Abstract
Retrotransposons are one type of mobile genetic element that abundantly reside in the genomes of nearly all animals. Their uncontrolled activation is linked to sterility, cancer, and other pathologies, thereby being largely considered detrimental. Here we report that, within a specific time window of development, retrotransposon activation can license the host’s immune system for future antiviral responses. We found the mdg4 (also known as Gypsy) retrotransposon selectively becomes active during metamorphosis at the Drosophila pupal stage. At this stage, mdg4 activation educates the host’s innate immune system by inducing the systemic antiviral function of the NF-κB protein, Relish, in a dSTING-dependent manner. Consequently, adult flies with mdg4, Relish, or dSTING silenced at the pupal stage are unable to clear exogenous viruses and succumb to viral infection. Altogether, our data reveal that hosts can establish a protective antiviral response that endows a long-term benefit in pathogen warfare due to the developmental activation of mobile genetic elements.
INTRODUCTION
Retrotransposons are the most abundant genetic elements in almost all animal genomes, comprising 38% of the human genome1–3. Retrotransposons use a self-replicating life cycle similar to retroviruses to propagate3–6. Upon activation, retrotransposons use their mRNAs as templates to make new DNA copies through reverse transcription. The newly synthesized DNA then integrates into the host genome to achieve the final mobilization step. To accomplish this cycle, two to three retrotransposon-encoded proteins are involved: Orf1/Gag for packaging the virus-like particle; Orf2/Pol for reverse transcription and integration; Envelope (Env) protein for mediating fusion between the virus-like particle and the cell membrane1–5,7.
Activation of retrotransposons is largely considered to be detrimental2,8–10. First, the products of retrotransposons, including mRNAs, proteins, and cDNAs, can potentially contribute to diseases, such as neurodegenerative disorders or cancer11,12. Second, their mobilization step generates DNA damage and mutations, causing sterility and possibly driving aging2,10,13–15. Thus, extensive efforts led to the identification of mechanisms that silence retrotransposons in both germline and somatic cells. However, even under strict regulation, it is possible that retrotransposons still can achieve activation in certain tissues during specific developmental stages16, with potential benefits and/or dangers to the hosts.
The innate immune system is essential for protecting a broad range of species, from fungi to plants and animals, against pathogen infection17–20. Upon infection, rapidly mounting the innate immune responses is crucial for host survival17,21,22. Notably, activation of innate immunity during a single event can protect hosts from future infection23,24. For example, Candida albicans infection renders mice protected from fungal reinfection in a monocyte-dependent manner25; mosquitoes invaded by the malaria parasite Plasmodium possess enhanced immunity upon reinfection26. These studies indicate that activating innate immunity once confers hosts a long-term protection from pathogen infection.
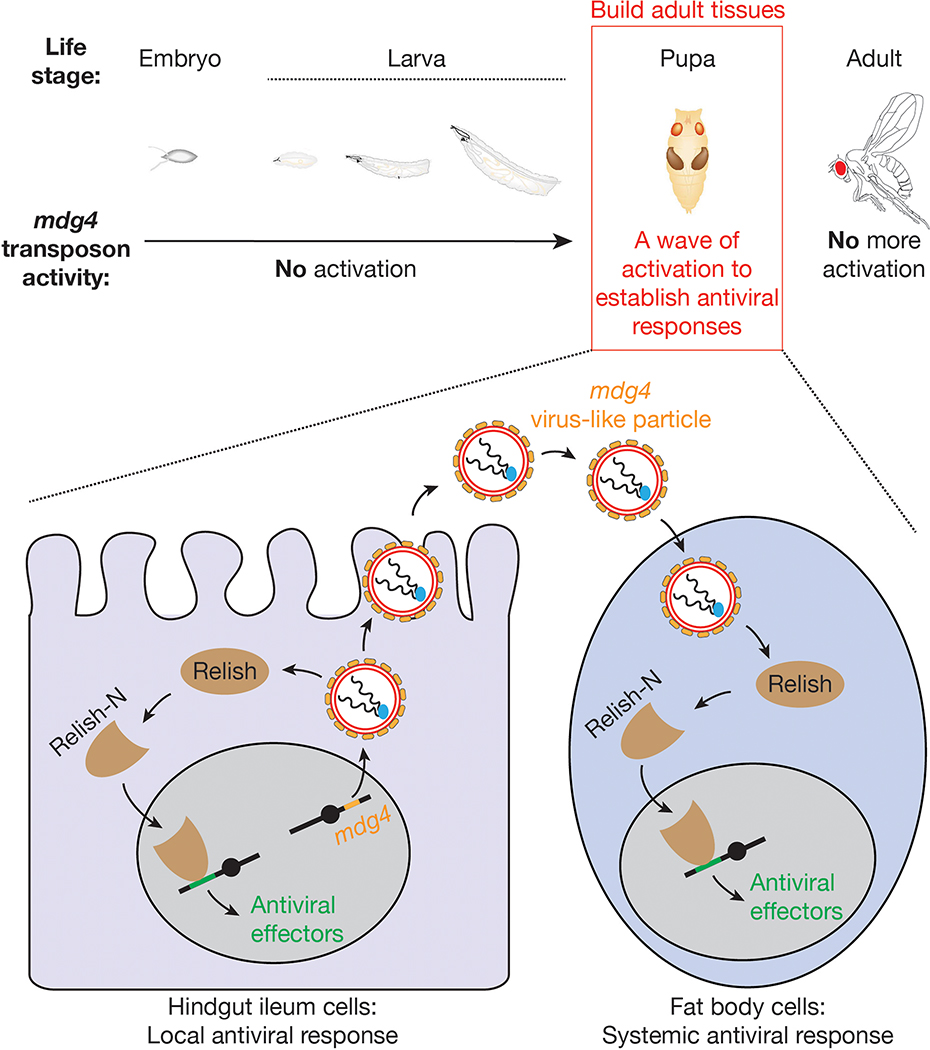
By using Drosophila as a model system, we systematically tracked retrotransposon activity at the mobilization level during animal development. We found that mdg4 retrotransposon selectively becomes active during metamorphosis, a stage when flies build new tissues for adult life. Notably, this wave of retrotransposon activation appears to be essential to induce innate immunity by activating the systemic antiviral function of an NF-κB factor, which protects the hosts from future viral infection. Our data therefore demonstrate a function on educating the immune system for pathogen warfare from the programmed activation of a class of mobile genetic elements.
RESULTS
mdg4 mobilizes in somatic tissues
The ultimate step of retrotransposon activation is mobilization–the insertion of a new copy of itself into the host genome3,5,8. Although retrotransposon activity has been examined at the transcription and translation levels, the spatiotemporal patterns of mobilization events during development are relatively unexplored27. Encouraged by our previous work monitoring their mobilization during oogenesis28, we sought to systematically characterize retrotransposon jumping events in somatic tissues of Drosophila, a powerful genetic model with well-curated retrotransposon annotation28.
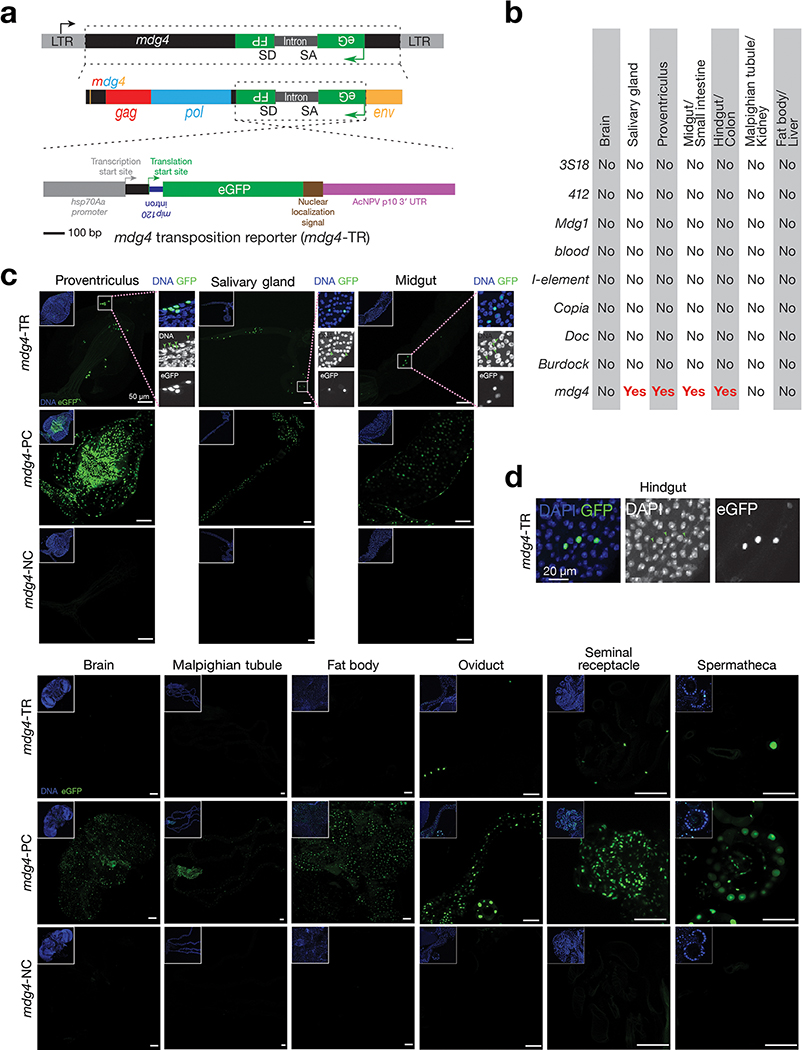
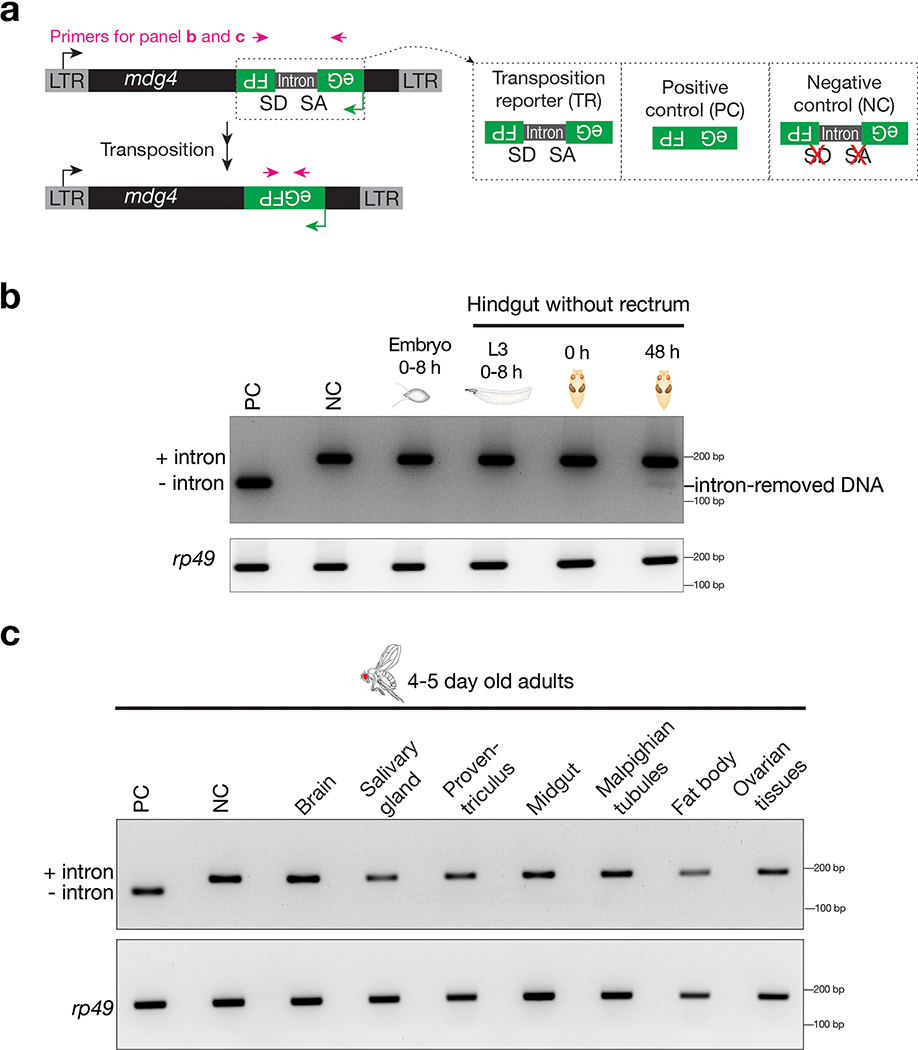
To achieve this, we examined nine retrotransposon families that maintain full-length copies in the genome and potentially can mobilize: 3S18, 412, Blood, Burdock, Copia, Mdg1, Doc, I-element, and mdg429–37. For each family, we engineered a corresponding eGFP mobilization reporter (Fig. 1a and Extended Data Fig. 1a). Similar to reporters designed previously5,29,38, an eGFP cassette was inserted into the retrotransposon in the antisense direction. This cassette contains a disruptive intron that can be spliced during transcription of the retrotransposon, but not of eGFP. As such, the reporter can only express eGFP after intron-spliced retrotransposon mRNAs are used as templates to make cDNA (Fig. 1a and Extended Data Fig. 1a). To minimize the positional effects from chromatin on retrotransposon activity, we generated fly alleles that carry each eGFP reporter at two different genomic loci and obtained the consistent findings reported below.
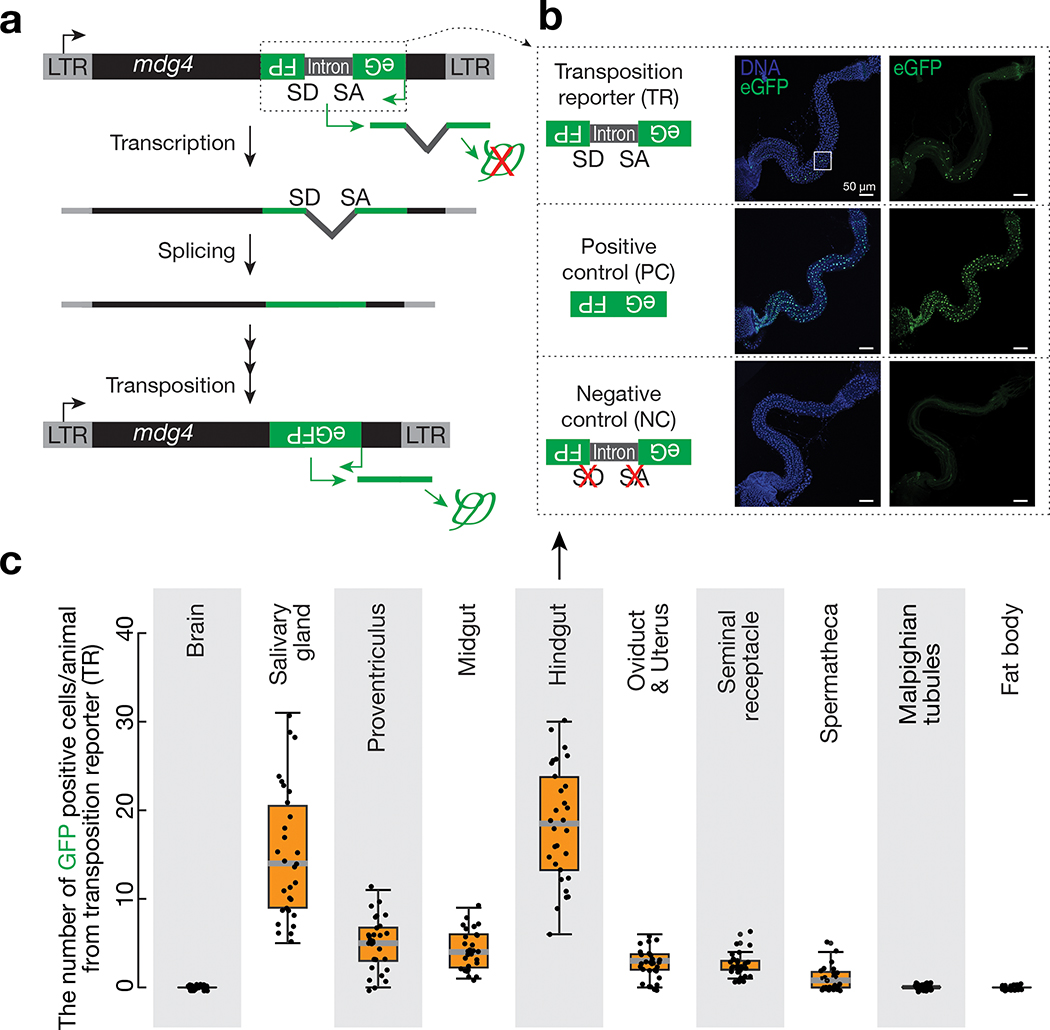
Fig. 1 |. Monitoring mdg4 mobilization in somatic cells via a transposition reporter.
a, Schematic design of an eGFP reporter to monitor mdg4 mobilization. The eGFP reporter is inserted in the non-coding sequence of mdg4 in the antisense orientation. eGFP is disrupted by an intron, which is in the same direction as mdg4 but opposite direction as eGFP. The intron cannot be spliced during eGFP transcription. mdg4 mobilization, which generates a new copy of the transgene without the disrupting intron, results in eGFP expression. b, mdg4 transposition reporter produces eGFP in hindgut cells in 2–4-day-old adult flies. An intronless construct is used as a positive control to test the potential sensitivity. A construct with mutated splicing acceptor and donor sites serves as a negative control. The boxed region is displayed in Extended Data Figure 1d to show eGFP signals are from nuclei (eGFP has nuclear localization signal sequences). c, The box plot shows the number of eGFP positive cells from mdg4 transposition reporter among different tissues from 2–4-day-old adult flies (N = 30). Box plots report the minimum, maximum, median, and interquartile ranges of the data.
We examined seven somatic tissues from 2–4-day-old flies to assess eGFP signals from the mobilization reporters: brain, salivary gland, proventriculus, midgut, hindgut, Malpighian tubule, and fat body. Among the nine retrotransposon families, eight displayed no detectable eGFP signals (Extended Data Fig. 1b). In contrast, the reporter for mdg4 produced eGFP signals in the salivary gland, proventriculus, midgut, and hindgut (Fig. 1b,c and Extended Data Fig. 1b,c). In summary, our data indicate that mdg4 still mobilizes in certain somatic tissues.
mdg4 only becomes active at the pupal stage
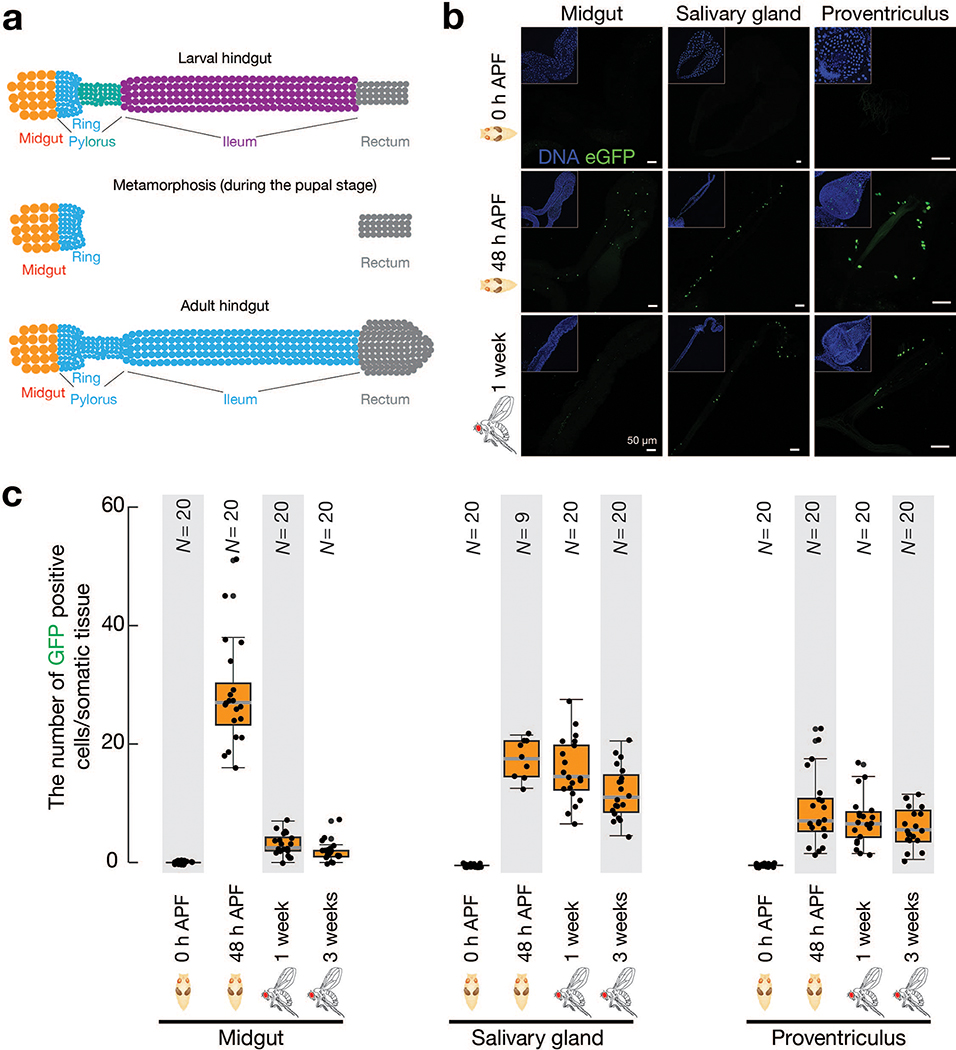
Based on the reporter design, both original cells harboring mobilization events and their progeny can produce eGFP. The mobilization events we detected could occur either in the adult cells we examined, or in their developmental precursors. As a holometabolous insect, Drosophila proceeds through four different life stages: embryo, larva, pupa and adult. To determine when mdg4 mobilizes, we first examined the most highly labelled tissue, the hindgut (Fig. 1c), at these stages. We discovered that mdg4 selectively becomes active at the pupal stage (Fig. 2 and Extended Data Fig. 2). Notably, metamorphosis is initiated in pupae, during which many larval tissues are first destroyed and then regenerated to produce adult structures, such as the salivary gland and the gut39. For the first 20 hours after puparium formation (APF), when the hindgut degenerates, we barely detected any mdg4 mobilization (Fig. 2a,b and Extended Data Fig. 2a). However, at 24 hours APF, we observed eGFP-positive cells in newly formed hindguts (Fig. 2a,b). As the hindgut grew during metamorphosis, we detected more cells harboring mobilization events (Fig. 2a,b). Similar to the hindgut, mdg4 does not appear to mobilize in the degenerating salivary gland, proventriculus, and midgut at the early pupal stage, but mobilizes in these tissues during their developmental regeneration (Extended Data Fig. 2b,c).
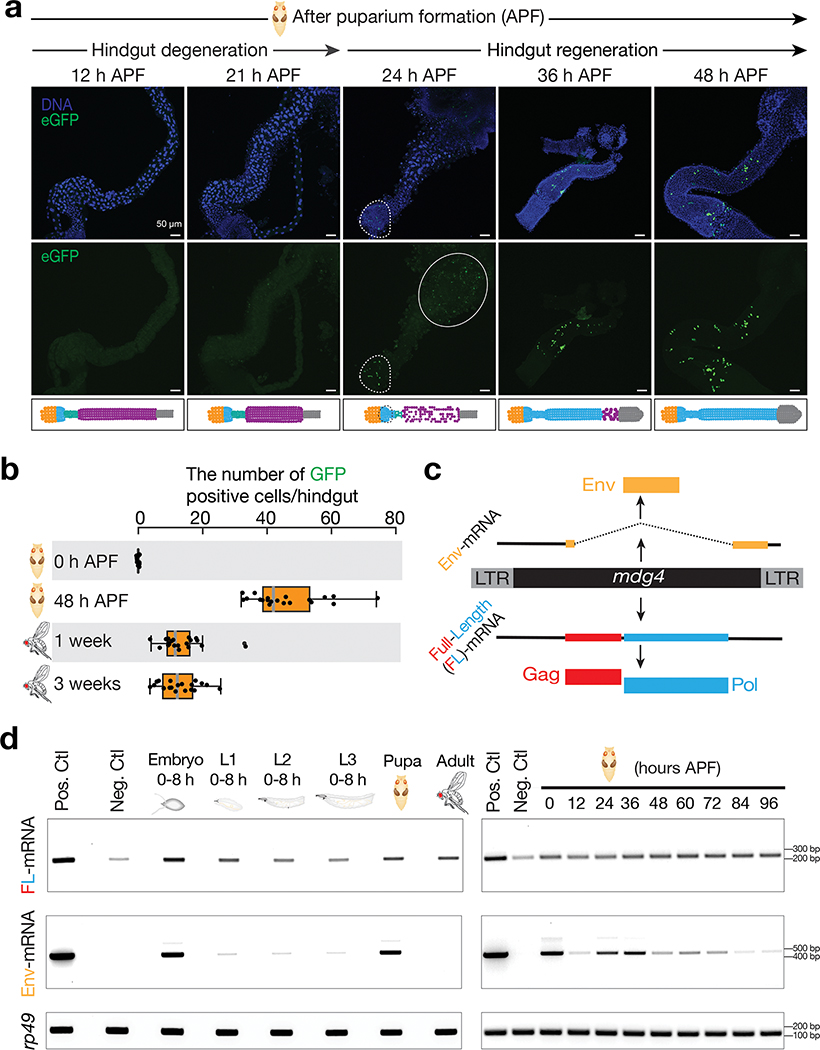
Fig. 2 |. mdg4 selectively mobilizes in the regenerating tissues during metamorphosis.
a, mdg4 mobilization in Drosophila during hindgut development at the pupal stage via eGFP transposition reporter. No eGFP is expressed during hindgut degeneration (12 h and 21 h APF). eGFP positive cells can be detected in the newly formed hindgut (24 h, 36 h, and 48 h APF). Dashed circle highlights eGFP signals. Solid circle depicts autofluorescence from the dying cells. Extended Data Figure 2a depicts the cell-type dynamics of hindgut during metamorphosis. b, The box plot shows the number of eGFP-positive cells per fly from mdg4 transposition reporter at different stages (N = 20). Box plots report the minimum, maximum, median, and interquartile ranges of the data. c, Diagram depicts the transcripts and proteins from mdg4. d, RT-PCR experiments to measure the expression of full-length and Env mRNAs from reporter-carrying flies. Pos. Ctl.: positive control, ovaries with Piwi being depleted in follicle cells. Neg. Ctl.: negative control, ovaries with mdg4 being depleted in both germ cells and follicle cells. APF: after puparium formation. Similar findings were made when using RNA-seq to quantify the full-length and Env mRNAs from endogenous mdg4 (Extended Data Fig. 4). Three independent biological replicates were performed.
As adult Drosophila hindguts possess no active stem cells and little cell turnover40, an unceasing flow of mdg4 mobilization is predicted to steadily increase the number of eGFP-positive cells as pupae develop into adulthood. However, we detected that the average number of eGFP-positive cells per hindgut decreased from an average of 46 in pupae to 13 in adults (Fig. 2b). Similarly, we also observed more eGFP-positive cells in pupal salivary glands, proventriculi, and midguts than in the corresponding adult structures (Extended Data Fig. 2c). Therefore, our data indicate that a proportion of cells harboring mdg4 mobilization events are eliminated during development, and that mdg4 only mobilizes at the pupal stage.
Since reporter cassettes can be epigenetically silenced after retrotransposition41, we further examined intron removal from the reporter by PCR to validate our findings from eGFP expression (Extended Data Fig. 3). Consistent with eGFP signals, we detected a clear intron-removed PCR product from the tissue that harbors the most eGFP-positive cells–hindguts from the 48-hour pupae (Extended Data Fig. 3b). For tissues without eGFP-positive cells, including embryos, hindguts from larvae or 0-hour pupae, the intron-removed PCR product was not detected (Extended Data Fig. 3b). Notably, this PCR-based method could not detect the intron-removed DNA from the adult tissues that harbor a few GFP-positive cells within a large population (Extended Data Fig. 3c), indicating that this method is less sensitive than monitoring eGFP fluorescence when probing infrequent mobilization events. Overall, these findings further support that mdg4 mobilizes during tissue regeneration, but not at other stages.
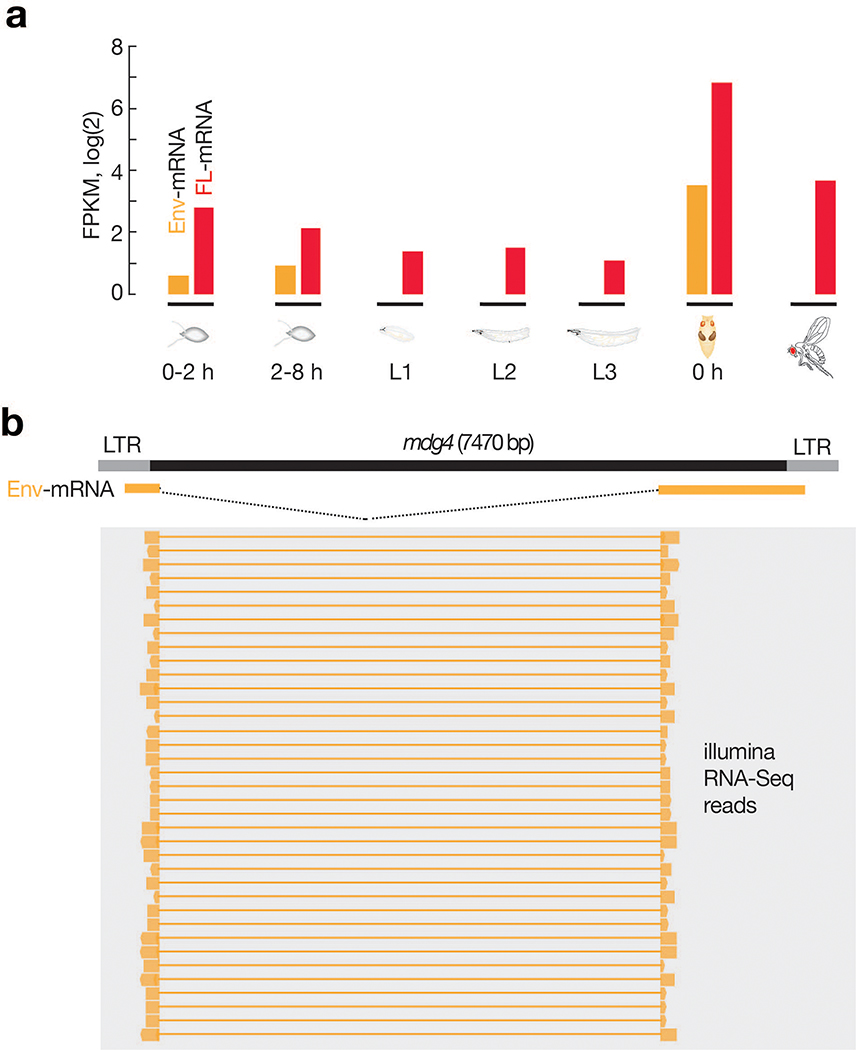
To further investigate the activity of mdg4 during development, we monitored its mRNA levels at different stages (Fig. 2c,d). In addition to transcribing full-length transcripts that encode Gag and Pol proteins, for its activation, mdg4 can produce spliced transcripts (Env mRNAs) encoding Env proteins (Fig. 2c)42. We found that full-length mdg4 mRNAs were transcribed throughout all life stages (Fig. 2d), arguing against transcriptional silencing. However, the Env mRNAs were primarily produced at the pupal stage (Fig. 2d). These results were confirmed by RNA sequencing (Extended Data Fig. 4). In summary, our data show that mdg4 selectively becomes active at the pupal stage to produce Env, potentially for infectious virus-like particle packaging and mobilization.
Flies harbor multiple full-length copies of endogenous mdg4
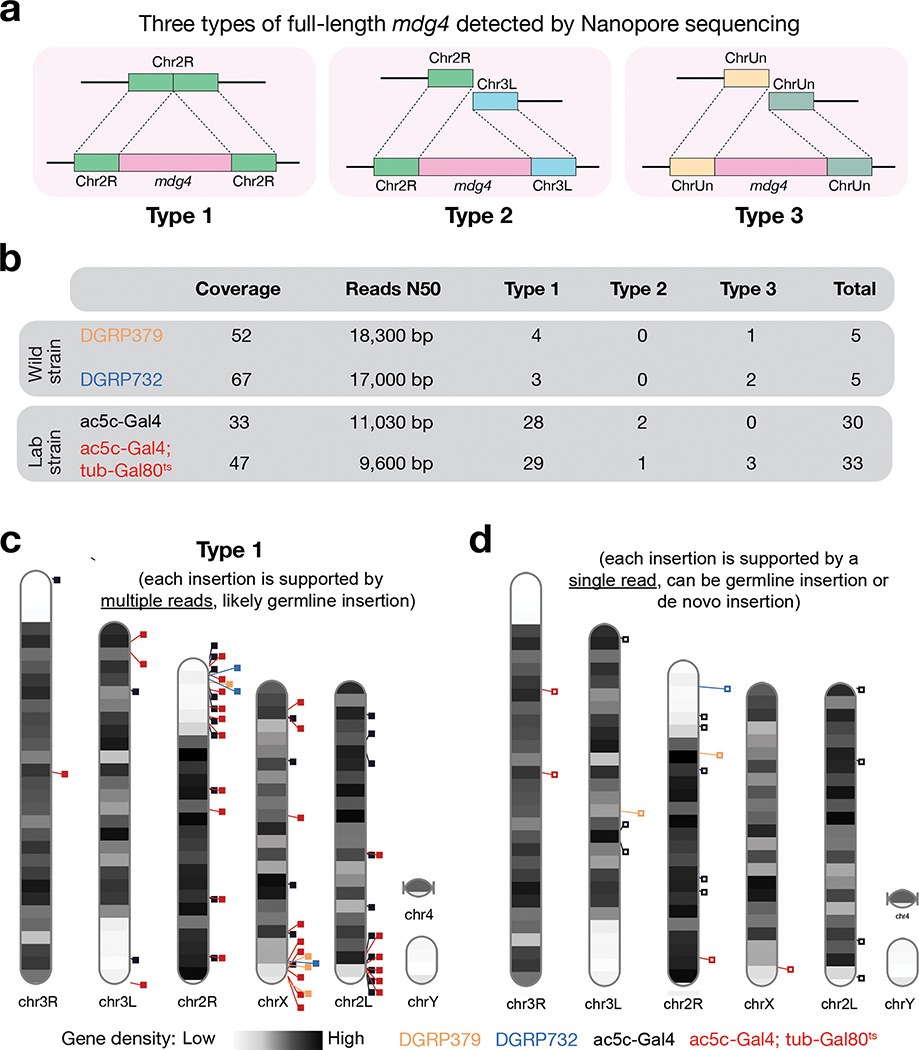
Given their repetitive nature, precisely quantifying transposon copy numbers has been challenging27. Nanopore sequencing technology, which can sequence DNA up to megabases without PCR amplification43, allows us to accurately examine the copy numbers of endogenous mdg4 in laboratory strains used in this study. From the laboratory strains we sequenced, we detected 30–33 copies of full-length endogenous mdg4 (Fig. 3). For most copies, we can unambiguously assign them to unique genomic regions (Type 1 from Fig. 3). Meanwhile, we also detected full-length mdg4 flanked by discordant sequences (Type 2 from Fig. 3) or unassembled genomic sequences (Type 3 from Fig. 3). These data indicate that the currently assembled fly genome still underestimates the full-length copies of mdg4. Furthermore, we analyzed published Nanopore sequencing data from two wild strains44, and found that both contain 6–7 copies of full-length mdg4. Similar to laboratory strains, while most of these full-length copies in wild strains can be uniquely assigned to the assembled fly genome (Type 1 from Fig. 3), there are one or two copies of full-length mdg4 flanked by unassembled DNA sequences (Type 3 from Fig. 3). Given that the laboratory strains were isolated ~100 years ago45,46, the heterogeneity we detected between laboratory and wild strains highlights that mdg4 has been actively mobilizing, potentially facilitating species evolution or adaptation.
Fig. 3 |. Nanopore sequencing detected full-length copies of mdg4 from both wild and laboratory strains.
a, Cartoon display of 3 types of full-length mdg4 detected. Type 1: Full-length mdg4 flanked by coordinated sequences from the reference genome. Type 2: Full-length mdg4 flanked by discordant sequences from the reference genome, likely reflecting the polymorphisms between the reference genome and the genome of individual strain. Type 3: Full-length mdg4 flanked unassembled genome sequences. b, A table to summarize the sequencing depth, N50 of the sequencing reads, and the number of full-length mdg4 detected from each strain. c, Chromosome ideogram to display the chromosomal positions of type 1 full-length mdg4. Each event reported in panels b and c is supported multiple reads, likely reporting germline mdg4 copies. d, Chromosome ideogram to display the chromosomal positions of full-length mdg4 detected by a single read, likely reporting de novo insertions or germline insertions with low regional coverage.
mdg4 protects adults from persistent viral infection
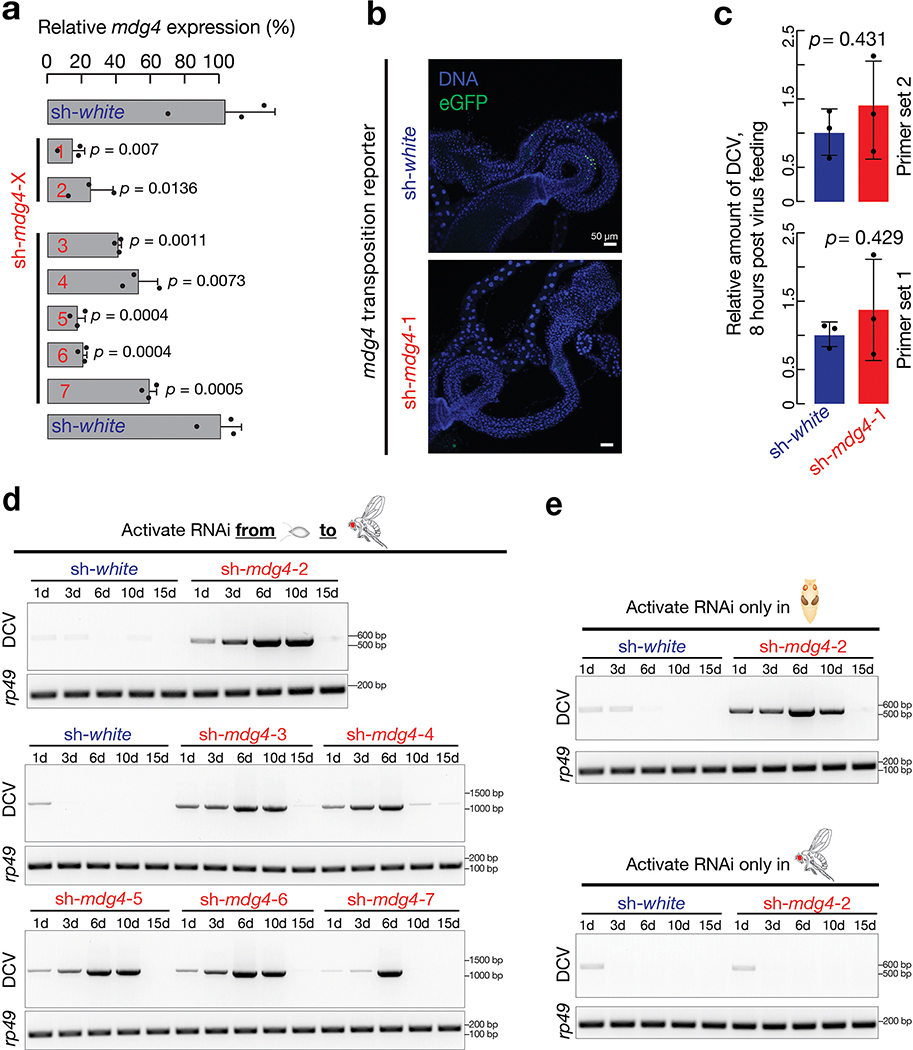
To determine the function of developmental mdg4 activation, we used RNAi constructs to ubiquitously suppress mdg4 in the laboratory strains. By depleting over 80% of mdg4 mRNAs and abolishing mdg4 mobilization during development (Extended Data Fig. 5a,b), we concluded that our RNAi constructs efficiently silenced mdg4. With mdg4 being suppressed (sh-mdg4), we found that flies still developed normally and exhibited no overt phenotypes.
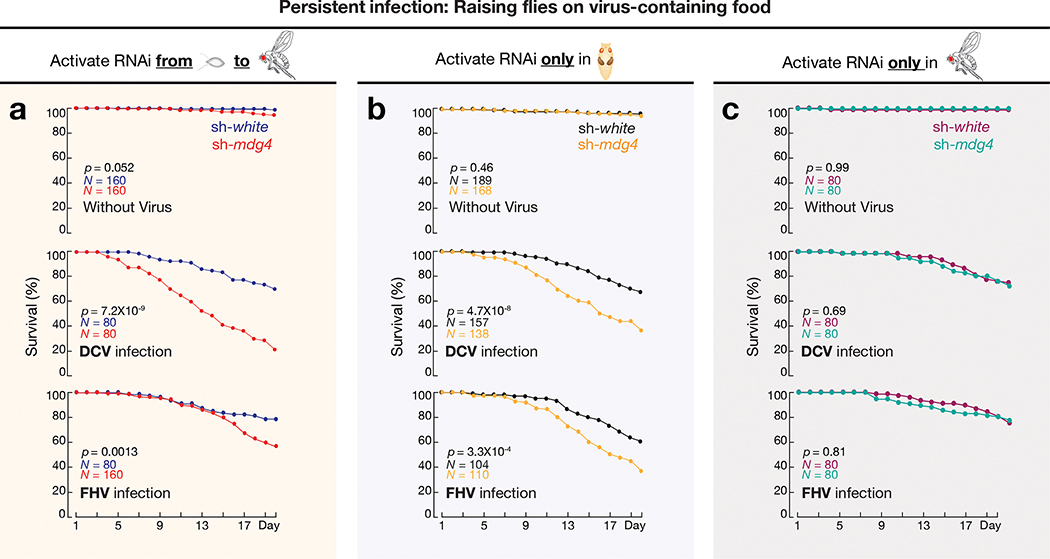
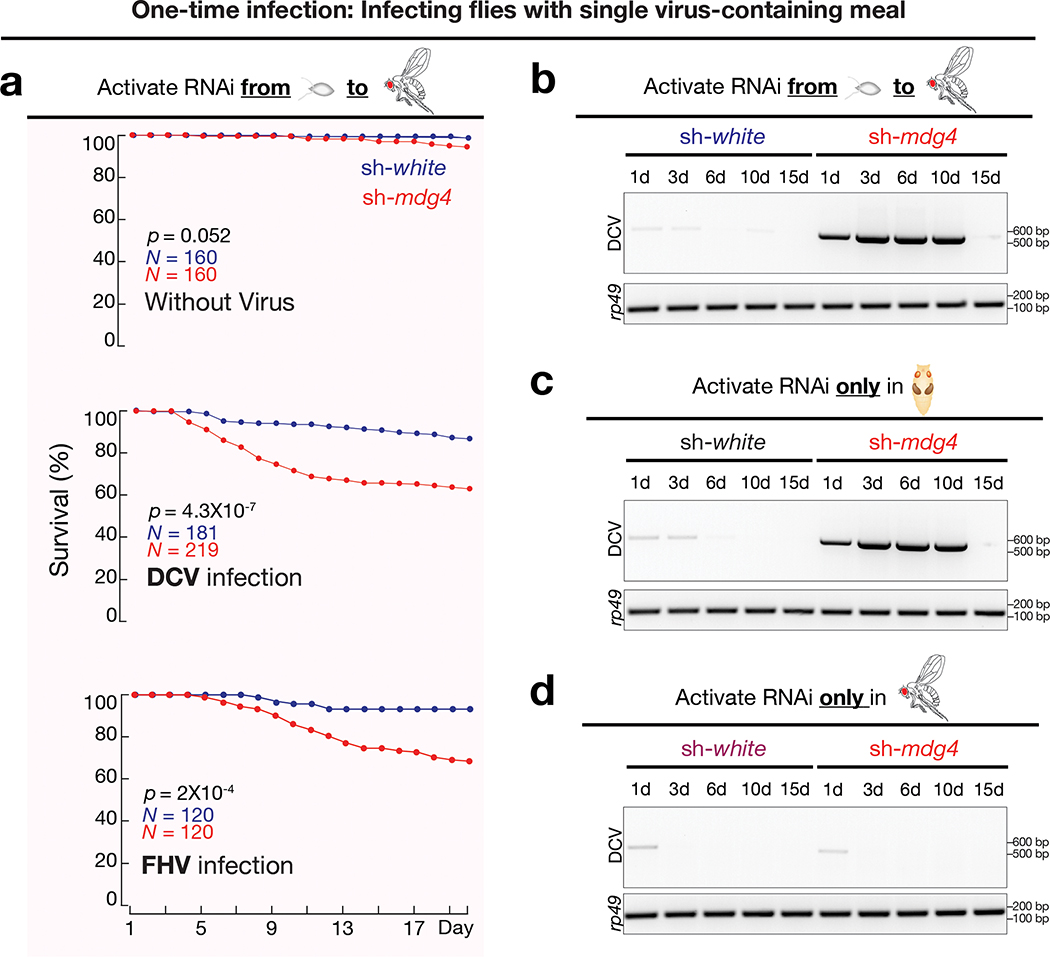
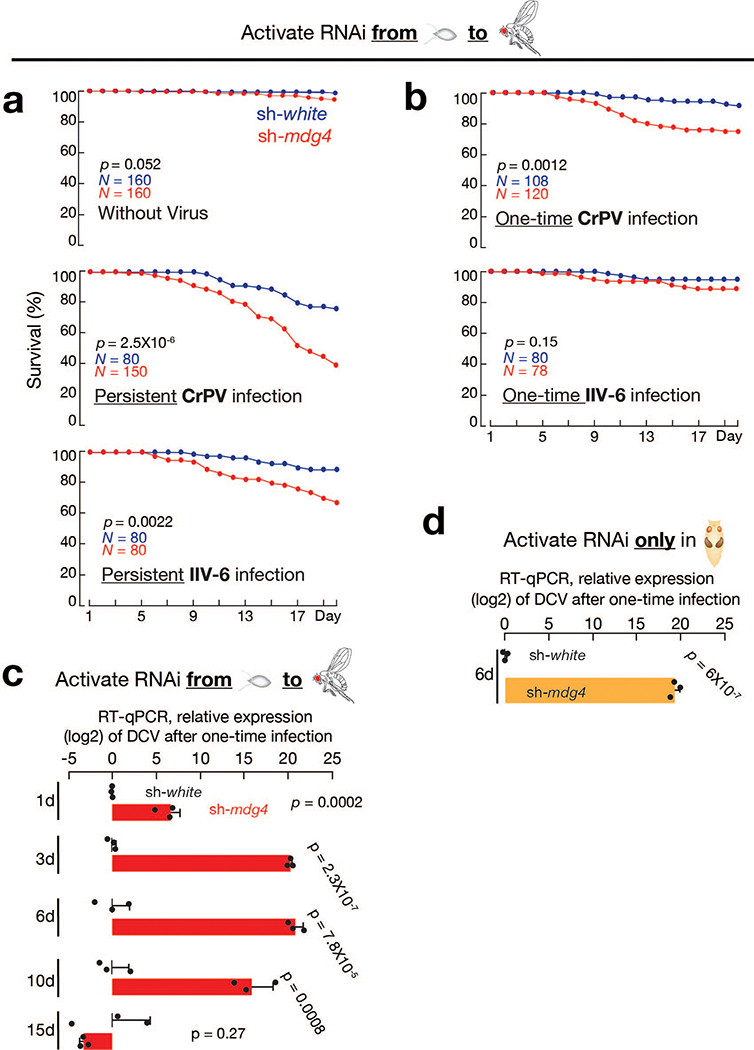
Given the retroviral origin of mdg4, we next tested whether flies can establish protective immunity from experiencing mdg4 activation for future viral defense. Since we observed most mdg4 mobilization events in digestive tissues (Fig. 1 and Extended Data Fig. 1), we utilized oral feeding to infect adult flies with multiple pathogenic viruses: Drosophila C virus (DCV), Cricket paralysis virus (CrPV), Flock House virus (FHV), and Invertebrate iridescent virus 6 (IIV-6). Among them, the first three are RNA viruses, and IIV-6 is a dsDNA virus. Upon housing adult flies on virus-containing food for 20 days, we observed that mdg4 activity optimally protects the hosts (Fig. 4a, Extended Data Fig. 6a,7a). For DCV infection, while only 30% of flies from the control group (sh-white) died (Fig. 4a), the lethality rate of flies with mdg4 silenced (sh-mdg4) significantly soared to 78% (P = 7.2 × 10−9, Cox proportional hazards regression; the same test applies to other P value calculations on lethality rates; Fig. 4a). For the other three viruses, the lethality rates were also significantly higher when mdg4 was silenced (Fig. 4a and Extended Data Fig. 7a): CrPV (from 24% in controls to 60% in sh-mdg4 flies, P = 2.5 × 10−6); FHV (from 21% to 43%, P = 0.0013); and IIV-6 (from 11% to 32%, P = 0.0022).
Fig. 4 |. mdg4 activation at pupal stage safeguards adult flies upon persistent viral infections.
a, By activating mdg4 RNAi from embryonic to adult stage, the survival rates were measured by raising flies on virus-containing food for 20 days. sh-white flies served as controls. All viral infection assays from this manuscript were performed with at least two biological replicates. Each replicate contains 20 males and 20 females. b, With mdg4 only silenced at pupal stage, the survival rates were measured by raising flies on virus-containing food for 20 days. c, With mdg4 only suppressed at adult stage, the survival rates were measured by raising flies on virus-containing food for 20 days. Flies used in panel a have one copy of the mdg4 reporter. Flies used in panels b and c only have endogenous copies of mdg4. Comparison of survival curves for all panels was completed using a Cox proportional-hazards model.
The aforementioned experiments were performed by activating the sh-mdg4 construct throughout the fly lifespan. While full-length transcripts are produced at all stages, Env mRNA production and mobilization only occur at the pupal stage (Fig. 2d). Thus, we next tested whether the protection from viral infection was gained from mdg4 activity specifically during the pupal stage. For the flies that only harbor the endogenous copies of mdg4, we suppressed mdg4 activity specifically at the pupal stage (Extended Data Fig. 6a–d). Compared to control flies (sh-white), adult flies with mdg4 silenced at the pupal stage showed significantly higher lethality rates upon viral infection (Fig. 4b). By treating flies with DCV or FHV for 20 days, the mortality rate significantly increased for sh-mdg4 flies compared to control flies (Fig. 4b). By contrast, specifically activating RNAi at the adult stage and raising the flies on food containing either DCV or FHV for 20 days led to similar survival rates between mdg4 depleted and control flies (Fig. 4c). Altogether, our data from stage specific suppression indicate that it is the mdg4 activity during the pupal stage, but not other stages, that bolsters the immune system for future antiviral defense.
mdg4 activity improves adult viral clearance efficiency
For fruit flies, natural viral infection also can occur from single-meal consumption. Instead of continuously housing flies on virus-containing food, we next tested the condition of one-time infection by feeding flies with viruses for only 16 hours (Extended Data Fig. 6a). Silencing mdg4 appears to have no impact on virus consumption (Extended Data Fig. 5c). Monitoring mortality rates thereafter revealed a similar function of mdg4 on protecting flies from the three RNA viruses (Fig. 5a and Extended Data Fig. 7b). For DCV, CrPV, and FHV, the lethality rates were all significantly higher when mdg4 was suppressed, compared with control animals (P = 4.3 × 10−7 for DCV, P = 0.0012 for CrPV, and P = 0.0002 for FHV; Fig. 5a and Extended Data Fig. 7b).
Fig. 5 |. mdg4 activation at pupal stage safeguards adult flies upon one-time viral infections.
a, By activating mdg4 RNAi from embryonic to adult stage, the survival rates were measured after infecting adult files with DCV or FHV from single meal. Comparison of survival curves was completed using a Cox proportional-hazards model. b, By activating mdg4 RNAi from embryonic to adult stage, RT-PCR experiments were performed to monitor the amount of DCV in fly bodies after one-time infection. c, With mdg4 only silenced at pupal stage, RT-PCR experiments were performed to monitor the amount of DCV in adult flies after one-time infection. d, With mdg4 only silenced at adult stage, RT-PCR experiments were performed to monitor the amount of DCV in fly bodies after one-time infection. Three independent biological replicates were performed for panels b, c, and d.
Noting a previous report that fruit flies are capable of clearing DCV upon one-time infection47, we next tested the potential function of mdg4 activation on DCV clearance. Control flies (sh-white) that experienced normal mdg4 activation at the pupal stage cleared the viruses within one day (Fig. 5b and Extended Data Fig. 5d). In contrast, flies with mdg4 silenced were unable to clear viruses (Fig. 5b and Extended Data Fig. 5d). Based on RT-qPCR experiments, there were > 62,557-fold more DCV RNA in sh-mdg4 flies than control animals (sh-white) for the first 10 days after infection (Extended Data Fig. 7c). During these first 10 days, sh-mdg4 flies showed a significantly 4.4-fold higher mortality rate than sh-white controls: 28.8% for sh-mdg4 flies vs. 6.6% for sh-white animals (P = 4.3 × 10−7; Fig. 5a). Subsequently, the mortality rates of sh-mdg4 flies were decreased to a level that is indistinguishable from sh-white control animals (Fig. 5a). Consistently, there was minimal, if any, DCV detectable in the sh-mdg4 survivors at day 15 after infection (Fig. 5b), indicating that the survivors eventually clear out DCV. In summary, our findings indicate that mdg4 activation renders a robust ability to rapidly clear invading DCV at the adult stage.
We also tested the necessity of having mdg4 activation at the pupal stage on adult DCV clearance with the one-time feeding assay (Extended Data Fig. 6a). When only silencing mdg4 during metamorphosis, adult flies lacked the ability to clear orally infected DCV (Fig. 5c and Extended Data Fig. 5e). For example, there was 787,514-fold more DCV in sh-mdg4 flies than controls at day 6 after infection (sh-mdg4: DCV ΔCt = −4.83; sh-white: DCV ΔCt = 14.76; and ΔΔCt = 19.59, Extended Data Fig. 7d). By contrast, flies with full-length transcripts suppressed at adulthood, but had experienced mdg4 activation at the pupal stage, had no defects on DCV clearance (Fig. 5d and Extended Data Fig. 5e). With the DCV one-time feeding assay, these flies cleared the invading viruses as efficiently as control animals (Fig. 5d). Our data thus suggest that having mdg4 activation at the pupal stage is essential for rapid clearance of invading viruses in adults.
Pupal Relish activation protects adults from viral infection
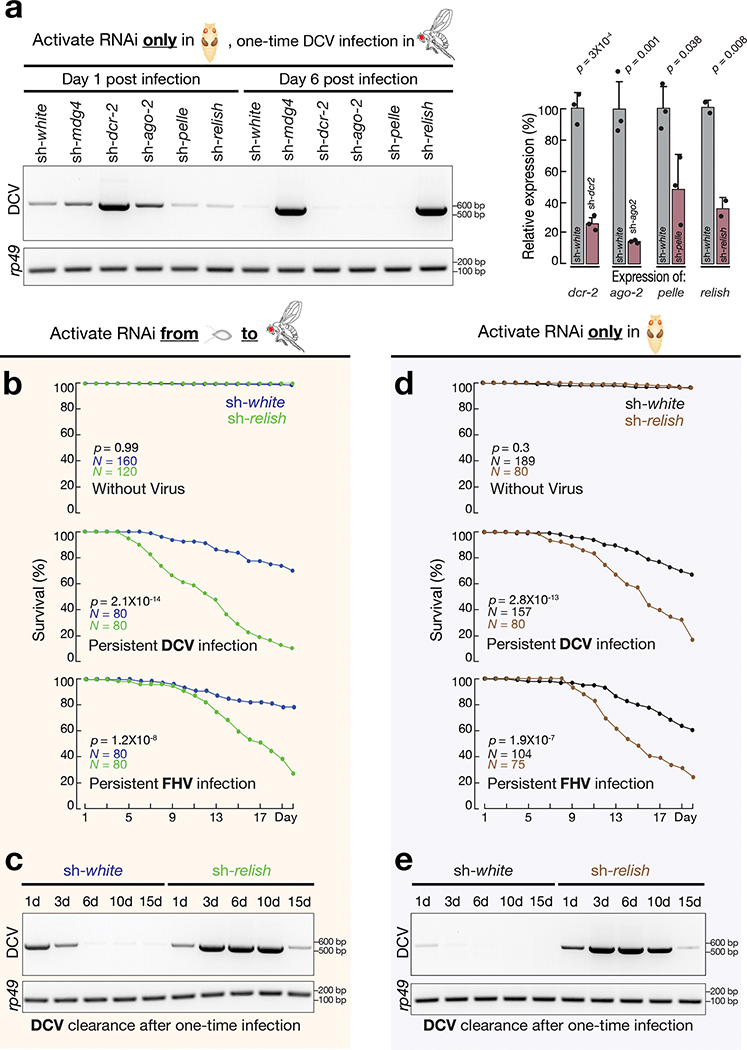
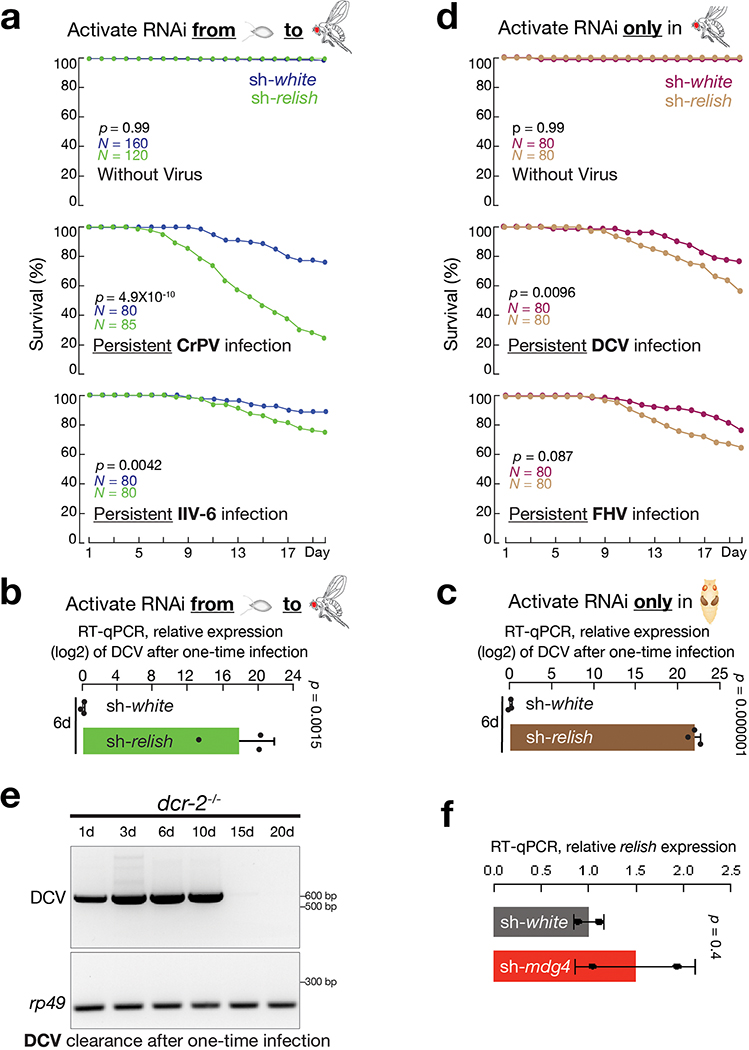
In Drosophila, multiple mechanisms have been reported to execute an antiviral function19. To screen key factors that potentially establish protective immunity at the pupal stage, we individually depleted the following four proteins only during metamorphosis and assayed DCV clearance in adult flies after one-time infection (Fig. 6a). These factors were: Dicer-2 and Argonaute-2 from the RNAi pathway, Pelle from the Toll pathway, and Relish from the IMD and dSTING pathways19,48–51. While suppressing Dicer-2, Argonaute-2, or Pelle at the pupal stage had no impact on DCV clearance in adult flies, silencing Relish led to similar DCV accumulation as observed from sh-mdg4 flies (Fig. 6a).
Fig. 6 |. Relish activation at pupal stage protects adult flies from viral infections.
a, With each key immune factor depleted only at pupal stage, adult flies were challenged with DCV by one-time feeding. The amount of DCV in fly bodies on day 1 and day 6 after infection was measured by RT-PCR. Right panel: the silencing efficiency of each RNAi construct was measured by RT-qPCR based on the two-tailed t-test. Newly eclosed flies were used to extract RNA. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for three biological replicates for sh-dcr2, sh-ago2, sh-pelle and two biological replicates for sh-relish. b, Upon activating Relish RNAi from embryonic to adult stage, the survival rates were measured by raising flies on virus-containing food for 20 days. sh-white flies served as controls. c, Upon activating Relish RNAi from embryonic to adult stage, RT-PCR experiments were performed to monitor the amount of DCV in fly bodies after one-time infection. d, With Relish only silenced at pupal stage, the survival rates were measured by raising flies on virus-containing food for 20 days. e, With Relish only silenced at pupal stage, RT-PCR experiments were performed to monitor the amount of DCV in adult flies after one-time infection. Three independent biological replicates were performed for panels c and e. Comparison of survival curves was completed using a Cox proportional-hazards model for panels b and d.
Although recent studies established the antiviral function of Relish when viruses were administrated via injection19, whether Relish possesses similar role upon oral-feeding is still unclear. By housing flies on virus-containing food for 20 days, we found Relish is essential to protect flies from oral infection (Fig. 6b and Extended Data Fig. 8a). Similar to mdg4 suppression, silencing Relish resulted in significantly higher mortality rates when flies were orally challenged by DCV, FHV, CrPV, and IIV-6 (Fig. 6b and Extended Data Fig. 8a). Similarly, sh-relish flies failed to efficiently clear DCV from the one-time feeding assay (Fig. 6c). At day six after infection, sh-relish flies had 292,659-fold more DCV than controls (sh-relish: DCV ΔCt = −3.25; sh-white: DCV ΔCt = 14.91; and ΔΔCt = 18.16; Extended Data Fig. 8b).
Given that mdg4 possibly induces the antiviral function of Relish at the pupal stage, we next tested whether Relish activation at this specific stage is essential for combating viruses in adults. Suppressing Relish solely at the pupal stage made adult flies significantly more vulnerable to viral infection (Fig. 6d,e). When raising these flies on DCV-containing food for 20 days, the mortality rate significantly increased from 33% in control flies to 82% in sh-relish group (P = 2.8 × 10−13; Fig. 6d). FHV infection led to similar findings: while control flies had a 39% mortality rate, the rate from sh-relish animals soared to 76% (P = 1.9 × 10−7; Fig. 6d). With the one-time feeding assay, flies with Relish silenced at the pupal stage failed to rapidly clear DCV (Fig. 6e and Extended Data Fig. 8c), resembling the findings from sh-mdg4 flies (Fig. 5c and Extended Data Fig. 7d). In contrast, the Relish activity at the adult stage is less prominent during viral warfare (Extended Data Fig. 8d). Raised on DCV-containing food, flies with Relish suppressed solely during adulthood increased the mortality from 24% in control animals to 44% (P = 0.0096; Extended Data Fig. 8d). For FHV infection, the changes in mortality rates between control and sh-relish animals were not statistically significant (from 24% in controls to 35% in sh-relish animals, P= 0.087; Extended Data Fig. 8d). It is worth noting that besides the protective response induced by mdg4/Relish, the RNAi pathway also contributes to antiviral responses upon oral infection in adults (Extended Data Fig. 8e)47. This pathway could directly degrade the viral RNA upon infection19,52, likely providing a compensatory mechanism on virus-silencing for the flies’ lack of adult Relish activity.
Altogether, our data indicate that inducing the antiviral function of Relish at the pupal stage provides flies optimal safeguard from future viral infection.
mdg4 triggers both local and systemic Relish activation
As an NF-κB factor, Relish activation requires an endoproteolytic step in the cytoplasm to release an N-terminal fragment (Relish-N), which contains the DNA-binding domain53. Upon translocating to the nucleus, Relish-N acts as a transcription factor to drive the expression of genes for antiviral responses53. Since mdg4 activation appears to have no impact on Relish expression (Extended Data Fig. 8f), we next sought to test whether mdg4 pupal activation affects Relish-N localization. Given that most tissues undergo degradation at the pupal stage, it is technically challenging to monitor Relish-N localization. Therefore, we focused our examination at the early pupal stage preceding tissue degeneration.
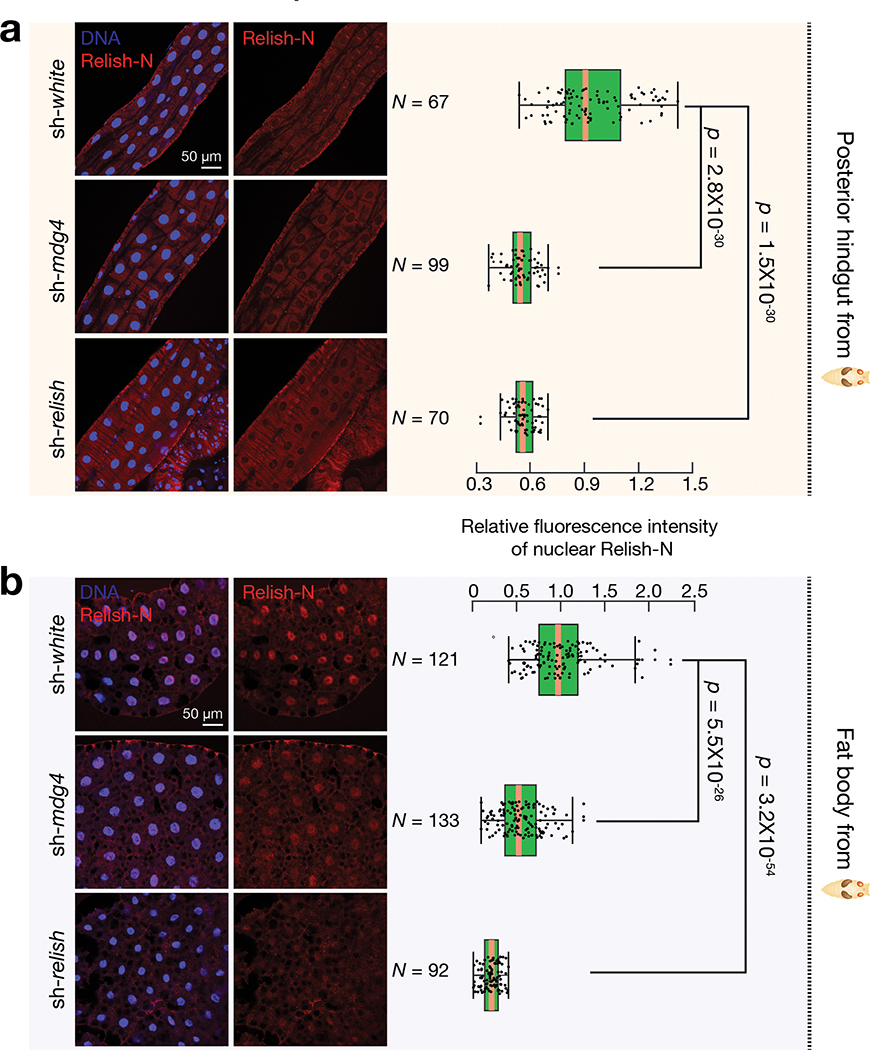
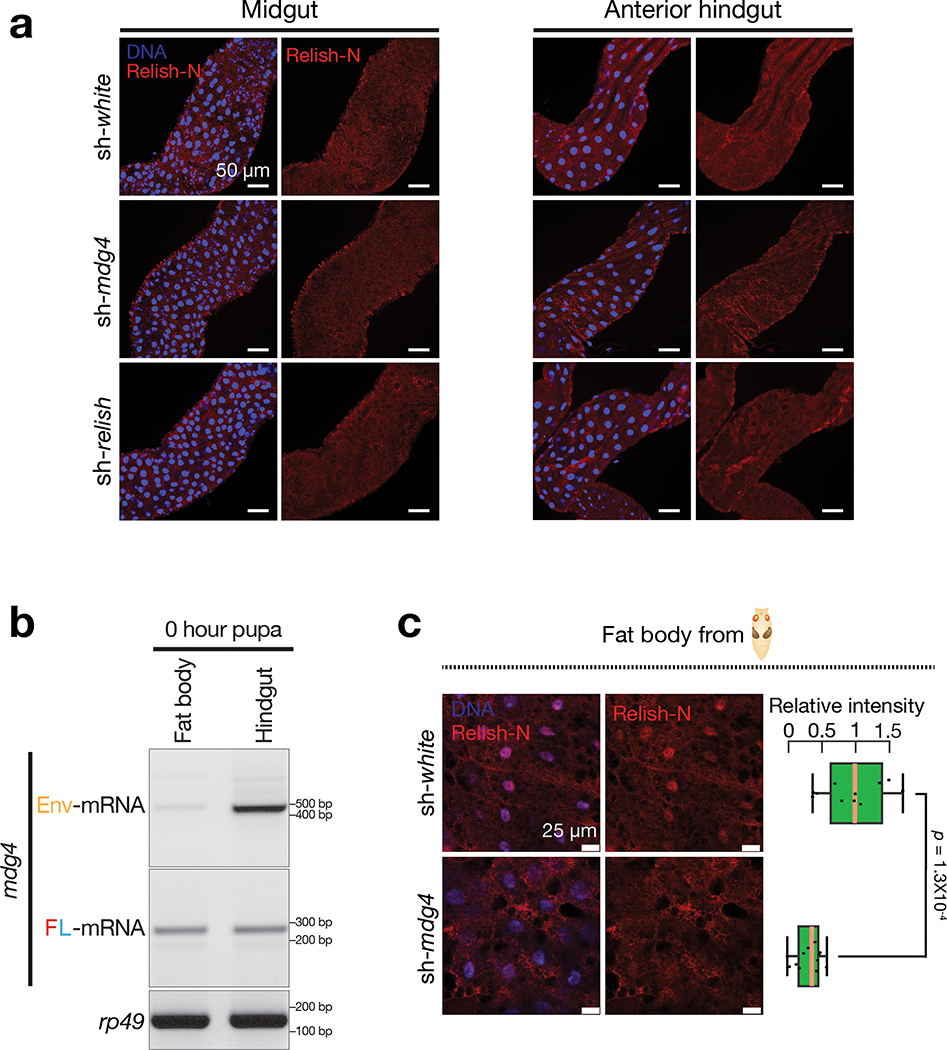
The antiviral response can be activated locally at epithelial barriers that directly encounter pathogens or in the fat body, which mediates a systemic response19. We first examined Relish localization in the gut, where we detected mobilization events and orally-acquired viruses likely invade the animals. Utilizing Relish-N antibodies that detect both full-length and N-terminal fragments of Relish, we observed very low, if any, signals in normal pupal midgut and the anterior hindgut (Extended Data Fig. 9a). However, at the posterior hindgut from control flies, Relish-N antibodies produced prominent signals in both cytoplasm and nucleus, with a slight enrichment at the nuclear periphery (Fig. 7a). Notably, upon mdg4 suppression, the nuclear Relish-N signal significantly decreased 1.6-fold (P = 1.2 × 10−23; Fig. 7a). These data suggest that mdg4 activation drives Relish translocation in the posterior part of the hindgut.
Fig. 7 |. mdg4 triggers Relish activation in both hindgut and fat body at pupal stage.
a, Immunostaining to detect the nuclear Relish-N signals in the posterior part of hindgut from sh-white, sh-mdg4, or sh-relish early pupae. The box plot shows the relative fluorescence intensity of nuclear Relish-N signals in the posterior part of hindgut. b, Immunostaining to detect the nuclear Relish-N signals in the fat body cells from sh-white, sh-mdg4, or sh-relish early pupae. Flies were raised on germ-free condition; Similar findings were made on non-germ-free condition (Extended Data Fig. 9c). The data were collected from 2 biological replicates with 3 animals per replicate. The box plot shows the relative fluorescence intensity of nuclear Relish-N signals in the fat body cells. Silencing mdg4 resulted in a significant decrease of the nuclear Relish-N in both hindgut and fat body cells. Box plots report the minimum, maximum, median, and interquartile ranges of the data. Two-tailed t-tests were used to evaluate the difference between each pair of groups.
Besides initiating a local antiviral response, fruit flies can also induce a systemic response by triggering the humoral immunity, which is mainly mediated by the fat body19. Although mdg4 mobilization events were nearly undetectable in fat body cells, mdg4 products produced within these cells or from the virus-like particles could potentially activate Relish in the fat body. To test this, we first measured mdg4 mRNA levels in fat body from early pupae and found both full-length and Env mRNAs (Extended Data Fig. 9b). Notably, in fat body cells from the control pupae, we detected strong nuclear localization of Relish-N (Fig. 7b and Extended Data Fig. 9c), indicating activation of the systemic antiviral response. However, in mdg4 silenced pupae, the amount of nuclear Relish-N significantly decreased 1.8-fold (P = 5.5 × 10−26; Fig. 7b and Extended Data Fig. 9c). Our data hence indicate that even without mobilization, mdg4 products can potentially license Relish activation in the fat body to systemically induce antiviral responses.
dSTING is also required for the protective benefit
In Drosophila, while the IMD pathway utilizes Relish-N for anti-bacterial/fungal responses, the antiviral function of Relish-N is activated through dSTING48–51. dSTING is the fly ortholog of mammalian STING, which can be activated through innate sensors that detect invading pathogens. We next asked whether dSTING is also required for licensing the Relish-mediated immunity at the pupal stage. Similar to mdg4 suppression, silencing dSTING significantly decreases nuclear Relish-N signals in both hindgut and fat body cells from early pupae (Fig. 8a,b). These data suggest a function of dSTING on triggering Relish activation when flies establish the protective immunity. Given that these flies were raised on germ-free conditions, dSTING is likely activated from recognition of endogenous products, like mRNA or cDNA from mdg4.
Fig. 8 |. dSTING triggers Relish activation at the pupal stage for adult anti-viral responses.
a, RT-qPCR to measure the RNAi silencing efficiency of dSTING based on the two-tailed t-test. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for three biological replicates. b, Immunostaining to detect the nuclear Relish-N signals in the posterior part of hindgut (top panel) and fat body cells (bottom panel) from sh-white and sh-dSTING early pupae. Silencing dSTING resulted in a significant decrease of the nuclear Relish-N in both hindgut and fat body cells. The flies were raised on germ-free condition. The data were collected from 2 biological replicates with 3 animals per replicate. Box plots report the minimum, maximum, median, and interquartile ranges of the data. A two-tailed t-test was used to compare the relative intensities of each group. c, With dSTING only silenced at pupal stage, the survival rates were measured by raising flies on virus-containing food for 20 days. Comparison of survival curves was completed using a Cox proportional-hazards model. d, With dSTING only silenced at pupal stage, RT-PCR experiments were performed to monitor the amount of DCV in adult flies after one-time infection. Three independent biological replicates were performed.
If the Relish activity triggered by dSTING establishes protective immunity, silencing dSTING at pupal stage should leave adult flies compromised. Indeed, upon persistent DCV infection, the adult flies that experienced normal dSTING expression at the pupal stage had a mortality rate of 33%. However, the adults with dSTING suppressed at the pupal stage showed a significantly higher mortality rate: 73%, P = 7.8 × 10−13 (Fig. 8c). Similarly, we observed a significant increase of mortality rate when files were raised on FHV for 20 days: from 39% for control flies to 83.5% in sh-dSTING animals (P = 1.9 × 10−14, Fig. 8c). With the one-time feeding assay, we found that adult flies lacking dSTING at pupal stage failed to clear orally acquired DCV (Fig. 8d), reminiscent of the findings when either mdg4 or Relish was silenced at the same stage (Fig. 5c and Fig. 6e).
Collectively, our data indicate that dSTING is an essential component to induce Relish-mediated immunity at the pupal stage to provide adult flies maximal protection against viral infection.
DISCUSSION
A current view posits that retrotransposons are largely suppressed during development to minimize their detrimental effects on their hosts. In contrast, our findings suggest a different paradigm for the function of developmental retrotransposon activation (Extended Data Fig. 10). By spatiotemporally tracking their activity at the mobilization level, we report that one retrotransposon family, mdg4, selectively becomes active during metamorphosis. Notably, compared with other somatic organs, digestive tissues harbor higher mdg4 activity. Given that these tissues likely encounter pathogens during natural infections, we tested whether mdg4 activation in them creates a protective response against oral virus infection. Our data indeed suggest that during the process of building adult tissues, hosts employ this wave of retrotransposon activation to prime their antiviral system, an effect that lasts well into adult stages. We hence propose that, at least in the animals we investigated, retrotransposon activation trains the immune system to provide long-term antiviral protection.
There are several mechanisms by which mdg4 might trigger a protective and long-lasting immune response. Given the restricted window of Env expression, its protein products may directly serve as ligands to activate immune signaling. Supporting this hypothesis, the Env proteins from vesicular stomatitis virus (VSV-G) could indeed directly trigger innate immune activation in Drosophila54,55. Alternatively, similar to how HIV DNA triggers cGAS/STING-mediated immune response in human cells56, mdg4 nucleic acid products may activate dSTING to drive an immune response. Our finding that suppressing dSTING or mdg4 at the pupal stage leads to similar defects on Relish location and adult antiviral responses indicates that mdg4 could induce Relish activation via dSTING. Rigorously testing this hypothesis first requires a thorough understanding of STING signaling in Drosophila.
Given that mounting a prompt response is essential for animal survival upon infection, priming of Relish-mediated viral-defense mechanism by mdg4 may enable adults to launch a swift response. Besides promoting antiviral signaling for infected cells, mdg4 activation might stimulate the activity or maturation of macrophages–the cells that engulf viruses and virus-infected cells. Supporting this hypothesis, evidence indicates that macrophages from fly embryos can engulf microbes only if they have previously been primed by encountering dead cells57.
Could mdg4 mobilization contribute to the antiviral response? As we discussed, it is likely the products from mdg4 that trigger Relish activation. Thereby, the integration step appears to be dispensable for inducing the host’s antiviral response. Consistently, we readily observed Relish activation when flies just enter the pupal stage, when Env production first peaks but mobilization is barely detectable from our reporter. Interestingly, a second wave of Env expression occurs later in pupation during tissue regeneration, and is accompanied by mdg4 integration events. It is possible that these integration events are just byproducts of mdg4 activation, which provides an antiviral benefit to the fly. As such, these mobilization events may be a concession that the host allows to gain the protection from future viral infections. However, increased mdg4 copies resulting from these mobilization events could manufacture additional products that further induce Relish activation. Therefore, it is also possible that allowing mobilization is a strategy employed by the hosts to maximize the benefits from mdg4.
In mammals, activation of LTR retrotransposons–the same class as mdg4–with epigenetic drugs can potentially stimulate interferon response58,59. Although the potential impact of retrotransposon activation to the host immunity during pathogenesis has been investigated11,58,59, less is known whether developmental activation of the mammalian retrotransposons can prepare the host to gain protective immunity. Interestingly, during human embryogenesis, there is also a wave of retrotransposon activation–from the 8-cell stage to the blastocyst stage–at which time the paternal genome is activated60. As maternal immunity is suppressed at these stages to avoid immune attack, future work will examine whether developing mammalian embryos employ an analogous mechanism to Drosophila to gain a long-term protection from pathogen infection. Although detailed mechanisms may differ, harnessing retrotransposons for pathogen warfare could potentially be a recurrent theme from flies to mammals.
METHODS
Fly husbandry and strains
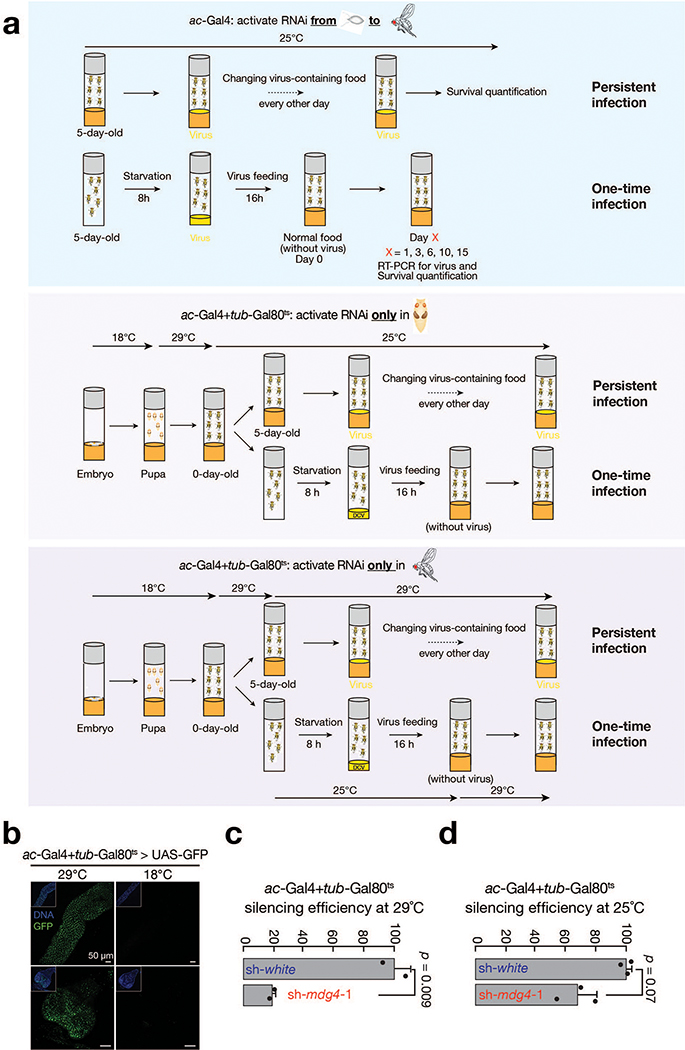
Generally, all flies were maintained at 25 °C and grown on standard agar-corn medium. Flies used to achieve gene silencing at all life stages carried one copy of the engineered mdg4 reporter. Flies used to achieve stage-specific gene silencing only had endogenous copies of mdg4. To silence white, mdg4, Relish, or dSTING at the pupal stage, embryos and larvae were raised at 18 °C. Wandering larvae were transferred to 29 °C and newly eclosed flies were transferred to 25 °C. To silence white, mdg4, or Relish in adult flies, embryos, larvae and pupae were raised in 18 °C and the newly eclosed flies were transferred to 29 °C. To minimize effects caused by genetic background, all of the sh-RNA flies were isogenized to w1118 isogenic background (w1118; iso2; Dr/TM6B,Tb,Hu). The fly alleles used in this study are listed in Supplementary Table 1. Supplementary Table 2 summarizes the fly genotypes for each figure. Animals were assigned to each biological groups in a randomized manner. Given that flies had to be sorted based on genotypes, data collection and analysis were not performed blind to the conditions of the experiments.
RT-PCR and RT-qPCR
Total RNA from 10 flies (5 females and 5 males) was extracted by using mirVana™ miRNA Isolation Kit (Thermo Fisher Scientific, Cat #AM1560). Ten μg of total RNA was treated with 2 μl of TURBO DNase (Thermo Fisher Scientific, Cat #AM2238) at 37 °C for 30 minutes. After DNase treatment, RNA was purified by RNA Clean & Concentrator-5 (Genesee Scientific, Cat #11-326), and the concentration was measured by NanoDrop. One μg of purified RNA was used for reverse transcription in 20 μl of reaction by using 1 μl of SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific, Cat #18080044) and 1 μl of random primers (Invitrogen, Cat #58875). To test the amount of DCV present after oral feeding, 1 μg of purified RNA was used in a 20 μl reaction with 4 μl of iScript reverse transcriptase supermix (BIO-RAD, Cat# 1708841). For each reaction, 1 μl of cDNA was used for either PCR by using GoTaq Green Master Mix or qPCR by using SsoFast EvaGreen Supermixes (BIO-RAD, Cat# 172-5204). PCR products were run on 1.5% agarose gel and scanned by BIO-RAD Gel Doc Universal Hood II. P values were calculated from at least three independent biological replicates using a two-tailed, two-sample unequal variance t-test. The error bars on the graphs report standard deviation for three biological replicates. qPCR for Extended Data Figure 8f was calculated from two biological replicates. Extended Data Figures 5c and 8f use the DART-PCR method of analysis61. Primers used for RT-PCR and RT-qPCR are listed in Supplementary Table 3a. P values were calculated by the Student’s t-test. The data met the assumptions of the statistical tests used. No animals or data points were excluded from these analyses.
Transgenic flies: eGFP reporter and sh- mdg4
The constructs of mdg4 transposition reporter (mdg4-TR) and negative control (NC), 412-TR, Blood-TR, Burdock-TR, Copia-TR and Mdg1-TR were generated by using Counter-Selection BAC Modification Kit (GENE BRIDGES, Cat# K002). The BAC clone used in this study to serve as template, which contains an intact copy of mdg4, is p[acman]-CH322-167N01. For other BAC clones: 412 is p[acman]-CH322-181C5, Blood is p[acman]-CH322-127N18, Burdock is p[acman]-CH322-187A21, Copia is p[acman]-CH322-186G13, and Mdg1 is p[acman]-CH322-86D10. Positive control of mdg4-TR (PC) and 3S18-TR was made by using Gibson Assembly and cloned into the BamHI site of attB-P[acman]-CmR-BW vector. mdg4 is 7,469 bp, Gag encoding sequence is from 1,080 to 2,435, Pol encoding sequence is from 2,438 to 5,470, and Env encoding sequence includes 567, 568 and from 5,551 to 7,000. The positive control, transposition reporter and negative control were inserted into mdg4 that is between 5,478 and 5,479, which will not disrupt any coding sequence. The transposition reporter for 3S18, 412, Mdg1, Blood, Copia and Burdock were constructed by landing the same GFP cassette (Extended Data Fig. 1a) into the following position respectively: 5,756–5,757, 6,914–6,915, 6,675–6,676, 6,823–6,824, 4,750–4,751, and 5,800–5,801.
To construct the plasmid for mdg4 silencing, the DNA fragments of sh-RNAs for depleting mdg4 were synthesized and cloned into the NheI and EcoRI sites of VALIUM20. All of the constructs were verified by colony PCR and sanger sequencing. All plasmids were site-specifically landed into fly genome at attP2 and attP40 sites. The 7 designed sh- mdg4 constructs suppressed mdg4 expression with different efficiencies (Extended Data Fig. 5a). However, all of them can silence mdg4 to a level that leads to consistent antiviral defects for the host flies (Extended Data Fig. 5). Notably, for the alleles that carry sh- mdg4-4 and −7 constructs that lead to lower RNAi efficiency, flies showed weaker virus-clearance defects. These data further suggest the antiviral response is specifically from mdg4. Primers used for constructing plasmids are listed in Supplementary Table 3b.
Quantifying mobilization events
To quantify eGFP-positive cells from mobilization reporter, pupae or adult flies were heat-shocked at 37 °C for 30 minutes, then recovered at room temperature for at least 3 hours. Corresponding tissues were dissected in cold PBS and fixed with 4% PFA. Samples were then washed 3 times with PBST, stained with DAPI and mounted with VECTASHIELD MOUNTING MEDIUM (VWR Cat #:101098-042). The cells that had GFP and DAPI co-localization were counted. Both adult male and female flies harbored similar number of GFP positive cells. For quantification, GFP fluorescence from female flies was directly visualized and the GFP-positive cells were counted by using ZEISS ApoTome fluorescence microscope. Representative images were taken by the Leica TCS SP5 confocal microscope.
Viral purification and titration
Drosophila S2 cells (Duke cell culture facility, Cat# CRL-1963) were cultured in T175 flask with 30 ml of complete medium (Schneider’s Drosophila Medium, Thermo Fisher Scientific, Cat# 21720024), 10% heat-inactivated FBS (Thermo Fisher Scientific, Cat# 10082139) and 1% penicillin & streptomycin (Thermo Fisher Scientific, Cat# 15140122)). For viral infection, S2 cells were expanded to 6 T175 flasks that contain complete medium plus 0.1% of Pluronic® F-68 (Thermo Fisher Scientific, Cat# 24040032) with a 1:6 split. Three days later, when cells reach about 75% confluence, 60 μl of viruses was thawed (DCV, CrPV, FHV and IIV-6) (3 × 1011 TCID50/ml) in 6 ml of Schneider’s Drosophila Medium and filtered with a 0.2 μm syringe filter (VWR, Cat# 28145-477). S2 cells were infected in each T175 flask with 1 ml of viruses diluted in medium.
For DCV, CrPV, and FHV purification: when more than 75% of cells were dead, cells with viruses were harvested and frozen at −20 °C for 1 hour to facilitate cell lysis. Then, viruses were thawed on the bench, transferred to ultracentrifuge tubes (BRCKMAN, Cat# 344058) and spun at 27,000 g in SW32Ti for 40 minutes at 4 °C to pellet cell debris. Supernatant was transferred to new ultracentrifuge tubes and spun at 110,000 g in SW32Ti for 3 hours at 4 °C to obtain viral pellet. Viral pellet was re-suspended in 1 ml of 50 mM Tris buffer (pH 7.4) and shaken in a cold room overnight. Suspended viruses were overlaid onto 10% sucrose, and centrifuged at 110,000 g in SW32Ti for 3 hours at 4 °C. Viruses were re-suspended in 1 ml of 50 mM Tris buffer (pH 7.4) and shaken at 4 °C for 20 hours. Viruses were aliquoted and stored at −70 °C.
For IIV-6 purification: the viruses were cultured with S2 cells for 7 days. After incubation, cells with viruses were harvested and frozen at −70 °C. The cells with IIV-6 were thawed and re-frozen for 3 cycles to facilitate cell lysis. Then the cells were transferred to 50 ml tubes and spun at 600 g for 15 minutes to remove the cell debris. This step was repeated once more. Supernatant was transferred to ultracentrifuge tubes and spun at 10,000 g in SW32Ti for 30 minutes at 4 °C to obtain viral pellet. IIV-6 pellet was re-suspended in 1 ml of ultrapure water and was laid onto 30% sucrose, and centrifuged at 30,000 g in SW32Ti for 1 hour at 4 °C. Viruses were then re-suspended in 1 ml of 50 mM Tris buffer (pH 7.4), aliquoted and stored at −70°C.
Viral titer was measured by endpoint dilution assay. Briefly, viral stocks were thawed on ice and made 10-fold serial dilution from 10−1 to 10−11. In 96-well plate, each well was filled with 100 μl of medium, 40,000 S2 cells, and 20 μl of medium with diluted viruses. Cells were incubated at 25 °C for 7 days. TCID50 was calculated by Spearman-Karber Method.
Viral infection
All flies used for DCV oral infection were dechorionated at the embryonic stage and treated with tetracycline for two generations. They were tested to be Wolbachia-negative by PCR (F: 5’-ACAGGGTCAGTAAAATATATTGCAG-3’, R: 5’-AGTTTTTCTGCTTTGATCACTACAC-3’). All viral infection assays were performed with at least two biological replicates. For each viral infection experiment: flies within 1 day old were collected (20 males and 20 females in each tube) and flipped to fresh food every day. These flies were used for the following experiments.
For one-time infection: five-day old flies were starved for 8 hours in an empty vial (horizontal). After starvation, flies were transferred to a vial containing a piece of sliced cotton plug in the bottom, which was soaked with 1 ml PBS with DCV at 3 × 109 TCID50/ml (DCV stock and 30% sucrose). After 16 hours, flies were transferred to normal food without DCV (day 0). Flies were transferred to clean food every other day. To extract RNA for RT-PCR and qPCR from these groups, 5 males and 5 females from each group were collected at day1, day3, day6, day10 and day15. To test the survival rate after one-time feeding, after starvation, flies were infected with DCV, CrPV, FHV and IIV-6 at 10 × 109 TCID50/ml. After infection, flies were transferred to fresh food every other day, and the number of dead flies were counted daily.
For persistent infection: 5 × 109 TCID50 of DCV, CrPV, FHV and IIV-6 were put on top of normal food and placed on bench until all liquid evaporated. Then 5-day old flies were transferred to the virus-containing food and counted as day 0. After that, we transferred flies to newly prepared virus-containing food every other day, and the number of dead flies were counted daily.
For survival analysis, we counted the number of dead flies from each group daily over 20 days. Comparison of the survival curves was completed in R 3.6.3 using Cox’s proportional hazards model.
Immunostaining
The Relish antibodies (Raybiotech, Cat# 130-10080) were first tested in S2 cells. S2 cells were treated with Tris buffer (pH 7.4) or infected with DCV (1 × 107 TCID50/ml) for 24 hours. After treatment, cells were fixed with 4% PFA for 15 minutes at room temperature. S2 cells with DCV infection showed strong Relish signals in nuclei (data not shown). For pupal tissue staining: to minimize Relish activation caused by bacteria, we made germ-free flies for immunostaining. Gut and fat body from white pupae were dissected in cold PBS and fixed with 4% PFA for 15 minutes at room temperature. After fixation, samples were washed 3 times with PBST (5 minutes each time). After washing, samples were blocked in blocking buffer (10% goat serum (G9023-10ML) in PBST) for 1 hour. Relish antibody (1:300 dilution) was diluted in blocking buffer and incubated with samples at 4 °C overnight. Next day, samples were washed with PBST for 3 times and then incubated with secondary antibody (Molecular Probes, Cat# A11011, 1:300) in blocking buffer for 1.5 hours at room temperature. Samples were then washed 3 times with PBST, stained with DAPI and mounted with VECTASHIELD MOUNTING MEDIUM.
For Relish signal quantification in pupal hindgut and fat body, single Z-stack images of individual sample were taken by the Leica TCS SP5 confocal microscope. The fluorescence signal from individual nucleus was determined by ImageJ. For each cell in fat body, the Relish signal in the nucleus was calculated by subtracting the brightness of the signal in cytoplasm from the brightness of the signal in nucleus determined by ImageJ. For each cell in hindgut, the Relish signal in nucleus was calculated by subtracting the brightness of the background (blank region within the same image) from the brightness of the signal in nucleus determined by ImageJ. Mean ImageJ value from the control animals was set as one and used for the calculation of relative signal sh- mdg4 and sh-Relish flies. P values were calculated by the Student’s t-test. The data met the assumptions of the statistical tests used. No animals or data points were excluded from these analyses.
RNA-Sequencing
Non-polyA selection strand-specific mRNA libraries were prepared by using TruSeq Stranded Total RNA Library Prep Gold (Illumina, Cat# 20020598). Briefly, 1 μg of total RNA from the 5 animals was diluted in nuclease-free ultrapure distilled water (Thermo Fisher Scientific, Cat #10977015) to a final volume of 10 μl, then rRNAs were removed by following the standard procedure from the kit. mRNAs were treated with 4 μl of Turbo DNase in 37 °C for 30 minutes and purified by RNA Clean & Concentrator-5. First strand cDNA was synthesized by using SuperScript™ III Reverse Transcriptase and cDNA was purified by using Ampure XP beads (BECKMAN COULTER, Cat# A63987). dUTP was incorporated after RNA template was removed and second strand cDNA was synthesized (RNase H, Thermo Fisher Scientific, Cat# 18021014; DNA Polymerase I, NEB, Cat# M0209L). dUTP incorporation was done in 16 °C for 2.5 hours and DNA was purified by using Ampure XP beads. Blunting of fragmented DNA and phosphorylation of DNA ends were achieved at 20 °C for 30 minutes. The enzymes used for end repair were T4 DNA polymerase (NEB, Cat# M0203L), Klenow DNA polymerase (NEB, Cat# M0210L) and T4 PNK (NEB, Cat# M0201L). After end repair, a single ‘A’ nucleotide was added to the 3’ ends of the blunt fragments by using Klenow Fragment (3’→5’ exo-) (NEB, Cat# M0212L). Adapter was then ligated to DNA fragments by using T4 DNA ligase (Enzymatics, Cat# L603-HC-L). After UDG (NEB, Cat# M0280S) treatment, DNA fragments were amplified by using Phusion HF DNA polymerase (NEB, Cat# M0530L). Libraries were sent to Novogene for quality control and sequencing.
Genomic DNA sequencing
Genomic DNA was isolated from adult flies using the Zymo Quick-DNA Microprep Kit (Zymo, Cat#D3021). Around 2 μg of DNA per sample was used as input for the Nanopore Ligation Sequencing Kit protocol (Oxford Nanopore Technologies, Cat# SQK-LSK109). Libraries were barcoded with the Nanopore Native Barcoding Expansion Kit (Oxford Nanopore Technologies, Cat# EXP-NBD104). Libraries were sequenced with R9.4 flow cells using the GridION system.
Full-length mdg4 copy number identification
The fast5 files generated by the Nanopore GridION machine were used as input in MinKNOW version 21.05.25 (MinKNOW core 4.3.12). Guppy 5.0.16 is integrated into the MinKNOW. Raw nanopore reads in fastq format were pre-processed by the software porechop with parameters: --extra_end_trim 0 --discard_middle. Clean reads were mapped to the mdg4 consensus sequence first. The mapped reads containing a full-length mdg4 were selected. To determine the flanking sequences, selected reads were mapped to the reference genome of Drosophila melanogaster version dm6 (GCA_000001215.4). Mapping was done by using minimap2 with the parameters: -ax map-ont -Y -t 16. Full-length mdg4 reads with continuous flanking sequences were classified as Type 1. Full-length mdg4 reads with flanking sequences that were mapped to two different chromosomes were classified as Type 2. Full-length mdg4 reads with flanking sequences that were not present in the major chromosomes of dm6 were classified as Type 3. For each type, individual reads were clustered based on the coordinates of the mdg4 and genome junctions. Drosophila chromosome ideogram is made by RIdeogram package. The heat map on the chromosomes represents gene density, which was calculated by 1 Mb windows.
Full-length mdg4 transcripts and Env transcripts identification and quantification
Raw Illumina reads were processed by the software trim_galore, with the parameter –paired –fastqc. The output files of trim_galore were mapped to both the reference genome of Drosophila melanogaster version dm6 (GCA_000001215.4) and the mdg4 consensus sequence using hisat2 with the default parameters except -p 16. To quantify the full-length mdg4 transcripts and the spliced Env transcripts, Illumina reads were assembled by stringtie with the default parameters except for -f 0. The full-length mdg4 mRNA was presented in the stringtie output as a transcript (1..7469) with a single exon (1..7469), while the Env transcript was presented in the stringtie output as an isoform with two exons (1..568; 5551..7469). The normalized transcript abundance values in FPKM were used for plotting Extended Data Fig. 4. Alignment visualization was achieved by using IGV version 2.11.1.
Extended Data
Extended Data Fig. 1 |. Monitoring retrotransposon mobilization in somatic cells via a transposition reporter.
a, Detailed schematic design of eGFP transposition reporter to monitor mdg4 mobilization. b, Summary of mobilization events from different somatic tissues for 9 retrotransposon families, as assayed by corresponding eGFP reporter. No: no eGFP positive cells are detected; Yes: eGFP positive cells can be detected. c, Detecting eGFP signals in somatic tissues from positive control, negative control, and mdg4 transposition reporter in 2–4-day-old adult flies. Note: Positive control construct gives low number of eGFP positive cells in brain and malpighian tubules, indicating that transcription of mdg4 is suppressed in these tissues. Three independent biological replicates were performed. d, Zoom-in display of the box region in Fig. 1b. In DAPI channels, green arrows point to the nuclei that have GFP expression.
Extended Data Fig. 2 |. mdg4 selectively mobilizes in the regenerating tissues during metamorphosis.
a, Schematic of Drosophila hindgut. Both larval and adult hindgut include the pylorus, ileum and rectum. During pupal stage metamorphosis, the pylorus and ileum from larval stage degenerate; the anterior part of pylorus (ring) regenerates to produce adult pylorus and ileum. b, Detecting eGFP positive cells from mdg4 transposition reporter in midgut, salivary gland and proventriculus at different stages. c, The box plot shows the number of eGFP positive cells from mdg4 transposition reporter in midgut, salivary gland and proventriculus.
Extended Data Fig. 3 |. Probing transposition events by PCR.
a, Transposition events generate intron-removed DNA, which produces a short PCR product. b, Probing mobilization events at different developmental stages. Only the DNA from 48 hours pupal hindguts can harbor enough mobilization events to be detected by this PCR assay. c, Probing mobilization events from different adult tissues. These tissues either have no––or too few––mobilization events to be detected by this method. Three independent biological replicates were performed for b and c.
Extended Data Fig. 4 |. RNA-Seq to measure transcripts from mdg4.
a, Bar graph to display the abundance of full-length and Env mRNAs from mdg4. Full-length mdg4 transcripts are constantly expressed at all stages. Env mRNAs can be detected from early stage embryos and pupal stage, but not adult stage. b, IGV browser screenshot to display the representative sequencing reads that support the expression of Env mRNAs.
Extended Data Fig. 5 |. Multiple RNAi constructs were designed to silence mdg4.
a, RT-qPCR to quantify the expression of mdg4 upon suppression by using one of the 7 RNAi constructs based on the two-tailed t-test. Each sh-RNA construct was driven by ac-Gal4. Flies were raised at 25°C and newly eclosed flies were used to extract RNA. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for three biological replicates (apply to all RT-qPCR data from this manuscript). b, Visualizing mdg4 transposition events (GFP positive cells) in hindgut from either white or mdg4 suppressed 2–4-day-old adults. Flies carrying sh-mdg4-5 construct were used for Fig. 6. sh-mdg4-1 was used in the rest of the experiments. The findings from it were validated by using other constructs (shown in Extended Data Fig. 11). c, RT-qPCR to measure the amount of DCV from fly bodies after feeding animals virus for 8 hours based on the two-tailed t-test. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for three biological replicates. d, By activating mdg4 RNAi from embryonic to adult stage, RT-PCR experiments were performed to monitor the amount of DCV in fly bodies after one-time infection. Together with Extended Data Fig. 5a, these data indicate RNAi efficiency negatively correlates with the robustness of antiviral response. e, By using sh-mdg4-2 to silence mdg4 at the stage specific manner, similar findings were made as Fig. 5. Three independent biological replicates were performed for sh-mdg4-2 in d and e, one time experiment was performed for sh-mdg4-3, 4, 5, 6 and 7 in d.
Extended Data Fig. 6 |. Schematic design of the virus-feeding assays used in this study.
a, Top panel: Schematic design to achieve RNAi from embryonic to adult stage. Middle panel: Schematic design to achieve RNAi only at pupal stage. At lower temperature, Gal80 inhibits Gal4 activity. At 29°C, Gal80 becomes inactive and cannot suppress Gal4. Bottom panel: Schematic design to achieve RNAi only at adult stage. b, Validating ac-Gal4+tub-Gal80ts system by driving UAS-GFP expression at high temperature (29°C) and low temperature (18°C) in larval midgut and proventriculus. This experiment was only performed once. c, RT-qPCR to measure the mdg4 silencing efficiency for the ac-Gal4+tub-Gal80ts system at 29°C based on the two-tailed t-test. Newly eclosed flies raised at 29°C during pupal stage were used to extract RNA. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for two biological replicates. d, RT-qPCR to measure the mdg4 silencing efficiency for the ac-Gal4+tub-Gal80ts system at 25°C based on the two-tailed t-test. Adult flies being shifted to 25°C for 5 days were used to extract RNA. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for three biological replicates.
Extended Data Fig. 7 |. mdg4 activation renders hosts protection from virus infection.
a, By activating mdg4 RNAi from embryonic to adult stage, the survival rates were measured by raising flies on CrPV- or IIV-6-containing food for 20 days. sh-white flies served as controls. b, By activating mdg4 RNAi from embryonic to adult stage, the survival rates were measured after infecting adult files with different viruses with a single meal. c, RT-qPCR to quantify the fold change of DCV mRNA in sh-mdg4 flies at different time points after one-time infection, relative to sh-white controls based on the two-tailed t-test. d, RT-qPCR to quantify the fold change of DCV mRNA in sh-mdg4 flies on day 6 after one-time infection, relative to sh-white controls based on the two-tailed t-test. The bars in panel c and d report mean ± standard deviation for three biological replicates. Comparison of survival curves was completed using a Cox proportional-hazards model for panels a and b.
Extended Data Fig. 8 |. Relish activation renders hosts protection from virus infection.
a, By activating Relish RNAi from embryonic to adult stage, the survival rates were measured by raising flies on CrPV- or IIV-6-containing food for 20 days. sh-white flies served as controls. b, RT-qPCR to quantify the fold changes of DCV mRNA in sh-relish flies on day 6 after one-time infection, relative to sh-white controls based on the two-tailed t-test. c, RT-qPCR to quantify the fold changes of DCV mRNA in sh-relish flies on day 6 after one-time infection, relative to sh-white controls based on the two-tailed t-test. The bars in panel B and C report standard deviation for three biological replicates. d, By activating Relish RNAi only at adult stage, the survival rates were measured by raising flies on DCV- or FHV-containing food for 20 days. sh-white flies served as controls. e, RT-PCR experiments to monitor the amount of DCV in adult dcr-2 mutant (dcr-2L811fsX) flies after one-time infection. Two independent biological replicates were performed. f, RT-qPCR to quantify relish expression upon mdg4 depletion in fly pupae. Data are normalized to rp49 (RpL32) expression; the bars report mean ± standard deviation for 2 biological replicates. Expression was compared using a two-tailed t-test. Comparison of survival curves was completed using a Cox proportional-hazards model for panels a and d.
Extended Data Fig. 9 |. mdg4 products promote the translocation of Relish-N into nucleus.
a, By performing immuno-staining with the Relish-N antibody, which can detect both full-length and N-terminal fragment of Relish, very low–if any–signals were detected in midgut and anterior part of hindgut from sh-white, sh-mdg4, or sh-relish early pupae. b, RT-PCR to examine the levels of mdg4 full-length and Env transcripts in fat body cells. Two independent biological replicates were performed for a and b. c, Immuno-staining to detect the nuclear Relish-N signals in the fat body cells from sh-white and sh-mdg4 early pupae. While the animals for Fig. 7 were raised in germ-free condition, the pupae examined for this figure were non-germ-free. In both conditions, silencing mdg4 resulted in a significant decrease of the nuclear Relish-N in fat body cells. Box plots report the minimum, maximum, median, and interquartile ranges of the data. A two-tailed t-test was used to compare the relative intensities of each genotype. The data were collected from 3 individual animals per genotype.
Extended Data Fig. 10 |. A model to depict the activity of transposon during animal development and how its activation prepares the host for antiviral responses.
Our data indicate that the mdg4 retrotransposon selectively becomes active during metamorphosis to prepare the antiviral responses.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Phillip Zamore for providing DCV, Maria-Carla Saleh for providing FHV and CrPV. We thank members from ZZ laboratory, Don Fox for critical suggestions, Brandy Kegeris for assistance on cloning, and Ken Poss for reading the manuscript. This work was supported by the grants to Z.Z. from the Pew Biomedical Scholars Program and the National Institutes of Health (DP5 OD021355 and R01 GM141018); and grant to N.S. from the National Institutes of Health (AI060025).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
CODE AVAILABILITY
Code from this manuscript is available at https://github.com/ZhaoZhangZZlab/2021_fly_mdg4
DATA AVAILABILITY
The sequencing data were deposited to the National Center for Biotechnology Information (NCBI) under accession number PRJNA7843705. The published datasets that were used in this manuscript under accession numbers: SRR1197325, SRR1197324, SRR1197326, SRR8627922, SRR8627923, and SRR8643355.
REFERENCES
- 1.Wells JN & Feschotte C A Field Guide to Eukaryotic Transposable Elements. Annu Rev Genet (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazazian HH Jr. & Moran JV Mobile DNA in Health and Disease. N Engl J Med 377, 361–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feschotte C & Gilbert C Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13, 283–96 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Garfinkel DJ, Boeke JD & Fink GR Ty element transposition: reverse transcriptase and virus-like particles. Cell 42, 507–17 (1985). [DOI] [PubMed] [Google Scholar]
- 5.Boeke JD, Garfinkel DJ, Styles CA & Fink GR Ty elements transpose through an RNA intermediate. Cell 40, 491–500 (1985). [DOI] [PubMed] [Google Scholar]
- 6.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF & Kazazian HH Jr. Isolation of an active human transposable element. Science 254, 1805–8 (1991). [DOI] [PubMed] [Google Scholar]
- 7.Feng Q, Moran JV, Kazazian HH Jr. & Boeke JD Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87, 905–16 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Kazazian HH Jr. et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature 332, 164–6 (1988). [DOI] [PubMed] [Google Scholar]
- 9.Miki Y et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 52, 643–5 (1992). [PubMed] [Google Scholar]
- 10.Burns KH Transposable elements in cancer. Nat Rev Cancer 17, 415–424 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Tam OH, Ostrow LW & Gale Hammell M Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob DNA 10, 32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodic N et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol 184, 1280–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosby RL, Chang NC & Feschotte C Host-transposon interactions: conflict, cooperation, and cooption. Genes Dev 33, 1098–1116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon M et al. LINE1 Derepression in Aged Wild-Type and SIRT6-Deficient Mice Drives Inflammation. Cell Metab 29, 871–885 e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Cecco M et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566, 73–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siudeja K et al. Unraveling the features of somatic transposition in the Drosophila intestine. EMBO J 40, e106388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medzhitov R & Janeway CA Jr. Innate immune recognition and control of adaptive immune responses. Semin Immunol 10, 351–3 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Akira S, Uematsu S & Takeuchi O Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Buchon N, Silverman N & Cherry S Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology. Nat Rev Immunol 14, 796–810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann JA & Reichhart JM Drosophila innate immunity: an evolutionary perspective. Nat Immunol 3, 121–6 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Mantovani A & Netea MG Trained Innate Immunity, Epigenetics, and Covid-19. N Engl J Med 383, 1078–1080 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Duneau D et al. Stochastic variation in the initial phase of bacterial infection predicts the probability of survival in D. melanogaster. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintin J, Cheng SC, van der Meer JW & Netea MG Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol 29, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Moorlag S et al. beta-Glucan Induces Protective Trained Immunity against Mycobacterium tuberculosis Infection: A Key Role for IL-1. Cell Rep 31, 107634 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintin J et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues J, Brayner FA, Alves LC, Dixit R & Barillas-Mury C Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 329, 1353–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faulkner GJ & Billon V L1 retrotransposition in the soma: a field jumping ahead. Mob DNA 9, 22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaminker JS et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3, RESEARCH0084 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Dou K, Moon S, Tan FJ & Zhang ZZ Hijacking Oogenesis Enables Massive Propagation of LINE and Retroviral Transposons. Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brookman JJ, Toosy AT, Shashidhara LS & White RA The 412 retrotransposon and the development of gonadal mesoderm in Drosophila. Development 116, 1185–92 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Potter SS, Brorein WJ Jr., Dunsmuir P & Rubin GM Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell 17, 415–27 (1979). [DOI] [PubMed] [Google Scholar]
- 32.Mevel-Ninio M, Mariol MC & Gans M Mobilization of the gypsy and copia retrotransposons in Drosophila melanogaster induces reversion of the ovo dominant female-sterile mutations: molecular analysis of revertant alleles. EMBO J 8, 1549–58 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunsmuir P, Brorein WJ Jr., Simon MA & Rubin GM Insertion of the Drosophila transposable element copia generates a 5 base pair duplication. Cell 21, 575–9 (1980). [DOI] [PubMed] [Google Scholar]
- 34.Zhao D & Bownes M The RNA product of the Doc retrotransposon is localized on the Drosophila oocyte cytoskeleton. Mol Gen Genet 257, 497–504 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Driver A, Lacey SF, Cullingford TE, Mitchelson A & O’Hare K Structural analysis of Doc transposable elements associated with mutations at the white and suppressor of forked loci of Drosophila melanogaster. Mol Gen Genet 220, 49–52 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Song SU, Kurkulos M, Boeke JD & Corces VG Infection of the germ line by retroviral particles produced in the follicle cells: a possible mechanism for the mobilization of the gypsy retroelement of Drosophila. Development 124, 2789–98 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Pelisson A et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13, 4401–11 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moran JV et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–27 (1996). [DOI] [PubMed] [Google Scholar]
- 39.Bainbridge SP & Bownes M Staging the metamorphosis of Drosophila melanogaster. J Embryol Exp Morphol 66, 57–80 (1981). [PubMed] [Google Scholar]
- 40.Fox DT & Spradling AC The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell 5, 290–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Perez JL et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466, 769–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song SU, Gerasimova T, Kurkulos M, Boeke JD & Corces VG An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes & Development 8, 2046–2057 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Ewing AD et al. Nanopore Sequencing Enables Comprehensive Transposable Element Epigenomic Profiling. Mol Cell 80, 915–928 e5 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Ellison CE & Cao W Nanopore sequencing and Hi-C scaffolding provide insight into the evolutionary dynamics of transposable elements and piRNA production in wild strains of Drosophila melanogaster. Nucleic Acids Res 48, 290–303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Painter TS & Reindorp EC Endomitosis in the nurse cells of the ovary of drosophila melanogaster. Chromosoma 1, 276–283 (1939). [Google Scholar]
- 46.Morgan TH Sex Limited Inheritance in Drosophila. Science 32, 120–2 (1910). [DOI] [PubMed] [Google Scholar]
- 47.Mondotte JA et al. Immune priming and clearance of orally acquired RNA viruses in Drosophila. Nat Microbiol 3, 1394–1403 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Cai H et al. 2’3’-cGAMP triggers a STING- and NF-kappaB-dependent broad antiviral response in Drosophila. Sci Signal 13(2020). [DOI] [PubMed] [Google Scholar]
- 49.Martin M, Hiroyasu A, Guzman RM, Roberts SA & Goodman AG Analysis of Drosophila STING Reveals an Evolutionarily Conserved Antimicrobial Function. Cell Rep 23, 3537–3550 e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goto A et al. The Kinase IKKbeta Regulates a STING- and NF-kappaB-Dependent Antiviral Response Pathway in Drosophila. Immunity 49, 225–234 e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y et al. Inflammation-Induced, STING-Dependent Autophagy Restricts Zika Virus Infection in the Drosophila Brain. Cell Host Microbe 24, 57–68 e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rij RP et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev 20, 2985–95 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoven S, Ando I, Kadalayil L, Engstrom Y & Hultmark D Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep 1, 347–52 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamoto M et al. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity 36, 658–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shelly S, Lukinova N, Bambina S, Berman A & Cherry S Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30, 588–98 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao D et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weavers H, Evans IR, Martin P & Wood W Corpse Engulfment Generates a Molecular Memory that Primes the Macrophage Inflammatory Response. Cell 165, 1658–1671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roulois D et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 162, 961–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiappinelli KB et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 162, 974–86 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grow EJ et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522, 221–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peirson SN, Butler JN & Foster RG Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res 31, e73 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data were deposited to the National Center for Biotechnology Information (NCBI) under accession number PRJNA7843705. The published datasets that were used in this manuscript under accession numbers: SRR1197325, SRR1197324, SRR1197326, SRR8627922, SRR8627923, and SRR8643355.