodich and Zoghbi, 2015). The Rett-iPSCs showed an absence of H3K27me3 foci, indicating the lack of XIST expression (Rett-iPSC:XISToff; Figure S4F), and RT-PCR showed the expression from the mutant allele of MECP2, indicating that MECP2 deregulation is protected in this line irrespective of XCI erosion (Figure S4G). Using the Rett-iPSC:XISToff line as the parent cell, we obtained the H3K27me3+ clone by puromycin selection and confirmed the heterogeneous clone with the Δ0-XIST allele (Figure 4B). The efficiency of the HDR approach using Δ0-XIST was more than 7-fold greater than that of the NHEJ approach (Figure 4C). We confirmed that Rett-iPSC lines with XIST re-expression (Rett-iPS:XISTre-on) maintain HUWE1 monoallelic expression with a normal karyotype (Figures 4D, 4E, and S4H), demonstrating that the use of Δ0-XIST for the HDR approach is the easiest method to generate an XIST re-expressed female hPSC line.
Figure 4.
An HDR-based efficient screening method for XIST-reactivated hPSCs and maintenance of XIST+ hPSCs
(A) Scheme of the XIST re-expressing line using an Δ0-XIST plasmid for HDR. hiPSC line obtained from a patient with Rett syndrome (Rett-iPSCs). Puromycin-resistant gene under the control of the EF1A promoter was inserted upstream of the XIST transcription starting site. The transfected cells were treated with puromycin before the H3K27me3 quantification experiment.
(B) Genotyping of Rett-iPSC:XISTre-on line generated byΔ0-XIST construct.
(C) Efficiency test of clones with H3K27me3 foci reacquisition. The efficiency of HDR using Δ0-XIST plasmid was compared with that of the NHEJ method using P region gRNA. Clones containing at least ten cells with H3K27me3 foci were counted as positive clones. n.c. refers to the number of colonies analyzed.
(D and E) XIST/XACT/HUWE1 RNA-FISH analysis. The representative images (D) and quantification results (E). n.c. refers to the number of cells analyzed. Scale bar represents 50 μm.
(F) Cell growth test. Cells with different passage numbers (different XIST expression statuses) were used for assays. After single-cell dissociation, the cells were cultured, and the viable cells were automatically counted by CountessII (Thermo Fisher Scientific, Waltham, MA, USA) after trypan blue staining at each time point.
(G) Quantification of viable cells. Each dot indicates an independent experiment. Error bars show standard deviations. p values were calculated using Student’s t test.
(H) Experimental scheme for maintaining XIST+ hPSCs using a Rho-kinase inhibitor (ROCK inhibitor [RI]). The RI inhibitor Y27632 was used transiently or continuously, and the XCI status was examined by immunofluorescence analysis using OCT4 and H3K27me3 at each time point.
(I) Representative image of the H3K27me3/OCT4 status at day 20.
(J) Quantification of the H3K27me3 foci at each time point. Each dot indicates an independent experiment. Error bars show standard deviations. p values were calculated using Student’s t test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Scale bar represents 50 μm.
Our strategy for XIST re-expression by gene editing is useful for the de-erosion of dosage compensation. However, given that DNMT3A/3B is active (Fukuda et al., 2021), the permanent prevention of XIST repression is inevitable. Thus, we next sought culture methods to maintain XIST+ female hPSCs. We noted that, compared with that observed in eroded female hPSC lines, during conventional culture, female hPSC lines with low passage, often retaining XIST expression, showed markedly poor growth after the removal of the Rho-kinase (ROCK) inhibitor Y27632 (Watanabe et al., 2007) (Figures 4F and 4G). We inferred that the presence of the ROCK inhibitor in the culture medium might reduce the loss of XIST+ cells. To test this possibility, we cultured the rescued XCI lines in the presence of the ROCK inhibitor Y27632 every day and assessed their XCI statuses (Figure 4H). We found that the daily use of the ROCK inhibitor significantly inhibited the loss of H3K27me3+ cells with OCT4 expression (Figures 4I and 4J). These results indicate that the continuous use of a ROCK inhibitor during culture is beneficial for the maintenance of XIST+ female hPSCs.
XCI reacquisition restores differentiation ability
A key question is if XCI reacquisition can rescue the differentiation potential of cells. Since the method for MN differentiation is useful for all studied hPSC lines, we generated MNs using the eroded and rescued XCI lines (Figure 5A). Immuno-FISH analysis showed that in the SEES1 line, the differentiation efficiency of the SE1-res-5a line was comparable to that of the parental eroded line (Figures 5B and 5C), suggesting that XCI is not a major determinant of MN differentiation in the SEES1 line. In contrast, a drastic improvement in the differentiation efficiency was observed in both the SE5-res-2C and -4C lines (>37% of cells) compared with that in the eroded line (3% of cells) (Figures 5B and 5C).
Figure 5.
XCI reacquisition restores the differentiation ability of female hPSCs
(A) Experimental schemes for evaluating the differentiation potential by XIST repression. XIST re-expressing hPSC lines and their parental lines without XIST expression were used. Immuno-FISH (for MN) or immunofluorescence (for cortical organoid) analysis was used for the evaluation.
(B and C) The efficiency of MN differentiation from XIST re-expressing hPSC lines. The representative images (B) and bar graphs are quantification results for ISL1+ cells (C). Each dot and n.c. indicate the percentage in the observed area and number of cells analyzed in total, respectively. Scale bar represents 50 μm. Error bars show the SD. p values were calculated using Student’s t test.
(D and E) The efficiency of neuronal differentiation from Rett-iPSC:XISTre-on. The representative images (D) and bar graphs are quantification results of TUJ-1 or ISL1+ cells (E). Each dot and n.c. indicate the percentage in the observed area and number of cells analyzed in total, respectively. Scale bar represents 50 μm. Error bars show the SD. p values were calculated using Student’s t test.
(F) Summary of female hPSC erosion and de-erosion. XIST is irreversibly repressed by DNMT3A/3B (Fukuda et al., 2021), leading to XCI erosion. XCI erosion affects various phenotypes, not only X-linked gene dysregulation but also the autosomal gene functional assay. XIST is reactivated by the editing of XIST promoter regions, resulting in the generation of de-eroded hPSCs by natural selection under normal culture conditions.
We further investigated whether the differentiation potential of Rett-iPSCs depends on XIST expression. Since Rett syndrome affects the CNS (Pohodich and Zoghbi, 2015), we generated cortical organoids in eroded Rett-iPS:XISToff and Rett-iPS:XISTre-on by HDR using Δ0-XIST (Figure 5A). Surprisingly, immunofluorescence analysis revealed that the Rett-iPS:XISToff line showed poor neuronal development, characterized by TUJ-1/ISL1, at day 20 from differentiation (Figures 5D and 5E). In contrast, the Rett-iPS:XISTre-on line exhibited steady neuronal differentiation (Figures 5D and 5E). Considering that neurogenesis is not markedly impaired in hPSCs with MECP2 mutations (Ohashi et al., 2018; Xiang et al., 2020), these results indicated that XIST silencing could lead to the misinterpretation of the experimental outcomes in disease iPSCs.
Taken together, erosion of dosage compensation causes underestimation of the intrinsic differentiation abilities of female hPSCs in some genetic backgrounds; however, this can be restored through XIST re-expression.
Relation of the XIST expression status with cell types in vivo
Finally, we focused on the effect of the XIST expression status in female cells in vivo. Using 61 published scRNA-seq datasets (Han et al., 2020), we first examined if the overexpression of X-linked genes is observed in specific cell types. Considering that the transcriptome status varies among cell types (Han et al., 2020; Thomsen et al., 2016), we focused on cells in the same cluster and further divided the cells into two groups based on XIST expression status (Figure S5A). Although we could not determine if the cells without XIST expression show lack of XIST transcripts, as observed in the RNA-FISH of female hPSCs (Figure 3D), to obtain insights into the X-linked gene-expression status, we calculated the expression ratio of X-linked genes based on the median expression levels between XIST+ and XIST– cells (Figure S5A). The median levels varied widely in each cluster of various tissues (Figures S5B and S5C), but some clusters, such as cluster 2 in the transverse colon and cluster 3 in the fetal intestine, showed a strong tendency of overexpression in XIST– cells (Figures S5B and S5C). Interestingly, ACE2, a gene encoding a protein with an important role in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (Lan et al., 2020), has been reported to show a heterogeneous expression status in vivo (Tukiainen et al., 2017) and is expressed at high levels in intestinal tissues (Figures S5B and S5C). To examine the relation of ACE2 expression with the XIST status, we focused on fetal intestinal and ileal cells (Martin et al., 2019) (Figure S5D). After the identification of the clusters marked by ACE2 (Figure S5D), we checked the ACE2 expression levels based on the XIST expression status in individual cells. The ACE2 expression levels in XIST– cells tended to be higher than those in XIST+ cells, and the differences were statistically significant in some clusters (Figure S5E), suggesting that XIST repression might increase the risk of SARS-CoV-2 infection in women.
We further investigated whether XIST expression status is linked with neuronal development, such as in ex vivo modeling using hPSCs (Figure S2). Using fetal brain tissue datasets, we examined the cell types based on the XIST expression status (Figure S5F). Although they were derived at different developmental stages (Han et al., 2020), various cell types, such as radial glial cells and post-mitotic neurons, were identified (Figure S8G). UMAP showed that XIST expression was widely distributed (Figure S5G), but the gene was identified as a positive marker in clusters of some excitatory neuron types, such as those with LHX5-AS1 expression, recently identified in developing brains (Eze et al., 2021) (Figure S5H). We further found that the percentages of neuronal cells tend to be higher among XIST+ cells (Figure S5I). In contrast, progenitors were more enriched in XIST– cells (Figure S5I). Although the differences in differentiating cell types between XIST+ and XIST– cells were not remarkable compared with the results obtained from ex vivo modeling (Figure S2K), these results suggested that XIST repression might affect neuronal differentiation in vivo, as observed in hPSC modeling.
Discussion
In the present study, we revealed that XIST repression is a key source of noise and one of the chief drivers of variation determining how and if female hPSCs differentiate effectively (Figure 5F). The scRNA-seq data revealed that among female hPSCs, the differentiation scores of XIST+ cells tend to be higher than those of XIST– cells (Figure 1D), consistent with findings from previous reports showing that XCI erosion results in a low degree of differentiation into the three germ layers (Anguera et al., 2012; Salomonis et al., 2016). We found that post-mitotic neurons from female hPSCs are enriched in XIST+ cells rather than in XIST– cells (Figures S2C and S2K), whereas progenitor cells show the opposite pattern (Figures S2C and S2K). Interestingly, a similar tendency was observed in in vivo development (Figure S5I). Although the causes leading to the differential differentiation status based on XIST expression remain unclear, some possibilities are that XIST transcripts might be more accumulated in neurons because they are post-mitotic cells and that overexpression of X-linked genes might be a roadblock in the differentiation of progenitors to neurons.
We also found that in primary and differentiated cells, representative heterochromatin modifications were lost in some cells even when XIST was expressed, suggesting that biallelic expression might occur even when XIST is expressed. Therefore, the situation of XCI erosion occurring in vivo might not be the same as that in female hPSCs.
Since XIST reactivation can reduce the side effects of XCI erosion, it is beneficial for the application of female hPSCs in different research areas, including the application of disease iPSCs with mutation in not only X-linked but also autosomal genes. Moreover, the methods can induce gene expression at physiological levels, eliminating the unexpected effects of the artificial transcription system with strong induction.
We also found that daily exposure to the ROCK inhibitor is beneficial for the maintenance of female hPSCs with XIST expression. Although the exact mechanisms underlying the development of the phenotype are unknown, the population of early-passage female hPSCs, which occasionally includes XIST+ cells, showed slower growth than the eroded lines (Figure 4G), consistent with the findings from a previous report (Anguera et al., 2012). Furthermore, among female hPSCs, the metabolism of cells in non-eroded states differed from that of cells in eroded states (Brenes et al., 2021). These observations indicate that female hPSCs with XIST expression might be vulnerable to single-cell dissociation and that the ROCK inhibitor might prevent cell death.
XIST is the only identified lncRNA that acts in cis at a chromosomal scale in humans, and its dysregulation affects broad aspects of female cells both in vitro and in vivo. With dosage compensation, when we revisit inexplicable biological phenomena, we might discover new frontiers in human biology and determine the extent to which phenotypes are attributed to sex differences.
Limitations of the study
Although the in vivo scRNA-seq data did not detect XIST transcripts, given that these data were not validated by other assays, such as RNA-FISH, and that the scRNA-seq platform used in the study mainly detected the 3′ region of transcripts, we cannot exclude the possibility that XIST was not detected because of technical limitations, such as drop-out events in the scRNA-seq analysis and/or low expression levels at the 3′ region.
As for the DNMT3A/3B dKO data, we observed that the XIST expression status greatly affected MN differentiation rather than DNMT3A/3B mutations; however, a previous study using male hPSC lines showed that these mutations impaired MN differentiation (Ziller et al., 2018). Thus, the effects of DNMT3A/3B on the differentiation potential of MNs may also depend on the genetic background.
For the SE5-res-2C line, we did not obtain evidence that recombination occurs at the 5′ region of the endogenous XIST gene. However, the affinity of the primer sequences used for the genotyping assay toward the genome may have been poor. Therefore, we could not ascertain whether or not the recombination occurred at the 5′ region.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-ISL1 | Abcam | Cat# ab109517; RRID: AB_10866454 |
| Rabbit monoclonal anti-PAX6 | Abcam | Cat# ab195045; RRID: AB_2750924 |
| Mouse monoclonal anti-SATB2 | Abcam | Cat# ab51502; RRID: AB_882455 |
| Rat monoclonal anti-Ctip2 | Abcam | Cat# ab18465; RRID: AB_2064130 |
| Rabbit monoclonal anti-TBR1 | Abcam | Cat# ab183032; RRID: AB_2313773 |
| Mouse monoclonal anti-β3-Tubulin | Cell Signaling Technology | Cat# 4466; RRID: AB_1904176 |
| Mouse monoclonal anti-Oct-3/4 (C-10) | Santa Cruz Biotechnology | Cat# sc-5279; RRID: AB_628051 |
| Rabbit monoclonal anti-H3K27me3 | Cell Signaling Technology | Cat# 9733; RRID: AB_2616029 |
| Rabbit polyclonal anti-Dnmt3b | Abcam | Cat# ab16049; RRID: AB_443299 |
| Rabbit polyclonal anti-Dnmt3a | Cell Signaling Technology | Cat# 2160S; RRID: AB_2263617 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling Technology | Cat# 2118; RRID: AB_561053 |
| Chemicals, peptides, and recombinant proteins | ||
| StemFlex | Thermo Fisher Scientific | Cat# A3349401 |
| Matrigel | Corning, Inc | Cat# 354277 |
| Y-27632 | Stemcell Technologies | Cat# ST-72308 |
| Neurobasal medium | Thermo Fisher Scientific | Cat# 21103049 |
| B-27 supplement | Thermo Fisher Scientific | Cat# 17504044 |
| N-2 supplement | Thermo Fisher Scientific | Cat# 17502048 |
| GlutaMAX | Thermo Fisher Scientific | Cat# 35050061 |
| SB431542 | Stemcell Technologies | Cat# ST-72234 |
| retinoic acid | Stemcell Technologies | Cat# R2625 |
| LDN-193189 | Stemcell Technologies | Cat# ST-72147 |
| Smoothend agonist | Stemcell Technologies | Cat# ST-73412 |
| DAPT | Stemcell Technologies | Cat# ST-72082 |
| SU-5402 | Stemcell Technologies | Cat# ST-73914 |
| STEMdiff Cerebral Organoid Kit | Stemcell Technologies | Cat# ST-08570 |
| Accutase | Stemcell Technologies | Cat# ST-07920 |
| Neural Tissue Dissociation kit (P) | Miltenyi Biotec | Cat# 130-092-628 |
| Poly-D-Lysine | Thermo Fisher Scientific | Cat# A3890401 |
| VECTASHIELD with DAPI | VECTOR LABORATORIES | Cat# H-1200 |
| RIPA lysis and extraction buffer | Thermo Fisher Scientific | Cat# 89900 |
| Critical commercial assays | ||
| Nick translation kit | Abbott | Cat# 32-801300 |
| EZ DNA Methylation Kit | Zymo Research | Cat# D5001 |
| RNeasy plus micro kit | Qiagen | Cat# 74034 |
| pGEM -T Easy Vector System | Promega | Cat# A137A |
| PUC118 cloning vector | Takara Bio Inc. | Cat# 3324 |
| HiFi DNA assembly cloning kit | New England Biolabs | Cat# E2621 |
| Deposited data | ||
| Raw and processed data | This paper | GEO: GSE186128 |
| human cell landscape dataset | Han et al., 2020 | GEO: GSE134355 |
| ileal uninvolved tissue samples | Martin et al., 2019 | GEO: GSE134809 |
| cortical organoid dataset from the HUES66 line | Broad Institute | portals.broadinstitute.org/single_cell/study/reproducible-brain-organoids |
| Gene expression data in scRNA-seq. | Mendeley | https://doi.org/10.17632/s3yyxhczs3.1 |
| Experimental models: Cell lines | ||
| Human: female ES cells; SEES1, SEES2, SEES4, SEES5 and SEES7 | National Center for Child Health and Development | N/A |
| Human: female iPS cells; ADSC and EDOM | National Center for Child Health and Development | N/A |
| Human: male ES cells; SEES3 and SEES6 | National Center for Child Health and Development | N/A |
| Human: male iPS cell; BJ | National Center for Child Health and Development | N/A |
| Human: male iPS cell; 2B | RIKEN BRC | HiPS-RIKEN-2B |
| Human: female Rett-iPS cell; HPS3049 | RIKEN BRC | HPS3049 |
| Human: DNMT3A/3B double-mutant knockout (dKO) line from SEES5 | This paper | N/A |
| Human: XIST-reactivate lines from SEES1, SEES5, ADSC and HPS3049 | This paper | N/A |
| Human: GTL2-reactivate lines from BJ and 2B | This paper | N/A |
| Human: EDOM; female primary cells | National Center for Child Health and Development | N/A |
| Oligonucleotides | ||
| Primers for TIDE5′, Genotyping, BS and qPCR see Table S2 | This paper | N/A |
| gRNAs of Target region see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| CAG-Cas9-T2A-EGFP-ires-puro | Saarimäki-Vire et al., 2017 | Addgene, #78311 |
| pSPgRNA | Perez-Pinera et al., 2013 | Addgene, #47108 |
| G1A, Plasmid for human XIST | Clemson et al., 1996 | Addgene, #24690; GenBank: NG_016172 |
| BAC clone: human XACT | Thermo Fisher Scientific | CTD 3063K22 |
| BAC clone: human HUWE1 | Thermo Fisher Scientific | PR11-975N19 |
| Software and algorithms | ||
| Benchling | Benchling | https://benchling.com; RRID:SCR_013955 |
| TIDE | Bas van Steensel lab. | http://shinyapps.datacurators.nl/tide/ |
| QUMA | RIKEN | http://quma.cdb.riken.jp/ |
| R | R Development Core Team | https://cran.r-project.org |
| Seurat package (version 3 or 4) | Hao et al., 2021 | https://satijalab.org/seurat/ |
| UCSC genome browser | UCSC Genome Bioinformatics Group | https://genome.ucsc.edu/; RRID:SCR_005780 |
| Monocle3 | Cao et al., 2019 | https://cole-trapnell-lab.github.io/monocle3/; RRID:SCR_018685 |
| edgeR package | McCarthy et al., 2012 | https://bioconductor.org/packages/release/bioc/html/edgeR.html; RRID:SCR_012802 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Atsushi Fukuda (fa972942@tsc.u-tokai.ac.jp).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All human ES cell lines in this study were used under the instruction on the use of human embryonic stem cell research guidelines of Tokai University, the National Center for Child Health and Development, and the Ministry of Education in Japan, and the cell lines used in this study were reported previously (Motosugi et al., 2021). The experiments on X-linked disease iPSCs were approved by the Institutional Review Board for Clinical Research, Tokai University.
Method details
hPSC culture
hPSCs were cultured in StemFlex (Thermo Fisher Scientific, Waltham, MA, USA) on a plate coated with hES cell-grade Matrigel (Corning Inc., Corning, NY, USA). For passaging hPSCs, 1 mM EDTA and 10 μM Y-27632 (StemCell Technologies, Vancouver, Canada) were added to the culture medium and maintained for 24 h, unless otherwise specified. We obtained Rett-iPSCs from the RIKEN bioresource center (HPS3049), and the hPSCs were cultured as described above. The normal chromosome status was confirmed previously (Akutsu et al., 2015). In high passage states, karyotype analysis using G-band staining was performed at Hokkaido Chromosome Science Laboratories, Hokkaido, Japan. Chromosomes were classified according to the International System for Human Cytogenetic Nomenclature. At least 20 metaphase chromosomes were analyzed per cell line.
MN differentiation
Spinal MN differentiation was performed based on methods described in previous reports (Klim et al., 2019; Motosugi et al., 2021). Briefly, the culture medium for confluent hPSCs was replaced with an MN induction medium. The differentiation medium contained ½ Neurobasal (Thermo Fisher Scientific) and ½ DMEM-F12 (Thermo Fisher Scientific) supplemented with Gibco B-27 supplement (×1; Thermo Fisher Scientific), Gibco N-2 supplement (×1; Thermo Fisher Scientific), Gibco GlutaMAX (×1; Thermo Fisher Scientific), and 100 μM non-essential amino acids. The time point at which the medium was changed was defined as day 0 of MN differentiation. We used the following small molecules: 10 μM SB431542 (StemCell Technologies), 1 μM retinoic acid (StemCell Technologies), 100 nM LDN-193189 (StemCell Technologies), and 1 μM Smoothened agonist (StemCell Technologies) on days 0–5; and 5 μM DAPT (StemCell Technologies), 4 μM SU-5402 (StemCell Technologies), 1 μM retinoic acid (StemCell Technologies), and 1 μM Smoothened agonist (StemCell Technologies) on days 6–14. On days 10 or 14, the differentiation efficiency of MNs was analyzed using immuno-FISH analysis. All cells, except naïve cells, were cultured at 37°C in the presence of 5% CO2.
Generation of cortical organoids
To generate cortical organoids from hPSCs, we used a cerebral organoid differentiation medium (STEMdiff Cerebral Organoid Kit, #08570; StemCell Technologies), in accordance with the manufacturer’s instructions. Approximately 70% of the confluent hPSCs were dissociated into single cells using Accutase (#ST-07920; StemCell Technologies), and 9,000 cells were plated in a 96-well plate with ultra-low attachment (#7007; Corning Inc.) and cultured for 5 days. On day 5, the embryoid bodies (EBs) were transferred to a 24-well plate with ultra-low attachment (#3473; Corning Inc.) in 500 μL of neural induction medium. After 48 h, the EBs were embedded in Matrigel and incubated for 30 min at 37°C in a CO2 incubator to allow the Matrigel to polymerize. After polymerization, 1 mL of expansion medium was added to each well. On day 10, the medium was replaced with 1 mL of maturation medium, and the culture plate was placed in a shaker at 37°C in a CO2 incubator. The maturation medium was replaced every 3–4 days until further analysis.
Immunofluorescence analysis
Immunofluorescence analysis was performed as previously described (Fukuda et al., 2021; Motosugi et al., 2021). Briefly, the cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 20 min. The samples were permeabilized using 0.1% Triton-X for 20 min. After washing with PBS, the samples were blocked with 1.5% bovine serum albumin (BSA) in PBS for 1 h and then subjected to the first antibody reaction. After washing with PBS, the samples were subjected to the second antibody reaction and used for microscopy analysis. Images were captured using an LSM770 microscope (Carl Zeiss, Oberkochen, Germany) or an Axio Imager 2 microscope (Carl Zeiss).
For the analysis of organoids developed from Rett-iPSCs, the organoids were dissociated using Neural Tissue Dissociation Kit (P) (Miltenyi Biotec) in accordance with manufacturer’s instruction and plated on a coverslip coated with poly-d-lysine (Thermo Fisher Scientific). After 24 h, the cells were fixed and subjected to immunofluorescence analysis, as described above. Detailed information on the antibodies used in this study is provided in Table S2.
RNA-FISH
Based on previous reports, RNA-FISH was performed as described previously (Fukuda et al., 2021; Motosugi et al., 2021). Briefly, the cells were cultured on a coverslip and fixed by treating with 4% PFA in PBS for 20 min. The samples were permeabilized using 0.1% Triton-X for 20 min and then subjected to RNA-FISH. The probes were prepared using a nick translation kit (Abbott Laboratories, Chicago, IL, USA), with G1A (#24690; Addgene, Cambridge, MA, USA) for XIST, and the BAC clones CTD 3063K22 and PR11-975N19 for XACT and HUWE1, respectively. VECTASHIELD with DAPI was used for nuclear staining (Vector Laboratories Inc., Burlingame, CA, USA). Images were acquired using an LSM770 microscope (Carl Zeiss) or an Axio Imager 2 microscope (Carl Zeiss).
Immunofluorescence analysis combined with RNA-FISH (Immuno-FISH)
Immuno-FISH analysis was conducted based on a method described in a previous report (Fukuda et al., 2021; Motosugi et al., 2021). Briefly, differentiating cells in a 2D culture were detached with Accutase (StemCell Technologies) and plated on a coverslip coated with Matrigel (Corning Inc.) for at least 1 h. After confirming the attachment of the cells under the microscope, the cells were fixed and permeabilized as described above. For immuno-FISH, nuclease-free BSA was used as the blocking buffer and for first antibody incubation. After secondary antibody treatment, the samples were subjected to post-fixation with 4% PFA in PBS for 20 min before XIST RNA-FISH. VECTASHIELD with DAPI was used for nuclear staining (Vector Laboratories Inc.).
On day 90, the cortical organoids were fixed overnight in 4% PFA. They were then washed in PBS and transferred to a 10%, 15%, or 20% sucrose solution every 24 h at 4°C. Subsequently, they were transferred into an embedding medium (Tissue-Tek OCT Compound 4583; Sakura Finetek Japan, Tokyo, Japan), frozen using an ultra-low-temperature freezing system (Histo-Tek PINO; Sakura Finetek Japan), and stored at −80°C. For immunohistochemistry, 5 μm-thick sections were obtained using a cryostat, and the specimens were placed onto glass slides and subjected to immuno-FISH, as described above. On day 90, the organoids were analyzed using immuno-FISH. Images were captured using an LSM700 microscope (Carl Zeiss) or an Axio Imager 2 microscope (Carl Zeiss).
DNA-FISH
The DNA-FISH procedures were based on a previous study (Fukuda et al., 2016). The fixed and permeabilised cells were treated with RNaseA and then incubated with 0.2N HCl containing 0.05% tween-20 solution on ice for 10 min. The samples were incubated at 85°C for 10 min and then for overnight at 37°C. Entire-hXIST and CTD 3063K22 were prepared by nick translation. Images were captured using an LSM700 microscope (Carl Zeiss).
Generation of XIST re-expressing female hPSC lines
For the XIST re-expression efficiency test, SEES1, SEES5, and ADSC-hiPSC lines (all of them containing >99% of cells without the H3K27me3 foci) were used. The XIST re-expressing SEES1 and SEES5 cell lines were used. For the gene targeting approach in XIST re-expression experiments, using bacterial recombineering technology, we generated a Targeting vector containing the whole human XIST gene (32 kb): Entire-hXIST contains exogenous sequences of a tetracycline response element upstream of the XIST transcription starting site and a SV40 polyA signal downstream of the XIST gene (Vector Builder, Chicago, IL, USA). Entire-hXIST was used for the transfection experiments. To induce recombination, Entire-hXIST was co-transfected with CAG-Cas9-T2A-EGFP-ires-puro (a gift from Timo Otonkoski, #78311; Addgene) and pSPgRNA (a gift from Charles Gersbach, #47108; Addgene). The gRNAs were designed to target the promoter (P-region) and 3′ regions of XIST. For the gRNA-mediated XIST or GTL2 reactivation experiments, CAG-Cas9-T2A-EGFP-ires-puro and pSPgRNA containing each target sequence were co-transfected. The gRNA sequences used in this study are listed in Table S2. For the GTL2 re-expression experiments, BJ- and 2B-hiPSC lines were used. The gRNAs were designed using Benchling (https://benchling.com).
At 24 h after transfection, 1.5–2 μg of puromycin (depending on the cell line used) was added to the culture medium and maintained for 24 h. Then, each colony was subjected to immunofluorescence staining against H3K27me3 to evaluate the efficiency of XCI reacquisition or sub-cloning into 96-well plates. After sub-cloning, we screened H3K27me3-positive clones (>85% of cells in each population).
For the generation of the Rett-iPSC: XISTre-on line, the Δ0-XIST plasmid was constructed using HiFi DNA assembly according to the manufacturer’s instruction (NEB). PUC118-HindIII (#3324; Takara Bio Inc., Siga, Japan) was used as a backbone plasmid. All fragments were generated by PCR using Entire-hXIST and genomic DNA, and original plasmids contained EF1a promoter sequences (available upon request). A part of Exon 1 in the XIST gene and upstream regions from TSS was used for the targeting arms. The gRNAs were generated as described above, and the sequences are listed in Table S2. After transfection, Rett-iPSCs were cultured till approximately 70% confluence was achieved and were then subjected to puromycin selection.
Generation of DNMT3A/3B double-mutant lines
The previously validated gRNAs for DNMT3A or 3B and the SEES5 line with or without XIST expression were used (Fukuda et al., 2021; Liao et al., 2015). gRNA was cloned into pSPgRNA, and two gRNAs targeting DNMT3A and 3B were co-transfected with CAG-Cas9-T2A-EGFP-ires-puro. The screening method is described above; the mutations were identified by genomic PCR, and deletion was confirmed by western blotting analysis.
Genotyping
XIST re-expressing hPSC lines were subjected to genomic PCR using primers spanning exogenous sequences when they were generated using TRE-hXIST. To check for scars in each gene editing scenario, TIDE (http://shinyapps.datacurators.nl/tide/) analysis was conducted around the gRNA targeting sequences. For Rett-iPSCs, the mutant region reported in the RIKEN bioresource center was confirmed by PCR and Sanger sequencing. All primer sequences are listed in Table S2.
Western blotting
Western blotting was performed using the conventional method (Fukuda et al., 2021; Liao et al., 2015). Briefly, proteins from hPSCs were extracted using RIPA lysis and extraction buffer (Thermo Fisher Scientific). The extracted proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis using 7.5% TGX gel (Bio-Rad, Hercules, CA, USA) for DNMT3B (ab16049; Abcam, Cambridge, UK) and 4%–20% TGX Gel (Bio-Rad) for DNMT3A (2160S; Cell Signaling Technology, Danvers, MA, USA) and GAPDH (2118; Cell Signaling Technology).
Bisulfite sequencing
Bisulfite sequencing was conducted based on a method described in a previous report (Fukuda et al., 2021). Briefly, genomic DNA was extracted using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). The amplified products were cloned into the pGEM-T Easy Vector System (Promega, Madison, WI, USA) by TA cloning or the PUC118 cloning vector (#3324; Takara Bio Inc., Siga, Japan) and sequenced. The results were analyzed using QUMA (http://quma.cdb.riken.jp/). The primer sequences are listed in Table S2.
Single-cell RNA-sequencing analysis
Sequencing using a 10× Chromium (10× Genomics, Pleasanton, CA, USA) platform was performed in Genewiz (Genewiz, South Plainfield, NJ, USA) based on the manufacturer’s instructions. The Cell Ranger 3.1.0 pipeline (10× Genomics) was used for the alignment of the RNA sequence with the GRCh38 human reference genome. The default parameters were used to generate filtered gene expression matrix datasets. The datasets were loaded into R (https://www.r-project.org/), and the Seurat package (version 3 or 4) was used for transcriptomic analysis (Hao et al., 2021; Stuart et al., 2019). Cells with unique feature counts greater than 200 and less than 25% of mitochondrial counts were filtered, and the genes expressed in at least ten cells were used for UMAP analysis in hPSCs. Based on Seurat’s default normalization methods, data from each hPSC line were merged using Seurat’s merge function to plot cells from all hPSC lines. The clustering analysis for UMAP was performed using 20 dims with Seurat’s RunUMAP function.
To calculate the X-linked gene dosage in hPSCs, the cells were filtered as described above. To exclude the cells showing potential initiation of differentiation, we removed the cells with less than 2 UMI (OCT4 expression), so that more than 93% of cells were used for the X-linked gene expression analysis (Figure S1B). In XIST expression, as 0.01 was the threshold for XIST detection, we used it as the cut-off value for XIST expression (XIST+ cells: ≥ 0.01 and XIST– cells: < 0.01). A gene expression matrix with gene names was extracted using Seurat’s GetAssayData function. The gene names were extracted from the matrix and annotated using the UCSC genome browser (https://genome.ucsc.edu/) to identify X-linked genes for the calculation. The extraction of expression data for X-linked genes and static analysis was performed using the default function of R. The raw and processed data were deposited in GEO (GSE186128).
In the public dataset analysis, the cortical organoid dataset from the HUES66 line was downloaded from Single Cell Portal (portals.broadinstitute.org/single_cell/study/reproducible-brain-organoids). The data were loaded in R and re-analyzed using Seurat. In the trajectory analysis, we used Monocle3(Cao et al., 2019) and calculated the pseudotime scores.
For in vivo data analysis, we used the human cell landscape dataset (GSE134355) and ileal uninvolved tissue samples (GSE134809). In the GSE134355 datasets, we obtained only female samples. The gene expression matrix was loaded into R, converted to a Seurat object, and normalized using Seurat’s default function. The cells were screened based on the above parameters, considering cells without OCT4 expression and mitochondrial gene selection, because OCT4 is a pluripotent cell marker and ACE2 expression affects mitochondrial activity (Shi et al., 2018). In cell type classifications, we used Seurat’s FindMarkers function with MAST to identify marker genes (Table S1). Based on findings from previous reports (Bhaduri et al., 2020; Thomsen et al., 2016; Velasco et al., 2019), the cells were separated into each cell type.
For the calculation of the median expression levels of each X-linked gene excluding the XIST gene, the cells in each cluster were divided into either the XIST+ or – group using Seurat’s subset function, and then the gene expression matrix was extracted using Seurat’s Getassaydata function. Using the extracted gene expression matrix in R, the median expression levels of X-linked genes in at least more than 20 cells (for HUES66 data) or more than ten cells (in vivo data) in both XIST+ and – groups were calculated. For the analysis of the cell-type status based on XIST expression, the percentages of XIST+ or – cells in each cluster were calculated, and the log2 ratios were shown in figures. The median expression data are shown as heatmaps in figures using R.
The gene expression data in all scRNA-seq analysis in the present study is deposited at Mendeley and the DOI is listed in the key resources table. The related statistical data is provided in Table S3.
Module score calculation
Each module score in female hPSCs was calculated using the AddModuleScore function in Seurat with default parameters. The genes used for the differentiation modules were reported previously (Bock et al., 2011), and OCT4, SOX2, and NANOG were used for the core pluripotency module.
Bulk RNA-sequencing
RNA-sequencing analysis using bulk samples has been reported previously (Fukuda et al., 2021). Briefly, total RNA from the hPSC line (SEES5 background: wild-type, DNMT3A/3B DKO mutants) was extracted using the RNeasy Plus Micro Kit (Qiagen, Hilden, Germany), and polyA mRNA selection was used to generate the sequence library. The sequencing and read count data alignment with the human reference genome (hg38) were performed in Genewiz (Genewiz). The read count data were normalized by the trimmed mean of M-values normalization using the edgeR package (McCarthy et al., 2012). Hierarchical clustering analysis was performed using R’s hclust function. The raw and processed data were deposited in GEO (GSE186128).
Quantification and statistical analysis
We used Seurat package in all statistical analyses in scRNA-seq data. For pair-wise comparison between XIST+ and - cells and multiple comparisons among the samples, we used Wilcoxon rank sum test and ANOVA with post-hoc test, respectively. In HUES66 and in vivo samples, FindMarkers function (MAST package) implemented in Seurat was used to identify the differentially expressed genes. All statistical data in the scRNA-seq analysis was shown in Table S3. For the data of neuronal differentiation assays, cell number analysis, and erosion induction tests, we performed paired, two-tailed t-tests to calculate p-values using R. p-values less than 0.05 were considered significant. The number of analyzed cells or genes was shown in figure legends.
Acknowledgments
We would like to thank the members of the Support Center for Medical Research and Education, Tokai University School of Medicine, for assisting with the various experiments. The study was supported by AMED under grant numbers JP21bm0704038 and 21bm0804030h0001 to A.F.
Author contributions
A.F. conceptualized the project. A.F., N.M., and A.S. designed the experiments. N.M. and A.S. conducted most of the experiments, with help from A.S., C.O., and A.F. N.M., A.S., C.O., A.O., S.H., and A.F. analyzed the data. N.M., A.S., and A.F. generated cell lines with gene manipulations. A.U. and H.A. generated wild-type ESCs and ADSC-, EDOM-, and BJ-hiPSC lines. All authors interpreted the data, and A.F. supervised the project. A.F. wrote the manuscript with input from all authors.
Declaration of interests
A.F. is applying for a patent for the method to generate XIST re-expressing female hPSCs by gRNA-mediated XIST demethylation (2022-062974).
Published: November 29, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100352.
Supplemental information
Data and code availability
-
•
Bulk RNA-seq and Single-cell RNA-seq data used in this study have been deposited at GEO and are publicly available. The gene expression data in scRNA-seq have been deposited at Mendeley and the DOI is listed in the key resources table and are publicly available as of the date of publication. Original western blot images have been shown in the figure. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Akutsu H., Machida M., Kanzaki S., Sugawara T., Ohkura T., Nakamura N., Yamazaki-Inoue M., Miura T., Vemuri M.C., Rao M.S., et al. Xenogeneic-free defined conditions for derivation and expansion of human embryonic stem cells with mesenchymal stem cells. Regen. Ther. 2015;1:18–29. doi: 10.1016/j.reth.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An C., Feng G., Zhang J., Cao S., Wang Y., Wang N., Lu F., Zhou Q., Wang H. Overcoming autocrine FGF signaling-induced heterogeneity in naive human ESCs enables modeling of random X chromosome inactivation. Cell Stem Cell. 2020;27:482–497.e84. doi: 10.1016/j.stem.2020.06.002. [DOI] [PubMed] [Google Scholar]
- Anguera M.C., Sadreyev R., Zhang Z., Szanto A., Payer B., Sheridan S.D., Kwok S., Haggarty S.J., Sur M., Alvarez J., et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augui S., Nora E.P., Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Bhaduri A., Andrews M.G., Mancia Leon W., Jung D., Shin D., Allen D., Jung D., Schmunk G., Haeussler M., Salma J., et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z.D., Ziller M., Croft G.F., Amoroso M.W., Oakley D.H., et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenes A.J., Yoshikawa H., Bensaddek D., Mirauta B., Seaton D., Hukelmann J.L., Jiang H., Stegle O., Lamond A.I. Erosion of human X chromosome inactivation causes major remodeling of the iPSC proteome. Cell Rep. 2021;35:109032. doi: 10.1016/j.celrep.2021.109032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali C.P., Park D.S., Lee E.B. Targeted DNA methylation of neurodegenerative disease genes via homology directed repair. Nucleic Acids Res. 2019;47:11609–11622. doi: 10.1093/nar/gkz979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A.G., Cotton A.M., Kelsey A.D., Brown C.J. Differentially methylated CpG island within human XIST mediates alternative P2 transcription and YY1 binding. BMC Genet. 2014;15:89. doi: 10.1186/s12863-014-0089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson C.M., McNeil J.A., Willard H.F., Lawrence J.B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha S.T., Edwards C.A., Ito M., Ogata T., Ferguson-Smith A.C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Charpentier E. Genome editing. The new Frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Eze U.C., Bhaduri A., Haeussler M., Nowakowski T.J., Kriegstein A.R. Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nat. Neurosci. 2021;24:584–594. doi: 10.1038/s41593-020-00794-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A., Hazelbaker D.Z., Motosugi N., Hao J., Limone F., Beccard A., Mazzucato P., Messana A., Okada C., San Juan I.G., et al. De novo DNA methyltransferases DNMT3A and DNMT3B are essential for XIST silencing for erosion of dosage compensation in pluripotent stem cells. Stem Cell Rep. 2021;16:2138–2148. doi: 10.1016/j.stemcr.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A., Mitani A., Miyashita T., Sado T., Umezawa A., Akutsu H. Maintenance of Xist imprinting depends on chromatin condensation state and Rnf12 dosage in mice. PLoS Genet. 2016;12:e1006375. doi: 10.1371/journal.pgen.1006375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., von Meyenn F., Rostovskaya M., Clarke J., Dietmann S., Baker D., Sahakyan A., Myers S., Bertone P., Reik W., et al. Epigenetic resetting of human pluripotency. Development. 2017;144:2748–2763. doi: 10.1242/dev.146811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W., et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klim J.R., Williams L.A., Limone F., Guerra San Juan I., Davis-Dusenbery B.N., Mordes D.A., Burberry A., Steinbaugh M.J., Gamage K.K., Kirchner R., et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 2019;22:167–179. doi: 10.1038/s41593-018-0300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liao J., Karnik R., Gu H., Ziller M.J., Clement K., Tsankov A.M., Akopian V., Gifford C.A., Donaghey J., Galonska C., et al. Targeted disruption of DNMT1, DNMT3A and DNMT3B in human embryonic stem cells. Nat. Genet. 2015;47:469–478. doi: 10.1038/ng.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.S., Nayar S., Greenstein A.J., et al. Single-cell analysis of Crohn's disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S., Bock C., de Boer A.S., Kiskinis E., Meissner A., Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motosugi N., Okada C., Sugiyama A., Kawasaki T., Kimura M., Shiina T., Umezawa A., Akutsu H., Fukuda A. Deletion of lncRNA XACT does not change expression dosage of X-linked genes, but affects differentiation potential in hPSCs. Cell Rep. 2021;35:109222. doi: 10.1016/j.celrep.2021.109222. [DOI] [PubMed] [Google Scholar]
- Ohashi M., Korsakova E., Allen D., Lee P., Fu K., Vargas B.S., Cinkornpumin J., Salas C., Park J.C., Germanguz I., et al. Loss of MECP2 leads to activation of P53 and neuronal senescence. Stem Cell Rep. 2018;10:1453–1463. doi: 10.1016/j.stemcr.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Bonora G., Sahakyan A., Kim R., Chronis C., Langerman J., Fitz-Gibbon S., Rubbi L., Skelton R.J.P., Ardehali R., et al. Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep. 2017;18:54–67. doi: 10.1016/j.celrep.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny G.D., Kay G.F., Sheardown S.A., Rastan S., Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Perez-Pinera P., Kocak D.D., Vockley C.M., et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohodich A.E., Zoghbi H.Y. Rett syndrome: disruption of epigenetic control of postnatal neurological functions. Hum. Mol. Genet. 2015;24:R10–R16. doi: 10.1093/hmg/ddv217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarimäki-Vire J., Balboa D., Russell M.A., et al. An Activating STAT3 Mutation Causes Neonatal Diabetes through Premature Induction of Pancreatic Differentiation. Cell Rep. 2017;19:281–294. doi: 10.1016/j.celrep.2017.03.055. [DOI] [PubMed] [Google Scholar]
- Saelens W., Cannoodt R., Saeys Y. A comprehensive evaluation of module detection methods for gene expression data. Nat. Commun. 2018;9:1090. doi: 10.1038/s41467-018-03424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W., Kuoy E., Langerman J., Clark A.T., Jaenisch R., Plath K. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N., Dexheimer P.J., Omberg L., Schroll R., Bush S., Huo J., Schriml L., Ho Sui S., Keddache M., Mayhew C., et al. Integrated genomic analysis of diverse induced pluripotent stem cells from the progenitor cell biology consortium. Stem Cell Rep. 2016;7:110–125. doi: 10.1016/j.stemcr.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T.T., Yang F.Y., Liu C., Cao X., Lu J., Zhang X.L., Yuan M.X., Chen C., Yang J.K. Angiotensin-converting enzyme 2 regulates mitochondrial function in pancreatic beta-cells. Biochem. Biophys. Res. Commun. 2018;495:860–866. doi: 10.1016/j.bbrc.2017.11.055. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tan S.M., Wang S.T., Hentze H., Dröge P. A UTF1-based selection system for stable homogeneously pluripotent human embryonic stem cell cultures. Nucleic Acids Res. 2007;35:e118. doi: 10.1093/nar/gkm704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen E.R., Mich J.K., Yao Z., Hodge R.D., Doyle A.M., Jang S., Shehata S.I., Nelson A.M., Shapovalova N.V., Levi B.P., Ramanathan S. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods. 2016;13:87–93. doi: 10.1038/nmeth.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tukiainen T., Villani A.C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot C., Ouimette J.F., Makhlouf M., Féraud O., Pontis J., Côme J., Martinat C., Bennaceur-Griscelli A., Lalande M., Rougeulle C. Erosion of X Chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell. 2015;16:533–546. doi: 10.1016/j.stem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Oron E., Nelson B., Razis S., Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S.i., Nishikawa S., Muguruma K., Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Patterson B., Hwang S.M., Hysolli E., Cakir B., Kim K.Y., Wang W., Kang Y.J., Clement E.M., et al. Dysregulation of BRD4 function underlies the functional abnormalities of MeCP2 mutant neurons. Mol. Cell. 2020;79:84–98.e9. doi: 10.1016/j.molcel.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Pluripotent stem cell-based cell therapy-promise and challenges. Cell Stem Cell. 2020;27:523–531. doi: 10.1016/j.stem.2020.09.014. [DOI] [PubMed] [Google Scholar]
- Ziller M.J., Ortega J.A., Quinlan K.A., Santos D.P., Gu H., Martin E.J., Galonska C., Pop R., Maidl S., Di Pardo A., et al. Dissecting the functional consequences of de novo DNA methylation dynamics in human motor neuron differentiation and physiology. Cell Stem Cell. 2018;22:559–574.e9. doi: 10.1016/j.stem.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Bulk RNA-seq and Single-cell RNA-seq data used in this study have been deposited at GEO and are publicly available. The gene expression data in scRNA-seq have been deposited at Mendeley and the DOI is listed in the key resources table and are publicly available as of the date of publication. Original western blot images have been shown in the figure. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.