Abstract
During pharmacotherapy, knowledge about the actual drug and metabolite concentrations in plasma is often critical. Individual dose adjustments can be performed based on pre‐emptive genotyping of certain absorption, distribution, metabolism, and excretion (ADME) genes but also using therapeutic drug monitoring (TDM). Analyses of liquid biopsies for tumor‐derived components are well‐established and have been found to be a good complement to biopsy examinations. Recently, liquid biopsy‐based quantification of cell‐free RNA (cfRNA) in plasma exosomes was proposed as a proxy measurement for the expression of different hepatic ADME genes and for the rate of drug metabolism, constituting an alternative to TDM. In this study, we validated these findings by examining the correlation between mRNA expression of eight different CYP genes in liver and the corresponding rate of enzyme‐specific drug metabolism in 96 donor‐matched liver samples. Analyses of CYP‐dependent drug metabolism in liver microsomes in comparison to the level of mRNA expression for the different CYP genes revealed a mean Pearson correlation coefficient of 0.28. The highest correlations (0.33–0.34) were obtained for CYP2D6 and CYP3A4 and the weakest correlations were observed for CYP1A2 and CYP2B6 (0.18–0.21). In all cases, the correlations obtained were too weak to demonstrate a predictive relationship, likely due to different regulatory and post‐translational events controlling the rate of enzyme activity. Our results reinforce the notion that, whilst liquid biopsy‐based approaches might have utility for prediction of hepatic CYP protein expression, they are not currently an important substitute for TDM.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Therapeutic drug monitoring (TDM) is clinically used to optimize patient treatment with drugs, many of which are metabolized by polymorphic enzymes. Two recent relatively small studies suggest that plasma exosomal cell free RNA (cfRNA) levels can be used as a substitute for TDM because a correlation between the cfRNA levels and hepatic protein expression of variant pharmacogenes as well a relationship between the cfRNA levels and CYP activity was found.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study examined whether hepatic CYP mRNA expression could predict hepatic CYP enzyme activity within liver pieces from 96 different donors.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study indicates that the correlation between hepatic mRNA expression and enzyme‐specific drug metabolism in eight different CYPs is poor, and that mRNA expression is not a useful predictor of hepatic CYP activity.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
The data indicate that cfRNA expression in plasma exosomes by liquid biopsy serum samples might not be a useful marker for the catalytic activity of different hepatic CYPs, which supports the use of TDM in a clinical setting.
Therapeutic drug monitoring (TDM) is a common clinical method for determination of drug and metabolite profiles in patients undergoing drug treatment. Efforts have been made to predict individualized drug therapy based on a patient’s ability for absorption, distribution, metabolism, and excretion (ADME). Such genetic predictability can be highly valuable, although the influence of factors, including drug–drug interactions, pathophysiology, and, importantly, the incomplete understanding of the heritability in pharmacokinetics and pharmacodynamics, underlines the importance of TDM for a successful clinical drug therapy optimization. However, it is true that TDM analyses can be difficult to successfully reproduce due to uncontrolled drug interactions, variations in physiological and pathophysiological factors, etc.
Anticancer therapies based on liquid biopsies are well‐established, largely because tumor cells export biomarkers with unique properties. Recently, attempts have been made to use similar liquid biopsies to quantify cell‐free RNA (cfRNA) in plasma exosomes as proxy measurements for the rate of hepatic drug metabolism. Achour et al. 1 used liquid biopsies and found a correlation between plasma exosomal cfRNA expression and hepatic protein expression of 8 CYPs and 4 UGTs in 29 patients (r = 0.6–0.89). In another relatively small study (n = 30) a correlation (r = 0.44–0.70; P < 0.05) between cfRNA and activity of four different CYPs in patients was phenotyped using the Geneva cocktail. 2
In this study, we intended to validate these finding by examining the correlation between mRNA expression of several different CYP genes in the liver and the enzymatic activity of the corresponding enzymes in donor‐matched liver samples.
MATERIALS AND METHODS
A biobank of snap frozen human liver pieces (n = 96) that was previously established and characterized 3 , 4 was used. Liver donors had provided written consent and the study was approved by the ethical committee at Karolinska Institutet, Stockholm (D:nr 97:112; 429‐01 03.6022010/541‐31/1; 2010/678‐31‐3).
Human liver microsomes
Microsomes were prepared by subcellular fractionation, as described elsewhere. 5 The frozen liver samples were thawed at 4°C in five volumes of homogenizing buffer (250 mM sucrose, 50 mM Tris–HCl, pH 7.4). The human liver microsomes were suspended in 100 mM potassium phosphate buffer (pH 7.4) containing 20% (v/v) glycerol, pearl frozen in liquid nitrogen and stored at −70°C.
Analysis of enzyme activities
Cytochrome P450 activities were analyzed using the liver microsomal fractions by Pharmacelsus GmbH (Saarbrücken, Germany). Determination of enzyme reactions was performed using microsomal samples incubated in phosphate buffer (pH 7.4, 20% (v/v) glycerol), 1 mM NADPH at 37°C, with two sampling time points within 0–120 minutes. The substrate concentrations were phenacetin (26 μM), bupropion (50 μM), diclofenac (9 μM), midazolam (3 μM), coumarin (5 μM), chlorzoxazone (50 μM), S‐mephenytoin (20 μM), and bufuralol (9 μM). Linearity of product formation with time was ensured in all cases. Samples were subjected to acetonitrile precipitation. Liquid chromatography‐high resolution mass spectrometry (LC/HRMS) analysis was then performed on a Q‐Exactive mass spectrometer (Orbitrap technology with accurate mass) applying simultaneous dual polarity measurement in a single run for substrate and metabolite quantification. Lower limits of detection were found in the range of 1–6 nM. Accuracy and precision were within the acceptance criteria of the US Food and Drug Administration (FDA) validation guidelines for quantitative bioanalytical methods.
Analysis of mRNA expression
The mRNA samples from the different livers were previously analyzed, as described elsewhere. 4 For the current study, the same mRNA data were used. As described, 4 RNA was isolated from the liver bank samples using AllPrep DNA/RNA/Protein Mini kit (Qiagen, Hilden, Germany, Cat. #80004). The RNA samples were quantified and assessed for integrity using an Agilent Bioanalyzer 2100 with the RNA 6000 Nano kit (Agilent Technologies, Santa Clara, California; Cat. #5067‐1511). Briefly, starting with 300 ng of RNA, the TargetAmp‐Nano Labeling Kit for Illumina Expression BeadChip (Illumina, San Diego, CA) was used to amplify and biotinylate the RNA samples from the livers according to the manufacturer’s instructions. For each sample, 750 ng of biotinylated cRNA was hybridized to Human HT‐12 BeadChips (Illumina, v4 arrays), according to the standard protocol. The BeadChips were scanned within 24 hours using a HiScanSQ scanner. The raw signals were exported using GenomeStudio (Illumina, San Diego, CA).
Statistical analysis
Analyses were performed using GraphPad 9. Correlations were determined using Pearson correlation coefficient. Outliers were detected and excluded using the Robust Regression followed by Outlier Identification (ROUT) method with a Q‐value of 0.1. 6
RESULTS
Human liver microsomes (n = 96) were analyzed for CYP enzyme‐specific metabolic activity by liquid chromatography‐mass spectrometry LC‐MS by Pharmacelsus GmbH (Saarbrücken, Germany) as described in the Materials and Methods section. In total, eight different CYP enzymes were examined by quantification of: phenacetin‐demethylation (CYP1A2), bupropion‐hydroxylation (CYP2B6), diclofenac‐4‐hydroxylation (CYP2C9), bufuralol‐1‐hydroxylation (CYP2D6), midazolam‐1‐hydroxylation (CYP3A4), coumarin‐7‐hydroxylation (CYP2A6), chlorzoxazone‐6‐hydroxylation (CYP2E1), and mephenytoin‐4‐hydroxylation (CYP2C19) using LC/HRMS. The relationship between mRNA expression and catalytic activity for each CYP enzyme was analyzed and is presented in Figure 1 . The mean Pearson correlation of these analyses was 0.285. The highest correlations (0.33–0.34) were obtained for CYP2D6 and CYP3A4 and the weakest correlations were observed for CYP1A2 and CYP2B6 (0.18–0.21). Outlier analysis, performed using the ROUT method applied with a Q value of 0.1, revealed that in most cases removal of outliers decreased the strength of the correlation between mRNA expression and activity causing a decrease of the mean r value from 0.313 to 0.285. In all cases, the correlations obtained were too weak to demonstrate a predictive relationship.
Figure 1.
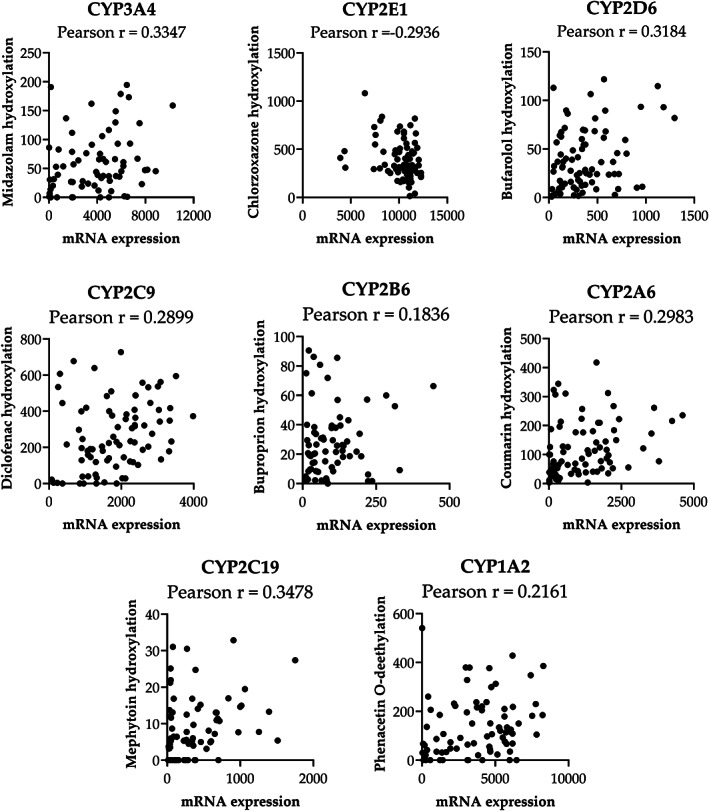
Correlation between CYP mRNA expression and enzyme activity in human liver tissue and microsomes. The mRNA values are presented as the raw signals from GenomeStudio. Activity values are presented as pmol/min/mg microsomal protein measured after 10 minutes of drug incubation. Outliers were detected using the Robust Regression followed by Outlier Identification (ROUT) method, Q value 0.1. Pearson correlation coefficient calculated using GraphPad Prism 9.
DISCUSSION
The use of a liquid biopsy based on plasma exosomes for enzyme activity prediction relies on the quantification of cfRNA and exosomal protein as proxy measurements of pharmacologically relevant gene products in the liver. Naturally, this approach does not provide the clinical levels of the drugs and metabolites in question. In addition, uncertainties in this extrapolation include the extent to which the analyzed exosomes originate from the liver and also the extent to which the cfRNA values relate to the true catalytic or transport activity of the corresponding gene products within the liver. The catalytic activities of hepatic drug metabolizing CYPs are regulated at several levels 7 and are influenced by the genetic variation in multiple genes. 8 Post‐transcriptional regulation occurs on several levels, including RNA splicing, regulation by miRNAs, phosphorylation determining enzyme activity and degradation, and regulation by substrate binding. Therefore, the critical issue is the extent to which the RNA expression in the liquid biopsies correlate to true hepatic drug metabolizing activity.
The data presented in Figure 1 indicate that the mechanisms discussed above manifest as difficulties in determining a simple link between hepatic mRNA expression and catalytic activity. Thus, post‐transcriptional and post‐translational events make it challenging to use circulating exosomal cfRNAs as true markers of hepatic P450‐mediated drug metabolism. This also includes the fact that the tissue origin of the cfRNA measured in the liquid biopsy represents a problem. Achour et al. demonstrated the correlation between cfRNA expression in plasma exosomes and hepatic protein expression for different CYPs 1 in liver samples from the same patients. However, the data presented here indicate that hepatic mRNA expression from donor‐matched liver tissue does not correlate significantly to CYP activity. This indicates a disconnect between hepatic protein expression and enzyme activity and a similar disconnect between plasma exosome cfRNA and hepatic enzyme activity would be anticipated. The authors also recently presented a relationship (R = 0.44–0.7; P < 0.05; n = 30) among plasma exosomal cfRNA levels of CYP1A2, CYP2B6, CYP2C9, and CYP3A and activities monitored using the Geneva cocktail. 2 The data are interesting and should be verified in a larger cohort, however, the results presented here do not support such a relationship. In conclusion, our results reinforce the notion that although liquid biopsy‐based approaches might have utility for prediction of hepatic CYP mRNA expression, in our opinion, this method is not currently an important substitute for TDM in a clinical setting.
FUNDING
This study was supported by grants from the European Research Council (ERC)–Advanced Grant (AdG) project HEPASPHER (grant agreement 742020), The Swedish Research Council (grants 2021‐02732 & 2018‐05766) and from European Union’s Horizon 2020 research and innovation program PSY‐PGx under grant agreement No. 94515.
CONFLICT OF INTEREST
M.I.‐S. is a co‐founder and co‐owner of HepaPredict AB. All other authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
C.S.P. and M.I.‐S. wrote the manuscript. M.I.‐S. and I.J. designed the research. I.J. performed the research. C.S.P. and I.J. analyzed the data.
References
- 1. Achour, B. et al. Liquid biopsy enables quantification of the abundance and Interindividual variability of hepatic enzymes and transporters. Clin Pharmacol Ther 109, 222–232 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Achour, B. et al. Liquid biopsy for patient characterization in cardiovascular disease: verification against markers of cytochrome P450 and P‐glycoprotein activities. Clin Pharmacol Ther 111, 1268–1277 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Westlind, A. et al. Interindividual differences in hepatic expression of CYP3A4: relationship to genetic polymorphism in the 5′‐upstream regulatory region. Biochem Biophys Res Commun 259, 201–205 (1999). [DOI] [PubMed] [Google Scholar]
- 4. Bonder, M.J. et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics 15, 860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ernster, L. , Siekevitz, P. & Palade, G.E. Enzyme structure relationship in endoplasmatic reticulum of rat liver: a morphological and biochemical study. Cell Biol 15, 541–562 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Motulsky, H. & Brown, R. Detecting outliers when fitting data with nonlinear regression – a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform 7, 123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zanger, U.M. & Schwab, M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138, 103–141 (2013). [DOI] [PubMed] [Google Scholar]
- 8. Ingelman‐Sundberg, M. , Sim, S.C. , Gomez, A. & Rodriguez‐Antona, C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116, 496–526 (2007). [DOI] [PubMed] [Google Scholar]


