Abstract
Tenacibaculum maritimum is a devastating bacterial pathogen affecting a large variety of marine fish species. It is responsible for significant economic losses in aquaculture farms worldwide. Different typing methods have been proposed to analyse bacterial diversity and population structure. Serological heterogeneity has been observed and up to four different serotypes have been described so far. However, the underlying molecular factors remain unknown. By combining conventional serotyping and genome‐wide association study, we identified the genomic loci likely involved in the O‐antigen biosynthesis. This finding allowed the development of a robust multiplex PCR‐based serotyping scheme able to detect subgroups within each serotype and therefore performs better than conventional serotyping. This scheme was successfully applied to a large number of isolates from worldwide origin and retrieved from a large variety of fish species. No obvious correlations were observed between the mPCR‐based serotype and the host species or the geographic origin of the isolates. Strikingly, the distribution of mPCR‐based serotypes does not follow the core genome phylogeny. Nevertheless, this simple and cost‐effective mPCR‐based serotyping method could be useful for different applications such as population structure analysis, disease surveillance, vaccine formulation and efficacy follow‐up.
Keywords: aquaculture, fish diseases, molecular serotyping, o‐antigen gene cluster, tenacibaculosis, Tenacibaculum maritimum
1. INTRODUCTION
The global fish production is estimated to have reached about 179 million tons in 2018. Aquaculture accounted for 52% of fish for human consumption and is currently the fastest growing food‐producing sector, playing an increasingly critical role in global food security (FAO, 2020). However, the fast development of aquaculture has been associated with a dramatic increase in infectious disease outbreaks, which can be catastrophic for the industry, causing large economic losses and impacting the environment and animal welfare (Lafferty et al., 2015; Stentiford et al., 2017).
Tenacibaculum species are Gram negative, filamentous and exclusively marine bacteria belonging to the family Flavobacteriaceae (phylum Bacteroidetes) (Bernardet, 2011). This genus encompasses several species pathogenic for marine fish, responsible for a condition referred to as tenacibaculosis (also known as marine flexibacteriosis) (see Avendaño‐Herrera et al. [2006], Fernández‐Álvarez & Santos [2018], and Nowlan et al. [2020] for recent reviews). Tenacibaculum maritimum (formerly Flexibacter maritimus) (Wakabayashi et al., 1986) was the first species to be described and constitutes the best‐known pathogen within the genus (Avendaño‐Herrera et al., 2006). It has originally been isolated in 1977 in Japan during mortality events on marine red seabream (Pagrus major) and black sea bream (Acanthopagrus schlegelii) (Masumura & Wakabayashi, 1977; Wakabayashi et al., 1984). Since then, it has been retrieved from a large variety of marine fish species worldwide including: Atlantic salmon (Salmo salar) in Australia (van Gelderen, 2007) and in Chile (Apablaza et al., 2017), rainbow trout (Oncorhynchus mykiss) in Australia (van Gelderen, 2007) and in Chile (Valdes et al., 2021), Chinook salmon (Oncorhynchus tshawytscha) in Canada (Ostland et al., 1999), turbot (Scophthalmus maximus), sole (Solea solea) and blackspot seabream (Pagellus bogaraveo) in Spain (Castro et al., 2007), batfish (Platax orbicularis) in French Polynesia (Lopez et al., 2022) and sea bass (Dicentrarchus labrax) and seabream (Sparus aurata) in Europe (Muniesa et al., 2020). Fish often display gross external lesions including eroded mouth, skin ulcers or necrosis, frayed fins and tail rot (Avendaño‐Herrera et al., 2006; Nowlan et al., 2020). So far, a unique vaccine1 is commercially available for a single fish species (turbot) and a single serotype (i.e. O2). Hence, in order to cope with the disease, the treatment broadly relies on disinfectants (e.g. formol and hydrogen peroxide) and a variety of antibiotics (e.g. amoxycillin, trimethoprim, oxytetracycline, enrofloxacin, flumequine, furazolidone and florfenicol) (Avendaño‐Herrera et al., 2005).
To improve the control of bacterial infections in fish farms, data at both the epidemiological and molecular levels are urgently needed (Bayliss et al., 2017). Indeed, different typing methods for studying the relationships between T. maritimum isolates have been proposed allowing to better understand the population structure, the spreading and the epidemiology of this pathogen (see Bridel et al. [2020] and reference therein). Among those, serotyping methods have been assessed and up to four serotypes have been described so far (Avendaño‐Herrera et al., 2004; Avendaño‐Herrera et al., 2005; Fernández‐Álvarez et al., 2018; Piñeiro‐Vidal et al., 2007). This serological diversity might have important consequences for the selection of appropriate candidate strain(s) for vaccine development, selective breeding for increased disease resistance, follow‐up studies, epidemiological surveillance, disease control as well as for a better understanding of virulence and host resistance traits. However, conventional serotyping methods suffer from several severe disadvantages such as the choice of the appropriate scheme, the need to raise antisera from animals and the bias of human interpretation (Gorski, 2021; Sloan et al., 2017). Therefore, molecular tools such as microarray or multiplex PCR (mPCR) serotyping assays have been developed for a number of bacteria (Howell et al., 2015; Ludwig et al., 2020; Nakaue et al., 2021), including fish pathogens (Rochat et al., 2017; Torres‐Corral & Santos, 2020).
Conventional serotyping is based on the immunogenicity of various surface‐exposed bacterial structures (e.g. lipopolysaccharide [LPS], capsular polysaccharide, flagella). LPS is one of the most pro‐inflammatory compounds of Gram‐negative bacteria, representing 75% of the total bacterial surface. Essential for the structure and function of the external membrane, LPS is considered as a determining factor during host–bacteria interactions (Tan & Kagan, 2014). This amphipathic glycoconjugate is composed in three different domains: the lipid A anchored in the outer membrane, the core oligosaccharide which connects the lipid A to the O‐antigen and the O‐antigen itself which is the hydrophilic and immunodominant portion, facing the external environment. The latter is composed of repeated carbohydrate units and its chemical composition and structure exhibit very high levels of variation, even within a single species (Mostowy & Holt, 2018). Genes coding for the biosynthesis of the O‐antigen are often located in a single genomic cluster (the O‐AGC), and fall into three classes: (i) the nucleotide sugar biosynthesis genes, (ii) the sugar transferase genes and (iii) those required for O‐units translocation, chain synthesis and chain length determination (see Bohl & Aihara [2018] and Whitfield et al. [2020] for recent reviews). The O‐antigen of T. maritimum is composed of a disaccharide repeated unit that has been resolved for a unique strain; nothing is known about the mechanisms and genes involved (Vinogradov et al., 2003).
In this study, we took advantage of a set of recently published T. maritimum genomes (Bridel et al., 2020; Pérez‐Pascual et al., 2017) to identify the O‐AGC in this species. We used conventional serotyping together with a genome‐wide association study to develop an mPCR‐based serotyping scheme (mPCR). Our study aims to provide an efficient, cost‐effective and reproducible molecular tool able to discriminate distinct gene combinations. This mPCR serotyping scheme was successfully applied to a collection of 124 T. maritimum isolates.
2. MATERIALS AND METHODS
2.1. Genome comparison
The complete or draft genome sequences (whole‐genome shotgun [WGS]) used in this study are listed in Table 1. Genome comparisons were performed using the MicroScope platform v3.15.3 (Vallenet et al., 2020), which allows graphic visualization enhanced by a synchronized representation of synteny groups. Analysis of gene organization was conducted using syntons (i.e. maximal set of orthologous gene pairs displaying a conserved organization) constructed with MicroScope default parameters (i.e. orthologous gene sets were computed using BLAST‐P Bidirectional Best Hit or displaying at least 30% identity on 80% of the shortest sequence and a genomic co‐localization with allowed gap set to five genes). In order to analyse the gene content of the T. maritimum type strain's O‐AGC, similarity searches were performed with each encoding proteins: (i) using BLAST‐P against the Swiss‐Prot database (release 2022_01) and (ii) using HHpred with default parameters in the PDB_mmCIF70 database (release 12 October 2021). HHpred is a sensitive protein homology detection, function and structure prediction based on HMM–HMM comparison (Zimmermann et al., 2018).
TABLE 1.
Tenacibaculum maritimum isolates used in this study
| Strain | Serotype | Type | Geographic origin | Host | Year | Genome c |
|---|---|---|---|---|---|---|
| NCIMB 2154T | O1, O2, O1/O2 a | 1‐0 | Japan | Pagrus major | 1977 | GCF_900119795.1 |
| NBRC 15946 b | O1 | 1‐0 | Japan | ND | ND | GCF_000509405.1 |
| P2‐27 | O1 | 1‐0 | Spain | Scophthalmus maximus | 2011 | GCF_902705465.1 |
| Aq 16–85 | O1 | 1‐0 | French Polynesia | Platax orbicularis | 2016 | GCF_902705305.1 |
| FC | O1 | 1‐0 | Chile | Scophthalmus maximus | 1998 | GCF_902705415.1 |
| 190605A03b | ND | 1‐0 | French Polynesia | Platax orbicularis | 2019 | pubmlst.org (id=151) |
| Tasmania_1513 | ND | 1‐0 | Australia | Latris lineata | 2003 | pubmlst.org (id=131) |
| FS08(1) | O1 | 1‐1 | Italy | Sparus aurata | 2006 | GCF_902705395.1 |
| NAC SLCC MFF | O1 | 1‐1 | Malta | Dicentrarchus labrax | ND | GCF_902705345.1 |
| P2‐48 | O1 | 1‐1 | France | Solea senegalensis | 2010 | GCF_902705555.1 |
| CVI1001048 | O1 | 2‐0 | The Netherlands | Solea solea | 2010 | GCF_902705265.1 |
| NCIMB 2158 | O2 | 2‐1 | Scotland | Solea solea | 1981 | GCF_902705425.1 |
| UCD SB2 | O2 | 2‐1 | USA | Atractoscion nobilis | 1995 | GCF_902705445.1 |
| JIP10/97 | O2 | 2‐1 | France | Scophthalmus maximus | 1997 | GCF_902705285.1 |
| JIP46/00 | O2 | 2‐1 | France | Scophthalmus maximus | 2000 | GCF_902705435.1 |
| P1‐39 | O2 | 2‐1 | France | Dicentrarchus labrax | 2010 | GCF_902705535.1 |
| 190115A05h | ND | 2‐1 | French Polynesia | Platax orbicularis | 2019 | pubmlst.org (id=150) |
| Tasmania_0663 | ND | 2‐1 | Australia | Salmo salar | 1996 | pubmlst.org (id=117) |
| Tasmania_0809 | ND | 2‐1 | Australia | Salmo salar | 2000 | pubmlst.org (id=127) |
| Tasmania_0811 | ND | 2‐1 | Australia | Salmo salar | 2000 | pubmlst.org (id=129) |
| Tasmania_2854 | ND | 2‐1 | Australia | Salmo salar | 2009 | pubmlst.org (id=132) |
| TM‐KORJJ | ND | 2‐1 | South Korea | Paralichthys olivaceus | 2016 | GCF_004803875.1 |
| 190628A06a | ND | 3‐0 | French Polynesia | Platax orbicularis | 2019 | pubmlst.org (id=153) |
| Baxa GBF‐8601 | ND | 3‐0 | Japan | Paralichthys olivaceus | 1986 | pubmlst.org (id=46) |
| USC RPM522.1 | ND | 3‐0 | Spain | Scophthalmus maximus | 1992 | pubmlst.org (id=61) |
| TFA4 | O3 | 3‐1 | French Polynesia | Platax orbicularis | 2013 | GCF_902705565.1 |
| 902 | O3 | 3‐1 | France | Dicentrarchus labrax | 2013 | GCF_902705365.1 |
| JIP32/91‐4 | O3 | 3‐1 | France | Dicentrarchus labrax | 1991 | GCF_902705385.1 |
| P4‐45 | O3 | 3‐1 | France | Dicentrarchus labrax | 2010 | GCF_902705495.1 |
| 190709D08d | ND | 3‐1 | French Polynesia | Platax orbicularis | 2019 | pubmlst.org (id=152) |
| Tasmania_3064 | ND | 3‐1 | Australia | Salmo salar | 2013 | pubmlst.org (id=120) |
| Tasmania_4635 | ND | 3‐1 | Australia | Salmo salar | 2018 | pubmlst.org (id=137) |
| DPIF 89/0239‐1 | O3 | 3‐2 | Australia | Salmo salar | 1989 | GCF_902705355.1 |
| DPIF 89/3001‐6.2 | O3 | 3‐2 | Australia | Latris lineata | 1989 | GCF_902705315.1 |
| Tasmania_0759 | ND | 3‐2 | Australia | Salmo salar | 1998 | pubmlst.org (id=126) |
| USC SE30.1 | O4 | 4‐0 | Spain | Oncorhynchus kisutch | 1993 | GCF_902705525.1 |
| USC SP9.1 | O4 | 4‐0 | Spain | Salmo salar | 1993 | GCF_902705515.1 |
| Tasmania_0814 | ND | 4‐0 | Australia | Oncorhynchus mykiss | 1991 | pubmlst.org (id=130) |
| Tasmania_4574 | ND | 4‐0 | Australia | Salmo salar | 2017 | pubmlst.org (id=135) |
| Tasmania_4579 | ND | 4‐0 | Australia | Salmo salar | 2017 | pubmlst.org (id=136) |
Note: The representative strains are indicated in bold.
Abbreviation: ND, not determined.
Different serotypes have been reported for the type strain NCIMB 2154T.
Strain NBRC 15946 is not the type strain according to Pérez‐Pascual et al. (2017).
NCBI genome accession number or genome available in PubMLST (https://pubmlst.org/bigsdb?db=pubmlst_tenacibaculum_isolates&page=query). Isolate id is given in parentheses.
For phylogenetic reconstruction, comparison of the gene content between strains was done by pairwise proteome similarity search using BLAST‐P Bidirectional Best Hit and the MicroScope default parameters (i.e. >80% protein identity, >80% coverage). A set of 2034 groups of core genome proteins was retained and multiple alignments on individual groups were performed using MUSCLE using an inhouse R script. The resulting alignments were concatenated using an inhouse script and tree phylogenetic reconstruction was inferred by approximately‐maximum‐likelihood using Fastree v2.1.10 with default parameters (Price et al., 2010). The tree was visualized using FigTree v1.1.4 and rooted using midpoint.
2.2. Bacterial isolates
The T. maritimum isolates used in this study are listed in Table 1 for those with complete or draft genome sequences and in Table S2 for those typed using mPCR. Additional information including country, year, host, MLST type (Habib et al., 2014) or MALDI‐TOF profile (Bridel et al., 2020) was included when available. Bacterial isolates were grown on Marine Agar 2216 (Difco) at 27°C for 48 h. All isolates were confirmed as belonging to the species T. maritimum by molecular techniques (PCR and/or MALDI‐TOF MS) and/or biochemistry depending on the way of identification.
2.3. Conventional serotyping
The serological characterization of the T. maritimum isolates was performed using both slide agglutination and dot blot assay with unabsorbed and absorbed antisera as described previously (Avendaño‐Herrera et al., 2005; Castro et al., 2007). Rabbit antisera raised against representative strains of the four serotypes described for this pathogen were used. Antisera raised against the sole (Solea senegalensis) isolates PC503.1 (O1) and ACC13.1(O3), the turbot (S. maximus) isolate PC424.1(O2) and the salmon (S. salar) isolate SNW20.2 (O4) were used. Only a reaction similar to that exhibited by the homologous control strain was scored as positive.
2.4. multiplex PCR
Primers (Table 2) were designed using the Benchling software2 according to the complete and draft genome sequences available and conservation of their sequences was verified. For mPCR optimization, reactions were performed using bacterial gDNA extracted using the genomic DNA purification kit (Macherey Nagel) according to the manufacturer's instructions. For this purpose, the type strain NCIMB 2154T and different representative strains (i.e. strains FS080(1), CVI100104801, NCIMB 2158, 190628A06a, TFA4 and DPIF 89/0239‐1) were used. mPCR reactions contained 0.25 μl DreamTaq DNA polymerase (Thermo Fisher Scientific), 5 μl of 10× DreamTaq buffer (Thermo fisher scientific), 5 μl of MgCl2 (at 25 mM), 1 μl dNTPs at 10 mM, 1.5 μl of each primer at 10 μM and 1 μl (about 100 ng) of gDNA template in a 50 μl final reaction volume according to the manufacturer's recommendations. The mPCR amplification mix was heated at 95°C for 2 min, followed by 30 cycles of 95°C for 30 s, 53°C for 30 s, 72°C for 90 s and a final extension at 72°C for 5 min. The amplified products were electrophoresed in 2.0% agarose gels run in 1× TBE buffer with the GeneRuler™ DNA ladder mix (Thermo Fisher Scientific) as a molecular size marker. For typing the large collection of isolates, cells from a bacterial colony (or a few depending on the size) were resuspended in 20 μl sterile water, vortexed and heated at 100°C for at least 5 min for full lysis, then briefly centrifuged to pellet the bacterial debris; 1 μl of the supernatant was used as DNA template for the mPCR reaction.
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′ → 3′) | Amplicon size in bp | Target gene (locus tag) in representative strains |
|---|---|---|---|
| P1F | TAGCCAGTATCTATTATTTGTTTACTG | 196 (Types 1 and 2) | MARIT_2530 |
| P1R | AGTTAGCGTGCCATTTATCA | ||
| P2/3F | TTTAGAATCTAGAGATATAAGGATTCC | 357 (Type 2) or 717 (Type 3) | CVI1001048_20215 or TM190628A06A_50037 |
| P2/3R | ATCAATTTTTGAAACATAATCTTCAT | ||
| P4F | GAGGCATTTCACATTCTGTAT | 985 (Type 4) | USCSE301_230029 |
| P4R | CCATGAGTCAATAATGTTGAGA | ||
| P5F | AACCTGAATATTGGTATTGGAG | 872 (Types 1‐1, 3‐1 and 3‐2) | FS0810_80069 |
| P5R | AATAGACATCAGAATCACTATAAGAA | ||
| P6F | GAGAGAAAATTACGCCTAAAAC | 476 (Types 2‐1 and 3‐2) | NCIMB2158_120032 |
| P6R | CAAAAATCTCCCCAACTTTAAC |
3. RESULTS
3.1. Conventional serotyping assay
Conventional serotyping was performed using both slide agglutination and dot blot assay on 22 T. maritimum strains, whose genomes were previously sequenced (Bridel et al., 2020). These strains were retrieved from various geographic locations and host fish species. They display a significant diversity as they encompass the four serotypes previously described (Table 1). Indeed, strains were originally selected to maximize the genomic diversity and therefore expected to display such a serotypic heterogeneity. Each strain reacted strongly with a unique serum, except strain NCIMB 2154T that was successively reported to belong to the intermediate minor serotype O1/O2 by Avendaño‐Herrera et al. (2004), to serotype O2 by Avendaño‐Herrera et al. (2005) and to serotype O1 by Fernández‐Álvarez et al. (2018). This is likely the consequence of typical drawbacks arising from conventional serotyping protocols. Nevertheless, the resulting data were used as a cornerstone for comparing serotypes with strains’ genomic organization.
3.2. Identification of the O‐antigen biosynthesis gene cluster of the T. maritimum type strain
Polysaccharide antigens, including the O‐antigen, are usually synthetized by a specialized group of enzymes which are encoded by genes located in an O‐AGC (Mostowy & Holt, 2018). Comparative genomics, including analysis of gene synteny, was performed between T. maritimum (strain NCIMB 2154T) and Flavobacterium psychrophilum (strain JIP02/86), a phylogenetically related species whose O‐AGC has been well characterized (Cisar et al., 2019; Rochat et al., 2017). Comparisons with Tenacibaculum dicentrarchi (strain 35/09T), whose genome is available and O‐antigen biosynthetic genes were recently described (Saldarriaga‐Córdoba et al., 2021), were also performed. This analysis allowed to identify a gene cluster very likely involved in the O‐antigen biosynthesis. This locus contains eight and 10 genes out of 16 that are conserved between T. maritimum NCIMB 2154T O‐AGC and T. dicentrarchi USC 35/09T (32%–78% protein identity) and F. psychrophilum JIP02/86 (26%–71% protein identity) O‐AGCs, respectively (Figure S1). Most of these orthologous genes are predicted to be involved in the nucleotide sugar precursors biosynthesis. Tenacibaculum maritimum NCIMB 2154T O‐AGC is located between the core genome genes folC and MARIT_2521 (encoding a conserved exported protein), the latter being immediately adjacent to gyrB. This locus is about 20 kb long and displays a 30.80% GC, slightly under the median guanine‐cytosine (GC) content (32.01%) of the complete genome. All genes of the cluster are transcribed in the same direction, some likely polycistronic.
3.3. Gene content of the O‐antigen biosynthesis gene cluster of the T. maritimum type strain's genome
Starting immediately downstream folC and two adjacent Val‐tRNA genes (and orientated 5′ to 3′), MARIT_2537, MARIT_2536 and MARIT_2535 display significant sequence similarities (35%–60% protein identity; >95% coverage) with genes wbpA, wbpB and udg of Pseudomonas aeruginosa, respectively (Figure S1 and additional details in Table S1). Their products are predicted to be involved in the biosynthesis of a nucleotide sugar precursor, probably an uridine diphosphate (UDP)‐glucose derivative. MARIT_2534 encodes a predicted periplasmic protein with poor and partial sequence similarities (below 25% identity) with proteins involved in exopolysaccharide export such as KpsD of Escherichia coli, Wza of Salmonella typhimurium and AmsH of Erwinia amylovora. MARIT_2533 encodes a predicted transmembrane protein with poor similarity (18.9% protein identity) with WzzE, the enterobacterial common antigen polysaccharide chain length modulation protein of E. coli (Rai & Mitchell, 2020). MARIT_2532 encodes a protein similar (56.5% identity, 98% coverage) to PseB of Campylobacter jejuni that possess both C6 dehydratase and C5 epimerase activities on UDP‐N‐acetyl‐α‐D‐glucosamine (Schoenhofen et al., 2006). MARIT_2531, MARIT_2530 and MARIT_2529 encode multi‐pass transmembrane proteins with very poor similarities with proteins having any experimental evidence. MARIT_2528 and MARIT_2527 encode proteins with poor similarities with predicted epimerases/dehydratases and acetyltransferases, respectively, but possibly involved in nucleotide sugar modifications. The MARIT_2526 gene product is highly similar (59.5% protein identity; 98% coverage) to PglC of C. jejuni and therefore likely encodes the undecaprenyl phosphate transferase that mediates the first step in the biosynthesis of the undecaprenyl‐linked nucleotide sugar, thus creating the first membrane‐associated intermediate for the O‐antigen unit synthesis (Glover et al., 2006). The MARIT_2525 gene product displays significant similarities (>50% identity) with different pyridoxal phosphate (PLP)‐dependent aminotransferases including EpsN of Bacillus subtilis and PglE of C. jejuni that modify amino sugars using L‐glutamate as an amino donor (Schoenhofen et al., 2006). The lysine, located at the amino acid position 200 in MARIT_2525 and required for covalent bond formation between the ε‐amino group of lysine and the aldehyde group of PLP in PLP‐containing proteins (Kaundinya et al., 2018), is well conserved. Finally, the MARIT_2524, MARIT_2523 and MARIT_2522 genes encode proteins highly similar to RmlA, RmlB and RmlC that are predicted to be involved in the biosynthesis of dTDP‐L‐rhamnose, a nucleotide sugar precursor of the O‐antigen. One might conclude that most if not all genes encompassed in this locus encode for proteins whose function are related to nucleotide sugar biosynthesis, transfer, chain synthesis and export and thus are very likely involved in O‐antigen biosynthesis of T. maritimum.
3.4. Comparison of the genomic organization of the O‐antigen biosynthesis gene cluster between T. maritimum strains
Extensive comparative genomic analysis of the O‐AGCs between T. maritimum strains revealed a globally well‐conserved structural backbone, located in the same chromosomal region between the core genome genes folC and gyrB. The loci encompass some genes (i.e. wbpA, wbpB, udg, epsN, rmlA, rmlB and rmlC) that encode proteins involved in nucleotide sugar metabolism and globally well‐conserved between strains. However, O‐AGCs display significant and striking differences among T. maritimum strains (Figure 1). By grouping the strains harbouring identical or nearly identical O‐AGC gene organization, different O‐AGCs, hereafter designated ‘Types’, were identified.
FIGURE 1.
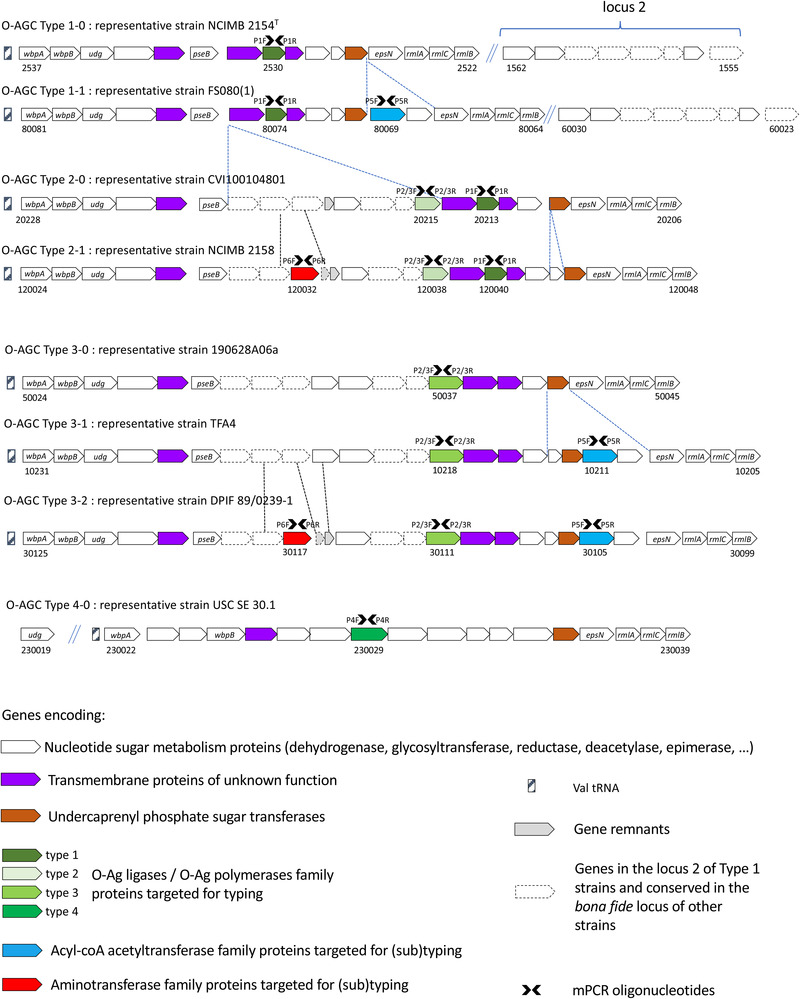
Genomic organization of the T. maritimum O‐AGCs. The loci were analysed using the Microscope platform. Eight distinct ‘Types’ were defined by grouping strains harbouring identical or nearly identical O‐AGC gene organization. Gene conservation was predicted using both homology and synteny criteria (see Section 2 for details). A second locus (locus 2) is present only in Type 1‐0 and Type 1‐1 strains. Genes conserved between the second locus and those encompassed in the bona fide O‐AGC are shown in dotted line. Oligonucleotides used in the mPCR are indicated by black arrows. The locus_tags for each representative strain are indicated for genes at both extremities of the O‐AGCs and for those targeted by the mPCR.
Genomic organization of O‐AGCs Type 1‐0 and Type 1‐1 displays obvious similarities with little variation between them. They differ from the other Types not only because of their intrinsic gene content composition but by harbouring a group of eight genes located elsewhere in their genome (locus 2 in Figure 1) and likely also involved in polysaccharide biosynthesis. This second locus contains genes encoding nucleotide sugar epimerase/dehydratase, aminotransferase, oxidoreductase and acetyltransferase, among others. Most of these genes (five out of eight) are encompassed in the bona fide, unique O‐AGC identified in Types 2‐0 to 3‐2. Types 2‐0 and Type 2‐1 display an overall conserved genomic structure with little variation among them, which is also the case for Types 3‐0, 3‐1 and 3‐2. Finally, a unique Type 4‐0 distantly related to the previously mentioned Types was identified. Therefore, the O‐AGCs of T. maritimum strains (not taking into account the locus 2 of O1 strains) vary from 19.8 to 27.5 kb in length and contain from 16 genes for strains belonging to Type 1‐0 up to 25 genes for strains belonging to Type 3‐1.
3.5. Comparative genomics of O‐AGC loci to identify key molecular determinant of T. maritimum serotypes
A very strong correlation was observed between the conventional serotypes and the structure of the locus. Indeed, with the exception of a single strain (CVI1001048), all O1 strains belong to Type 1 (Type 1‐0 or 1‐1), all O2 strains belong to Type 2 (Type 2‐0 or 2‐1), all O3 strains belong to Type 3 (Type 3‐0, 3‐1 or 3‐2) and all O4 strains belong to Type 4‐0 (Table 1). Even though strain CVI1001048 was serotyped O1 using conventional serotyping, the structure of its O‐AGC (named Type 2‐0) is closely related to the structure of Type 2‐1 O‐AGCs. In addition, and contrary to Type 1 strains, strain CVI1001048 and all other Type 2 strains do not possess the second locus elsewhere in the genome (Figure 1). The type strain NCIMB 2154T, which was typed O1 or O2 and even O1/O2 by conventional methods, typically belongs to the Type 1 group and accordingly contains the second locus.
3.6. Development of an mPCR serotyping scheme
Based on these observations and on the grouping of strains according to the Type they belong to, we noticed that the presence of distinct genes coding transmembrane proteins was the best marker able to discriminate the Types and displaying obvious correlations with the results of conventional serotyping. Indeed, all strains belonging to Type 1 and Type 2 have a gene (MARIT_2530 in strain NCIMB 2154T and orthologs) that is absent in strains belonging to Type 3 and Type 4. This gene encodes a protein with predicted 11 transmembrane segments but with very low BLAST‐P similarities with proteins having any known function. However, using HHpred to detect remote homologies, eight out of the 10 best hits (probability ranging from 94.84% to 99.94% that the database match is a true positive) belong to O‐antigen ligases and O‐antigen polymerases family proteins. One might conclude that MARIT_2530 and its orthologs likely encode a main actor of the O‐antigen biosynthesis with oligosaccharyltransferase activity (Whitfield et al., 2020). This gene was chosen as the first target (using the oligonucleotide pair P1F–P1R) for the development of an mPCR able to discriminate Type 1 and Type 2 from the other Types. In order to distinguish between Type 1 and Type 2 strains, we identified a gene (NCIMB2158_120038 in strain NCIMB 2158 and its orthologs) present in Type 2 strains and absent in Type 1 strains. This gene encodes an eight‐segment transmembrane protein with no significant homologies (even using HHpred) and was chosen as the second target. We also took advantage of a gene (TFA4_10218 in strain TFA4 and its orthologs) only present in Type 3 strains; this gene displays significant similarities to NCIMB2158_120038 though containing a 360‐bp in‐frame insertion. Therefore, this second oligonucleotide pair (P2/3F ‐ P2/3R), encompassing this insertion, not only distinguishes Type 1 from Type 2 strains but also discriminates between Type 2 and Type 3 strains, by amplifying a 357‐ or 717‐bp fragment, respectively. The divergent Type 4 O‐AGC also encompass a gene (USCSE301_v1_230029 in strain USC SE 301.1) encoding a predicted 12‐segment transmembrane protein without any BLAST‐P similarities with proteins having any known function but with significant structural homologies (>99% probability HHpred) with O‐antigen ligases and O‐antigen polymerases. A third oligonucleotide pair (P4F–P4R) targeting USCSE301_v1_230029 and its orthologs was designed to distinguish Type 4 strains (see Figure 1 for genes targeted for mPCR).
Because the different O‐AGCs also contain genes likely involved in nucleotide‐sugar metabolism and unevenly distributed among the strains, we selected two genes (FS0810_80069 in strain FS08(1) and its orthologs encoding an acyl‐CoA acyltransferase family protein and NCIMB2158_120032 in strain NCIMB 2158 and its orthologs encoding an aminotransferase family protein) to be included as targets (using the oligonucleotide pairs P5F–P5R and P6F–P6R, respectively) to increase the sensitivity of the proposed mPCR serotyping scheme. Therefore, the final scheme uses six primer pairs (Table 2) and is able to distinguish up to eight different O‐AGCs: the four Types (Types 1–4) that correspond to the classical serotypes delineated using conventional serum‐based methods) and additional variations (i.e. Types 1‐0 and 1‐1, Types 2‐0 and 2‐1 and Types 3‐0, 3‐1 and 3‐2). Migration of the PCR products after mPCR amplification can virtually provide six bands ranging from 196 to 985 bp. Their presence/absence and combinations allow the unambiguous assignation of any isolate to its Type (Figure 2). Using the strains whose genomes have been sequenced and available in our laboratory, a 100% correlation was observed between the in silico‐predicted Type and the banding pattern obtained by mPCR.
FIGURE 2.
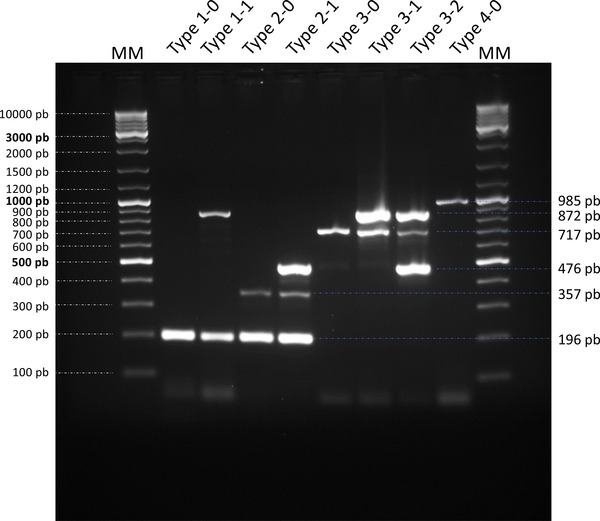
Multiplex PCR. Migration of the PCR products after amplification of a representative strain of each O‐AGC Type. MM, molecular marker; Type 1‐0, representative strain NCIMB 2154T; Type 1‐1, representative strain FS080(1); Type 2‐0, representative strain CVI100104801; Type 2‐1, representative strain NCIMB 2158; Type 3‐0, representative strain 190628A06a; Type 3‐1, representative strain TFA4; Type 3‐2, representative strain DPIF 89/0239‐1; and Type 4‐0, representative strain USC SE 30.1.
3.7. Screening a collection of T. maritimum isolates using mPCR
In addition to the 40 strains mentioned in Table 1, we used the proposed mPCR serotyping scheme to screen a collection of 124 T. maritimum isolates retrieved from a wide variety of geographic areas and host fish species during the last 50 years. Most of these isolates have been previously typed by various methods including MLST (Habib et al., 2014) and MALDI–TOF MS (Bridel et al., 2020). All amplifications were successful and the results are show in Table S2. Strikingly, and taking into account the whole dataset (i.e. the 40 WGS‐types strains and the 124 mPCR‐types isolates), the isolates are unevenly distributed among the Types: isolates belonging to Types 1‐0, 2‐1 and 3‐1 are widely predominant, whereas isolates belonging to Types 1‐1, 2‐0, 3‐2 and 4‐0 are scarce (Figure 3). In addition, our results revealed no clear association between mPCR Type, host fish species or geographic origin of the isolates. When many representatives retrieved from a single fish species were available, some tendencies can be noticed (Figure S2). For example, among the 57 isolates retrieved from sea bass, 25 and 23 belong to Types 2‐1 and 3‐1, respectively. Among the 45 isolates retrieved from batfish, about half belong to Type 1‐0. On the other hand, isolates originating from Tasmania, most of which isolated from Atlantic salmon, belong to Types 2‐1, 3‐0, 3‐1, 3‐2 or 4‐0, while isolates retrieved from batfish in French Polynesia belong to Types 1‐0, 2‐1, 3‐0 or 3‐1. It is also interesting to note that a major serological diversity is present in the isolates from sea bass (six different types were detected), while sea bream isolates only belong to Types 1‐0 and 1‐1. On the other hand, no isolates retrieved from Atlantic salmon belong to Types 1‐0 and 1‐1. Although a limited number of isolates from turbot is available, representatives of four diverse serotypes (Types 1‐0, 2‐1, 3‐0 and 4‐0) were detected. Of importance, no clear correlation was observed between the mPCR Types and the MALDI groups, but tendencies could be observed between the mPCR Types and the MLST types, as most isolates belonging to a specific ST display the same mPCR Type. However, the distribution of Types does not follow the phylogeny based on core genome proteins (Figure 4). For instance, strains Tasmania_0759 and Tasmania_0809, both retrieved from Atlantic salmon and tightly clustered in the phylogenetic tree, belong to Type 3‐2 and 2‐1, respectively. The same lack of correlation between strain position in the phylogenetic tree and the Type they belong to can be observed in many branches of the tree.
FIGURE 3.
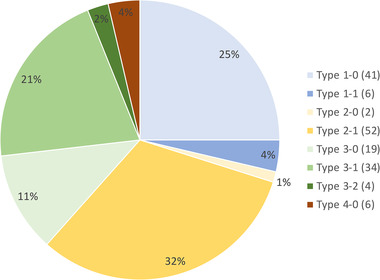
Distribution of the isolates according to their Types. The number of isolates is given in parenthesis.
FIGURE 4.
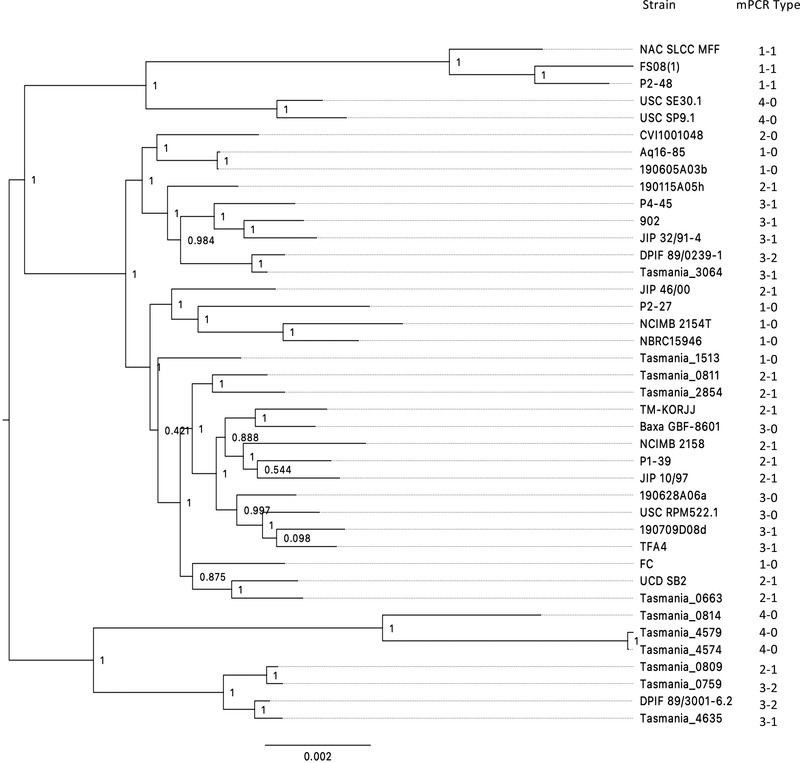
O‐AGC organization (Types) and evolutionary relationships between the T. maritimum strains. Types are shown on a tree attempting to capture the phylogenetic relationships between the 40 T. maritimum strains used in this study for which complete genomes were available. The tree was constructed with 2034 core genome proteins (see Section 2 for details). Local support values are indicated at nodes. The bar at the bottom provides the scale of branch lengths and the unit corresponds to the number of amino acid change per sites.
4. DISCUSSION
Serological techniques are important for diagnosis and epidemiological studies as well as for antigenic characterization to select strains for vaccine development and efficiency follow‐up. Tenacibaculum maritimum was initially described as a very homogeneous group of bacteria (Wakabayashi et al., 1984), but different serological studies have revealed the existence of several serotypes, thus revealing some antigenic diversity (Avendaño‐Herrera et al., 2004; Avendaño‐Herrera et al., 2005; Fernández‐Álvarez et al., 2018; Piñeiro‐Vidal et al., 2007). In addition, the antigenic differences are mainly due to differences in the ‘O’ chains of the LPS (Avendaño‐Herrera et al., 2004). Moreover, it has been suggested that a vaccine produced against one T. maritimum serotype may not be effective against another (Romalde et al., 2005). Indeed, the unique vaccine commercially available so far exclusively targets the O2 serotype affecting turbot1. However, conventional protocols for the serotyping of T. maritimum isolates suffer several drawbacks. For instance, different methods have been proposed: microagglutination, slide agglutination, immunodiffusion, indirect immunofluorescence, enzyme‐linked immunosorbent assay and dot blot assay (see Fernández‐Álvarez & Santos [2018] for a recent and exhaustive review). The choice of reference strains and the procedure used to extract antigens (from whole cells to purified O‐antigen) in order to raise antisera greatly varied among studies. Furthermore, the use of unabsorbed or cross‐absorbed sera (and the choice of the strain(s) used to perform reciprocal absorption) has also important consequences. For example, the type strain NCIMB 2154T was typed O1 or O2 and even O1/O2 by conventional methods. Therefore, these discrepancies probably originated either from limits of the conventional serotyping method or, less plausibly, from subtle variations within the coding sequences or altered regulation of some key genes. In addition, conventional serotyping protocols require to raise antisera from animals (sometimes with variable immune responses) and some isolates display strong, weak or lack of reactivity, or even cross‐reactivity with multiple antisera. Hence, because traditional serotyping is costly, labour‐intensive and requires significant technical expertise, molecular serotyping relying of the presence/absence of relevant genes is a promising alternative. In full expansion for the molecular serotyping of human pathogens (Howell et al., 2015; Ludwig et al., 2020; Nakaue et al., 2021), such schemes were recently developed for animal and especially fish pathogenic bacteria (Rochat et al., 2017; Torres‐Corral & Santos, 2020). The strategy is based on the genomic identification of molecular markers that determine the serotype. In this study, we first identified a locus in the T. maritimum type strain genome with similarities with the O‐AGC of F. psychrophilum and T. dicentrarchi (Figure S1). We noticed that the metabolic pathway of dTDP‐L‐rhamnose biosynthesis also requires the product of rmlD, which is not part of the O‐AGC and is located elsewhere in the T. maritimum genome (MARIT_0966). This organization was also observed in the F. psychrophilum genome in which rmlC and rmlD genes are not located in the O‐AGC. As the rmlABC genes are located immediately downstream to MARIT_2525 and MARIT_2526, it is tempting to speculate that an amino‐modified dTDP‐L‐rhamnose is the first nucleotide sugar to be loaded on the undecaprenyl carrier (C55‐P) on the cytoplasmic face of the inner membrane. It was also noteworthy that most of the genes encompassed in the T. maritimum O‐AGC encode for proteins that display similarities with proteins from phylogenetically distantly related bacteria (e.g. P. aeruginosa, C. jejuni and B. subtilis) (Table S1). Therefore, and as previously reported in diverse bacterial species (Holt et al., 2020; Wang et al., 2010), horizontal gene transfer and homologous recombination likely play instrumental roles in the evolution of T. maritimum O‐AGCs.
We took advantage of the availability of the whole‐genome sequences of 40 T. maritimum strains to perform genomic comparisons of the O‐AGCs. By grouping the strains harbouring identical or nearly identical O‐AGC gene organization, we identified different ‘Types’, with obvious links with the serotypes obtained using conventional procedures. This genome‐based association study allowed us to develop a robust mPCR serotyping scheme able to discriminate up to eight different allelic combinations. This scheme is able to detect subgroups within each serotype and therefore performs better than conventional serotyping. We successfully applied our typing scheme to an additional collection of 124 isolates. In the pioneering works using conventional serotyping methods, some correlations were observed between serotype and host fish species: O1 isolates were retrieved from sole, O2 isolates from sea bream and sea bass and O3 isolates from turbot (Avendaño‐Herrera et al., 2003). However, further studies revealed no strict association between serotype and host fish species. For instance, isolates retrieved from sea bass belong to serotype O1, O2 or O3 (Avendaño‐Herrera et al.,2005; Yardimci & Timur, 2016), isolates retrieved from turbot belong to serotype O1, O2 or O3 (Avendaño‐Herrera et al., 2003; Avendaño‐Herrera et al., 2004; Castro et al., 2007; Fernández‐Álvarez et al., 2018; Piñeiro‐Vidal et al., 2007) and sole isolates belong to serotype O1, O3 or O4 (Avendaño‐Herrera et al., 2003; Avendaño‐Herrera et al., 2005; Fernández‐Álvarez et al., 2018). In addition, Fernández‐Álvarez et al. (2018) stated that serotype O1 is the dominant one regardless of the source of the isolates (i.e. host fish species or geographic location). The same conclusions were drawn when isolate genotypes (determined using either MLST, MALDI–TOF or complete genome‐based typing approaches) were compared to the background information on the isolation source (host fish, year and geographic origin) that could account for only a limited amount of the total genetic variance. Indeed, only trends were observed between the geographic origin of the isolates and the genotypes (Bridel et al., 2020; Habib et al., 2014). Therefore, the lack of any obvious correlation between the mPCR Type and the host fish species was not surprising and only trends were noticed. Yet, it is difficult to state if these trends are the consequences of sampling bias or if they arise from hidden functional links (e.g. bacterial fitness, virulence, colonization of a specific host). Anyhow, this situation is in sharp contrast with the conclusions obtained with F. psychrophilum where obvious links between the serotype and the host fish species were noticed (Li et al., 2021; Rochat et al., 2017). Strikingly, in T. maritimum the Types distribution does not follow the core genome‐based phylogeny (Figure 4). This result is probably the consequence of allelic exchanges mediated by horizontal gene transfer in relation to the high recombination rate observed in this bacterium (Bridel et al., 2020; Habib et al., 2014).
In conclusion, our data revealed a significant diversity of the O‐AGCs in T. maritimum and provided a valuable mPCR‐based serotyping scheme for this important fish pathogen. This scheme should help to improve our knowledge of the serotype distribution at different scales in order to facilitate future epidemiological studies and to help the development of vaccines, including the selection of appropriate strains for autogenous vaccination. If necessary, the proposed mPCR scheme could be enriched with additional targets for a more accurate discrimination of isolates. The O‐AGCs genomic structure described in this study may also serve as a milestone for future complete genome‐based typing approaches (Bentley & Lo, 2021; Lee et al., 2021; Mostowy & Holt, 2018; Zhang et al., 2019).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Pierre Lopez performed the mPCR development and isolate typing and contributed to data analysis and to the writing of the manuscript. Sébastien Bridel performed genome comparisons and O‐AGCs identification. Denis Saulnier contributed to the writing of the manuscript. Beatriz Magariños and Beatriz S. Torres performed the serotyping. Jean François Bernardet managed the T. maritimum bacterial strains collection and contributed to the writing of the manuscript. Eric Duchaud wrote the manuscript, contributed to data analysis and coordinated the work. All authors have read and agreed to the published version of the manuscript.
Supporting information
FIGURE S1 Comparison of the genomic organization of the O‐AGC between T. maritimum strain NCIMB 2154T and F. psychrophilum strain JIP02/86 and T. dicentrarchi strain USC 35/09T. The O‐AGCs were analysed and compared using the Microscope platform. Gene conservation was predicted using both homology and synteny criteria using default parameters. Red arrows represent conserved genes, white arrows not conserved genes and the hatched box the Val‐tRNA. The locus_tag numbers are indicated for strain NCIMB 2154T and at both extremities of the O‐AGC for strains JIP02/86 and USC 35/09T.
FIGURE S2 Distribution of the ‘Types’ for isolates with many representatives retrieved from a single fish species. The number of isolates is given in parenthesis.
TABLE S1 Genes encompassed in T. maritimum NCIMB 2154T O‐AGC. Functional predictions were performed using BLAST‐P on the SwissProt database. Predictions of protein localization was manually verified by using the online TMHMM Server v.2.0 software (http://www.cbs.dtu.dk/services/TMHMM‐2.0/) and the PSORTb v3.0.2 software (https://www.psort.org/psortb/). HHpred was run online (https://toolkit.tuebingen.mpg.de/tools/hhpred) using default parameters on the on PDB_mmCIF70 (release 12 October 2021).
TABLE S2 Tenacibaculum maritimum isolates typed using the mPCR serotyping scheme
ACKNOWLEDGEMENTS
The authors are very grateful to Maelenn Le Roy (IFREMER) and Benjamin Fradet (INRAe) for skillful technical assistance, to Alicia Estévez Toranzo (USC) and Tatiana Rochat (INRAe) for helpful discussions and critical reading of the manuscript, to the many individuals listed in Table S2 for kindly providing bacterial strains and to Zoé Rouy from the MicroScope Platform for her kind help with genome data management. This work has benefited from the facilities and expertise of the high‑throughput sequencing facility of I2BC. We also wish to thank the INRAE MIGALE bioinformatics platform, the LABGeM and the National Infrastructure ‘France Génomique’ for providing computational resources.
Lopez, P. , Bridel, S. , Saulnier, D. , David, R. , Magariños, B. , Torres, B. S. , Bernardet, J. F. , & Duchaud, E. (2022). Genomic characterization of Tenacibaculum maritimum O‐antigen gene cluster and development of a multiplex PCR‐based serotyping scheme. Transboundary and Emerging Diseases, 69, e2876–e2888. 10.1111/tbed.14637
Footnotes
DATA AVAILABILITY STATEMENT
New sequences used in this study have been deposited in the PubMLST database and are freely available in the PubMLST web site (https://pubmlst.org/bigsdb?db=pubmlst_tenacibaculum_isolates&page=query).
REFERENCES
- Apablaza, P. , Frisch, K. , Brevik, Ø. J. , Småge, S. B. , Vallestad, C. , Duesund, H. , Mendoza, J. , & Nylund, A. (2017). Primary isolation and characterization of Tenacibaculum maritimum from Chilean Atlantic salmon mortalities associated with a Pseudochattonella spp. algal bloom. Journal of Aquatic Animal Health, 29(3), 143–149. 10.1080/08997659.2017.1339643 [DOI] [PubMed] [Google Scholar]
- Avendaño‐Herrera, R. , Magariños, B. , López‐Romalde, S. , Romalde, J. L. , & Toranzo, A. E. (2004). Phenotypic characterization and description of two major O‐serotypes in Tenacibaculum maritimum strains from marine fishes. Diseases of Aquatic Organisms, 58(1), 1–8. 10.3354/dao058001 [DOI] [PubMed] [Google Scholar]
- Avendaño‐Herrera, R. , Magariños, B. , Moriñigo, M. A. , Romalde, J. , & Toranzo, A. E. (2005). A novel O‐serotype in Tenacibaculum maritimum strains isolated from cultured sole (Solea senegalensis). Bulletin of the European Association of Fish Pathologists, 25(2), 70–74. [Google Scholar]
- Avendaño‐Herrera, R. , Magariños, B. , Romalde, J. L. , & Toranzo, A. E. (2003). An update on the antigenic diversity of Tenacibaculum maritimum strains isolated from marine fishes. FHS/AFS Newsletter, 31(2), 24–26. [Google Scholar]
- Avendaño‐Herrera, R. , Toranzo, A. E. , & Magariños, B. (2006). Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: A review. Diseases of Aquatic Organisms, 71(3), 255–266. 10.3354/dao071255 [DOI] [PubMed] [Google Scholar]
- Avendaño‐Herrera, R. , Toranzo, A. E. , Romalde, J. L. , Lemos, M. L. , & Magariños, B. (2005). Iron uptake mechanisms in the fish pathogen Tenacibaculum maritimum . Applied and Environmental Microbiology, 71(11), 6947–6953. 10.1128/AEM.71.11.6947-6953.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss, S. C. , Verner‐Jeffreys, D. W. , Bartie, K. L. , Aanensen, D. M. , Sheppard, S. K. , Adams, A. , & Feil, E. J. (2017). The promise of whole genome pathogen sequencing for the molecular epidemiology of emerging aquaculture pathogens. Frontiers in Microbiology, 8, 121. 10.3389/fmicb.2017.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, S. D. , & Lo, S. W. (2021). Global genomic pathogen surveillance to inform vaccine strategies: A decade‐long expedition in pneumococcal genomics. Genome Medicine, 13(1), 84. 10.1186/s13073-021-00901-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardet, J. F. (2011). Family I. Flavobacteriaceae Reichenbach 1992. In Krieg N. R., Ludwig W., Whitman W. B., Hedlund B. P., Paster B. J., Stanley J. T., Ward N. L., Brown D. R., & Parte A. C. (Eds.), Bergey's manual of systematic bacteriology (2nd ed., Vol. 4, pp. 106–111). Springer. 10.1007/978-0-387-68572-4_3 [DOI] [Google Scholar]
- Bohl, T. E. , & Aihara, H. (2018). Current progress in the structural and biochemical characterization of proteins involved in the assembly of lipopolysaccharide. International Journal of Microbiology, 2018, 5319146. 10.1155/2018/5319146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridel, S. , Bourgeon, F. , Marie, A. , Saulnier, D. , Pasek, S. , Nicolas, P. , Bernardet, J.‐F. , & Duchaud, E. (2020). Genetic diversity and population structure of Tenacibaculum maritimum, a serious bacterial pathogen of marine fish: From genome comparisons to high throughput MALDI‐TOF typing. Veterinary Research, 51(1), 60. 10.1186/s13567-020-00782-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, N. , Magariños, B. , Nunez, S. , & Toranzo, A. E. (2007). Reassessment of the Tenacibaculum maritimum serotypes causing mortalities in cultured marine fish. Bulletin of the European Association of Fish Pathologists, 27(6), 229–233. [Google Scholar]
- Cisar, J. O. , Bush, C. A. , & Wiens, G. D. (2019). Comparative structural and antigenic characterization of genetically distinct Flavobacterium psychrophilum o‐polysaccharides. Frontiers in Microbiology, 10, 1041. 10.3389/fmicb.2019.01041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Álvarez, C. , & Santos, Y. (2018). Identification and typing of fish pathogenic species of the genus Tenacibaculum . Applied Microbiology and Biotechnology, 102(23), 9973–9989. 10.1007/s00253-018-9370-1 [DOI] [PubMed] [Google Scholar]
- Fernández‐Álvarez, C. , Torres‐Corral, Y. , & Santos, Y. (2018). Comparison of serological and molecular typing methods for epidemiological investigation of Tenacibaculum species pathogenic for fish. Applied Microbiology and Biotechnology, 102(6), 2779–2789. 10.1007/s00253-018-8825-8 [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization (FAO) . (2020). The state of world fisheries and aquaculture 2020: Sustainability in action. 10.4060/ca9229en [DOI]
- Glover, K. J. , Weerapana, E. , Chen, M. M. , & Imperiali, B. (2006). Direct biochemical evidence for the utilization of UDP‐bacillosamine by PglC, an essential glycosyl‐1‐phosphate transferase in the Campylobacter jejuni N‐linked glycosylation pathway. Biochemistry, 25(6), 5343–5350. 10.1021/bi0602056 [DOI] [PubMed] [Google Scholar]
- Gorski, L. (2021). Serotype assignment by sero‐agglutination, ELISA, and PCR. In Fox E. M., Bierne H., & Stessl B. (Eds.), Listeria monocytogenes: Methods and protocols (pp. 57–78). Springer US. 10.1007/978-1-0716-0982-8_5 [DOI] [PubMed] [Google Scholar]
- Habib, C. , Houel, A. , Lunazzi, A. , Bernardet, J.‐F. , Olsen, A. B. , Nilsen, H. , Toranzo, A. E. , Castro, N. , Nicolas, P. , & Duchaud, E. (2014). Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Applied and Environmental Microbiology, 80(17), 5503–5514. 10.1128/AEM.01177-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, K. E. , Lassalle, F. , Wyres, K. L. , Wick, R. , & Mostowy, R. J. (2020). Diversity and evolution of surface polysaccharide synthesis loci in Enterobacteriales. The ISME Journal, 14(7), 1713–1730. 10.1038/s41396-020-0628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, K. J. , Peters, S. E. , Wang, J. , Hernandez‐Garcia, J. , Weinert, L. A. , Luan, S.‐L. , Chaudhuri, R. R. , Angen, Ø. , Aragon, V. , Williamson, S. M. , Parkhill, J. , Langford, P. R. , Rycroft, A. N. , Wren, B. W. , Maskell, D. J. , Tucker, A. W. , & BRaDP1T Consortium . (2015). Development of a multiplex PCR assay for rapid molecular serotyping of Haemophilus parasuis . Journal of Clinical Microbiology, 53(12), 3812–3821. 10.1128/JCM.01991-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundinya, C. R. , Savithri, H. S. , Rao, K. K. , & Balaji, P. V. (2018). EpsN from Bacillus subtilis 168 has UDP‐2,6‐dideoxy 2‐acetamido 4‐keto glucose aminotransferase activity in vitro. Glycobiology, 28(10), 802–812. 10.1093/glycob/cwy063 [DOI] [PubMed] [Google Scholar]
- Lafferty, K. D. , Harvell, C. D. , Conrad, J. M. , Friedman, C. S. , Kent, M. L. , Kuris, A. M. , Powell, E. N. , Rondeau, D. , & Saksida, S. M. (2015). Infectious diseases affect marine fisheries and aquaculture economics. Annual Review of Marine Science, 7(1), 471–496. 10.1146/annurev-marine-010814-015646 [DOI] [PubMed] [Google Scholar]
- Lee, I. , Ha, S.‐M. , Baek, M. , Kim, D. W. , Yi, H. , & Chun, J. (2021). VicPred: A vibrio cholerae genotype prediction tool. Frontiers in Microbiology, 12, 691895. 10.3389/fmicb.2021.691895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Chai, J. , Knupp, C. , Nicolas, P. , Wang, D. , Cao, Y. , Deng, F. , Chen, F. , Lu, T. , & Loch, T. P. (2021). Phenotypic and genetic characterization of Flavobacterium psychrophilum recovered from diseased salmonids in China. Microbiology Spectrum, 9(2), e00330–21. 10.1128/Spectrum.00330-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, P. , Saulnier, D. , Swarup‐Gaucher, S. , David, R. , Lau, C. , Taputuarai, R. , Belliard, C. , Basset, C. , Labrune, V. , Marie, A. , Bernardet, J. F. , & Duchaud, E. (2022). First isolation of virulent tenacibaculum maritimum isolates from diseased orbicular batfish (Platax orbicularis) farmed in Tahiti Island. Pathogens, 11(2), 131. 10.3390/pathogens11020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, J. B. , Shi, X. , Shridhar, P. B. , Roberts, E. L. , DebRoy, C. , Phebus, R. K. , Bai, J. , & Nagaraja, T. G. (2020). Multiplex PCR assays for the detection of one hundred and thirty seven serogroups of shiga toxin‐producing Escherichia coli associated with cattle. Frontiers in Cellular and Infection Microbiology, 10, 378. 10.3389/fcimb.2020.00378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumura, K. , & Wakabayashi, H. (1977). An outbreak of gliding bacterial disease in hatchery‐born red seabream (Pagrus major) and gilthead (Acanthopagrus schlegeli) fry in Hiroshima. Fish Pathology, 12(3), 171–177. 10.3147/jsfp.12.171 [DOI] [Google Scholar]
- Mostowy, R. J. , & Holt, K. E. (2018). Diversity‐generating machines: Genetics of bacterial sugar‐coating. Trends in Microbiology, 26(12), 1008–1021. 10.1016/j.tim.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa, A. , Basurco, B. , Aguilera, C. , Furones, D. , Reverte, C. , Sanjuan‐Vilaplana, A. , Jansen, M. D. , Brun, E. , & Tavornpanich, S. (2020). Mapping the knowledge of the main diseases affecting sea bass and sea bream in Mediterranean. Transboundary and Emerging Diseases, 67(3), 1089–1100. 10.1111/tbed.13482 [DOI] [PubMed] [Google Scholar]
- Nakaue, R. , Qin, T. , Morita, M. , Ren, H. , Chang, B. , Murai, M. , Amemura‐Maekawa, J. , & Ohnishi, M. (2021). Development of a multiplex‐PCR serotyping assay for characterizing Legionella pneumophila serogroups based on the diversity of lipopolysaccharide biosynthetic loci. Journal of Clinical Microbiology, 59(11), e0015721. 10.1128/JCM.00157-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlan, J. P. , Lumsden, J. S. , & Russell, S. (2020). Advancements in characterizing Tenacibaculum infections in Canada. Pathogens, 9(12), 1029. 10.3390/pathogens9121029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostland, V. E. , Morrison, D. , & Ferguson, H. W. (1999). Flexibacter maritimus associated with a bacterial stomatitis in Atlantic salmon smolts reared in net‐pens in British Columbia. Journal of Aquatic Animal Health, 11(1), 35–44. 10.1577/1548-8667(1999)011<0035:FMAWAB>2.0.CO;2 [DOI] [Google Scholar]
- Pérez‐Pascual, D. , Lunazzi, A. , Magdelenat, G. , Rouy, Z. , Roulet, A. , Lopez‐Roques, C. , Larocque, R. , Barbeyron, T. , Gobet, A. , Michel, G. , Bernardet, J.‐F. , & Duchaud, E. (2017). The complete genome sequence of the fish pathogen Tenacibaculum maritimum provides insights into virulence mechanisms. Frontiers in Microbiology, 8, 1542. 10.3389/fmicb.2017.01542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro‐Vidal, M. , Centeno‐Sestelo, G. , Riaza, A. , & Santos, Y. (2007). Isolation of pathogenic Tenacibaculum maritimum‐related organisms from diseased turbot and sole cultured in the Northwest of Spain. Bulletin of the European Association of Fish Pathologists, 27(1), 29–35. [Google Scholar]
- Price, M. N. , Dehal, P. S. , & Arkin, A. P. (2010). FastTree 2—Approximately maximum‐likelihood trees for large alignments. PLoS ONE, 5(3), e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, A. K. , & Mitchell, A. M. (2020). Enterobacterial common antigen: Synthesis and function of an enigmatic molecule. MBio, 11(4), e01914–e01920. 10.1128/mBio.01914-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci, R. E. , & Timur, G. (2016). Antigenic characterisation of Tenacibaculum maritimum isolates from sea bass (Dicentrarchus labrax, L.) farmed on the Aegean Sea coasts of Turkey. Journal of Aquaculture Research & Development, 7(2), 1000408. 10.4172/2155-9546.1000408 [DOI] [Google Scholar]
- Rochat, T. , Fujiwara‐Nagata, E. , Calvez, S. , Dalsgaard, I. , Madsen, L. , Calteau, A. , Lunazzi, A. , Nicolas, P. , Wiklund, T. , Bernardet, J.‐F. , & Duchaud, E. (2017). Genomic characterization of Flavobacterium psychrophilum serotypes and development of a multiplex PCR‐based serotyping scheme. Frontiers in Microbiology, 8, 1752. 10.3389/fmicb.2017.01752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romalde, J. L. , Ravelo, C. , López‐Romalde, S. , Avendaño‐Herrera, R. , Magariños, B. , & Toranzo, A. E. (2005). Vaccination strategies to prevent emerging diseases for Spanish aquaculture. Developments in Biologicals, 121, 85–95. [PubMed] [Google Scholar]
- Saldarriaga‐Córdoba, M. , Irgang, R. , & Avendaño‐Herrera, R. (2021). Comparison between genome sequences of Chilean Tenacibaculum dicentrarchi isolated from red conger eel (Genypterus chilensis) and Atlantic salmon (Salmo salar) focusing on bacterial virulence determinants. Journal of Fish Diseases, 44(11), 1843–1860. 10.1111/jfd.13503 [DOI] [PubMed] [Google Scholar]
- Schoenhofen, I. C. , McNally, D. J. , Vinogradov, E. , Whitfield, D. , Young, N. M. , Dick, S. , Wakarchuk, W. W. , Brisson, J.‐R. , & Logan, S. M. (2006). Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: Enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. Journal of Biological Chemistry, 281(2), 723–732. 10.1074/jbc.M511021200 [DOI] [PubMed] [Google Scholar]
- Sloan, A. , Wang, G. , & Cheng, K. (2017). Traditional approaches versus mass spectrometry in bacterial identification and typing. Clinica Chimica Acta, 473, 180–185. 10.1016/j.cca.2017.08.035 [DOI] [PubMed] [Google Scholar]
- Stentiford, G. D. , Sritunyalucksana, K. , Flegel, T. W. , Williams, B. A. P. , Withyachumnarnkul, B. , Itsathitphaisarn, O. , & Bass, D. (2017). New paradigms to help solve the global aquaculture disease crisis. PLoS Pathogens, 13(2), e1006160. 10.1371/journal.ppat.1006160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Y. , & Kagan, J. C. (2014). A cross‐disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Molecular Cell, 54(2), 212–223. 10.1016/j.molcel.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres‐Corral, Y. , & Santos, Y. (2020). Comparative genomics of Streptococcus parauberis: New target for molecular identification of serotype III. Applied Microbiology and Biotechnology, 104(14), 6211–6222. 10.1007/s00253-020-10683-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes, S. , Irgang, R. , Barros, M. C. , Ilardi, P. , Saldarriaga‐Córdoba, M. , Rivera–Bohle, J. , Madrid, E. , Gajardo–Córdova, J. , & Avendaño‐Herrera, R. (2021). First report and characterization of Tenacibaculum maritimum isolates recovered from rainbow trout (Oncorhynchus mykiss) farmed in Chile. Journal of Fish Diseases, 44(10), 1481–1490. 10.1111/jfd.13466 [DOI] [PubMed] [Google Scholar]
- Vallenet, D. , Calteau, A. , Dubois, M. , Amours, P. , Bazin, A. , Beuvin, M. , Burlot, L. , Bussell, X. , Fouteau, S. , Gautreau, G. , Lajus, A. , Langlois, J. , Planel, R. , Roche, D. , Rollin, J. , Rouy, Z. , Sabatet, V. , & Médigue, C. (2020). MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Research, 48, D579–D589. 10.1093/nar/gkz926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen, R. (2007). Vaccination of Atlantic salmon (Salmo salar) against marine flexibacteriosis (PhD thesis). University of Tasmania. https://eprints.utas.edu.au/22196/ [Google Scholar]
- Vinogradov, E. , MacLean, L. L. , Crump, E. M. , Perry, M. B. , & Kay, W. W. (2003). Structure of the polysaccharide chain of the lipopolysaccharide from Flexibacter maritimus . European Journal of Biochemistry, 270(8), 1810–1815. 10.1046/j.1432-1033.2003.03543.x [DOI] [PubMed] [Google Scholar]
- Wakabayashi, H. , Hikida, M. , & Masumura, K. (1984). Flexibacter infection in cultured marine fish in Japan. Helgol Meeresunters, 37, 587–593. [Google Scholar]
- Wakabayashi, H. , Hikida, M. , & Masumura, K. (1986). Flexibacter maritimus sp. nov., a pathogen of marine fishes. International Journal of Systematic and Evolutionary Microbiology, 36(3), 396–398. 10.1099/00207713-36-3-396 [DOI] [Google Scholar]
- Wang, L. , Wang, Q. , & Reeves, P. R. (2010). The variation of o antigens in gram‐negative bacteria. In Wang X. & Quinn P. J. (Eds.), Endotoxins: Structure, function and recognition (pp. 123–152). Springer. 10.1007/978-90-481-9078-2_6 [DOI] [PubMed] [Google Scholar]
- Whitfield, C. , Williams, D. M. , & Kelly, S. D. (2020). Lipopolysaccharide O‐antigens—Bacterial glycans made to measure. The Journal of Biological Chemistry, 295(31), 10593–10609. 10.1074/jbc.REV120.009402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , den Bakker, H. C. , Li, S. , Chen, J. , Dinsmore, B. A. , Lane, C. , Lauer, A. C. , Fields, P. I. , & Deng, X. (2019). SeqSero2: Rapid and improved salmonella serotype determination using whole‐genome sequencing data. Applied and Environmental Microbiology, 85(23), e01746–19. 10.1128/AEM.01746-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, L. , Stephens, A. , Nam, S.‐Z. , Rau, D. , Kübler, J. , Lozajic, M. , Gabler, F. , Söding, J. , Lupas, A. N. , & Alva, V. (2018). A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. Journal of Molecular Biology, 430(15), 2237–2243. 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Comparison of the genomic organization of the O‐AGC between T. maritimum strain NCIMB 2154T and F. psychrophilum strain JIP02/86 and T. dicentrarchi strain USC 35/09T. The O‐AGCs were analysed and compared using the Microscope platform. Gene conservation was predicted using both homology and synteny criteria using default parameters. Red arrows represent conserved genes, white arrows not conserved genes and the hatched box the Val‐tRNA. The locus_tag numbers are indicated for strain NCIMB 2154T and at both extremities of the O‐AGC for strains JIP02/86 and USC 35/09T.
FIGURE S2 Distribution of the ‘Types’ for isolates with many representatives retrieved from a single fish species. The number of isolates is given in parenthesis.
TABLE S1 Genes encompassed in T. maritimum NCIMB 2154T O‐AGC. Functional predictions were performed using BLAST‐P on the SwissProt database. Predictions of protein localization was manually verified by using the online TMHMM Server v.2.0 software (http://www.cbs.dtu.dk/services/TMHMM‐2.0/) and the PSORTb v3.0.2 software (https://www.psort.org/psortb/). HHpred was run online (https://toolkit.tuebingen.mpg.de/tools/hhpred) using default parameters on the on PDB_mmCIF70 (release 12 October 2021).
TABLE S2 Tenacibaculum maritimum isolates typed using the mPCR serotyping scheme
Data Availability Statement
New sequences used in this study have been deposited in the PubMLST database and are freely available in the PubMLST web site (https://pubmlst.org/bigsdb?db=pubmlst_tenacibaculum_isolates&page=query).


