Abstract
The olfactory placode (OP) of vertebrates generates several classes of migrating cells, including hypothalamic gonadotropin‐releasing hormone (GnRH)‐producing neurons, which play essential roles in the reproduction system. Previous studies using OP cell labeling have demonstrated that OP‐derived non‐GnRH cells enter the developing forebrain; however, their final fates and phenotypes are less well understood. In chick embryos, a subpopulation of migratory cells from the OP that is distinct from GnRH neurons transiently expresses somatostatin (SS). We postulated that these cells are destined to develop into brain neurons. In this study, we examined the expression pattern of SS mRNA in the olfactory‐forebrain region during development, as well as the destination of OP‐derived migratory cells, including SS mRNA‐expressing cells. Utilizing the Tol2 genomic integration system to induce long‐term fluorescent protein expression in OP cells, we found that OP‐derived migratory cells labeled at embryonic day (E) 3 resided in the olfactory nerve and medial forebrain at E17–19. A subpopulation of green fluorescent protein (GFP)‐labeled GnRH neurons that remained in the olfactory nerve was considered to comprise terminal nerve neurons. In the forebrain, GFP‐labeled cells showed a distribution pattern similar to that of GnRH neurons. A large proportion of GFP‐labeled cells expressed the mature neuronal marker NeuN. Among the GFP‐labeled cells, the percentage of GnRH neurons was low, while the remaining GnRH‐negative neurons either expressed SS mRNA, neuropeptide Y, or calbindin D‐28k or did not express any of them. These results indicate that a diverse population of OP‐derived neuronal cells, other than GnRH neurons, integrates into the chick medial forebrain.
Keywords: forebrain, GnRH neurons, neuronal migration, olfactory placode, somatostatin mRNA, terminal nerve, Tol2‐transposon
This long‐lasting early OP cell labeling study, utilizing the Tol2 genomic integration system, shows that OP‐derived neurons in the forebrain just before hatching are not specific to hypothalamic GnRH neurons. They express somatostatin mRNA, neuropeptide Y, calbindin D‐28k, or none of these substances. Our results indicate that diverse OP‐derived neurons are differentiated in the chick forebrain.
1. INTRODUCTION
Epithelial cells migrate from the olfactory placode (OP) or olfactory epithelium toward the forebrain during development in various vertebrates (De Carlos et al., 1995; Mendoza et al., 1982; Pellier et al., 1994; Quintana‐Urzainqui et al., 2014; Tarrozo et al., 1995). These migratory cells are heterogeneous populations, including neurons and olfactory ensheathing cells (Forni & Wray, 2015; Miller et al., 2010). Among them, gonadotropin‐releasing hormone (GnRH)‐producing neurons typically migrate from the OP and enter the forebrain, settling in the hypothalamus (Schwanzel‐Fukuda & Pfaff, 1989; Wray, 2010; Wray et al., 1989). They play an essential role in reproductive functions after maturation (Silverman et al., 1994; Forni & Wray, 2015).
Various immunohistochemical studies have revealed the presence of other cells associated with migrating GnRH neurons along olfactory, terminal, and/or vomeronasal nerves. These cells express gamma‐aminobutyric acid (GABA) in rodents and human embryos (Tobet et al., 1996), somatostatin (SS) (Murakami & Arai, 1994b) or neuropeptide Y (NPY) (Hilal et al., 1996) in chicks, olfactory marker protein (Valverde et al., 1993) or calcium binding protein, calbindin D‐28k (CB) (Toba et al., 2001) in rats, and tyrosine hydroxylase (TH) in human embryos (Verney et al., 1996). More recent studies have shown that early migratory cells from the OP express the LIM homeodomain transcription factor Islet1/2 in mice, chicks, and zebrafish (Aguillon et al., 2018; Palaniappan et al., 2019; Shan et al., 2020; Taroc et al., 2020). Expression of these molecules and GnRH immunoreactivity are mostly observed in separate cell populations; however, GABA, NPY, TH, and Islet1/2 are coexpressed in a subpopulation of GnRH neurons. It has therefore been proposed that a subpopulation of OP‐derived cells migrate in association with GnRH neurons to enter the forebrain in a variety of species. However, cell lineage tracing techniques have not been applied to the olfactory system of mammals, and their final destinations remain unknown. In contrast, analysis of the OP cell lineage in chicks further provided evidence supporting the possibility that OP‐derived GnRH‐negative cells enter the forebrain. Studies using carbocyanine dye, DiI, labeling, and retrovirus infection in chick embryos have reported that OP‐derived non‐GnRH cells were observed in the forebrain at embryonic day (E) 7−13 (Murakami & Arai, 1994a; Silverman et al., 2002). In a quail‐chick transplantation study, OP‐derived cells expressing molluscan cardioexcitatory peptide (FMRFamide), which appear to comprise a different population from GnRH neurons, were found in the olfactory bulb at E9 (Yamamoto et al., 1996). Moreover, a study using OP labeling with green fluorescent protein (GFP) demonstrated that early OP‐labeled cells expressing GnRH, Lhx2, or Islet1 entered the medial forebrain at E6−6.5 in chicks (Palaniappan et al., 2019). These observations raise the possibility that OP‐derived cells differentiate into distinct brain neurons, although the final fate of these cells largely remains unknown, except for GnRH neurons, in both mammals and birds.
In chick embryos, SS expression has been observed in the olfactory‐forebrain region during early embryonic development (Blähser & Heinrichs, 1982; Murakami & Arai, 1994b). A small number of SS‐immunoreactive (‐ir) cells were found in the olfactory nerve and rostral forebrain during active migration of GnRH neurons (Murakami & Arai, 1994b). Several immunohistochemical studies have demonstrated that SS is a neuropeptide present in neurons throughout the brain, including in the hypothalamus, cerebral cortex, and olfactory bulb of vertebrates (Blähser, 1984; Epelbaum, 1986; Mikami & Yamada, 1984; Takatsuki et al., 1981; Viollet et al., 2008). Although SS‐ir neurons in these regions are believed to be generated in the brain, the close relationship between SS‐ir cells and GnRH neurons during their migratory course indicates that a subpopulation of SS‐ir cells in the olfactory nerve might be a candidate for OP‐derived brain neurons. However, weak and transient expression of the SS protein in migrating cells is not useful for tracing their destination at late embryonic stages. To address this issue, we examined the expression of SS mRNA in the developing chick olfactory‐forebrain region using in situ hybridization histochemistry. We found that SS mRNA was expressed in the olfactory epithelium, migrating cells from the OP, and forebrain during early to late embryonic development, indicating that the expression of the SS gene in the olfactory‐forebrain region can identify a subset of OP‐derived migrating cells.
In light of these findings, we aimed to determine the final destination of OP‐derived SS mRNA‐expressing cells at late embryonic stages. Cells that express the SS protein form a small population in the migratory tract of GnRH neurons in the olfactory nerve and forebrain (Murakami & Arai, 1994b); therefore, we also focused on the fate and properties of other OP‐derived migratory cells. We performed an OP cell labeling experiment using a transposon‐mediated genomic integration system. The Tol2 transposable element was isolated from the genome of medaka fish (Kawakami et al., 1998). The Tol2 transposon is frequently integrated into intragenetic regions (Kawakami & Noda, 2004; Kotani et al., 2006) and is functional in a wide range of host species, including chickens (Sato et al., 2007). The epithelium of the OP on E3, when cells expressing SS mRNA migrate from the OP, was selectively and stably labeled with GFP using the Tol2 transposon system combined with in ovo electroporation, and the labeled cells were then analyzed in the olfactory‐forebrain region at E17−19. Here, we describe the expression pattern of SS mRNA in the developing chick olfactory‐forebrain region and provide evidence that a few OP‐derived cells expressing SS mRNA reside in the medial forebrain. Moreover, a long‐lasting OP cell labeling study clearly showed that OP‐derived GnRH‐negative neurons expressing NPY, CB, or OP‐derived GnRH‐, NPY‐, and CB‐negative neurons integrate into the forebrain and that OP‐derived GnRH neurons remain in the olfactory nerve at the late embryonic stage.
2. MATERIALS AND METHODS
2.1. Animals and tissue preparations
All animal experiments were approved by the Animal Care Committee of the Juntendo University School of Medicine. Fertilized White Leghorn eggs were purchased locally and incubated in a humidified incubator at 37.6°C. Embryonic stage was determined according to Hamburger and Hamilton (1951). For histological examinations, embryos were fixed overnight in 4% paraformaldehyde in 0.1 M phosphate buffer (PB, pH 7.4) at 4°C and were cryoprotected with 20% sucrose in PB. The heads were cut into 16 μm thick serial coronal or sagittal sections.
2.2. In situ hybridization
In situ hybridization was performed as previously described (Ding et al., 2005; Murakami et al., 2010), with minor modifications. A fragment of chick somatostatin cDNA (nt 183–481, GenBank accession number NM_205336) was amplified by reverse transcription polymerase chain reaction (RT‐PCR) and cloned into the pBluescriptⅡSK(‐) (Agilent Technologies/Stratagene, CA, USA) vector to produce the riboprobe. A digoxigenin (DIG)‐labeled RNA probe was generated using a DIG RNA‐labeling kit (Roche Diagnostics, Switzerland) and hybridized to 16 μm thick cryostat sections. The hybridized DIG‐labeled probes were detected with an alkaline phosphatase‐conjugated anti‐DIG antibody (Roche Diagnostics) and visualized by the addition of nitrobluetetrazodium/5‐bromo‐4‐chloro‐3‐indolyl phosphate substrate (NBT/BCIP, Roche Diagnostics).
2.3. Antibody characterization and specificity
The antibodies used in this study are listed in Table 1. Mouse monoclonal anti‐HuC/D (clone 16A11; Molecular Probes, Thermo Fisher Scientific, OR, USA) was generated against the human HuC/HuD neuronal protein. This antibody recognizes the embryonic lethal abnormal visual family members HuC, HuD, and Hel‐N1 neuronal proteins. Mouse monoclonal anti‐highly polysialylated neural cell adhesion molecule (PSA‐NCAM) (generously provided by Dr. T. Seki, Tokyo Medical University) was generated against the forebrain of E18 Wistar rats (Seki & Arai, 1991). This antibody recognizes a broad 180–280 kD band, and the minimum chain length required for antigen‐antibody binding is five N‐acetylneuraminic acid residues (Sato et al., 1995). Mouse monoclonal anti‐GnRH (Gunma University, Japan) was generated against the synthetic peptide EHWSYGLRPG (C‐terminal free mammalian LHRH) conjugated with bovine thyroglobulin. This antibody recognizes the region around Ser4‐Tyr5, which is a common amino acid sequence of GnRH in a variety of animal species. Immunostaining was eliminated by preincubation of the diluted antiserum with 1 μg/ml synthetic LHRH. Rabbit polyclonal anti‐GFP (Molecular Probes) was generated against GFP isolated directly from Aequorea victoria. Goat polyclonal anti‐GFP (Frontier Institute, Japan) was generated against the full GFP sequence (YP_002302326). Mouse monoclonal anti‐neuronal nuclei (NeuN) clone A60 (Millipore, MA, USA) was generated against purified cell nuclei from the mouse brain. According to the manufacturer, this antibody recognizes the DNA‐binding neuron‐specific protein NeuN in most neuronal cell types of all vertebrates, with some exceptions, such as olfactory bulb mitral cells and cerebellar Purkinje cells (Mullen et al., 1992). Rabbit polyclonal anti‐NPY (Abcam, UK) was generated against a synthetic peptide corresponding to pig NPY aa 1 at the C‐terminus conjugated to keyhole limpet hemocyanin. This antibody stained the soma and processes of neurons and was consistent with the expected distribution of NPY in the chick brain. Rabbit polyclonal calbindin D‐28k (Swant, Switzerland) was generated against recombinant rat calbindin D‐28k, and no staining was detected with this antibody in the brains of calbindin D‐28k knockout mice (Airaksinen et al., 1997). Control staining was performed by omitting the primary or secondary antibodies to ensure that the combination of primary and secondary antisera was free of cross‐reactivity.
TABLE 1.
List of primary antibodies
| Antibody | Antigen | Source, Cat. # | Host and clonality | Dilution | RRID |
|---|---|---|---|---|---|
| HuC/D | Human HuC/HuD neuronal protein | Molecular Probes, Thermo Fisher Scientific, Cat# A‐21271 | Mouse, monoclonal IgG (clone 16A11) | 1:100 | AB_221448 |
| PSA‐NCAM | Forebrain of embryonic day 18 Wistar rat | T. Seki, Tokyo Medical University | Mouse, monoclonal IgM | 1:2000 | AB_2315216 |
| GnRH | Mammalian LHRH epitope: pEHWSYGLRPG‐NH2 | Gunma University, Cat# LRH‐13 | Mouse, monoclonal IgG | 1:2000 | AB_2490075 |
| GFP | GFP directly isolated from Aequorea victoria | Molecular Probes, Cat# A‐11122 | Rabbit, polyclonal | 1:1000 | AB_221569 |
| GFP | Full sequence of GFP (YP_002302326) | Frontier Institute, Cat# GFP‐Go‐Af1480 | Goat, polyclonal | 1:400 | AB_2571574 |
| NeuN | Purified cell nuclei from mouse brain | Millipore, Cat# MAB377 | Mouse, monoclonal IgG (clone A60) | 1:100 | AB_2298772 |
| Neuropeptide Y | Synthetic peptide corresponding to pig Neuropeptide Y aa 1 to the C‐terminus | Abcam, Cat# ab30914 | Rabbit, polyclonal | 1:1000 | AB_1566510 |
| CB | Reconbinant rat calbindin D‐28k | Swant, Cat# CB38 | Rabbit, polyclonal | 1:2000 | AB_10000340 |
Abbreviations: CB, calbindin; GFP, green fluorescent protein; GnRH, gonadotropin‐releasing hormone; NeuN, neuronal nuclei; PSA‐NCAM, polysialylated neural cell adhesion molecule.
2.4. Combined fluorescent in situ hybridization and immunohistochemistry
For the simultaneous detection of SS mRNA and HuC/D or GnRH peptide on migratory cells in early embryonic stages or SS mRNA on GFP‐labeled cells in the olfactory‐forebrain region of late chick embryos, a combination of fluorescent in situ hybridization and immunohistochemistry was performed. Immunodetection with an anti‐DIG antibody conjugated with horseradish peroxidase (Roche Diagnostics) was enhanced using the TSA plus DNP system (PerkinElmer Life Sciences, MA, USA). Fluoro‐detection was performed using an anti‐DNP antibody conjugated with Alexa Fluor 488 (Molecular Probes). The sections were further processed with HuC/D, GnRH (LRH13, Gunma University), and PSA‐NCAM antibodies or both GnRH antibody and GFP antibody (Frontiers Institute). The intensity of GFP fluorescence in Tol2‐GFP‐positive cells was not easy to detect in the brain except in the olfactory epithelium; therefore, GFP antibody was used for GFP‐fluorescence detection. The secondary antibodies used were Cy3‐conjugated donkey anti‐mouse IgG or a mixture of Cy3‐conjugated donkey anti‐goat IgG and Cy5‐conjugated donkey anti‐mouse IgG (Jackson ImmunoResearch, PA, USA). To detect double staining for monoclonal antibodies (HuC/D, GnRH, and PSA), Cy3‐ or Cy5‐conjugated anti‐mouse IgG (gamma) and Cy3‐conjugated (Jackson Immunoresearch Laboratories, PA, USA) or Alexa Fluor 647‐conjugated (Molecular Probes) anti‐mouse IgM were used.
2.5. Immunohistochemistry
Immunostaining was performed as previously described (Murakami et al., 2010). Sections were incubated with rabbit anti‐GFP antibody (Molecular Probes), followed by incubation in biotinylated anti‐rabbit IgG and avidin‐biotin peroxidase complex (Vector Laboratories, CA, USA). The peroxidase complex was visualized with 3‐3′diaminobenzidine. Sections were then treated in a 0.01 M citric acid solution (pH 6.0) using microwaves for 1–3 min to enhance immunoreactivity, processed for GnRH immunostaining, and finally detected with Vector SG (Vector Laboratories). Sections were counterstained with methyl green. Triple immunofluorescence was performed to characterize GFP‐labeled and GnRH‐negative cells. After antigen retrieval treatment, sections were incubated with a mixture of the following primary antibodies: goat anti‐GFP (Frontiers Institute), mouse anti‐GnRH (LRH13), mouse anti‐NeuN (Millipore), rabbit anti‐neuropeptide Y (Abcam), or rabbit anti‐calbindin D‐28k (Swant) for 24−48 h at 4°C and subsequently with a species‐specific secondary antibody conjugated to fluorescent dyes. Cell nuclei were visualized with 4′, 6′‐diamindino‐2‐phenylindole (DAPI, Sigma‒Aldrich, MO, USA).
2.6. OP ablation
The OP invaginates to form the olfactory pit at E3 (Hamburger and Hamilton stages [HH] 18). At E3.5 (HH20−23), the epithelium of the olfactory pit on the right side was excised using a finely sharpened steel needle under a dissecting microscope. Four days after surgery, the embryos were fixed to detect SS mRNA‐expressing cells and GnRH neurons in the olfactory‐forebrain region.
2.7. In ovo electroporation
E3 (HH18) chick embryos were electroporated in the olfactory placode epithelial region to transfer a Tol2‐based vector containing the EGFP gene (pT2AL200R175G‐CAGGS‐EGFP) with CAGGS‐transposase (pCAGGS‐T2TP) (Urasaki et al., 2006; kindly provided by Dr. K. Kawakami, National Institute of Genetics) at 1 μg/μl in sterile physiological saline containing 0.01% Fast Green. Three electric pulses of 10 V, 50 ms, and 100 ms intervals were applied using a square‐pulse electroporator (CUY 21EDIT, Nepa Gene, Japan). Embryos were then incubated until E17−19, anesthetized with sodium pentobarbital, and transcardially perfused with 4% paraformaldehyde in 0.1 M PB. The heads were removed, postfixed overnight in 4% paraformaldehyde at 4°C, and processed for cryostat sectioning.
2.8. Image acquisition and analysis
Bright‐field images were obtained with a Zeiss Axiocam camera (Carl Zeiss, Germany) on a Zeiss Axioplan microscope using Axiovision software (Version 4.5; Carl Zeiss). Confocal images were taken using a confocal laser scanning microscope (Zeiss LSM 780 or LSM 880, Carl Zeiss) and merged with the ZEN 2.3 (Carl Zeiss), and images were adjusted for brightness and contrast or changed colors. All images were stored and assembled into figures using Adobe Photoshop 5.5 (Adobe Systems, Mountain View, CA, USA), and when necessary, brightness and contrast were adjusted. In most bright‐field images, the figure backgrounds were adjusted to show a white background by using white level correction of Adobe Photoshop software. The number of SS mRNA‐expressing cells, HuC/D‐positive cells, and GnRH neurons was counted in the confocal stacked images of serial sections at the second to fourth intervals of E3 (HH17, HH19) and E6.5–7 embryos. To map the distribution of GFP‐labeled cells and GnRH neurons at E17–19, camera lucida drawings were made of every fifth coronal serial section, and the positions of all GFP‐labeled cells and GnRH neurons were recorded on the drawings using an Olympus BH2 microscope (Olympus, Japan) with 5–20× objectives. For the figures, the cell locations were approximated to their locations on the representative drawings. For quantitative analysis, every fifth serial section of the forebrain immunolabeled for GFP and GnRH, NeuN, NPY, or CB (each 41 or 47 sections from two E17–19 embryos) was examined under a confocal laser scanning microscope, and the number of GFP‐labeled cells and percentage of cells colocalized with NeuN, NPY, CB, or GnRH were determined. Brain areas were identified according to the report by García‐Calero and Puelles (2009) and the chick brain atlas (Kuenzel & Masson, 1988; Puelles et al., 2007).
3. RESULTS
3.1. Expression of SS mRNA in the developing olfactory‐forebrain region
The SS peptide is expressed in a small population of migrating cells from the chick olfactory epithelium during E5−8 (Murakami & Arai, 1994b). SS mRNA expression in the chick hypothalamic area, which is an SS expression site in the adult brain, has been detected at E3 (Caqueret et al., 2005), whereas SS gene expression in the peripheral olfactory region has not been reported. To ascertain whether the migrating cells along the olfactory nerve express SS mRNA from early developmental stages, SS mRNA expression in the olfactory‐forebrain region was examined by in situ hybridization using a DIG‐coupled mRNA probe.
At E2.5 (HH16), early migratory cells from the OP that express HuC/D have been detected in the mesenchymal tissue adjacent to the OP (Fornaro et al., 2001, 2003). SS mRNA expression was observed in a subpopulation of these early migratory cells from the OP (Figure 1a). At E3 (HH17), SS mRNA‐expressing cells were observed in the epithelium of the OP and cluster of HuC/D‐positive cells but not in the telencephalon (Figure 1b,c). SS mRNA‐expressing cells coexpressed HuC/D (Figure 1, c 1, 2, 3), and the proportion of SS mRNA‐expressing cells to the total number of HuC/D‐positive cells in the mesenchymal tissue was 38.7% (n = 1 embryo). At E3.5 (HH18−20), it has been demonstrated that early migratory cells from the OP form a cellular cord between the epithelium of the olfactory pit and the rostral telencephalon (Drapkin & Silverman, 1999; Miyakawa et al., 1997). The number of SS mRNA‐expressing cells increased not only in the epithelium of the olfactory pit but also in the HuC/D‐positive cellular cord (Figure 1d, e1–3). A similar percentage of SS mRNA‐expressing cells to the total number of HuC/D‐positive migratory cells was observed (41.5%, n = 1 embryo). SS mRNA expression was apparent in the superficial layer of the telencephalon (Figure 1d). At E5.5−7, many SS mRNA‐expressing cells were found throughout the olfactory epithelium but not in the prospective respiratory epithelium (Figure 1f,i). SS mRNA‐expressing cells and HuC/D‐positive cells were continuously found along the developing olfactory nerve, which was immunoreactive to PSA‐NCAM (Figures 1g,h, and 2a). A subpopulation of SS mRNA‐expressing cells was observed in close proximity to migrating GnRH neurons (Figure 1h). At approximately E6.5−7, a small number of SS mRNA‐expressing cells were found in the medial wall of the forebrain from the prospective olfactory bulb to the septum region, corresponding to the migratory pathway of GnRH neurons (Figures 1i and 2c). Many SS mRNA‐expressing cells were observed in the ventral hypothalamic area at E7 (Figure 1j).
FIGURE 1.
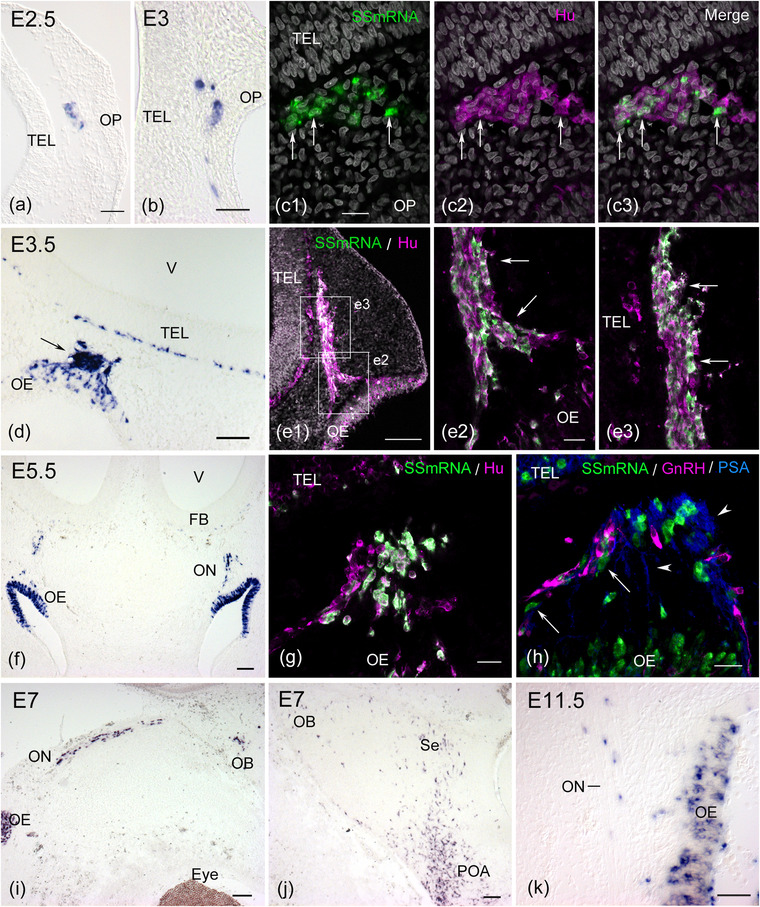
Somatostatin (SS) mRNA expression in the developing chick olfactory‐forebrain region. (a) Cross section through the head of an E2.5 (HH16) chick embryo. Few SS mRNA‐positive cells are seen in the mescenchymal tissue beneath the olfactory placode (OP). (b) At E3 (HH17), SS mRNA‐expressing cells are found in the epithelium of the OP and mesenchymal tissue. (c) In situ hybridization on a cross section followed by immunostaining for HuC/D labels a cluster of migratory cells from the OP at HH17. SS mRNA‐expressing cells that coexpress HuC/D are found in the HuC/D‐positive cellular mass‐like structure (arrows in c1–c3). (d) Cross section through the olfactory‐forebrain region of an E3.5 (HH21) chick embryo. A cluster of migratory cells from the olfactory epithelium (OE) strongly expresses SS mRNA (arrow). (e) Sagittal section through the head of an E3.5 (HH19) embryo. HuC/D‐positive migratory cells form the cellular cord between the OE and telencephalon (TEL). Many SS mRNA‐expressing cells are found in the cellular cord (e1). Regions similar to those outlined by the squares show that SS mRNA and HuC/D double‐positive cells are located in a thick cellular cord (arrows in e2 and e3). (f–h) Cross sections through the olfactory‐forebrain region of E5.5 chick embryos. Intense expression of SS mRNA is seen in the OE and migratory cells along the olfactory nerve (ON) (f). SS mRNA‐expressing cells that coexpress HuC/D migrate from the OE (g). Some SS mRNA‐expressing cells comigrated with gonadotropin‐releasing hormone (GnRH) neurons (arrows in h). These migratory cells are associated with olfactory fibers that express highly polysialylated neural cell adhesion molecule (PSA‐NCAM, arrowheads). (i–j) Sagittal section through the head of an E7 chick embryo. SS mRNA‐positive cells are seen in the OE, ON, and olfactory bulb (OB). SS mRNA‐expressing cells are sparsely distributed in the dorsal forebrain, including the septum region (Se), while many SS mRNA‐expressing cells are found in the preoptic area (POA) of the ventral forebrain. (k) Sagittal section through the olfactory region of an E11.5 chick embryo. SS mRNA is still expressed in the OE and the cells associated with the ON. (a), (b), (d), and (k): Viewed with Nomarski optics. FB, forebrain. Scale bars: 20 μm in c1, e2, g, and h; 50 μm in a, b, and k; 100 μm in d, e1, f, i, and j
FIGURE 2.
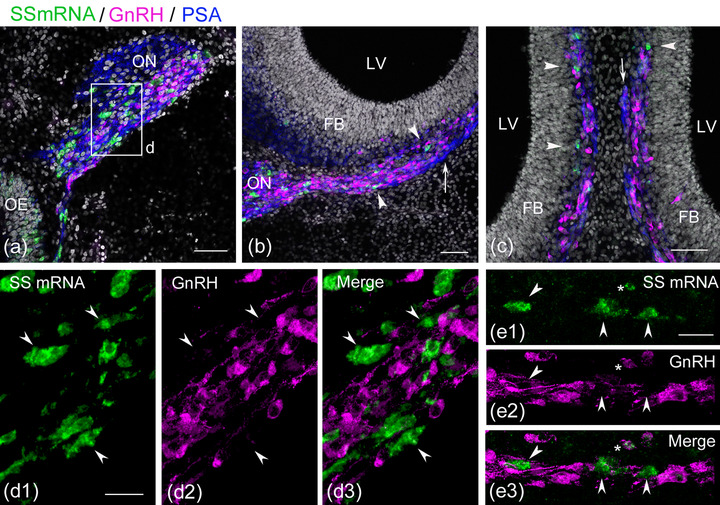
Somatostatin (SS) mRNA‐expressing cells are accompanied by gonadotropin‐releasing hormone (GnRH) neurons in the olfactory‐forebrain region. Coronal sections through the olfactory‐forebrain region of E6.5−7 embryos. (a) SS mRNA‐expressing cells (green) and GnRH neurons (magenta) migrate along the medial portion of the olfactory nerve (ON, blue) that express highly polysialylated neural cell adhesion molecule (PSA‐NCAM). (b) A small subset of PSA‐NCAM‐immunoreactive (‐ir) fibers branch medially from the ON and extend to the medial forebrain (arrow). SS mRNA‐expressing cells and GnRH neurons migrate along these fibers into the medial forebrain (arrowheads). (c) At a more caudal level of panel (b), a small number of SS mRNA‐expressing cells are seen in the parenchyma of the medial forebrain (arrowheads), where migrating GnRH neurons are aligned continuously along the PSA‐NCAM‐ir fibers. A small fascicle of the medial branch of the ON is seen to extend along the medial forebrain (arrow). (d) A confocal stacked image of an olfactory region of an E6.5 chick embryo shows that SS mRNA‐expressing cells are immunonegative for GnRH (arrowheads in d1–d3). (e) A confocal stacked image of the medial forebrain of an E7 chick embryo shows that almost none of the SS mRNA‐expressing cells express GnRH. In this region, three SS mRNA‐expressing cells adjacent to GnRH neurons form a distinct cell population from GnRH neurons (arrowheads in e1–e3), while only one SS mRNA‐expressing cell exhibits very weak GnRH immunoreactivity (asterisk in e1–e3). Scale bars: 50 μm in a, b, and c; 20 μm in d1 and e1. Abbreviations: FB, forebrain; LV, lateral ventricle; OE, olfactory epithelium
FIGURE 3.
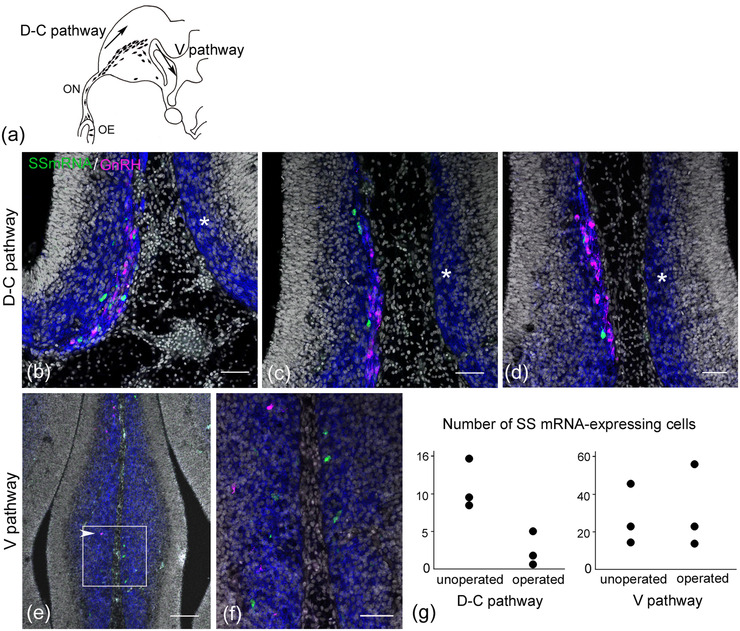
Unilateral olfactory placodectomy induces a loss of somatostatin (SS) mRNA‐expressing cells and gonadotropin‐releasing hormone (GnRH) neurons in the migratory course of the forebrain. Coronal section through the forebrain of an E7.5 embryo after unilateral placodectomy at E3.5. (a) Schematic figure shows the proposed migration pathway of GnRH neurons in the forebrain (Murakami et al., 2010). (b–d) Coronal sections through the forebrain are aligned rostrally (b) to caudally (d) at different intervals. On the unoperated side, a small number of SS mRNA‐expressing cells (green) migrate in association with GnRH neurons (magenta) into the medial forebrain (b). Highly polysialylated neural cell adhesion molecule (PSA‐NCAM) immunoreactivity (blue) highlights immature neurons in the developing forebrain. SS mRNA‐expressing cells are found within a cluster of migratory GnRH neurons in the medial forebrain surface (c–d). On the operated side, no SS mRNA‐expressing cells or GnRH neurons are seen in the equivalent region on the unoperated side (asterisk in b–d). (e–f) At the level of the V pathway of GnRH neurons, scattered GnRH neurons migrate ventrally on the unoperated side (arrowhead in e) but not on the operated side. An enlarged image of panel (e) demonstrates that SS mRNA‐expressing cells are detected in the V pathway of GnRH neurons on both sides (f). (g) Quantification of the number of SS mRNA‐expressing cells in the D‐C and V pathways of GnRH neurons (n = 3 embryos), which were counted in every fifth coronal section. The D‐C pathway is characterized by a small cluster of GnRH neurons that are aligned along the medial forebrain surface in coronal sections. Thus, the counting of SS mRNA‐expressing cells in the D‐C pathway was performed within this restricted region. Ablation of the olfactory placode induces a remarkable reduction in the number of SS mRNA‐expressing cells in the D‐C pathway on the operated side. Scale bars, 50 μm in b, c, d, and f; 100 μm in e. Abbreviations: OE, olfactory epithelium; ON, olfactory nerve
The immunoreactivity for SS peptides was remarkably reduced in the olfactory region at E11.5 (Murakami & Arai, 1994b). However, the expression of SS mRNA was consistently observed in the olfactory epithelium and olfactory nerve at E11.5 (Figure 1k) and was also detected in the olfactory epithelium at E18 just before hatching (data not shown). The developmental expression pattern of SS mRNA in the olfactory‐forebrain region raises the possibility that a subpopulation of SS mRNA‐expressing cells migrates from the OP and enters the forebrain.
3.2. SS mRNA‐expressing cells are different from GnRH neurons
Because the time of appearance of SS mRNA‐expressing cells in the olfactory region largely corresponded with that of GnRH neurons migrating from the olfactory epithelium to the forebrain, the possibility that GnRH neurons coexpress SS mRNA was examined using a combination of fluorescent in situ hybridization and immunohistochemistry. Previous studies have demonstrated that GnRH neurons enter the medial forebrain near the prospective olfactory bulb at E6 and travel dorsocaudally along the medial forebrain surface beneath the pia matter (Murakami et al., 2010, 2000; Sullivan & Silverman, 1993). A subset of olfactory fibers branching medially from the olfactory nerve also extends in association with migrating GnRH neurons along the medial forebrain surface (Murakami et al, 2010, 2000; Norgren & Branckenbury, 1993). At E6.5−7, a subpopulation of cells expressing SS mRNA was usually found near migrating GnRH neurons, extending the olfactory nerve and medial forebrain surface (Figure 2a–c). A confocal stacked image of the olfactory nerve showed that SS mRNA‐expressing cells were immunonegative for GnRH (Figure 2, d 1, 2, 3). The total number of SS mRNA‐expressing cells counted in the olfactory nerve was 103 (n = 2 embryos), and all were immunonegative for GnRH. Within the medial forebrain surface, the proportion of SS mRNA‐expressing cells to the total number of SS mRNA‐expressing cells and GnRH neurons was 23.2% (n = 2 embryos). The majority of SS mRNA‐expressing cells did not show colocalization with GnRH, whereas a small percentage of SS mRNA‐expressing cells exhibiting very weak or dot‐like GnRH immunoreactivity was observed (2.5%, Figure 2, e 1, 2, 3). These results indicate that migratory cells expressing SS mRNA are largely different from the cell population of GnRH neurons.
3.3. Olfactory placodectomy induces a loss of SS mRNA‐expressing cells associated with migrating GnRH neurons
Unilateral olfactory placodectomy was performed at E3.5 to determine the precise origin of the SS mRNA‐expressing cells associated with GnRH neurons in the olfactory‐forebrain region. Four days after the complete unilateral placodectomy, the olfactory components were ablated on the operated side, and GnRH neurons were absent in the olfactory‐forebrain region, consistent with the results of a previous study (Akutsu et al., 1992). It has been demonstrated that two different modes of GnRH neuron migration exist in the chick forebrain: they initially undergo axophilic migration along a subset of olfactory fibers in a dorsocaudal direction and subsequently undergo ventrally directed migration (Murakami et al., 2010; Figure 3a). At E7.5, GnRH neurons disappeared completely in the axophilic dorsocaudal migratory course (D‐C pathway) and ventral migratory course (V pathway) on the operated side (Figure 3b–f), while the loss of SS mRNA‐expressing cells was restricted to the D‐C pathway along the medial forebrain surface (Figure 3b–d). At the level of the ventral pathway of GnRH neurons, SS mRNA‐expressing cells were found on both the unoperated and operated sides (Figure 3e,f). The number of SS mRNA‐expressing cells in the D‐C pathway was markedly decreased on the operated side compared to that on the unoperated side (Figure 3g). In contrast, there was no difference in the number of SS mRNA‐expressing cells on the unoperated and operated sides of the V pathway of GnRH neurons (Figure 3g). These results indicate that migratory cells expressing SS mRNA are generated in the OP, and some of them that are associated with GnRH neurons invade the medial forebrain.
3.4. Distribution patterns of OP‐derived migrating cells labeled with GFP using the Tol2‐transposon‐mediated gene transfer system
Previous cell tracing studies could not characterize the properties of labeled cells that were immunonegative for GnRH (Murakami & Arai, 1994a; Silverman et al., 2002), although one study demonstrated that non‐GnRH OP‐derived cells entering the medial forebrain express Islet1 at E6−6.5 (Palaniappan et al., 2019). The epithelial cells of the OP at E3 (HH18) were labeled with GFP using Tol2 transposon‐mediated gene genomic integration, and the final destination of cells expressing SS mRNA and other OP‐derived migratory cells was determined at E17−19. The long‐term survival of early labeled OP‐derived cells has not been studied previously; therefore, we first examined the distribution patterns of GFP‐labeled cells in the olfactory‐forebrain region.
In five successfully electroporated embryos at E17−19, GFP‐labeled cells were found in the peripheral olfactory region and in various regions of the medial forebrain. In the peripheral olfactory region, GFP expression was found in a part of the olfactory epithelium and olfactory nerve, indicating that a subpopulation of migratory cells was labeled (Figure 4a). GFP‐labeled olfactory fibers were observed to terminate in the olfactory bulb, indicating persistent labeling of olfactory sensory neurons (Figure 4b). GFP‐labeled cells and GnRH neurons were observed in the medial portion of the olfactory nerve layer (Figure 4c). A small fascicle of GFP‐labeled fibers was separated from the GFP‐labeled olfactory nerve in the caudal region of the olfactory bulb (Figure 4c) and extended along the medial forebrain surface to the dorsocaudal region of the septum, where some labeled fibers were dispersed into the deep layer (Figure 4d). In the nasal cavity, a small cluster of GFP‐labeled cells was detected in the olfactory nerve near the olfactory epithelium (Figure 4, e1, e4), indicating that these OP‐derived cells had ceased to migrate into the forebrain during development. We analyzed the expression of calbindin D‐28k (CB) in GFP‐labeled cells in the olfactory nerve, which has been previously examined in the developing rat olfactory system (Toba et al., 2001). A portion of GFP‐labeled cells expressed CB (Figure 4, e1, e2, e4, arrow), while some GFP‐labeled cells expressed both CB and GnRH (Figure 4, 1, 2, 3, 4, arrowhead). GFP‐labeled OP‐derived cells showed a neuronal phenotype in the olfactory nerve.
FIGURE 4.
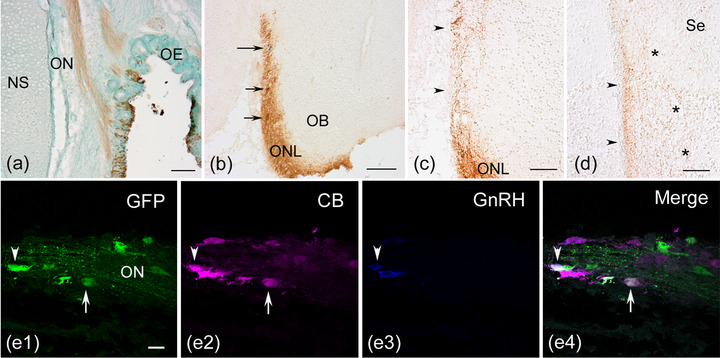
Persistent green fluorescent protein (GFP) expression in the peripheral olfactory region and neuronal phenotypes of GFP‐labeled cells. The Tol2‐mediated GFP construct was electroporated into the olfactory placode at HH18 (E3), and horizontal (a and e) or coronal (b–d) sections were obtained at E17−19. (a) A part of the olfactory epithelium (OE) and the olfactory nerve (ON) are labeled by transfer to the Tol2‐GFP construct. (b) The GFP‐labeled ON fibers terminate in the olfactory nerve layer (ONL) of the olfactory bulb (OB). GFP‐labeled cells (short arrows) and a gonadotropin‐releasing hormone (GnRH) neuron (long arrow) are seen in the GFP‐positive ONL. (c) A small number of GFP‐labeled fibers apart from the ON extend to the medial forebrain surface caudally (arrowheads). (d) A small fascicle of GFP‐labeled fibers is seen to terminate in the medial edge of the dorsocaudal portion of the septum (arrowheads). Some GFP‐labeled fibers are seen to disperse into the deep layer of the septum (asterisks). (e) A cluster of GFP‐labeled cells is observed in the ON, and two labeled cells coexpress calbindin D‐28k (CB). One GFP‐labeled cell coexpresses CB but not GnRH (arrow in e1, e2, and e4), while another GFP‐labeled cell coexpresses both CB and GnRH (arrowhead in e1–e4). (c) and (d): Viewed with Nomarski optics. Scale bars: 100 μm in a and b; 50 μm in c and d; 20 μm in e. Abbreviations: NS, nasal septum; Se, septum
Within the brain, the total number of GFP‐labeled cells in the two cases determined by counting every fifth serial section was 35 and 70, respectively. Tol2 transposon‐mediated GFP labeling showed a specific distribution of OP‐derived cells restricted to the medial wall of the subpallium and the preoptic area. GFP‐labeled cells were located throughout the rostro‐bulbar olfactory area, including the olfactory bulb, septal regions from the rostral to caudal level, vertical limb of the diagonal band, and preoptic area lying ventral to the anterior commissure (Figure 5a). The distribution pattern of GFP‐labeled cells was similar to that of GnRH neurons (Figure 5a). GFP‐labeled cells tended to be located near the GnRH neurons (Figure 5a–c). Specifically, a cluster of GFP‐labeled cells and GnRH neurons was found at the dorsocaudal level of the septum region, including the septofimbrial nucleus (Figure 5a,b). This region, which has a large number of GnRH neurons, corresponds to the bed nucleus of the pallial commissure (nCPa), a major group of GnRH neurons in the adult chick brain, as previously described (Kuenzel & Blähser, 1991). Many GFP‐labeled cells were negative for GnRH (Figure 5b,c, arrows), whereas a small number of GFP‐labeled cells coexpressed GnRH (Figure 5b, arrowheads). GFP‐labeled GnRH‐negative cells had multipolar processes, suggesting that they differentiated into mature neurons (Figure 5d,e).
FIGURE 5.
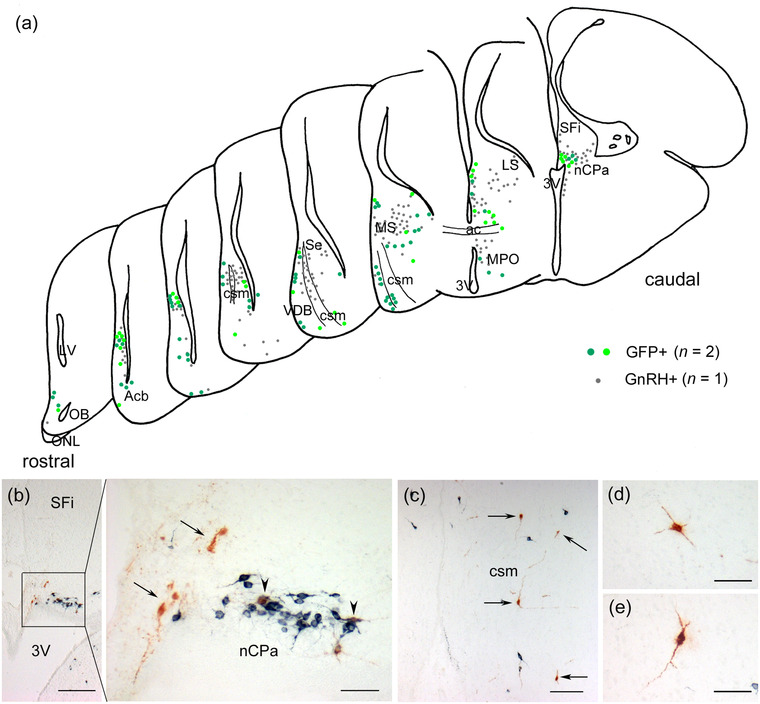
Distribution patterns of green fluorescent protein (GFP)‐labeled cells and gonadotropin‐releasing hormone (GnRH) neurons in the forebrain of E17−19 embryos. Coronal sections through the forebrain electroporated with the Tol2‐mediated GFP plasmid into the olfactory placode at HH18 (E3) and processed at E17−19. (a) Schematic drawings of hemisections through the forebrain of E18 chick embryos showing the distribution of GFP‐labeled cells and GnRH neurons. They are arranged from rostral to caudal at 80−400 μm intervals. The location of GFP‐labeled cells is represented by the data of two embryos (green and yellow‒green circles, 94 cells), while that of GnRH neurons is the data of one embryo (gray circles, 207 neurons). The distribution pattern was largely similar between the two cell groups. GFP‐labeled cells that coexpressed GnRH were excluded. (b) GFP‐labeled cells are seen in and around the bed nucleus of the pallial commissure (nCPa) at the dorsocaudal level of the septum, including the septofimbrial nucleus (SFi). GFP‐labeled GnRH‐negative cells are found near the nCPa (arrows). A cluster of GnRH neurons is seen in the nCPa, where two GnRH neurons coexpress GFP (arrowheads). (c) GFP‐labeled cells are located in the vertical limb of the diagonal band (VDB) at the ventral part of the subpallium (arrows). (d and e) GFP‐labeled cells have multipolar processes. Scale bars: 200 μm in b; 100 μm in c; 50 μm in the large panels of b, d, and e. Abbreviations: ac, anterior commissure; Acb, accumbens nucleus; csm, corticoseptomesencephalic tract; LS, lateral septum; MPO, medial preoptic area; MS, medial septum; OB, olfactory bulb; ONL, olfactory nerve layer; Se, septum; 3V, third ventricle
3.5. Phenotypes of long‐lasting GFP‐labeled OP‐derived cells in the forebrain
Although early migratory cells from the OP at E3 (HH17−18) express immature and mature neuronal markers (Drapkin & Silverman, 1999; Miyakawa et al., 1997), we examined whether OP‐derived cells that were labeled at E3 were part of the neuronal population. We analyzed an E17 (HH43) embryo with a relatively large number of GFP‐labeled cells in the brain and found that a large proportion of GFP‐labeled cells expressed the mature neuronal marker NeuN (73.3%, n = 1 embryo, Figure 6a,b). Next, we examined the molecular properties of GFP‐labeled GnRH‐negative neurons in the brain. The detection of SS mRNA in GFP‐labeled cells was performed by combining fluorescent in situ hybridization and immunohistochemistry for GFP and GnRH. Thirty‐four GFP‐labeled cells were found in serial sections collected at different intervals throughout the forebrain of three E17−19 embryos. Six GFP‐labeled cells coexpressed SS mRNA at the level of the rostral forebrain, ventral portion of the diagonal band, and caudal septum region, including the nCPa (Figure 6c,d). Three GFP‐labeled cells expressing SS mRNA did not coexpress GnRH in the forebrain of two embryos (Figure 6, d 1, 2, 3, 4). Other GFP‐labeled cells expressing SS mRNA exhibited very weak GnRH immunoreactivity (data not shown). These results clearly show that a subpopulation of SS mRNA‐expressing cells in the medial forebrain is derived from the OP and resides in the brain.
FIGURE 6.
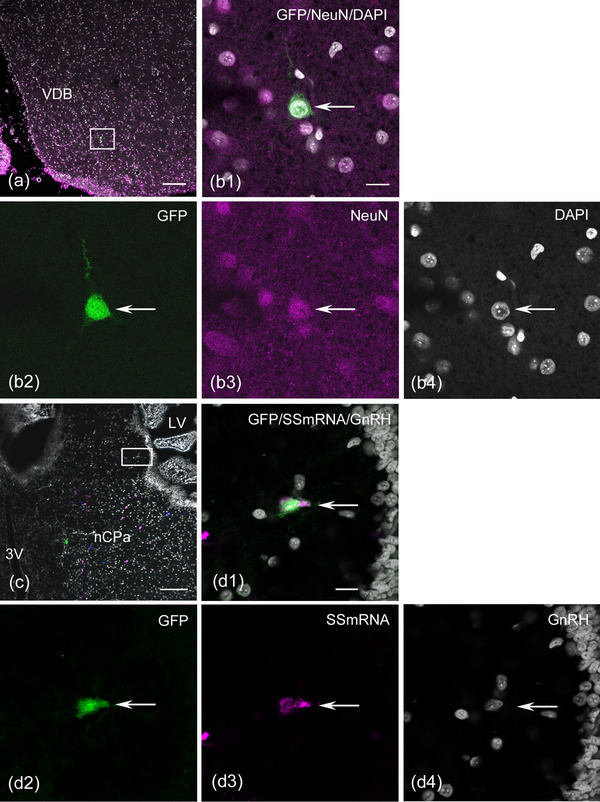
Green fluorescent protein (GFP)‐labeled olfactory placode (OP)‐derived cells coexpress neuronal nuclei (NeuN) or somatostatin (SS) mRNA in the medial forebrain. Coronal sections through the forebrain of E17 or E19 embryos electroporated with the Tlo2‐mediated EGFP plasmid into the OP at HH18 (E3). (a and b) Confocal single image at the ventral portion of the rostral‐medial forebrain. An enlarged image of panel (a) demonstrates that a GFP‐labeled cell expresses NeuN (arrow in b1–b4). (c and d) The combination of fluorescent in situ hybridization and immunohistochemistry reveals that GFP‐labeled cells coexpress SS mRNA in the bed nucleus of the pallial commissure (nCPa). Pseudocolored confocal images for GFP and SS mRNA were produced using Zeiss ZEN. GFP‐labeled cells (green), SS mRNA‐expressing cells (magenta), and gonadotropin‐releasing hormone (GnRH) neurons (blue) are seen in the nCPa (c). An enlarged confocal single image of panel (c) shows a GFP‐labeled cell expressing SS mRNA negative for GnRH (arrow in d1–d4). Scale bars: 100 μm in a and c; 10 μm in b1 and d1. Abbreviations: LV, lateral ventricle; 3V, third ventricle; VDB, vertical limb of the diagonal band
We further examined the molecular properties of GFP‐labeled GnRH‐negative cells in the forebrain using triple immunofluorescence staining with GFP, GnRH, and NPY or with CB antibodies. NPY and CB are widely expressed in neurons of the mature brain, including the telencephalon and hypothalamus (Celio, 1990; Chronwall et al., 1985; Esposito et al., 2001; Rogers & Resibois, 1992; Roth et al., 1981), and in migrating cells from the OP during development (Hilal et al., 1996; Toba et al., 2001). In two E17−19 embryos, every fifth serial section was analyzed. GFP‐labeled NPY‐positive cells were found in the ventral telencephalon, including in the diagonal band nucleus, preoptic area, and nCPa at the caudal level of the septum, whereas GFP‐labeled CB‐positive cells were uniformly found in the telencephalon in the rostral to caudal direction. A modest ratio of GFP‐labeled GnRH‐negative cells colabeled with NPY to the total number of GFP‐labeled cells was detected in the forebrain (31.8%, n = 1 embryo, Figure 7a, b1–4). A similar percentage of GFP‐labeled GnRH‐negative cells colocalized with CB (24.6%, n = 2 embryos, Figure 7c, d1–4) was observed. The proportion of GFP‐labeled GnRH neurons that were immunonegative for NPY or CB was 13.6% (Figure 7, b 1, b2, b4, arrowhead, n = 1 embryo) and 15.3% (n = 2 embryos), respectively. The identities of the remaining GFP‐labeled GnRH‐negative cells could not be determined (54.5%, n = 1 embryo and 60.1%, n = 2 embryos). These findings indicate that diverse populations of OP‐derived neurons, other than GnRH neurons, exist in the medial forebrain at late embryonic stages.
FIGURE 7.
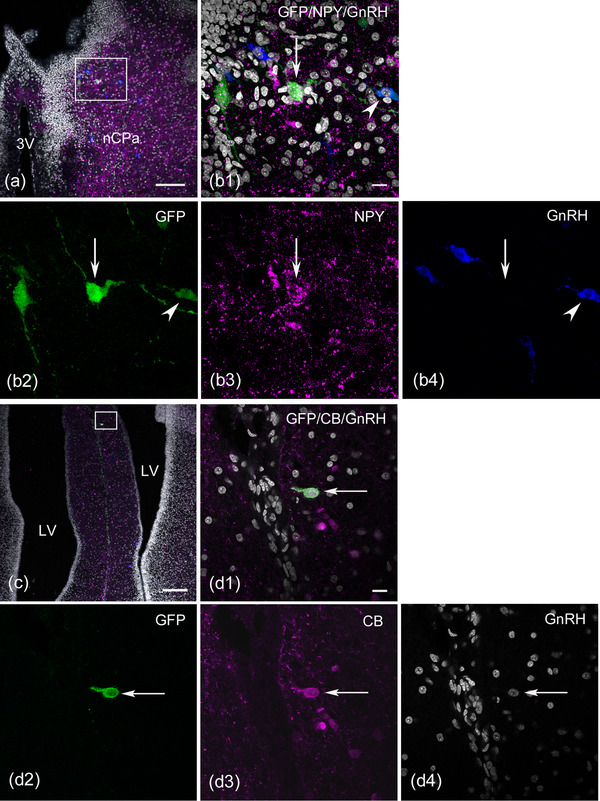
Green fluorescent protein (GFP)‐labeled olfactory placode‐derived cells also coexpress neuropeptide Y (NPY) or calbindin D‐28k (CB) in the medial forebrain at E17 or E19. (a and b) Confocal single image at the dorsocaudal level of the septum region, including the bed nucleus of the pallial commissure (nCPa). An enlarged image of panel (a) shows the colocalization of NPY in the GFP‐labeled cell. The stacked image (z‐projection image stacks) reveals that many GFP‐labeled cells are observed in the nCPa (b1 and b2). A GFP‐labeled cell expresses NPY (arrow in b1‐ b3) but does not express gonadotropin‐releasing hormone (GnRH) (arrow in b4), while another labeled cell expresses GnRH (arrowhead in b1, b2, and b4), and the remaining labeled cell does not express either NPY or GnRH. (c and d) Confocal single image of triple immunostaining with GFP, CB, and GnRH antibodies shows a GFP‐labeled CB‐expressing neuron in the rostral medial forebrain caudal to the olfactory bulb. An enlarged image of panel (c) demonstrates that a GFP‐labeled cell expresses CB (arrow in d1‐ d3) but does not express GnRH (arrow in d4). Scale bars: 100 μm in a and c; 10 μm in b1 and d1. Abbreviations: LV, lateral ventricle; 3V, third ventricle
4. DISCUSSION
In the present study, we showed the following: (1) SS mRNA‐expressing cells were found in the olfactory‐forebrain region from E2.5 (HH16) until E11.5; (2) a subpopulation of migratory cells expressing SS mRNA was associated with GnRH neurons; (3) the long‐lasting labeling experiment of early OP cells demonstrated that GFP‐labeled cells resided in regions from the olfactory nerve to the restricted region of the medial forebrain at E17−19; (4) a large proportion of GFP‐labeled cells in the medial forebrain expressed NeuN. GFP‐labeled GnRH‐negative cells expressed SS mRNA, NPY, and CB. GFP‐labeled cells immunonegative for these substances were also found; and (5) GFP‐labeled cells were detected in the olfactory nerve even at later stages, and they also expressed GnRH and CB. These results indicate that OP‐derived brain neurons are not specific to GnRH neurons and that OP‐derived neurons reside in the olfactory nerve as terminal nerve neurons.
4.1. Fate of SS mRNA‐expressing cells derived from the OP
Early expression of SS mRNA in the peripheral olfactory region was histologically identified in this study. The expression pattern was consistent with the microarray analysis data of chick OP at HH16−20, in which the SS gene was one of the upregulated transcripts in early OP (Poopalasundaram et al., 2016). The cells expressing SS mRNA identified in the mesenchyme at E2.5−3 correspond to a population of early migratory cells from the OP, which form a cellular cord between the OP and telencephalon at approximately HH18−20 (Drapkin & Silverman, 1999; Miyakawa et al., 1997). Previous studies in which GnRH neurons migrate from the OP at HH20−21 (Mulrenin et al., 1999; Murakami et al., 1991) have raised the possibility that the early migratory cells expressing SS mRNA remain in the cellular cord prior to the migration of GnRH neurons and early extension of the olfactory nerve and may contribute to a scaffold for migrating cells and extending olfactory axons, such as the migratory mass in the mesenchyme of embryonic mice (Miller et al., 2010).
As development progresses, SS mRNA‐expressing cells increase in number and are situated adjacent to GnRH neurons in their migratory pathway, suggesting that cells expressing SS mRNA and GnRH neurons migrate in association with each other toward the forebrain. It has been demonstrated that heterogeneous cell populations originate in the OP and follow the migratory pathway of GnRH neurons (Hilal et al., 1996; Toba et al., 2001; Tobet et al., 1996; Valverde et al., 1993), but their entry into the forebrain has not yet been determined. A recent study demonstrated that non‐GnRH OP‐derived cells expressing Islet1 entered the medial forebrain at E6−6.5 (Palaniappan et al., 2019); however, their final fates at later stages have not been studied. In contrast, our OP ablation study suggests that SS mRNA‐expressing cells in the medial forebrain surface, which correspond to the migratory course of GnRH neurons, are generated from the OP. Moreover, an OP long‐lasting labeling study provided direct evidence that OP‐derived cells expressing SS mRNA reside in the medial forebrain, distinct from GnRH neurons, at E17−19. A ratio of 3:25 for GFP‐labeled GnRH‐negative cells expressing SS mRNA to GFP‐labeled cells was found in the medial forebrain of two E17−19 embryos, suggesting that a relatively small number of OP‐derived cells expressing SS mRNA integrate into the brain.
4.2. A diverse population of OP‐derived neurons in the medial forebrain
The most significant finding of the present study was the existence of different cell types of OP‐derived neurons, other than GnRH neurons, in the medial forebrain at the late embryonic stage. A large proportion of GFP‐labeled cells in the forebrain showed neuronal properties, which were identified by the expression of NeuN, a mature neuronal marker. GFP‐labeled GnRH neurons accounted for approximately 15% of all GFP‐labeled cells. In contrast, 25−30% of GFP‐labeled cells that had GnRH‐negative properties expressed the neurochemical markers NPY or CB. Overall, the proportion of GFP‐labeled neurons expressing NPY or CB was higher than that of GFP‐labeled GnRH neurons. It has previously been shown in chick embryos that cells expressing NPY and NPY/GnRH double‐labeled cells are found in the migratory pathway of GnRH neurons at approximately E4.5−16 (Hilal et al., 1996). One possibility for this observation is that NPY‐positive cells change their phenotype, switching to expressing GnRH or other neuropeptides during development. However, there is no evidence of adult GnRH neurons coexpressing NPY in mammals and birds. Thus, GFP‐labeled NPY‐positive cells in the forebrain are thought to comprise a distinct subpopulation. They migrate from the OP along the olfactory nerve and/or terminal nerve and integrate into the forebrain. GFP‐labeled CB‐positive cells have also been suggested to be generated in the OP and migrate into the forebrain, where they reside. During late development, CB appears when neurons make synaptic contacts or become functional (Baimbridge et al., 1992). GFP/CB double‐labeled cells that have reached their destination may join the neuronal network.
The remaining GFP‐labeled neurons that had GnRH‐negative properties (approximately 30%) could not be identified as specific neuronal subtypes. Several cell populations have been reported to be generated from the OP and/or olfactory epithelium and then migrate toward the forebrain (Tarrozo et al., 1995; von Bartheld, 2004). Candidate substrates for identifying unknown OP‐derived neurons may be present in these migratory cells. In contrast, OP‐derived GnRH‐negative cells that express Islet1 in the medial forebrain at E6−6.5 (Palaniappan et al., 2019) may be a distinct population of OP‐derived brain neurons. The identity of GFP‐labeled cells with NeuN‐negative properties remains unclear. NeuN expression is observed in most mature neuronal populations in mammals (Mullen et al., 1992) and chicks (Mezey et al., 2012). However, its expression level may depend on the physiological state of neurons (Weyer & Schilling, 2003). In adult neurogenesis, immature neurons do not express NeuN during the early differentiation phase (Seki, 2002; von Bohlen und Halbach, 2007). Since GFP‐labeled cells did not express GFAP, a glial marker for astrocytes, in some sections (data not shown), it is possible that GFP‐labeled NeuN‐negative cells are immature neurons.
It was believed that adult brain neurons in mammals and birds, which originate outside the brain and subsequently enter the brain, are derived from two neuronal populations: (i) hypothalamic GnRH neurons, which are derived from the OP (Schwanzel‐Fukuda & Pfaff, 1989; Wray et al., 1989), and (ii) mesencephalic trigeminal nucleus (Me5) neurons, which are thought to be embryologically derived from neural crest cells (Narayanan & Narayanan, 1978; Stainier et al., 1991). However, recent studies have demonstrated that Me5 neurons have a central nervous system origin (Hunter et al., 2001; Louvi et al., 2007). Thus, OP‐derived neurons uniquely contribute to forebrain development in vertebrates. To the best of our knowledge, our study is the first to show that additional OP‐derived neurons reside in the medial forebrain, some of which are neuropeptidergic neurons at the late embryonic stage. The postnatal development of novel populations of OP‐derived neurons is unknown; however, our results raise the possibility that a more diverse population of OP‐derived neurons constitutes the medial forebrain than previously thought. Immunohistochemical studies have demonstrated the comigration of OP‐derived neurons expressing GABA, OMP, CB, or TH with GnRH neurons along the olfactory‐related fibers in rodents and human embryos, without showing their final destinies (Toba et al., 2001; Tobet et al., 1996; Valverde et al., 1993; Verney et al., 1996). An OP origin for hypothalamic GnRH neurons has been conserved among most vertebrates; therefore, these observations reinforce the possibility that OP‐derived non‐GnRH neurons in rodents and other mammals have a similar destination in the forebrain to that in chicks.
4.3. OP‐derived GnRH neurons in the olfactory nerve in relation to the terminal nerve
Two distinct types of OP‐derived cells were distinguished in the present study: (i) cells that migrated into the brain and (ii) cells that resided in the olfactory nerve. The latter population is thought to settle in the migratory route outside of the brain during development. Subpopulations of these cells expressed GnRH or CB, suggesting that nervus terminalis (terminal nerve [TN]) neurons exist in the chick olfactory region at the late embryonic stage. TN has been identified in the nasal cavity and forebrain of various vertebrates (Vilensky, 2014; von Bartheld, 2004; Wirsig‐Wiechmann et al., 2002). The distribution of TN neurons differs between species, and subpopulations of TN neurons and fibers contain GnRH, thus providing a selective marker for TN (von Bartheld, 2004). In postnatal chicks, subpopulations of TN neurons that express FMRFamide and acetylcholinesterase, but not GnRH, are distributed in ganglia or singly along the medial half of the olfactory nerve bundle (Wirsig‐Wiechmann, 1990). The location of GFP‐labeled GnRH‐ or CB‐positive neurons in the nasal region was consistent with that of chick TN demonstrated in this report. Our results indicate that TN GnRH neurons exist in the peripheral olfactory region, which has been previously shown, but are not identified as TN in birds (Mulrenin et al., 1999). In addition, the location of GFP‐labeled cells in the olfactory nerve layer and olfactory bulb regions is similar to that of TN GnRH neurons reported in adult pigeons (Norgren et al., 1992). Moreover, the trajectory of a small number of GFP‐labeled fibers extending to the caudal septum is in accordance with the observation of quail‐chick chimeras with OP transplants at E9.5 (Yamamoto et al., 1996). The OP‐derived fiber tract in the medial forebrain surface corresponds to the central projections of the TN, as described in other species (von Bartheld, 2004; Wirsig‐Wiechmann et al., 2002). Although the developmental origin of the TN is still debated, possibly either OP cells or neural crest‐derived cells (Forni et al., 2011; von Bartheld, 2004; Whitlock, 2004), our findings revealed that chick TN GnRH neurons are derived from the OP.
In summary, this study reports that cells migrating from the OP express SS mRNA, which is mainly associated with GnRH neurons. The long‐lasting OP cell labeling system provides evidence that OP‐derived neurons other than GnRH neurons settle in the medial forebrain. They represent a distribution pattern similar to that of GnRH neurons and express SS mRNA, NPY, CB, or none of these substances, indicating that a diverse population of OP‐derived brain neurons contributes to the neuronal network of the medial forebrain. Additionally, OP‐derived GnRH neurons develop in the peripheral olfactory region. Further studies on the molecular identities and postnatal development of diverse populations of OP‐derived neurons in the forebrain are required to ascertain their possible functions in the adult brain.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/cne.25389.
ACKNOWLEDGMENTS
We thank Dr. Koichi Kawakami for the Tol2‐based vector and Dr. Tatsunori Seki for the PSA‐NCAM antibody. This research was supported by JSPS KAKENHI Grant Number 16K08454 to Shizuko Murakami and the MEXT‐supported Program for the Strategic Research Foundation at Private Universities to Yasuo Uchiyama.
Murakami, S. , Ohki‐Hamazaki, H. , & Uchiyama, Y. (2022). Olfactory placode generates a diverse population of neurons expressing GnRH, somatostatin mRNA, neuropeptide Y, or calbindin in the chick forebrain. Journal of Comparative Neurology, 530, 2977–2993. 10.1002/cne.25389
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Aguillon, R. , Batut, J. , Subramanian, A. , Madelaine, R. , Dufourcq, P. , Schilling, T. F. , & Blader, P. (2018). Cell‐type heterogeneity in the early zebrafish olfactory epithelium is generated from progenitors within preplacodal ectoderm. eLife, 7, e32041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akutsu, S. , Takada, M. , Ohki‐Hamazaki, H. , Murakami, S. , & Arai, Y. (1992). Origin of luteinizing hormone‐releasing hormone (LHRH) neurons in the chick embryo: Effect of the olfactory placode ablation. Neuroscience Letters, 142, 241–244. [DOI] [PubMed] [Google Scholar]
- Airaksinen, M. S. , Eilers, J. , Garaschuk, O. , Thoenen, H. , Konnerth, A. , & Meyer, M. (1997). Ataxia and altered dendritic calcium signaling in mice carrying a targeted null mutation of the calbindin D28k gene. Proceedings of the National Academy of Sciences of the United State of America, 94, 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baimbridge, K. G. , Celio, M. R. , & Rogers, J. H. (1992). Calcium‐binding proteins in the nervous system. Trends in Neuroscience, 15, 303–308. [DOI] [PubMed] [Google Scholar]
- Blähser, S. (1984). Peptidergic pathways in the avian brain. The Journal of Experimental Zoology, 232, 397–403. [DOI] [PubMed] [Google Scholar]
- Blähser, S. , & Heinrichs, M. (1982). Immunoreactive neuropeptide system in avian embryos (domestic mallard, domestic fowl, Japanese quail). Cell and Tissue Research, 223, 287–303. [DOI] [PubMed] [Google Scholar]
- Caqueret, A. , Coumilleau, P. , & Michaud, J. L. (2005). Regionalization of the anterior hypothalamus in the chick embryo. Developmental Dynamics, 233, 652–658. [DOI] [PubMed] [Google Scholar]
- Celio, M. R. (1990). Calbindin D‐28k and parvalbumin in the rat nervous system. Neuroscience, 35, 375–475. [DOI] [PubMed] [Google Scholar]
- Chronwall, B. M. , DiMaggio, D. A. , Massari, V. J. , Pickel, V. M. , Ruggiero, D. A. , & O'Donohue, T. L. (1985). The anatomy of neuropeptide‐Y‐containing neurons in the rat brain. Neuroscience, 15, 1159–1181. [DOI] [PubMed] [Google Scholar]
- De Carlos, J. A. , Lopez‐Mascaraque, L. , & Valverde, F. (1995). The telencephalic vesicles are innervated by olfactory placode‐derived cells: A possible mechanism to induce neocortical development. Neuroscience, 68, 1167–1178. [DOI] [PubMed] [Google Scholar]
- Ding, L. , Takabayashi, H. , Watanabe, K. , Ohtsuki, T. , Tanaka, K. F. , Nabeshima, Y. , Chisaka, O. , Ikenaka, K. , & Ono, K. (2005). Short‐term lineage analysis of dorsally derived Olig3 cells in the developing spinal cord. Developmental Dynamics, 234, 622–632. [DOI] [PubMed] [Google Scholar]
- Drapkin, P. T. , & Silverman, A.‐J. (1999). Development of the chick olfactory nerve. Developmental Dynamics, 214, 349–360. [DOI] [PubMed] [Google Scholar]
- Epelbaum, J. (1986). Somatostatin in the central nervous system: Physiology and pathological modifications. Progress in Neurobiology, 27, 63–100. [DOI] [PubMed] [Google Scholar]
- Esposito, V. , Pelagalli, G. V. , De Girolamo, P. , & Gargiulo, G. (2001). Anatomical distribution of NPY‐like immunoreactivity in the domestic chick brain (Gallus domesticus). The Anatomical Record, 263, 186–201. [DOI] [PubMed] [Google Scholar]
- Fornaro, M. , Geuna, S. , Fasolo, A. , & Giacobini‐Robecchi, M. G. (2001). Evidence of very early neuronal migration from the olfactory placode of the chick embryo. Neuroscience, 10, 191–197. [DOI] [PubMed] [Google Scholar]
- Fornaro, M. , Geuna, S. , Fasolo, A. , & Giacobini‐Robecchi, M. G. (2003). HuC/D confocal imaging points to olfactory migratory cells as the first cell population that expresses a post‐mitotic neuronal phenotype in the chick embryo. Neuroscience, 122, 123–128. [DOI] [PubMed] [Google Scholar]
- Forni, P. E. , & Wray, S. (2015). GnRH, anosmia and hypogonadotropic hypogonadism—Where are we? Frontiers in Neuroendocrinology, 36, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni, P. E. , Taylor‐Burds, C. , Melvin, V. S. , Williams, T. , & Wray, S. (2011). Neural crest and ectodermal cells intermix in the nasal placode to give rise to GnRH‐1 neurons, sensory neurons, and olfactory ensheathing cells. The Journal of Neuroscience, 31, 6915–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Calero, E. , & Puelles, L. (2009). Enc1 expression in the chick telencephalon at intermediate and late stages of development. The Journal of Comparative Neurology, 517, 564–580. [DOI] [PubMed] [Google Scholar]
- Hamburger, C. , & Hamilton, H. L. (1951). A series of normal stages in the development of chick embryo. Journal of Morphology, 88, 49–92. [PubMed] [Google Scholar]
- Hilal, E. M. , Chen, J. H. , & Silverman, A. J. (1996). Joint migration of gonadotropin‐releasing hormone (GnRH) and neuropeptide Y (NPY) neurons from olfactory placode to central nervous system. Journal of Neurobiology, 31, 487–502. [DOI] [PubMed] [Google Scholar]
- Hunter, E. , Begbie, J. , Mason, I. , & Graham, A. (2001). Early development of the mesencephalic trigeminal nucleus. Developmental Dynamics, 222, 484–493. [DOI] [PubMed] [Google Scholar]
- Kawakami, K. , Koga, A. , Hori, H. , & Shima, A. (1998). Excision of the Tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio . Gene, 225, 17–22. [DOI] [PubMed] [Google Scholar]
- Kawakami, K. , & Noda, T. (2004). Trasnposition of the Tol2 element, an Ac‐like element from the Japanese medaka fish Oryzias latipes, in mouse embryonic stem cells. Genetics, 166, 895–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani, T. , Nagayoshi, S. , Urasaki, A. , & Kawakami, K. (2006). Transposon‐mediated gene trapping in zebrafish. Methods, 39, 199–206. [DOI] [PubMed] [Google Scholar]
- Kuenzel, W. J. , & Blähser, S. (1991). The distribution of gonadotropin‐releasing hormone (GnRH) neurons and fibers throughout the chick brain (Gallus domesticus). Cell and Tissue Research, 264, 481–495. [DOI] [PubMed] [Google Scholar]
- Kuenzel, W. J. , & Masson, M. (1988). A stereotaxic atlas of the brain of the chick (Gallus domesticus). Johns Hopkins University Press. [Google Scholar]
- Louvi, A. , Yoshida, M. , & Grove, E. A. (2007). The derivatives of the Wnt3a lineage in the central nervous system. The Journal of Comparative Neurology, 504, 550–569. [DOI] [PubMed] [Google Scholar]
- Mendoza, A. S. , Breipohl, W. , & Miragall, F. (1982). Cell migration from the chick olfactory placode: A light and electron microscopic study. Journal of Embryology and Experimental Morphology, 69, 47–59. [PubMed] [Google Scholar]
- Mezey, S. , Krivokuca, D. , Balint, E. , Adorjan, A. , Zachar, G. , & Csillag, A. (2012). Postnatal changes in the distribution and density of neuronal nuclei and doublecortin antigens in domestic chicks (Gallus domesticus). The Journal of Comparative Neurology, 520, 100–116. [DOI] [PubMed] [Google Scholar]
- Mikami, S. , & Yamada, S. (1984). Immunohistochemistry of the hypothalamic neuropeptides and anterior pituitary cells in the Japanese quail. The Journal of Experimental Zoology, 232, 405–417. [DOI] [PubMed] [Google Scholar]
- Miller, A. M. , Treloar, H. B. , & Greer, C. A. (2010). Composition of the migratory mass during development of the olfactory nerve. The Journal of Comparative Neurology, 518, 4825–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa, M. , Seki, T. , & Arai, Y. (1997). Supportive role of cellular bridge of neurons expressing a highly polysialylated form of NCAM (NCAM‐H) at the initial stage of migration of LHRH neurons. Zoological Science, 14, 489–495. [Google Scholar]
- Mullen, R. J. , Buck, C. R. , & Smith, A. M. (1992). NeuN, a neuronal specific nuclear protein in vertebrates. Development, 116, 201–211. [DOI] [PubMed] [Google Scholar]
- Mulrenin, E. M. , Witkin, J. W. , & Silverman, A. J. (1999). Embryonic development of the gonadotropin‐releasing hormone (GnRH) system in the chick: A spatio‐temporal analysis of GnRH neuronal generation, site of origin, and migration. Endocrinology, 140, 422–433. [DOI] [PubMed] [Google Scholar]
- Murakami, S. , & Arai, Y. (1994a). Direct evidence for the migration of LHRH neurons from the nasal region to the forebrain in the chick embryo: A carbocyanine dye analysis. Neuroscience Research, 19, 331–338. [DOI] [PubMed] [Google Scholar]
- Murakami, S. , & Arai, Y. (1994b). Transient expression of somatostatin immunoreactivity in the olfactory‐forebrain region in the chic embryo. Brain Research, Developmental Brain Research, 82, 277–285. [DOI] [PubMed] [Google Scholar]
- Murakami, S. , Ohki‐Hamazaki, H. , Watanabe, K. , Ikenaka, K. , & Ono, K. (2010). Netrin 1 provides a chemoattractive cue for the ventral migration of GnRH neurons in the chick forebrain. The Journal of Comparative Neurology, 518, 2019–2034. [DOI] [PubMed] [Google Scholar]
- Murakami, S. , Seki, T. , Rutishauser, U. , & Arai, Y. (2000). Enzymatic removal of polysialic acid from neural cell adhesion molecule perturbs the migration route of luteinizing hormone‐releasing hormone neurons in the developing chick forebrain. The Journal of Comparative Neurology, 420, 171–181. [DOI] [PubMed] [Google Scholar]
- Murakami, S. , Seki, T. , Wakabayashi, K. , & Arai, Y. (1991). The ontogeny of luteinizing hormone‐releasing hormone (LHRH) producing neurons in the chick embryo: Possible evidence for migrating LHRH neurons from the olfactory epithelium expressing a highly polysialylated neural cell adhesion molecule. Neuroscience Research, 12, 421–431. [DOI] [PubMed] [Google Scholar]
- Narayanan, C. H. , & Narayanan, Y. (1978). Determination of the embryonic origin of the mesencephalic nucleus of the trigeminal nerve in birds. Journal of Embryology and Experimental Morphology, 43, 85–105. [PubMed] [Google Scholar]
- Norgren, R. B. , & Branckenbury, R. (1993). Cell adhesion molecules and the migration of LHRH neurons during development. Developmental Biology, 160, 377–387. [DOI] [PubMed] [Google Scholar]
- Norgren, R. B. , Lippert, J. , & Nehman, M. N. (1992). Luteinizing hormone‐releasing hormone in the pigeon terminal nerve and olfactory bulb. Neuroscience Letters, 135, 201–204. [DOI] [PubMed] [Google Scholar]
- Palaniappan, T. K. , Sleikiene, L. , Gunhaga, L. , & Pattey, C. (2019). Extensive apoptosis during the formation of the terminal nerve ganglion by olfactory placode‐derived cells with distinct molecular markers. Differentiation, 110, 8–16. [DOI] [PubMed] [Google Scholar]
- Pellier, V. , Astic, L. , Oestreicher, A. B. , & Saucier, D. (1994). B50/GAP‐43 expression by the olfactory receptor cells and the neurons migrating from the olfactory placode in embryonic rats. Brain Research, Developmental Brain Research, 80, 63–72. [DOI] [PubMed] [Google Scholar]
- Poopalasundaram, S. , Chambers, D. , Graham, A. , & Bouloux, P. M. (2016). Serotonin receptor 1A (HTR1A), a novel regulator of GnRH neuronal migration in chick embryo. Endocrinology, 157, 4632–4640. [DOI] [PubMed] [Google Scholar]
- Puelles, L. , Martínez‐de‐la‐Torre, M. , Paxinos, G. , Watson, C. , & Martínez, S. (2007). The chick brain in stereotaxic coordinates. Academic Press. [Google Scholar]
- Quintana‐Urzainqui, I. , Rodriguez‐Moldes, I. , & Candal, E. (2014). Developmental, tract‐tracing and immunohistochemical study of the peripheral olfactory system in a basal vertebrate: Insights on Pax6 neurons migrating along the olfactory nerve. Brain Structure & Function, 219, 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J. H. , & Resibois, A. (1992). Calretinin and calbindin‐D28k in rat brain: Patterns of partial co‐localization. Neuroscience, 51, 843–865. [DOI] [PubMed] [Google Scholar]
- Roth, J. , Baetens, D. , Norman, A. W. , & Garcia‐Segura, L. (1981). Specific neurons in chick central nervous system stain with an antibody against chick intestinal vitamin D‐dependent calcium‐binding protein. Brain Research, 222, 452–457. [DOI] [PubMed] [Google Scholar]
- Sato, C. , Kitajima, K. , Inoue, S. , Seki, T. , Troy, F. A. , & Inoue, Y. (1995). Characterization of the antigenic specificity of four different anti‐(alpha 2→8‐linked polysialic acid) antibodies using lipid‐conjugated oligo/polysialic acids. Journal of Biological Chemistry, 270, 18923–18928. [DOI] [PubMed] [Google Scholar]
- Sato, Y. , Kasai, T. , Nakagawa, S. , Tanabe, K. , Watanabe, T. , Kawakami, K. , & Takahashi, Y. (2007). Stable integration and conditional expression of electroporated transgene in chicken embryos. Developmental Biology, 305, 616–624. [DOI] [PubMed] [Google Scholar]
- Schwanzel‐Fukuda, M. , & Pfaff, D. W. (1989). Origin of luteinizing hormone‐releasing hormone neurons. Nature, 338, 161–164. [DOI] [PubMed] [Google Scholar]
- Seki, T. (2002). Expression patterns of immature neuronal markers PSA‐NCAM, CRMP‐4 and NeuroD in the hippocampus of young adult and aged rodents. Journal of Neuroscience Research, 70, 327–334. [DOI] [PubMed] [Google Scholar]
- Seki, T. , & Arai, Y. (1991). The persistent expression of a highly polysialylated NCAM in the dentate gyrus of the adult rat. Neuroscience Research, 12, 503–513. [DOI] [PubMed] [Google Scholar]
- Shan, Y. , Saadi, H. , & Wray, S. (2020). Heterogeneous origin of gonadotropin releasing hormone‐1 neurons in mouse embryos detected by Islet‐1/2 expression. Frontiers in Cell and Developmental Biology, 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman, A. J. , Cserjesi, P. , & Kanter, E. (2002). Distribution of gonadotropin‐releasing hormone neurons in the chick forebrain is independent of lineage relationships among cells of the early nasal placode. Journal of Neuroendocrinology, 14, 207–212. [DOI] [PubMed] [Google Scholar]
- Silverman, A. J. , Livne, I. , & Witkin, J. W. (1994). The gonadotropin‐releasing hormone (GnRH) neuronal systems: Immunocytochemistry and in situ hybridization. In Knobil, E. & Neil, J. D. (Eds.). The physiology of reproduction (2nd ed., pp. 1683–1709). Raven Press Ltd. [Google Scholar]
- Stainier, D. Y. R. , Rilder, D. H. , & Gilbert, W. (1991). The B30 ganglioside is a cell surface marker for neural crest‐derived neurons in the developing mouse. Developmental Biology, 144, 177–188. [DOI] [PubMed] [Google Scholar]
- Sullivan, K. A. , & Silverman, A.‐J. (1993). The ontogeny of gonadotropin‐releasing hormone neurons in the chick. Neuroendocrinology, 58, 597–608. [DOI] [PubMed] [Google Scholar]
- Takatsuki, K. , Shiosaka, S. , Inagaki, S. , Sakanaka, M. , Takagi, H. , Senba, E. , Matsuzaki, T. , & Tohyama, M. (1981). Topographic atlas of somatostatin‐containing neurons system in the avian brain in relation to catecholamine‐containing neurons system. I. Telencephalon and diencephalon. The Journal of Comparative Neurology, 202, 103–113. [DOI] [PubMed] [Google Scholar]
- Taroc, E. G. M. , Katreddi, R. R. , & Foni, P. E. (2020). Identifying Isl1 genetic lineage in the developing olfactory system and in GnRH‐1 neurons. Frontiers in Physiology, 11, 601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrozo, G. , Peretto, P. , & Fasolo, A. (1995). Cell migration from the olfactory placode and the ontogeny of the neuroendocrine compartments. Zoological Science, 12, 367–383. [DOI] [PubMed] [Google Scholar]
- Toba, Y. , Ajiki, K. , Horie, M. , Sango, K. , & Kawano, H. (2001). Immunohistochemical localization of calbindin D‐28k in the migratory pathway from the rat olfactory placode. Journal of Neuroendocrinology, 13, 683–694. [DOI] [PubMed] [Google Scholar]
- Tobet, S. A. , Chickering, T. W. , King, J. C. , Stopa, E. G. , Kim, K. , Kuo‐Leblank, V. , & Schwarting, G. A. (1996). Expression of gamma‐aminobutyric acid and gonadotropin‐releasing hormone during neuronal migration through the olfactory system. Endocrinology, 137, 5415–5420. [DOI] [PubMed] [Google Scholar]
- Urasaki, A. , Morvan, G. , & Kawakami, K. (2006). Functional dissection of the Tol2 transposable element identified the minimal cis‐sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics, 174, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, F. , Heredia, M. , & Santacana, M. (1993). Characterization of neuronal cell varieties migrating from the olfactory epithelium during prenatal development in the rat: Immunocytochemical study using antibodies against olfactory marker protein (OMP) and luteinizing hormone‐releasing hormone (LHRH). Brain Research, Developmental Brain Research, 71, 209–220. [DOI] [PubMed] [Google Scholar]
- Verney, C. , Amraoui, A. I. , & Zecevic, N. (1996). Comigration of tyrosine hydroxylase‐ and gonadotropin‐releasing hormone‐immunoreactive neurons in the nasal area of human embryos. Brain Research, Developmental Brain Research, 97, 251–259. [DOI] [PubMed] [Google Scholar]
- Vilensky, J. A. (2014). The neglected cranial nerve: Nervus terminalis (cranial nerve N). Clinical Anatomy, 27, 46–53. [DOI] [PubMed] [Google Scholar]
- Viollet, C. , Lepousez, G. , Loudes, C. , Videau, C. , Simon, A. , & Epelbaum, J. (2008). Somatostainergic system in brain: Networks and functions. Molecular and Cellular Endocrinology, 286, 75–87. [DOI] [PubMed] [Google Scholar]
- von Bartheld, C. S. (2004). The terminal nerve and its relation with extrabulbar “olfactory” projections: Lessons from lampreys and lungfish. Microscopy Research and Technique, 65, 13–24. [DOI] [PubMed] [Google Scholar]
- von Bohlen und Halbach, O. (2007). Immunohistological markers for staging neurogenesis in adult hippocampus. Cell and Tissue Research, 329, 409–420. [DOI] [PubMed] [Google Scholar]
- Weyer, A. , & Schilling, K. (2003). Developmental and cell type‐specific expression of the neuronal marker NeuN in the murine cerebellum. Journal of Neuroscience Research, 73, 400–409. [DOI] [PubMed] [Google Scholar]
- Whitlock, K. E. (2004). Development of the nervus terminalis: Origin and migration. Microscopy Research and Technique, 65, 2–12. [DOI] [PubMed] [Google Scholar]
- Wirsig‐Wiechmann, C. R. (1990). The nervus terminalis in the chick: A FMRFamide‐immunoreactive and AChE‐positive nerve. Brain Research, 523, 175–179. [DOI] [PubMed] [Google Scholar]
- Wirsig‐Wiechmann, C. R. , Wiechmann, A. F. , & Eisthen, H. L. (2002). What defines the nervus terminalis? Neurochemical, developmental, and anatomical criteria. Progress in Brain Research, 141, 45–58. [DOI] [PubMed] [Google Scholar]
- Wray, S. (2010). From nose to brain: Development of gonadotropin‐releasing hormone‐1 neurons. Journal of Neuroendocrinology, 22, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, S. , Grant, P. , & Gainer, F. (1989). Evidence that cells expressing luteinizing hormone‐releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proceedings of the National Academy of Sciences of the United State of America, 86, 8132–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, N. , Uchiyama, H. , Ohki‐Hamazaki, H. , Tanaka, H. , & Ito, H. (1996). Migration of GnRH‐immunoreactive neurons from the olfactory placode to the brain: A study using avian embryonic chimeras. Brain Research, Developmental Brain Research, 95, 234–244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


