Abstract
The design and implementation of systematic reviews and meta-analyses are often hampered by high financial costs, significant time commitment, and biases due to researchers' familiarity with studies. We proposed and implemented a fast and standardized method for search term selection using Natural Language Processing (NLP) and co-occurrence networks to identify relevant search terms to reduce biases in conducting systematic reviews and meta-analyses.
-
•
The method was implemented using Python packaged dubbed Ananse, which is benchmarked on the search terms strategy for naïve search proposed by Grames et al. (2019) written in “R”. Ananse was applied to a case example towards finding search terms to implement a systematic literature review on cumulative effect studies on forest ecosystems.
-
•
The software automatically corrected and classified 100% of the duplicate articles identified by manual deduplication. Ananse was applied to the cumulative effects assessment case study, but it can serve as a general-purpose, open-source software system that can support extensive systematic reviews within a relatively short period with reduced biases.
-
•
Besides generating keywords, Ananse can act as middleware or a data converter for integrating multiple datasets into a database.
Keywords: Search Strategy, Search Terms, Data Deduplication, Software Implementation, Evidence Synthesis, Systematic Literature Review
Graphical abstract
Specifications table
| Subject Area | Environmental Science |
| More specific subject area | Evidence synthesis in environmental and biological sciences |
| Method name | Text mining and keyword co‐occurrence networks to identify the most important terms for a review |
| Name and reference of original method | Grames, E. M., Stillman, A. N., Tingley, M. W., & Elphick, C. S. (2019). An automated approach to identifying search terms for systematic reviews using keyword co‐occurrence networks. Methods in Ecology and Evolution, 10(10), 1645-1654. |
| Resource availability |
Documentation:https://baasare.github.io/ananse/_build/html/index.html Software:GitHub - baasare/ananse Method description: ananse · PyPI |
Background
Historically, summaries of scientific evidence have helped discover patterns of phenomena, develop theories or concepts, and inform practice. Although common with editors and readers alike, this approach is less rigorous since evidence summarized this way is less likely to answer specific clinical questions and more likely to contain literature selected by the authors and recommendations prejudiced strongly by opinion. With exponential growth in scientific literature, the search for a structured and effective evidence synthesis has become a critical scientific endeavor. Evidence synthesis involves combining information from multiple studies or research that have investigated the same or similar issue to come to a conclusive understanding of a specific topic [1]. It often involves summarizing trends, identifying emerging questions, and clarifying disagreements and conflicting results [2,3].
Since 1753 when James Lind published the first evidence synthesis to provide a concise and unbiased summary of evidence on scurvy, improvement in the state of evidence synthesis has grown [4,5]. In the past two decades, advances in computer-aided technology have enabled the growth and development of various forms of evidence synthesis. The two central techniques known to have originated from the medical sciences and are commonly used today to synthesize evidence are systematic reviews (SRs) – which search available literature for evidence that addresses the research question, - and meta-analyses – which quantitatively assess statistical evidence found through systematic reviews [5]. Evolutionary and behavioral ecologists started adopting meta-analyses in the mid-1990s and became fully embraced since 2010 [6]. Meta-analysis has since become the gold standard for combining information from multiple studies across disciplines. However, a good meta-analysis is dependent on a good sampling of the core universe of studies, thus requiring a careful and comprehensive SR. A SR involves the review of an articulated research question using systematic and testable methods to help to identify, select and evaluate all pertinent research [7], and collect and analyze data from the studies that are included in the review [8]. An excellent SR assembles and presents an impartial and objective summary of findings, assesses all results for inclusion/exclusion and quality, and minimizes bias at all stages of the process [7].
However, the process of evidence synthesis is very tedious and often involves experienced methodologists and disciplinary experts combing through all relevant studies, both published and unpublished, through a guided methodological process. As such, it tends to be costly and tedious as it can take months, or even years, to complete, making it practically challenging [9]. According to some estimates, conducting a SR can take up to 2 years to complete. [10] also suggest that the time needed to complete a SR with meta-analyses ranges from 216 to 2,518 hours. According to [11], conducting an effective systematic search requires an information specialist's expertise and time, who need an average aggregated time of 26.9 hours when developing a search strategy. Thus, the design and implementation of evidence-based synthesis are hampered by high financial costs [3] and significant time commitment [2].
To overcome time and resource constraints required to synthesize evidence, scholars have adopted automation of the laborious tasks in SR [12]. Advances in computer-aided technology have helped automate aspects of the evidence synthesis process to improve efficiency and cut costs and time while still maintaining the standards of conventional search methods [13]. Automation occurs in different forms; from the most basic of tasks to complicated ones [13], such as removing duplicate articles, prioritizing articles for screening, and extracting data from tables and figures [14,15]. Research on different approaches for automating systematic reviews via technologies such as machine learning, text mining, and natural language processing exists [12]. Text mining is the process of discovering knowledge and structure from unstructured data [16], while Natural Language Processing (NLP) supports human analysts to carry out various linguistic analytical tasks on textual documents [17], such as identifying potential keywords in systematic literature reviews [18], [19], [20]. Using NLP to extract information from text automatically leads to decreased labor of manual extraction from a large volume of text material and saves time [21].
However, automation in SR has focused chiefly on extracting data or results after a literature search, while methods or strategies to find or assemble all relevant evidence, including developing a search strategy, have received little attention [22]. According to [22], search strategies for SR should be able to return all the studies relevant to the review (‘recall’) without retrieving irrelevant studies (‘precision’). Unfortunately, not all fields of study have a structured or standardized ontology for search strategy development. The field of public health has institutionalized support and standardized ontology (i.e., Medical Subject Headers, or MeSH) for search strategy development [23]. However, ecology or environmental sciences, generally, does not have standardized ontologies. Thus, researchers tend to use broad, non‐specific keywords in their search (Pullin & Stewart, 2006), leading to low precision of search results (0.473%; [2]). With low precision, more time and cost are spent on screening articles. Thus, enhanced standardization in search strategy development is critical to improving the specificity, objectivity, and reproducibility of SRs [24]. Two primary approaches for automating search strategy development are citation networks and text mining [22]; both use a set of predetermined articles that researchers deem relevant to the review. Thus, both approaches require researchers to select a starting set of articles with which they are already familiar. This predisposes citation networks and text mining towards familiar articles. Although this approach has high precision, it has a low recall, and the risk of selection, citation, and publication bias is increased as the initial set of articles influences what is eventually retrieved [25,22,26].
In this research, we mediate the problems associated with search strategy development in systematic literature reviews by developing a method that uses NLP and keyword co-occurrence networks to identify potential keywords to support SR. We adapted the search terms strategy for naïve search proposed by [22] written in R. To facilitate reproducibility and transparency; we created the python package dubbed ‘Ananse’ (a Ghanaian vernacular translated as a spider) to aid the implementation of the method in a user-friendly format. The software and documentation are publicly available via Github [27] and PyPI [28], [29], [30], respectively. We tested our approach by applying it to selecting keywords for a systematic literature review of cumulative effect assessment of disturbance on forest ecosystems (see [30]).
The remainder of the study is structured as follows. Materials and methods are presented in section 2, where the process flow of Ananse in finding search terms are described. Using Ananse to perform a search tailored to a SR of cumulative effect studies is described in section 3. In section 4, we discuss the outcomes of using Ananse to perform cumulative effect search terms [30] and compare our results with other related works. Finally, in section 5, we draw conclusions based on our findings and forecast future work.
Methods details
We developed a Python package to partially automate search term selection and write search strategies for SRs. We refer to this Python package as Ananse (a Ghanaian vernacular translated as a spider). We adapted the search strategy for black-backed woodpecker occupancy of post-fire forest systems ( [22] and [31]) written in R. Our search term selection strategy focuses on cumulative effect and seeks to create an open-source search software in Python.
Software design
Software design describes the structure of the software to be implemented, the data models used by the system, the interfaces, and, sometimes, the algorithms used [32]. Requirements usually precede the design. We present the following design considerations during the creation of Ananse: functional requirements, use case diagram, and data flow diagram. We do not intend to offer a technical software engineering perspective but to guide the user to appreciate the design concepts which gave birth to Ananse.
Functional requirements
The functional requirements for a software system describe what the system should do [33,34]. We considered the SR process from the NLP perspective and specified the requirements for Ananse. Ananse is able to:
-
1.
Import results of a naïve search from a literature database such as JSTOR, Web of Science, and Scopus just to mention a few.
-
2.
Deduplicate combined search results.
-
3.
Extract terms using Rapid Automatic Keyword Extraction (RAKE) algorithm
-
4.
Create document term matrix.
-
5.
Convert document term matrix into data frames.
-
6.
Create document network from data frames.
-
7.
Generate node strength and final cut-off.
-
8.
Generate keywords.
These eight requirements were used to formulate a use case diagram.
Use case diagram
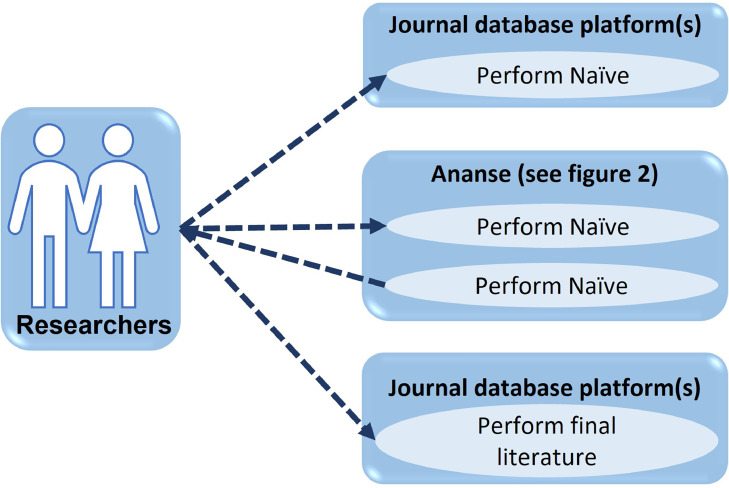
Use cases are documented using a high-level use case diagram. The set of use cases represents all of the possible interactions described in the system requirements. Actors in the process, who may be human or other systems, are represented as stick figures. Each class of interaction is represented as a named ellipse. Lines link the actors with the interaction; arrowheads show how the interaction is initiated.
Figure 1 is the use case diagram for Ananse. A researcher performs naïve a search from a journal database platform such as Web of Science, Scopus, or JSTOR.
Fig. 1.
Use case diagram for Ananse.
Flow diagram
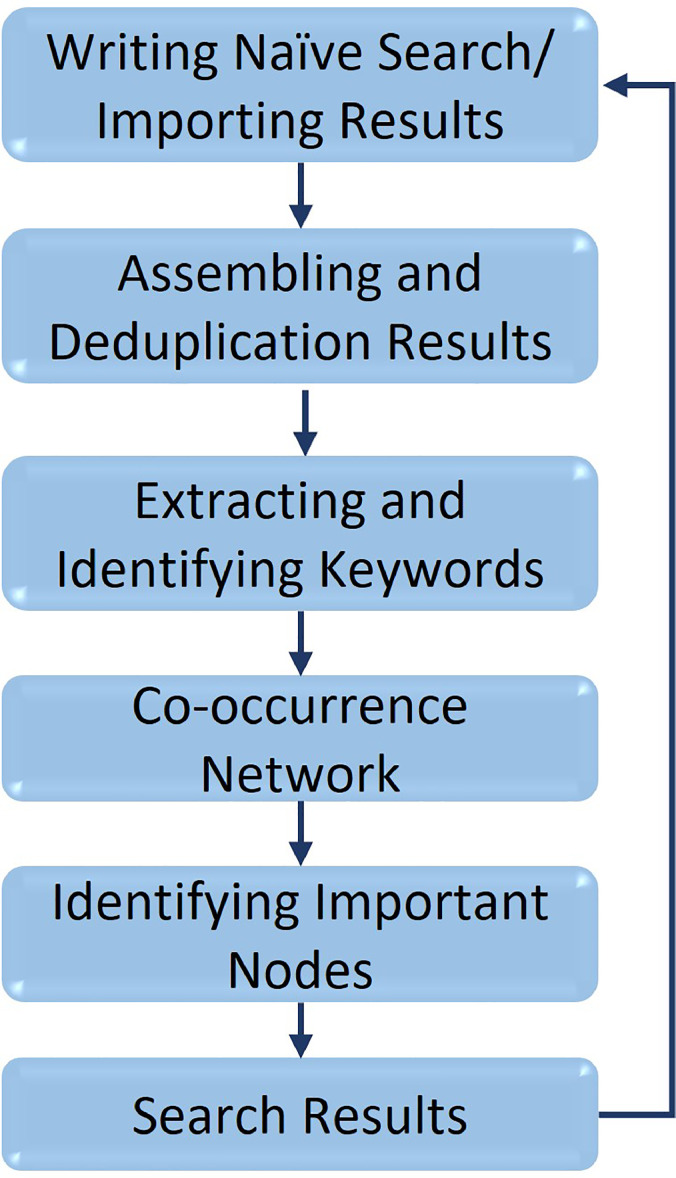
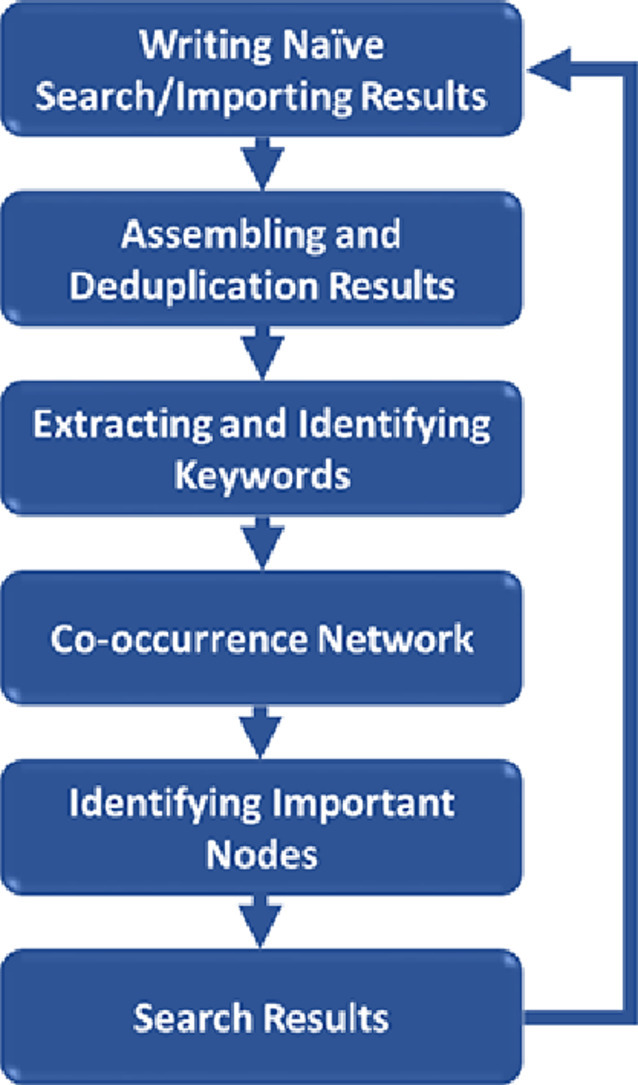
Figure 2 shows the process flow used in implementing Ananse.
Fig. 2.
Ananse process flow
Naïve search is written and imported. Results are assembled and deduplicated, followed by keyword extraction, creating a co-occurrence network, and identifying important nodes. After getting results, the process can be initiated for other searches.
Software implementation and results
Writing the naïve search and exporting the results
When writing a naive search, the first step is to clearly articulate the research question (Grames et al., 2020). The naїve search must be precise; otherwise, it will return several unrelated articles, weakening the subsequent keyword selection [22]. The authors, who are experts in the domain of cumulative effect assessment, developed the initial search terms (76 search terms) under different concept categories to guide the identification of studies for the naïve search. We grouped the search terms into three concept categories and combined them into a Boolean search (see Table 1). Using the initial search terms of 76, we conducted a naïve literature search in three sample databases: JSTOR, Scopus, and Web of Science. These three databases were chosen to broaden the available pool of search terms on the topic as their coverage differs substantially [45].
Table 1.
Search terms grouped under different concept categories.
| Concept category 1:(Cumulative effect* OR Cumulative impart* OR Cumulative disturbance* OR Cumulative environmental effect*) OR (Environmental effect* OR Environmental impact*) OR (Strategic environmental assessment* OR Impact assessment*) OR (Social effect* OR Economic effects* OR Strategic effects* OR Economic Effects*) OR (Human health* OR Human Health Effects*) OR Regulatory drive OR Risk assessment* OR Systematic approach* |
| Concept category 2:forest* OR forest ecosystem* OR forest management* OR forest disturbance OR forest dynamics* OR forest growth* OR forest community* OR forest bird* OR forest land* OR forest policy* OR forest sustainability* OR forest cover OR forest carbon* OR forest soil* OR forest soil nutrients* OR forest biodiversity* OR forest conservation* OR forest structure* OR understory vegetation* OR Indigenous people livelihoods* OR Electricity generation OR forest stream* OR silviculture* OR ecosystem* OR population* OR community* OR land use/cover conversion* OR water quality* OR water quantity* OR species composition* OR endangered species* |
| Concept category 3:Mining OR Minerals and metal OR Oil and gas OR Oil sands development OR Peat mining OR Storm (wind) OR Pulp and paper industry OR Barriers OR Wildfire OR Planting OR Forest disease OR Forest health OR Forest pest OR Deforestation OR Linear features OR Electricity generation OR Roads OR Power lines OR Seismic lines OR Urbanization OR Land reclamation /restoration OR Global change OR Climate change OR Defoliation OR Insect outbreak OR water and wetlands OR Logging OR Wells OR Flood OR Drought OR Hydro development |
Importing naïve search results into Ananse
Ananse is a package and is provoked through a file. The naïve search results from Jstor, Scopus, and Web of Science databases were exported as an ris file, csv file, and txt file, respectively; s; Jstor with a .ris, Web of Science with a .csv file extension, and Scopus with a .txt file extension. Due to the different formats in the exportation of results from the databases, this manual process takes more time. All these three files were fed into Ananse at the same time. Using these files as input, Ananse merges all the different file formats into a single Pandas data frame. The merging resulted in a csv file containing 129,407 articles. Figure 3 shows the results of the naïve search and the file “ananse_test.py” that provokes Ananse to perform the search.
Fig. 3.
Naïve search file and results from the three databases
Assembling and deduplicating results
Many articles indexed in multiple databases may pop up more than once searching for information, resulting in an overrepresentation of terms. The naïve search results were assembled and deduplicated to prevent over-representation. Provided that the path to the directory of search results is given, the import_naive_results function in Ananse automatically finds each file's database and file type, selects analogous columns, and joins them to form a single dataset. This function imports the search results from a specified path. If the parameters clean_dataset and save_dataset are set to TRUE, the function deduplicates search results after importing and saves the full search results to a csv file. The parameter save_directory contains the path to a directory where search results will be saved. If save_dataset is set to TRUE while the parameter save_directory is set to the directory of choice, the merged file is saved to that directory path containing the naive search results files. After the results are obtained, a pandas data frame consisting of assembled search results is returned. After the merging, Ananse performs deduplication based on the article titles and abstracts and returns different articles. In this instance, Ananse removes the exact title duplicates; titles that are over 95% similar or abstract that are more than 90% similar are removed. The user can change these similarity levels. Ananse returned 6,786 distinct articles out of the 7,809 articles fed into it and created a csv file, a screenshot of it is as shown in Fig. 4 (the content of the csv file is available in the appendix). Ananse automatically corrected and classified 100% of the 1023 duplicate articles identified by manual deduplication.
Fig. 4.
Screenshot of deduplicated files in csv format
Extracting and identifying keywords
Ananse uses the Rapid Automatic Keyword Extraction (RAKE) [35], a keyword extraction method, to extract potential keywords from the titles, keywords and abstracts of articles in the deduplicated dataset. The RAKE is designed to identify keywords in scientific literature by selecting strings of words uninterrupted using a list of stopwords (6+) and phrase delimiters (punctuation) to detect the most relevant words or phrases in a piece of text [36]. The function extract_terms call the RAKE algorithm and eliminates keywords that only appear in a single article and excludes phrases with only one word from the list of potential keywords resulting in a more precise search. Ananse then combines the author- and database-tagged keywords with the search terms. The author and database tagged keywords are combined as dictionary objects created with extract_terms to define all possible keywords. All the possible keywords are then passed to a function create_dtm for function wrapping, which generates a document-feature matrix using the potential keywords as features and the combined titles, abstracts, and keywords of each article (also referred to as noted) as the documents.
Co-occurrence network
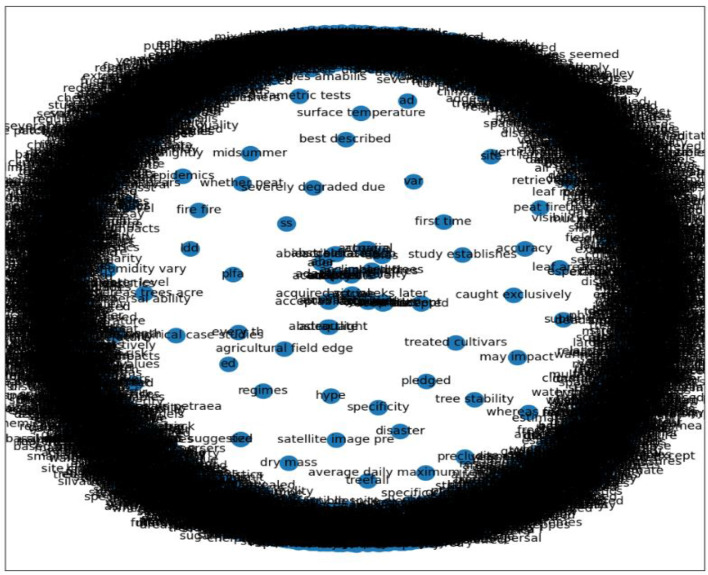
The selection of keywords using the frequency of occurrence can be a good indicator of the relevance of a word/term to a search strategy. However, we moved beyond this and generated a keyword co‐occurrence network. The co-occurrence network creates and measures each term's importance and influence in relation to the topic being reviewed [37]. Using the document matrix containing the potential keywords, we generated a keyword co-occurrence network. Each keyword is represented by a point referred to as the node, and an edge also represents a link between the keywords. Each node represents a potential search term, and the edges are co-occurrences of two terms in a study's title, abstract, or tagged keywords [37]. In Ananse, the co-occurrence network is implemented with the function create_network, which measures the importance of each term in relation to the selected topic being reviewed. The function get_centrality is used to evaluate the node importance of a graph and returns a dictionary containing nodes with their importance.
Figure 5 shows a co-occurrence network with important keywords closely grouped. The dense region shows keywords that are closely related.
Fig. 5.
Co-occurrence Network from the case study
Identifying important nodes using a full network
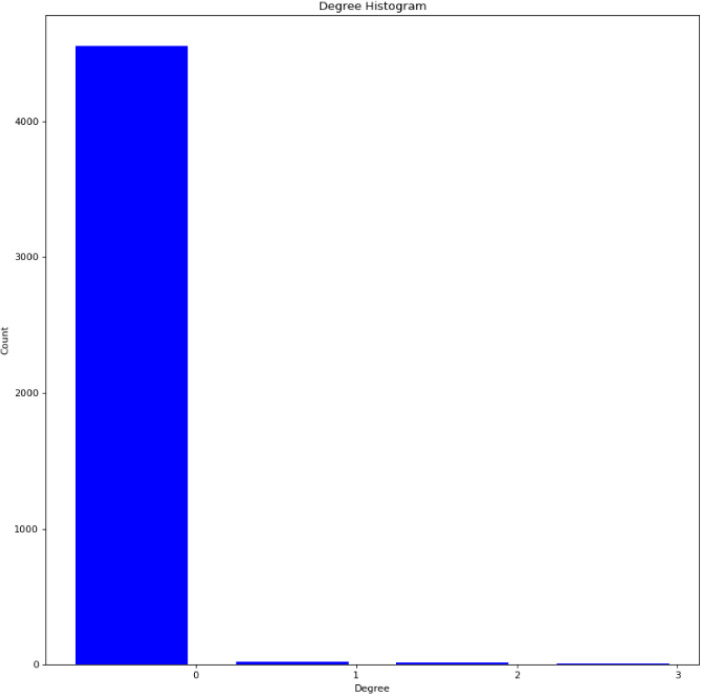
Important nodes represent keywords to be used to generate final search terms. Two methods to identify important nodes were explored in Ananse: fitting a spline model to the node importance to select tipping points and cumulative approach, which finds the minimum number of nodes to capture a large percentage of the total importance of the network. One can decide which method to use depending on the distribution and preference. In choosing a method, the first thing to do is to look at the distribution of node importance. In Ananse, the distribution was plotted with the function plot_degree_distribution, plot_rank_degree_distribution, or plot_degree_histogram as shown in Fig. 6
Fig. 6.
Degree Histogram of degree and counts
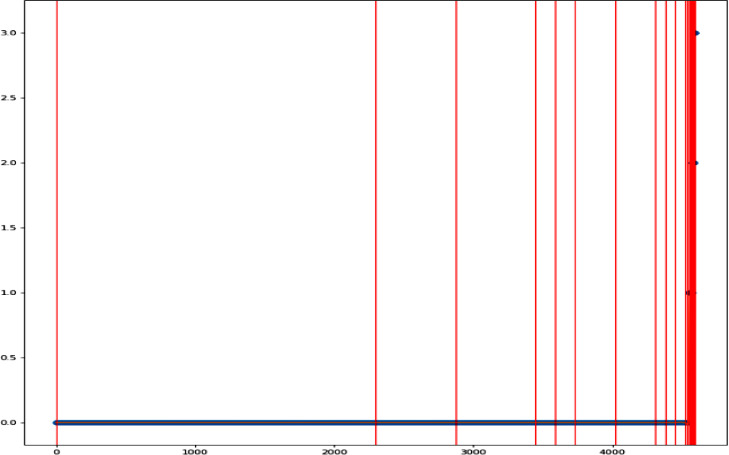
A spline model for finding cut-off is an appropriate method to identify the cut-off threshold for keyword importance if the rank distribution plot has a lot of weak nodes with a long tail. On the other hand, the cumulative approach is more appropriate when there are no clear breaks in the data. In Ananse, the find_cutoff function finds the cut-off for a graph network using either cumulative or spline method of cutting the degree distribution, as shown in Fig. 7. The reduce_graph function then generates a graph consisting of only important nodes, after which the get_keyword function extracts the keywords from the reduced network.
Fig. 7.
(Ranked Node Strength with cut-off points)
Ananse uses the node strength to generate relevant keywords from which the experts can now select their final keywords. In this research, Ananse generated 4,596 keywords. A screenshot of it is shown in Fig. 8 (the content of the csv file is available in the appendix). Afterward, the researchers manually reviewed each word or phrase using their expert knowledge to arrive at the final keywords.
Fig. 8.
A section of relevant keywords.
The final list of search terms (listed as search strings) was grouped under three concepts, as shown in Table 2. These concepts (and terminology) are cumulative effects, forests and forest ecosystems, and types and forms related to forest disturbance [30].
Table 2.
Final list of search terms.
| Concept A: Cumulative effets terminologies |
Concept B: Resource development/disturbance |
Concept C: Forest landscape dynamics |
|---|---|---|
| Cumulative effect | Mining | forest |
| Cumulative impact | Minerals and metal | forest ecosystem |
| Environmental effect | Oil and gas | forest management |
| Environmental impact | Oil sands development | forest disturbance |
| Cumulative disturbance | Peat mining | forest dynamics |
| Impact assessment | Storm (wind) | forest growth |
| Cumulative environmental effect | Pulp and paper industry | understory vegetation |
| Social effects | Barriers | forest community |
| Economic effects | Wildfire | forest bird |
| Strategic environmental assessment | Planting | forest land |
| Risk assessment | Forest disease | Indigenous people livelihoods |
| Systematic approach | Forest health | forest policy |
| Human health | Forest pest | forest sustainability |
| Human Health Effects | Deforestation | forest cover |
| Regulatory drive | Linear features | forest carbon |
| Electricity generation | landscape | |
| Roads | forest stream | |
| Power lines | silviculture | |
| Seismic lines | ecosystem | |
| Urbanization | population | |
| Land reclamation /restoration | community | |
| Global change | land cover conversion | |
| Climate change | water quality | |
| Defoliation | water quantity | |
| Insect outbreak | forest soil | |
| water and wetlands | forest soil nutrients | |
| Logging | forest biodiversity | |
| Wells | forest conservation | |
| Flood | forest structure | |
| Drought | species composition | |
| Hydro development | endangered species | |
| Habitat fragmentation | forest habitat | |
| Landscape fragmentation | wildlife | |
| Species invasion | soil compaction | |
| Urban expansion | soil porosity | |
| Habitat alteration | soil quality | |
| Loss of biological diversity | functional traits | |
| Soil acidification | Forest soil biodiversity | |
| Forest harvesting | ||
| Air Pollution | ||
| Water pollution |
Discussion
Evidence synthesis has become an essential feature of the current academic landscape, although a lack of transparency often hampers the process. This research reports on the methods used to select search terms that form the building block for performing evidence synthesis and offers a transparent approach to understand underlying assumptions. In systematic reviews, the selection of key search terms is considered the basic building block for the successful assemblage of knowledge in a particular field. However, this process is often left to researchers' discretion, leaving room for biases and a subjective selection process, affecting the outcomes of effective evidence synthesis. In this research, we designed and implemented a partially automated keyword search software package using Python for SR to enhance efficiency, maximize transparency and comprehensiveness while minimizing subjectivity and bias. Dubbed Ananse, our tool provides an efficient and standardized method for developing search strategies using NLP and co-occurrence networks to identify relevant search terms.
Our approach combines expert knowledge with a quasi‐automated method which enhances search recall. This is very important for fields such as ecology, where non‐standardized or nuanced terminology or a lack of formal ontologies exist for conducting SRs [22]. Most importantly, Ananse significantly reduces the time required to conduct a SR by decreasing time spent on search strategy development and tedious tasks like assembling and deduplication. Compared with the manual process of assembling results, Ananse reduced by more than half the time required to assembly results. Similarly, while it took two of the co-authors two days of full-time work to remove duplicates, Ananse removed the duplicates efficiently in about a minute or less and achieved 100% accuracy. With the reduction in time needed to develop a search strategy and assemble and deduplicate the results, our approach makes extensive systematic reviews and meta‐analyses more efficient and effective compared with conventional approaches. Our research contributes to the emergence and application of an ever-growing set of tools and software that can be used to facilitate transparent, reproducible reviews and develop reproducible synthesis workflows such as metaDigitise [38], litsearchr [22] in R, and revtools [39]. These efforts should help facilitate the reproducibility of ecological reviews, enhance transparency, and improve the rigor of evidence used to guide policy decisions [40].
In its current implementation, Ananse, a Python package, contains a suite of functions to improve the efficiency of keywords selection for systematic reviews. For instance, by automatically deduplicating and assembling results from separate databases, Ananse provides a systematic approach to facilitate knowledge synthesis through SR. Also, apart from generating keywords, it can act as middleware or a data converter for integrating multiple datasets into a database. Done manually, this is a time‐intensive process because platforms and databases export results in different formats [2]. Furthermore, we used the agile method of software engineering with open-source software development, thereby making Ananse easily customizable and improved upon as researchers use it beyond the application to cumulative effects assessments. Currently, Ananse has a popularity of 131 downloads per week on the Python Package Index (https://snyk.io/advisor/python/ananse). Ananse contributes to the development of open-source software systems needed to speed up systematic review. In its current state, Ananse provides a means to merge and deduplicate keywords for experts programmatically. By its design and implementation, Ananse allows researchers to modify their requirements without creating new software. Even though Ananse has been used for a cumulative effect use case [30], it is general-purpose software for a systematic review of any kind. It can be applied broadly in ecology and evolutionary biology as well as other fields.
Conclusion
Compared to conventional approaches for developing keywords for systematic review, our method is far effective and efficient by significantly reducing the time and resources needed to develop search strategies to conduct systematic reviews. Ananse substantially reduces the time spent on the systematic review by automating time-consuming tasks such as assembling and deduplicating large search results. Ananse saves time and enhances effective keyword generation compared to traditional methods by automating the tedious and bias-prone aspect of systematic review tasks. Therefore, Ananse presents an approach to performing large systematic reviews within a short period of time.
Our results can be used as a starting point to frame future studies according to well-defined terminology. Future research would enhance the front-end of Ananse through a user-friendly graphical interface. Currently, Ananse allows one user per time; this functionality can be improved by making Ananse a server-type software with capabilities to permit concurrent and multi-user interaction. The requirements would be modified as we get feedback from the research community.
Software, data, and documentation availability
The source of this software is publicly available via Github [41] and also via PyPI [42]. Documentation is accessible via [43] and [44].
Acknowledgments
Acknowledgements
The CFS Cumulative Effects Program supported this work. We want to thank Sonja Kosuta, Tracey Cook, and Danny Galarneau from Natural Resources Canada for their instrumental roles in moving the study forward.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This research was funded by Natural Resources Canada (Canadian Forest Service, Forest Ecosystem Integrity and Cumulative Effects Programs). Natural Resources Canada (Canadian Forest Service, Forest Ecosystem Integrity, and Cumulative Effects Programs) did not play any role in preparing this manuscript.
References
- 1.Cochrane . Cochrane; 2021. Evidence Synthesis - What is it and why do we need it?https://www.cochrane.org/news/evidence-synthesis-what-it-and-why-do-we-need-it (accessed May 06, 2021) [Google Scholar]
- 2.Haddaway N.R., Westgate M.J. Predicting the time needed for environmental systematic reviews and systematic maps. Conservation Biology. 2019;33:2. doi: 10.1111/cobi.13231. Blackwell Publishing Inc., pp. 434–443, Apr. 01, 2019. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland W.J., Wordley C.F. A fresh approach to evidence synthesis. Nature. 2018;558:364–366. doi: 10.1038/d41586-018-05472-8. [DOI] [PubMed] [Google Scholar]
- 4.Lind J. Cambridge University Press; 2014. A Treatise of the Scurvy, in Three Parts. [Google Scholar]
- 5.Peričić, T.P. and Tanveer, S. (2019). “Why systematic reviews matter.” https://www.elsevier.com/connect/authors-update/why-systematic-reviews-matter (accessed May 06, 2021).
- 6.Berger-Tal O., Greggor A.L., Macura B., Adams C.A., Blumenthal A., Bouskila A.…Blumstein D.T. Systematic reviews and maps as tools for applying behavioral ecology to management and policy. Behavioral Ecology. 2019;30(1):1–8. [Google Scholar]
- 7.Mengist W., Soromessa T., Legese G. Method for conducting systematic literature review and meta-analysis for environmental science research. MethodsX. 2020;7 doi: 10.1016/j.mex.2019.100777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtin University (2021) “What is a systematic review? - Systematic Reviews in the Health Sciences - LibGuides at Curtin University.” https://libguides.library.curtin.edu.au/systematic-reviews (accessed May 06, 2021).
- 9.Marshall I.J., Wallace B.C. Toward systematic review automation: a practical guide to using machine learning tools in research synthesis. Systematic reviews. 2019;8(1):1–10. doi: 10.1186/s13643-019-1074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nussbaumer-Streit B., Ellen M., Klerings I., Sfetcu R., Riva N., Mahmić-Kaknjo M.…Gartlehner G. Resource use during systematic review production varies widely: a scoping review. Journal of Clinical Epidemiology. 2021;139:287–296. doi: 10.1016/j.jclinepi.2021.05.019. [DOI] [PubMed] [Google Scholar]
- 11.Bullers K., Howard A.M., Hanson A., Kearns W.D., Orriola J.J., Polo R.L., Sakmar K.A. It takes longer than you think: librarian time spent on systematic review tasks. Journal of the Medical Library Association: JMLA. 2018;106(2):198. doi: 10.5195/jmla.2018.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Mara-Eves, A., Thomas, J., McNaught, J., Miwa, M., & Ananiadou, S. (2015). Using text mining for study identification in systematic reviews: a systematic review of current approaches. Systematic reviews, 4(1), 1-22. doi: 10.1186/2046-4053-4-5. [DOI] [PMC free article] [PubMed]
- 13.Tsafnat G., Glasziou P., Choong M.K., Dunn A., Galgani F., Coiera E. Systematic review automation technologies. Systematic reviews. 2014;3(1):1–15. doi: 10.1186/2046-4053-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rathbone J., Hoffmann T., Glasziou P. Faster title and abstract screening? Evaluating Abstrackr, a semi-automated online screening program for systematic reviewers. Systematic reviews. 2015;4(1):1–7. doi: 10.1186/s13643-015-0067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shemilt I., Simon A., Hollands G.J., Marteau T.M., Ogilvie D., O'Mara-Eves A.…Thomas J. Pinpointing needles in giant haystacks: use of text mining to reduce impractical screening workload in extremely large scoping reviews. Research Synthesis Methods. 2014;5(1):31–49. doi: 10.1002/jrsm.1093. [DOI] [PubMed] [Google Scholar]
- 16.Ananiadou S., McNaught J. 2006. Text mining for biology and biomedicine. Boston, MA. [Google Scholar]
- 17.Zhao L., Alhoshan W., Ferrari A., Letsholo K.J., Ajagbe M., Chioasca E.V., Batista-Navarro R.T. Natural Language Processing (NLP) for Requirements Engineering (RE): A Systematic Mapping Study. ACM Computing Surveys. 2020 [Google Scholar]
- 18.Basyal, G. P., Rimal, B. P., & Zeng, D. (2020). A Systematic Review of Natural Language Processing for Knowledge Management in Healthcare. arXiv preprint arXiv:2007.09134
- 19.Turchin A., Florez Builes L.F. Using Natural Language Processing to Measure and Improve Quality of Diabetes Care: A Systematic Review. Journal of Diabetes Science and Technology. 2021;15(3):553–560. doi: 10.1177/19322968211000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J., Deng H., Liu B., Hu A., Liang J., Fan L.…Lei J. Systematic evaluation of research progress on natural language processing in medicine over the past 20 years: Bibliometric study on PubMed. Journal of medical Internet research. 2020;22(1):e16816. doi: 10.2196/16816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montazeri M., Afraz A., Farimani R.M., Ghasemian F. Natural Language Processing Systems for Diagnosing and Determining Level of Lung Cancer: A Systematic Review. Frontiers in Health Informatics. 2021;10(1):68. [Google Scholar]
- 22.Grames E.M., Stillman A.N., Tingley M.W., Elphick C.S. An automated approach to identifying search terms for systematic reviews using keyword co-occurrence networks. Methods in Ecology and Evolution. 2019;10(10):1645–1654. [Google Scholar]
- 23.Bramer Wichor M., De Jonge Gerdien B., Rethlefsen Melissa L., Mast Frans, Kleijnen Jos. A systematic approach to searching: an efficient and complete method to develop literature searches. Journal of the Medical Library Association: JMLA. 2018;106(4):531. doi: 10.5195/jmla.2018.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stansfield C., O'Mara-Eves A., Thomas J. Text mining for search term development in systematic reviewing: A discussion of some methods and challenges. Research synthesis methods. 2017;8(3):355–365. doi: 10.1002/jrsm.1250. [DOI] [PubMed] [Google Scholar]
- 25.Belter C.W. Citation analysis as a literature search method for systematic reviews. Journal of the Association for Information Science and Technology. 2016;67(11):2766–2777. [Google Scholar]
- 26.Sarol, M. J., Liu, L., & Schneider, J. (2018, January). Testing a citation and text-based framework for retrieving publications for literature reviews. In BIR@ECIR.
- 27.Antwi, E, Owusu-Banahene W., Boakye-Danquah J., Asare, B. A and Frimpong-Boateng A. F (2020a), Ananse: https://github.com/baasare/ananse
- 28.Antwi, E, Owusu-Banahene W., Boakye-Danquah J., Asare, B. A and Frimpong-Boateng A. F (2020b), Ananse 1.1.2.: https://pypi.org/project/ananse
- 29.Antwi, E, Owusu-Banahene W., Boakye-Danquah J., Asare, B. A and Frimpong-Boateng A. F (2020c), Ananse Documentation: https://ananse.readthedocs.io/en/latest/
- 30.Antwi E.K., Boakye-Danquah J., Owusu-Banahene W., Webster K., Dabros A., Wiebe P.…Sarfo A.K. A Global review of cumulative effects assessments of disturbances on forest ecosystems. Journal of Environmental Management. 2022;317 doi: 10.1016/j.jenvman.2022.115277. [DOI] [PubMed] [Google Scholar]
- 31.Grames, Stillman Introduction to litsearchr with an example of writing a systematic review search strategy for black-backed woodpecker occupancy of post-fire forest systems. Elizagrames.github.io. 2020 https://elizagrames.github.io/litsearchr/introduction_vignette_v010.html 2020. [Online]. Available. [Google Scholar]
- 32.Sommerville I., Fowler M., Beck K., Brant J., Opdyke W., Roberts D. 2019. Edition: Software Engineering. Instructor. [Google Scholar]
- 33.Sommerville I. Pearson; 2020. Engineering Software Products. [Google Scholar]
- 34.Stol K.J., Fitzgerald B. The ABC of software engineering research. ACM Transactions on Software Engineering and Methodology (TOSEM) 2018;27(3):1–51. [Google Scholar]
- 35.MonkeyLearn (2020) "Keyword Extraction". Available: https://monkeylearn.com/keywordextraction/.
- 36.Rose S., Engel D., Cramer N., Cowley W. Automatic keyword extraction from individual documents. Text mining: applications and theory. 2010;1:1–20. [Google Scholar]
- 37.Lee P.C., Su H.N. Investigating the structure of regional innovation system research through keyword co-occurrence and social network analysis. Innovation. 2010;12(1):26–40. [Google Scholar]
- 38.Pick J.L., Nakagawa S., Noble D.W. Reproducible, flexible and high-throughput data extraction from primary literature: The metaDigitise r package. Methods in Ecology and Evolution. 2019;10(3):426–431. [Google Scholar]
- 39.Westgate M.J. revtools: An R package to support article screening for evidence synthesis. Research synthesis methods. 2019;10(4):606–614. doi: 10.1002/jrsm.1374. [DOI] [PubMed] [Google Scholar]
- 40.Grames E.M., Elphick C.S. Use of study design principles would increase the reproducibility of reviews in conservation biology. Biological Conservation. 2020;241 [Google Scholar]
- 41.Antwi, E, Owusu-Banahene W., Boakye-Danquah J., Asare, B. A. and Frimpong-Boateng AF. (2020)(d),Ananse documentation: https://baasare.github.io/ananse/_build/html/index.html
- 42.Cornell University (2021). “Types of Evidence Synthesis - A Guide to Evidence Synthesis - LibGuides at Cornell University.” https://guides.library.cornell.edu/evidence-synthesis/types (accessed May 06, 2021).
- 43.Hausner E., Waffenschmidt S., Kaiser T., Simon M. Routine development of objectively derived search strategies. Systematic reviews. 2012;1(1):1–10. doi: 10.1186/2046-4053-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su Hsin-ning, Lee Pei-Chun. Mapping knowledge structure by keyword co-occurrence: A first look at journal papers in Technology Foresight. Scientometrics. 2020;85(1):65–79. [Google Scholar]
- 45.Mongeon P., Paul-Hus A. The journal coverage of Web of Science and Scopus: a comparative analysis. Scientometrics. 2016;106(1):213–228. [Google Scholar]