Highlights
-
•
The new informatic pipeline VICOS revealed 2% of SARS-CoV-2 coinfections during the first COVID-19 wave in Argentina.
-
•
VICOS output together with additional metadata help to differentiate between coinfection by 2 SARS-CoV-2, intra-host evolution or sequencing contamination.
-
•
SARS-CoV-2 inter- and intra-lineage coinfections need to be monitored as the driving force of recombination and evolution.
Keywords: SARS-CoV-2, Coinfection, Within-host, Intra-host, Evolution, Virus
Abstract
Introduction
Coinfection with two SARS-CoV-2 viruses is still a very understudied phenomenon. Although next generation sequencing methods are very sensitive to detect heterogeneous viral populations in a sample, there is no standardized method for their characterization, so their clinical and epidemiological importance is unknown.
Material and methods
We developed VICOS (Viral COinfection Surveillance), a new bioinformatic algorithm for variant calling, filtering and statistical analysis to identify samples suspected of being mixed SARS-CoV-2 populations from a large dataset in the framework of a community genomic surveillance. VICOS was used to detect SARS-CoV-2 coinfections in a dataset of 1,097 complete genomes collected between March 2020 and August 2021 in Argentina.
Results
We detected 23 cases (2%) of SARS-CoV-2 coinfections. Detailed study of VICOS's results together with additional phylogenetic analysis revealed 3 cases of coinfections by two viruses of the same lineage, 2 cases by viruses of different genetic lineages, 13 were compatible with both coinfection and intra-host evolution, and 5 cases were likely a product of laboratory contamination.
Discussion
Intra-sample viral diversity provides important information to understand the transmission dynamics of SARS-CoV-2. Advanced bioinformatics tools, such as VICOS, are a necessary resource to help unveil the hidden diversity of SARS-CoV-2.
Key points.
The new informatic pipeline VICOS revealed 2% of SARS-CoV-2 coinfections during the first COVID-19 wave in Argentina.
VICOS output together with additional metadata help to differentiate between coinfection by 2 SARS-CoV-2, intra-host evolution or sequencing contamination.
SARS-CoV-2 inter- and intra-lineage coinfections need to be monitored as the driving force of recombination and evolution.
Alt-text: Unlabelled box
Introduction
Genomic sequencing of SARS-CoV-2 has proven to be fundamental for detection and response to the pandemic by providing valuable information on the biology and evolution of the causal agent of COVID-19. According to the GISAID database (www.gisaid.org), more than 13 million full-length SARS-CoV-2 sequences have been generated up to October 1st, 2022, allowing real-time monitoring of the genomic diversity of SARS-CoV-2 around the world. The rapid implementation and sharing of laboratory protocols and bioinformatic analysis pipelines among the scientific community leveraged the use of high-throughput sequencing technologies for SARS-CoV-2 full-length sequencing. Of the different next-generation sequencing (NGS) technologies, Illumina was the most commonly adopted, accounting for 80% of the SARS-CoV-2 genomes [1]. While highly sensitive, NGS is usually analyzed using simple bioinformatic platforms that can readily provide a single consensus sequence but fail to detect and inform polymorphisms that occur at low frequency in the samples (also called sub-consensus or intra-host single nucleotide variations, iSNVs). In some cases, consensus sequences contain IUPAC letters that account for a “mixture” of nucleotides at any given position along the viral genome but population abundance and haplotypes are lost. Thus, the lack of easy-to-use bioinformatic pipelines applicable to laboratories in charge of genomic surveillance hinders the potential of NGS methods to unveil SARS-CoV-2 diversity and evolution. In addition, there is no consensus as to what the minor allele frequency threshold should be for identification of mixed populations of SARS-CoV-2 in clinical samples.
In COVID-19 infected patients, iSNVs can occur as a consequence of: i) coinfection or superinfection by more than one SARS-CoV-2 virus; ii) intra-host viral evolution during infection or; iii) laboratory cross-contamination or PCR amplification artifacts prior to sequencing. The simultaneous infection by two different lineages of SARS-CoV-2 was first proven soon after the start of the pandemic in a previously healthy 17-year-old female from Portugal that developed severe COVID-19 disease [2]. As opposed to coinfections, that may be facilitated by an environment of high exposure to multiple viruses, superinfections by SARS-CoV-2 are commonly associated to prolonged viral shedding and underlying conditions of the host such as immunosuppression [3]. In both cases, identification of the mixed SARS-CoV-2 populations is easier when viral divergence is high, but can be cumbersome to detect in settings of low genetic heterogeneity. Therefore, real frequency and factors conditioning coinfections/ superinfections by multiple SARS-CoV-2 in any given population remain largely unknown, and more studies are needed to determine their occurrence in the population.
SARS-CoV-2 is a positive-sense single-stranded RNA virus of the Coronaviridae family with a low evolutionary rate, estimated around 1.8×10−3 substitutions per site per year [4]. Because SARS-CoV-2 triggers an acute and usually self-limited respiratory infection with a median viral shedding of 16 days, within-host viral evolution is expected to be minimal. Importantly, a number of studies have shown presence of viral quasispecies in COVID-19 patients 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, and high intra-host SARS-CoV-2 evolution, [17] underscoring the role of iSNVs as molecular markers of viral population dynamics during SARS-CoV-2 infection. Identification of SARS-CoV-2 coinfections is more important considering the emergence of viral variants with potential immune escape and greater transmissibility, which the WHO has called variants of concern (VOC) or variants of interest (VOI), and the possibility of viral recombination between them [18]. Thus, the identification and study of iSNVs in patients with SARS-CoV-2 has great potential to identify mutations associated with the antibody escape as well as for monitoring viral spread, identifying susceptible populations and guiding evidence-based policies in public health.
In this work we present VICOS (Viral COinfection Surveillance), an integrated bioinformatic pipeline that allows the characterization of iSNVs in SARS-CoV-2 sequences obtained by Illumina technology. Through the analysis of 1,097 SARS-CoV-2 sequences from clinical samples obtained between March 2020 and August 2021 for genomic surveillance by the Argentine Inter-Institutional SARS-CoV-2 Genomic Consortium (Proyecto PAIS), VICOS allowed the identification and detailed characterization of mixed SARS-CoV-2 populations in 2% of the samples. Our results not only add a new powerful bioinformatic tool for viral genomic analysis but also provide important evidence of SARS-CoV-2 diversity and intra-host evolution.
Methods
VICOS: a novel bioinformatic pipeline for iSNV identification from Illumina NGS outputs
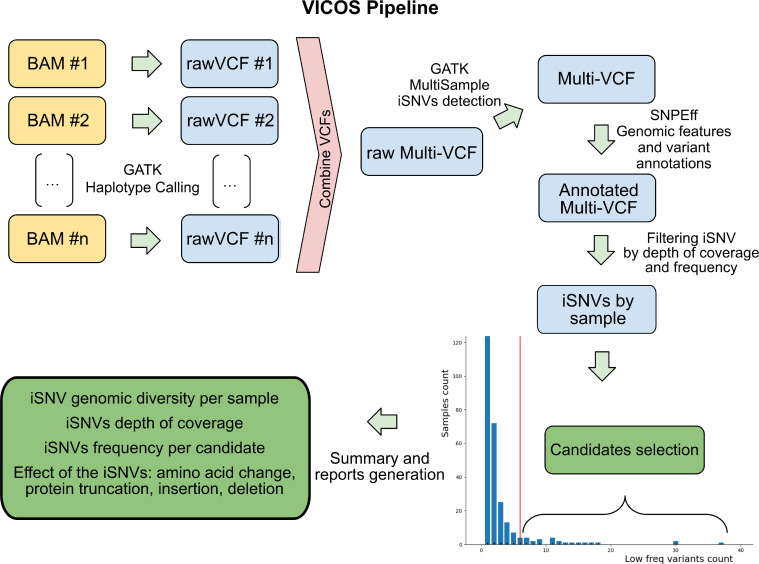
A new bioinformatic pipeline called VICOS was developed for the identification of viral genomic positions with iSNV in sequences obtained by Illumina technology. The input for the analysis is a set of BAM files generated from the mapping against a reference sequence. The output includes the generation of tables and graphics, helping the user to identify cases of suspected mixed viral populations (https://github.com/ProyectoPAIS/VICOS). VICOS analysis starts with the haplotype calling and VCF (Variant Call Format) files generation using GATK software [19] (Fig. 1). Then, the VCFs of all samples are joined into one single multi-sample VCF file, followed by a joint genotyping call considering the detection of more than one possible “allele”. Variant annotation and amino acid change annotation, when applicable, is performed with SNPEff [20]. Then, genome positions with more than one “allele” are kept considering a minimum depth of coverage of 10X and a minimum frequency of 20% for each nucleotide detected. Finally, in surveillance mode, VICOS calculates the distribution of samples by the amount of detected iSNVs and fits a Poisson distribution. That fitted distribution's lambda (λ) parameter, which represents its median value and standard deviation will be used as threshold to mark a sample for further analysis. This means that all samples with more iSNVs than this lambda value will be included as “candidates for mixed viral populations”. Subsequently, VICOS builds a set of charts and report files for every candidate with the genome position, frequency and depth of coverage of each iSNV (Fig. 1). When working with data that does not meet the assumption that most samples do not have mixed viral populations, the threshold can be set manually in the script.
Figure 1.
Algorithm of work used by VICOS to identify and report the number, frequency, and position of iSNVs in SARS-CoV-2 genomes obtained using Illumina technology. The pipeline requires a set of BAM files as input. The details on the algorithm of work are described in the article.
Evaluation of VICOS performance
In silico positive controls were generated using pooled ratios of FASTQ reads from the following SARS-CoV-2 sequences (GISAID accession numbers in Supplementary Material 1, GenBank SRA PRJEB56223): PAIS-A0830: lineage B.1.1.7, Alpha variant, 99.9% coverage, 1,611X mean depth; PAIS-A0910: lineage B.1.1.7, Alpha variant, 99.9% coverage, 2,446X mean depth; and PAIS-A0856: lineage P.1, Gamma variant, 99.9% coverage, 1,832X mean depth.
Genetic inter-lineage (PAIS-A0830 and PAIS-A0856 mixture) and intra-lineage (PAIS-A0830 and PAIS-A0910 mixture) coinfections were generated in silico from the FASTQ read ratio mixtures 10:90, 20:80, 30:70, 40:60 and 50:50. Each mixture was performed by 60 replicates for analyzing both the detection of iSNVs and the accuracy on the iSNV frequency. In addition, an in vitro control was generated from a 60:40 mixture of ARTIC v3 PCR products from two different clinical samples previously identified as lineage B.1.1.7 or Alpha variant (PAIS-A0828) and lineage P.1 or Gamma variant (PAIS-A0833) (GISAID accession numbers in Supplementary Material 1, GenBank SRA PRJEB56223). The mixed library was sequenced in an Illumina NextSeq 500 sequencer.
Mutations characterizing different lineages were obtained from cross-reference nucleotide markers using outbreak.info [21].
Clinical samples and SARS-CoV-2 sequencing
A total of 1,097 nasopharyngeal swabs from symptomatic patients testing positive for COVID-19 from March 2020 to August 2021 in Argentina underwent SARS-CoV-2 genome sequencing using ARTIC v3 protocol and Illumina technology (MiSeq and NextSeq 500 platforms). The FASTQ files were filtered by quality >Q30 and read length >50nt using Trimmomatic [22], and then mapped against the WIV04 reference (GISAID accession number EPI_ISL_402124) with BWA [23] software generating the BAM files.
The coverage range of the viral genomes was between 57% and 100% referred to the WIV04 reference sequence (Supplementary Fig. 1). The average depth of coverage was 812X for samples sequenced in the MiSeq and 2,077X for those sequenced in the NextSeq 500. Majority consensus genomes of all analyzed samples are shared in GISAID (Supplementary Material 1).
All samples analyzed in this work were produced by the SARS-CoV-2 genomic community surveillance in Argentina by the Argentine Inter-Institutional SARS-CoV-2 Genomic Consortium (PAIS Project) created by the Ministry of Science and Technology in 2020 (http://pais.qb.fcen.uba.ar/). All samples were anonymized prior to sampling procedure.
Phylogenetic analysis
Minority and majority consensus nucleotide sequences were reconstructed considering low-frequency (<50%) iSNVs reported by VICOS for the minority consensus, and high-frequency (>50%) for the majority consensus. A phylogenetic analysis was performed on a dataset of SARS-CoV-2 sequences including the majority and minority consensus sequences for the VICOS candidates, the majority consensus sequences of the non-candidate samples, the reference sequences according to the PANGO lineages and the ten sequences with the lowest number of SNPs for each majority/minority consensus sequence retrieved using AudacityInstant (GISAID on to June 1st, 2022).
The alignment was built with MAFFT v7.486 [24], inspected and edited with BioEdit (available at: https://bioedit.software.informer.com/), and the maximum likelihood tree was built with IQ-TREE v.2.1 [25]. The molecular evolution model was estimated with ModelFinder [26] and the reliability of sequences clusters was evaluated using UFBoot2 method (1000 replicates) [27] and the SH-approximate likelihood ratio test (1000 replicates) [28].
Results
VICOS performance
To evaluate the performance of VICOS when a minor viral population is present at different frequencies, we first performed in-silico mixtures of two SARS-CoV-2 genomes from different lineages (B.1.1.7 or Alpha variant, and P.1 or Gamma variant) that differ at 67 positions along the viral genome. The analysis of B.1.1.7:P.1 mixtures showed that only 1% of iSNVs could be detected by VICOS when the minority viral population represents 10% of the total sequences. Instead, while minority viral population frequency raises to 20% the average detection improved to 52.22% of the iSNVs (Supplementary Fig. 2a). When the minority viral frequency is at 30%, an average detection of 99.25% iSNVs was already obtained, and finally, viral frequencies at 40% and 50% resulted in a detection of 100% iSNVs. In addition to the iSNVs detection, the accuracy in the correct assignment of the minority and majority nucleotide was analyzed. As expected, 100% certainty in the iSNVs detected at viral ratios of 10:90, 20:80 and 30:70 was found, and slightly decreased for the 40:60 ratio (average certainty 94.77%) (Supplementary Fig. 2b). The assignment for the 50:50 ratio was not analyzed since no majority and minority nucleotide can be defined.
Next, we analyzed the performance of VICOS using two SARS-CoV-2 genomes of the same lineage B.1.1.7 or Alpha variant. In this case, the genomes differed at 16 nucleotide positions. As in the previous case, an average of 53.33% of iSNVs could be detected when present at a 20% frequency, and 93.75% of the iSNVs were detected when present at a 30% frequency (Supplementary Fig. 2c). Minor viral frequency of 40% resulted in 96.87% detection and a viral frequency of 50% had 100% of the iSNVs detected. The correct assignment of the minority and majority nucleotide decreased when the minor viral population frequency increased from 10% to 40%, as it was also seen in the inter-lineage in-silico coinfection (Supplementary Fig. 2d). Mixture ratios of 10:90 and 20:80 resulted in 100% correct assignment on the detected iSNVs; 30:70 ratio had an iSNVs detection certainty of 93.33% of the and 40:60 ratio, an 87.08%.
Last, we performed an invitro control by mixing PCR products from clinical samples previously known to carry an Alpha or Gamma SARS-CoV-2 variant. To study stochasticity of the laboratory work during the routine sample preparation, PCR products were mixed at a 60:40 ratio. From a total of 58 iSNVs expected from the genetic differences between the genomes, VICOS detected 54 (93%) and the correct assignment of iSNV frequencies was 59% (32/54 iSNVs detected).
Detection of mixed SARS-CoV-2 intra-sample population during the COVID-19 pandemic in Argentina
In order to investigate the presence of mixed SARS-CoV-2 intra-sample populations during the first 18 months of COVID-19 pandemic in Argentina, we used VICOS to analyze a total of 1,097 BAMs generated using Illumina sequencing platforms for genomic surveillance of SARS-CoV-2 in Argentina.
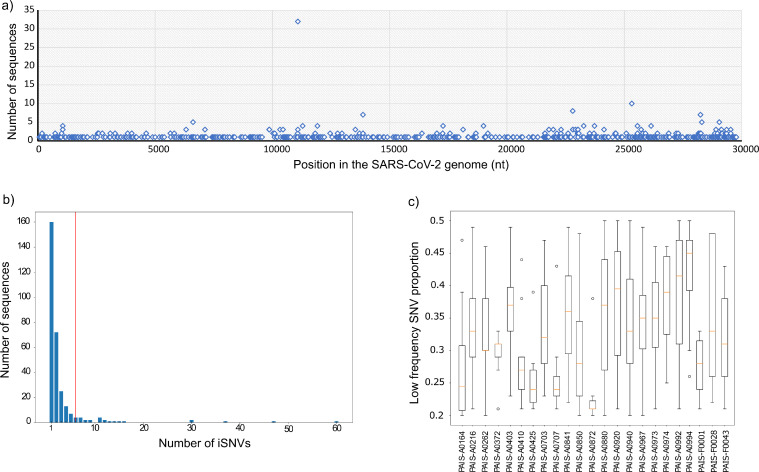
Results showed that 302 of the 1,097 analyzed samples (27.5%) presented at least 1 iSNV at any of the 567 positions along the SARS-CoV-2 genome in which they were detected (Supplementary Table 1, Fig. 2a). In most cases, iSNVs were detected in a single sequence. However, 100 iSNVs were shared between two to ten sequences. Interestingly, the same iSNV was found at the genomic position 11,083 (mixture of Thymine “t” and a nucleotide deletion) in 32 samples sequenced at different times and using different Illumina equipment.
Figure 2.
VICOS result for the analysis of genomic surveillance in Argentina. For a dataset of 1,097 whole genome sequences by Illumina collected from March 2020 to August 2021 the results on the number, frequency and position of iSNVs are shown. a) Number of sequences containing an iSNVs in a specific location at the SARS-CoV-2 genome. b) Distribution of sequences containing at least one iSNV in the dataset. The red line identifies the cutoff value defined by VICOS according to a Poisson distribution. C) Dispersion of the frequency values of iSNVs per sample, the boxes highlight 95% HDP and the red line indicates the mean.
Analysis of the iSNVs genomic location showed that while 25 cases were found in the intergenic region, 542 iSNV were located in the viral genes ORF1ab, S, ORF3a, E, M, ORF6, ORF7a, ORF7b, ORF8 and N. From those, 299 cases (55% of iSNV in viral genes) the iSNV was also reflected in an amino acid change. At the same time, 44 iSNVs comprised a nucleotide insertion or deletion generating a frameshift (located in viral genes ORF1ab, S, ORF3a, E, M, ORF8 and N), while in other 27 cases the insertion or deletion did not affect the open reading frame (viral genes ORF1ab, S, ORF3a, ORF7b and ORF8) (Supplementary Table 1). In addition, iSNVs in single sequences involving protein truncation were also found at genes ORF1ab (Gln283*, Glu1801*), S (Cys1254*), ORF3a (Gly254*) and ORF8 (Gln18*, Gln27*, Glu110*).
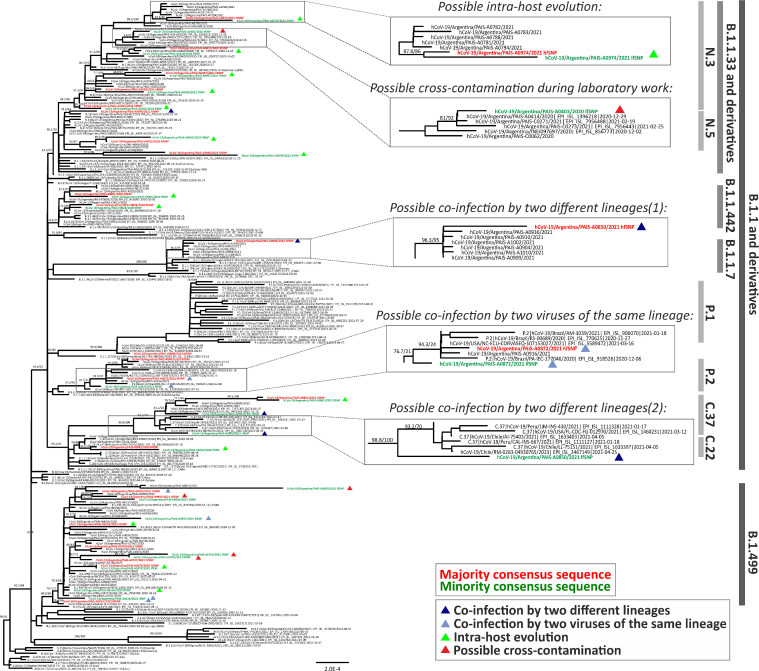
As expected, distribution of sequences according to the number of iSNVs showed that only a few sequences have a high number of iSNVs. After adjusting the data to a Poisson distribution, we used the VICOS calculated cut-off value to consider the candidates of mixed SARS-CoV-2 populations. For our dataset, VICOS determined a threshold of six iSNVs. Thus, 23 samples (2%) were considered to contain mixed SARS-CoV-2 populations. The number of detected iSNVs in the selected cases ranged from seven to 60 per sample (Fig. 2b and Table 1), and the mean iSNVs frequencies per sample ranged from 23% to 43% (Fig. 2c). To further characterize the mixed SARS-CoV-2 genomes, we reconstructed the majority and minority consensus viral sequences according to VICOS information and compared the phylogenetic relationship between them and to a set of 1,336 reference sequences downloaded from GISAID. Of the 23 candidates, 13 cases showed a high degree of similarity between majority SARS-CoV-2 genome sequence (with high frequency polymorphism, hfSNP) and minority sequence (with low frequency polymorphism, lfSNP) clustering monophyletically and suggesting possible within-host evolution (Table 1 and Fig. 3). At the remaining ten cases, the majority and minority consensus sequences clustered independently in the phylogenetic tree, suggesting possible coinfections of distantly related viruses. Further analysis of these ten cases revealed that the minority consensus sequence at five of them (PAIS-A0403, PAIS-A0703, PAIS-A0707, PAIS-A0880, PAIS-A0992) was closely related to another sequence from the genomic surveillance of the PAIS Project (Fig. 3). Internal laboratory records about which samples were processed simultaneously during the wet-lab experiments suggested that those five cases may represent possible laboratory contaminations. Thus, the remaining five cases with mixed SARS-CoV-2 populations were considered possible coinfections (PAIS-A0216, PAIS-A0850, PAIS-A0872, PAIS-0973, and PAIS-A0994). Two of them (PAIS-0850 and PAIS-0994) involved different SARS-CoV-2 lineages (C.37 + B.1.1.7, and C.37 + N.3, respectively), while the remaining three cases (PAIS-A0216, PAIS-A0872, and PAIS-0973) involved the same genetic lineage or a divergent sub-lineage (B.1.499 + B.1, P.2 + P.2, and B.1.499 + B.1.499, respectively) (Table 1).
Table 1.
VICOS candidates to SARS-CoV-2 coinfections. The 23 candidate samples identified by VICOS, including the number of intra-sample nucleotide variations (iSNV) detected, the proportion and average of depth of coverage for the low frequency iSNVs and the sample collection date. Based on phylogenetic analysis of the majority and minority consensus sequences reconstruction, the lineage assignment is informed as well as the classification of the samples when the majority and minority consensus sequences were monophyletic.
| Candidate Sample | Number of iSNVs | Mean proportion of Low-freqiSNVs | Depth of coverage of Low-freqiSNVs | Sample Collection Date (MM-YYYY) | High-freq lineage (VOC/VOI) | Low-freq lineage (VOC/VOI) | |
|---|---|---|---|---|---|---|---|
| Monophyletic consensus sequences | PAIS-A0164/2020 | 8 | 28% | 65.38X | 05-2020 | B.1.499 | B.1.499 |
| PAIS-A0262/2020 | 9 | 34% | 116.67X | 05-2020 | N.5 | N.5 | |
| PAIS-A0372/2020 | 7 | 29% | 140.57X | 08-2020 | B.1.499 | B.1.499 | |
| PAIS-A0410/2020 | 9 | 28% | 166.89X | 12-2020 | N.5 | N.5 | |
| PAIS-A0425/2020 | 11 | 26% | 279.09X | 11-2020 | N.3 | N.3 | |
| PAIS-A0841/2021 | 7 | 36% | 29.57X | 06-2021 | C.37 (Lambda) | C.37 (Lambda) | |
| PAIS-A0920/2021 | 12 | 37% | 588.5X | 01-2021 | N.5 | N.5 | |
| PAIS-A0940/2021 | 37 | 34% | 472.89X | 04-2021 | N.5 | N.5 | |
| PAIS-A0967/2021 | 14 | 35% | 360.71X | 12-2020 | N.5 | N.5 | |
| PAIS-A0974/2021 | 11 | 38% | 291.36X | 12-2020 | N.5 | N.5 | |
| PAIS-F0001/2020 | 11 | 27% | 103.91X | 07-2020 | B.1.1.442 | B.1.1.442 | |
| PAIS-F0028/2020 | 7 | 36% | 117.71X | 08-2020 | B.1.1.442 | B.1.1.442 | |
| PAIS-F0043/2020 | 13 | 32% | 122.85X | 04-2020 | C.22 | C.22 | |
| Non-monophyletic consensus sequences | Inter- SARS-CoV-2 lineage divergence | ||||||
| PAIS-A0403/2020* | 30 | 37% | 328.97X | 12-2020 | P.2 (Zeta) | N.5 | |
| PAIS-A0850/2021 | 47 | 29% | 251.26X | 06-2021 | B.1.1.7 (Alpha) | C.37 (Lambda) | |
| PAIS-A0880/2021* | 60 | 36% | 413.18X | 06-2021 | B.1.1 | B.1.499 | |
| PAIS-A0994/2021 | 30 | 43% | 799.67X | 05-2021 | N.3 | C.37 (Lambda) | |
| Intra-SARS-CoV-2 lineage divergence | |||||||
| PAIS-A0216/2020 | 7 | 34% | 66.57X | 05-2020 | B.1 | B.1.499 | |
| PAIS-A0703/2021* | 12 | 34% | 383.33X | 08-2020 | B.1.499 | B.1.499 | |
| PAIS-A0707/2021* | 11 | 26% | 266.45X | 09-2020 | B.1.499 | B.1.499 | |
| PAIS-A0872/2021 | 8 | 23% | 284.25X | 02-2021 | P.2 (Zeta) | P.2 (Zeta) | |
| PAIS-A0973/2021 | 15 | 35% | 339X | 11-2020 | B.1.499 | B.1.499 | |
| PAIS-A0992/2021* | 16 | 38% | 292.75X | 01-2021 | B.1.499 | B.1.499 | |
(*) possible laboratory cross-contaminations
Figure 3.
Phylogenetic analysis of SARS-CoV-2 suspected coinfection cases. The names of the sequences of the 23 “candidate” cases by VICOS are shown in red (majority consensus reconstruction, hfSNP) and green (minority consensus reconstruction, lfSNP). In black are shown the reference sequences downloaded from GISAID and the majority consensus sequences of the non-candidate samples analyzed (details in text). The support value (SH-aLRT/UFB) is shown only for selected groups of relevance. The color of the triangles next to the sequence name indicates blue, coinfection with two viruses of different SARS-CoV-2 lineage; light blue, coinfection with two viruses of the same SARS-CoV-2 lineage; green, possible intra-host evolution; red, possible laboratory cross-contamination. Examples of cases of each category found are shown in larger size, where in the case of coinfection by two different lineages, the pair of sequences is reported in different boxes with the numbers (1) and (2). The names of the various lineages and sub-lineages are described to the right of the phylogenetic tree.
The presence of iSNVs, including VOC/VOI amino acid markers defined by the WHO, were analyzed in all 23 candidates. Seven candidates with iSNVs involving VOC/VOI amino acid markers were detected, in three of them a significant number of markers was related with a particular VOC/VOI (Supplementary Table 1). In PAIS-A0850 sequence, 14 Alpha amino acid markers were detected at high frequency out of 19 reported for this VOC, and 10 Lambda markers were found at low frequency out of 19 reported for this VOI. The results were in agreement with the lineage classification of the majority (lineage B.1.1.7 – Alpha variant) and minority (C.37 – Lambda variant) consensus sequences of this sample (Table 1). Similarly, 12 out of 19 Lambda markers were detected at a low frequency in PAIS-A0994 sequence, in which the minority consensus sequence phylogenetically classified as C.37 lineage. Interestingly, despite monophyly of majority and minority consensus genomes in PAIS-A0940 sequence associated with N.5 lineage, 13 out of 22 Gamma amino acid markers were found, mainly at low frequency. It is important to note that this candidate has a remarkable amount of iSNVs (37 iSNVs) comparing with the other samples of the same monophyly category, which showed ≤14 iSNVs (Table 1).
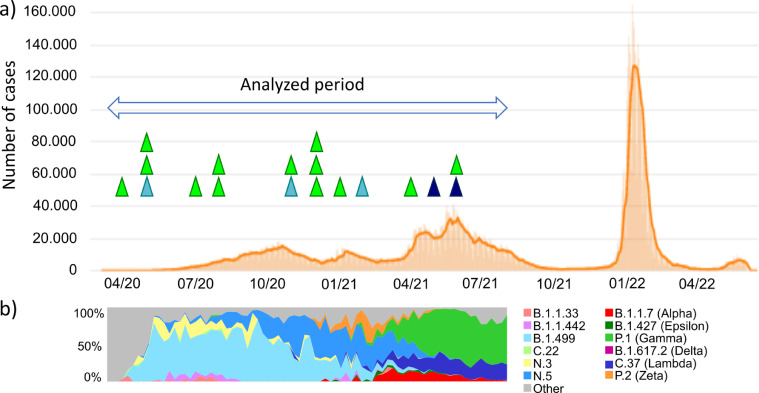
Lastly, we assessed the correlation between SARS-CoV-2 lineages detected as intra-sample mixed populations in the 23 sample candidates and the prevalence of the different lineages that circulated in Argentina during the analyzed period (Fig. 4). As in other countries, the pandemic in Argentina occurred as outbreaks or “waves” (Fig. 4a). The first wave occurred from 2020 to early 2021 when the largest number of cases of mixed populations with viruses of the same lineage were detected, together with most of the cases of mixed populations whose majority and minority consensuses were monophyletic in the phylogenetic tree. Finally, all lineages found in majority and minority consensus sequences matched with the lineages circulating at the time and place of clinical sample collection, discarding further motif of suspected contamination (Fig. 4b).
Figure 4.
Seasonality of SARS-CoV-2 coinfection cases in Argentina. a) Seasonality of SARS-CoV-2 cases from the beginning of the pandemic to June 2022, the arrow indicates the sample collection period analyzed with VICOS. Triangles mark the temporality of the suspected cases identified with VICOS: in blue the coinfections by two different lineages, in light blue the cases of intra-lineage coinfections, in green the undetermined cases categorized as intra-host evolution. b) Frequency of the SARS-CoV-2 lineages in which the reconstructed majority and minority sequences were associated for each suspected VICOS case. The seasonality of all the VOC/VOI circulating in the analyzed period is also indicated. Details on phylogenetic results are mentioned in the manuscript and Table 1.
Discussion
The detection of low frequency mutations in clinical samples has potential relevance both at the epidemiological level and in public health. However, there is no agreed method for their identification from NGS, thus low frequency mutations have been underestimated in comparison to the use of majority consensus viral genomes. In this study, we present VICOS, a novel and powerful open-source tool for the analysis, interpretation and quality control of SARS-CoV-2 sequences obtained by Illumina NGS platforms. This bioinformatic pipeline not only allows the detection and characterization of iSNVs but, most importantly, provides a list of suspected cases with mixed SARS-CoV-2 viral populations. We used VICOS to assess 1,097 SARS-CoV-2 sequences obtained by Illumina technology during a community epidemiological surveillance framework in Argentina over an 18-months period covering the first two waves of SARS-CoV-2 pandemic and the introduction of the first VOC/VOIs in the country. Detection of mixed SARS-CoV-2 populations in 2% of the cases highlights the importance of further characterizing mutations occurring at a sub-consensus level to further understand their role in SARS-CoV-2 evolution and adaptation within hosts and in the population.
Previous studies performed in-depth analysis of iSNVs on SARS-CoV-2 full-length genomic data during genomic surveillance to reveal important evolutionary aspects of SARS-CoV-2 associated with transmission bottlenecks [29,30], evolutionary dynamics of variant spectra in COVID-19 patients [7], and also to identify coinfections by SARS-CoV-2 viruses of different lineages [31]. While the basis of these analyses shared the same logic as VICOS, in most of the cases the workflow was not automated in a single pipeline, and a validated criterion to define the iSNV threshold was lacking. One of VICOS´s main features rely on the massive analysis of datasets of thousands of samples at the same time, the selection of candidate cases based on a statistical parameter, and an easy-to-interpret table and graphical output that contain all relevant details of iSNVs detected in candidate sequences -such as genome location, depth of coverage and amino acid change in the protein sequence-. Recently, a bioinformatic pipeline called ViralFlow was developed by Dezordi et al for identification of contaminations and coinfections by SARS-CoV-2 [32]. A few key differences distinguish VICOS from ViralFlow. First, VICOS was designed to work with samples a depth of 50, since a lot of our samples (and of those around the world) do not reach a depth of 100 per position (2000 in the sample) required by ViralFlow, but there is a trade off with the minor allele frequency, 20% in our case, and 5% in ViralFlow. According to our in vitro calibration, VICOS allows the detection of iSNVs minority variants when present at a frequency of 20% or higher, while minority variants occurring at a frequency of 10% or less are undetectable. The performance of VICOS for minority variants between 10% and 20% was not evaluated. Differences in the percentage of detection according to the frequency of the minority population mostly rely on the use of read depth as cutoff by the variant caller GATK for detection of iSNVs in SARS-CoV-2 genomes, an adaptation that produced a small sacrifice in sensitivity, but that did not alter the expected purpose of the tool. Second, ViralFlow uses an in-house script for iSNV calling, while VICOS uses GATK providing a more complex and widely tested probabilistic framework. Third, ViralFlow output directly generates minority and majority consensus sequences together with PANGO lineage classification while VICOS focuses only on the detection and detailed description of iSNVs present in the samples. As shown in our study, also in a study of 1.462 SARS-CoV-2 Brazilian sequences using ViralFlow [33], inter-lineage coinfections still require confirmation by phylogenetic challenge analysis and visual inspection.
VICOS can be applied to monitor and quantify the presence of iSNVs of interest in a population, to evaluate the frequency of iSNVs in longitudinal samples, detect laboratory cross-contamination, or quantitatively compare the distribution of iSNVs between different epidemiological outbreaks, among other uses. The sensitivity assessment of VICOS pipeline showed that low frequency iSNVs of minor viral populations have low probability of being detected but, at the same time, those iSNVs have higher probability of correct assignment as minority or majority variant. Thus, VICOS criteria of considering a minimum frequency of 20% and depth of coverage higher than 10X ensures to identify the majority of the iSNVs in a sample with the minimum frequency possible. As expected, within-host viral populations frequencies close to 50% may mix up majority and minority iSNVs. VICOS generates reports per sample indicating the individual iSNVs frequencies, providing the user all the information associated to sequence diversity either to be used directly, or to facilitate further ad-hoc studies. We hypothesize that the errors in iSNVs detection and frequency assignment are related with either low sequencing depth of coverage or “dropout” during sequencing, and GATK variant calling interpreting the position as an error. In addition, during the wet-lab experiments the sample treatment may also bias the original frequency of iSNVs, for example due to differences in the annealing of specific primers in within-host viral populations during genome amplification.
In our study of 1,097 samples collected in Argentina from March 2020 to August 2021 we detected 23 cases (2%) carrying more than 6iSNVs - the number of iSNVs calculated by VICOS from the input sequence data-. This frequency of mixed SARS-CoV-2 populations is in agreement with a previous report conducted on 1,313 samples obtained during the first wave of COVID-19 in the United Kingdom that estimated a SARS-CoV-2 coinfection rate of 1-2% [29]. In that study, the authors were unable to distinguish between real coinfections and laboratory cross-contaminations, leading to a possible overestimation of the coinfection rate. Due to the availability of internal experimental reports during sequencing, and phylogenetic analysis of minority or majority consensus sequences with SARS-CoV-2 sequences obtained by PAIS Project in the same day or sequencing run, we were able to distinguish possible cross-contaminations in five of the 23 samples with high number of iSNVs, thus restricting the coinfections to 1.6%. Of the remaining 18 cases with significant intra-sample viral diversity, 2 represented coinfections with viruses of different lineages, and 3 represented coinfections with viruses of the same lineage. We were unable to distinguish between same-lineage coinfections and intra-host viral evolution in the remaining 13 cases that presented a monophylogenetic relationship between minority and majority consensus sequences. Unfortunately, information related to the patient´s clinical and immune status and time of symptoms onset was unavailable to us in these cases.
The epidemiological information about COVID-19 pandemic in Argentina supported the lineage assignment of the VICOS candidates, i.e., the lineages detected coincided with the lineages circulating in the population at the time of sample collection. The first wave in Argentina occurred prior to the massive availability of vaccines (between March 2020 and December 2020) affecting 1.25 million people. During this period, the molecular epidemiology of SARS-CoV-2 was characterized by a low genetic diversification and with circulation of lineages highly similar to the index virus. In this setting, new mutations are expected to accumulate at a rate of approximately 25–30 mutations per lineage, per year [34]. However, at the end of 2020 new lineages of SARS-CoV-2 emerged worldwide, including the VOC/VOI variants that were introduced to Argentina during the first months of 2021 [35]. These drove thesecond wave of COVID-19, which affected 2.75 million people. During this period, we found two SARS-CoV-2 coinfections involving at least one VOC/VOI (Alpha + Lambda, N.3 + Lambda). As expected, these cases showed a high number of iSNVs (47 and 30, respectively) and are usually easier to identify even by non-sequencing methods such as real time qPCR specific for SARS-CoV-2 VOCs [36]. In light of recent findings that show that 2.7% of SARS-CoV-2 genomes have detectable recombinant ancestry [37], our results further highlight the importance of recognizing SARS-CoV-2 coinfections in real-time to monitor the spread of emerging recombinant variants with improved transmission efficiency and ability to evade immune response.
In summary, detection of low frequency SARS-CoV-2 viral populations is of increasing epidemiological and clinical importance. With VICOS development we demonstrated that the power of NGS has much more to offer than the usual majority consensus genomes. Here we easily showed the detection of SARS-CoV-2 coinfections in the genomic surveillance framework. Comprehensive analysis with bioinformatic tools such as VICOS provides insights into the evolution and adaptation of SARS-CoV-2.
Author statements
We declare all authors made substantial contributions to the work.
Supplementary Material
Supplementary Figure 1. Coverage depth profiles of the 23 candidate coinfection samples selected by VICOS. The light blue bars show the depth of coverage of each position of the SARS-CoV-2 genome per sample. The red lines below each profile indicate the regions with a depth of coverage less than 50X. The organization of the viral genome is indicated at the top of the figure.
Supplementary Figure 2. Sensitivity analysis of VICOS pipeline.In silicocoinfection was created from a mixture of FASTQ files of two SARS-CoV-2 whole genome sequencing by Illumina. Mixture ratios (10:90, 20:80, 30:70, 40:60 and 50:50) were replicated 60 times for the assessment. Analysis of percentage of iSNVs detected in an inter-lineage and intra-lineage coinfection are shown in a) and c) respectively. Analysis of correct assignment of majority and minority iSNV when detected for inter-lineage and intra-lineage coinfections are shown in b) and d) respectively. In each case the diameter of the circle informs the number of replicas.
Supplementary Table 1. Complete iSNVs detected by VICOS. The location in the genome of each iSNV is reported together with the nucleotide change, the detection of the iSNV in any of the candidate samples by VICOS, the detection of the iSNV in any of the non-candidate samples by VICOS, the location in a SARS-CoV-2 viral gene, the nucleotide, and amino acid change where appropriate.
Supplementary Material 1. GISAID accession numbers of the majority consensus sequences analyzed by VICOS.
Declaration of Competing Interest
The authors declare no competing interest or personal relationship that might have affected the work reported in this paper.
Acknowledgments
Funding
This work was supported by Ministry of Science, Technology and Innovation, Argentina [IP COVID-19 N°08]; and MERCOSUR Structural Convergence Fund [Focem COF 03/11 Covid-19]
Acknowledgement
We would like to thank all healthcare workers for their hard work, especially during the COVID-19 pandemic. We would especially thank the laboratories that have been and continue to be part of the Argentine Inter-Institutional SARS-CoV-2 Genomic Consortium(PAIS Project) for their dedicated effort. We gratefully acknowledge the authors from the Originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which part of this research is based (a list of accession numbers of the sequences used, with information of the contributions of the submitting and the originating laboratories, can be retrieved through the Data Acknowledgement Locator at https://www.gisaid.org with ID EPI_SET_20220602zx).
Footnotes
(http://pais.qb.fcen.uba.ar/)
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2022.199035.
Contributor Information
Paula Aulicino, Email: paulicino@garrahan.gov.ar.
Mariana Viegas, Email: viegasmariana@conicet.gov.ar.
Appendix. Supplementary materials
Data availability
Data will be publicly available in EBI/NCBI accession PRJEB56223 upon when the publication is released. Also the code and scripts used in the project are in: https://github.com/ProyectoPAIS/VICOS
References
- 1.Chen Z, Azman AS, Chen X, et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet. 2022;54:499–507. doi: 10.1038/s41588-022-01033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedro N, Silva CN, Magalhães AC, et al. Dynamics of a Dual SARS-CoV-2 Lineage Co-Infection on a Prolonged Viral Shedding COVID-19 Case: Insights into Clinical Severity and Disease Duration Dynamics of a Dual SARS-CoV-2. Microorganisms. 2021;9:300. doi: 10.3390/microorganisms9020300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarhini H, Recoing A, Bridier-Nahmias A, et al. Long-Term Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infectiousness Among Three Immunocompromised Patients: From Prolonged Viral Shedding to SARS-CoV-2 Superinfection. J Infect Dis. 2021;223:1522–1529. doi: 10.1093/infdis/jiab075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Wang W, Zhao X, et al. Transmission dynamics and evolutionary history of 2019-nCoV. J Med Virol. 2020;92:501–511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.al Khatib HA, Benslimane FM, Elbashir IE, et al. Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities. Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.575613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen Z, Xiao Y, Kang L, et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin Infect Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Wang D, Zhang L, et al. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021;13:30. doi: 10.1186/s13073-021-00847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andres A, Garcia-Cehic D, Gregori J, et al. Naturally occurring SARS-CoV-2 gene deletions close to the spike S1/S2 cleavage site in the viral quasispecies of COVID19 patients. Emerg Microbes Infect. 2020;9:1900–1911. doi: 10.1080/22221751.2020.1806735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jary A, Leducq V, Malet I, et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin Microbiol Infect. 2020;26(1560):e1–1560. doi: 10.1016/j.cmi.2020.07.032. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capobianchi MR, Rueca M, Messina F, et al. Molecular characterization of SARS-CoV-2 from the first case of COVID-19 in Italy. Clin Microbiol Infect. 2020;26:954–956. doi: 10.1016/j.cmi.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rueca M, Bartolini B, Gruber CE, et al. Compartmentalized Replication of SARS-Cov-2 in Upper vs. Lower Respiratory Tract Assessed by Whole Genome Quasispecies Analysis. Microorganisms. 2020;8:1302. doi: 10.3390/microorganisms8091302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karamitros T, Papadopoulou G, Bousali M, et al. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies. J Clin Virol. 2020;131 doi: 10.1016/j.jcv.2020.104585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Gonzalez B, Vazquez-Sirvent L, Soria ME, et al. Vaccine breakthrough infections with SARS-CoV-2 Alpha mirror mutations in Delta Plus, Iota, and Omicron. J Clin Investig. 2022;132 doi: 10.1172/JCI157700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Gonzalez B, Soria ME, Vazquez-Sirvent L, et al. SARS-CoV-2 Point Mutation and Deletion Spectra and Their Association with Different Disease Outcomes. Microbiol Spectr. 2022;10 doi: 10.1128/spectrum.00221-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Gonzalez B, Soria ME, Vazquez-Sirvent L, et al. SARS-CoV-2 Mutant Spectra at Different Depth Levels Reveal an Overwhelming Abundance of Low Frequency Mutations. Pathogens. 2022;11:662. doi: 10.3390/pathogens11060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun F, Wang X, Tan S, et al. SARS-CoV-2 Quasispecies Provides an Advantage Mutation Pool for the Epidemic Variants. Microbiol Spectr. 2021;9 doi: 10.1128/spectrum.00261-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baang JH, Smith C, Mirabelli C, et al. Prolonged Severe Acute Respiratory Syndrome Coronavirus 2 Replication in an Immunocompromised Patient. J Infect Dis. 2021;223:23–27. doi: 10.1093/infdis/jiaa666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson B, Boni MF, Bull MJ, et al. Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic. Cell. 2021;184:5179–5188. doi: 10.1016/j.cell.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsueng G, Mullen JL, Alkuzweny M, et al. Outbreak.info Research Library: A standardized, searchable platform to discover and explore COVID-19 resources. bioRxiv. 2022 doi: 10.1101/2022.01.20.477133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minh BQ, Schmidt HA, Chernomor O, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoang DT, Chernomor O, von Haeseler A, et al. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guindon S, Dufayard J-F, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 29.Lythgoe KA, Hall M, Ferretti L, et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372:eabg0821. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonkin-Hill G, Martincorena I, Amato R, et al. Patterns of within-host genetic diversity in SARS-CoV-2. Elife. 2021;10:e66857. doi: 10.7554/eLife.66857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francisco R da S, Benites LF, Lamarca AP, et al. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2021;296 doi: 10.1016/j.virusres.2021.198345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dezordi FZ, Neto AM da S, Campos T de L, et al. ViralFlow: A Versatile Automated Workflow for SARS-CoV-2 Genome Assembly, Lineage Assignment, Mutations and Intrahost Variant Detection. Viruses. 2022;14:217. doi: 10.3390/v14020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dezordi FZ, Resende PC, Naveca FG, et al. Unusual SARS-CoV-2 intrahost diversity reveals lineage superinfection. MicrobGenom. 2022;8 doi: 10.1099/mgen.0.000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balloux F, Tan C, Swadling L, et al. The past, current and future epidemiological dynamic of SARS-CoV-2. Oxf Open Immunol. 2022;3:iqac003. doi: 10.1093/oxfimm/iqac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torres C, Mojsiejczuk L, Acuña D, et al. Cost-Effective Method to Perform SARS-CoV-2 Variant Surveillance: Detection of Alpha, Gamma, Lambda, Delta, Epsilon, and Zeta in Argentina. Front Med (Lausanne) 2021;8 doi: 10.3389/fmed.2021.755463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pisano MB, Sicilia P, Zeballos M, et al. SARS-CoV-2 genomic surveillance enables the identification of Delta/Omicron coinfections in Argentina. Front Virol (Lausanne) 2022;2 [Google Scholar]
- 37.Turakhia Y, Thornlow B, Hinrichs A, et al. Pandemic-scale phylogenomics reveals the SARS-CoV-2 recombination landscape. Nature. 2022;609:994–997. doi: 10.1038/s41586-022-05189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be publicly available in EBI/NCBI accession PRJEB56223 upon when the publication is released. Also the code and scripts used in the project are in: https://github.com/ProyectoPAIS/VICOS