Abstract
Antibodies specific for capsular polysaccharides play a central role in immunity to encapsulated Streptococcus pneumoniae, but little is known about their genetics or the variable (V) region polymorphisms that affect their protective function. To begin to address these issues, we used combinatorial library cloning to isolate pneumococcal polysaccharide (PPS)-specific Fab fragments from two vaccinated adults. We determined complete V region primary structures and performed antigen binding analyses of seven Fab fragments specific for PPS serotype 6B, 14, or 23F. Fabs were of the immunoglobulin G2 or A isotype. Several VHIII gene segments (HV 3-7, 3-15, 3-23, and 3-11) were identified. VL regions were encoded by several κ genes (KV 4-1, 3-15, 2-24, and 2D-29) and a λ gene (LV 1-51). Deviation of the VH and VL regions from their assigned germ line counterparts indicated that they were somatically mutated. Fabs of the same serotype specificity isolated from a single individual differed in affinity, and these differences could be accounted for either by the extent of mutation among clonal relatives or by usage of different V-region genes. Thus, functionally disparate anti-PPS antibodies can arise within individuals both by activation of independent clones and by intraclonal somatic mutation. For one pair of clonally related Fabs, the more extensively mutated VH was associated with lower affinity for PPS 14, a result suggesting that somatic mutation could lead to diminished protective efficacy. These findings indicate that the PPS repertoire in the adult derives from memory B-cell populations that have class switched and undergone extensive hypermutation.
Streptococcus pneumoniae is a serious human bacterial pathogen causing pneumonia, bacteremia, meningitis, and acute otitis media (7). Encapsulated pneumococci are considered one of the leading causes of death worldwide (4), and in the United States approximately 500,000 cases of invasive pneumococcal disease occur per year, resulting in 40,000 deaths. Ninety or more different pneumococcal capsular polysaccharide (PPS) serotypes have been identified, but only a subset of these are responsible for the majority of invasive disease (7). Immunity to pneumococcal infection is mediated principally by opsonic PPS-specific antibodies (Abs) (17). Accordingly, efforts to develop effective pneumococcal vaccines have focused upon induction of these Ab specificities.
The vaccine presently licensed in the United States consists of a mixture of 23 purified PPS capsular serotypes (4, 51). The young and the elderly are particularly susceptible to developing pneumococcal infection and comprise the principal target groups for vaccination. The polyvalent vaccine is generally immunogenic in healthy young adults and the elderly, although efficacy estimates vary considerably (18). In contrast, the majority of the PPS serotypes are poorly immunogenic in infants, and therefore, the polyvalent pneumococcal vaccine does not provide uniform protection against invasive pneumococcal disease in this age group. The lack of an effective pediatric vaccine and the emergence of antibiotic-resistant pneumococci have prompted the development of new vaccines in which protein carriers are covalently coupled to the PPS (28, 30, 58). This design is based on that used for the development of efficacious pediatric vaccines against Haemophilus influenzae type b (Hib) (24). Unlike plain polyvalent PPS vaccines, the protein-conjugated forms of PPS are immunogenic in infants, and a recent clinical trial of a heptavalent PPS conjugate vaccine in infants has demonstrated high efficacy in preventing invasive diseases caused by pneumococci expressing the capsular serotypes contained in the vaccine (13).
Renewed interest in the serological and functional characterization of anti-PPS Ab responses has accompanied these vaccine development efforts. Although this interest stems primarily from the need to evaluate vaccine immunogenicity and to establish reliable surrogates of protection, the Ab response to PPS antigens (Ags) represents an ideal opportunity to examine the inheritance and development of protective immunity in humans. Ab responses to PPS Ags are markedly oligoclonal within individuals (31, 34, 46), and consequently variable (V) region diversity is limited. This property leads to individual variation in PPS-specific Ab fine specificity (41), avidity, and protective efficacy (52, 66). While V region polymorphism undoubtedly affects antibody protective function, little is known about the V regions encoding PPS antibodies or the structural determinants of PPS binding.
In this study we describe our initial efforts aimed at the molecular definition of the human Ab repertoire to PPS Ags. We used combinatorial library cloning to isolate Fab fragments specific for PPS serotypes 6B, 14, and 23F. We focused on these particular serotypes because they are structurally disparate, they are components of both licensed and experimental conjugate vaccines, and the respective pneumococci are significant pathogens.
MATERIALS AND METHODS
Human subjects and vaccination.
Two healthy adults, a 45-year-old Caucasian female (002) and a 24-year-old African-American male (018), received an intramuscular injection of 0.5 ml of Pneumovax vaccine (Merck & Co., Inc., West Point, Pa.). Peripheral blood samples were taken before, 7 days after, and 30 days after vaccination. The protocols were reviewed and approved by the Children's Hospital Oakland Research Committee and Institutional Review Board.
Preparation of PPS paramagnetic beads and enrichment of PPS-binding B cells.
Lyophilized PPS 6B, 14, and 23F were purchased from the American Type Culture Collection, Rockville, Md. Ten milligrams of PPS was dissolved in 1.0 ml of 0.2 M sodium bicarbonate (pH 10). Cyanogen bromide (2.5 mg dissolved in 50 μl of dimethylformamide) was added, and the mixture was stirred on ice for 10 min. Another 2.5 mg of cyanogen bromide was added, and the reaction proceeded for an additional 10 min. Biotin hydrazide (Pierce Chemical Co., St. Louis, Mo.) was dissolved in dimethyl sulfoxide, and 14.3 mg was added to the solution of activated PPS, giving a final biotin hydrazide concentration of 5 mM. The solution was stirred at room temperature for 2 h, after which it was dialyzed extensively at 4°C against phosphate-buffered saline (PBS). The PPS-biotin was sterilized by filtration and stored at −80°C.
PPS-coated paramagnetic beads were prepared by adding 50 μg of PPS-biotin to 1.0 mg of washed avidin paramagnetic beads (Immunotech Inc., Marseille, France) in a total volume of 0.5 ml of PBS–0.2% bovine serum albumin (BSA). Beads were mixed, incubated at room temperature for 15 min, isolated with a magnet and washed several times with PBS-BSA.
Ficoll-Hypaque was used to isolate mononuclear cells (MNC) from the heparinized peripheral blood sample obtained 7 days after vaccination. After a wash with RPMI 1640 medium, MNC were suspended to a concentration of 107/ml in PBS containing 30% fetal calf serum (FCS) and 20 μg of pneumococcal common cell wall polysaccharide (CPS; Danish Staten Seruminstitut, Copenhagen, Denmark). To enrich for PPS-specific B cells, 1.0 mg of PPS-coated paramagnetic beads was added to 2 × 107 MNC, and the mixture was incubated for 15 min on ice. Cells binding to beads were isolated by magnetic separation. The beads were washed two times with PBS-BSA, and RNA was extracted from the beads using RNA-STAT-60 (Tel-Test “B,” Inc., Friendswood, Tex.). RNA was stored at −80°C as an ethanol precipitate. cDNA was prepared from RNA using oligo(dT)18 as a primer. Reverse transcriptase, nucleotides, and buffers were purchased from Pharmacia, Inc. (Piscataway, N.J.) and were used according to instructions provided by the manufacturer.
Preparation of L chain and Fd chain libraries.
The general procedures used for construction of Fab libraries have been described elsewhere (9, 50). Fd and light (L) chain cDNAs were amplified by PCR using appropriate primers. The L chain primers were those described previously (50) plus VK6a (gaaattgagctcactcagtctcc [all sequences are 5′-3′]), VK6b (gatgttgagctcacacagtctcc), VLAM1 (aattttgagctcactcagccccac), VLAM2 (tctgccgagctccagcctgcgtccgtg), VLAM3 (tctgtggagctccagccgccctcagtg), VLAM4 (tctgaagagctccaggaccctgttgtgtctgtg), VLAM5 (cagtctgagctcacgcagccgccc), VLAM6 (cagactgagctcactcaggagccc), VL1/b5 (cagtctgagctcactcagccacc), VL3′ (tcctatgagctcactcagccaccc), VL8′ (cagcttgagctcactcaatcgccc), VL7′ (caggttgagctcactcaaccgccc), VL2.1 (cagtctgagctcactcagcctgcc), and lambdacon (cgccgtctagaactatgaacattctgtagg). The Fd primers were those described previously (50) plus VH4.21 (5′-caggtgcagctactcgagtggggc-3′). L chain and Fd chain PCR products were separately inserted into the pComb3H vector (kindly provided by Carlos Barbas and The Scripp's Research Institute [9]), using the appropriate restriction sites. Phagemid DNA was electroporated into XL1-Blue cells, and transformants were selected. Combinatorial Fab libraries were prepared by inserting bulk Fd chain DNA into bulk phagemid DNA containing L chains. Purified clonal phagemid DNA was sequenced directly with Sequenase 2.0 (U.S. Biochemical Corp., Cleveland, Ohio) as previously described (50).
Identification of PPS-specific Fabs.
Fab libraries were screened for the presence of PPS-specific Fab fragments by either autoradiography of blots of clones from agar plates or radioantigen binding assay (RABA) of individual randomly picked clonal isolates. For screening by the blotting method, Fab library phagemid clones were plated on LB-carbenicillin plates and incubated overnight at 37°C. Nitrocellulose circles were coated overnight with either anti-human kappa or anti-human lambda Ab (Biosource International, Camarillo, Calif.) at a concentration of 5 μg/ml in PBS. The nitrocellulose was blocked with PBS–1% BSA and washed five times with PBS–0.1% Tween 20. The nitrocellulose was placed directly onto the surface of the agar plate having bacterial colonies, followed by two pieces of Whatman filter paper. After incubation for 1 to 2 h at 37°C, the nitrocellulose blots were lifted from the plates, washed several times with PBS-Tween, and then incubated with rotation for 1 h at room temperature with 125I-PPS (5 × 105 cpm/ml) diluted in PBS–1% BSA containing 10 μg of CPS/ml. The blots were washed with PBS-Tween, air dried, and exposed to X-ray film. Following blotting, the agar plates were incubated for 4 to 8 h at 37°C to regenerate the colonies and then stored at 4°C. PPS-binding bacterial clones identified by autoradiography were picked from the agar plate and isolated by streaking onto agar plates, followed by liquid culture.
Random screening of clones was performed on supernatants of clonal bacterial lysates obtained by repetitive freezing and thawing of bacteria harvested from overnight cultures. The presence of PPS binding Fab in the supernatant was determined by RABA.
Fab purification.
To facilitate purification, the carboxy-terminal region of each Fab CH1 domain was engineered to contain a polyhistidine region. A primer (5′-cctcctgactagtatgatgatggtgatggtgacaagatttgggc-3′) spanning the SpeI site at the CH1/gIII junction and encoding six histidine residues was used in PCR with an upstream VH3 primer to generate the appropriate Fd fragment. Subsequent enzymatic digestion and ligation generated a Fab with an internal polyhistidine tag. SpeI/NheI digestion, gel purification, and religation of the resulting phagemid produced a Fab lacking the gIII component of pComb3H and having a six-histidine tag at the carboxy terminus of the Fd chain. Enzymatic digestion with AocI and NotI produces a cassette containing the polyhistidine region that can be subsequently used to modify other Fabs to contain the polyhistidine region. Correct insertion and lack of PCR errors was determined by sequencing the CH1 region, and binding studies verified that the introduced modification did not affect antigen binding.
Cultures of Fab clones were prepared by seeding 500 ml of LB broth with an overnight suspension culture of bacteria carrying the appropriate phagemid. The culture was shaken at 300 rpm at 37°C for ∼8 h followed by overnight induction with 1 mM isopropyl β-d-thiogalactoside. Bacteria were then harvested by centrifugation. The bacterial pellet was resuspended in PBS containing protease inhibitors and was subjected to three cycles of freezing and thawing. Debris was removed by centrifugation, and the supernatant was used for immobilized metal affinity chromatography. Ni-nitrilotriacetic acid-agarose (Pierce Chemical Co., St. Louis, Mo.) chromatography was performed as recommended by the manufacturer. Fabs were eluted from the absorbent with 0.2 M imidazole (pH 8.0) and were dialyzed extensively against PBS. Following dialysis, Fabs were spun at 100,000 × g for 1 h, sterilized by filtration, and stored at 4°C. Fab concentration was determined either by absorbance at 280 nm using extinction coefficients calculated from amino acid composition or by a previously described enzyme-linked immunosorbent assay (ELISA) where Fabs were captured on wells coated with an anti-Fd Ab and detected using alkaline phosphatase-conjugated anti-L chain Ab (36).
RABA and PPS ELISA.
The preparation of tyraminated and iodinated PPS and the RABA have been described in a previous report (34). RABA was used to determine anti-PPS Ab concentrations in pre- and postvaccination sera and to evaluate Fab binding specificity. Serum anti-PPS levels were calculated from a standard curve generated by the reference serum 89-SF as previously described (34). Prior to assay, Fab fragments were spun for 1 h at 100,000 × g. Fab fragments or sera, diluted in PBS containing 10% FCS and 10 μg of CPS/ml, were mixed with ∼300,000 cpm of 125I-PPS in a total volume of 100 μl. After incubation overnight at 4°C, 100 μl of 100% saturated ammonium sulfate was added for 4 h at 4°C. Precipitates were harvested by centrifugation and washed with 200 μl of 50% saturated ammonium sulfate, and bound radioactivity was determined by counting in a gamma counter.
The PPS ELISA was used to determine the isotypes of anti-PPS Abs in sera and in supernatants from MNC cultures. The ELISA and calibration standards are described in reference 34.
Determination of Fab affinity.
The affinity of Fab binding to PPS was determined by RABA. 125I-PPS binding was evaluated at various Fab concentrations, and the concentration of Fab binding 50% of added 125I-PPS was calculated. Affinity is expressed as the inverse of the Fab molar concentration binding 50% of the added 125I-PPS.
GenBank accession numbers.
GenBank accession numbers for VH cDNA sequences of Fabs 6B-1, 14-1, 14-2, 14-3, 14-4, 23F-1, and 23F-2 are AF165100, AF165107, AF165109, AF165105, AF165112, AF165104, and AF165102, respectively. GenBank accession numbers for VL cDNA sequences of Fabs 6B-1, 14-1, 14-2, 14-3, 14-4, 23F-1, and 23F-2 are AF165099, AF165108, AF165110, AF106, AF165111, AF165103, and AF165101, respectively.
RESULTS
Response to vaccination.
Two adult subjects were vaccinated with polyvalent pneumococcal vaccine. Blood samples were obtained before, 7 days after, and 30 days after vaccination. Prevaccination and 30-day postvaccination serum samples were analyzed for total anti-PPS Ab, and PPS-specific κ/λ ratios were determined on the 30-day-postvaccination samples (Table 1). Also, MNC isolated from the 7-day-postvaccination blood sample were cultured for 7 days, and the anti-PPS Abs present in the culture supernatants were analyzed for heavy (H) and L chain isotypes (Table 1).
TABLE 1.
Serological characterization of subjects' responses to pneumococcal vaccinationa
| Subject | In vitro Ab H/L chain isotype(s)b
|
Serum Abc concn (μg/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Pre
|
Post
|
||||||||
| 6B | 14 | 23F | 6B | 14 | 23F | 6B | 14 | 23F | |
| 002 | G/A/κ | G/λ | G/κ | 17 | 5.0 | 3.8 | 33 (1.3) | 307 (0.1) | 34 (2.7) |
| 018 | G/κ | 0.8 | 3.0 (4.2) | ||||||
Two adult subjects were vaccinated with polyvalent pneumococcal vaccine. Blood was taken before and then 7 and 30 days after vaccination.
MNC were isolated from heparinized blood obtained 7 days following vaccination and were cultured (106/ml) in RPMI 1640 medium containing 10% FCS at 37°C in 5% CO2 atmosphere. Supernatants were harvested after 7 days and assayed for the presence of PPS-specific Ab by ELISA using isotype-specific secondary Abs (34).
Serum samples obtained before (Pre) and 30 days after (Post) vaccination were assayed for total anti-PPS Abs using RABA. κ/λ ratios were calculated by ELISA as previously described (34) and are shown in parentheses.
Both subjects had detectable serum PPS-specific Ab prior to vaccination, and both responded to vaccination with increased Ab levels to the PPS. In subject 002, vaccination elicited predominantly κ Abs to PPS 23F, approximately equal representation of κ and λ Abs to PPS 6B, and a λ-dominated response to PPS 14. κ Abs dominated the response of subject 018 to PPS 14. PPS-specific Abs were detectable in MNC culture supernatants, a result indicating that specific B cells were present in the peripheral circulation. The Abs secreted from MNC had L chain representations resembling the 30-day-postvaccination serum Abs. With the exception of PPS 6B Abs in subject 002, which were immunoglobulin G (IgG) and IgA, IgG was the predominant H chain isotype of the PPS Abs secreted from the MNC.
Cloning and isolation of PPS-specific Fab fragments.
To isolate PPS-specific Fab fragments, we prepared (Fd × L) chain combinatorial libraries using RNA obtained from 7-day-postvaccination MNC populations from the two subjects described in Table 1. Prior to library construction, we enriched for specific B cells using PPS-coated paramagnetic beads. Three separate (Fd × L) combinatorial libraries were prepared from subject 002: one each for PPS 6B (Fd × κ), PPS 14 (Fd × κ, λ), and PPS 23F (Fd × κ, λ). One (Fd × κ, λ) library was prepared from subject 018 using B cells enriched for PPS 14-binding cells.
Bacterial colonies carrying the relevant phagemids were screened for expression of PPS-binding Fab fragments as described in Materials and Methods. PPS-binding Fab clones were present in a frequency of approximately 1% in these libraries. For subject 002, five distinct Fabs were isolated: one specific for PPS 6B, two specific for PPS 14, and two specific for PPS 23F (Table 2). For subject 018, two distinct PPS 14-specific Fabs were isolated. Six of the Fabs were of the IgG2 isotype, and one was IgA. The L chain isotypes of the Fab fragments paralleled that observed by analysis of both culture-derived and serum Abs.
TABLE 2.
Summary of PPS-specific Fabsa
| Subject | Fab | PPS specificity | CH-CL | Affinityb (M−1, 106) | H
|
L
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V | % Identityc
|
CDR-3d | J | V | % Identityc
|
CDR-3 | J | |||||||
| Nucleotide | Amino acid | Nucleotide | Amino acid | |||||||||||
| 002 | 6B-1 | 6B | α1-κ | 0.5 | HV 3-7 | 89 | 82 | 9 | 4b | KV 4-1 | 95 | 98 | 8 | κ2 |
| 14-1 | 14 | γ2-λ | 33 | HV 3-15 | 94 | 89 | 10 | 4b | LV 1-51 | 97 | 97 | 11 | λ2/3 | |
| 14-2 | 14 | γ2-λ | 2.5 | HV 3-15 | 92 | 84 | 10 | 4b | LV 1-51 | 97 | 97 | 11 | λ2/3 | |
| 018 | 14-3 | 14 | γ2-κ | 1.3 | HV 3-23 | 93 | 88 | 11 | 4b | KV 3-15 | 93 | 84 | 9 | κ4 |
| 14-4 | 14 | γ2-κ | 1.3 | HV 3-23 | 93 | 89 | 11 | 4b | KV 3-15 | 94 | 84 | 9 | κ4 | |
| 002 | 23F-1 | 23F | γ2-κ | 17 | HV 3-23 | 92 | 82 | 7 | 5a | KV 2-24 | 96 | 94 | 9 | κ2 |
| 23F-2 | 23F | γ2-κ | 3.3 | HV 3-11 | 95 | 91 | 6 | 4b | KV 2D-29 | 98 | 94 | 8 | κ2 | |
Fab fragments were isolated from (Fd × L) combinatorial libraries prepared from peripheral blood MNC of subjects who were vaccinated 7 days previously with polyvalent pneumococcal vaccine.
Inverse of the Fab concentration required to bind 50% of added 125I-PPS in the RABA.
Percent nucleotide and amino acid sequence identity of the Fab V gene sequence to the assigned germ line gene and its formal translation product, respectively. H, kappa, and lambda V gene designations are according to references 45, 10, and 44, respectively.
Length in amino acid residues.
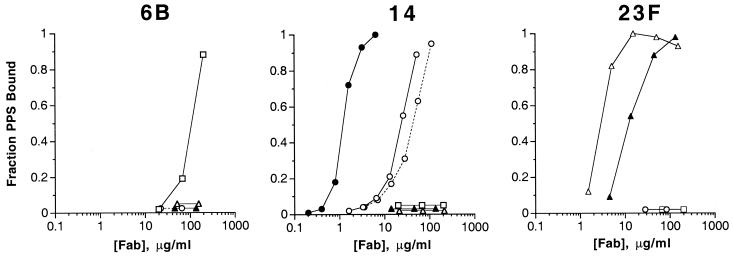
Fab specificity and affinity were analyzed using RABA. As shown in Fig. 1, the Fabs gave specific and concentration-dependent binding to their PPS Ags. Different affinities were observed for Fabs of the same serotype specificity (Table 2). For example Fabs 14-1 and 14-2, both isolated from subject 002, differed 13.2-fold in their affinity for PPS 14. They used similar V regions and appeared to be intraclonal variants (see below). Fabs 23F-1 and 23F-2, also isolated from subject 002, differed fivefold in their affinity for PPS 23F. These two Fabs, however, appeared to be derived from two distinct clones, as they utilized different VH and VL regions (see below).
FIG. 1.
RABA of purified Fab fragments 6B-1 (□), 14-1 (●), 14-2 (○——○), 14-3 and 14-4 (○––○), 23F-1 (▵), and 23F-2 (▴). Fab concentration was determined either by absorbance at 280 nm using the extinction coefficient calculated from the amino acid sequence or by ELISA as described in Materials and Methods.
V region sequences.
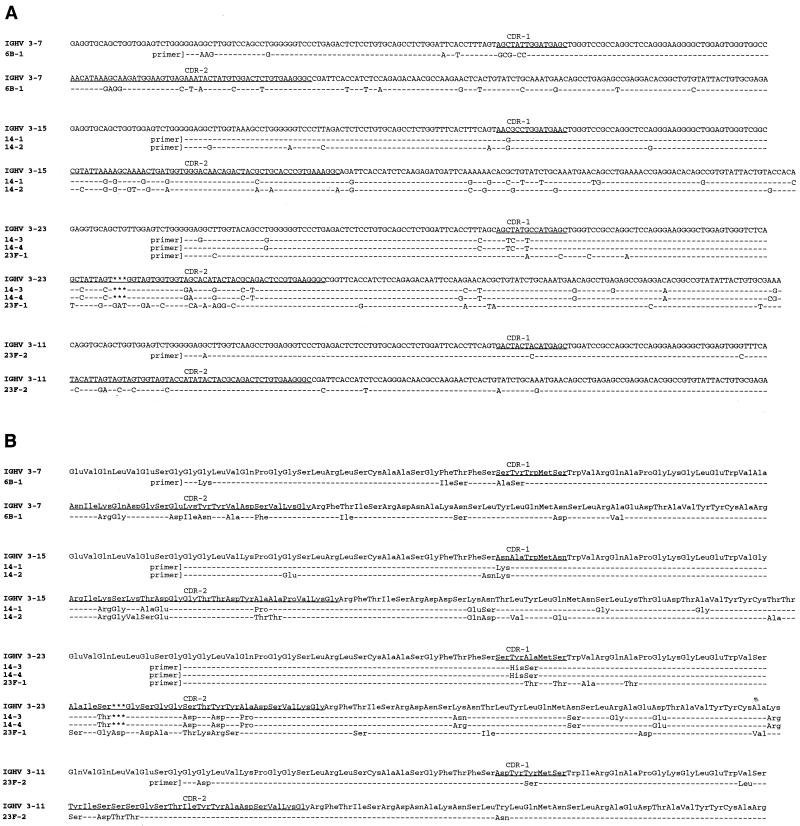
Complete VH (Fig. 2) and VL (Fig. 3) cDNA sequences were determined for all Fabs. The likely germ line V gene segments encoding these V regions were identified by a search of the databases, and assignments were made based on the germ line gene segment having the closest sequence identity to the PPS-specific Fab V sequence. Four distinct VH gene segments were used by these Fabs (Fig. 2 and Table 2). Fab 6B.1 utilized the HV 3-7 gene. HV 3-23 (also commonly known as VH26 and DP-47) was used by the PPS 14-specific Fabs 14-3 and 14-4 and also by the PPS 23F-specific Fab 23F-1. An amino acid insertion (aspartate) was present in the VH complementarity determining region 2 (CDR-2) of Fab 23F-1. In keeping with previously observed insertion patterns (23, 43, 70), the aspartate codon (GAT) appeared to be related to the immediately adjacent sequence (GGT). Similar insertions in the CDR-2 of the HV 3-23 gene have been described and are thought to arise during the course of somatic hypermutation (23, 43). Fabs 14-1 and 14-2 used the VH 3-15 gene or its close relative, known as LSG6.1 (2). Fab 23F-2 used the VH 3-11 gene. All of these gene segments are of the VHIII family. The 3-15 gene is of the IIIb subfamily and belongs to the 1-U canonical class, whereas the 3-11, 3-23, and 3-7 genes are of the IIIa subfamily and belong to the 1-3 canonical structure class (20, 65). The increased length of VH CDR-2 in Fab 23F-1 resulting from the inserted residue is likely to alter the loop configuration and thereby change the canonical structure class (23).
FIG. 2.
cDNA nucleotide sequences (A) and formal translation products (B) of Fab VH segments. Shown for comparison are the assigned germ line VH genes. CDR locations are according to Kabat et al. (27).
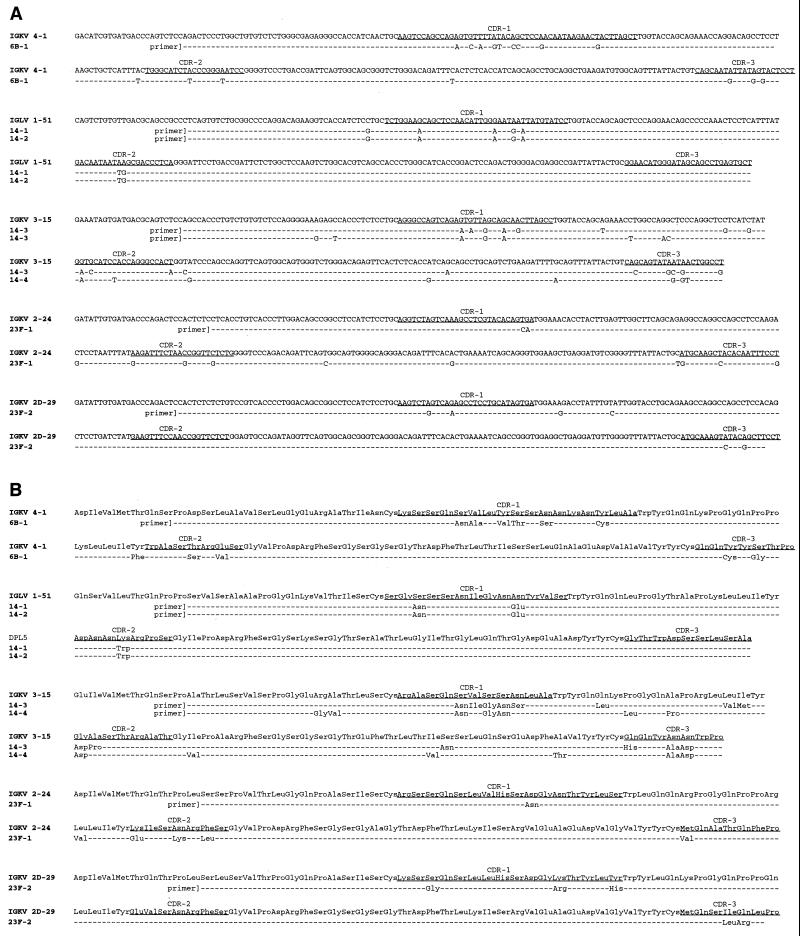
FIG. 3.
cDNA nucleotide sequences (A) and formal translation products (B) of Fab VL segments. Shown for comparison are the assigned germ line VL genes. CDR locations are according to Kabat et al. (27).
The VH segments of the Fabs appeared to be mutated, as their identity to the candidate germ line nucleotide sequence ranged from 82 to 95%. As shown in Fig. 2 and summarized in Table 2, many of the nucleotide differences between the Fab VH sequence and the respective germ line gene resulted in amino acid replacements. In the absence of germ line gene sequences from the donors, the attribution of these polymorphisms to somatic hypermutation cannot be made with certainty. However, the human VH locus has been studied extensively, and the sequence of the entire VH locus was published recently (39). While some of the differences between the Fab VH sequence and the assigned germ line gene could be due to PCR errors or to individual genomic polymorphisms, we think it is likely that most of the observed differences resulted from hypermutation of germ line gene segments that are either identical or closely related to the assigned germ line gene.
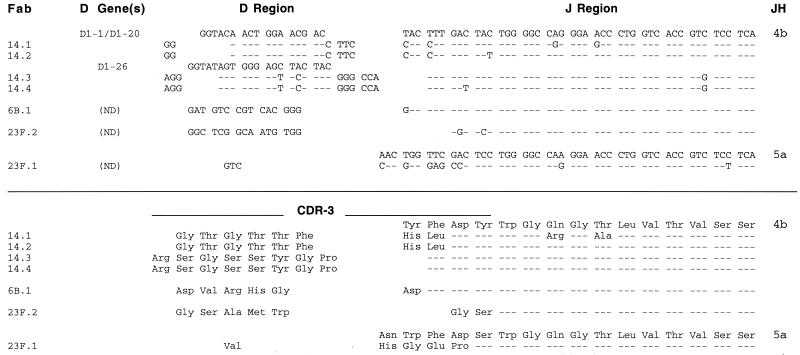
VH CDR-3 regions of the PPS-specific Fab fragments are shown in Fig. 4. Candidate germ line D segments were identified for the PPS 14-specific Fabs. Lengths of the D regions varied, as did the overall length of CDR-3 (6 to 11 amino acid residues). Six Fabs used JH4b and one used JH5a. CDR-3 length was not strictly conserved between Fabs of the same serotype specificity. Fabs 14-1 and 14-2 had a CDR-3 of 10 residues, whereas Fabs 14-3 and 14-4 had a CDR-3 of 11 residues. PPS 14-specific Fabs tended to have longer CDR-3s (10 or 11 residues) compared to PPS 23F-specific Fabs (6 or 7 residues). No identical CDR-3 motifs recurred in unrelated Fabs of the same serotype specificity, although the two pairs of PPS 14-specific Fabs, which used different V gene segments and D regions, had either Ser-Gly-Ser-Ser-Tyr or Thr-Gly-Thr-Thr-Phe in VH CDR-3.
FIG. 4.
Fab VH CDR-3 regions. CDR locations are according to Kabat et al. (27).
VL cDNA sequences and formal translation products of the PPS-specific Fabs are shown in Fig. 3 and summarized in Table 2. The Vλ gene known as LV 1-51 was used by PPS 14.1 and 14.2 in combination with the λ2 or λ3 J region. The remainder of the Fabs used genes derived from either the κII, κIII, or κIV subgroup in association with either Jκ2 or Jκ4. The PPS 23F-specific Fabs used two different Vκ genes, 2-24 and 2D-29. Both are from the κII subgroup and are of the 4-1-1 canonical structure class (64). With the exception of the 2D-29 gene, which is located on the portion of the κ locus distal to the telomere, the Vκ genes used by the Fabs derived from the proximal cluster. The KV 4-1 gene used by the PPS 6B-specific Fab, the most proximal gene segment, is located immediately adjacent to the J regions but in opposite transcriptional orientation (32). The 2D-29 gene (also known as A2 and DPK-12) used by Fab 23F.2 is the most common Vκ gene used in the human Ab response to the Hib polysaccharide (PS) (35, 55).
Similar to the VH genes, VL regions appeared to be somatically mutated. The sequence identity of the L chain V gene segments to their assigned germ line counterparts ranged from 93 to 98% (Table 2 and Fig. 3). L chain CDR-3 lengths were 8, 9, and 11 residues. PPS Fabs 14-3 and 14-4 have a 9-amino-acid CDR-3 resulting from the direct joining of the KV 3-15 gene to Jκ4, and Fabs 14-1 and 14-2 have a CDR-3 11 amino acids in length resulting from direct joining of the LV 1-51 gene segment to Jλ2/3 (Fig. 5). Fabs 6B-1 and 23F-2 have a CDR-3 of eight amino acids that resulted from the deletion of the proline codon at position 95. The PPS 23F-specific Fabs had CDR-3s either eight or nine residues in length. The truncation of the 2D-29 V gene of Fab 23F-2 is of note, as this gene segment is used by Abs to Hib PS, where the CDR-3 is 10 amino acids in length and contains an insertional arginine at position 95a, the V-J joint (35, 55). All the Fab L chain CDR-3s have amino acid substitutions compared to the germ line assignments. Some of these substitutions could result from mutation, while others, such as the truncations of Pro at position 95, may occur during V-J joining.
FIG. 5.
Fab VL CDR-3 regions. CDR locations are according to Kabat et al. (27).
The sequence data indicate that some of the Fabs isolated from the same subject were likely clonally related. For example, Fab 14-1 and 14-2 used the same VL region, but their VH regions, although derived from the same rearrangement, differed at 13 amino acid residues (4 in CDR-1 and CDR-2 and 9 in the framework regions). These sequence differences, which presumably arose as a consequence of differential somatic mutation between daughter clones, conferred a 13-fold difference in affinity for PPS 14 (Table 2 and Fig. 1). The more extensively mutated Fab, 14-2, had lower affinity for PPS 14 than its clonal relative 14-1. The PPS 14-specific Fabs isolated from subject 018 also appeared to be clonal variants, but the primary sequence differences between these Fabs (2 in VH and 15 in VL [Fig. 2 and 4]) did not confer a measurable difference in affinity (Table 2 and Fig. 1).
Independently derived clones present in an individual also can have disparate functions, as shown by Fabs 23F-1 and 23F-2. These Fabs used different VH and VL genes, and differed fivefold their affinity for PPS 23F. Thus, affinity differences can arise in PPS Ab responses by interclonal and intraclonal variation.
DISCUSSION
In this study, we used combinatorial library cloning to isolate PPS-specific Fab fragments from two vaccinated adults. This approach has permitted us to identify V genes contributing to the repertoire, to assess their mutation, and to analyze the relationship between V region polymorphism and PPS binding affinity.
Before discussing the implications of these findings, it is important to consider the cloning methodology as a potential caveat to repertoire analysis. Unlike hybridomas which retain the chain combinations present in the native B cell, Fabs from combinatorial libraries derive from the recombination of bulk Fd and L chains, a process that scrambles the H-L pairs present in vivo. PPS-binding Fab fragments may therefore not represent physiological pairing configurations. Although this possibility cannot be discounted with certainty, several observations point to the likelihood that native VH-VL configurations are being reassembled. First, only one or two distinct Fab fragments of any single serotype specificity were isolated from an individual donor, and in some cases these appeared to be intraclonal variants. This result is consistent with previous studies showing that Ab responses to PPS Ags are markedly oligoclonal and can be dominated by the products of a single clone (31, 34, 46). Second, the MNC populations used for library construction were enriched for specific B cells using PPS-coated paramagnetic beads and were obtained 7 days after vaccination, a time when there is an increased frequency of PPS-specific B cells in the peripheral circulation (38). This increased representation of PPS-specific B cells and the relevant mRNA would increase the probability of reassembling native pairs with the appropriate PPS binding specificity. Although the use of PPS-coated beads could potentially result in affinity biases in the resulting libraries, the isolation from the same library of Fabs having greater than 10-fold differences in binding affinity for PPS14 suggests that this does not represent a significant limitation. Third, Fabs of differing serotype specificity were isolated from a single donor, and they used distinctive gene rearrangements, CDR-3 configurations, and chain pairing combinations. These features are not consistent with the promiscuous assembly of irrelevant chain combinations. Fourth, the H and L chain isotypes of the Fab fragments obtained from the combinatorial libraries resembled the isotypes expressed by serum- and MNC culture-derived PPS Abs of the donor. Fifth, previous studies of the human Ab repertoire to Hib PS have shown that combinatorial library-derived Fab fragments recapitulate V gene configurations and chain pairing of native antibodies (11, 26, 50). Thus, this collective evidence indicates that the PPS-specific Fabs studied here represent bona fide products of Ag-driven Ab responses.
We determined the complete primary structures of seven Fab fragments representing three PPS serotype specificities. A common pattern to emerge from this sequence analysis was the preferential usage of VHIII gene segments, a result in agreement with previous studies (1, 12, 19, 34, 57, 62, 71). Biased usage of VHIII genes appears to be a feature common not only to PPS Abs but to other human anti-PS Ab specificities, including Hib PS (3, 56, 59), Cryptococcus neoformans (49), and α-galactosyl (68). The mechanism underlying the preferential usage of VHIII gene segments is not understood. Structural constraints in formation of the combining site (29) and induction by VHIII-specific superantigens such as staphylococcal protein A (33, 60) have been proposed as explanations. However, the demonstration of preferential expression of VHIII genes in both productive and nonproductive rearrangements in peripheral blood B cells (16) suggests that this phenomenon does not involve selection at the level of the combining site but is determined by intrinsic molecular properties of VHIII genes (47).
The data presented here show that the same VH gene segment can be used by Fabs of different PPS specificity. The Fab pair 14-3–14-4 and Fab 23F-1 used HV 3-23, but their CDR-3s were distinctive, and they paired with different L chains. This promiscuity of HV 3-23 is perhaps not too surprising in light of the fact that this gene segment is commonly expressed in the peripheral repertoire (61), is used by a plethora of Abs having reactivities with either self or foreign Ags (47), and is a prominent member of the Hib PS repertoire, where it pairs with the KV 2D-29 V region to form the canonical combining site (37, 48). Table 3 summarizes our Fab results and the results of previous studies of V region gene segment usage by PPS-specific monoclonal Abs (MAbs). The recurrence of the same VH gene segments among different PPS specificities is apparent. For example, in addition to encoding PPS 14 and PPS 23F Abs, HV 3-23 is used by PPS 6B-specific Abs; HV 3-74 is used by Abs to either PPS 3 or 6B; HV 3-15 is used by PPS 6B, 8, and 14 Abs; and the HV 3-48 gene is used by Abs to PPS 9V, 18C, and 23F. The promiscuity of VH genes extends to other PS specificities. The HV 3-15 pairs, which pairs with the LV 1-51 region to form a PPS 14 combining site, also is found in association with another Vλ region, LV 7-43, to form a Hib PS combining site (3, 25). Similar patterns are seen with VL usage. VK 4-1 is associated with PPS 3 and PPS 6B Abs, VK 3-20 is associated with PPS 6B and PPS 14 Abs, and VK 2(D)-28 is associated with PPS 6B and PPS 9V Abs. The usage of the 2D-29 Vκ gene by the 23F-2 Fab is notable since as mentioned above this gene in conjunction with HV 3-23 is the most commonly expressed VL gene in the Hib PS repertoire. Furthermore, the LV 1-51 gene used by PPS 14-specific Fabs is also found in association with either HV 3-23 or HV 3-7 to form a combining site specific for the capsular PS of C. neoformans (49). From these data, it is apparent that chain pairing and CDR-3 diversity serve as critical determinants of PPS binding specificity. Thus, it is problematic to assign a germ line specificity to any particular V gene segment.
TABLE 3.
V gene segments encoding human anti-PPS Absa
| PPS specificity | VH | Vκ | Vλ |
|---|---|---|---|
| 3 | 3-74,b 3-73b | 4-1b | 7-46b |
| 4 | 1-46,h 3-7h | 2-30,h 3-20h | |
| 6B | 1-3,ch 1-e,h 3-7,cd 3-15,eh 3-23,c 3-30,h 3-66,c 3-74c | 1(D)-39,h 2(D)-28,h 2(D)-30,c 3-20,ch 4-1d | 1-51,h 2-8,c (2-11),c 2-14,e 10-54c |
| 8 | 3-15f | 1-27f | |
| 9V | 1-02,h 3-48h | 2(D)-28h | |
| 14 | 3-15,d 3-23d | 3-15,d 3-20g | 1-51d |
| 18C | 3-7,h 3-48h | 2-30,h 7-3h | |
| 23F | 3-23,d 3-11,d 3-48h | 2-24,d 2D-29d |
Compiled from published reports and this study. Succeeding footnotes refer to method of V gene identification, Ab source, and reference; nomenclature is according to references 10, 44, and 45.
cDNA sequence of hybridoma-derived MAb (57). GenBank sequences were reported by authors, but V gene assignments were made by us.
cDNA sequence of combinatorial library Fab (this report).
cDNA of hybridoma-derived MAb (62).
cDNA of EBV transformant-derived MAb (71).
Partial amino acid sequence of purified serum Ab (34).
cDNA sequence of hybridoma-derived MAb (12).
Our study was restricted to two adult donors; therefore, this limited scope does not permit estimates of V repertoire size for any single PPS specificity. However, our findings taken with previous studies (Table 3) suggest that at the population level, the number of V gene segments contributing to a particular PPS serotype specificity may be quite large. From the present analysis we identified LV 1-51 and KV 3-15 genes as contributors to the PPS-14 VL repertoire, and in a previous study we showed usage of the KV 3-20 gene (34). Thus, 3 different VL regions are used by 3 unrelated individuals to encode PPS-14 Abs. The PPS 6B repertoire is encoded by at least 10 VL gene segments and 8 VH gene segments (Table 3). This diversity contrasts to the human Ab repertoire to Hib PS, which although encoded by 3 to 4 VH genes and as many as 12 VL genes, is dominated by Abs using a single VH-VL canonical configuration (37, 48, 55). The capacity to generate potentially complex PPS Ab repertoires at the population level suggests that unlike the Hib PS repertoire, the occurrence of canonical combining sites may be infrequent within a particular PPS serotype specificity. Despite this diversity, however, there are likely to be structural constraints on the formation of PPS-specific combining sites as indicated by the recurrence of a VH CDR-3 motif among the PPS14-specific Fabs. Furthermore, VH gene segments belonging to the 1-3 canonical class are prominent among our Fabs and the panel of PPS-specific MAbs described by Baxendale et al. (12).
We found evidence for both interclonal and intraclonal mechanisms for generating PPS-specific combining site diversity within an individual. The two PPS 23F-specific Fabs isolated from subject 002 represent two independent clones that used different VH and VL combinations and whose affinities for PPS 23F differed fivefold. Since both of these Fabs had accumulated mutations, the attribution of higher affinity or fitness to one or the other germ line configuration cannot be made at present. Nonetheless, clones of independent origin coexist within an individual, and affinity differences between them could affect their ability to be selected by Ag, to be maintained in the memory pool, or to mediate protection.
It is interesting that one of the PPS 23F-specific Fabs had acquired an insertion in VH CDR-2. The insertion or deletion of residues into V gene segments during somatic maturation is becoming increasingly recognized as an important mechanism for generating combining site diversity (23, 43, 70). Changes in CDR length would generate a canonical loop structure entirely different from that of the original V gene (23) and could be expected to have a substantial impact on Ab affinity. The finding of an insertionally modified V region in this relatively small survey of PPS-specific V regions suggests that this mechanism may not be uncommon.
The potential for combining site functional diversity is present even when the products of a single clone dominate the expressed repertoire of an individual, as shown by the isolation of two PPS 14-specific Fab pairs from both subjects. The pair isolated from subject 018 (14-3 and 14-4) had differentially accumulated mutations, but these sequence polymorphisms did not confer measurable differences in affinity. However, the pair of clonally related Fabs isolated from subject 002 (14-1 and 14-2) varied in the extent of mutation in their VH regions sufficiently to confer a 13-fold difference in affinity. The more heavily mutated VH was associated with lower affinity for PPS 14, a result suggesting that hypermutation could lead to diminished anti-PPS Ab protective function. Parallel findings have been obtained in the murine response to the capsular PS of C. neoformans, where it has been shown that Abs originating from a single clone can differ in antigenic fine specificity and protective efficacy as a result of somatic mutation (40). Since the primary structures of Fabs 14-1 and 14-2 differed only in the VH region (13 positions: 4 in the CDRs and 9 in the frameworks), the disparities in affinity between these Fabs can be attributed to the VH region. The extent of the sequence differences precludes any precise assignment of the critical positions, but based upon our mutagenesis studies of Hib PS-specific Fabs showing that a single amino acid replacement can ablate binding (36), we might expect that only a small subset of the observed substitutions are responsible for the differences in PPS 14 binding affinity. VH CDR-3 is likely to play a critical role in PPS 14 binding as the homologous sequences Ser-Gly-Ser-Ser-Tyr and Thr-Gly-Thr-Thr-Phe were present in the CDR-3s of the two PPS 14-specific Fab pairs even though they used different canonical class VH gene segments and different D regions.
The affinity variation observed among the Fabs likely has consequence with respect to their potential to mediate protective immunity in vivo. Avidity functions as a determinant of anti-PPS Ab protective efficacy, as assessed in an in vitro model of opsonophagocytosis and in a mouse model of bacteremia (52, 66). Avidity variation is present among anti-PPS Ab populations elicited in adults by the polyvalent vaccine (52, 66) and in infants following vaccination with PPS-protein conjugates (5, 6). Irrespective of whether they are generated interclonally or intraclonally, the coexistence of affinity variants implies that Ab functional capability is not equivalent between responding clones and could in principle differ within a clone as expansion generates new variants with changed affinity.
While we cannot exclude the possibility that some of the observed deviations from the assigned candidate V gene segment could be generated by PCR artifacts or by unknown germ line polymorphisms, we assume that the majority of the observed sequence polymorphisms result from the process of somatic hypermutation. The mutated nature of all of the PPS-specific Fab fragments and their expression of non-IgM isotypes (IgG2 and IgA) lead us to conclude that these Fabs originated from memory B cells. Baxendale and colleagues reached the same conclusion in their recently published sequence analysis of human PPS-specific hybridomas generated from five adults (12). Their MAb panel, representing specificities to PPS serotypes 4, 6B, 9V, 18C, and 23F, showed a consistent pattern of mutation, and the majority were isotype switched.
Although PPSs as well as other purified PS Ags are thought to elicit minimal memory owing to their T-cell-independent nature, studies in mice have shown that some PS Ags have the capability to generate germinal centers (63, 67), regulatory T cells (8), and avidity maturation associated with hypermutation (14). Therefore, it is possible that immunization with PPS vaccine could directly induce class switch and hypermutation in primary B cells. However, we think it more likely that the B cells responding to PPS vaccination in the adult derive from a preexisting memory population. The presence of PPS-specific serum Abs to all three serotypes in the subjects prior to vaccination indicates their primed status. Furthermore, it is known that vaccination with PPS can activate memory B cells generated by prior vaccination with protein-conjugated PPS (15, 22, 42). The antigens responsible for natural priming could be the homologous pneumococci or other bacteria or food substances expressing antigenic determinants cross-reactive with PPS. Unlike purified PPS, these natural antigenic stimuli may occur on cell surfaces in a milieu of proteins or lipids, and this form of antigen might elicit T-cell-dependent activation and promote memory generation and somatic hypermutation. This process may apply generally to anti-PS repertoires in the adult, as somatically mutated V regions have been observed among a variety of human anti-PS Ab specificities, including Hib PS (3, 11, 26, 37), C. neoformans (49), α-galactosyl (68), and gangliosides (69).
Our findings together with results of previous studies indicate that Abs of a particular PPS serotype specificity can be encoded by a potentially large number of V genes, and conversely, a single V gene segment can encode combining sites of different specificity. Such degeneracy and promiscuity suggest that humoral immunity to encapsulated pneumococci has evolved not by investing a germ line specificity in a single V gene segment (21) but rather by distributing specificity (fitness) potential over a variety of genes. The high capacity for creating combining site diversity at the population level contrasts to what is seen in the individual where only one or a few V gene combinations dominate the expressed PPS repertoire. While V gene germ line content can vary among individuals (53, 54) and could limit diversity, we think it more likely, especially given the properties of V gene degeneracy and promiscuity, that repertoire restriction is determined primarily by somatic events. These events could include Ag-independent processes that control the pace of V gene assembly leading to establishment of the precursor B cell pool as well as Ag-driven processes involving memory generation, mutation, and clonal competition. For the PPS Ab repertoire in the adult, the outcome of these processes is an oligoclonal memory population that has matured under the influence of natural antigenic exposure and that has acquired a substantial mutational load. The impact this mutation exerts on the maintenance of memory and protective immunity remains to be elucidated.
ACKNOWLEDGMENTS
We thank Nancy Sweeters and Julie Simon for performing the vaccinations and phlebotomy, Charles Connolly and Adam O'Connor for technical assistance, and the volunteers for their participation in this study.
This work was supported by grants AI25008, AI45250, and RR01271 from the National Institutes of Health.
REFERENCES
- 1.Abadi J J, Friedman R, Jefferis R, Mageed R A, Pirofski L. Human antibodies elicited by a pneumococcal vaccine express idiotypic determinants indicative of VH3 gene segment usage. J Infect Dis. 1998;178:707–716. doi: 10.1086/515369. [DOI] [PubMed] [Google Scholar]
- 2.Adderson E E, Azmi F H, Wilson P M, Shackelford P G, Carroll W L. The human VH3b gene subfamily is highly polymorphic. J Immunol. 1993;151:800–809. [PubMed] [Google Scholar]
- 3.Adderson E E, Shackelford P G, Quinn A, Carroll W L. Restricted Ig H chain V gene usage in the human response to Haemophilus influenzae type b capsular polysaccharide. J Immunol. 1991;147:1667–1674. [PubMed] [Google Scholar]
- 4.AlonsoDeVelasco E, Verheul A F, Verhoef J, Snippe H. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev. 1995;59:591–603. doi: 10.1128/mr.59.4.591-603.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anttila M, Eskola J, Ähman H, Käyhty H. Avidity of IgG for Streptococcus pneumoniae type 6B and 23F polysaccharides in infants primed with pneumococcal conjugates and boosted with polysaccharide or conjugate vaccines. J Infect Dis. 1998;177:1614–1621. doi: 10.1086/515298. [DOI] [PubMed] [Google Scholar]
- 6.Anttila M, Eskola J, Käyhty H. Differences in the avidity of antibodies evoked by four different pneumococcal conjugate vaccines in early childhood. Vaccine. 1999;17:1970–1977. doi: 10.1016/s0264-410x(98)00458-7. [DOI] [PubMed] [Google Scholar]
- 7.Austrian R. Pneumococcal infections. In: Germanier R, editor. Bacterial vaccines. Orlando, Fla: Academic Press, Inc.; 1984. pp. 257–288. [Google Scholar]
- 8.Baker P J. T cell regulation of the antibody response to bacterial polysaccharide antigens: an examination of some general characteristics and their implications. J Infect Dis. 1992;165:S44–S48. doi: 10.1093/infdis/165-supplement_1-s44. [DOI] [PubMed] [Google Scholar]
- 9.Barbas C F, Kang A S, Lerner R A, Benkovic S J. Assembly of combinatorial libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbié V, Lefranc M-P. The human immunoglobulin kappa variable (IGKV) genes and joining (IGKJ) segments. Exp Clin Immunogenet. 1998;15:171–183. doi: 10.1159/000019068. [DOI] [PubMed] [Google Scholar]
- 11.Barington T, Hougs L, Juul L, Madsen H O, Ruder L P, Heilmann C, Svejgaard A. The progeny of a single virgin B cell predominates the human recall B cell response to the capsular polysaccharide of Haemophilus influenzae type b. J Immunol. 1996;157:4016–4027. [PubMed] [Google Scholar]
- 12.Baxendale H E, Davis Z, White H N, Spellerberg M B, Stevenson F K, Goldblatt D. Immunogenetic analysis of the immune response to pneumococcal polysaccharide. Eur J Immunol. 2000;30:1214–1223. doi: 10.1002/(SICI)1521-4141(200004)30:4<1214::AID-IMMU1214>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 13.Black S, Shinefield H, Pay P, Fireman B, Austrian R, et al. Efficacy of heptavalent conjugate pneumococcal vaccine (Wyeth Lederle) in 7000 infants and children: results of the Northern California Kaiser Permanente efficacy trial. Pediatr Res. 1999;45:157A. [Google Scholar]
- 14.Boswell C M, Stein K E. Avidity maturation, repertoire shift, and strain differences in antibodies to bacterial levan, a type 2 thymus-independent polysaccharide antigen. J Immunol. 1996;157:1996–2005. [PubMed] [Google Scholar]
- 15.Breukels M A, Rijkers G T, Voorhorst-Ogink M M, Zegers B M, Sanders L A M. Pneumococcal conjugate vaccine primes for polysaccharide-inducible IgG2 antibody responses in children with recurrent otitis media acuta. J Infect Dis. 1999;179:1152–1156. doi: 10.1086/314705. [DOI] [PubMed] [Google Scholar]
- 16.Brezinschek H P, Brezinschek R I, Lipsky P E. Analysis of heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 17.Bruyn G A, Zegers B J M, van Furth R. Mechanisms of host defense against infection with Streptococcus pneumoniae. Clin Infect Dis. 1992;14:251–262. doi: 10.1093/clinids/14.1.251. [DOI] [PubMed] [Google Scholar]
- 18.Butler J C, Breiman R F, Campbell J F, Lipman H B, Broome C V, Facklam R R. Pneumococcal polysaccharide vaccine efficacy: an evaluation of current recommendations. JAMA. 1993;270:1826–1831. [PubMed] [Google Scholar]
- 19.Chang Q, Abadi J, Alpert P, Pirofski L. A pneumococcal capsular polysaccharide vaccine induces a repertoire shift with increased VH3 expression in peripheral blood B cells from human immunodeficiency virus (HIV)-uninfected but not HIV-infected persons. J Infect Dis. 2000;181:1313–1321. doi: 10.1086/315405. [DOI] [PubMed] [Google Scholar]
- 20.Chothia C, Lesk A M, Gherardi E, Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. Structural repertoire of the human VH segments. J Mol Biol. 1992;227:799–817. doi: 10.1016/0022-2836(92)90224-8. [DOI] [PubMed] [Google Scholar]
- 21.Cohn M, Langman R E. The protection: the unit of humoral immunity selected by evolution. Immunol Rev. 1990;115:11–147. doi: 10.1111/j.1600-065x.1990.tb00783.x. [DOI] [PubMed] [Google Scholar]
- 22.Dagan R, Melamed R, Zamir O, Leroy O. Safety and immunogenicity of tetravalent pneumococcal vaccines containing 6B, 14, 19F and 23F polysaccharides coupled to either tetanus toxoid or diphtheria toxoid in young infants and their boosterability by native polysaccharide antigens. Pediatr Infect Dis J. 1997;16:1053–1059. doi: 10.1097/00006454-199711000-00010. [DOI] [PubMed] [Google Scholar]
- 23.De Wildt R M, van Venrooij W J, Winter G, Hoet R M A, Tomlinson I M. Somatic insertions and deletions shape the human antibody repertoire. J Mol Biol. 1999;294:701–710. doi: 10.1006/jmbi.1999.3289. [DOI] [PubMed] [Google Scholar]
- 24.Ellis R W, Granoff D M. Development and clinical uses of Haemophilus b conjugate vaccines. New York, N.Y: Marcel Dekker; 1994. [Google Scholar]
- 25.Granoff D M, Shackelford P G, Holmes S J, Lucas A H The Collaborative Study Group. Variable region expression in the antibody responses of infants vaccinated with Haemophilus influenzae type b polysaccharide-protein conjugates: description of a new λ light chain-associated idiotype and the relation between idiotype expression, avidity, and vaccine formulation. J Clin Investig. 1993;91:788–796. doi: 10.1172/JCI116298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hougs L, Juul L, Ditzel H J, Heilman C, Svejgaard A, Barington T. The first dose of a Haemophilus influenzae type b conjugate vaccine reactivates memory B cells: evidence for extensive clonal selection, intraclonal affinity maturation and multiple isotype switches to IgA2. J Immunol. 1999;162:224–237. [PubMed] [Google Scholar]
- 27.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of proteins of immunological interest, 5th ed. NIH publication no. 91-3242. Washington, D.C.: U.S. Department of Health and Human Services; 1991. [Google Scholar]
- 28.Käyhty H, Eskola J. New vaccines for the prevention of pneumococcal infections. Emerg Infect Dis. 1996;2:289–298. doi: 10.3201/eid0204.960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkham P M, Mortari F, Newton J A, Schroeder H W., Jr Immunoglobulin VH clan and family predicts variable domain structure and may influence antigen binding. EMBO J. 1992;11:603–609. doi: 10.1002/j.1460-2075.1992.tb05092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein D L, Ellis R W. Conjugate vaccines against Streptococcus pneumoniae. In: Levine M M, Woodrow G C, Kaper J C, Cobon G S, editors. New generation vaccines. 2nd ed. New York, N.Y: Marcel Dekker; 1997. pp. 503–525. [Google Scholar]
- 31.Konradsen H B, Hahn-Zoric M, Hanson L Å. Differences within mono- and dizygotic twin-pairs in spectrotypes and clones of IgG2 antibodies to pneumococcal polysaccharide type-1 and C-polysaccharide after vaccination. Scand J Immunol. 1994;40:423–428. doi: 10.1111/j.1365-3083.1994.tb03484.x. [DOI] [PubMed] [Google Scholar]
- 32.Lautner-Rieske A, Huber C, Meindl A, Pargent W, Schäble K F, Thiebe R, Zocher I, Zachau H G. The human immunoglobulin kappa locus. Characterization of the duplicated A regions. Eur J Immunol. 1992;22:1023–1029. doi: 10.1002/eji.1830220422. [DOI] [PubMed] [Google Scholar]
- 33.Lucas A H. Genetic basis and somatic evolution of a human polysaccharide-specific antibody repertoire. In: Zouali M, editor. Human B cell superantigens. R. G. Austin, Tex: Landes Co.; 1996. pp. 121–143. [Google Scholar]
- 34.Lucas A H, Granoff D M, Mandrell R E, Connolly C C, Shan A S, Powers D C. Oligoclonality of serum immunoglobulin G antibody responses to Streptococcus pneumoniae capsular polysaccharide serotypes 6B, 14, and 23F. Infect Immun. 1997;65:5103–5109. doi: 10.1128/iai.65.12.5103-5109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucas A H, Langley R J, Granoff D M, Nahm M H, Kitamura M Y, Scott M G. An idiotypic marker associated with a germ-line encoded κ light chain variable region that predominates the vaccine-induced human antibody response to Haemophilus influenzae b polysaccharide. J Clin Investig. 1991;88:1811–1818. doi: 10.1172/JCI115502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucas A H, Moulton K D, Reason D C. Role of κII-A2 L chain CDR-3 junctional residues in human antibody binding to the Haemophilus influenzae type b polysaccharide. J Immunol. 1998;161:3776–3780. [PubMed] [Google Scholar]
- 37.Lucas A H, Reason D C. Polysaccharide vaccines as probes of antibody repertoires in man. Immunol Rev. 1999;171:89–104. doi: 10.1111/j.1600-065x.1999.tb01343.x. [DOI] [PubMed] [Google Scholar]
- 38.Lue C, Tarkowski A, Mestecky J. Systemic immunization with pneumococcal vaccine induces a predominant IgA2 response of peripheral blood lymphocytes and increases of both serum and secretory anti-pneumococcal antibodies. J Immunol. 1988;140:3793–3800. [PubMed] [Google Scholar]
- 39.Matsuda F, Ishii K, Bourvagnet P, Kuma K, Hayashida H, Miyata T, Honjo T. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J Exp Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee J, Nussbaum G, Scharff M D, Casadevall A. Protective and nonprotective monoclonal antibodies to Cryptococcus neoformans originating from one B cell. J Exp Med. 1995;181:405–409. doi: 10.1084/jem.181.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nahm M H, Olander J V, Magyarlaki M. Identification of cross-reactive antibodies with low opsonophagocytic activity for Streptococcus pneumoniae. J Infect Dis. 1997;176:698–703. doi: 10.1086/514093. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien K L, Steinhoff M C, Edwards K, Keyserling H, Thoms M L, Madore D. Immunologic priming of young children by pneumococcal glycoprotein conjugate, but not polysaccharide vaccines. Pediatr Infect Dis J. 1996;15:425–430. doi: 10.1097/00006454-199605000-00009. [DOI] [PubMed] [Google Scholar]
- 43.Ohlin M, Borrebaeck C A K. Insertions and deletions in hypervariable loops of antibody heavy chains contribute to molecular diversity. Mol Immunol. 1998;35:233–238. doi: 10.1016/s0161-5890(98)00030-3. [DOI] [PubMed] [Google Scholar]
- 44.Pallarès N, Frippiat J-P, Giudicelli V, Lefranc M P. The human immunoglobulin lambda variable (IGLV) genes and joining (IGLJ) segments. Exp Clin Immunogenet. 1998;15:8–18. doi: 10.1159/000019054. [DOI] [PubMed] [Google Scholar]
- 45.Pallarès N, Lefebvre S, Content V, Matsuda F, Lefranc M-P. The human immunoglobulin heavy variable genes. Exp Clin Immunogenet. 1999;16:36–60. doi: 10.1159/000019095. [DOI] [PubMed] [Google Scholar]
- 46.Park M K, Sun Y, Olander J V, Hoffmann J W, Nahm M H. The repertoire of human antibodies to the carbohydrate capsule of Streptococcus pneumoniae 6B. J Infect Dis. 1996;174:75–82. doi: 10.1093/infdis/174.1.75. [DOI] [PubMed] [Google Scholar]
- 47.Pascual V, Capra D J. Human immunoglobulin heavy-chain variable region genes: organization, polymorphism, and expression. Adv Immunol. 1991;49:1–74. doi: 10.1016/s0065-2776(08)60774-9. [DOI] [PubMed] [Google Scholar]
- 48.Pinchuk G V, Nottenburg C, Milner E C B. Predominant V-region gene configurations in the human antibody response to Haemophilus influenzae capsule polysaccharide. Scand J Immunol. 1995;41:324–330. doi: 10.1111/j.1365-3083.1995.tb03574.x. [DOI] [PubMed] [Google Scholar]
- 49.Pirofski L, Lui R, DeShaw M, Kressel A B, Zhong Z. Analysis of human monoclonal antibodies elicited by vaccination with a Cryptococcus neoformans glucuronooxylomannan capsular polysaccharide vaccine. Infect Immun. 1995;63:3005–3014. doi: 10.1128/iai.63.8.3005-3014.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reason D C, Wagner T C, Lucas A H. Human Fab fragments specific for the Haemophilus influenzae b polysaccharide isolated from a bacteriophage combinatorial library use variable region gene combinations and express idiotype that mirror in vivo expression. Infect Immun. 1997;65:261–266. doi: 10.1128/iai.65.1.261-266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robbins J B, Austrian R, Lee C-J, Rastogi S C, Schiffman G, Henrichsen J, Mäkela P H, Broome C V, Facklam R R, Tiesjema R H, Parke J C. Considerations for formulating the second-generation pneumococcal capsular polysaccharide vaccine with emphasis on the cross-reactive types within groups. J Infect Dis. 1983;148:1136–1159. doi: 10.1093/infdis/148.6.1136. [DOI] [PubMed] [Google Scholar]
- 52.Romero-Steiner S, Musher D M, Cetron M S, Pais L B, Groover J E, Fiore A E, Plikaytis B D, Carlone G M. Reduction in functional antibody activity against Streptococcus pneumoniae in vaccinated elderly individuals highly correlates with decreased IgG antibody avidity. Clin Infect Dis. 1999;29:281–288. doi: 10.1086/520200. [DOI] [PubMed] [Google Scholar]
- 53.Sasso E H, Van Dijk K W, Milner E C B. Prevalence and polymorphism of human VH3 genes. J Immunol. 1990;145:2751–2757. [PubMed] [Google Scholar]
- 54.Schaible G, Rappold G A, Pargent W, Zachau H G. The human immunoglobulin kappa locus: polymorphism and haplotypes of Caucasoid and non-Caucasoid individuals. Hum Genet. 1993;91:261–267. doi: 10.1007/BF00218268. [DOI] [PubMed] [Google Scholar]
- 55.Scott M G, Crimmins D L, McCourt D W, Zocher I, Thiebe R, Zachau H G, Nahm M H. Clonal characterization of the human IgG antibody repertoire to Haemophilus influenzae type b polysaccharide. III. A single VκII gene and one of several J genes are joined by an invariant arginine to form the most common L chain V region. J Immunol. 1989;143:4110–4116. [PubMed] [Google Scholar]
- 56.Scott M G, Tarrand J F, Crimmins D L, McCourt D W, Siegel N R, Smith C E, Nahm M H. Clonal characterization of the human antibody repertoire to Haemophilus influenzae b polysaccharide: IgG antibodies contain VH genes from a single VH family and VL genes from at least 4 VL families. J Immunol. 1989;143:293–298. [PubMed] [Google Scholar]
- 57.Shaw D R, Kirkham P, Schroeder H W, Jr, Roben P, Silverman G J. Structure-function studies of human monoclonal antibodies to pneumococcus type 3 polysaccharide. Ann N Y Acad Sci. 1995;764:370–373. doi: 10.1111/j.1749-6632.1995.tb55849.x. [DOI] [PubMed] [Google Scholar]
- 58.Siber G R. Pneumococcal disease: prospects for a new generation of vaccines. Science. 1994;265:1385–1387. doi: 10.1126/science.8073278. [DOI] [PubMed] [Google Scholar]
- 59.Silverman G J, Lucas A H. Variable region diversity in circulating human antibodies specific for the capsular polysaccharide of Haemophilus influenzae type b. J Clin Investig. 1991;88:911–920. doi: 10.1172/JCI115394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silverman G J, Nayak J V, Warnatz K, Hajjar F F, Cary S, Tighe H, Curtiss V E. The dual phases of the response to neonatal exposure to a VH family-restricted staphylococcal B cell superantigen. J Immunol. 1998;161:5720–5732. [PubMed] [Google Scholar]
- 61.Stewart A K, Huang C, Stollar B D, Schwartz R S. High-frequency representation of a single VH gene in the expressed human B cell repertoire. J Exp Med. 1993;177:409–418. doi: 10.1084/jem.177.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun Y, Park M K, Kim J, Diamond B, Solomon A, Nahm M H. Repertoire of human antibodies against the polysaccharide capsule of Streptococcus pneumoniae serotype 6B. Infect Immun. 1999;67:1172–1179. doi: 10.1128/iai.67.3.1172-1179.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sverremark E, Fernandez C. Role of T cells and germinal center formation in the generation of immune responses to the thymus-independent carbohydrate dextran B512. J Immunol. 1998;161:4646–4651. [PubMed] [Google Scholar]
- 64.Tomlinson I M, Cox J P L, Gherardi E, Lesk A M, Chothia C. The structural repertoire of the human Vκ domain. EMBO J. 1995;14:4628–4638. doi: 10.1002/j.1460-2075.1995.tb00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomlinson I M, Walter G, Marks J D, Llewelyn M B, Winter G. The repertoire of human germline VH sequences reveals about fifty groups of VH segments with different hypervariable loops. J Mol Biol. 1992;227:776–798. doi: 10.1016/0022-2836(92)90223-7. [DOI] [PubMed] [Google Scholar]
- 66.Usinger W R, Lucas A H. Avidity as a determinant of the protective efficacy of human antibodies to pneumococcal capsular polysaccharides. Infect Immun. 1999;67:2366–2370. doi: 10.1128/iai.67.5.2366-2370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang D, Wells S M, Stall A M, Kabat E A. Reaction of germinal centers in the T-cell-independent response to the bacterial polysaccharide α(1-6)dextran. Proc Natl Acad Sci USA. 1994;91:2502–2506. doi: 10.1073/pnas.91.7.2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Radic M Z, Galili U. Human anti-gal heavy chains: preferential use of VHIII and the presence of somatic mutations. J Immunol. 1995;155:1276–1285. [PubMed] [Google Scholar]
- 69.Weng N-P, Yu-Lee L-Y, Sanz I, Patten B M, Marcus D M. Structure and specificity of anti-ganglioside autoantibodies associated with motor neuropathies. J Immunol. 1992;149:2518–2529. [PubMed] [Google Scholar]
- 70.Wilson P C, de Boutellier O, Liu Y-J, Potter K, Banchereau J, Capra J D, Pascual V. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J Exp Med. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong Z, Burns T, Chang Q, Carroll M, Pirofski L. Molecular and functional characteristics of a protective human monoclonal antibody to serotype 8 Streptococcus pneumoniae capsular polysaccharide. Infect Immun. 1999;67:4119–4127. doi: 10.1128/iai.67.8.4119-4127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]