Abstract
The high organ specification of the human heart is inversely proportional to its functional recovery after damage. The discovery of induced pluripotent stem cell‐derived cardiomyocytes (iPSC‐CMs) has accelerated research in human heart regeneration and physiology. Nevertheless, due to the immaturity of iPSC‐CMs, they are far from being an representative model of the adult heart physiology. Therefore, number of laboratories strive to obtain a heart tissues by engineering methods by structuring iPSC‐CMs into complex and advanced platforms. By using the iPSC‐CMs and arranging them in 3D cultures it is possible to obtain a human heart muscle with physiological capabilities potentially similar to the adult heart, while remaining in vitro. Here, we attempt to describe existing examples of heart muscle either in vitro or ex vivo models and discuss potential options for the further development of such structures. This will be a crucial step for ultimate derivation of complete heart tissue‐mimicking organs and their future use in drug development, therapeutic approaches testing, pre‐clinical studies, and clinical applications. This review particularly aims to compile available models of advanced human heart tissue for scientists considering which model would best fit their research needs.
Keywords: cardiac tissue engineering, engineered heart tissue, human cardiomyocytes, induced pluripotent stem cells, iPSCs based tissue modeling
1. INTRODUCTION
Cardiovascular diseases (CVDs) are the leading cause of death globally. According to the WHO report, it is estimated that CVDs take approximately 17.9 million lives each year (https://www.who.int/health‐topics/cardiovascular‐diseases/#tab=tab_1]). They are a group of heart and blood vessels disorders and include coronary heart disease, cerebrovascular disease, and rheumatic heart disease. Most heart failures are connected with impaired activity of cardiomyocytes (CMs). Although amphibians and fish are able to regenerate their heart after infarction (Poss et al., 2002), human adult heart is terminally differentiated organ, consequently, the potential for repair through cardiomyocyte proliferation is extremely low. Thus adult cardiomyocytes lose their ability to undergo cell division and proliferation after a narrow proliferative window at the neonatal stages (Payan et al., 2020; Wu et al., 2021). To study the biology of CMs and the development of new therapeutic strategies for CVDs, reliable and advanced heart models are needed. For needs of the manuscript we define a model as a complex structure that have a very close physiological functions, mimic the environment and may respond to medical test or treatment in similar way as the organ/tissue of interest. In our opinion the perfect heart model should:
-
‐
be patient‐specific
-
‐
carry the genetical background (in case of genetic diseases)
-
‐
recreate the environment of the tissue in physiological or pathological conditions.
Such models allow for an improved understanding of the healthy heart's molecular processes as well as disease mechanisms and therapeutic strategies.
During the development and testing of new therapeutic compounds, cardiotoxicity remains a major cause of failure. It is estimated that only 8% of new drugs will successfully transit from clinical trials to market launch, and even then, several will be withdrawn after preliminary approval (Ferri et al., 2013; McNaughton et al., 2014). This is in part due to the fact that conventional in vitro cell cultures and in vivo animal models of the heart failure are ineffective in modeling human diseases and predicting drug responses, which is caused by limited relevance and scalability. Moreover development of new heart models may solve the problem regarding maturation of the iPSC‐CMs. This will be a milestone to obtain the perfect environment for testing the new therapeutic approaches in the field of regenerative medicine. Heart tissue engineering may also provide models for treatment genetic diseases that involve myocardial pathologies. Certainly creation of patient‐specific fully matured and vascularized patched will push the personalized medicine forward.
However, human cardiac conditions may be more precisely reflected by developing engineered, physiologically relevant 3D models. Human pluripotent stem cells, and their potential for differentiation into CMs, provided an appropriate starting point and reasonable background for the development of human cardiac models that allow improved recapitulation of the human heart physiology and pathology.
In this review, we focus on innovative techniques for cardiac tissues engineering in both two and three dimensions that closely replicate the human heart. We discuss why reliable cardiac models are needed, which cells have been already used for model generation, and how iPSC‐derived cardiomyocytes could be maturated. Next, we provide an overview of the selected models, including cell‐based models, such as organoids, muscular thin films and cell sheets, followed by engineered heart tissues, heart‐on‐a‐chip systems, and decellularized heart approaches. For each system, we describe its usefulness, as well as its advantages and disadvantages, possible application, and future directions for technology development. Moreover, we also show a comparison of the selected models in terms of cardiac muscle features, metabolic supply, and readiness for use.
2. WHY DO WE NEED HUMAN HEART IN VITRO MODEL
An ideal model of cardiac tissue should mimic the physiological conditions of cardiomyocytes in adult human heart such as: morphology; contractility and hemodynamics properties of whole heart that is associated with proper electrophysiology of cells; calcium distribution within the contractions. Unfortunately, animal models do not fully fulfill these criteria. In models of small animals, the resting heart beats approximately tenfold faster, while the QT interval is fourfold shorter than in a typical human heart (Passier, van Laake, & Mummery, 2008). In the case of models of large animal, they more closely resemble the human heart. For example, the ratio of heart weight to body weight in 20‐ to 30‐kg pigs, which are used frequently in cardiovascular studies, is identical (5 g/kg) to that of adult humans whereas in dogs it is approximately 7 g/kg. The porcine heart anatomy is more similar to humans then dogs: the number of orifices for the left atrium‐in pigs the left atrium receives oxygenated blood from two pulmonary veins, in dogs from five or six, and in humans from four or five; the tricuspid valve of swine has three leaflets as in humans, whereas dogs typically have two. Therefore during past couple of decades, pigs have gained favor over dogs, mostly because of marked differences in coronary anatomy between dogs and humans and in part due to ethical and social concerns regarding their use in biomedical research (Lelovas et al., 2014). On the other hand, the adolescent human heart is similar to a porcine heart, while the older heart with ischemic heart disease is considered to have more dog‐like characteristics. However, for example, ischemia‐reperfusion induced arrhythmias are still more frequent in animals than in humans (Camacho et al., 2016). Models of large animals are also less suitable for the genetic selection and production of transgenic strains and disease modeling. Finally, models of large animals are difficult to manage and require specialized infrastructure and trained personnel, which vastly increases the already enormous costs of pre‐clinical studies, putting additional strains on the already decreasing effectiveness of the drug discovery field (Camacho et al., 2016).
In terms of newly developed drug testing, animal models do not allow for a full prediction of cardiotoxicity due to differences in ion channels expression, biological pathways as well as in pharmacokinetic properties. In addition to heart function, CVD modeling often requires additional stimulation, such as mimicking the immune system or hormone response mechanisms, thus defining the complicated nature of CVDs. Although a fully mature human heart model still does not exist, no species or animal model will present an accurate simulation of human cardiac disease; this will remain an approximation. However, the development of a reliable in vitro models (either 2D or 3D) using iPSC‐CMs may be crucial to study some important parameter that reflect the physiology of cardiomyocytes such as: morphology, calcium handling and contractility. Moreover generation of adequate pathological model that mimic some abnormalities in physiology of the cardiomyocytes (genetic disease, ischemia, hypertropia etc.) will allow to investigate alterations in cells for example‐abnormal sarcomere organization, cellular hypertrophy, and altered calcium handling.
So far there are a few models based on iPSC‐CMs that enable studies on the disease and the physiology of the cardiomyocytes. Lan et al. presented an elegant model regarding hypertrophy of cardiomyocytes. This model enable to investigate the impairment in the calcium transient caused by reorganization in contractile apparatus (Lan et al., 2013). Some models allow the study of the molecular pathways underlying cardiomyocyte physiology. For example, the genetic disorder such as Duchenne Muscular Dystrophy affects contractile dysfunction due to the degeneration of cardiac muscle (Kamdar et al., 2020). iPSC‐CMs based models are also convenient and reliable tool to describe aberrant features in arrhythmic diseases and electrophysiological properties of cardiomyocytes. The generation of such models enable to recapitulated the disease phenotype, including sodium or potassium current abnormalities (Shinnawi et al., 2019).
Presented studies are commonly based on patient‐specific models what is important in developing personalized treatment systems.
Taking all together, hPSC‐CMs system could play a pivotal role in discovering new therapeutic approaches by drug testing, gene therapy application and significantly facilitate the study of the CMs biology under physiological and pathological conditions.
3. FACTORS UNDERLYING THE UNIQUENESS OF CARDIAC TISSUE MODELS
To construct the human adult heart muscle, several components are necessary. The proper organization of specific cell types and maturation factors are among them. A brief summary of currently exploited materials and methods to reconstruct the human heart model is presented below.
3.1. Cells currently used for cardiac tissue models
An adult human heart is a complex organ, composed of four morphologically and functionally distinct chambers. The cellular landscape of the human heart contains many cell types, such as atrial and ventricular cardiomyocytes, fibroblasts, endothelial cells, pericytes and smooth muscle cells (mural cells), immune cells, adipocytes, and mesothelial cells. Moreover, the distributions of these main cell types differ between heart chambers, including atrial and ventricular tissues. In the atrium, there are fewer CMs and mural cells, but more fibroblasts, endothelial and immune cells compared to the ventricle (Litviňuková et al., 2020).
CMs, being the major element of heart force production machinery, account for approximately 75% of the heart volume yet represent only 33% of the total cell number, as estimated from nucleus numbers. The remaining 60% comprises fibroblasts and approximately 10%, pacemaker cells, endothelial cell, pericytes, and others (Litviňuková et al., 2020). Fibroblasts, as the most common cell type in the heart, play a pivotal role in maintaining the heart's structure as well as its mechanical and electrical functions (Doppler et al., 2017). The last group of different cell types, although the smallest, is also essential for normal cardiac function, such as vascular formation, stabilization, remodeling, and signal transduction (Armulik, Abramsson, & Betsholtz, 2005). Of note depending on the context, the ratio and percentage of cell types may wary. When considering the heart structure we only provide the information about cells that are exactly involve in heart itself, without the ones that are associated to the vasculature structure. When we will consider the whole structure of the organ along with the blood vessels obviously the endothelial cells or pericytes will also be the major groups of the non‐myocyte cell type (Pinto et al., 2016).
The derivation of CMs in vitro is important in the context of developing cardiac models. There is a need for robust and renewable cell sources. Previously, common sources of CMs were animals; however, the differences in physiology have prompted scientists to search for more reliable cells to build human‐like cardiac tissue. Human CMs can be derived directly from myocardium. Coppini et al. optimized the protocol for the isolation of human ventricular CMs from patients undergoing cardiac surgery. They showed that isolated cells stay viable for the time needed for analysis, display an adaptation of action potential duration and electrophysiological responses (calcium transient, sarcomere organization). This suggests the preservation of the functionality and overall structural integrity of ion channels. Using patient samples may provide important information on disease‐related changes in cardiomyocyte function. The main disadvantage of this approach, however, is that during isolation, cell to cell connections as well as association with cardiac ECM are lost. It is difficult to keep cells in culture for long time. As the heart is postmitotic organ, the cardiomyocytes proliferation potential is very limited. During the time they rather increase their size then number. Till now the process that drives cardiomyocytes into cell cycle arrest and switch them from proliferative state into hypertrophy remains unknown (van Amerongen and Engel, 2008). As it was mentioned, CMs undergo significant remodeling with time, which may affect the relevance of the studies (Coppini et al., 2014). Therefore, currently, the main approach for obtaining a relatively homogenous CM population is focused on human pluripotent stem cells, including hESCs and hiPSCs (Dell’Era et al., 2015). Studies show that hiPSC‐CM‐based models are reliable and seem to be the most relevant to human physiology for pharmacological purposes (Mathur et al., 2015; Takasuna et al., 2017). Their differentiation process resembles that occurring under in vivo conditions.
During gastrulation, CMs emerge from mesodermal tissue. Their fate is influenced by proteins produced by the adjacent endoderm, whereas WNT‐mediated signals from the underlying neural tube and notochord act against CM specification. The addition of molecules that may suppress or activate the aforementioned signaling pathways drive hiPSC differentiation into mesoderm, cardiomyogenic mesoderm, cardiac precursors, and cardiomyocytes. In widely employed protocols, the initial step relies on mesodermal induction by the initial addition of Wnt pathway agonists, including CHIR99021 and CHIR98014, which leads to an increase in beta‐cathenin accumulation in cell cytosol. The second step focuses on early cardiac progenitor cell differentiation by subsequent WNT pathway inhibition (IWP2 or IWR1). Finally, cardiac fitted medium leads to the maturation of sarcomeres thanks to the insulin‐induced Akt pathway, by exploiting RepSox, forskolin, and valproic acid signaling (Kolanowski, Antos, & Guan, 2017). iPSCs‐derived cardiomyocytes are immature and have prenatal or newborn‐like phenotype (Feric & Radisic, 2016). The maturation process drives cardiomyocytes into two subtypes‐atrial (aCMs) and ventricular (vCMs). Starting from the morphology of both subtypes the aCMs are smaller in size and surface area than vCMs. Transcriptomic analysis conducted by Cyganek et al. reveal some crucial differences in gene expression profile. Based on NGS studies ventricular CMs found to have a higher expression of following genes: HAND1, HEY2, and IRX4; MYL2, MYH7, and GJA1; as well as KCNJ2 and KCNJ4 for instance atrial CMs were expressed HEY1, TBX5, and NR2F2; MYL7, MYH6, and GJA5; as well as KCNA5, KCNJ3, and KCNK3. Presented key transcription factors, structural proteins, and ion channels, are considered as a major determinants of these two subpopulations. Due to the distinction in expression profile these two subtypes are characterized by different electrophysiological properties. In aCMs predominantly Kv1.5 (KCNA5), Kir3.1 (KCNJ3), and K2p3.1 (KCNK3) ion channels are expressed. In turn for vCMs Kir2.1 (KCNJ2) ion channel is characteristic (Cyganek et al., 2018).
Cardiac tissue models, regardless of type, often follow heart muscle complexity in terms of cell types and organization. It was shown that the combination of non‐CMs and CMs enhance tissue formation and maturity processes (Fennema, Rivron, Rouwkema, van Blitterswijk, & de Boer, 2013; Giacomelli et al., 2017). In the organoid model of Buono et al., three types of cells, hiPSC‐CMs, human cardiac microvascular endothelial cells (HCMECs), and human cardiac fibroblast (HCFs), were used to improve organoid characteristics (Filippo Buono et al., 2020). Cell sheets designed as a mixture of iPSC‐CMs, endothelial cells, and vascular smooth muscle cells (VSMCs) show increased vascularization and thus survival of the tissue after grafting into the heart in vivo (Ishigami et al., 2018). Engineered heart tissues (EHTs) consisting of CMs, endothelial cells, and fibroblasts develop stabilized microstructures that align with CM orientation, with fibroblasts being essential for proper extracellular matrix organization (Caspi et al., 2007). It is important to bear in mind that only the addition of non‐CMs in physiological ratios and within structured tissue organization allows proper CM–fibroblast coupling and avoids arrhythmic events often occurring in cardiac models (Sridhar, Vandersickel, & Panfilov, 2017; V. Balashov et al., 2018).
3.2. Maturation factors
CM maturity can be assessed by analysis of the expression of characteristic genes (RyR2, SERCA, Myh6, Myh7, NCX, Hey1, Hey2, Mlc2v, Mlc2a, etc.), as well as analysis of the composition and organization of the cytoskeleton and contractile apparatus, oxidative metabolism, and electrophysiological features (Kolanowski et al., 2017).
Currently 2D protocols of in vitro iPSC‐CMs differentiation generates the immature fetal‐like cells. It is probably due to the absence of physical and environmental stimuli. Number of approaches have been proposed to advance iPSC‐CMs toward a mature stage (Zhao, Ye et al., 2020 and; Zhao, Rafatian et al., 2020). An optimal in vitro tissue model should incorporate iPSC‐CMs into in vivo‐like tissue structures with an ECM composition and appropriate cell types that resemble cell–cell interaction as well as microenvironment geometry. Nowadays the most efficient method to obtain more mature cardiomyocytes is based on generation of 3D EHT (engineered heart tissue). It appears that the protocols for 3D cultures reconstruct the biophysical stimuli and intercellular crosstalk which are essential for the physiological hypertrophy of postnatal cardiomyocytes (Feric & Radisic, 2016; Scuderi & Butcher, 2017). A great advancement in the cell–ECM interaction field was achieved by the group of Agladze. They showed that fibroblasts interact with ECM elements by creating focal adhesion clusters interconnecting piles and polymers of ECM. Simultaneously, CMs “envelop” single polymers, forming “sheathed” structures. Based on this work, it is clear that only by applying the appropriate ECM composition and CM:fibroblast ratio can a tissue mimicking model resemble the original heart tissue (V. Balashov et al., 2018). According to Ronaldson‐Bouchard et al., 2018 supporting iPSC‐CMs by fibroblasts can advance them to an adult myocardial‐like state after a few weeks of 3D culture under physical conditioning and electrical stimulation (Ronaldson‐Bouchard et al., 2018). They show adult‐like gene expression profiles, well‐organized sarcomere structures, presence of transverse tubules, a positive force‐frequency relationship and high‐density mitochondria. Moreover, using a Biowire chip seeded with cell‐hydrogel mixture, Zhao et al. (2019) have constructed a platform which can generate atrial‐ and ventricular‐specific cardiac tissue by combining the directed cell differentiation and electric field conditioning, which is very close to the simulation of distinct electric signaling in adult heart chambers (Zhao et al., 2019).
The spontaneous contractions that appear as a result of the differentiation process are considered as a functional effect of cardiac ion channels and transporter presence, with many related mainly to action potential generation. However, hiPSC‐CMs generated with available protocols are still immature due to the lack of humoral factors as well as mechanical and electrical stimuli (Tu, Chao, & Wu, 2018 Tu et al., 2018). In the literature, there are many approaches stimulating maturity of cardiac tissue. They can be divided into biochemical and physical stimulation. Biochemical stimuli include adrenergic signaling agonists, such as isoproterenol, norepinephrine and phenylephrine, and are used in a time‐ and dose‐dependent manner. For instance, the addition of triiodo‐L‐thyronine is crucial for later stages of maturation and increases force output (Yang, Rodriguez et al., 2014). In the case of physical stimulation, mechanical stretching (Liaw & Zimmermann, 2016), flow stimulation (dynamic culture conditions) (Jackman, Carlson, & Bursac, 2016), electrical stimulation (Baumgartner et al., 2015), and magnetic fields (Sapir, Polyak, & Cohen, 2014 Sapir et al., 2014), have been successfully employed as maturation factors. The relative hiPSC‐CM immaturity is a general challenge in the field, and many newly developed approaches are currently being tested to improve this issue, such as molecular modifications, including miRNAs or selected gene overexpression (Yang, Pabon et al., 2014).
4. CURRENTLY DEVELOPED CARDIAC TISSUE MODELS
Although each model consists of specific components, they still need to be adequately organized. Cardiac tissue engineering methods rely mainly on the use of heart cells together with the synthetic or biological matrix to mimic a cardiac‐like tissue environment. These approaches have been developed in order to regenerate or replace diseased myocardia in vivo as well as a reliable model for drug testing. Engineered cardiac tissue should mimic a tissue‐like structure, show efficient contractility, remain electrophysiologically stable, be flexible (in terms of size expansion), and be prone to vascularization. To ensure reliability, systems should also recapitulate conditions of the human heart, such as the 3D anisotropic structure, extracellular matrix (ECM) network, vascularization, and circulation (Mathur, Ma, Loskill, Jeeawoody, & Healy, 2016). Thus, an optimal approach would include the incorporation of iPSC‐CMs into an vivo‐like tissue structure with an organized ECM and required cell types that resemble cell–cell interaction as well as microenvironment geometry. Cardiac tissue engineering methods can be divided into matrix based and non‐matrix based. The first group can be further subdivided based on matrix constitution of solid (gelatin mesh, collagen mesh, polystyrene beads, and polyglycolic acid) and liquid/gel (collagen/matrigel). Moreover, different types of equipment for environment control are used in order to provide metabolic advancement and mechanical stimulation (Zimmermann, Melnychenko, & Eschenhagen, 2004).
Thus far, cardiac engineering techniques have been focused on three main aims: (1) development of advanced cardiac muscle features (e.g., Frank–Starling mechanism; force–frequency relationship) to model heart tissue development and diseases; (2) improvement of nutrition supply (microfluidic devices, heart‐on‐chip systems, etc.); (3) development of application‐oriented strategies, such as tissue patches for heart regeneration, decellularized hearts scaffolds for engineering of artificial heart, and muscular thin films or microstructures for pharmacological screening. However, these aims are often overlapping; thus, many models can be used interchangeably, for example, heart‐on‐chip systems are integrated with matrix‐based approaches to improve the positive force–frequency relationship for detecting a precise pharmacological drug interaction with the heart muscle.
Defining a comprehensive classification for cardiac tissue models, due to a number of existing approaches, is challenging. Nevertheless, for the purpose of this manuscript, we proposed two crucial factors that are often used to make an initial decision to develop a particular tissue technology for a defined application. These are as follows: (1) the potential for increased throughput in conducted research within a reasonable cost and (2) the system complexity that defines structural potential for mimicking the physiological characteristics of the human heart (see Tables 1 and 2; Figures 1 and 2). Potential for scaling varies between models and depends highly on the costs and effort needed to create a model unit (machines, controlling units, and manufacturing efforts) and availability of the substrates (e.g., heart tissue for decellularization). On the other hand, the potential for mimicking physiological characteristics is directly connected to the purposes for which the system is developed. Although lab‐on‐a‐chip‐based approaches are often highly complex and could allow formation of mature tissues, they often do not have a clinical approach as their first goal (e.g., preferably small sizes that increase throughput), whereas cardiac de‐/re‐cellularization is particularly designed for heart mimicking and in vivo applications.
TABLE 1.
Comparison of maturation state of selected cardiac tissue models
| Model | Calcium dynamics | Action potential (AP) | Action potential duration (APD90) | Metabolism/Maturation | References |
|---|---|---|---|---|---|
| Human heart in vivo | • Myofilament calcium sensitivity | • 4 phases: Upstroke (0); early fast repolarization (1); plateau (2); repolarization (3), diastole (4); | ∼250 ms (atrium) | • Reliance on oxidative phosphorylation (80% of cardiac ATP production fatty acid β‐oxidation); | Zhao et al., 2019 EL‐Armouche & Eschenhagen, 2009 |
| EC50 = 701 nM. | • Atrial AP is shorter than ventricular AP. | ∼350–400 ms (ventricle). | • Lactic acid metabolism; | Lopaschuk & Jaswal, 2010 | |
| • β ‐adrenergic stimulation by β‐adrenergic receptors (β1‐ is the most abundant subtype). | Torres, Varian, Canan, Davis, & Janssen, 2013 | ||||
| Organoids | • Changes in the gene expression of calcium‐handling‐related genes (e.g., decrease in: ATP2A2, RYR2, CACNA1C, and SLC8A1, and increase in ITPR3). | • The atrium‐like regions exhibited significantly shorter APD90 than the ventricle‐like region. | 75 ms (atrial‐like) and 140 ms (ventricle‐like). | • Physiologically relevant metabolism–basal respiration ∼100 pmol/min; | J Et al., 2020 |
| • Presence of the K+ channel Kir2.1. | Richards et al., 2020 | ||||
| Muscular thin films (MTFs) | • Faster cycling of Ca2+ in comparison with 2D cultures. | • Contractile wave through MTFs, takes 160 ms, and back to its diastolic state‐360 ms; | Not presented. | • Increase in parallel registration of the sarcomeres at the Z‐disc. | Nishimura et al., 2004 |
| • Under isometric conditions, the duration of contraction shortened by 10‐fold; | Alford, Feinberg, Sheehy, & Parker, 2010 | ||||
| • Increase in peak systolic stress in comparison with 2D cultures. | Denning et al., 2016 | ||||
| Feinberg et al., 2012 | |||||
| Cell sheets | • Regular calcium transients proven. | • Conduction velocities of up to 25 cm/s (mESC‐CM/fibroblast); | 247 ms. | • Basal respiration‐46.1 pmol/min; | P. Lee et al., 2012 |
| • Contractile forces of up to 2 mN/tissue (mESC‐CM/fibroblast); | • Increase in inward sodium current density and a decrease in funny current densities; | Liau, Christoforou, Leong, & Bursac, 2011 | |||
| • Unidirectional action potential propagation, coupling with neighboring cell sheet; | • Presence of IKr and Na + channel (∼60% of the Na + current is inactivated); | Yoshida et al., 2018 | |||
| • A positive force–length and a negative isometric force–frequency relationship. | • Presence of gap junctions with connexin 43. | Shaheen et al., 2018 | |||
| Laksman et al., 2017 | |||||
| Wong et al., 2020 | |||||
| Engineered heart tissue (EHTs) | • Calcium‐handling proteins (L‐type calcium channels, LTCC; Na+/Ca2+−exchanger, NCX1; Na+/K+ ‐ATPase; Na+/H+ ‐exchanger, NHE1; SR Ca2+‐ATPase, SERCA; PLN are detected); | • Possibility to compare APD between atrial and ventricular EHTs. | 230 ms (atrial) and 420 ms (ventricular). | • More oxidative metabolism of glucose, lactate, and fatty acid and less glycolysis, and generated 2.3‐fold more ATP by oxidation than in 2D models; | Saleem et al., 2020 |
| Mannhardt et al., 2016 | |||||
| Ulmer et al., 2018 | |||||
| Lemme et al., 2018 | |||||
| Heart‐on‐a‐chip | • Contraction of CMs is strongly related to the transition of the intracellular calcium ion concentration; | • Possibility to mimic hypoxia influence on APD (a substantial reduction in mean APD50 (−46%) and APD90 (−34%)). | 236 ms. | • Basal respiration‐373.38 pmol/min and ATP production‐225.27 pmol/min; | Sakamiya et al., 2020 |
| • Spontaneous and synchronous calcium transient. | • Elongated cardiomyocytes with well‐developed sarcomeric structure and connexin‐43 positive gap junctions. | Pasqualini et al., 2018 | |||
| Sidorov et al., 2017 | |||||
| De/Recellularized heart | • Reduction in the duration of CaT90 and calcium upstroke time; | • Improved cardiac commitment; | 408 ms. | • Ion channel formation of CMs in cECM; | Garreta et al., 2016) |
| • Expression of calcium‐handling genes. | • Reduction of the conduction velocity. | • Sarcomere formation; | |||
| • Sarcomeric α‐actinin, cardiac troponin T, connexin‐43, N‐cadherin and myosin heavy chain expression; | Guyette et al., 2016 | ||||
| • Increases in the expression of different cardiac channels, such as SNCA5, KCNJ2, KCNA4, CACNA1C, SERCA2, KCNQ1, and KCNQ2. | |||||
| In vivo mouse heart model | • Full functional development of calcium storage system; | • Differences in ion channels affects APD90: both KV4.2 and KV4.3 are responsible for a fast transient outward current Ito,f in mouse; in human, only KV4.3. | 20–50 ms (atrial) and 52–54 ms (ventricular); almost ten‐fold shorter than in human heart in vivo. | • Basal respiration in young mice (2–3 months old)‐450 625 pmol/min; in old mice (22–28 months old)‐625 pmol/min. | Das & Muniyappa, 2013 |
| • Lack of voltage‐gated calcium channel CaV3.3 in mouse. | Lomax, Kondo, & Giles, 2003 | ||||
| Tanner & Beeton, 2018 | |||||
| Ying et al., 2016 |
TABLE 2.
Advantages and disadvantages of the selected cardiac tissue models and their possible applications
| Model | Advantages | Disadvantages | Application |
|---|---|---|---|
| Organoids | Robust; high throughput | Difficult to control, many cardiac lineages, low perfusability; limited maturity and pharmaceutical responses; no organotypic behavior | Screening for cardiotoxicity in early drug development phases |
| Muscular thin films (MTFs) | Organized structures, robust, high‐throughput; well‐developed and real‐time response measurement system | Medium‐to‐low maturity; limited tissue organization; limited possibility for ECM development | Screening of drug effects and cardiotoxicity |
| Cell sheets | Well established and standardized manufacturing procedures; unlimited tissue source; easy to handle in the laboratory | Difficult for application for inexperienced medical staff; rely on proper homing and adhesion; limited complexity; use limited to the size of the tissue injury; relatively low throughput | Regenerative medicine; heart infarction treatment |
| Engineered heart tissue (EHT) | Advanced maturation due to the mechanical stimulation in 3D environment; ECM allows for modeling of the larger than cellular scale disease effects; reproduces physiological features and complicated structural mechanisms of the heart tissue; response to the stimuli similar to mature human heart tissue; possibility to employ several cell types | Complicated culture system; difficult to handle and measure (requires specific equipment); hard to standardize; low throughput | Disease modeling; drug screening in preclinical phases; regenerative medicine (limited use) |
| Heart‐on‐a‐chip | Higher CM maturity; direct experimental access to the cell/tissue; controlled environment for cell development; medium–high throughput; automated cultures | Complicated system requiring installation of the specific equipment and supply of additional gases and media; relatively low flexibility (requires changes in chip design, specific expertize, and equipment). High chip prices | Drug screening for functionality and toxicity; studies of paracrine effect of the cells (combined media systems) |
| De‐/Recellularized heart | Mimic the properties of native heart matrix, maintain cell–cell contact and stiffness, mechanical anisotropy | Difficult to reproduce, hard to recellularize in terms of cells compartmentalization; species/personal differences; require access to the source of human heart tissues in good shape—limits the accessibility of the technique (the only model to still have this limit); currently low physiological functionality; lowest throughput | Heart transplantation procedures, replacement of the viable human heart organs |
| In vivo mouse heart model | Possibility to study hormonal regulation and organ‐to‐organ interactions in the whole organism | Difference in physiology (ex. Action potential duration, heat rate), differences in ion channels expression (ex.) | Basic research, preclinical studies (currently major model) |
FIGURE 1.
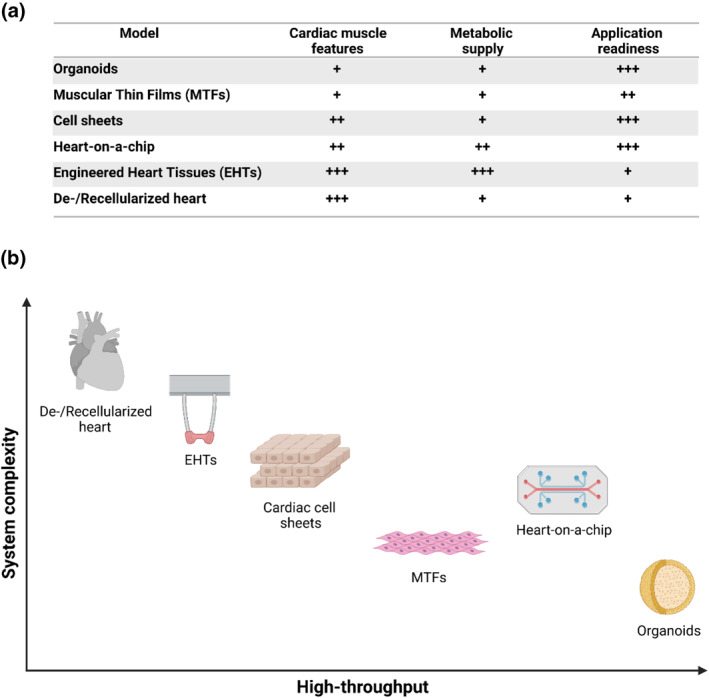
Characteristics of the described cardiac tissue models (a). Comparison of selected fea‐tures of models: + represents the level of recapitulation of selected heart tissue features, that is, +: low; ++ moderate; +++ high (b). Graph discriminating the models based on their complexity and high throughput (created with BioRender.com)
FIGURE 2.
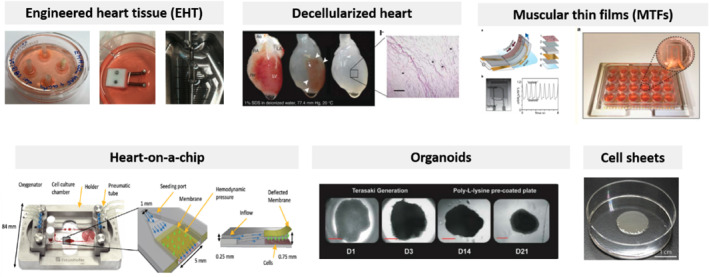
Images showing each described cardiac tissue model in vitro. Modified from MTFs (Brandenburger et al., 2012), decellularized heart (Bejleri & Davis, 2019), heart‐on‐a‐chip (Kolanowski et al., 2020), organoids (Filippo Buono et al., 2020), and cell sheets (Inui et al., 2019). License no. 5025300023207 (heart‐on‐a‐chip) and 5033880066840 (decellularized heart)
The attention should also be paid on a novel techniques involving tissue and organs 3D bioprinting. This novel technology still requires refinement and effort, but there are some studies that prove the method is forward looking. The group of Tal Davir were able to obtain personalized patient‐specific hydrogels that were used as a bioink to further studies on bioprinting (Edri et al., 2019). The group developed personalized strategy to prepare patch that fully match the immunological, biochemical and anatomical properties of the patient. They prepared 3D‐print thick, vascularized, and perfusable cardiac patches using patient‐specific iPS‐CMs. The result of their study demonstrate a great potential of this novel approach (Noor et al., 2019).
In this section, we provide an overview of available in vitro cardiac models along with their advantages and disadvantages and possible applications. We stratified the presented approaches on the basis of cell‐oriented model formation complexity (Figure 1 a, b).
4.1. Human cardiac cellular models
4.1.1. Organoids
Human cardiac organoids (hCOs) were proposed as a physiologically relevant model of the human heart, which has several applications, from drug screening to studying heart biology, including its regenerative potential. hCOs are created by the self‐organization of pluripotent cells into structures that should resemble the organogenesis of native cardiac tissue. Usually, they are small in size, approximately 100 μm, and contain multiple cardiac lineages (Filippo Buono et al., 2020; Voges et al., 2017).
Richards et al. showed that hCOs are promising models of myocardial infraction (MI) and drug cardiotoxicity. To model the organotypic response of the myocardium after infraction, they designed 3D microtissues with oxygen gradient diffusion and chronic adrenergic stimulation. Transcriptomic meta‐analysis of human control and infract organoids together with data on ischemic hearts from multiple species revealed the relevance of organoids, especially regarding acute post‐infarct ventricular tissue. Moreover, performed analyses (such as calcium dynamics inside 3D cardiac microtissues, gene expression analysis, immunofluorescence staining, and micropipette aspiration tests) showed that this model recapitulates certain aspects of the metabolism observed in the human infracted myocardium and may display pathological fibrosis. Taken together, the model proposed by the authors seems, to an extent, to recapitulate major hallmarks of the acute post‐MI cardiac state at the transcriptomic, structural, and functional level (Richards et al., 2020). Nevertheless, several important limitations of the model have been also reported. These limitations include the immature state of hiPSC‐CMs, lack of developed extracellular matrix, and limited possibilities of mimicking organ physiological characteristics. Finally, the lack of inflammatory cells, which play an important role in the activation of fibrotic response in terms of MI modeling, is a factor that limits hCO implementation (Forte, Furtado, & Rosenthal, 2018).
Voges et al. developed hCOs in order to provide insight into the endogenous repair processes of the immature human heart, which cannot be investigated in vitro. Cardiac tissue was obtained from human ESCs differentiated into cardiomyocytes and stromal cells. They showed that myocytes in presented in vitro model have an endogenous ability to recover contractile force after injury, not being impacted by surrounding cells. The designed hCO model in many aspects corresponds to neonatal heart regeneration in vivo, for example, lack of fibrosis and hypertrophy, and high rate of proliferation. Based on the obtained results, authors confirmed that immature human heart tissue has an intrinsic capacity to regenerate in response to injury (Voges et al., 2017).
Buono et al., in turn, adapted hCOs for the modeling of genetic hypertrophic cardiomyopathy, caused by heterozygous missense mutation (G‐ > A in position 2156) in the β‐myosin heavy chain (MYH7) gene. To mimic adult human heart tissue, their model consisted of three types of cells: hiPSC‐CMs, human cardiac microvascular endothelial cells (HCMECs), and human cardiac fibroblast (HCFs). HiPSCs for the generation of hCOs were derived from healthy human donors and patients with hypertrophic cardiomyopathy. Authors showed that hCOs represent significant phenotypical features of the healthy as well as hypertrophic human heart in vitro. They observed different beating behavior between the two groups studied, suggesting that hCOs may serve as a platform for the design and testing of patient‐tailored treatment as well as for drug development, particularly in the case of diseases caused by mutations in major CM contractile proteins (Filippo Buono et al., 2020).
Taken together, hCO models may, to an extent, mimic human tissue and bring significant advances for the establishment of innovative therapies. Organoids may be used for studies that cannot be performed in animal models or humans. However, despite the aforementioned advances, many challenges remain. The main limitation of hCOs is the relatively low maturity of CMs derived from pluripotent cells as well as other cells used for co‐culture. The maturity of CMs is pivotal for studies on drug toxicity and heart regeneration potential. The small number of cells and analyte volume (need for more sensitive biosensors) as well as the limitation of imaging depth (small size and compact structure) should also be taken into account when hCOs are considered. Another important issue is the minimal control over the arrangement of organoids in terms of their structure and function. External stimuli may be considered to enhance cell maturation and improve control over hCO development. Studies suggest that hCO models are still relatively immature, and they do not resemble the characteristics of the adult heart, but still show potential to expand our knowledge on human biology (Mills et al., 2017).
4.1.2. Muscular thin films and microstructures
Muscular thin film (MTF) models have been developed to recapitulate the ventricular microarchitecture in vitro. They can be engineered using both CMs (cMTFs) and vascular smooth muscle cells (vMTFs). Feinberg et al. showed the generation of MTFs, which included the seeding of dissociated muscle cells on a multilayer construct consisting of ECM proteins on multilayer cantilevers substrates. At a temperature below 35°C, MTFs are released from the surface and free float (Feinberg et al., 2007). During muscle contraction, the cantilever bends proportionally to the stress generated by the tissue. Changes in contractile stress can be quantified, enabling drug testing and disease modeling (G. Wang et al., 2014). Nishimura et al. showed that MTFs allow local changes in the radius of curvature, together with its length during the cardiac cycle, to be tracked. Cardiac MTFs, generated from neonatal rat ventricular myocytes, were developed to mimic ventricular lamellae, which recapitulated the anisotropic alignment of normal cardiac muscle and systolic stress levels (Nishimura et al., 2004).
Balashov et al. designed MTFs suitable for the dye‐free mapping of excitation waves in cardiac tissue cultures with little technical effort. This approach can be used for the study of the influence of chemical compounds on cardiac tissue, mechanisms underlying arrhythmias as well as maturation processes of hiPSC‐derived cardiac tissue. The main novelty of the approach is the use of a flexible cell/tissue film that generates off‐axis illumination during CM contraction. Inflection of the membrane creates bright/dark areas that could be used for consistent contraction wave tracking. System flexibility facilitates the larger displacements of cells during their contraction, which is beneficial for detection purposes (V. A. Balashov, Gorbunov, Guria, & Agladze, 2020).
Mathur et al. presented a cardiac microphysiological system generated from hiPSC‐CMs, which can be used for drug cardiotoxicity prediction, general drug testing, and disease modeling. In this system, the reported half maximal inhibitory/effective concentration values (IC50/EC50) were more consistent with the data on tissue‐scale references, in comparison to cellular‐scale studies. In the system, cells were aligned in the microtissue, which allowed efficient and continuous nutrient exchange, and cultured in a serum‐free medium. It was shown that the use of serum in culture medium may cause variations in the result due to variations in the serum composition as well as the possibility of serum contamination (Mathur et al., 2015; van der Valk et al., 2010). Moreover, authors reported that treatment with isoproterenol increased the beat rate with an EC50 of 315 nM. This value is in line with values observed for isolated human tissues, and higher than values observed for 2D hiPSC‐CMs. Similarly, good concordance with the contractility measurements of human ventricular heart tissue slices was also found (Brandenburger et al., 2012). Taken together, the aforementioned system seems to exhibit several advantages over conventional 2D systems. The possibility to combine it with microfluidic devices makes it a promising tool with a wide range of applications in pharmaceutical and biological studies. The microfluidic system ensures drug supply in a precise concentration continuously throughout the analysis (Mathur et al., 2015).
Expanding the MTF system, Lind et al. designed a multiwell cMTF platform with integrated flexible strain gauges. The platform allows for the continuous electronic readout of the contractile stress and beat rate of cardiac tissues. Considering that the platform has an open‐well format and provides simultaneous measurement for 24 different tissues, it seems to be a simple and convenient tool for drug testing as well as analyzing temporal drug transport across the barriers (Lind et al., 2017). Undoubtedly, the system developed by Parkers lab paves the way for high‐throughput preclinical research, based on human stem cell‐derived cardiac tissues.
Distinct in vitro model of myocardium was presented by Watson et al. They developed a robust and reproducible method to obtain thin ventricular myocardial slices from the myocardia of human and small mammals. Slices may be used as in vitro models allowing for the analysis of the myocardial structure and function at the cellular level. This model represents a hybrid approach between CM 2D culture after isolation from patients and advanced engineered heart tissues (EHTs). Within the slices, all cardiac cell populations are present in a proper ratio, with cell‐to‐cell interactions maintained within intact cells. Thus, the main advantage of slices is retaining the native multicellularity and microphysiology of the heart. Watson et al. demonstrated that slices obtained with their protocol are viable, and have maximum contractility, Ca2+ handling, and structure. They can be used for pharmacological studies, such as drug efficacy and toxicity screening (Watson et al., 2017, 2019). Miller et al. proposed a slice model of 300 μm thick that could be maintained as viable for up to 6 days. The major problem of native tissue explants is, however, their long‐term viability and low availability of the human tissue (Miller et al., 2020).
4.1.3. Cell sheets
Stem cell‐derived cell sheets have been developed as a treatment strategy for tissue repair. In the case of heart tissue, this approach was used for MI treatment and reported to have many advantages compared to direct stem cell transplantation as well as conventional, scaffold tissue engineering methods. Many studies have reported that cell sheets improve cardiac function in patients after MI, including perfusion, angiogenesis, and enhanced regenerative potential (Xu, Ding, Zhao, Pu, & He, 2014). Moreover, it was also shown that stem cell transplantation may increase the chance of graft survival, by induction of blood vessel formation (Segers & Lee, 2008). Whereas the direct injection may have a drawbacks. For example, this method is proved to be less efficient, there are also some concerns over the graft survival within the site of intervention. Therefore application of polymers and preparation of cell sheets takes advantages over classical injection of the cells (Augustine et al., 2021).
Cell sheets can be obtained from different cell types: skeletal myoblast (SkMbs), mesenchymal stem cells (MSCs), autologous marrow mononuclear cells (MNCs), cardiac progenitor/stem cells (CPCs/CSCs), hiPSC‐CMs and hESC‐CMs, etc. (Guo et al., 2020). To obtain cell sheets, temperature‐responsive culture dishes covered with polymers are used. One of the commonly used temperature‐responsive substance is poly (N‐isopropylacrylamide)‐ PNIPAm. The polymer is in a hydrophobic state at 37°C and becomes hydrophilic at temperatures below 32°C. Therefore, at room temperature, cells detach from the surface and can be collected without losing cell to cell contact and the extracellular matrix. This feature of cell sheets provides an advantage over techniques based on scaffolds, for example, solves the problem of scaffold degradation during cell or tissue construct transfer [47] (J. Yang et al., 2007).
Increasing evidence has demonstrated that cell sheet treatment improves the ejection fraction and thickness of the heart wall, enhances angiogenesis, and decreases fibrosis (Miyagawa et al., 2017; Sawa et al., 2012; Sawa & Miyagawa, 2013; Yamamoto et al., 2019; Yoshikawa et al., 2018).
For MI treatment, cell sheets can be used in both the acute and chronic phases. It was shown that neonatal rat cardiomyocyte cell sheets can contract synchronously after transplantation, with host (nude rats) myocardium (Matsuura, Masuda, & Shimizu, 2014). In the chronic phase of MI, cell sheets significantly improve systolic and diastolic function in animal models. In addition, in rats, they ameliorate cardiac regenerative potential by enhancing paracrine effects and neovascularization (Hoashi et al., 2009; Matsuura et al., 2014; Matsuura, Haraguchi, Shimizu, & Okano, 2013; Sawa & Miyagawa, 2013). Stimulation of angiogenesis occurs via increasing cytokine secretion (Hoashi et al., 2009). Several studies have demonstrated that cell sheets composed solely of CMs derived from pluripotent cells are able to enhance heart regenerative potential and show synchronous beating in small animal models of acute MI. However, sheets composed of mixed stem cell‐derived cells (CMs, endothelial cells, and VSMCs) have a superior impact on cardiac function due to neovascularization and longer retention in MI animal models, especially in the chronic phase (Kawamura et al., 2013; Masumoto et al., 2012; Sekine et al., 2011). Another function of cell sheets is an attenuating inflammatory response in MI. Inflammatory modulation is a significant element of the healing process but also ventricular remodeling, leading to alterations in the heart's structure and function. It was shown that cell sheets can decrease inflammation and increase the expression of anti‐inflammatory genes (Imanishi et al., 2011; Park et al., 2019).
Although there are many promising results on the usage of cell sheets, several important limitations and obstacles should be also considered. In the case of SkMbs, fatal arrhythmias were reported. CMs sheets, in turn, were shown to create gap junctions with connexin 43 and contract synchronously with the host heart (Matsuura et al., 2014). To avoid immunological rejection the stem cell source should be host specific‐it could be either autologous or obtained from the homozygous (best possible donor) human leukocyte antigen or autologous cells. The approach to obtain autologous hiPSCs, although justified from an immunological point of view, generates higher costs, then preparation of the stable hiPSC cell line for allogenic transplantation (Matsuura et al., 2014).
4.2. Engineered heart tissue (EHT)
The first system, which can now be considered as the earliest version of engineered heart tissue (EHT) was developed by Eschenhagen et al., in 1997 (Eschenhagen et al., 1997). They created a cardiac 3D model consisting of mixed embryonic chicken CMs and collagen, which was placed between two Velcro‐coated glass tubes. This group was the first that used the collagen I in cardiac tissue formation. The designed platform made it possible to anchor constructs and generate static force during tissue formation (Eschenhagen et al., 1997).
In general, EHTs are composed of heart cells together with a liquid matrix, including collagen type I or fibrin, which can be supplemented with extracellular basement membrane proteins (e.g., Matrigel®) and serum‐containing medium. The mixture is further loaded into circular molds, and after 5–7 days, spontaneously and synchronously contractile EHTs are transferred into metal spacers to impose mechanical stimuli for an‐other 10 days. Stretching plays an important role in maturation of EHTs and improves their functional and mechanical properties (Fink et al., 2000). The cell composition of EHTs includes cardiac and non‐cardiac cells in order to mimic the in vivo environment. It was reported that EHTs generated from unpurified heart–cell mixtures exhibit advanced structures as well as increased contractile and passive forces in comparison to EHTs containing only the cardiac myocyte population (Zimmermann et al., 2001). The shape and size of EHTs can be modified by adjusted casting molds, and 3D structures may be generated by the stacking and merging of multiple EHTs in vitro (Yoshikawa et al., 2018).
Regarding functional properties, studies from Eschenhagen and Zimmermann showed that EHTs respond to mechanical and pharmacological treatments in an organotypic manner (Matsuura et al., 2014; Yamamoto et al., 2019; Yoshikawa et al., 2018). Constructs were generated by mixing cardiac myocytes from neonatal Fisher 344 rats with collagen type I, Matrigel, and serum‐free media. The implantation of developed EHTs in healthy rats showed that this approach requires immunosuppression. Under immunosuppression, EHTs were viable and contractile for at least 8 weeks, becoming vascularized and innervated (Zimmermann et al., 2002). For clinical testing, the source of cells used for EHT formation should be autologous, possible to obtain in a large quantity, and high quality (differentiated and functional). The most important limitation of using myocytes for EHTs is their inability to proliferate (Pasumarthi & Field, 2002). On the contrary, hESC and certain adult stem cells show proliferation capacity, and could be useful cell sources for cardiac replacement therapy; however, they give rise to ethical issues together with required long‐term immunosuppression (Fändrich et al., 2002). Therefore, iPSCs have fewer ethical issues and enable autologous treatment options. Without a doubt, autologous hiPSCs are a highly promising cell source for the development of patient‐specific cell‐based organ repair strategies. However, as mentioned before, this approach is relatively time consuming due to the need for cell generation and then differentiation into the desired cell type. This obstacle can be omitted by using prefabricated allogeneic cell or tissue products. However, in the case of allogenic cells, immunosuppression treatment must also be implemented. Considering short immunosuppression, the main drawback is the short period of cell survival, but for long‐term suppression, there is a risk of renal toxicity, allograft vasculopathy, etc (Jabbour, Owen, Pandey, & Harding, 2020). Recently, Deuse et al. reported interesting data on hiPSC‐derived EHTs after the activation of certain MHC class I and II markers and overexpression of CD47, using the CRISPR‐Cas9 method. After inactivation, cells still showed pluripotency and differentiation potential as well as proliferation capacity. After implantation in mice, constructs were able to evade immune rejection, in fully MHC‐mismatched allogeneic recipients, and were retained long term without the use of immunosuppression (Deuse et al., 2019).
Goldfracht et al. developed a novel, combined EHT model, generated with hiPSC‐CMs and chitosan‐enhanced ECM hydrogel, derived from decellularized pig hearts. The model showed an anisotropic muscle structure with a more mature CM state than 2D‐cultured hiPSC‐CMs. Authors used fluorescent reporters and unique mapping technologies to perform detailed molecular and ultrastructural characterization of the model at the cellular and tissue resolution. Moreover, the generated model also allowed the observation of drug‐related changes in morphology, contraction rate, tissue conduction properties, and cell arrhythmogenicity. The ECM‐EHT model can be subjected to high‐resolution, long‐term serial functional phenotyping with possible application for cardiac disease modeling and drug testing (Goldfracht et al., 2019). The same group also presented ring‐shaped EHTs derived from hPSC‐CMs. The model included differentiated CMs into atrial and ventricular subtypes, which were further mixed with a collagen‐hydrogel matrix. Authors revealed that aCMs and vCMs display significant differences, confirmed by immunostaining, gene expression analysis, the evaluation of conduction velocity, mechanical force, and the response to pharmacological treatments. They also developed an atrial EHT‐based arrhythmia model, the usefulness of which was confirmed by pharmacological testing (Goldfracht et al., 2020). Here, it is worth emphasizing that in order to develop a reliable cardiac tissue platform for drug screening, physiological differences in atrial and ventricular CMs need to be considered. These differences include gene and protein expression, electrophysiology, and ion channel activities. For instance, atrial CMs show differences in potassium current, and are smaller in size and surface area (Asp, Synnergren, Jonsson, Dellgren, & Jeppsson, 2012Asp et al., 2012). Moreover, they are distinguished by the presence of the potassium channels which are major determinants of electrophysiological differences between atrial and ventricular CMs—Kv1.5 and Kir3.1/3.4, respectively (Schram, Pourrier, Melnyk, & Nattel, 2002). Therefore, to study the cardiotoxicity and off‐target effect of drugs in humans, chamber‐specific tissue platforms are needed. The limited progress on obtaining atrial CMs has been caused by the low efficiency of available differentiation protocols. Several groups are working on the improvement of generation, culture, and maturation engineered atrial tissues, but to date, it remains a challenge (Zhao, Rafatian et al., 2020; Zhao, Ye et al., 2020).
EHTs seem to be promising and reliable models for cardiac disease modeling and regeneration medicine. Thus far, EHTs are one of the most advanced models in terms of mimicking heart structure and physiological properties in vitro. Development of new 3D models certainly is beneficial for the preclinical cardiology and regenerative therapies. Tissue heart models are crucial for studying the physiology and pathology of designated organs thereby brings us closer to recreate the perfect environment for testing new approaches. So far it is impossible to obtain the fully functional bioartificial heart, however MacQueen et al. presented an elegant tissue‐engineered scale models of the human left ventricle, combined of nanofibrous scaffolds that promote native‐like anisotropic myocardial tissue genesis and stimulate chamber‐level contractions (MacQueen et al., 2018). This proves that scientist are able to recreate step by step the functional parts of the heart. As a general cardiac tissue model problem, for EHTs, the main limitation is still the inadequate the level of CM maturity regarding the subcellular structure, however this is significantly improved compared to other systems. This immaturity implicates limited mechanical forces and relatively slower conduction velocity in comparison to the adult heart. Thus, the inclusion of reliable references of fresh human tissue patches is of great importance. Chamber‐specific human tissue explants would provide the extensive control of maturation state and set reference for each particular functional parameter. This would clearly provide further EHT development pathways. To an extent, maturation issues can be addressed by EHT incorporation into microdevices, which would then generate another cardiac in vitro system—heart‐on‐a‐chip, described in the next section.
4.3. Heart‐on‐a‐chip
Organ‐on‐a‐chip (OOC) technology combines tissue models with microfluidic platforms that provides a circulation system and mimics the physiology of a heart/organ system. This group of models encompasses a wide variety of systems that differ in complexity, throughput, and physiology‐mimicking abilities. Nunes et al. presented an elegant model of Biowire platform that combines architectural and electrical cues. Construction was generated by seeding the human iPSC‐CMs into template polydimethylsiloxane (PDMS) channel, around a sterile surgical suture in type I collagen gels. The platform was also electrically stimulated. Such construction enabled to obtain more mature cardiomyocytes. Biowire improved their morphology (induced physiological hypertrophy), architecture of contractile apparatus (induced sarcomere maturation) and electrophysiological properties in a stimulation frequency dependent manner (Nunes et al., 2013). On the other hand Xiao et al. generated cardiac constructs by mixing neonatal rat CMs and hESC‐CMs and seeding cells into a polytetrafluorethylene (PTFE) tubing template, integrated with electrical stimulation using a carbon rod. The designed Biowire platform enables the assessment of a drug's pharmacological effects on cardiac tissue in vitro by perfusion that provides high physiological relevance (Xiao et al., 2014). This effort was more recently updated by Zhao et al. who created an updated version called Biowire‐2 (Zhao et al., 2019). Biowire‐2 is one of the first attempts to combine atrial and ventricular CMs in one organized tissue. With the ECM matrix involved and more environmental parameters controlled (e.g., electrical stimulation), the system combines both ECM and organ‐on‐chip, which is an example of the remaining problems associated with cardiac tissue system classification attempts (Zhao et al., 2019).
Chen et al., 2013. Attempted to create a model that reflects cell to cell and cell to ECM interactions and mimics vascular/valvular 3D environments observed in vivo. To this end, they developed a bilayer microfluidic device that incorporates different cells seeded on a hydrogel. The system included fluid flow over an endothelial monolayer and a porous membrane that provided cell interactions as well as simultaneously maintained their compartmentalization. Authors proved that their model is a useful tool for mechanistic vascular and valvular biology studies and may also be used for the assessment of drug effects on cardiac tissue in physiologically relevant 3D microenvironments (Chen, Srigunapalan, Wheeler, & Simmons, 2013).
Kim et al. designed and investigated a biosensor that allows for the real‐time monitoring of cardiotoxicity. They inserted a specific device in the bottom of the chamber in order to monitor the behavior of ESC‐CMs under different drug treatments. This approach is cost effective, does not need a lens, has a compact size, and seems to be highly useful for high‐throughput drug screening (Kim et al., 2011). Cardiotoxicity examination is also important during the anti‐cancer drug screening. Platforms such an OOC are reliable models for physiological studies, therefore can be use during the development of new strategies for cancer treatment. Chramiec et al. conducted studies with bone Ewing Sarcoma tumor and heart muscle‐both using OOC platforms and subjected to a clinically used linsitinib dosage. Then measured anti‐tumor efficacy and cardiotoxicity were compared with the results obtained from the clinical trial (Chramiec et al., 2020). Kolanowski et al. proposed a microfluidic system that allows for the control of the oxygen concentration in the cell chamber, ensuring both physiological and hypoxic O2 amounts suitable for heart development and disease modeling. This chip combined with video‐based CM activity monitoring and analysis delivers a complete system for drug screening purposes (Kolanowski et al., 2020). Sakamiya et al. developed and validated a heart‐on‐a‐chip platform that enables the online monitoring and analysis of 3D cardiac tissue features, such as contraction force, frequency, and synchronization. Authors concluded that their system is reliable and can be used in cardio‐related drug screening applications (Sakamiya, Fang, Mo, Shen, & Zhang, 2020). Van Dijk et al. presented a new microfluidic (heart) vessel‐on‐a‐chip system that allows for the monitoring of vascular structure and cell interactions in a 3D biological matrix. The model included endothelial cells, pericytes and an ECM under controlled, continuous, and unidirectional flow. The interaction of the mentioned cells allowed for the creation of a human cell‐derived blood vessel. Authors revealed that new vessels show natural vascular response under thrombin stimulation and can be used for assessing the interaction between the cells in response to TNFα. The developed model seems to be biologically functional and could be a useful tool for the in vitro analysis of the human microvasculature in physiological and pathological conditions (van Dijk et al., 2020). Moreover Parsa et al. invented a microfluidic model that enables monitoring of cardiomyocytes hypertrophy induced by volume overload. This platform provides reliable and feasible method for mechanical stimulation of 3D cardiac tissues (Parsa et al., 2017). Therefore, it opens new possibilities to study the pathological processes that lead to heart failure. Additionally tissue engineered models promotes maturation of iPSC‐CMs therefore are suitable for physiological studies. For example, examination of ischemia‐reperfusion injury was conducted mainly on animal models, however nowadays the 3D EHT may perfectly replace the in vivo studies (Chen & Vunjak‐Novakovic, 2018).
4.4. Decellularized heart
A decellularized extracellular matrix (dECM) of the heart has been developed and used as a natural scaffold for cardiac tissue. It is a relatively new approach aimed to increase the global supply for transplantation organs. The first protocol for the whole heart decellularization process was published in 2008 and concerned the rat heart. Two years later, a whole porcine heart was decellularized. In 2013, the first recellularization of a mouse heart with hiPSC‐derived cardiac progenitor cells was described. Concerning the human heart, the first decellularization occurred in 2015, and one year later, recellularization was reported (Kc, Hong, & Zhang, 2019).
The main purpose of human heart decellularization is to mimic the native microenvironment for cardiac cells and stimulate iPSC‐CMs to maturation. Although there are some evidences that maturation of iPSC‐CMs may be driven by heart itself. In the literature there are studies that involves transplantation of fetal‐like differentiated cardiomyocytes at the early stages if differentiation. The results clearly indicates that the transplanted cardiomyocytes are mature following the seeding in heart (Cho et al., 2017; Funakoshi, et al., 2016). However, scaffold, which is obtained after cell removal, offers many mechanical and biochemical advantages that ameliorate cell attachment, integrity, growth, and proliferation as well as cardiovascular differentiation during subsequent recellularization (Gilbert, Sellaro, & Badylak, 2006; Iop, Dal Sasso, Menabò, Di Lisa, & Gerosa, 2017). It was shown that the decellularized heart consists of major cardiac ECM components, such as collagen, laminin, elastin, and glycosaminoglycans (Johnson et al., 2016). Methe et al. reported that the dECM of the porcine heart also includes cytokines and growth factors, such as fibroblast growth factor, endoglin, interleukin‐8, etc. These molecules play an important role in cardiac regeneration and remodeling and can significantly modulate inflammation and heart healing (Methe et al., 2014). The dECM was also shown to be a robust platform for cardiac stem cell differentiation and maturation, which can be used for the rapid production of human heart grafts (K. M. Lee et al., 2015; Garreta et al., 2016). Wang B. et al. Generated 3D porcine myocardial scaffolds that were seeded with mesenchymal stem cells and then treated with biochemical and electrical stimuli. By immunofluorescence staining, authors proved that the differentiated cells possessed a cardiomyocyte‐like phenotype and expressed sarcomeric α‐actinin, myosin heavy chain, cardiac troponin T, connexin‐43, and N‐cadherin (B. Wang et al., 2013). Wang Q. et al. Developed functional human cardiac patches using hiPSC‐CMs and a decellularized natural rat heart ECM as scaffolds. They showed that the generated patches had similar to normal contractile and electrical physiology in vitro. Moreover, they improved the heart function of rats with acute myocardial infarction in vivo after patching on the infarct area. Improvement was shown as reduction in infract size and recovery of the left‐ventricular ejection fraction (Q. Wang et al., 2016).
The dECM derived from the heart gained a large amount of interest, especially in the context of cardiac engineering, regeneration, and repair. However, it is important to emphasize that obtaining a dECM is extremely complicated and needs to be optimized (method, agents, and time) in order to maintain a balance between cell removal and ECM preservation. Achieving an optimal dECM scaffold is crucial for minimizing the immunogenicity and risk of rejection. Another important issue is the recellularization of the dECM. Scaffolds must be fully populated with suitable cell numbers and distribution. The non‐cardiac dECM has been also studied in clinical trials for myocardium regeneration (Yanagawa, Rao, Yau, & Cusimano, 2013). It seems that the cardiac scaffold will be a more appropriate choice in terms of heart failure treatment, but several problems still need to be solved, for example, batch to batch variations (individual differences), and assessment of long‐term efficacy and safety. Other important issues include the in situ cell differentiation procedure, which is challenging and must be performed precisely, likewise further CM maturation within the model. The human heart is a complex and multilayered structure; thus, it is a great challenge to train and maturate the whole heart ex vivo. The many heart layers also make it difficult to recapitulate cell to cell interactions, vascular and nervous systems, proper channel distribution, and the generation of contractile force. Nevertheless, the cardiac dECM, after recellularization, is an interesting and complex model, which in the long term may be promising candidates for clinical trials. Details on cardiac model maturation states are shown in Table 1, and advantages and pitfalls are summarized in Table 2.
5. CONCLUSIONS AND FUTURE PROSPECTS
Cardiac tissue engineering is a promising and evolving field that aims to develop a model for drug testing as well as regeneration and replacement strategies for the human heart. During the last decade, significant progress in the development of relevant and functional models of cardiac tissue has been made. Different approaches have been implemented to increase CM maturity, and iPSC‐differentiated cells have been used to obtain functional cardiac tissue, surpassing troublesome human cardiac cell supply shortages. Introducing structural scaffolds and microfluidic systems further shifted researchers toward more physiologically relevant human heart model development. Another important milestone in cardiac tissue engineering was the generation of CMs in high purity from hiPSCs. Here, it should be emphasized that generated CMs are mainly ventricular, and obtaining atrial CM populations requires a large amount of effort and, additionally, the optimization of the protocol. It is important to develop models of chamber‐specific tissues in order to achieve proper disease modeling. Thus, refining efficient methods for the direct differentiation of hiPSCs toward atrial‐, ventricular‐, and pacemaker‐like cells within the tissue model remains a major challenge in the field.
Due to the extremely widespread work currently being performed by a number of groups worldwide, this review was targeted to be only an introduction to help readers distinguish between different model‐specific functionalities. Thus, we focused on summarizing the models that have been already established. Nevertheless, there are several important differences among the described model systems (Tables 1 and 2, Figures 1 and 2), with advantages and disadvantages that are often dependent on the physiological or pathophysiological condition being studied. These differences should be taken into account when deciding on a model type suitable for a particular application. Of particular interest are physiological data that could support clinical output comparisons between models; thus, this remains extremely difficult due to their different functions. From these data, the action potential duration (APD) is shown in Table 1. It is worth mentioning that although more robust methods, such as MTFs and organoids, provide the highest throughput, their physiological relevance might be reversely proportional when compared with systems such as EHTs and decellularized hearts. On the other hand, EHTs not only allow for atrial and ventricular chamber specification but also present highly similar results to the referential human heart chambers. Nevertheless, each model is designed for its desired functionality (image of each model is shown in Figure 2); which one should be picked depends on the application. For instance, in cardiotoxicity studies, MTFs are now used rather than organoids as they offer more reliable results and robust data output, despite the simplicity of hCO excel formation. In more advanced drug effect screening, organ‐ and body‐on‐a‐chip are favored due to their automatization techniques. For disease modeling, EHTs seem to be the most reliable source of data, whereas in heart regeneration, the most intriguing sources are cell sheets and decellularized hearts, although this is the specific scope of other reviews (Zhang, Zhu, Radisic, & Vunjak‐Novakovic, 2018).
Future directions of cardiac tissue engineering field development will undoubtedly include advances in 3D bioprinting technologies that have a huge potential for improving many systems, such as EHTs and decellularized scaffolds. The incorporation of this technology may help to combine the atrial/ventricular cells with the relevant chamber‐specific structures. The development of more capable subtype differentiation protocols would have a tremendous effect on chamber‐specific disease modeling. Regulator and funding agency support for the development of in vitro organ models that is currently more often observed, and should lead to the popularization of cardiac tissue models among culture systems in research laboratories. This would put more strain on animal use in preclinical studies (over 30 million of laboratory animals used yearly in the EU and US only), ultimately altering drug development processes.
To date, the EHT models are certainly the most advanced platforms for studying the physiology, pathology of the cardiomyocytes. However preparation of the classical engineering heart tissue require extremely large number of cells, what may be considered as some limitation. Due to this also the cost of the EHT preparation may be high. Approaches to miniaturize the EHT models are promising to overcome such inconvenience (Huebsch et al., 2016).
Advanced method of tissue engineering are promising for personalized/targeted medicine development. The iPSC technology generates unlimited source of patient‐specific cells. That enables to preparation of engineered cardiac tissues for autologous transplantation in treatment of heart failure. Moreover, generation of heart models affected with genetic alterations will be prospective for testing new possible gene therapies. Additionally, preparation of engineered tissues opens a new perspectives for screening of new drugs and testing the cardiotoxicity for example, during application of new chemotherapeutics in oncology. In vitro cell cultures are not sufficient for mentioned aspects. Verification of new drugs and therapies should be conducted in a complex models, that can mimic the accurate environment of selected organ/tissue. Therefore by using the advanced platforms it is possible to recreate the physiological or pathological states. Obtained results will be more reliable for further clinical application. It is also worth to point out that tissue engineering methods omits the ethical issues considered during in vivo studies using animal models.
Taken together, organ models are on the intensive development track that, each year, achieves improved results, and heart tissue models are at the forefront of these advances, leading to tremendous changes in many biomedical research applications.
AUTHOR CONTRIBUTIONS
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Ewelina Kałużna, Agnieszka Nadel, Agnieszka Zimna drafted the work, designed and prepared the figures. Natalia Rozwadowska critically revised manuscript and contributed to manuscript concept, Tomasz Kolanowski defined the manuscript concept, helped with design and research interpretation. All authors prepared, read, and approved the final manuscript.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest. The funders had no role in interpretation of data or the writing of the manuscript.
ACKNOWLEDGMENTS
This manuscript was supported by National Science Centre Poland, grant number 2018/31/D/NZ3/01719.
Kałużna, E. , Nadel, A. , Zimna, A. , Rozwadowska, N. , & Kolanowski, T. (2022). Modeling the human heart ex vivo—current possibilities and strive for future applications. Journal of Tissue Engineering and Regenerative Medicine, 16(10), 853–874. 10.1002/term.3335
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- Alford, P. W. , Feinberg, A. W. , Sheehy, S. P. , & Parker, K. K. (2010). Biohybrid thin films for measuring contractility in engineered cardiovascular muscle. Biomaterials, 31(13), 3613–3621. 10.1016/j.biomaterials.2010.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik, A. , Abramsson, A. , & Betsholtz, C. (2005). Endothelial/pericyte interactions. Circulation Research, 97(6), 512–523. 10.1161/01.RES.0000182903.16652.d7 [DOI] [PubMed] [Google Scholar]
- Asp, J. , Synnergren, J. , Jonsson, M. , Dellgren, G. , & Jeppsson, A. (2012). Comparison of human cardiac gene expression profiles in paired samples of right atrium and left ventricle collected in vivo. Physiological Genomics, 44(1), 89–98. 10.1152/physiolgenomics.00137.2011 [DOI] [PubMed] [Google Scholar]
- Augustine, R. , Dan, P. , Hasan, A. , Khalaf, I. M. , Prasad, P. , Ghosal, K. , Gentile, C. , McClements, L. , & Maureira, P. (2021). Stem cell‐based approaches in cardiac tissue engineering: Controlling the microenvironment for autologous cells. Biomed. Pharmacotheraphy, 138, 111425. 10.1016/j.biopha.2021.111425 [DOI] [PubMed] [Google Scholar]
- Balashov, V. , Efimov, A. , Agapova, O. , Pogorelov, A. , Agapov, I. , & Agladze, K. (2018). High resolution 3D microscopy study of cardiomyocytes on polymer scaffold nanofibers reveals formation of unusual sheathed structure. Acta Biomaterialia, 68, 214–222. 10.1016/j.actbio.2017.12.031 [DOI] [PubMed] [Google Scholar]
- Balashov, V. A. , Gorbunov, V. S. , Guria, K. G. , & Agladze, K. I. (2020). Muscular thin films for label‐free mapping of excitation propagation in cardiac tissue. Annals of Biomedical Engineering, 48(10), 2425–2437. 10.1007/s10439-020-02513-0 [DOI] [PubMed] [Google Scholar]
- Baumgartner, S. , Halbach, M. , Krausgrill, B. , Maass, M. , Srinivasan, S. P. , Sahito, R. G. A. , Peinkofer, G. , Nguemo, F. , Muller‐Ehmsen, J. , & Hescheler, J. (2015). Electrophysiological and morphological maturation of murine fetal cardiomyocytes during electrical stimulation in vitro. Journal of Cardiovascular Pharmacology and Therapeutics, 20(1), 104–112. 10.1177/1074248414536273 [DOI] [PubMed] [Google Scholar]
- Bejleri, D. , & Davis, M. E. (2019). Decellularized extracellular matrix materials for cardiac repair and regeneration. Advanced Healthcare Materials, 8(5), 1801217. 10.1002/adhm.201801217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburger, M. , Wenzel, J. , Bogdan, R. , Richardt, D. , Nguemo, F. , Reppel, M. , Hescheler, J. , Terlau, H. , & Dendorfer, A. (2012). Organotypic slice culture from human adult ventricular myocardium. Cardiovascular Research, 93(1), 50–59. 10.1093/cvr/cvr259 [DOI] [PubMed] [Google Scholar]
- Camacho, P. , Fan, H. , Liu, Z. , & He, J.‐Q. (2016). Large mammalian animal models of heart disease. Journal of Cardiovascular Development and Disease, 3(4). 10.3390/jcdd3040030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi, O. , Lesman, A. , Basevitch, Y. , Gepstein, A. , Arbel, G. , Habib, I. H. M. , Gepstein, L. , & Levenberg, S. (2007). Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circulation Research, 100(2), 263–272. 10.1161/01.RES.0000257776.05673.ff [DOI] [PubMed] [Google Scholar]
- Chen, M. B. , Srigunapalan, S. , Wheeler, A. R. , & Simmons, C. A. (2013). A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell‐cell interactions. Lab on a Chip, 13(13), 2591–2598. 10.1039/c3lc00051f [DOI] [PubMed] [Google Scholar]
- Chen, T. , & Vunjak‐Novakovic, G. (2018). Vitro models of ischemia‐reperfusion injury. Regenerative engineering and translational medicine, 4(3), 142–153. 10.1007/s40883-018-0056-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, G. S. , Lee, D. I. , Tampakakis, E. , Murphy, S. , Andersen, P. , Uosaki, H. , Chelko, S. , Chakir, K. , Hong, I. , Seo, K. , Chen, H. S. V. , Chen, X. , Basso, C. , Houser, S. R. , Tomaselli, G. F. , O’Rourke, B. , Judge, D. P. , Kass, D. A. , & Kwon, C. (2017). Neonatal transplantation confers maturation of PSC‐derived cardiomyocytes conducive to modeling cardiomyopathy. Cell Reports, 18(2), 571–582. 10.1016/j.celrep.2016.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chramiec, A. , Teles, D. , Yeager, K. , Marturano‐Kruik, A. , Pak, J. , Chen, T. , Hao, L. , Wang, M. , Lock, R. , Tavakol, D. N. , Lee, M. B. , Kim, J. , Ronaldson‐Bouchard, K. , & Vunjak‐Novakovic, G. (2020). Integrated human organ‐on‐a‐chip model for predictive studies of anti‐tumor drug efficacy and cardiac safety. Lab on a Chip, 20(23), 4357–4372. 10.1039/d0lc00424c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppini, R. , Ferrantini, C. , Aiazzi, A. , Mazzoni, L. , Sartiani, L. , Mugelli, A. , Poggesi, C. , & Cerbai, E. (2014). Isolation and functional characterization of human ventricular cardiomyocytes from fresh surgical samples. Journal of Visualized Experiments : Journal of Visualized Experiments(86). 10.3791/51116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyganek, L. , Tiburcy, M. , Sekeres, K. , Gerstenberg, K. , Bohnenberger, H. , Lenz, C. , Henze, S. , Stauske, M. , Salinas, G. , Zimmermann, W. H. , Hasenfuss, G. , & Guan, K. (2018). Deep phenotyping of human induced pluripotent stem cell‐derived atrial and ventricular cardiomyocytes. JCI Insight, 3(12), e99941. 10.1172/jci.insight.99941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, K. C. , & Muniyappa, H. (2013). Age‐dependent mitochondrial energy dynamics in the mice heart: Role of superoxide dismutase‐2. Experimental Gerontology, 48(9), 947–959. 10.1016/j.exger.2013.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Era, P. , Benzoni, P. , Crescini, E. , Valle, M. , Xia, E. , Consiglio, A. , & Memo, M. (2015). Cardiac disease modeling using induced pluripotent stem cell‐derived human cardiomyocytes. World Journal of Stem Cells, 7(2), 329–342. 10.4252/wjsc.v7.i2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, C. , Borgdorff, V. , Crutchley, J. , Firth, K. S. A. , George, V. , Kalra, S. , Kondrashov, A. , Hoang, M. D. , Mosqueira, D. , Patel, A. , Prodanov, L. , Rajamohan, D. , Skarnes, W. C. , Smith, J. G. , & Young, L. E. (2016). Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochimica et Biophysica Acta, 1863(7 Pt B), 1728–1748. 10.1016/j.bbamcr.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuse, T. , Hu, X. , Gravina, A. , Wang, D. , Tediashvili, G. , De, C. , Thayer, W. O. , Wahl, A. , Garcia, J. V. , Reichenspurner, H. , Davis, M. M. , Lanier, L. L. , & Schrepfer, S. (2019). Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nature Biotechnology, 37(3), 252–258. 10.1038/s41587-019-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doppler, S. A. , Carvalho, C. , Lahm, H. , Deutsch, M.‐A. , Dreßen, M. , Puluca, N. , Lange, R. , & Krane, M. (2017). Cardiac fibroblasts: More than mechanical support. Journal of Thoracic Disease, 9(Suppl 1), S36–S51. 10.21037/jtd.2017.03.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edri, R. , Gal, I. , Noor, N. , Harel, T. , Fleischer, S. , Adadi, N. , Green, O. , Shabat, D. , Heller, L. , Shapira, A. , Gat‐Viks, I. , Peer, D. , & Dvir, T. (2019). Personalized hydrogels for engineering diverse fully autologous tissue implants. Advanced Materials, 31(1), e1803895. 10.1002/adma.201803895 [DOI] [PubMed] [Google Scholar]
- El‐Armouche, A. , & Eschenhagen, T. (2009). Beta‐adrenergic stimulation and myocardial function in the failing heart. Heart Failure Reviews, 14(4), 225–241. 10.1007/s10741-008-9132-8 [DOI] [PubMed] [Google Scholar]
- Eschenhagen, T. , Fink, C. , Remmers, U. , Scholz, H. , Wattchow, J. , Weil, J. , Zimmermann, W. , Dohmen, H. H. , Schafer, H. , Bishopric, N. , Wakatsuki, T. , & Elson, E. L. (1997). Three‐dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: A new heart muscle model system. Federation of American Societies for Experimental Biology Journal : Official Publication of the Federation of American Societies for Experimental Biology, 11(8), 683–694. 10.1096/fasebj.11.8.9240969 [DOI] [PubMed] [Google Scholar]
- Fändrich, F. , Lin, X. , Chai, G. X. , Schulze, M. , Ganten, D. , Bader, M. , Holle, J. , Huang, D. S. , Parwaresch, R. , Zavazava, N. , & Binas, B. (2002). Preimplantation‐stage stem cells induce long‐term allogeneic graft acceptance without supplementary host conditioning. Nature Medicine, 8(2), 171–178. 10.1038/nm0202-171 [DOI] [PubMed] [Google Scholar]
- Feinberg, A. W. , Alford, P. W. , Jin, H. , Ripplinger, C. M. , Werdich, A. A. , Sheehy, S. P. , Grosberg, A. , & Parker, K. K. (2012). Controlling the contractile strength of engineered cardiac muscle by hierarchal tissue architecture. Biomaterials, 33(23), 5732–5741. 10.1016/j.biomaterials.2012.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A. W. , Feigel, A. , Shevkoplyas, S. S. , Sheehy, S. , Whitesides, G. M. , & Parker, K. K. (2007). Muscular thin films for building actuators and powering devices. Science (New York, N.Y.), 317(5843), 1366–1370. 10.1126/science.1146885 [DOI] [PubMed] [Google Scholar]
- Fennema, E. , Rivron, N. , Rouwkema, J. , vanBlitterswijk, C. , & deBoer, J. (2013). Spheroid culture as a tool for creating 3D complex tissues. Trends in Biotechnology, 31(2), 108–115. 10.1016/j.tibtech.2012.12.003 [DOI] [PubMed] [Google Scholar]
- Feric, N. T. , & Radisic, M. (2016). Maturing human pluripotent stem cell‐derived cardiomyocytes in human engineered cardiac tissues. Advanced Drug Delivery Reviews, 15(96), 110–134. 10.1016/j.addr.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri, N. , Siegl, P. , Corsini, A. , Herrmann, J. , Lerman, A. , & Benghozi, R. (2013). Drug attrition during pre‐clinical and clinical development: Understanding and managing drug‐induced cardiotoxicity. Pharmacology & Therapeutics, 138(3), 470–484. 10.1016/j.pharmthera.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Filippo Buono, M. , von Boehmer, L. , Strang, J. , Hoerstrup, S. P. , Emmert, M. Y. , & Nugraha, B. (2020). Human cardiac organoids for modeling genetic cardiomyopathy. Cells, 9(7). 10.3390/cells9071733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, C. , Ergün, S. , Kralisch, D. , Remmers, U. , Weil, J. , & Eschenhagen, T. (2000). Chronic stretch of engineered heart tissue induces hypertrophy and functional improvement. Federation of American Societies for Experimental Biology Journal : Official Publication of the Federation of American Societies for Experimental Biology, 14(5), 669–679. 10.1096/fasebj.14.5.669 [DOI] [PubMed] [Google Scholar]
- Forte, E. , Furtado, M. B. , & Rosenthal, N. (2018). The interstitium in cardiac repair: Role of the immune‐stromal cell interplay. Nature Reviews Cardiology, 15(10), 601–616. 10.1038/s41569-018-0077-x [DOI] [PubMed] [Google Scholar]
- Funakoshi, S. , Miki, K. , Takaki, T. , Okubo, C. , Hatani, T. , Chonabayashi, K. , Nishikawa, M. , Takei, I. , Oishi, A. , Narita, M. , Hoshijima, M. , Kimura, T. , Yamanaka, S. , & Yoshida, Y. (2016). Enhanced engraftment, proliferation, and therapeutic potential in heart using optimized human iPSC‐derived cardiomyocytes. Scientific Reports, 6, 19111. 10.1038/srep19111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreta, E. , de Oñate, L. , Fernández‐Santos, M. E. , Oria, R. , Tarantino, C. , Climent, A. M. , Marco, A. , Samitier, M. , Martinez, E. , Valls‐Margarit, M. , Matesanz, R. , Taylor, D. A. , Fernandez‐Aviles, F. , Izpisua Belmonte, J. C. , & Montserrat, N. (2016). Myocardial commitment from human pluripotent stem cells: Rapid production of human heart grafts. Biomaterials, 98, 64–78. 10.1016/j.biomaterials.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Giacomelli, E. , Bellin, M. , Sala, L. , vanMeer, B. J. , Tertoolen, L. G. J. , Orlova, V. V. , & Mummery, C. L. (2017). Three‐dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co‐differentiated from human pluripotent stem cells. Development (Cambridge, England), 144(6), 1008–1017. 10.1242/dev.143438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, T. W. , Sellaro, T. L. , & Badylak, S. F. (2006). Decellularization of tissues and organs. Biomaterials, 27(19), 3675–3683. 10.1016/j.biomaterials.2006.02.014 [DOI] [PubMed] [Google Scholar]
- Goldfracht, I. , Efraim, Y. , Shinnawi, R. , Kovalev, E. , Huber, I. , Gepstein, A. , Arbel, G. , Shaheen, N. , Tiburcy, M. , Zimmermann, W. H. , Machluf, M. , & Gepstein, L. (2019). Engineered heart tissue models from hiPSC‐derived cardiomyocytes and cardiac ECM for disease modeling and drug testing applications. Acta Biomaterialia, 92, 145–159. 10.1016/j.actbio.2019.05.016 [DOI] [PubMed] [Google Scholar]
- Goldfracht, I. , Protze, S. , Shiti, A. , Setter, N. , Gruber, A. , Shaheen, N. , Nartiss, Y. , Keller, G. , & Gepstein, L. (2020). Generating ring‐shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell‐derived cardiomyocytes. Nature Communications, 11(1), 75. 10.1038/s41467-019-13868-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, R. , Morimatsu, M. , Feng, T. , Lan, F. , Chang, D. , Wan, F. , & Ling, Y. (2020). Stem cell‐derived cell sheet transplantation for heart tissue repair in myocardial infarction. Stem Cell Research & Therapy, 11(1), 19. 10.1186/s13287-019-1536-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyette, J. P. , Charest, J. M. , Mills, R. W. , Jank, B. J. , Moser, P. T. , Gilpin, S. E. , Gershlak, J. R. , Okamoto, T. , Gonzalez, G. , Milan, D. J. , Gaudette, G. R. , & Ott, H. C. (2016). Bioengineering human myocardium on native extracellular matrix. Circulation Research, 118(1), 56–72. 10.1161/CIRCRESAHA.115.306874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoashi, T. , Matsumiya, G. , Miyagawa, S. , Ichikawa, H. , Ueno, T. , Ono, M. , Saito, A. , Shimizu, T. , Okano, T. , Kawaguchi, N. , Matsuura, N. , & Sawa, Y. (2009). Skeletal myoblast sheet transplantation improves the diastolic function of a pressure‐overloaded right heart. The Journal of Thoracic and Cardiovascular Surgery, 138(2), 460–467. 10.1016/j.jtcvs.2009.02.018 [DOI] [PubMed] [Google Scholar]
- Huebsch, N. , Loskill, P. , Deveshwar, N. , Spencer, C. I. , Judge, L. M. , Mandegar, M. A. , B. Fox, C. , Mohamed, T. M. , Ma, Z. , Mathur, A. , Sheehan, A. M. , Truong, A. , Saxton, M. , Yoo, J. , Srivastava, D. , Desai, T. A. , So, P. L. , Healy, K. E. , & Conklin, B. R. (2016). Miniaturized iPS‐cell‐derived cardiac muscles for physiologically relevant drug response analyses. Scientific Reports, 6, 24726. 10.1038/srep24726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi, Y. , Miyagawa, S. , Maeda, N. , Fukushima, S. , Kitagawa‐Sakakida, S. , Daimon, T. , Hirata, A. , Shimizu, T. , Okano, T. , Shimomura, I. , & Sawa, Y. (2011). Induced adipocyte cell‐sheet ameliorates cardiac dysfunction in a mouse myocardial infarction model: A novel drug delivery system for heart failure. Circulation, 124(11 Suppl), S10–S17. 10.1161/CIRCULATIONAHA.110.009993 [DOI] [PubMed] [Google Scholar]
- Inui, A. , Sekine, H. , Sano, K. , Dobashi, I. , Yoshida, A. , Matsuura, K. , Kobayashi, E. , Ono, M. , & Shimizu, T. (2019). Generation of a large‐scale vascular bed for the in vitro creation of three‐dimensional cardiac tissue. Regenerative Therapy, 11, 316–323. 10.1016/j.reth.2019.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iop, L. , Dal Sasso, E. , Menabò, R. , Di Lisa, F. , & Gerosa, G. (2017). The rapidly evolving concept of whole heart engineering. Stem Cells International, 18, 8920940. 10.1155/2017/8920940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigami, M. , Masumoto, H. , Ikuno, T. , Aoki, T. , Kawatou, M. , Minakata, K. , Ikeda, T. , Sakata, R. , Yamashita, J. K. , & Minatoya, K. (2018). Human iPS cell‐derived cardiac tissue sheets for functional restoration of infarcted porcine hearts. PLoS One, 13(8), e0201650. 10.1371/journal.pone.0201650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbour, R. J. , Owen, T. J. , Pandey, P. , & Harding, S. E. (2020). Future potential of engineered heart tissue patches for repairing the damage caused by heart attacks. Expert Review of Medical Devices, 17(1), 1–3. 10.1080/17434440.2020.1700793 [DOI] [PubMed] [Google Scholar]
- Jackman, C. P. , Carlson, A. L. , & Bursac, N. (2016). Dynamic culture yields engineered myocardium with near‐adult functional output. Biomaterials, 111, 66–79. 10.1016/j.biomaterials.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, T. D. , Hill, R. C. , Dzieciatkowska, M. , Nigam, V. , Behfar, A. , Christman, K. L. , & Hansen, K. C. (2016). Quantification of decellularized human myocardial matrix: A comparison of six patients. Proteomics ‐ Clinical Applications, 10(1), 75–83. 10.1002/prca.201500048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdar, F. , Das, S. , Gong, W. , Klaassen‐Kamdar, A. , Meyers, T. A. , Shah, P. , Ervasti, J. M. , Townsend, D. , Kamp, T. J. , Wu, J. C. , Garry, M. G. , Zhang, J. , & Garry, D. J. (2020). Stem cell‐derived cardiomyocytes and beta‐adrenergic receptor blockade in Duchenne muscular dystrophy cardiomyopathy. Journal of the American College of Cardiology, 75, 1159–1174. 10.1016/j.jacc.2019.12.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura, M. , Miyagawa, S. , Fukushima, S. , Saito, A. , Miki, K. , Ito, E. , Sougawa, N. , Kawamura, T. , Daimon, T. , Shimizu, T. , Okano, T. , Toda, K. , & Sawa, Y. (2013). Enhanced survival of transplanted human induced pluripotent stem cell‐derived cardiomyocytes by the combination of cell sheets with the pedicled omental flap technique in a porcine heart. Circulation, 128(11 Suppl 1), S87–S94. 10.1161/CIRCULATIONAHA.112.000366 [DOI] [PubMed] [Google Scholar]
- Kc, P. , Hong, Y. , & Zhang, G. (2019). Cardiac tissue‐derived extracellular matrix scaffolds for myocardial repair: Advantages and challenges. Regenerative Biomaterials, 6(4), 185–199. 10.1093/rb/rbz017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. B. , Bae, H. , Cha, J. M. , Moon, S. J. , Dokmeci, M. R. , Cropek, D. M. , & Khademhosseini, A. (2011). A cell‐based biosensor for real‐time detection of cardiotoxicity using lensfree imaging. Lab on a Chip, 11(10), 1801–1807. 10.1039/c1lc20098d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolanowski, T. J. , Antos, C. L. , & Guan, K. (2017). Making human cardiomyocytes up to date: Derivation, maturation state and perspectives. International Journal of Cardiology, 241, 379–386. 10.1016/j.ijcard.2017.03.099 [DOI] [PubMed] [Google Scholar]
- Kolanowski, T. J. , Busek, M. , Schubert, M. , Dmitrieva, A. , Binnewerg, B. , Pöche, J. , Fisher, K. , Schmieder, F. , Grunzner, S. , Hansen, S. , Richter, A. , El‐Armouche, A. , Sonntag, F. , & Guan, K. (2020). Enhanced structural maturation of human induced pluripotent stem cell‐derived cardiomyocytes under a controlled microenvironment in a microfluidic system. Acta Biomaterialia, 102, 273–286. 10.1016/j.actbio.2019.11.044 [DOI] [PubMed] [Google Scholar]
- Laksman, Z. , Wauchop, M. , Lin, E. , Protze, S. , Lee, J. , Yang, W. , Izaddoustdar, F. , Shafaattalab, S. , Gepstein, L. , Tibbits, G. F. , Keller, G. , & Backx, P. H. (2017). Modeling atrial fibrillation using human embryonic stem cell‐derived atrial tissue. Scientific Reports, 7(1), 5268. 10.1038/s41598-017-05652-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, F. , Lee, A. S. , Liang, P. , Sanchez‐Freire, V. , Nguyen, P. K. , Wang, L. , Han, L. , Yen, M. , Wang, Y. , Sun, N. , Abilez, O. J. , Hu, S. , Ebert, A. D. , Navarrete, E. G. , Simmons, C. S. , Wheeler, M. , Pruitt, B. , Lewis, R. , Yamaguchi, Y. , … Wu, J. C. (2013). Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient‐specific induced pluripotent stem cells. Cell Stem Cell, 12(1), 101–113. PMID: 23290139; PMCID: PMC3638033. 10.1016/j.stem.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. M. , Kim, H. , Nemeno, J. G. , Yang, W. , Yoon, J. , Lee, S. , & Lee, J. I. (2015). Natural cardiac extracellular matrix sheet as a biomaterial for cardiomyocyte transplantation. Transplantation Proceedings, 47(3), 751–756. 10.1016/j.transproceed.2014.12.030 [DOI] [PubMed] [Google Scholar]
- Lee, P. , Klos, M. , Bollensdorff, C. , Hou, L. , Ewart, P. , Kamp, T. J. , Zhang, J. , Bizy, A. , Guerrero‐Serna, G. , Kohl, P. , Jalife, J. , & Herron, T. J. (2012). Simultaneous voltage and calcium mapping of genetically purified human induced pluripotent stem cell‐derived cardiac myocyte monolayers. Circulation Research, 110(12), 1556–1563. 10.1161/CIRCRESAHA.111.262535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelovas, P. P. , Kostomitsopoulos, N. G. , & Xanthos, T. T. (2014). A comparative anatomic and physiologic overview of the porcine heart. Journal of the American Association for Laboratory Animal Science, 53(5), 432–438. [PMC free article] [PubMed] [Google Scholar]
- Lemme, M. , Ulmer, B. M. , Lemoine, M. D. , Zech, A. T. L. , Flenner, F. , Ravens, U. , Reichenspurner, H. , Rol‐Garcia, M. , Smith, G. , Hansen, A. , Christ, T. , & Eschenhagen, T. (2018). Atrial‐like engineered heart tissue: An in vitro model of the human atrium. Stem Cell Reports, 11(6), 1378–1390. 10.1016/j.stemcr.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau, B. , Christoforou, N. , Leong, K. W. , & Bursac, N. (2011). Pluripotent stem cell‐derived cardiac tissue patch with advanced structure and function. Biomaterials, 32(35), 9180–9187. 10.1016/j.biomaterials.2011.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw, N. Y. , & Zimmermann, W.‐H. (2016). Mechanical stimulation in the engineering of heart muscle. Advanced Drug Delivery Reviews, 96, 156–160. 10.1016/j.addr.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Lind, J. U. , Yadid, M. , Perkins, I. , O’Connor, B. B. , Eweje, F. , Chantre, C. O. , Hemphill, M. A. , Yuan, H. , Campbell, P. H. , Vlassak, J. J. , & Parker, K. K. (2017). Cardiac microphysiological devices with flexible thin‐film sensors for higher‐throughput drug screening. Lab on a Chip, 17(21), 3692–3703. 10.1039/c7lc00740j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litviňuková, M. , Talavera‐López, C. , Maatz, H. , Reichart, D. , Worth, C. L. , Lindberg, E. L. , Kanda, M. , Polanski, K. , Heinig, M. , Lee, M. , Nadelmann, E. R. , Roberts, K. , Tuck, L. , Fasouli, E. S. , DeLaughter, D. M. , McDonough, B. , Wakimoto, H. , Gorham, J. M. , Samari, S. , & Teichmann, S. A. (2020). Cells of the adult human heart. Nature, 588(7838), 466–472. 10.1038/s41586-020-2797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax, A. E. , Kondo, C. S. , & Giles, W. R. (2003). Comparison of time‐ and voltage‐dependent K+ currents in myocytes from left and right atria of adult mice. American Journal of Physiology ‐ Heart and Circulatory Physiology, 285(5), H1837–H1848. 10.1152/ajpheart.00386.2003 [DOI] [PubMed] [Google Scholar]
- Lopaschuk, G. D. , & Jaswal, J. S. (2010). Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. Journal of Cardiovascular Pharmacology, 56(2), 130–140. 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- MacQueen, L. A. , Sheehy, S. P. , Chantre, C. O. , Zimmerman, J. F. , Pasqualini, F. S. , Liu, X. , Goss, J. A. , Campbell, P. H. , Gonzalez, G. M. , Park, S. J. , Capulli, A. K. , Ferrier, J. P. , Kosar, T. F. , Mahadevan, L. , Pu, W. T. , & Parker, K. K. (2018). A tissue‐engineered scale model of the heart ventricle. Nature Biomedical Engineering, 2(12), 930–941. 10.1038/s41551-018-0271-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannhardt, I. , Breckwoldt, K. , Letuffe‐Brenière, D. , Schaaf, S. , Schulz, H. , Neuber, C. , Benzin, A. , Werner, T. , Eder, A. , Schulze, T. , Klampe, B. , Christ, T. , Hirt, M. , Huebner, N. , Moretti, A. , Eschenhagen, T. , & Hansen, A. (2016). Human engineered heart tissue: Analysis of contractile force. Stem Cell Reports, 7(1), 29–42. 10.1016/j.stemcr.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto, H. , Matsuo, T. , Yamamizu, K. , Uosaki, H. , Narazaki, G. , Katayama, S. , Marui, A. , Shimizu, T. , Ikeda, T. , Okano, T. , Sakata, R. , & Yamashita, J. K. (2012). Pluripotent stem cell‐engineered cell sheets reassembled with defined cardiovascular populations ameliorate reduction in infarct heart function through cardiomyocyte‐mediated neovascularization. Stem Cells (Dayton, Ohio), 30(6), 1196–1205. 10.1002/stem.1089 [DOI] [PubMed] [Google Scholar]
- Mathur, A. , Loskill, P. , Shao, K. , Huebsch, N. , Hong, S. , Marcus, S. G. , Marks, N. , Mandegar, M. , Conklin, B. R. , Lee, L. P. , & Healy, K. E. (2015). Human iPSC‐based cardiac microphysiological system for drug screening applications. Scientific Reports, 5, 8883. 10.1038/srep08883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, A. , Ma, Z. , Loskill, P. , Jeeawoody, S. , & Healy, K. E. (2016). In vitro cardiac tissue models: Current status and future prospects. Advanced Drug Delivery Reviews, 96, 203–213. 10.1016/j.addr.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura, K. , Haraguchi, Y. , Shimizu, T. , & Okano, T. (2013). Cell sheet transplantation for heart tissue repair. Journal of Controlled Release : Official Journal of the Controlled Release Society, 169(3), 336–340. 10.1016/j.jconrel.2013.03.003 [DOI] [PubMed] [Google Scholar]
- Matsuura, K. , Masuda, S. , & Shimizu, T. (2014). Cell sheet‐based cardiac tissue engineering. Anatomical Record (Hoboken, N.J.), 297(1), 65–72. 10.1002/ar.22834 [DOI] [PubMed] [Google Scholar]
- McNaughton, R. , Huet, G. , & Shakir, S. (2014). An investigation into drug products withdrawn from the EU market between 2002 and 2011 for safety reasons and the evidence used to support the decision‐making. BMJ Open, 4(1), e004221. 10.1136/bmjopen-2013-004221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methe, K. , Bäckdahl, H. , Johansson, B. R. , Nayakawde, N. , Dellgren, G. , & Sumitran‐Holgersson, S. (2014). An alternative approach to decellularize whole porcine heart. BioResearch Open Access, 3(6), 327–338. 10.1089/biores.2014.0046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. M. , Meki, M. H. , Ou, Q. , George, S. A. , Gams, A. , Abouleisa, R. R. E. , Tang, X. L. , Ahern, B. M. , Giridharan, G. A. , El‐Baz, A. , Hill, B. G. , Satin, J. , Conklin, D. J. , Moslehi, J. , Bolli, R. , Ribeiro, A. J. , Efimov, I. R. , & Mohamed, T. M. A. (2020). Heart slice culture system reliably demonstrates clinical drug‐related cardiotoxicity. Toxicology and Applied Pharmacology, 406, 115213. 10.1016/j.taap.2020.115213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, R. J. , Titmarsh, D. M. , Koenig, X. , Parker, B. L. , Ryall, J. G. , Quaife‐Ryan, G. A. , Voges, H. K. , Hodson, M. P. , Ferguson, C. , Drowley, L. , Plowright, A. T. , Needham, E. J. , Wang, Q. D. , Gregorevic, P. , Xin, M. , Thomas, W. G. , Parton, R. G. , Nielsen, L. K. , Launikonis, B. S. , & Hudson, J. E. (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proceedings of the National Academy of Sciences of the United States of America, 114(40), E8372–E8381. 10.1073/pnas.1707316114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa, S. , Domae, K. , Yoshikawa, Y. , Fukushima, S. , Nakamura, T. , Saito, A. , Sakata, Y. , Hamada, S. , Toda, K. , Pak, K. , Takeuchi, M. , & Sawa, Y. (2017). Phase I clinical trial of autologous stem cell‐sheet transplantation therapy for treating cardiomyopathy. Journal of American Heart Association, 6(4). 10.1161/JAHA.116.003918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, S. , Yasuda, S. , Katoh, M. , Yamada, K. P. , Yamashita, H. , Saeki, Y. , Sunagawa, K. , Nagai, R. , Hisada, T. , & Sugiura, S. (2004). Single cell mechanics of rat cardiomyocytes under isometric, unloaded, and physiologically loaded conditions. American Journal of Physiology ‐ Heart and Circulatory Physiology, 287(1), H196–H202. 10.1152/ajpheart.00948.2003 [DOI] [PubMed] [Google Scholar]
- Noor, N. , Shapira, A. , Edri, R. , Gal, I. , Wertheim, L. , & Dvir, T. (2019). 3D printing of personalized thick and perfusable cardiac patches and hearts. Advanced Science, 6(11), 1900344. 10.1002/advs.201900344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes, S. S. , Miklas, J. W. , Liu, J. , Aschar‐Sobbi, R. , Xiao, Y. , Zhang, B. , Jiang, J. , Masse, S. , Gagliardi, M. , Hsieh, A. , Thavandiran, N. , Laflamme, M. A. , Nanthakumar, K. , Gross, G. J. , Backx, P. H. , Keller, G. , & Radisic, M. (2013). Biowire: A platform for maturation of human pluripotent stem cell‐derived cardiomyocytes. Nature Methods, 10(8), 781–787. 10.1038/nmeth.2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.‐J. , Kim, R. Y. , Park, B.‐W. , Lee, S. , Choi, S. W. , Park, J.‐H. , Choi, J. J. , Kim, S. W. , Jang, J. , Cho, D. W. , Chung, H. M. , Moon, S. H. , Ban, K. , & Park, H.‐J. (2019). Dual stem cell therapy synergistically improves cardiac function and vascular regeneration following myocardial infarction. Nature Communications, 10(1), 3123. 10.1038/s41467-019-11091-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa, H. , Wang, B. Z. , & Vunjak‐Novakovic, G. (2017). A microfluidic platform for the high‐throughput study of pathological cardiac hypertrophy. Lab on a Chip, 17(19), 3264–3271. 10.1039/c7lc00415j [DOI] [PubMed] [Google Scholar]
- Pasqualini, F. S. , Agarwal, A. , O’Connor, B. B. , Liu, Q. , Sheehy, S. P. , & Parker, K. K. (2018). Traction force microscopy of engineered cardiac tissues. PLoS One, 13(3), e0194706. 10.1371/journal.pone.0194706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passier, R. , vanLaake, L. W. , & Mummery, C. L. (2008). Stem‐cell‐based therapy and lessons from the heart. Nature, 453(7193), 322–329. 10.1038/nature07040 [DOI] [PubMed] [Google Scholar]
- Pasumarthi, K. B. S. , & Field, L. J. (2002). Cardiomyocyte cell cycle regulation. Circulation Research, 90(10), 1044–1054. 10.1161/01.res.0000020201.44772.67 [DOI] [PubMed] [Google Scholar]
- Payan, S. M. , Hubert, F. , & Rochais, F. (2020). Cardiomyocyte proliferation, a target for cardiac regeneration. Biochimica et Biophysica Acta Molecular Cell Research, 1867(3), 118461. 10.1016/j.bbamcr.2019.03.008 [DOI] [PubMed] [Google Scholar]
- Pinto, A. R. , Ilinykh, A. , Ivey, M. J. , Kuwabara, J. T. , D'Antoni, M. L. , Debuque, R. , Chandran, A. , Wang, L. , Arora, K. , Rosenthal, N. A. , & Tallquist, M. D. (2016). Revisiting cardiac cellular composition. Circulation Research, 118(3), 400–409. 10.1161/CIRCRESAHA.115.307778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss, K. D. , Wilson, L. G. , & Keating, M. T. (2002). Heart regeneration in zebrafish. Science, 298(5601), 2188–2190. 10.1126/science.1077857. PMID: 12481136 [DOI] [PubMed] [Google Scholar]
- Richards, D. J. , Li, Y. , Kerr, C. M. , Yao, J. , Beeson, G. C. , Coyle, R. C. , Chen, X. , Jia, J. , Damon, B. , Wilson, R. , Starr Hazard, E. , Hardiman, G. , Menick, D. R. , Beeson, C. C. , Yao, H. , Ye, T. , & Mei, Y. (2020). Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nature Biomedical Engineering, 4(4), 446–462. 10.1038/s41551-020-0539-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson‐Bouchard, K. , Ma, S. P. , Yeager, K. , Chen, T. , Song, L. , Sirabella, D. , Morikawa, K. , Teles, D. , Yazawa, M. , & Vunjak‐Novakovic, G. (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature, 556, 239–243. 10.1038/s41586-018-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamiya, M. , Fang, Y. , Mo, X. , Shen, J. , & Zhang, T. (2020). A heart‐on‐a‐chip platform for online monitoring of contractile behavior via digital image processing and piezoelectric sensing technique. Medical Engineering & Physics, 75, 36–44. 10.1016/j.medengphy.2019.10.001 [DOI] [PubMed] [Google Scholar]
- Saleem, U. , Mannhardt, I. , Braren, I. , Denning, C. , Eschenhagen, T. , & Hansen, A. (2020). Force and calcium transients analysis in human engineered heart tissues reveals positive force‐frequency relation at physiological frequency. Stem Cell Reports, 14(2), 312–324. 10.1016/j.stemcr.2019.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir, Y. , Polyak, B. , & Cohen, S. (2014). Cardiac tissue engineering in magnetically actuated scaffolds. Nanotechnology, 25(1), 014009. 10.1088/0957-4484/25/1/014009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, Y. , & Miyagawa, S. (2013). Present and future perspectives on cell sheet‐based myocardial regeneration therapy. BioMed Research International. 6, 583912. 10.1155/2013/583912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, Y. , Miyagawa, S. , Sakaguchi, T. , Fujita, T. , Matsuyama, A. , Saito, A. , Shimizu, T. , & Okano, T. (2012). Tissue engineered myoblast sheets improved cardiac function sufficiently to discontinue LVAS in a patient with DCM: Report of a case. Surgery Today, 42(2), 181–184. 10.1007/s00595-011-0106-4 [DOI] [PubMed] [Google Scholar]
- Schram, G. , Pourrier, M. , Melnyk, P. , & Nattel, S. (2002). Differential distribution of cardiac ion channel expression as a basis for regional specialization in electrical function. Circulation Research, 90(9), 939–950. 10.1161/01.res.0000018627.89528.6f [DOI] [PubMed] [Google Scholar]
- Scuderi, G. J. , & Butcher, J. (2017). Naturally engineered maturation of cardiomyocytes. Frontiers in Cell and Developmental Biology, 5, 50. 10.3389/fcell.2017.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers, V. F. M. , & Lee, R. T. (2008). Stem‐cell therapy for cardiac disease. Nature, 451(7181), 937–942. 10.1038/nature06800 [DOI] [PubMed] [Google Scholar]
- Sekine, H. , Shimizu, T. , Dobashi, I. , Matsuura, K. , Hagiwara, N. , Takahashi, M. , Kobayashi, E. , Yamato, M. , & Okano, T. (2011). Cardiac cell sheet transplantation improves damaged heart function via superior cell survival in comparison with dissociated cell injection. Tissue Engineering Part A, 17(23–24), 2973–2980. 10.1089/ten.tea.2010.0659 [DOI] [PubMed] [Google Scholar]
- Shaheen, N. , Shiti, A. , Huber, I. , Shinnawi, R. , Arbel, G. , Gepstein, A. , Setter, N. , Goldfracht, I. , Gruber, A. , Chorna, S. V. , & Gepstein, L. (2018). Human induced pluripotent stem cell‐derived cardiac cell sheets expressing genetically encoded voltage indicator for pharmacological and arrhythmia studies. Stem Cell Reports, 10(6), 1879–1894. 10.1016/j.stemcr.2018.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnawi, R. , Shaheen, N. , Huber, I. , Shiti, A. , Arbel, G. , Gepstein, A. , Ballan, N. , Setter, N. , Tijsen, A. J. , Borggrefe, M. , & Gepstein, L. (2019). Modeling reentry in the short QT syndrome with human‐induced pluripotent stem cell‐derived cardiac cell sheets. Journal of the American College of Cardiology, 73, 2310–2324. 10.1016/j.jacc.2019.02.055 [DOI] [PubMed] [Google Scholar]
- Sidorov, V. Y. , Samson, P. C. , Sidorova, T. N. , Davidson, J. M. , Lim, C. C. , & Wikswo, J. P. (2017). I‐Wire Heart‐on‐a‐Chip I: Three‐dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomaterialia, 48, 68–78. 10.1016/j.actbio.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar, S. , Vandersickel, N. , & Panfilov, A. V. (2017). Effect of myocyte‐fibroblast coupling on the onset of pathological dynamics in a model of ventricular tissue. Scientific Reports, 7, 40985. 10.1038/srep40985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasuna, K. , Asakura, K. , Araki, S. , Ando, H. , Kazusa, K. , Kitaguchi, T. , Kunimatsu, T. , Suzuki, S. , & Miyamoto, N. (2017). Comprehensive in vitro cardiac safety assessment using human stem cell technology: Overview of CSAHi HEART initiative. Journal of Pharmacological and Toxicological Methods, 83, 42–54. 10.1016/j.vascn.2016.09.004 [DOI] [PubMed] [Google Scholar]
- Tanner, M. R. , & Beeton, C. (2018). Differences in ion channel phenotype and function between humans and animal models. Frontiers in Bioscience, 23, 43–64. 10.2741/4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, C. A. A. , Varian, K. D. , Canan, C. H. , Davis, J. P. , & Janssen, P. M. L. (2013). The positive inotropic effect of pyruvate involves an increase in myofilament calcium sensitivity. PLoS One, 8(5), e63608. 10.1371/journal.pone.0063608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C. , Chao, B. S. , & Wu, J. C. (2018). Strategies for improving the maturity of human induced pluripotent stem cell‐derived cardiomyocytes. Circulation Research, 123(5), 512–514. 10.1161/CIRCRESAHA.118.313472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer, B. M. , Stoehr, A. , Schulze, M. L. , Patel, S. , Gucek, M. , Mannhardt, I. , Funcke, S. , Murphy, E. , Eschenhagen, T. , & Hansen, A. (2018). Contractile work contributes to maturation of energy metabolism in hiPSC‐derived cardiomyocytes. Stem Cell Reports, 10(3), 834–847. 10.1016/j.stemcr.2018.01.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen, M. J. , & Engel, F. B. (2008). Features of cardiomyocyte proliferation and its potential for cardiac regeneration. Journal of Cellular and Molecular Medicine, 12(6A), 2233–2244. 10.1111/j.1582-4934.2008.00439.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Valk, J. , Brunner, D. , De Smet, K. , Fex Svenningsen, A. , Honegger, P. , Knudsen, L. E. , Lindl, T. , Noraberg, J. , Price, A. , Scarino, M. , & Gstraunthaler, G. (2010). Optimization of chemically defined cell culture media‐‐replacing fetal bovine serum in mammalian in vitro methods. Toxicology in Vitro : An International Journal Published in Association with BIBRA, 24(4), 1053–1063. 10.1016/j.tiv.2010.03.016 [DOI] [PubMed] [Google Scholar]
- van Dijk, C. G. M. , Brandt, M. M. , Poulis, N. , Anten, J. , van der Moolen, M. , Kramer, L. , Homburg, E. F. G. A. , Louzao‐Martinez, L. , Pei, J. , Krebber, M. M. , van Balkom, B. W. M. , de Graaf, P. , Duncker, D. J. , Verhaar, M. C. , Luttge, R. , & Cheng, C. (2020). A new microfluidic model that allows monitoring of complex vascular structures and cell interactions in a 3D biological matrix. Lab on a Chip, 20(10), 1827–1844. 10.1039/d0lc00059k [DOI] [PubMed] [Google Scholar]
- Voges, H. K. , Mills, R. J. , Elliott, D. A. , Parton, R. G. , Porrello, E. R. , & Hudson, J. E. (2017). Development of a human cardiac organoid injury model reveals innate regenerative potential. Development (Cambridge, England), 144(6), 1118–1127. 10.1242/dev.143966 [DOI] [PubMed] [Google Scholar]
- Wang, B. , Wang, G. , To, F. , Butler, J. R. , Claude, A. , McLaughlin, R. M. , Williams, L. N. , de Jongh Curry, A. L. , & Liao, J. (2013). Myocardial scaffold‐based cardiac tissue engineering: Application of coordinated mechanical and electrical stimulations. Langmuir : The ACS Journal of Surfaces and Colloids, 29(35), 11109–11117. 10.1021/la401702w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. , McCain, M. L. , Yang, L. , He, A. , Pasqualini, F. S. , Agarwal, A. , Yuan, H. , Jiang, D. , Zhang, D. , Zangi, L. , Geva, J. , Roberts, A. E. , Ma, Q. , Ding, J. , Chen, J. , Wang, D. Z. , Li, K. , Wang, J. , Wanders, R. J. A. , & Pu, W. T. (2014). Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart‐on‐chip technologies. Nature Medicine, 20(6), 616–623. 10.1038/nm.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Yang, H. , Bai, A. , Jiang, W. , Li, X. , Wang, X. , Mao, Y. , Lu, C. , Qian, R. , Guo, F. , Ding, T. , Chen, H. , Chen, S. , Zhang, J. , Liu, C. , & Sun, N. (2016). Functional engineered human cardiac patches prepared from nature’s platform improve heart function after acute myocardial infarction. Biomaterials, 105, 52–65. 10.1016/j.biomaterials.2016.07.035 [DOI] [PubMed] [Google Scholar]
- Watson, S. A. , Duff, J. , Bardi, I. , Zabielska, M. , Atanur, S. S. , Jabbour, R. J. , Simon, A. , Tomas, A. , Smolenski, R. T. , Harding, S. E. , Perbellini, F. , & Terracciano, C. M. (2019). Biomimetic electromechanical stimulation to maintain adult myocardial slices in vitro. Nature Communications, 10(1). 10.1038/S41467-019-10175-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, S. A. , Scigliano, M. , Bardi, I. , Ascione, R. , Terracciano, C. M. , & Perbellini, F. (2017). Preparation of viable adult ventricular myocardial slices from large and small mammals. Nature Protocols, 12(12), 2623–2639. 10.1038/nprot.2017.139 [DOI] [PubMed] [Google Scholar]
- Wong, A. O.‐T. , Wong, N. , Geng, L. , Chow, M. Z. , Lee, E. K. , Wu, H. , Khine, M. , Kong, C. W. , Costa, K. D. , Keung, W. , Cheung, Y. F. , & Li, R. A. (2020). Combinatorial treatment of human cardiac engineered tissues with biomimetic cues induces functional maturation as revealed by optical mapping of action potentials and calcium transients. Frontiers in Physiology, 11, 165. 10.3389/fphys.2020.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Zhou, L. , Liu, H. , Duan, R. , Zhou, H. , Zhang, F. , He, X. , Lu, D. , Xiong, K. , Xiong, M. , Zhuang, J. , Liu, Y. , Li, L. , Liang, D. , & Chen, Y. H. (2021). LRP6 downregulation promotes cardiomyocyte proliferation and heart regeneration. Cell Research, 31, 450–462. 10.1038/s41422-020-00411-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Zhang, B. , Liu, H. , Miklas, J. W. , Gagliardi, M. , Pahnke, A. , Thavandiran, N. , Sun, Y. , Simmons, C. , Keller, G. , & Radisic, M. (2014). Microfabricated perfusable cardiac biowire: A platform that mimics native cardiac bundle. Lab on a Chip, 14(5), 869–882. 10.1039/c3lc51123e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , Ding, S. , Zhao, Y. , Pu, J. , & He, B. (2014). Autologous transplantation of bone marrow/blood‐derived cells for chronic ischemic heart disease: A systematic review and meta‐analysis. Canadian Journal of Cardiology, 30(11), 1370–1377. 10.1016/j.cjca.2014.01.013 [DOI] [PubMed] [Google Scholar]
- Yamamoto, R. , Miyagawa, S. , Toda, K. , Kainuma, S. , Yoshioka, D. , Yoshikawa, Y. , Hata, H. , Ueno, T. , Kuratani, T. , & Sawa, Y. (2019). Long‐term outcome of ischemic cardiomyopathy after autologous myoblast cell‐sheet implantation. The Annals of Thoracic Surgery, 108(5), e303–e306. 10.1016/j.athoracsur.2019.03.028 [DOI] [PubMed] [Google Scholar]
- Yanagawa, B. , Rao, V. , Yau, T. M. , & Cusimano, R. J. (2013). Initial experience with intraventricular repair using CorMatrix extracellular matrix. Innovations, 8(5), 348–352. 10.1097/IMI.0000000000000014 [DOI] [PubMed] [Google Scholar]
- Yang, J. , Yamato, M. , Shimizu, T. , Sekine, H. , Ohashi, K. , Kanzaki, M. , Ohki, T. , Nishida, K. , & Okano, T. (2007). Reconstruction of functional tissues with cell sheet engineering. Biomaterials, 28(34), 5033–5043. 10.1016/j.biomaterials.2007.07.052 [DOI] [PubMed] [Google Scholar]
- Yang, X. , Pabon, L. , & Murry, C. E. (2014a). Engineering adolescence: Maturation of human pluripotent stem cell‐derived cardiomyocytes. Circulation Research, 114(3), 511–523. 10.1161/CIRCRESAHA.114.300558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Rodriguez, M. , Pabon, L. , Fischer, K. A. , Reinecke, H. , Regnier, M. , Sniadecki, N. J. , Ruohola‐Baker, H. , & Murry, C. E. (2014b). Tri‐iodo‐l‐thyronine promotes the maturation of human cardiomyocytes‐derived from induced pluripotent stem cells. Journal of Molecular and Cellular Cardiology, 72, 296–304. 10.1016/j.yjmcc.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, S. , Cao, H. , Hu, H. , Wang, X. , Tang, Y. , & Huang, C. (2016). Alk7 depleted mice exhibit prolonged cardiac repolarization and are predisposed to ventricular arrhythmia. PLoS One, 11(2), e0149205. 10.1371/journal.pone.0149205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S. , Miyagawa, S. , Fukushima, S. , Kawamura, T. , Kashiyama, N. , Ohashi, F. , Toyofuku, T. , Toda, K. , & Sawa, Y. (2018). Maturation of human induced pluripotent stem cell‐derived cardiomyocytes by soluble factors from human mesenchymal stem cells. Molecular Therapy : The Journal of the American Society of Gene Therapy, 26(11), 2681–2695. 10.1016/j.ymthe.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa, Y. , Miyagawa, S. , Toda, K. , Saito, A. , Sakata, Y. , & Sawa, Y. (2018). Myocardial regenerative therapy using a scaffold‐free skeletal‐muscle‐derived cell sheet in patients with dilated cardiomyopathy even under a left ventricular assist device: A safety and feasibility study. Surgery Today, 48(2), 200–210. 10.1007/s00595-017-1571-1 [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Zhu, W. , Radisic, M. , & Vunjak‐Novakovic, G. (2018). Can we engineer a human cardiac patch for therapy? Circulation Research, 123(2), 244–265. 10.1161/CIRCRESAHA.118.311213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, M. T. , Ye, S. , Su, J. , & Garg, V. (2020a). Cardiomyocyte proliferation and maturation: Two sides of the same coin for heart regeneration. Frontiers in Cell and Developmental Biology, 8, 594226. 10.3389/fcell.2020.594226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Rafatian, N. , Feric, N. T. , Cox, B. J. , Aschar‐Sobbi, R. , Wang, E. Y. , Aggarwal, P. , Zhang, B. , Conant, G. , Ronaldson‐Bouchard, K. , Pahnke, A. , Protze, S. , Lee, J. H. , Davenport Huyer, L. , Jekic, D. , Wickeler, A. , Naguib, H. E. , Keller, G. M. , Vunjak‐Novakovic, G. , & Radisic, M. (2019). A platform for generation of chamber‐specific cardiac tissues and disease modeling. Cell, 176(4), 913–927. e18. 10.1016/j.cell.2018.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Rafatian, N. , Wang, E. Y. , Wu, Q. , Lai, B. F. L. , Lu, R. X. , Savoji, H. , & Radisic, M. (2020b). Towards chamber specific heart‐on‐a‐chip for drug testing applications. Advanced Drug Delivery Reviews, 165–166, 60–76. 10.1016/j.addr.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, W. H. , Didié, M. , Wasmeier, G. H. , Nixdorff, U. , Hess, A. , Melnychenko, I. , Boy, O. , Neuhuber, W. L. , Weyand, M. , & Eschenhagen, T. (2002). Cardiac grafting of engineered heart tissue in syngenic rats. Circulation, 106(12 Suppl 1), I151–I157. [PubMed] [Google Scholar]
- Zimmermann, W. H. , Melnychenko, I. , & Eschenhagen, T. (2004). Engineered heart tissue for regeneration of diseased hearts. Biomaterials, 25(9), 1639–1647. 10.1016/s0142-9612(03)00521-0 [DOI] [PubMed] [Google Scholar]
- Zimmermann, W. H. , Schneiderbanger, K. , Schubert, P. , Didie, M. , El‐ Armouche, A. , & Eschenhagenm, T. (2001). Engineering of an organoid cardiac tissue equivalent in vitro. Circulation, 104. :II–129 [abstract]. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


