Abstract
Streptococcus pyogenes secretes many proteins that influence host-pathogen interactions. Despite their importance, relatively little is known about the regulation of these proteins. The rgg gene (also known as ropB) is required for the expression of streptococcal erythrogenic toxin B (SPE B), an extracellular cysteine protease that contributes to virulence. Proteomics was used to determine if rgg regulates the expression of additional exoproteins. Exponential- and stationary-phase culture supernatant proteins made by S. pyogenes NZ131 rgg and NZ131 speB were separated by two-dimensional electrophoresis. Differences were identified in supernatant proteins from both exponential- and stationary-phase cultures, although considerably more differences were detected among stationary-phase supernatant proteins. Forty-two proteins were identified by peptide fingerprinting with matrix-assisted laser desorption mass spectrometry. Mitogenic factor, DNA entry nuclease (open reading frame [ORF 226]), and ORF 953, which has no known function, were more abundant in the culture supernatants of the rgg mutant compared to the speB mutant. ClpB, lysozyme, and autolysin were detected in the culture supernatant of the speB mutant but not the rgg mutant. To determine if Rgg affected protein expression at the transcriptional level, real-time (TaqMan) reverse transcription (RT)-PCR was used to quantitate Rgg-regulated transcripts from NZ131 wild-type and speB and rgg mutant strains. The results obtained with RT-PCR correlated with the proteomic data. We conclude that Rgg regulates the transcription of several genes expressed primarily during the stationary phase of growth.
Infection with Streptococcus pyogenes (group A streptococcus) typically results in pharyngitis and is self-limiting. Rarely, severe infections such as necrotizing fasciitis and toxic shock syndrome occur. Although the molecular mechanisms of severe streptococcal infections are poorly understood, S. pyogenes secretes to the extracellular environment many proteins that may contribute to disease. For example, the extracellular cysteine protease streptococcal erythrogenic toxin B (SPE B) degrades human extracellular matrix proteins (18) and activates human enzymes involved in host tissue remodeling (4). In this manner, SPE B may contribute to the massive tissue destruction and concomitant dissemination of infection that is characteristic of necrotizing fasciitis and myositis. In addition, extracellular superantigens, in conjunction with cell-associated components, undoubtedly contribute to the systemic effects that characterize streptococcal toxic shock syndrome (10). Genetic studies support the idea that extracellular proteins (ECPs) contribute to severe streptococcal disease. Inactivation of speB (28, 30) and the sic gene (29), encoding the extracellular streptococcal inhibitor of complement, reduced virulence compared to the isogenic wild-type strains in animal models of infection. In addition, the extracellular plasminogen activator streptokinase A (SKA) has been linked to the development of acute post-streptococcal glomerulonephritis in animal models (33, 34). Thus, ECPs are important determinants of host-pathogen interactions and are potential targets for chemotherapeutic intervention designed to prevent or treat severe disease.
Several loci that influence the expression of streptococcal ECPs have been identified. Mga (multiple gene activator) regulates the expression of several cell-associated proteins including M protein, M-like proteins (Mrp, Enn, and FcR), and C5a peptidase, in addition to the ECPs SIC and serum opacity factor (10). Mga-regulated genes are expressed primarily during the exponential phase of growth (32). A two-component regulatory system designated csrRS (2, 14, 25), also known as covRS (11), regulates the expression of several proteins, including ECPs. Specifically, nonpolar inactivation of csrR enhanced expression of the has operon responsible for capsule formation and of the ECPs SKA, SagA (streptolysin S-associated gene A), SPE B, and mitogenic factor (MF) (11, 14). Inactivation of the pleiotropic effect locus (pel) altered the expression of genes encoding SPE B, SKA, and streptolysin S (26). Despite recent progress, much remains to be learned about the regulation of ECP expression and how expression is coordinated with additional regulatory networks.
The expression of speB is dependent on the rgg gene (6), also known as ropB (31), which is located proximal to speB in the chromosome. Rgg is homologous to the transcriptional regulatory factor Rgg of Streptococcus gordonii (41, 42), which is required for the transcription of the gene encoding glucosyltransferase G (gtfG). GtfG is a secreted enzyme responsible for the polymerization of glucose to form water-soluble and insoluble glucans important in bacterial adherence to tooth enamel (27). Rgg of S. pyogenes is also similar to GadR of Lactococcus lactis, which is required for expression of the GadABCD regulon (37). GadC is an antiporter that transports glutamate into the cytoplasm and exports glutamate-γ-aminobutyrate, formed by GadA and GadB-mediated decarboxylation of glutamate. The GadABCD regulon is required for glutamate-dependent acid resistance and is maximally expressed in the stationary phase of growth (37). In addition, Rgg of S. pyogenes is similar to MutR of Streptococcus mutans (36), which is required for the expression of an operon encoding the lantibiotic mutacin II (MutA), the modifying enzyme MutM, a transport protein (MutT), and three polypeptides required for immunity to the lantibiotic (MutF, MutE, and MutG) (36). Mutacin activity is maximal in the stationary phase of growth, although the level of mutA transcripts does not vary between the exponential and stationary phases of growth (36). Although rgg and speB are physically linked, it is unclear if rgg acts directly to influence speB expression. Moreover, the influence of rgg on the expression of additional ECPs of S. pyogenes is unknown.
The determination of the complete nucleotide sequence of an S. pyogenes serotype M1 genome (B. Roe, S. P. Linn, L. Song, X. Yuan, S. Clifton, R. E. McLaughlin, M. McShan, and J. J. Ferretti, streptococcal genome sequencing project, online [http://www.genome.ou.edu/strep.html]) facilitates the use of functional genomic methods to study global changes in gene expression. Proteomics involves separating and identifying proteins composing a defined proteome, such as the ECPs of S. pyogenes. High-resolution separation of complex protein mixtures is typically done by two-dimensional electrophoresis (2-DE). Identification of proteins of interest can be accomplished by determining the masses of peptides after in-gel trypsinization. The masses represent a fingerprint of the protein and are used to identify the corresponding gene in a genomic database. Criteria used to describe the quality of protein identifications include the number of tryptic peptides detected, the coverage of the identified protein with detected peptides, and the accuracy of peptide mass determinations. As few as three peptides are sufficient to identify a protein; however, confidence in the identification increases with the detection of additional peptides (15). Although coverage values greater than 35% are typical of unambiguous identifications, posttranslational modifications such as proteolytic removal of the signal peptide or proteolytic modification of proproteins or zymogens are not considered when calculating the coverage because the precise modification is typically not defined. Finally, differences in the calculated mass and observed mass of a peptide that are less than 10 ppm approach the technical limitations of many mass spectrometers. Protein identification by mass spectrometry (MS) offers a relatively rapid method to identify proteins of interest, such as those whose expression is altered by inactivation of a regulatory gene.
The objective of this study was to determine if the rgg gene influences expression of ECPs other than SPE B. To achieve this objective, proteomics was used to identify differences in ECP expression between NZ131 speB and rgg mutants. Real-time (TaqMan) reverse transcription-PCR (RT-PCR) showed that the differences in protein expression were due to changes in the level or stability of the corresponding transcripts. The results indicate that Rgg regulates the expression of several genes in the stationary phase of growth.
MATERIALS AND METHODS
Strains and media.
S. pyogenes NZ131 (serotype M49) and isogenic mutant derivatives NZ131 speB and NZ131 rgg have been previously described (6, 8). Strains were grown on Trypticase soy agar containing 5% sheep blood (Becton Dickinson, Cockeysville, Md.) overnight at 37°C in 5% CO2. Todd-Hewitt broth containing 0.2% (wt/vol) yeast extract (THY; Difco Laboratories, Detroit, Mich.) was passed through a 10,000-molecular-weight cutoff (MWCO) filter using a Millipore ProFlux M12 tangential flow filtration system to prepare protein-free THY.
Preparation of extracellular proteins.
Plate-grown bacteria were used to inoculate 10 ml of protein-free THY in 15-ml polypropylene tubes (Corning, New York, N.Y.), and the cultures were incubated for 8 h at 37°C in 5% CO2. Each 10-ml culture was added to 40 ml of prewarmed protein-free THY and incubated for approximately 14 h prior to inoculation into 1-liter bottles containing 950 ml of protein-free THY equilibrated overnight in a 37°C incubator containing 5% CO2. The cultures were incubated at 37°C with 5% CO2 with no agitation. Exponential-phase cultures had an A600 of 0.2 to 0.3 and corresponded to 2 to 3 h of growth. Stationary-phase cultures had an A600 of 0.5 to 0.6 and were grown for approximately 18 h. Following growth in protein-free THY broth, bacteria were centrifuged for 15 min at 13,679 × g at 4°C, and the supernatant fluids were sterilized with a 0.2-μm-pore-size filter (NalgeNunc, Rochester, N.Y.). Culture supernatant proteins were concentrated approximately 10-fold with a Millipore ProFlux M12 tangential flow concentrator fitted with an Amicon S3Y10 spiral cartridge with a 10,000-MWCO filter. Culture supernatant proteins were precipitated by adding 85% (wt/vol) ammonium sulfate (Sigma Chemical Co., St. Louis, Mo.). Proteins were resuspended in 3 ml of water and dialyzed extensively with a Slide-A-Lyzer 10,000-MWCO cartridge (Pierce Chemical Co., Rockford, Ill.). When necessary, protein preparations were further concentrated by lyophilization with a Savant (Hicksville, N.Y.) SpeedVac concentrator. Total protein concentration was determined with an ESL protein determination kit, as described by the manufacturer (Boehringer Mannheim, Mannheim, Germany).
2-DE.
First-dimension isoelectric focusing was done with an IPGphor isoelectric focusing system as described by the manufacturer (Amersham Pharmacia Biotech, Piscataway, N.J.). Immobiline dry strips (18 cm) with a 3 to 10 linear separation range were rehydrated with 350 μl of protein sample in rehydration buffer consisting of 8 M urea (Amersham Pharmacia Biotech), 2% (wt/vol) 3-[3-(cholamidopropyl)-dimethyl-ammonio]-1-propane sulfonate (CHAPS; Amersham Pharmacia Biotech), 0.5% (vol/vol) IPG buffer (Amersham Pharmacia Biotech), and 2.8 mg of dithiothreitol (Amersham Pharmacia Biotech) per ml at 20°C for 14 h. Isoelectric focusing was done with 500 V for 500 V·h, 1,000 V for 1,000 V·h, and 8,000 V for 32,000 V·h. The strips were then incubated in sodium dodecyl sulfate (SDS) equilibration buffer (50 mM Tris-Cl [pH 8.8], 6 M urea, 30% [vol/vol] glycerol, 2% SDS, bromophenol blue) for 10 min. SDS-polyacrylamide gel electrophoresis separation was done with a DALT electrophoresis system (Amersham Pharmacia Biotech) and a 12% acrylamide resolving gel (1.5 by 23.4 by 19.5 mm; Bio-Rad, Hercules, Calif.) containing 1% SDS for approximately 18 h at 115 V. Following staining with Coomassie colloidal blue (Bio-Rad), the gels were scanned with a calibrated UMAX transmission scanner (Amersham Pharmacia Biotech). Spot volume was determined with ImageMaster 2D Elite software (Amersham Pharmacia Biotech) and is defined as the sum of the pixel values comprising the protein spot minus the sum of background pixel values.
In-gel tryptic digestion of proteins.
Proteins of interest were excised from the SDS-polyacrylamide gels and washed three times for 15 min in 400 μl of 25 mM NH4HCO3–50% acetonitrile (ACN; Aldrich, Milwaukee, Wis.). The proteins were incubated in 100% ACN for 5 min and lyophilized in a SpeedVac for 30 min. The dried gel plugs were rehydrated with 25 mM NH4HCO3 containing 10 μg of sequencing-grade trypsin (Sigma) per ml. Following incubation at 37°C for approximately 16 h, the trypsin solution was aspirated to a microcentrifuge tube, and additional peptides were recovered from the gel plugs by extraction twice with 50% ACN–5% trifluoroacetic acid (TFA; Applied Biosystems, Foster City, Calif.) for 1 h. The extracted peptides were lyophilized in a SpeedVac, resuspended in 10 μl of 0.1% TFA, and purified with ZipTip Microcolumns (Millipore, Bedford, Mass.). The peptides were recovered from the ZipTip columns by elution with 30, 50, and 80% ACN in 0.1% TFA, lyophilized, and resuspended in 3 μl of 50% ACN–0.1% TFA.
MALDI-TOF MS.
The mass of each extracted peptide was determined with a Voyager STR MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) mass spectrometer (PE Biosystems, Framingham, Mass.). Peptides were crystallized on a stainless steel MALDI plate by using a dry-drop method with an equal volume of 10.0 mg/ml α-cyano-4-hydroxycinnamic acid matrix (Aldrich) in 50% ACN–0.1% TFA. The masses of the peptides were determined in positive reflector mode with internal calibrants obtained from PE Biosystems (des-Arg1-bradykinin [Mr 904.56] and adrenocorticotropin [clip 18–39; Mr 2,465.20]).
Database searches and protein identification.
Protein Prospector (University of California, San Francisco, Mass Spectrometry Facility) was used to search a genomic database of S. pyogenes containing 2,241 open reading frames (ORFs). The database included ORFs identified by WIT2 analysis (www.genome.ou.edu) of S. pyogenes strain SF370 (serotype M1), by genome sequencing projects in the Laboratory of Human Bacterial Pathogenesis, and by contigs of genomic sequences of S. pyogenes Manfredo strain (serotype M5) assembled by the Sanger Institute (www.sanger.ac.uk).
RNA isolation.
S. pyogenes was grown in 10 ml of protein-free THY broth in 15-ml tubes (Corning) for approximately 10 h (A600 of 0.5 to 0.6). Cultures were centrifuged; the bacteria were resuspended in 200 μl of diethyl pyrocarbonate (DEPC; Sigma)-treated water and frozen in liquid nitrogen. Bacterial pellets (200 μl) were thawed on ice and added to 2-ml FastPrep Blue tubes containing ceramic matrices, 500 μl of acid phenol, and 500 μl of CPRS-Blue, as described by the manufacturer (Bio 101, La Jolla, Calif.). The bacteria were lysed with a FastPrep instrument (Bio 101) at setting 6 for 11 s and immediately placed on ice for 1 min. Samples were incubated at 65°C for 10 min and centrifuged at 10,000 × g for 5 min at 4°C. The upper aqueous phase was aspirated to a 2-ml phase-lock microcentrifuge tube (Eppendorf Scientific, Westbury, N.Y.) and extracted with an equal volume of acid-phenol heated to 65°C. The phases were separated by centrifugation at 10,000 × g for 4 min at 4°C, and the extraction was repeated with acid-phenol:chloroform (1:1, vol/vol) and chloroform:isoamyl alcohol (24:1, vol/vol). The aqueous phase was treated with 100 U of DNase I (Roche Molecular Biochemicals, Mannheim, Germany) for 2 h at 37°C and then extracted three times with acid-phenol and chloroform; the nucleic acid was precipitated by adding an equal volume of isopropanol. Samples were centrifuged at 10,000 × g for 15 min at 4°C, and the pellets were washed with 75% DEPC-treated ethanol. RNA was suspended in 50 μl of DEPC-treated water and stored at −80°C. The quality of the RNA was assessed by agarose gel electrophoresis and spectrophotometry.
Real-time RT-PCR.
Oligonucleotide primers and probes (Table 1) were designed with Primer Express 1.0 software (ABI Prism; PE Biosystems) and purchased from MegaBases Inc. (Evanston, Ill.). The probes consisted of an oligonucleotide labeled at the 5′ end with the reporter dye 5-carboxyfluorescein and at the 3′ end with the quencher N,N,′N′-tetramethyl-6-carboxyrhodamine. RT-PCR was done with the TaqMan One-Step RT-PCR Master Mix Reagents kit (PE Applied Biosystems) as described by the manufacturer. The RT-PCR mixture (25 μl) contained 6.25 U of Multiscribe reverse transcriptase, 10.0 U of RNase inhibitor, 500 nM each gene-specific primer, 100 nM each probe, and 25 ng of total RNA template. Amplification and detection of specific products was performed with the ABI Prism 7700 sequence detection system (PE Applied Biosystems) with the following cycle profile: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. The critical threshold cycle (Ct) is defined as the cycle at which the fluorescence becomes detectable above background and is inversely proportional to the logarithm of the initial number of template molecules. A standard curve was plotted for each primer-probe set with Ct values obtained from amplification of known quantities of genomic DNA isolated from strain NZ131. The standard curves were used to transform Ct values to the relative number of DNA molecules. The amount of contaminating chromosomal DNA in each sample was determined with control reactions that did not contain reverse transcriptase. The amount of contaminating DNA was subtracted from each experimental value to give the quantity of cDNA. The quantity of cDNA for each experimental gene was normalized to the quantity of gyrA cDNA in each sample.
TABLE 1.
Oligonucleotide primers and fluorescent probes used to quantitate cDNA
| Gene | Forward primer (5′ - 3′) | Reverse primer (5′ - 3′) | Fluorescent probea(5′ - 3′) |
|---|---|---|---|
| mf | CCCAAAATGTAGGAGGTCGTG | CATTCTTGAGCTCTTTGTTCGGT | CCAAAAAGGCGGCATGCGCT |
| orf226 | TCCTCCTGGCTGGCATAATT | CTAAATGGCCACGGTCCATT | AAATTGACTGACGCTAATGGAAAAACAACTTGG |
| orf953 | TTGGTTAGGAATGGTATCAGTCTTTTT | AATTTGCGAGCTAGAGTTATTATGATTG | CGATTCTCCTTTTTTTAACTGCAGCATCGA |
| orf204 | GGCTGTCGCAGCAGTAGCA | GAACCTATTGGACGTTTACCATCATC | TGCTATTCGACGAAATCGGGCAGGTT |
| orf1669 | TTAAGGGTGCCTATAACGGTTCTT | TGATCAATCGTATAGGTATTGCCATTA | TGTGACGATGTCAACTTGGGAAGATGATG |
| orf1324 | TTCAAATGGCAATGCTTACGAT | GTCTGCGTAGCCATCCCAA | TTGATGGATCGCTTGGTGCGCAAT |
| gyrA | CGACTTGTCTGAACGCCAAA | TTATCACGTTCCAAACCAGTCAA | CGACGCAAACGCATATCCAAAATAGCTTG |
Covalently linked at the 5′ end to 5-carboxyfluorescein and at the 3′ end to N,N,N-tetramethyl-6-carboxyrhodamine.
RESULTS
Comparison of supernatant proteins of NZ131 speB and NZ131 rgg grown to the mid-exponential phase.
The role of rgg in the regulation of ECP expression was assessed by comparing 2-DE patterns of ECPs from NZ131 speB and NZ131 rgg. NZ131 speB was used rather than the wild-type strain because SPE B accounts for nearly 95% of culture supernatant protein when strain NZ131 is grown in protein-free THY broth (7). The abundance of SPE B would interfere with the detection of other ECPs potentially influenced by Rgg. In addition, SPE B, which is not detected in strain NZ131 rgg (6), degrades a variety of human and streptococcal proteins (1, 7, 18).
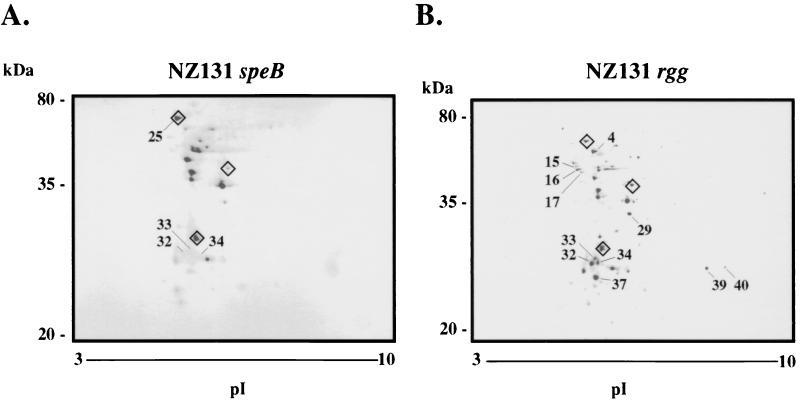
NZ131 speB and NZ131 rgg were grown in protein-free THY broth medium to mid-exponential phase. The concentrated ECPs were separated by 2-DE and stained with Coomassie colloidal blue. Representative gels from two independent protein isolations are shown in Fig. 1. The majority of ECPs from each strain had an isoelectric point (pI) between 4 and 6. The protein spots were designated on the basis of the source strain (NZ131 rgg or NZ131 speB) and arbitrarily numbered. Differences were identified in the protein composition of exponential-phase culture supernatant fluids from NZ131 speB and NZ131 rgg. For example, proteins (designated Rgg-15 to Rgg-17) present in supernatant fluid from NZ131 rgg were not detected in the analogous region of the 2-DE gels of the speB mutant (Fig. 1). Protein Rgg-16 was identified as DNA K and migrated similarly to the inferred molecular weight and pI (Table 2). Proteins designated Rgg-32, Rgg-33, and Rgg-34 were detected in the supernatant fluid of NZ131 rgg but not NZ131 speB (SpeB-32, SpeB-33, and SpeB-34 [Fig. 1]). Finally, Rgg-39 and Rgg-40 were detected in the ECP obtained from the rgg mutant but not the speB mutant.
FIG. 1.
Coomassie colloidal blue-stained 2-DE gels of supernatant proteins from exponential-phase cultures of NZ131 speB (A) and NZ131 rgg (B). Proteins identified by peptide mass fingerprinting are summarized in Table 3. Diamonds around selected proteins are used to orient the gels to each other. The migration of molecular mass standards is indicated. The gels (oriented with acidic proteins to the left) are representative of two independent experiments.
TABLE 2.
Exponential-phase supernatant proteins identified by peptide mass fingerprinting
| Designation | Name | Accession no.a | Mr/pIb | No. of peptides (coverage)c | Δppmd |
|---|---|---|---|---|---|
| SpeB-25 | Heat shock protein | U72721 | 64,920/4.62 | 10 (18) | 2.9 ± 3.2 |
| Rgg-4 | Heat shock protein | X89236, RST0771 | 50,497/4.68 | 14 (37) | 4.0 ± 1.8 |
| Rgg-16 | DNA K | RST00473 | 58,144/4.58 | 6 (16) | 9.9 ± 5.4 |
| Rgg-29 | Enolase | RST00465 | 37,330/4.83 | 8 (34) | 12.2 ± 2.3 |
| Rgg-37 | Enolase | RST00465 | 37,330/4.83 | 7 (24) | 0.5 ± 2.3 |
NCBI accession number or WIT2 RST designation.
Inferred from the nucleotide sequence.
Number of tryptic peptides of the identified protein detected by MALDI-TOF MS (percentage of the protein covered with detected peptides).
Mean difference ± standard error between masses detected by MALDI-TOF MS and calculated masses of the tryptic peptides.
Comparison of supernatant fluid proteins of NZ131 speB and NZ131 rgg grown to stationary phase.
The Rgg-regulated exoprotein SPE B is expressed by strain NZ131 primarily in the stationary phase of growth (9). Thus, it was of interest to characterize stationary-phase supernatant proteins from NZ131 speB and NZ131 rgg. NZ131 speB and NZ131 rgg were grown in protein-free THY broth medium for approximately 18 h, and the concentrated culture supernatant proteins were analyzed by 2-DE (Fig. 2).
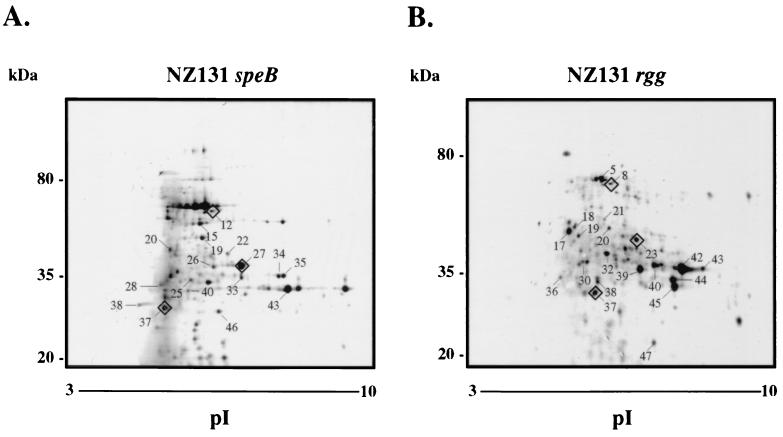
FIG. 2.
Coomassie colloidal blue-stained 2-DE gels of supernatant proteins from stationary-phase cultures of NZ131 speB (A) and NZ131 rgg (B). Proteins identified by peptide mass fingerprinting are summarized in Table 2. Diamonds around selected proteins are used to orient the gels to each other. The migration of molecular mass standards is indicated. The gels (oriented with acidic proteins to the left) are representative of two independent experiments.
Considerable differences were detected in the protein composition of stationary-phase supernatant fluids between the two strains (Fig. 2). The proteins identified by peptide mass fingerprinting in stationary-phase supernatant fluids are summarized in Table 3. Approximately 4.8-fold more MF was detected in the supernatant of NZ131 rgg cultures than in NZ131 speB culture supernatant (total spot volumes of 2,606 and 540, respectively) (Fig. 2; Table 4). Five positional variants of MF were detected in the NZ131 rgg supernatant fluid (Rgg-39, Rgg-40, Rgg-42, Rgg-43, and Rgg-45), whereas only one was identified in the supernatant fluid from NZ131 speB (SpeB-43). In addition, proteins designated Rgg-39 and Rgg-40 in the exponential phase 2-DE map (Fig. 1) migrated similarly to protein spots identified as MF in the stationary phase (Fig. 2, Rgg-42 and Rgg-43), which suggested that NZ131 rgg produced detectable MF in the exponential phase of growth whereas the speB mutant did not (cf. Fig. 1 and 2). In addition, DNA entry nuclease (ORF 226) was highly expressed by the rgg mutant compared to the speB mutant (Table 4). Proteins Rgg-17, Rgg-19, Rgg-36, Rgg-44, and Rgg-47 were identified as DNA entry nuclease in NZ131 rgg supernatant fluid (Table 3); however, only SpeB-26 was identified as DNA entry nuclease in ECPs from the speB mutant. Protein Rgg-20, uniquely expressed by the rgg mutant, was identified as ORF 953 and has no known function (Fig. 2; Table 3).
TABLE 3.
Stationary-phase supernatant proteins identified by peptide mass fingerprinting
| Designation | Name | Accession no.a | Mr/pIb | No. of peptides (coverage)c | Δppmd |
|---|---|---|---|---|---|
| SpeB-12 | Streptokinase | M19347 | 50,084/5.54 | 8 (13) | 2.8 ± 6.0 |
| SpeB-15 | Glyceraldehyde-3-phosphate dehydrogenase/plasmin receptor | M95569 | 35,958/5.34 | 8 (30) | 2.9 ± 4.5 |
| SpeB-19 | 6-Phosphofructokinase | RST00803 | 35,748/5.33 | 7 (23) | 0.8 ± 2.6 |
| SpeB-20 | Immunogenic secreted protein | RST00809, U31811 | 53,292/5.5 | 5 (10) | 8.2 ± 7.2 |
| SpeB-22 | Streptokinase | M19347 | 50,084/5.54 | 5 (14) | 1.3 ± 3.6 |
| SpeB-25 | Streptokinase | M19347 | 50,084/5.54 | 5 (18) | 3.3 ± 9.6 |
| SpeB-26 | DNA entry nuclease | RST00226 | 30,398/4.93 | 4 (16) | 0.3 ± 5.8 |
| SpeB-27 | CAMP factor | RST0342, AF079502 | 28,480/6.25 | 4 (15) | 6.9 ± 1.8 |
| SpeB-28 | ClpB | RST00204 | 63,349/5.41 | 5 (9) | 10.4 ± 17.8 |
| SpeB-33 | CAMP factor | RST0342, AF079502 | 28,480/6.25 | 5 (26) | 2.8 ± 2.7 |
| SpeB-34 | CAMP factor | RST0342, AF079502 | 28,480/6.25 | 6 (29) | 0.1 ± 4.9 |
| SpeB-35 | CAMP factor | RST0342, AF079502 | 28,480/6.25 | 6 (29) | 0.1 ± 4.9 |
| SpeB-37 | Enolase | RST00465 | 37,330/4.83 | 6 (23) | 7.1 ± 1.2 |
| SpeB-38 | Autolysin | RST01669 | 23,020/4.66 | 4 (15) | 11.7 ± 2.7 |
| SpeB-40 | Lysozyme | RST01324 | 49,729/5.04 | 5 (15) | 1.5 ± 16.5 |
| SpeB-43 | Mitogenic factor | D13428 | 30,061/9.24 | 7 (37) | 7.6 ± 1.8 |
| SpeB-46 | Ribosome recycling factor | RST00322 | 20,572/5.68 | 8 (45) | 11.7 ± 4.7 |
| Rgg-5 | Streptokinase | M19347 | 50,084/5.54 | 8 (23) | 1.7 ± 3.2 |
| Rgg-8 | Streptokinase | M19347 | 50,084/5.54 | 7 (17) | 5.4 ± 6.7 |
| Rgg-17 | DNA entry nuclease | RST00226 | 30,398/4.93 | 8 (23) | 4.2 ± 3.5 |
| Rgg-18 | Immunogenic secreted protein | RST00809, U31811 | 53,292/5.50 | 5 (37) | 4.7 ± 5.5 |
| Rgg-19 | DNA entry nuclease | RST00226 | 30,398/4.93 | 6 (27) | 6.2 ± 3.1 |
| Rgg-20 | Unknown | RST00953 | 32,053/5.79 | 4 (22) | 2.6 ± 6.2 |
| Rgg-21 | Streptokinase | M19347 | 50,084/5.54 | 9 (20) | 1.7 ± 1.7 |
| Rgg-23 | CAMP factor | RST0342, AF079502 | 28,480/6.25 | 10 (38) | 6.0 ± 2.6 |
| Rgg-30 | Immunogenic secreted protein | RST00809, U31811 | 53,292/5.50 | 4 (11) | 15.2 ± 13.4 |
| Rgg-32 | Streptokinase | M19347 | 50,084/5.54 | 7 (18) | 18.7 ± 6.1 |
| Rgg-36 | DNA entry nuclease | RST00226 | 30,398/4.93 | 5 (24) | 1.0 ± 4.1 |
| Rgg-37 | Streptokinase | M19347 | 50,084/5.54 | 8 (13) | 1.7 ± 7.3 |
| Rgg-38 | Phosphoglycerate kinase | RST00513 | 42,130/4.82 | 5 (17) | 4.9 ± 11.8 |
| Rgg-39 | Mitogenic factor | D13428 | 30,061/9.24 | 5 (28) | 18.3 ± 9.1 |
| Rgg-40 | Mitogenic factor | D13428 | 30,061/9.24 | 8 (37) | 9.1 ± 5.0 |
| Rgg-42 | Mitogenic factor | D13428 | 30,061/9.24 | 9 (45) | 3.1 ± 15.9 |
| Rgg-43 | Mitogenic factor | D13428 | 30,061/9.24 | 4 (19) | 11.4 ± 10.6 |
| Rgg-44 | DNA entry nuclease | RST00226 | 30,398/4.93 | 6 (27) | 11.2 ± 1.9 |
| Rgg-45 | Mitogenic factor | D13428 | 30,061/9.24 | 5 (26) | 18.0 ± 7.6 |
| Rgg-47 | DNA entry nuclease | RST00226 | 30,398/4.93 | 6 (14) | 11.2 ± 1.9 |
NCBI accession number or WIT2 RST designation.
Inferred from the nucleotide sequence.
Number of tryptic peptides of the identified protein detected by MALDI-TOF MS (percentage of the protein covered with detected peptides).
Mean difference ± standard error between masses detected by MALDI-TOF MS and calculated masses of the tryptic peptides.
TABLE 4.
Quantitation of Rgg-regulated exoproteins
| Name | NZ131 speB
|
NZ131 rgg
|
||
|---|---|---|---|---|
| Designation | Vola | Designation | Vol | |
| Mitogenic factor | SpeB-43 | 540.2 | Rgg-39 | 430.4 |
| Rgg-43 | 200.3 | |||
| Rgg-45 | 465.5 | |||
| Rgg-42 | 1,171.5 | |||
| Rgg-40 | 338.3 | |||
| DNA entry nuclease (ORF 226) | SpeB-26 | 102.1 | Rgg-17 | 318.8 |
| Rgg-19 | 91.2 | |||
| Rgg-36 | 50.6 | |||
| Rgg-44 | 334.7 | |||
| Rgg-47 | 149.9 | |||
| ORF 953 | NDb | Rgg-20 | 90.3 | |
| Lysozyme (RST1324) | SpeB-40 | 46.4 | ND | |
| Autolysin (RST1669) | SpeB-38 | 89.8 | ND | |
| ClpB (RST204) | SpeB-28 | 55.4 | ND | |
Sum of pixel values comprising the protein spot minus background pixel values.
ND, not detected.
Several proteins were detected in the supernatant from NZ131 speB cultures but not in the supernatant of the rgg mutant (Fig. 2). Peptide mass fingerprinting indicated that SpeB-28 had peptides that corresponded to ClpB (ORF 204); SpeB-38 and SpeB-40 were identified as autolysin (ORF 1669) and lysozyme (ORF 1324), respectively (Table 3). The results indicate that the rgg mutant did not synthesize detectable levels of autolysin, lysozyme, and ClpB.
Proteins whose expression was not significantly altered by rgg inactivation were also identified. For example, proteins identified as CAMP factor included SpeB-27, SpeB-33, SpeB-34, and SpeB-35 from NZ131 speB supernatant and protein Rgg-23 from the supernatant of NZ131 rgg. Proteins identified as streptokinase included SpeB-12, SpeB-22, and SpeB-25 from NZ131 speB supernatant and Rgg-5, Rgg-8, Rgg-21, Rgg-32, and Rgg-37 from NZ131 rgg supernatant.
Real-time quantitative RT-PCR.
The results of the proteomic analysis of ECPs from NZ131 speB and NZ131 rgg indicated that the quantity of several proteins in culture supernatants was affected by rgg inactivation. Inasmuch as Rgg is similar to known transcriptional regulatory proteins, the results suggested that Rgg influenced ECP expression at the level of transcription. To determine if this was the case, the following six genes were analyzed by real-time RT-PCR: mf, orf 226 (encoding DNA entry nuclease), orf953, orf204 (clpB), orf1669 (encoding autolysin), and orf1324 (encoding lysozyme). Standard curves were generated for each gene with genomic DNA isolated from the wild-type strain of NZ131 to determine the relative quantity of amplified cDNA. The quantity of cDNA for each gene was then normalized to the quantity of gyrA cDNA present in each RNA preparation. To confirm that gyrA was constitutively expressed, the amount of gyrA mRNA was measured in three separate RT-PCR experiments with two independent RNA samples isolated from both stationary- and exponential-phase cultures of NZ131. The results confirmed that gyrA is constitutively transcribed in NZ131 wild-type, speB, and rgg strains in both growth phases (data not shown).
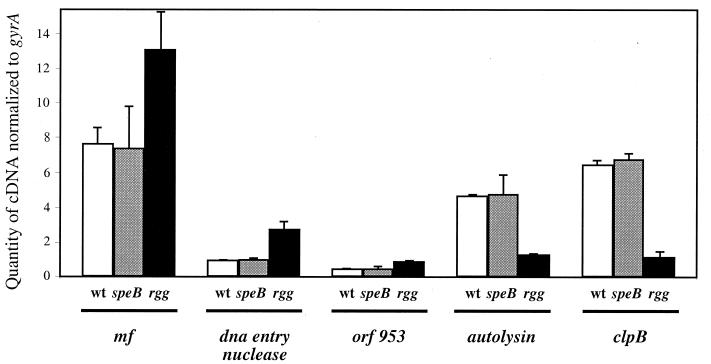
Equal amounts of total RNA from stationary-phase (10-h) cultures of wild-type, speB, and rgg strains were used to quantify the transcript levels of the Rgg-regulated exoproteins. Representative results, confirmed by analysis of two or three independently isolated sets of RNA, are shown in Fig. 3. Importantly, the RT-PCR and proteomic data were qualitatively cognate. The mf, orf226, and orf953 transcript levels were higher in the rgg mutant than in the wild-type strain and the speB mutant (Fig. 3). Levels of autolysin and clpB transcripts were lower in the rgg mutant than in the wild-type strain and the speB mutant (Fig. 3). Primers and probes based on an M1 nucleotide sequence (Roe et al., online) did not amplify lysozyme cDNA or the lysozyme structural gene when purified genomic DNA from NZ131 was used a control. However, the primers did amplify the lysozyme gene when genomic DNA from an M1 serotype (SF370) was used (data not shown).
FIG. 3.
Relative quantities of Rgg-regulated gene transcripts assessed by TaqMan assays. cDNA detected from stationary-phase cultures of NZ131 wild-type (wt), speB, and rgg strains was quantified for mf, the DNA entry nuclease gene (orf226), orf953, the autolysin gene (orf1669), and clpB (orf204). The cDNA values were normalized to the quantity of gyrA cDNA in each sample. The experiments were repeated using at least two independently isolated RNA preparations, and representative results are shown.
To determine if rgg transcription correlated with the influence of Rgg primarily on stationary-phase gene expression, total RNA was isolated from NZ131 wild-type and speB cultures grown to exponential (2 h) and stationary (10 h) phases. The rgg transcript level was higher in the stationary phase than in the exponential phase for both strains (Fig. 4). Interestingly, the rgg transcript was more abundant in the speB mutant than in the wild-type strain (Fig. 4).
FIG. 4.
Relative quantities of rgg cDNA in the exponential and stationary phases of growth assessed by TaqMan assays. The amount of rgg cDNA was determined and normalized to the amount of gyrA cDNA following reverse transcription of total RNA isolated from NZ131 wild-type (wt) and speB strains. The results shown are representative of those obtained with two independently isolated RNA preparations.
DISCUSSION
Several ECPs of S. pyogenes have been extensively characterized, but the function and contribution to virulence of many others are not well understood. Insight into protein function is often gained by determining the conditions under which a gene is expressed. Rgg is required for the expression of SPE B (6, 31), an extracellular cysteine protease that contributes to virulence (28, 30). In this study, we used proteomics and real time RT-PCR to identify additional Rgg-regulated exproteins. Comparative analysis of the extracellular proteomes of NZ131 speB and NZ131 rgg showed that the rgg mutant expressed less lysozyme (ORF 1324), autolysin (ORF 1669), and ClpB (ORF 204) in the stationary phase of growth. In addition, the rgg mutant expressed considerably more MF (ORF 1835), DNA entry nuclease (ORF 226), and ORF 953. The results show that Rgg regulates the expression of several genes expressed primarily in the stationary phase of growth.
Transcriptional regulation by Rgg.
Real time RT-PCR analysis showed that Rgg influenced the expression of MF, DNA entry nuclease, ORF 953, autolysin, and ClpB by altering the quantity or stability of the corresponding transcripts (Fig. 3). This finding is consistent with transcriptional regulation of speB by Rgg, as previously described (31). The results are also consistent with the observation that the amino acid sequence of Rgg is similar to those of several other gram-positive transcriptional regulatory proteins, including Rgg of S. gordonii (41, 42), GadR of L. lactis (37), and MutR of S. mutans (36). The gadR gene expressed primarily in the stationary phase of growth (37). Mutacin II activity, which is regulated by MutR, is also maximally expressed in the stationary phase (36). Similarly, the majority of S. pyogenes Rgg-regulated exoproteins were detected in the stationary phase of growth, which correlated with increased rgg expression (Fig. 4). No information is available regarding growth phase-dependent expression of rgg in S. gordonii. Several potential regulatory elements were identified in the promoter regions of S. pyogenes Rgg-regulated genes, but a common motif was not apparent.
Interestingly, more rgg transcript was detected in NZ131 speB than in the wild-type strain (Fig. 4), suggesting that SPE B represses the expression of rgg. The possibility that speB inactivation influenced the expression of orf204 (clpB), orf1669 (encoding autolysin), orf953, mf, and orf226 (encoding DNA entry nuclease) was excluded by RT-PCR, which showed that the transcript levels for each gene were similar between the wild-type strain and the isogenic speB mutant (Fig. 3). Additional experiments are required to determine if the speB transcript, SPE B protease, or SPE B degradation products are responsible for the inhibition of rgg expression.
Proteomics was used to identify several Rgg-regulated genes. It remains possible that additional exoproteins, not detected in this study, are regulated by Rgg. Although variation was periodically observed upon repeated analysis of culture supernatant proteins, MF, ORF 226, ORF 953, ClpB, autolysin, and lysozyme were selected for further study because their expression consistently differed between NZ131 speB and NZ131 rgg. Several exoproteins, including four that were detected in NZ131 speB supernatants but not in supernatants from NZ131 rgg, were not identified by peptide fingerprinting. The genomic database used to identify proteins by peptide fingerprinting was constructed primarily with nucleotide sequences from a serotype M1 genome sequencing project prior to its completion (Roe et al., online). Thus, the database did not contain all M1 ORFs. Moreover, NZ131 (serotype M49) may possess exoproteins not present in the M1 genome used to construct the database.
MF was more abundant in the stationary-phase supernatant of the rgg mutant compared to the speB mutant (Fig. 2). The mf gene is located proximal to rgg in the streptococcal chromosome. The intergenic region between mf and rgg is 240 bp in strain MGAS 8232 (serotype M18) (J. C. Smoot, unpublished data) and 241 bp in strain SF370 (serotype M1) (R. Overbeek, G. D. Pusch, M. Dsouza, N. Larsen, and E. Selkov, Functional overview of Streptococcus pyogenes, online [http://129.15.12.51:8080/WIT2/CGI/index.cgi?user=]); in both strains, the genes are divergently oriented. Promoter activity potentially contained within the heterologous DNA used to insertionally inactivate rgg is unlikely to have enhanced mf expression, because the insertion is downstream of the mf gene. Nonetheless, we cannot formally exclude the possibility that mf expression was altered by insertion of heterologous DNA into the rgg locus.
Function of Rgg-regulated proteins.
Four immunologically and electrophoretically distinct nucleases, designated DNases A, B, C, and D, have been previously identified in S. pyogenes supernatant fluids (45, 46). DNase D is encoded by the sdaD gene (35), and MF is thought to be identical to the protein previously described as DNase B (16). It is unclear if DNA entry nuclease (ORF 226) is identical to enzymes previously designated DNase A or C. ORF 226 is approximately 30% identical to several nucleases, including streptodornase (accession number X84793), EndA of Streptococcus pneumoniae (23), and MF (accession number D13428). In addition to ORF 226, RST00413 and RST0049 were also designated as DNA entry nucleases in the WIT2 analysis of an M1 streptococcal genome (Overbeek et al., online). However, in contrast to ORF 226, neither RST00413 nor RST00491 has an apparent signal sequence. Rgg thus regulates the expression of at least two of four extracellular nucleases described, suggesting that control of extracellular nuclease activity is an important component of the Rgg regulon.
Many gram-positive and gram-negative bacteria secrete nucleases. Although their function is unclear, it has been hypothesized that extracellular nucleases (i) provide phosphate, nitrogen, and carbon for catabolism following the transport of oligonucleotides and nucleotides to the cytoplasm, (ii) protect the bacterial cell against potentially mutagenic heterologous DNA, and (iii) contribute to host-pathogen interactions. Secreted nuclease activity among streptococci is primarily associated with S. pyogenes (44). This observation suggests that this enzyme activity is not an important component of streptococcal metabolism, since the activity is not conserved among related groups of streptococci that share many metabolic features. In addition, in the absence of evidence for natural DNA transformation, it seems unlikely that secreted nucleases are necessary to protect S. pyogenes from heterologous DNA, since the cell wall is an efficient barrier against DNA entry. Host mucus may contain significant amounts of DNA that can inhibit the adherence of microorganisms to epithelial cells. DNA is also present in pus, and secreted DNase may decrease the viscosity of pus and facilitate bacterial dissemination. As noted by Wannamaker (44), all strains of S. pyogenes have extracellular nuclease activity and produce significantly more activity compared to other groups of streptococci, suggesting that the activity may contribute to virulence. In this regard, the toxicity of the cytolethal distending toxin of Campylobacter jejuni was found to be dependent on its DNase activity (24). Nonetheless, the function and potential contribution to virulence of streptococcal exonucleases remain to be determined.
Autolysin and lysozyme were detected in the supernatant from stationary-phase cultures of NZ131 speB but not NZ131 rgg (Fig. 2). Bacterial peptidoglycan hydrolases are typically involved in cell wall turnover, cell separation, competence, and sporulation. Interestingly, their activity is often posttranslationally regulated by proteases. For example, in Enterococcus hirae, muramidase activity is activated by an extracellular protease (19). Similarly, an extracellular protease activates the ATL peptidoglycan hydrolase of Staphylococcus aureus (20). Alternatively, proteases may down regulate autolysin activity, as described for Bacillus subtilis (17) and exemplified by the degradation of the autolysin AcmA by the serine protease PrtP of L. lactis (3). The coordinate regulation of the extracellular protease SPE B and peptidoglycan hydrolases (autolysin and lysozyme) suggests a functional relationship.
Function of the Rgg regulon.
Exoproteins are often referred to as accessory gene products that are not essential for growth in nutrient-rich conditions. Typically expressed under conditions of stress, exoproteins are likely to be critical for survival in hostile environments, such as those encountered during infection. The comparison of ECPs in exponential- and stationary-phase culture supernatant fluids of NZ131 speB and NZ131 rgg showed that significantly more ECPs were produced in the stationary phase of growth (cf. Fig. 1 and 2). In addition, NZ131 produces SPE B in the stationary phase of growth in response to nutrient starvation (9), consistent with the general theme that many secreted proteins comprise a bacterial response to nutritional stress.
The expression of extracellular nuclease by Serratia marcescens is enhanced by induction of the SOS stress response (13). In addition, the extracellular thermonuclease of S. aureus is secreted at a higher level in a sigB mutant (22). SigB is a stationary-phase sigma factor involved in the cellular response to stress (5, 21). Similarly, we observed increased expression of MF (DNase B) and the putative DNase ORF 226 in the rgg mutant in the stationary phase of growth. Inactivation of rgg also resulted in decreased expression of autolysin, lysozyme, and ClpB. The ClpB heat shock proteins of Saccharomyces cerevisiae and Escherichia coli are necessary for survival at elevated temperatures (38, 39, 43). Peptidoglycan hydrolases such as autolysin and lysozyme are required for sporulation in B. subtilis (12), a developmental response induced, at least in part, by nutritional stress (40). Thus, Rgg regulates a variety of genes whose products potentially comprise a response to stress. It remains to be determined if the rgg mutant is deficient in responding to stress or in the detection and signaling of stressful conditions.
In conclusion, rgg was initially described as being required for speB expression (6, 31). The results described in the present study show that inactivation of the gene affects the expression of several extracellular proteins, some of which are likely to influence host-pathogen interactions. It remains unclear if Rgg acts directly or indirectly on each gene to alter expression. In addition, the influence of Rgg on the expression of a variety of secreted proteins may be due, at least in part, to the interaction of the Rgg regulon with additional regulatory networks. This hypothesis is currently being tested by DNA microarray analysis.
ACKNOWLEDGMENTS
We thank J. A. Carroll and J. R. Fitzgerald for critical review of the manuscript.
REFERENCES
- 1.Berge A, Björck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 2.Bernish B, van de Rijn I. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J Biol Chem. 1999;274:4786–4793. doi: 10.1074/jbc.274.8.4786. [DOI] [PubMed] [Google Scholar]
- 3.Buist G, Venema G, Kok J. Autolysis of Lactococcus lactis is influenced by proteolysis. J Bacteriol. 1998;180:5947–5953. doi: 10.1128/jb.180.22.5947-5953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns E H, Jr, Marciel A M, Musser J M. Activation of a 66-kilodalton human endothelial cell matrix metalloprotease by Streptococcus pyogenes extracellular cysteine protease. Infect Immun. 1996;64:4744–4750. doi: 10.1128/iai.64.11.4744-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan P F, Foster S J, Ingham E, Clements M O. The Staphylococcus aureus alternative sigma factor ςB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee M S, Ajdic D, Ferretti J J. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect Immun. 1999;67:1715–1722. doi: 10.1128/iai.67.4.1715-1722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaussee M S, Cole R L, van Putten J P M. Streptococcal erythrogenic toxin B abrogates fibronectin-dependent internalization of Streptococcus pyogenes by cultured mammalian cells. Infect Immun. 2000;68:3226–3232. doi: 10.1128/iai.68.6.3226-3232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee M S, Gerlach D, Yu C-E, Ferretti J J. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect Immun. 1993;61:3719–3723. doi: 10.1128/iai.61.9.3719-3723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussee M S, Phillips E R, Ferretti J J. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65:1956–1959. doi: 10.1128/iai.65.5.1956-1959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham M W. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Federle M J, McIver K S, Scott J R. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster S J. The role and regulation of cell wall structural dynamics during differentiation of endospore-forming bacteria. Soc Appl Bacteriol Symp Ser. 1994;23:25S–39S. doi: 10.1111/j.1365-2672.1994.tb04355.x. [DOI] [PubMed] [Google Scholar]
- 13.Guynn L J, Dai W, Benedik M J. Nuclease overexpression mutants of Serratia marcescens. J Bacteriol. 1998;180:2262–2264. doi: 10.1128/jb.180.8.2262-2264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heath A, DiRita V J, Barg N L, Engleberg N C. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henzel W J, Billeci T M, Stults J T, Wong S C, Grimley C, Watanbe C. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc Natl Acad Sci USA. 1993;90:5011–5015. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki M, Igarashi H, Yutsudo T. Mitogenic factor secreted by Streptococcus pyogenes is a heat-stable nuclease requiring His122 for activity. Microbiology. 1997;143:2449–2455. doi: 10.1099/00221287-143-7-2449. [DOI] [PubMed] [Google Scholar]
- 17.Jolliffe L K, Doyle R J, Streips U N. Extracellular proteases modify cell wall turnover in Bacillus subtilis. J Bacteriol. 1980;141:1199–1208. doi: 10.1128/jb.141.3.1199-1208.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapur V, Topouzis S, Majesky M W, Li L-L, Hamrick M R, Hamill R J, Patti J M, Musser J M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 19.Kariyama R, Shockman G D. Extracellular and cellular distribution of muramidase-2 and muramidase-1 of Enterococcus hirae ATCC 9790. J Bacteriol. 1992;174:3236–3241. doi: 10.1128/jb.174.10.3236-3241.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komatsuzawa H, Sugai M, Nakashima S, Yamada S, Matsumoto A, Oshida T, Suginaka H. Subcellular localization of the major autolysin, ATL and its processed proteins in Staphylococcus aureus. Microbiol Immunol. 1997;41:469–479. doi: 10.1111/j.1348-0421.1997.tb01880.x. [DOI] [PubMed] [Google Scholar]
- 21.Kullik I, Giachino P. The alternative sigma factor ςB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 22.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacks S, Neuberger M. Membrane location of a deoxyribonuclease implicated in the genetic transformation of Diplococcus pneumoniae. J Bacteriol. 1975;124:1321–1329. doi: 10.1128/jb.124.3.1321-1329.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lara-Tejero M, Galán J E. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290:354–357. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 25.Levin J C, Wessels M R. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Sledjeski D D, Kreikemeyer B, Podbielski A, Boyle M D P. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J Bacteriol. 1999;181:6019–6027. doi: 10.1128/jb.181.19.6019-6027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukomski S, Burns E H, Jr, Wyde P R, Podbielski A, Rurangirwa J, Moore-Poveda D K, Musser J M. Genetic inactivation of an extracellular cysteine protease (SpeB) expressed by Streptococcus pyogenes decreases resistance to phagocytosis and dissemination to organs. Infect Immun. 1998;66:771–776. doi: 10.1128/iai.66.2.771-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukomski S, Hoe N P, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou S J, Adams G G, Musser J M. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Investig. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon W R, Gibson C M, Caparon M G. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McIver K S, Scott J R. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordstrand A, McShan W M, Ferretti J J, Holm S E, Norgren M. Allele substitution of the streptokinase gene reduces the nephritogenic capacity of group A streptococcal strain NZ131. Infect Immun. 2000;68:1019–1025. doi: 10.1128/iai.68.3.1019-1025.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nordstrand A, Norgren M, Ferretti J J, Holm S E. Streptokinase as a mediator of acute post-streptococcal glomerulonephritis in an experimental mouse model. Infect Immun. 1998;66:315–321. doi: 10.1128/iai.66.1.315-321.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Podbielski A, Zarges I, Flosdorff A, Weber-Heynemann J. Molecular characterization of a major serotype M49 group A streptococcal DNase gene (sdaD) Infect Immun. 1996;64:5349–5356. doi: 10.1128/iai.64.12.5349-5356.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi F, Chen P, Caufield P W. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl Environ Microbiol. 1999;65:652–658. doi: 10.1128/aem.65.2.652-658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders J W, Leenhouts K, Burghoorn J, Brands J R, Venema G, Kok J. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol. 1998;27:299–310. doi: 10.1046/j.1365-2958.1998.00676.x. [DOI] [PubMed] [Google Scholar]
- 38.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 39.Squires C, Squires C L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauch M A, Hoch J A. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 41.Sulavik M C, Clewell D B. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J Bacteriol. 1996;178:5826–5830. doi: 10.1128/jb.178.19.5826-5830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sulavik M C, Tardif G, Clewell D B. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J Bacteriol. 1992;174:3577–3586. doi: 10.1128/jb.174.11.3577-3586.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki C K, Rep M, van Dijl J M, Suda K, Grivell L A, Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- 44.Wannamaker L W. Streptococcal deoxynucleases. In: Uhr J, editor. The streptococcus, rheumatic fever, and glomerulonephritis. Baltimore, Md: The Williams and Wilkins Co.; 1964. pp. 140–165. [Google Scholar]
- 45.Wannamaker L W, Hayes B, Yasmineh W. Streptococcal nucleases. II. Characterization of DNAse D. J Exp Med. 1967;126:497–508. doi: 10.1084/jem.126.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wannamaker L W, Yasmineh W. Streptococcal nucleases. I. Further studies on the A, B, and C enzymes. J Exp Med. 1967;126:475–496. doi: 10.1084/jem.126.3.475. [DOI] [PMC free article] [PubMed] [Google Scholar]