Abstract
Introduction
Nitrogen (N) is essential for plant growth and yield production and can be taken up from soil in the form of nitrate or peptides. The NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) genes play important roles in the uptake and transportation of these two forms of N.
Methods
Bioinformatic analysis was used to identify and characterize the NPF genes in Setaria. RNA-seq was employed to analyze time-series low nitrate stress response of the SiNPF genes. Yeast and Arabidopsis mutant complementation were used to test the nitrate transport ability of SiNRT1.1B1 and SiNRT1.1B2.
Results
We identified 92 and 88 putative NPF genes from foxtail millet (Setaria italica L.) and its wild ancestor green foxtail (Setaria viridis L.), respectively. These NPF genes were divided into eight groups according to their sequence characteristics and phylogenetic relationship, with similar intron-exon structure and motifs in the same subfamily. Twenty-six tandem duplication and 13 segmental duplication events promoted the expansion of SiNPF gene family. Interestingly, we found that the tandem duplication of the SiNRT1.1B gene might contribute to low nitrogen tolerance of foxtail millet. The gene expression atlas showed that the SiNPFs were divided into two major clusters, which were mainly expressed in root and the above ground tissues, respectively. Time series transcriptomic analysis further revealed the response of these SiNPF genes to short- and long- time low nitrate stress. To provide natural variation of gene information, we carried out a haplotype analysis of these SiNPFs and identified 2,924 SNPs and 400 InDels based on the re-sequence data of 398 foxtail millet accessions. We also predicted the three-dimensional structure of the 92 SiNPFs and found that the conserved proline 492 residues were not in the substrate binding pocket. The interactions of SiNPF proteins with were analyzed using molecular docking and the pockets were then identified. We found that the SiNPFs NO3 − binding energy ranged from-3.4 to -2.1 kcal/mol.
Discussion
Taken together, our study provides a comprehensive understanding of the NPF gene family in Setaria and will contribute to function dissection of these genes for crop breeding aimed at improving high nitrogen use efficiency.
Keywords: Setaria, nitrate/peptide transporter, expression profile, natural variation, three-dimensional structure, NRT1.1, low nitrogen tolerance
Introduction
Nitrogen (N) is an essential macronutrient for plant growth and crop production. Over the past half-century, global food production has increased considerably, which relies heavily on the application of N fertilizer (Liu et al., 2013; Li et al., 2017). It is estimated that over 120 Mt of N fertilizer is used annually and only approximately 30-40% of the applied N fertilizer can be used by crop plants (Raun and Johnson, 1999; Li et al., 2017). The excessive use of N fertilizers coupled with the low use efficiency of crops not only increases the cost of farmers but also causes severe environmental and ecological pollution (Guo et al., 2010; Xu et al., 2012). A better understanding of N uptake and transportation within the plant is crucial for molecular breeding for crops with high nitrogen use efficiency (NUE) and the development of sustainable agriculture (Xu et al., 2012).
In aerated soils, plants take up N mainly in the form of nitrate due to nitrification (Xu et al., 2012). To ensure an efficient up take over a wide range of external nitrate concentrations, plants have evolved two different nitrate transport systems, the low-affinity transport system (LATS) and the high-affinity transport system (HATS) (Edith Laugier et al., 2012). The LATS enables nitrate uptake in high (> 0.5 mM) external nitrate concentration, while the HATS allows nitrate absorption at low (< 0.5 mM) external nitrate concentrations (Crawford and Glass, 1998). The NITRATE TRANSPOTER 1 (NRT1)/PTR FAMILY (NPF) and NRT2 are responsible for LATS and HATS, respectively, except for AtNPF6.3 (also known as CHL1 or NRT1.1) that displays a dual affinity property (Liu et al., 1999). In addition to NPF and NRT2 genes, chloride channel (CLC) and slowly activating anion channel/homologues (SLAC1/SLAH) family genes also participate in nitrate transport (Krapp et al., 2014). Among these nitrate transporter families, NPF is the largest one and plays multifunctional roles in nitrate uptake and transport throughout the plant body (Wang et al., 2012).
During the last three decades, a great number of NPF genes were identified and extensively characterized in the model plants, especially in Arabidopsis and rice (Léran et al., 2014; Drechsler et al., 2018). Up to now, at least 53 NPF genes were identified in Arabidopsis, of which 19 were demonstrated to be involved in nitrate uptake and transport ( Supplementary Table 1 ). The AtNRT1.1 and AtNRT1.2 transporters localized on the plasma membrane of the root epidermis, cortex or endothelial cell absorb nitrate from the outside (Tsay et al., 1993; Huang et al., 1999). AtNRT1.1 was the first identified nitrate transport gene in Arabidopsis, which functions as a dual-affinity nitrate transport in the high and low affinity ranges (Tsay et al., 1993; Liu et al., 1999). The dual-affinity nitrate transport activity is achieved through the phosphorylation and dephosphorylation of threonine at position 101 (Parker and Newstead, 2014). This precise regulation mechanism enables plants to quickly switch between high-affinity and low-affinity systems to adapt to the varying levels of nitrate supply in the soil. While Nitrate Excretion Transport 1 (NAXT1), another member of the NRT1 family, can induce nitrate to flow out of the root under acidic conditions, thereby removing excess nitrate from the plants (Segonzac et al., 2007). The nitrate absorbed by the root system is further transported to the plant stem by AtNRT1.5, AtNRT1.8 and AtNRT1.9 (Lin et al., 2008; Li et al., 2010b; Wang and Tsay, 2011). In the later stages of plant growth and development, AtNRT1.7, AtNRT1.11 and AtNRT1.12 are responsible for the transport of nitrate from mature organs to young tissues and reproductive organs for reuse (Hsu and Tsay, 2013; Liu et al., 2017). The nitrate redistribution mediated by these transporters plays an important role in improving nitrogen use efficiency under low nitrogen conditions. In contrast to Arabidopsis, there are more NRT/PTR members in rice indicating that nitrate uptake and transport in rice are regulated more precisely. There are three putative homologs of AtNRT1.1 were identified in rice: OsNRT1.1A (OsNPF6.3), OsNRT1.1B (OsNPF6.5), and OsNRT1.1C (OsNPF6.4) (Plett et al., 2010). Overexpression of OsNRT1.1A conferred high NUE, high yield and early maturation in rice, thus providing a target to produce high yield and early maturation simultaneously (Wang et al., 2018b). Another NRT1.1 gene, OsNRT1.1B, also plays an important role in nitrate utilization in rice. A single polymorphism in this gene contributes to the long-noted divergence in NUE between indica and japonica subspecies of Asian cultivated rice (Hu et al., 2015). In addition to the function of nitrate transporter and sensor, OSNRT1.1B is also involved in the establishment of the rice root microbiota and contributes to the divergence of the root microbiota between indica and japonica subspecies (Zhang et al., 2019). Recently, a rare variant of OsNPF6.1, OsNPF6.1HapB was identified that contributes to high NUE and can be trans-activated by OsNAC42, another NUE-related gene (Tang et al., 2019). In addition to the model plants, the NPF genes were also identified and characterized in several other species including sugarcane and coffee (Santos et al., 2017; Wang et al., 2019). These NPF genes provides valuable resources for high NUE and yield breeding.
The Setaria system contains two important C4 Panicoid grass species, namely wild green foxtail (S Setaria viridis L.) and its cultivated cousin foxtail millet (Setaria italica L.) (Kellogg, 2017). These two species demonstrate a wide capacity for adaptation. Green foxtail is one of the most widespread weeds on the planet; while foxtail millet is one of the most resilient cereal crops with high NUE that can grow on marginal land with minimal agricultural inputs. However, the molecular mechanisms underlying the barren tolerance in Setaria remain largely unexplored. Although a unified nomenclature of NPF genes in plants has been reported (Léran et al., 2014), this gene family has not been studied thoroughly in the barren-tolerant crop foxtail millet and its wild ancestor green foxtail. Recently, we developed a mini-plant (dubbed xiaomi) of foxtail millet with an extremely short life cycle of two months and produced a high-quality genome sequence by combining PacBio single molecule real-time sequencing chromosome conformation capture sequencing technology (http://sky.sxau.edu.cn/MDSi.htm) (Yang et al., 2020b). In this study, we carried out a BLAST search using Arabidopsis and rice NPFs against the newly available xiaomi genome and the A10 green foxtail genome (Bennetzen et al., 2012), and identified 92 and 88 NPF genes from Xiaomi and A10, respectively. We further performed a comprehensive analysis of these NPF genes including expression pattern, response to low nitrate, and natural variation and domestication. As the first systematic study of the NPF genes in foxtail millet, our results will provide a valuable information for selecting candidate genes to improve crop NUE and further investigating the function of NPFs in foxtail millet.
Materials and methods
Plant materials and low nitrate treatment
For low nitrate treatment, seeds of xiaomi, a fast life cycle mini-foxtail millet, were germinated on moist sponge in the auto-controlled growth chamber under 28°C/22°C day/night cycle with a 14 h photoperiod. Seven-day-old, uniform size and healthy seedlings were selected and transferred to a tank containing full nutrient solution. The full nutrient contained 2 mM KNO3, 0.25 mM KH2PO4, 0.75 mM K2SO4, 0.65 mM MgSO4, 2.0 mM CaCl2, 0.2 mM Fe-EDTA, 1 μM MnSO4, 1 μM ZnSO4, 0.1 μM CuSO4, 0.005 μM Na2MO7O4 and 1 μM H3BO4. The solution was renewed every 3 days. On the tenth day, seedlings were transferred to continuous light condition to avoid the effect of photoperiod on gene expression. After 5-day culture, the seedlings were treated with low nitrate solution containing 0.2 mM KNO3. To avoid potential potassium deficiency due to the low concentration of KNO3 solution, KCl was added to the solution to maintain K level same as those under the higher KNO3. Each sample had three biological replications. The roots and shoots were separately harvested at the designated time point.
Identification of NPF genes and analysis of their physical and chemical properties
The NPF protein sequences of Arabidopsis and rice (Léran et al., 2014) were retrieved from TAIR (https://www.arabidopsis.org/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), respectively. These protein sequences were then blasted against the xiaomi genome database (http://sky.sxau.edu.cn/MDSi.htm) using BLASTP algorithm with an E value< 10-10 to retrieve putative NPFs. In addition, HMMER program was used to identify NPF genes in xiaomi. The Hidden Markov Model (HMM) profile of PTR (PF00854), a conserved domain of NPF proteins, were obtained from pfam 32.0 (http://pfam.xfam.org/). Then the hmmsearch program in HMMERv3.1b2 package (http://hmmer.org/) was used to search for proteins containing PTR domain with “trusted cutoff” as threshold (E-value<10-10) based on the local xiaomi protein database. The members of NPF proteins identified using BLAST and HMMER were merged as candidate NPF proteins in xiaomi. The conserved PTR domain of these candidate NPF proteins was further detected using SMART (http://smart.embl.de/smart/set_mode.cgi?NORMAL=1) and NCBI-CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and proteins without PTR domain were removed. The genomic and CDS sequences of NPF genes were downloaded from our MDSi database (http://sky.sxau.edu.cn/MDSi.htm). These NPF genes were named using the previous nomenclature (Wang et al., 2018a): SiNPFX.Y and SvNPFX.Y for foxtail millet and green foxtail millet, respectively. Si and Sv were the Latin abbreviation for species; X represented for the subfamily, Y for a specific number within the subfamily.
Homology analysis and phylogenetic tree construction
The MCScanX program was used to analyze homology genes among foxtail millet (xiaomi), green foxtail, rice and Arabidopsis, and the result was visualized using CIRCOS software (http://circos.ca/). To investigate the evolutionary relationship of NPF proteins between different species, the full-length NPF amino acid sequences of Arabidopsis, rice, and foxtail millet were aligned by ClustalW in MEGA X with default parameters (https://www.megasoftware.net/). Then, an unrooted neighbor-joining phylogenetic tree was constructed using Treeview.
Chromosome location, gene duplication and synteny analysis
The chromosome location data of NPF genes were obtained from xiaomi genomic database (http://sky.sxau.edu.cn/MDSi.htm) and then mapped to the chromosomes with Mapchart (https://www.wur.nl/en/show/Mapchart.htm). The genes failed to be mapped to chromosomes were not shown in the image. Gene duplication event was analyzed using the Multicollinearity Scanning Toolkit (McScanx) server with default parameters. KAKS_Calculator 2.0 was used to calculate the nonsynonymous substitution (ka) and synonymous substitution (ks) for each repeat NPF gene. In order to show the homologous relationship of homologous NPF genes, the homologous analysis map was drawn by Dual Synteny Plotter software (https://github.com/cj-chen/tbtools).
Gene structure, motif and cis-acting elements analysis
The NPF exon-intron structures were analyzed using the web-based gene structure display server 2.0 (http://gsds.cbi.pku.edu.cn/). The conserved motifs were analyzed using the online MEME tool (http://meme-suite.org/tools/meme) with the default parameters except that the maximum number of motifs was 10. For promoter analysis, 2.0 kb upstream sequence from the initiation codon (ATG) of each NPF was truncated, and then submitted to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict cis-acting elements.
Expression analysis of SiNPF genes in xiaomi
The expression data of 11 different tissues covering the entire lifecycle of xiaomi were used to monitor the expression of the above NPF genes, including the 3 d imbibed seeds (seed), 2-week-old whole seedling (seedling), root, stem, the top first fully extended leaf of 2-week-old seedling (leaf 1), the top second leaf of 30-day-old plants (leaf 2), flag leaf (leaf 3), the fourth leaf (leaf 4), immature panicle (panicle 1), panicle at pollination stage (panicle 2) and panicle at grain-filling stage (panicle 3). MeV software was used to generate and hierarchical cluster of heatmaps.
RNA isolation, library construction and sequencing
Total RNA was extracted from root and shoot samples using Plant RNA kit (OMEGA, USA) and RNAprep Pure Plant Kit (Tiangen Biotech Co.,Ltd., Beijing, China), respectively, according to the manufacturer’s protocols. RNA-Seq libraries were constructed using NEBNext Ultra RNA Library Prep Kit for Illumina ((#E7770, New England BioLabs, USA) following the manufacture’s instruction. Briefly, mRNA was purified and fragmented into 200 nt fragments. The fragmented mRNAs were then reverse transcribed to synthesize the first-strand cDNA and the second-strand cDNA. After end repair and adaptor ligation, the products were selected by Agencourt AMPure XP beads (Beckman Coulter, Inc.) and enriched by PCR amplification to create a cDNA library. Finally, the cDNA libraries were sequenced on an Illumina HiSeq X-ten platform.
RNA-seq reads processing and identification of differential expressed genes
RNA-seq reads were firstly cleaned using Trimmomatic (Bolger et al., 2014) with the following parameters: ILLUMINACLIP : TruSeq3-PE.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:30 HEADCROP:10. The clean reads were then mapped to the foxtail millet reference genome (http://sky.sxau.edu.cn/MDSi.htm) using hisat2 (Kim et al., 2015) with default parameters. Gene expression levels were calculated using R with the transcripts per million (TPM) (Li et al., 2010a). Fold changes were calculated using the log2 ratio of TPM. Average log values of three biological replications for each sample were then computed and used for further analysis. The cutoff of log2-fold changes ≥1 (2-fold absolute value) and padj value ≤ 0.05 were used for selecting significant DEGs.
Haplotype analysis and verification
Haplotype analysis was performed using the independently developed Perl script CandiHap.pl by our group (https://github.com/xukaili/CandiHap).
Three-dimensional structure prediction and molecular docking
Three-dimensional (3D) structure of the SiNPFs were predicted using the AlphaFold2 software (Jumper et al., 2021) following the instructions on the website https://github.com/deepmind/alphafold. The obtained 3D structures (pdb files) were then imported into AutoDockTools 1.5.6 for molecular docking analysis (Morris et al., 2009). The 2D and 3D NPF-nitrate interaction model were visualized by Ligplot 2.2.4 (Laskowski and Swindells, 2011) and PyMOL (http://www.pymol.org/), respectively.
Plasmid construction and complementation of yeast and Arabidopsis mutants
For yeast complementation analysis, the CDS fragments of SiNRT1.1B1 and SiNRT1.1B2 were synthesized and cloned into the Sal I-Spe I sites of the integrative pYNR-EX vector. The constructs were linearized at BstEII in LEU2 and transformed into Δynt1, a high-affinity nitrate transporter mutant-deficient strain. Finally, the yeast growth assay was performed as previously described (Martin et al., 2008).
For Arabidopsis complementation analysis, the genomic DNA fragments of SiNRT1.1B1 and SiNRT1.1B2 were cloned into pCAMBIA1300 vector, respectively. Due to the large size of the SiNRT1.1B1 gene, we first amplified two partially overlapped DNA fragments with primer pairs NRT1.1B1F1- NRT1.1B1R1 (fragment 1A) and NRT1.1B1F2- NRT1.2B1R2 (fragment 1B) using KOD-plus DNA polymerase (Toyobo, Osaka, Japan). The fragment 1A was then digested with EcoR I-Xho I and cloned into EcoR I-Sal I sites of pCAMBIA1300 to construct the intermediate vector. Finally, the resultant intermediate vector and fragment 1B were digested with Nru I and Nco I, and ligated to generate the pC1300-SiNRT1.1B1 vector. Similarly, the SiNRT1.1B2 gene was amplified and inserted into the pCAMBIA1300 vector using appropriate primer pairs and restriction sites to generate pC1300-SiNRT1.1B2 vector. After verification by extensive restriction digestion and DNA sequencing analysis, these constructs were transformed into the Arabidopsis atnrt1.1 mutant (SALK_097431C) via Agrobacterium-mediated floral dip method (Clough and Bent, 1998). All primers used for vector constructions are listed in Supplementary Table 2 .
Results
Genome-wide identification of the NPF genes in Setaria
To identify the entire NPF genes in Setaria, we performed a BLAST search against xiaomi (foxtail millet, http://sky.sxau.edu.cn/MDSi.htm) and A10 (green foxtail) (Bennetzen et al., 2012) genomes using the full-length amino acid sequences of 53 Arabidopsis and 93 rice, repectively. After removing redundant sequences manually, a total of 92 SiNPF genes and 88 SvNPF genes were retrieved from foxtail millet and green foxtail, respectively ( Supplementary Datasheet 1 ).
The length of the SiNPF proteins ranged from 191 amino acid (Si6g12910/SiNPF4.7) to 765 amino acid (Si2g07520/SiNPF2.3), with molecular weight ranging from 21035.05 to 82541.31 Da. Among these SiNPFs, 88 genes were unevenly distributed on 9 chromosomes of xiaomi except four genes (Si0g05510, Si0g08150, Si0g08180 and Si0g08230) not assembled on the genome ( Supplementary Figure 1 ). There were only two SiNPF genes (Si2g07520 and Si2g38080) on chromosome 2, which had the least number of NPF genes. There were 26 SiNPF genes on chromosome 9, which contained the largest number of SiNPF genes. Further analysis revealed that the SvNPF genes showed a similar chromosome distribution pattern to the SiNPF genes ( Supplementary Figure 2 ).
Gene duplication is a main driving force during evolution, which creates the raw genetic materials for natural selection. Of the 92 SiNPF genes, 56 genes were involved in duplication events, including 26 tandem gene pairs and 13 segmental duplications gene pairs ( Supplementary Datasheet 2 ). The Ka/Ks values of all gene pairs were less than 1, suggesting that these genes were subjected to different levels of purifying selection. Similarly, twenty-three SvNPF gene pairs underwent tandem duplication, and 12 gene pairs underwent segmental duplication. The Ka/Ks values were also less than 1 ( Supplementary Datasheet 2 ).
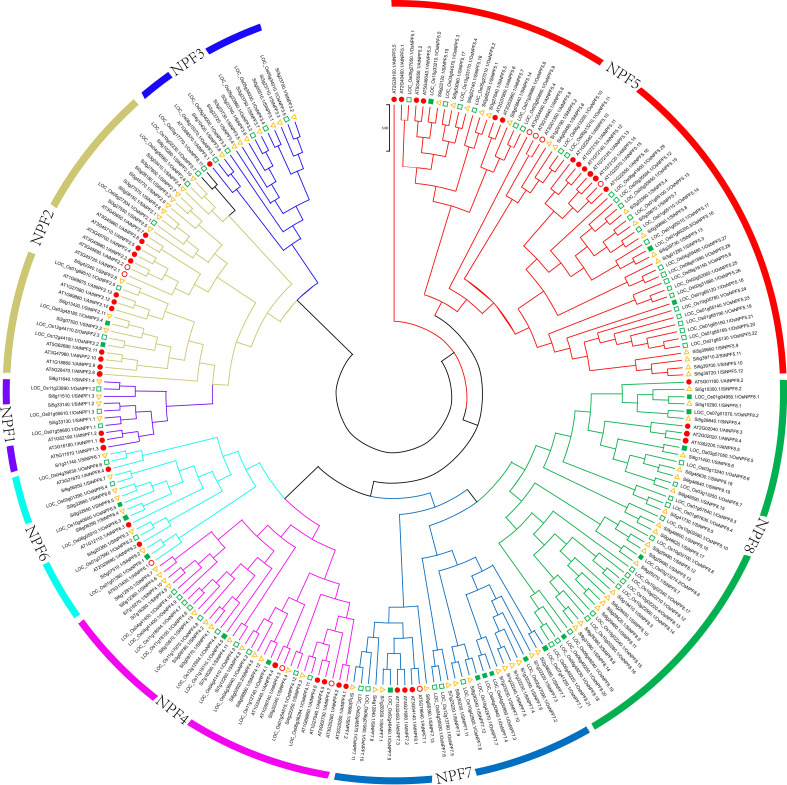
These NPF proteins were well-aligned with NPFs in Arabidopsis and rice, and separated into eight clades ( Figure 1 and Supplementary Figure 3 ). Group eight contains 19 SiNPF proteins, which is the largest group, followed by Group five, which contained 17 SiNPF proteins. There were 38 SiNPFs and 39 SvNPFs in the same branch of rice NPF proteins. The collinear relationship between different species was clearly distinguishable ( Figure 2 ). There were three, 59 and 68 orthologous genes of SiNPFs were found in Arabidopsis, rice and green foxtail ( Supplementary Datasheet 3 ). There were four and 58 orthologous genes of SvNPF genes in Arabidopsis and rice. Each orthologous gene pair belonged to the same subfamily. The number of homologous genes of SiNPF and SvNPF in rice was significantly higher than that in Arabidopsis.
Figure 1.
Phylogenetic relationship of NPF proteins among foxtail millet, rice and Arabidopsis. Different subfamilies are color-coded as illustrated in the figure. The red filled circle and the green filled square indicates these NPFs have been characterized in Arabidopsis (red filled circle) or rice (green filled square). The detailed information of these characterized NPF genes were summarized in Table S1 .
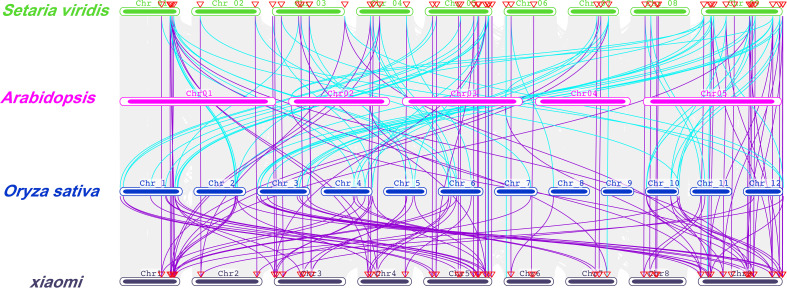
Figure 2.
Collinearity of the orthologous NPF genes in foxtail millet, green foxtail, rice and Arabidopsis. The genome of each species is shown in one row, and the NPF genes of foxtail millet and green foxtail are shown with different colored lines. The collinear relationship of all orthologous genes in different species was shown with the gray lines.
Gene structure and protein motif analysis of the NPF genes in Setaria
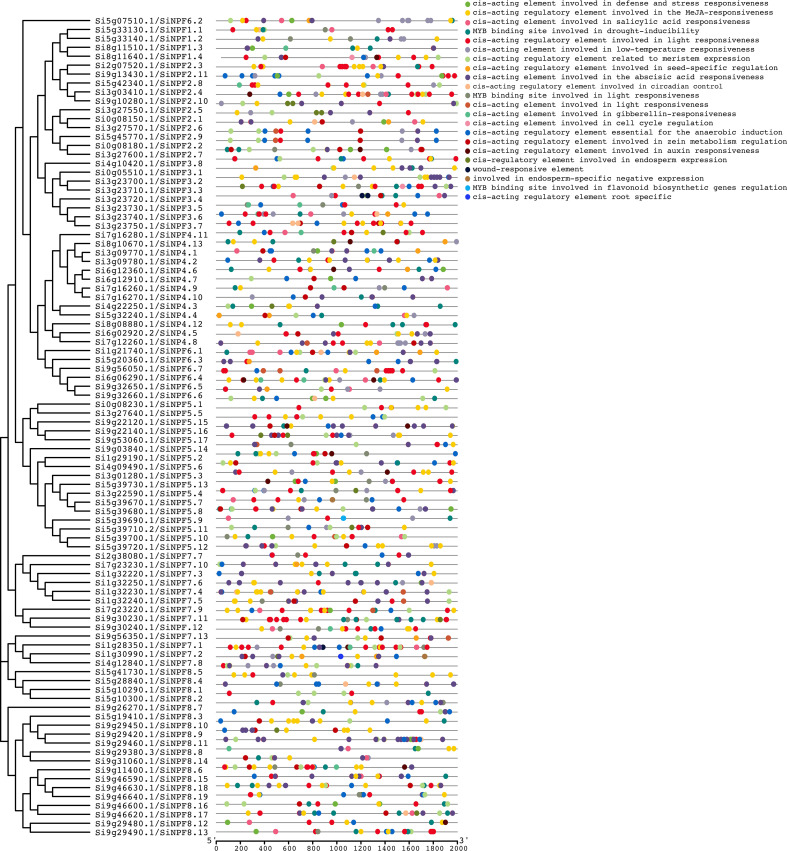
To further investigate the structural diversity of NPF genes in Setaria, the exon-intron organization of these genes were generated based on their coding sequences and corresponding genome sequences ( Figure 3 and Supplementary Figure 4 ). We found that the closely related NPFs tended to have similar gene structures with same number of exons and introns. Their differences mainly occurred in the length of UTR and intron regions ( Figure 3 ). The number of exons ranged from one to seven. Four genes, Si3g01280, Si5g07510, Si5g28840 and Si6g12910, had one exon, while Si4g12840 contained seven exons.
Figure 3.
Phylogenetic relationship, gene structure analysis, and motif distributions of SiNPF genes. Exon-intron structures are customized in each subfamily (with customized scale bar), the green bar represents exons, the black line represents introns, and the yellow bar represents UTR (untranslated region). The different motifs are color-coded as illustrated in the figure. The scale represents amino-acid length.
The MEME program was used to search the conserved motifs in these NPF proteins ( Figure 3 ). In total, we identified ten conserved motifs. The number of motifs in each NPF varied from three to 11. Approximately two-thirds of the SiNPF (65 out of 92) contained all of the 10 motifs. Although the number of motifs was different, the order of motif 1-10 in NPFs was similar. For instance, motif 7 was located at the C-terminus of all NPF proteins, whereas motif 4 was located at the N-terminus.
cis- regulatory elements and expression atlas of the SiNPFs
The cis-regulatory elements (CREs) in the promoter regions provide insights into gene functions. In total, 22 and 21 types of CREs were found in promoter regions of SiNPF and SvNPF genes, respectively ( Figure 4 and Supplementary Figure 5 ). These elements are involved in growth and development, stress and hormonal responses (ethylene, abscisic acid, auxin, gibberellin, methyl jasmonate, and salicylic acid). A total of 463 light-responsive elements were identified in the promoter regions of all SiNPF genes except for Si0g08230, which is the most abundant CREs in the SiNPF promoters. Among the hormone-responsive elements, the CREs related to the response to methyl jasmonate was the most numerous, followed by abscisic acid. These two hormones have important functions in plant stress response, so the function of SiNPF gene may be closely related to these two hormones under low nitrogen stress.
Figure 4.
cis-acting regulatory elements in the SiNPF promoters. Different colored circles represent different types of cis-elements.
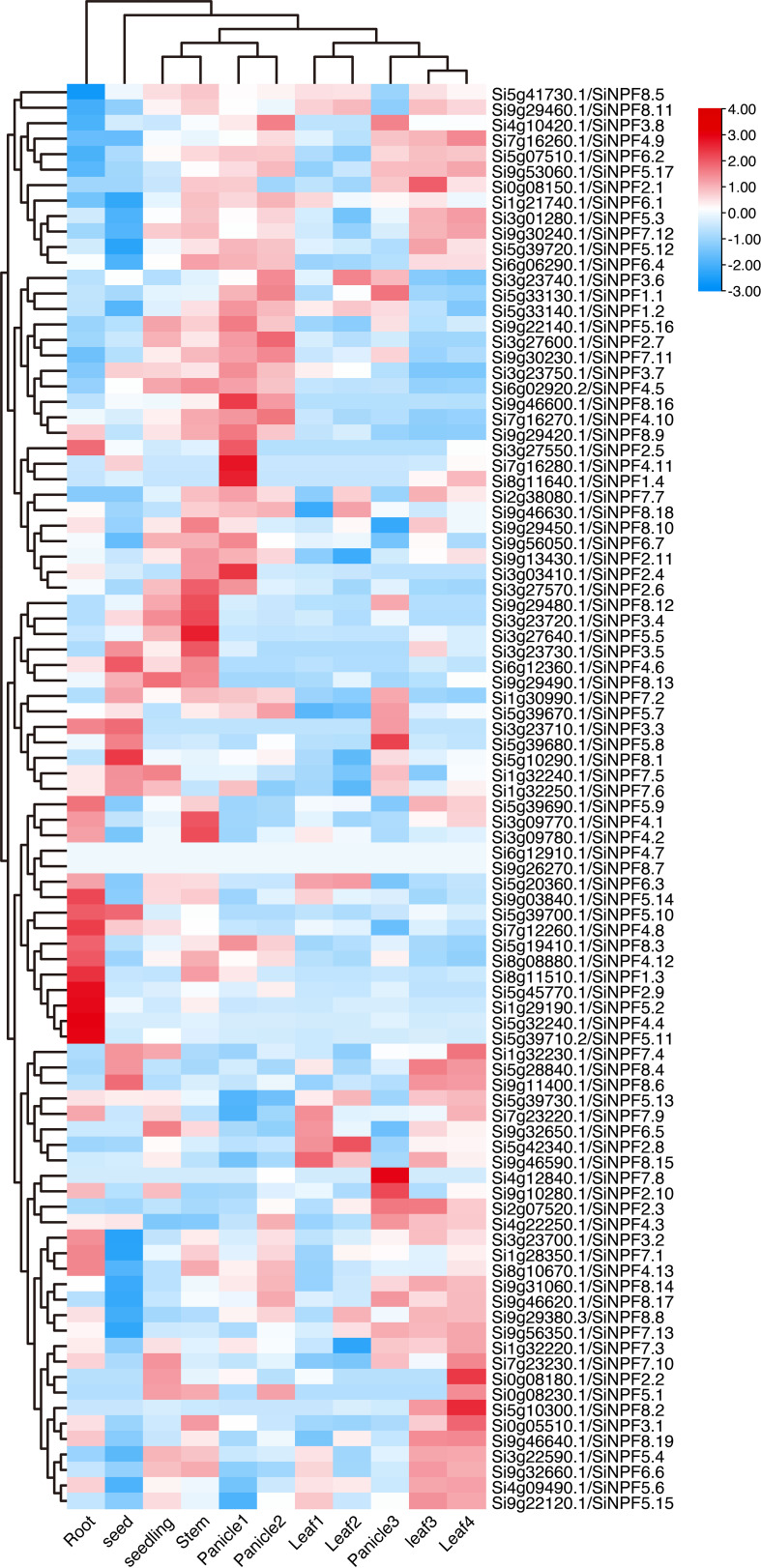
To provide a dynamic expression atlas for gene function dissection, we analyzed the gene transcript levels of the SiNPFs in eleven diverse tissues representing the major organs over various developmental stages. We found that the SiNPF genes expressed in a spatial and temporal manner ( Figure 5 and Supplementary Datasheet 4 ). These SiNPFs were divided into two major clusters based on their expression profiles. The first cluster SiNPFs were mainly expressed in root, while genes of another cluster were mainly expressed in above ground tissues (including seed, leaf, panicle and stem). Additionally, the young reproductive tissues (panicle 1 and panicle 2), the young leaves (leaf 1 and leaf 2) and the mature leaves (leaf 3 and leaf 4) clustered together strongly, respectively. In particular, Si1g29190, Si5g39710, Si5g45770 and Si9g03840 were highly and preferentially expressed in root, indicating that they might be involved in uptake of nitrate/peptide from soil. Thirty-six SiNPFs were highly expressed in stem, which might translocate nitrate from root to shoot. Si4g12840 was highly expressed in Panicle3, but hardly detected in other tissues. The expression of Si5g10290 in seed was significantly higher than that in other tissues, while the expression of Si5g42340 in leaf2 was higher than that in other tissues.
Figure 5.
Gene expression atlas of the SiNPFs across eleven diverse tissues. 3 d imbibed seeds (seed), 2-week-old whole seedling (seedling), root, stem, the top first fully extended leaf of 2-week-old seedling (leaf 1), the top second leaf of 30-day-old plants (leaf 2), flag leaf (leaf 3), the fourth leaf (leaf 4), immature panicle (panicle 1), panicle at pollination stage (panicle 2) and panicle at grain-filling stage (panicle 3).
Time-series low nitrate stress response of the SiNPF genes
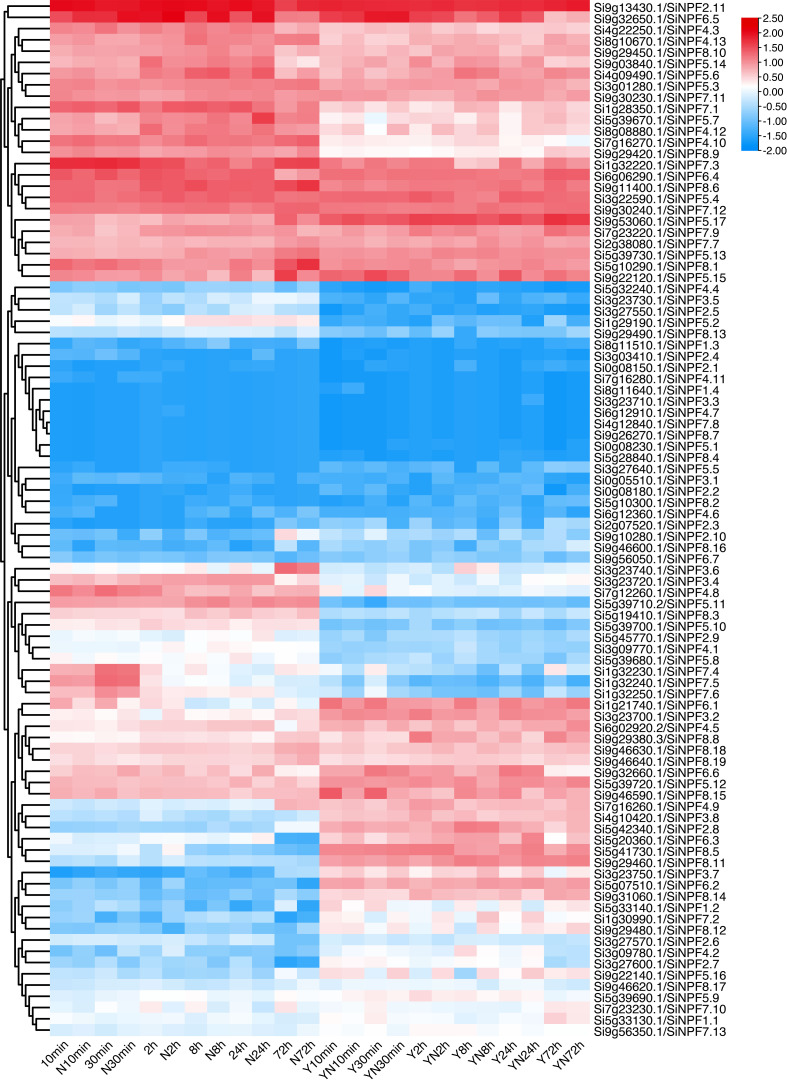
To gain a better view of the low nitrate response of these SiNPFs, we detected the gene expression profile under 10 min, 30 min, 2 h, 8 h, 24 h and 72 h of low nitrate stress. Transcriptome analysis showed that SiNPF genes exhibited abundant expression patterns under low nitrogen stress ( Figure 6 and Supplementary Datasheet 5 ). The expression patterns of these SiNPF genes were clustered into two broad classes (root and shoots). Eight genes (Si7g16280, Si8g11640, Si3g23710, Si6g12910, Si0g08230, Si5g28840, Si4g12840 and Si9g26270) were hardly expressed under normal N or low N stress. After low nitrogen stress, the number of genes with a foldchange greater than 2 in the shoot (38) was more than that in the root (24). Indeed, more differentially expressed genes (DEGs) were identified at 8h (11), 24h (12) and 72h (12) than at 10min (1), 30min (16) and 2h (10) under LN stress, suggesting the more SiNPF genes responded to a longer period of low nitrogen stress. The Si5g33140 gene was significantly up-regulated in roots after 72 hours of nitrogen stress, predominantly expressed in panicles.
Figure 6.
Time-series low nitrate stress response of the SiNPF genes.
Natural variations of the NPF family genes in foxtail millet
Natural variation is central to understanding gene function, evolution, and which in turn improves breeding in foxtail millet. To provide more information about SiNPF genes and facilitate the use of natural accessions, we performed a haplotype analysis based on the re-sequence data of 398 foxtail millet accessions including 162 cultivars, 198 landraces and 38 wild S. viridis (Li et al., 2022). A total of 2924 SNPs and 400 InDels were detected in the 92 SiNPF genes ( Supplementary Datasheets 6, 7 ). These variations exhibited an uneven distribution: the Si8g11510 contained the highest number of SNPs/InDels, whereas no SNPs/InDels were found in Si0g05510, Si0g08150, Si0g08180, Si0g08230, Si1g32240 and Si9g26270. Two SNPs (Si3g03410 and Si5g10300) and 1 InDel (Si9g29460) located in splicing site and may cause abnormal splicing of the intron. 245 SNPs/InDels were non-synonymous and were distributed in the coding regions of 53 SiNPF genes. Out of the 400 InDels, 22 were found in the coding sequencing and caused frameshift or nonframeshift deletion (Si1g29190, Si2g07520, Si3g03410, Si7g16280, Si8g08880, Si9g13430, Si9g56350, Si5g39700 and Si8g11640), insertion (Si1g29190, Si5g10300 and Si9g13430) or stop-gain (Si1g29190). These SiNPF haplotype data provided valuable information for further gene function dissection and molecular design breeding.
Three-dimensional structure of the SiNPFs and their interaction with nitrate
Protein 3D structure can provide invaluable information to predict its biological function. Thus, we predicted the three-dimensional structure of the NPFs in foxtail millet using AlphaFold2 (Jumper et al., 2021). The SiNPFs members shared a canonical major facilitator superfamily (MFS) fold structure, which was characterized by 12 transmembrane helices (TMs) with a central linker connecting the N-domain (TM1-TM6) and C-domain (TM7-TM12) ( Supplementary Data 1 , Supplementary Datasheet 8 ). Only 15 SiNPFs have few TMs, which are distributed in all except subfamily III ( Supplementary Datasheet 8 ). Further analysis revealed that both of the N- and C-terminal structure of the SiNPFs are not conserved, and their exact function in nitrate transport remains to be further elucidated.
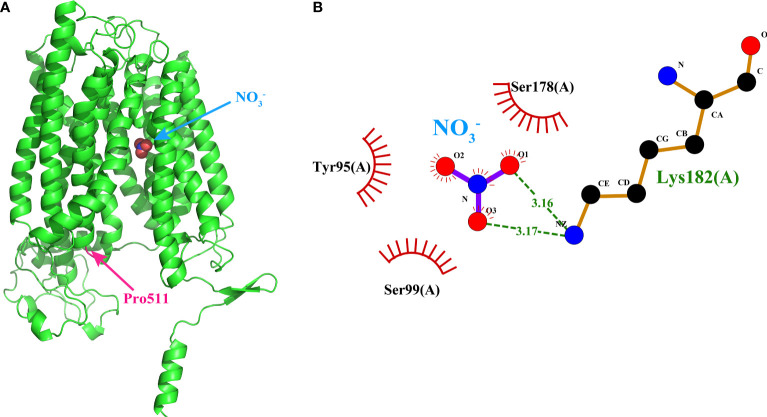
As nitrate is the major substrate of NPFs, we evaluate theaffinity of the 92 SiNPFs with NO3 − by molecular docking. Thebinding energy of the SiNPFs to NO3 − ranged from -3.4 to -2.1 kcal/mol ( Supplementary Datasheet 9 ). Among them, Si5g32240 had thelowest binding energy of -3.4 kcal/mol, indicating highly stablebinding. Previously, Ho et al. (2009) reported that the Pro 492 residue of NRT1.1 is important for the nitrate transport activity inArabidopsis. We found that only 10 members do not have theproline residue at the corresponding position, indicating that thisresidue is highly conserved in the SiNPFs ( Supplementary Figure 6 ). In the Si5g32240 structure, the conserved proline (Pro511) islocated at the short TMH10-TMH11 loop, but not in thesubstrate binding pocket ( Figure 7A ), indicated that variation ofthe conserved proline might not affect the nitrate binding ability. The 2D Si5g32240- NO3 − interaction analysis revealed that Tyr95, Ser99, Ser178, and Lys182 might interact with NO3− ( Figure 7B ). These amino acids might play important role in nitrate bind and transport for Si5g32240.
Figure 7.
Binding model of Si5g32240 to nitrate by moleculardocking. (A) The 3D Si5g32240-nitrate interaction model. The conserved Pro511 was marked in pink. (B) The 2D Si5g32240-nitrate interaction model. Dashed lines indicated potential covalent bonds between Lys182 and NO3 −, and the eyelash indicated the non-covalent bonds between Tyr95, Ser99, Ser178 and NO3 −. The3D and 2D interaction models of the other 91 NPFs could be obtained from Supplementary Data 2 and 3 , respectively.
Tandem duplication of the NRT1.1 gene may contribute to low nitrogen tolerance in foxtail millet
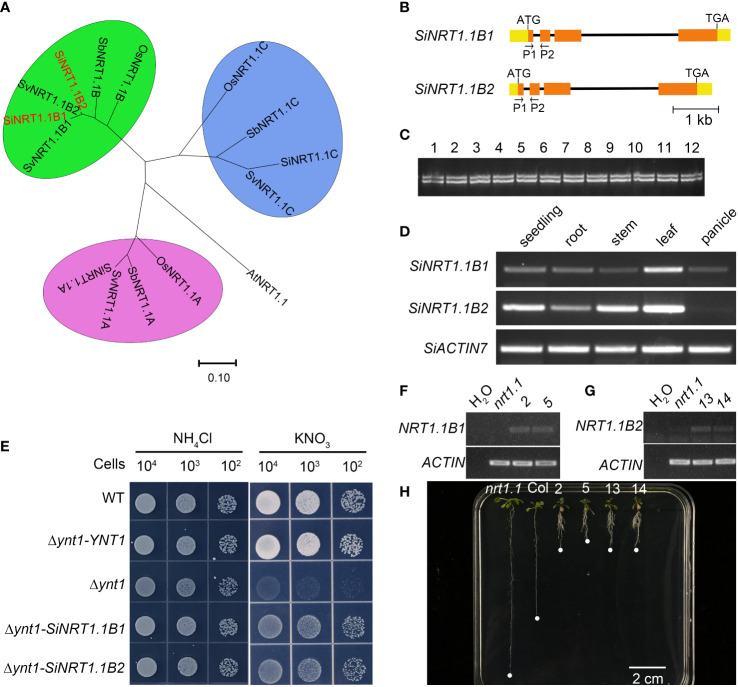
To dissect the possible molecular mechanisms underlying low nitrogen tolerance of foxtail millet, we performed a synteny analyses of the NPF genes between foxtail millet and Arabidopsis and rice ( Figure 2 and Supplementary Datasheet 3 ). Interestingly, we found there were two NRT1.1B gene copies in a tail-to-tail orientation in both foxtail millet (Si9g32650/SiNRT1.1B1 and Si9g32660/SiNRT1.1B2) and green foxtail (SvNRT1.1B1/Sevir.9g333900 and SvNRT1.1B2/Sevir.9g334100), while there was only one copy in rice and sorghum ( Figure 8A ). Despite of the very high amino acid sequence identity (96.27%) between the two SiNRT1.1Bs, we found the first intron of SiNRT1.1B1 was 26 bp longer than that of SiNRT1.1B2 ( Figure 8B ). To determine if there was copy number variation at the SiNRT1.1Bs locus in foxtail millet, we performed a PCR analysis using primers spanning the first intron. However, all the germplasm resources detected, including 360 foxtail millets and 38 green foxtails, harbored two copies of NRT1.1B gene ( Figure 8C , and data not shown). This result indicated that the gene duplication events might occur before the divergence of foxtail millet and green foxtail, and after the divergence of the Setaria and sorghum.
Figure 8.
Duplication of the SiNRT1.1B gene might confers tolerance to low nitrate in foxtail millet. (A) The phylogenetic relationship of the NRT1.1 subfamily in Arabidopsis, rice, sorghum, foxtail millet and green foxtail. AtNRT1.1 (At1G12110), OsNRT1.1A (LOC_Os08g05910), OsNRT1.1B (LOC_Os10g40600), OsNRT1.C (LOC_Os03g01290), SiNRT1.1A (Si6g06290), SiNRT1.1B1 (Si9G32650), SiNRT1.1B2 (Si9G32660), SiNRT1.1C (Si9g56050), SbNRT1.1A (Sobic.007G044300), SbNRT1.1B (Sobic.001G302800), SbNRT1.1C (Sobic.001G541900). (B) Gene structure of the SiNRT1.1B1 and SiNRT1.1B1 genes. P1 and P2 are primers used for copy number analysis in Panel C (C) Copy number analysis of the SiNRT1.1B1 gene in Setaria. 1-8 are foxtail millet, and 9-12 are green foxtail. (D) The SiNRT1.1B1 and SiNRT1.1B2 expression pattern in various organs. (E) SiNRT1.1B1 and SiNRT1.1B2 rescued the yeast Δynt1 mutant. Δynt1-YNT1, Δynt1-SiNRT1.1B1 and Δynt1-SiNRT1.1B2 are Δynt1 mutant strain transformed with the yeast YNT1 gene, SiNRT1.1B1 and SiNRT1.1B2 gene, respectively. (F) RT-PCR analysis of SiNRT1.B1 expression in atnrt1.1 mutant (nrt1.1), wild type (Col) and two independent atnrt1.1 transgenic lines carrying a pSiNRT1.1B1::SiNRT1.1B1 gene (2 and 5). (G) RT-PCR analysis of SiNRT1.B1 expression in atnrt1.1 mutant (nrt1.1), wild type (Col) and two independent atnrt1.1 transgenic lines carrying a pSiNRT1.1B2::SiNRT1.1B2 gene (13 and 14). (H) SiNRT1.1B1 and SiNRT1.1B2 enhanced chlorate sensitivity of the atnrt1.1 mutant. nrt1.1 was atnrt1.1 mutant; Col was the wildtype, 2, 5, 13 and 14 were transgenic lines showed in Panels (F, G).
Duplicated genes usually diverge at the level of gene expression, protein function or both. Thus, it is interesting to know whether the two copies of NRT1.1B have similar function or not. As shown in Figure 5 , SiNRT1.1B1 and SiNRT1.1B2 exhibited a similar expression pattern, with highly expressed in vegetative tissues, especially in leaves, but low in reproductive organs. These results were further confirmed by RT-PCR ( Figure 8D ). To further determine whether these two copies could transport nitrate, we transformed them into Δynt1, a high affinity nitrate transporter mutant deficient yeast strain (Martin et al., 2008). Unlike wildtype strain, the Δynt1 mutant could not grow under low nitrate condition. This growth defect could be partially completed by SiNRT1.1B1 and SiNRT1.1B2 ( Figure 8E ). To further confirm this result, we transformed the SiNRT1.1Bs to the Arabidopsis nrt1.1 mutant and performed a chlorate-sensitivity assay Figures 8F–H . As shown in Figure 8H , the transgenic lines of SiNRT1.1B1 and SiNRT1.1B2 exhibited higher chlorate sensitivity than that of the atnrt1.1 mutant. These results clearly demonstrated that both SiNRT1.1B1 and SiNRT1.1B2 had nitrate transport activity. Since NRT1.1B gene plays an important role in nitrate absorption and transport (Hu et al., 2015), the duplication of the NRT1.1B gene may contribute to low nitrogen tolerance of foxtail millet.
Due to the relatively short read length, large InDels could not be identified based on the genome re-sequencing data. To provide more genetic variation information, we randomly selected 114 foxtail millet accessions to identify large InDels. Interestingly, we found a 349 bp insertion in the promoter region at nucleotide -398 (the first nucleotide of the putative translation start codon of the genomic sequence is referred to as +1) ( Figure 9 ). The effects of this large InDel on gene expression and nitrateuptake need to be further studied.
Figure 9.
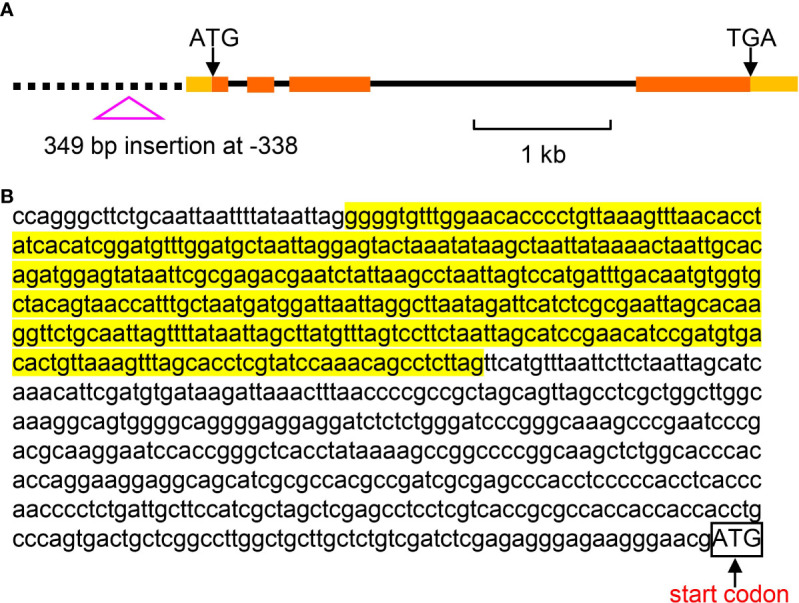
The large InDel in the promoter region of SiNRT1.1B2. (A) The genomic structure of SiNRT1.1B2 gene and the large InDel in the promoter. Exons, introns and promoter are denoted by filled boxes, lines and dash line, respectively. (B) The nucleotide sequence of the large InDel in the promoter of SiNRT1.1B2. The highlighted fragment indicates the 349 bp InDel. The box refers to the start codon.
Discussion
Genome-wide identification of NPF genes have been extensively studied in diverse plant species, including Arabidopsis (Léran et al., 2014), rice (Yang et al., 2020a), Populus tomentosa (Zhao et al., 2021), Spirodela polyrhiza (Lv et al., 2022) and Triticum aestivum (Kumar et al., 2022). However, there has been little information about NPF family in the barren tolerant species Setaria, which greatly limits our understanding of the molecular mechanisms underlying high NUE in this species. In this study, 92 and 88 NPF genes were identified in foxtail millet and its wild type ancestor green foxtail, respectively ( Supplementary Datasheet 1 ). Further natural variation analysis revealed that there were abundant genetic variations in the SiNPF genes ( Supplementary Datasheet 6, 7 ). Undoubtedly, our findings provide fundamental information for further research on the biological functions of NPFs and regulation mechanism of barren tolerance in Setaria. We also acknowledge that limitations still remain for identification and analysis of the natural variation based on a single reference genome, since a single-reference assembled genome represents only a small fraction information of the entire Setaria species. Thus, a species-representative pan-genome is necessary to better capture the full genetic diversity of the NPF gene family in Setaria.
The gene number of NPF family varies greatly among species, which is mainly caused by gene duplication including tandem duplication, segmental duplication and transposition events. Our results revealed that tandem duplication and segmental duplication were the driving forces of the SiNPF gene expansion ( Supplementary Datasheet 2 ). Further analysis revealed that all the SiNPF gene pairs underwent strong purifying selection, as evidenced by Ka/Ks ratios lower than 1.0, indicating that function of these genes was highly preserved. Notably, despite the similar number of NPF genes in foxtail millet (92) and rice (93), the duplicated events were not exactly the same. We found there were three clades of closely related AtNRT1.1 genes, OsNRT1.1A, OsNRT1.1B and OsNRT1.1C in rice, while four clades in foxtail millet, namely SiNRT1.1A, SiNRT1.1B1, SiNRT1.1B2 and SiNRT1.1C ( Figures 1 , 8A ). Among these gene, the SiNRT1.1B1 and SiNRT1.1B2 were the tandem duplicated gene pair of AtNRT1.1, while there only contains one linear homolog in rice. Interestingly, there is also only one linear homolog of OsNRT1.1B in Sorghum bicolor L, another Panicoideae species closely to foxtail millet. Therefore, there are at least two genes potentially fill the same functional role of OsNRT1.1B in foxtail millet, and this might contribute to the low nitrogen tolerance in Setaria. Therefore, it is possible to improve crop NUE by increasing the copy number of NRT1.1 gene in the future.
The demand for nitrate varies greatly in different tissues or at different growth stages in foxtail millet, therefore nitrate must reach to each of them by different routes (Dechorgnat et al., 2011). Our data showed that approximately one third of the SiNPF genes were highly expressed in root, which might play important role in uptake nitrate/peptide or other substrate from soil ( Figure 5 , Supplementary Datasheet 4 ). In additions, NPFs that are highly expressed in germinated seed, stem, leaf and panicle were also identified. Since gene express pattern reflects specific tissue functions, these expression data provide valuable clues to understanding their biological function in the future. Notably, we found that many SiNPF genes showed similar expression pattern to their orthologous and phylogenetic close Arabidopsis or rice NPF genes. For example, the SiNPF6.3 was predominately expressed in leaves. Similar expression pattern was also reported for the Arabidopsis AtNPF6.2/AtNRT1:4 (Chiu et al., 2004). Therefore, SiNPF6.3 is likely to be a key player in regulating leaf nitrate homeostasis and leaf development as AtNPF6.2. Differences in expression patterns were also found between SiNPFs and their orthologous. Previously, Li et al. (2009) reported that rice OsNPF4.1/OsSP1 was predominantly expressed in young panicle and with a unique function in panicle development. In contrast to OsNPF4.1, the foxtail millet homologous SiNPF4.12 was highly expressed in roots other than panicles, indicating an additional root function. The functional divergence between orthologues suggests that it is unreliable to predict the NPF function based on their phylogenetic relationships.
The plant NPFs are well known for their essential roles in nitrate uptake, transport and allocation. To date, at least 43 NPFs in Arabidopsis have been characterized, and 23 of them were able to transport nitrate ( Table S1 ). Moreover, several NPFs were also been extensively characterized in rice ( Table S1 ). However, the substrate(s) and biological functions of NPFs are largely unknown in Seteria, a barren-tolerant species. Here, we provide a solid foundation for further functional dissection of these NPF genes. Conventional, gene function could be predicted according to their sequence homology. The collinear analysis showed that there were 3 and 59 linear orthologous genes of SiNPFs in Arabidopsis and rice, respectively ( Supplementary Datasheet 3 ), which might have similar functions. The Pro492 residue of AtNPF6.3 is highly conserved in Arabidopsis, and is crucial for the nitrate transport activity in Arabidopsis (Ho et al., 2009; Chen et al., 2021). Consistently, there were only 10 SiNPFs that were not proline residue at the corresponding position ( Supplementary Figure 6 ). Whether these SiNPFs can transport nitrate remains to be further verified. The AtNRT1.1 (AtNPF6.3) is the most deeply studied NPF genes which play important roles in nitrate uptake and signal transduction. The AtNPF6.3- NO3 − binding energy was -2.4 kcal/ mol. We found that a total 87 SiNPFs had binding energy lower than or equal to -2.4 kcal/mol ( Supplementary Datasheet 9 ). These SiNPFs might involve in nitrate uptake and transport in foxtail millet, and should be the focus in future research. In addition to nitrate, some NPFs can also transport phytohormones including auxin, abscisic acid, jasmonates and gibberellins (Corratge-Faillie and Lacombe, 2017). On the other hand, several NPF genes involved in phytohormone transport demonstrated hormone- and/or stress- response characteristics (Saito et al., 2015; Tal et al., 2016). In this study, many cis-acting element involved in hormone responses were detected in the promoter region of SiNPF genes, which suggested their potential hormone-inducing characteristics of these genes ( Figure 3 ). Natural variations in NPF genes may greatly alter the N uptake and ability. For example, a single-nucleotide polymorphism (SNP) in OsNRT1.1B contributed to the NUE divergence between the two main rice subspecies, indica and japonica (Hu et al., 2015). Here, we identified 2,924 SNPs and 400 InDels in 92 SiNPF genes ( Supplementary Datasheet 6, 7 ). Undoubtedly, nonsynonymous or nonsense mutation in SiNPFs will be useful for gene function dissection. It is worth mentioning that most synonymous mutations in yeast are extremely harmful, instead of being neutral as generally believed (Shen et al., 2022). If it holds true for genes in foxtail millet, the large amount of synonymous mutation information provided here will be of great value for the future gene function analysis and crop breeding for high NUE. Finally, the accurate 3D structure of the SiNPFs and were also predicted with AlphaFold2. To our knowledge, it is the first NPF family that have 3D structure information.
Conclusion
In this study, 92 and 88 putative NPF genes were identified in foxtail millet and its wild ancestor green foxtail, respectively. These NPFs could be divided into eight subfamilies based on sequence similarity and phylogenetic relationship. Among the 92 SiNPF genes, about one fourth are highly expressed in root, suggesting that these genes might play roles in nitrate uptake. The time series of transcriptomes provided insight into response of these SiNPFs to short- and long- time low nitrate treatments. Interestingly, we found that the NRT1.1B gene might contribute to low nitrogen tolerance in Setaria. Coupled with the natural variation, 3D information and SiNPF-nitrate interaction models, these results provided the basis for comprehensive understanding of NPF genes for low nitrogen tolerance in Setaria.
Data availability statement
The raw RNA-seq data of foxtail millet treated with low nitrate have been deposited in the National Genomics Data Center (https://ngdc.cncb.ac.cn/) under the BioProject accession PRJCA012843. The other sequencing data were obtained from our Multi-omics Database for Setaria italica (MDSi) (http://sky.sxau.edu.cn/MDSi.htm) and JGI Phytozome (https://phytozome-next.jgi.doe.gov/).
Author contributions
XCW, ZRY conceived and designed the experiments. JJC performed bioinformatics analysis, low nitrate treatment of seedlings. HLT performed 3D structure and natural variation analysis. MS, MMD and YLY participated in data collection. LH and HMS performed molecular docking analysis. XCW and ZRY wrote the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We are very grateful to Yiwei Jiang of Purdue University for his critical reading of the manuscript, and Xingyu Jiang of Hainan University for presenting the Δynt1 mutant and pYNR-EX plasmid.
Funding Statement
This work was supported by the Central government Guides the Local Science and Technology Development Fund Project (YDZJSX2021B010), the Fundamental Research Program of Shanxi Province (20210302123385), Shanxi Province Science and Technology Major Special Project (202101140601027), Shanxi Breeding Innovation Program (2022XCZX04-10). The funding bodies had no role in the design of the study, collection, analysis, or interpretation of data or in the writing of the manuscript.
Abbreviations
DEG, differential expressed gene; HATS, high-affinity transport system; LATS, low-affinity transport system; N, nitrogen; NPF, nitrate transporter 1/peptide transporter family; NRT, nitrate transporter; TPM,s transcripts per million.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.1043832/full#supplementary-material
Chromosome location and distribution analysis of the SiNPF genes. Tandem duplicated genes are linked by a red curve.
Chromosome location and distribution analysis of the SvNPF genes. Tandem duplicated genes are linked by a red curve.
Phylogenetic relationship of NPF proteins among green foxtail, rice and Arabidopsis. Different subfamilies are color-coded as illustrated in the figure. The red filled circle and the green filled square indicates these NPFs have been characterized in Arabidopsis (red filled circle) or rice (green filled square). The detailed information of these characterized NPF genes were summarized in Table S1 .
Phylogenetic relationship, gene-structure analysis, and motif distributions of SiNPF genes. Exon-intron structures are customized in each subfamily (with customized scale bar), the green bar represents exons, the black line represents introns, and the yellow bar represents UTR (untranslated region). The different motifs are color-coded as illustrated in the figure. The scale represents amino-acid length.
cis-acting regulatory elements in the SvNPF promoters. Different colored circles represent different types of cis-elements.
Amino acid sequence alignment of AtNRT1.1 and SiNPFs. The red star indicates Pro492 in AtNRT1.1.
Summary of the characterized NPF genes in Arabidopsis and rice
Primers used in this study.
References
- Bennetzen J. L., Schmutz J., Wang H., Percifield R., Hawkins J., Pontaroli A. C., et al. (2012). Reference genome sequence of the model plant Setaria . Nat. Biotechnol. 30 (6), 555–561. doi: 10.1038/nbt.2196 [DOI] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30 (15), 2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Lin S. H., Cheng L. H., Wu J. J., Lin Y. C., Tsay Y. F. (2021). Potential transceptor AtNRT1.13 modulates shoot architecture and flowering time in a nitrate-dependent manner. Plant Cell 33 (5), 1492–1505. doi: 10.1093/plcell/koab051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. C., Lin C. S., Hsia A. P., Su R. C., Lin H. L., Tsay Y. F. (2004). Mutation of a nitrate transporter, AtNRT1:4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45 (9), 1139–1148. doi: 10.1093/pcp/pch143 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Corratge-Faillie C., Lacombe B. (2017). Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 68 (12), 3107–3113. doi: 10.1093/jxb/erw499 [DOI] [PubMed] [Google Scholar]
- Crawford N. M., Glass A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3 (10), 389–395. doi: 10.1016/S1360-1385(98)01311-9 [DOI] [Google Scholar]
- Dechorgnat J., Nguyen C. T., Armengaud P., Jossier M., Diatloff E., Filleur S., et al. (2011). From the soil to the seeds: the long journey of nitrate in plants. J. Exp. Bot. 62 (4), 1349–1359. doi: 10.1093/jxb/erq409 [DOI] [PubMed] [Google Scholar]
- Drechsler N., Courty P. E., Brule D., Kunze R. (2018). Identification of arbuscular mycorrhiza-inducible Nitrate transporter 1/Peptide transporter family (NPF) genes in rice. Mycorrhiza 28 (1), 93–100. doi: 10.1007/s00572-017-0802-z [DOI] [PubMed] [Google Scholar]
- Edith Laugier E. B., Mauriès A., Tillard P., Gojon A., Lejay L. (2012). Regulation of high-affinity nitrate uptake in roots of arabidopsis depends predominantly on posttranscriptional control of the NRT2.1/NAR2.1 transport system. Plant Physiol. 158 (2), 1067–1078. doi: 10.1104/pp.lll.l [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. H., Liu X. J., Zhang Y., Shen J. L., Han W. X., Zhang W. F., et al. (2010). Significant acidification in major Chinese croplands. Science 327 (5968), 1008–1010. doi: 10.1126/science.1182570 [DOI] [PubMed] [Google Scholar]
- Ho C. H., Lin S. H., Hu H. C., Tsay Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138 (6), 1184–1194. doi: 10.1016/j.cell.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Hsu P. K., Tsay Y. F. (2013). Two phloem nitrate transporters, NRT1.11 and NRT1.12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 163 (2), 844–856. doi: 10.1104/pp.113.226563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N.-C., Liu K.-H., Lo H.-J., Tsay Y.-F. (1999). Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11 (8), 1381–1392. doi: 10.1105/tpc.11.8.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Wang W., Ou S., Tang J., Li H., Che R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47 (7), 834–838. doi: 10.1038/ng.3337 [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E. A. (2017). “Evolution of setaria,” in Genetics and genomics of setaria. Eds. Dous A., Diao X. (Cham: Springer International Publishing; ), 3–27. [Google Scholar]
- Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., David L. C., Chardin C., Girin T., Marmagne A., Leprince A.-S., et al. (2014). Nitrate transport and signalling in Arabidopsis . J. Exp. Bot. 65 (3), 789–798. doi: 10.1093/jxb/eru001 [DOI] [PubMed] [Google Scholar]
- Kumar A., Sandhu N., Kumar P., Pruthi G., Singh J., Kaur S., et al. (2022). Genome-wide identification and in silico analysis of NPF, NRT2, CLC and SLAC1/SLAH nitrate transporters in hexaploid wheat (Triticum aestivum). Sci. Rep. 12 (1), 11227. doi: 10.1038/s41598-022-15202-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A., Swindells M. B. (2011). LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf Model. 51 (10), 2778–2786. doi: 10.1021/ci200227u [DOI] [PubMed] [Google Scholar]
- Léran S., Varala K., Boyer J.-C., Chiurazzi M., Crawford N., Daniel-Vedele F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19 (1), 5–9. doi: 10.1016/j.tplants.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Li J. Y., Fu Y. L., Pike S. M., Bao J., Tian W., Zhang Y., et al. (2010. b). The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22 (5), 1633–1646. doi: 10.1105/tpc.110.075242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Gao J., Song J., Guo K., Hou S., Wang X., et al. (2022). Multi-omics analyses of 398 foxtail millet accessions reveal genomic regions associated with domestication, metabolite traits, and anti-inflammatory effects. Mol. Plant 15 (8), 1367–1383. doi: 10.1016/j.molp.2022.07.003 [DOI] [PubMed] [Google Scholar]
- Li H., Hu B., Chu C. (2017). Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot. 68 (10), 2477–2488. doi: 10.1093/jxb/erx101 [DOI] [PubMed] [Google Scholar]
- Lin S. H., Kuo H. F., Canivenc G., Lin C. S., Lepetit M., Hsu P. K., et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20 (9), 2514–2528. doi: 10.1105/tpc.108.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Qian Q., Fu Z., Zeng D., Meng X., Kyozuka J., et al. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58 (4), 592–605. doi: 10.1111/j.1365-313X.2009.03799.x [DOI] [PubMed] [Google Scholar]
- Li B., Ruotti V., Stewart R. M., Thomson J. A., Dewey C. N. (2010. a). RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26 (4), 493–500. doi: 10.1093/bioinformatics/btp692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. H., Huang C. Y., Tsay Y. F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11 (5), 865–874. doi: 10.1105/tpc.11.5.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Sun Q., Wang K., Du Q., Li W. X. (2017). Nitrogen limitation adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis . New Phytol. 214 (2), 734–744. doi: 10.1111/nph.14396 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhang Y., Han W., Tang A., Shen J., Cui Z., et al. (2013). Enhanced nitrogen deposition over China. Nature 494 (7438), 459–462. doi: 10.1038/nature11917 [DOI] [PubMed] [Google Scholar]
- Lv M., Dong T., Wang J., Zuo K. (2022). Genome-wide identification of nitrate transporter genes from Spirodela polyrhiza and characterization of SpNRT1.1 function in plant development. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.945470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Y., Navarro F. J., Siverio J. M. (2008). Functional characterization of the Arabidopsis thaliana nitrate transporter CHL1 in the yeast Hansenula polymorpha . Plant Mol. Biol. 68 (3), 215–224. doi: 10.1007/s11103-008-9363-z [DOI] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30 (16), 2785–2791. doi: 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. L., Newstead S. (2014). Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507 (7490), 68–72. doi: 10.1038/nature13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plett D., Toubia J., Garnett T., Tester M., Kaiser B. N., Baumann U. (2010). Dichotomy in the NRT gene families of dicots and grass species. PloS One 5 (12), e15289. doi: 10.1371/journal.pone.0015289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raun W. R., Johnson G. V. (1999). Improving nitrogen use efficiency for cereal production. Agron. J. 91 (3), 357–363. doi: 10.2134/agronj1999.00021962009100030001x [DOI] [Google Scholar]
- Saito H., Oikawa T., Hamamoto S., Ishimaru Y., Kanamori-Sato M., Sasaki-Sekimoto Y., et al. (2015). The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis . Nat. Commun. 6, 6095. doi: 10.1038/ncomms7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T. B., Lima J. E., Felicio M. S., Soares J. D. M., Domingues D. S. (2017). Genome-wide identification, classification and transcriptional analysis of nitrate and ammonium transporters in Coffea . Genet. Mol. Biol. 40 (1 suppl 1), 346–359. doi: 10.1590/1678-4685-GMB-2016-0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C., Boyer J. C., Ipotesi E., Szponarski W., Tillard P., Touraine B., et al. (2007). Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter. Plant Cell 19 (11), 3760–3777. doi: 10.1105/tpc.106.048173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Song S., Li C., Zhang J. (2022). Synonymous mutations in representative yeast genes are mostly strongly non-neutral. Nature 606 (7915), 725–731. doi: 10.1038/s41586-022-04823-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal I., Zhang Y., Jorgensen M. E., Pisanty O., Barbosa I. C. R., Zourelidou M., et al. (2016). The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7, 11486. doi: 10.1038/ncomms11486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Ye J., Yao X., Zhao P., Xuan W., Tian Y., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10 (1), 5279. doi: 10.1038/s41467-019-13187-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y. F., Schroeder J. I., Feldmann K. A., Crawford N. M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72 (5), 705–713. doi: 10.1016/0092-8674(93)90399-b [DOI] [PubMed] [Google Scholar]
- Wang Y. Y., Hsu P. K., Tsay Y. F. (2012). Uptake, allocation and signaling of nitrate. Trends Plant Sci. 17 (8), 458–467. doi: 10.1016/j.tplants.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Wang W., Hu B., Yuan D., Liu Y., Che R., Hu Y., et al. (2018. b). Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell 30 (3), 638–651. doi: 10.1105/tpc.17.00809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Liu C., Dong Q., Huang D., Li C., Li P., et al. (2018. a). Genome-wide identification and analysis of apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) genes reveals MdNPF6.5 confers high capacity for nitrogen uptake under low-nitrogen conditions. Int. J. Mol. Sci. 19 (9), 2761. doi: 10.3390/ijms19092761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li Y., Zhu F., Ming R., Chen L.-Q. (2019). Genome-wide analysis of nitrate transporter (NRT/NPF) family in sugarcane Saccharum spontaneum l. Trop. Plant Biol. 12 (3), 133–149. doi: 10.1007/s12042-019-09220-8 [DOI] [Google Scholar]
- Wang Y. Y., Tsay Y. F. (2011). Arabidopsis nitrate transporter NRT1.9 is important in phloem nitrate transport. Plant Cell 23 (5), 1945–1957. doi: 10.1105/tpc.111.083618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Fan X., Miller A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63 (1), 153–182. doi: 10.1146/annurev-arplant-042811-105532 [DOI] [PubMed] [Google Scholar]
- Yang X., Xia X., Zeng Y., Nong B., Zhang Z., Wu Y., et al. (2020. a). Genome-wide identification of the peptide transporter family in rice and analysis of the PTR expression modulation in two near-isogenic lines with different nitrogen use efficiency. BMC Plant Biol. 20 (1), 193. doi: 10.1186/s12870-020-02419-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhang H., Li X., Shen H., Gao J., Hou S., et al. (2020. b). A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system. Nat. Plants. 6 (9), 1167–1178. doi: 10.1038/s41477-020-0747-7 [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu Y.-X., Zhang N., Hu B., Jin T., Xu H., et al. (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37 (6), 676–684. doi: 10.1038/s41587-019-0104-4 [DOI] [PubMed] [Google Scholar]
- Zhao L., Chen P. F., Liu P., Song Y. P., Zhang D. Q. (2021). Genetic effects and expression patterns of the Nitrate transporter (NRT) gene family in Populus tomentosa . Front. Plant Sci. 12. doi: 10.3389/fpls.2021.661635 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosome location and distribution analysis of the SiNPF genes. Tandem duplicated genes are linked by a red curve.
Chromosome location and distribution analysis of the SvNPF genes. Tandem duplicated genes are linked by a red curve.
Phylogenetic relationship of NPF proteins among green foxtail, rice and Arabidopsis. Different subfamilies are color-coded as illustrated in the figure. The red filled circle and the green filled square indicates these NPFs have been characterized in Arabidopsis (red filled circle) or rice (green filled square). The detailed information of these characterized NPF genes were summarized in Table S1 .
Phylogenetic relationship, gene-structure analysis, and motif distributions of SiNPF genes. Exon-intron structures are customized in each subfamily (with customized scale bar), the green bar represents exons, the black line represents introns, and the yellow bar represents UTR (untranslated region). The different motifs are color-coded as illustrated in the figure. The scale represents amino-acid length.
cis-acting regulatory elements in the SvNPF promoters. Different colored circles represent different types of cis-elements.
Amino acid sequence alignment of AtNRT1.1 and SiNPFs. The red star indicates Pro492 in AtNRT1.1.
Summary of the characterized NPF genes in Arabidopsis and rice
Primers used in this study.
Data Availability Statement
The raw RNA-seq data of foxtail millet treated with low nitrate have been deposited in the National Genomics Data Center (https://ngdc.cncb.ac.cn/) under the BioProject accession PRJCA012843. The other sequencing data were obtained from our Multi-omics Database for Setaria italica (MDSi) (http://sky.sxau.edu.cn/MDSi.htm) and JGI Phytozome (https://phytozome-next.jgi.doe.gov/).