Abstract
While still in its infancy, the application of deep convolutional neural networks in veterinary diagnostic imaging is a rapidly growing field. The preferred deep learning architecture to be employed is convolutional neural networks, as these provide the structure preferably used for the analysis of medical images. With this retrospective exploratory study, the applicability of such networks for the task of delineating certain organs with respect to their surrounding tissues was tested. More precisely, a deep convolutional neural network was trained to segment medial retropharyngeal lymph nodes in a study dataset consisting of CT scans of canine heads. With a limited dataset of 40 patients, the network in conjunction with image augmentation techniques achieved an intersection‐overunion of overall fair performance (median 39%, 25 percentiles at 22%, 75 percentiles at 51%). The results indicate that these architectures can indeed be trained to segment anatomic structures in anatomically complicated and breed‐related variating areas such as the head, possibly even using just small training sets. As these conditions are quite common in veterinary medical imaging, all routines were published as an open‐source Python package with the hope of simplifying future research projects in the community.
Keywords: artificial intelligence, automated segmentation, canine head
Abbreviations
- DCNN
deep convolutional neural network
- ECVDI
European College of Veterinary Diagnostic and Imaging
1. INTRODUCTION
Machine learning algorithms employing deep neural networks can solve problems that previously have been out of reach for any other computer system and as such have profoundly changed the technical world in the past decade. These techniques are starting to enter the medical world as well, and veterinary science is no exception in that. Of special importance for applications in medical radiology are so‐called deep convolutional neural networks (DCNNs). Compared to standard deep neural networks, these architectures explicitly use the fact that images are depicting spatial objects and thus pixels close to each other typically describe similar objects. Mathematically speaking, convolutions are a technique to extract “features”—more abstract descriptions of elements—out of an image, which basically contain the relevant information about the image. A convolutional neural network is a composition of several of these mathematical algorithms, interchanged with steps to reduce the information even more to the relevant parts, typically some kind of averaging over certain regions in the image. The final steps depend on the target application: “classification” or “segmentation.” In a “classification” task, the images shall be assigned a “label” such as deciding between healthy or diseased patients and ordering such as a tumor grade or even a differential diagnosis out of a finite set of possibilities.
In “segmentation” mode, a DCNN can identify regions of special importance in an image and provide this information back to the user. The associated architecture of these networks varies. A commonly used network is made from two convolutional neural networks: one for extracting the “features” and a “reversed” network for detecting the regions of interest based on those features. A particular example of such an algorithm is the U‐Net architecture. 1 A prototypical manual “segmentation” task in medical imaging is to contour tumor volumes and organs‐at‐risk, with the aim of correct differentiation between normal organ(s) and lesion(s). 2 In the head and neck area, anatomically close relationships among normal organs, organs at risk and tumor tissue are routinely encountered. Tumors of the head and neck are a commonly encountered disease in canine patients and are known for their high metastatic potential to the locoregional lymphnodes. 3 , 4 , 5 , 6 Radiation therapy is often the treatment of choice for these types of tumors, and the challenges while creating treatment plans are the same as in human medicine. Precise contouring of the lesion(s) and the organs‐at‐risk is a time‐consuming task and prone to intra‐ and interobserver variability and is currently manually performed with the help of commercially available treatment planning systems. 7 , 8 Studies in human medicine are exploring the possibility of deep learning‐based technology to be used in fully automated delineation of anatomic structures in the head and neck region in medical images and are showing promising results. 9 , 10 , 11 , 12 , 13 While the classification task of convolutional neural networks has already been used in some fields of veterinary imaging in the near past, veterinary literature dealing with the segmentation ability of DCNNs is scarce. 14 , 15 , 16 , 17 , 18 , 19 , 20 Only one other recently published study describes the implementation of deep learning‐based approaches to segment organs such as the eye or salivary glands in canine patients. 21
The authors of the current study considered contour delineation of the medial retropharyngeal lymph nodes in contrast‐enhanced CT studies of canine heads as a well‐suited topic for examining the potential of “a fully automated” algorithm‐based approach for segmentation of anatomic structures in the head and neck region. The objective of the current study was therefore to determine the diagnostic performance of a deep convolutional neural network in segmenting the medial retropharyngeal lymph nodes in dogs. The working hypothesis was that the network's segmentation performance would be able to identify the contours of the medial retropharyngeal lymph nodes similar to manually placed contours from veterinary radiologists.
2. MATERIALS AND METHODS
2.1. Study design
The current study was designed as a retrospective exploratory study. The aim was to show that automated, deep learning‐based image segmentation is possible with Veterinary Data given the constraints of limited availability of labeled original data (lymph nodes in this case) and varying head sizes. If the network would be successfully delineating and segmenting the medial retropharyngeal lymph nodes from their surroundings, the future role of this automated approach would have potential for its use in organs at risk delineation for radiation therapy planning.
2.2. Data collection
The data collection for this project was performed in two steps. The first step consisted of reviewing the imaging database of the Vetsuisse Faculty Zurich for CT examinations of the head of canine patients with the help of a keyword search termed “medial retropharyngeal lymph node(s)” in the years 2017–2020. Taking the results of this database query, two of the authors (D.S. and I.L.) randomly selected 30 radiology reports of dogs in which a European College of Veterinary Diagnostic and Imaging (ECVDI)–certified veterinary radiologist had described both medial retropharyngeal lymph nodes as small and normal in size in accordance with available literature. 22 An additional 10 reports of dogs were selected by the same two authors in which at least one of the medial retropharyngeal lymph nodes had been described as moderately enlarged. The database query provided only basic information about the species of the listed animals, which helped to differentiate dogs and cats for patient selection. Detailed information about the signalment was not included in the initial database query. Query results did, however, include the patient clinical identification number. With the help of this number, additional information about age and breed could be identified at the end of the data collection process. In the second step, the contrast‐enhanced soft tissue series of the 40 patients was separated from the native soft tissue and bone window reconstruction series and manually imported into free‐available DICOM viewer software (HOROS; version 2.1.1; free, open‐source medical image viewer; www.horosproject.org). As the data used were generated during routine patient workup, approval from the animal welfare committee was not needed. The use of data was in accordance with the hospital director.
2.3. Ground truth determination
The same two authors who had performed the report selection proceeded with building the ground truth dataset. One of the evaluators was an ECVDI imaging resident (D.S.), and the other evaluator was an ECVDI‐certified veterinary radiologist (I.L.). Both evaluators were familiar with the DICOM viewer software used. Ten of the normal series were evaluated by the veterinary radiologist in a first step to test the applicability of the contouring tools provided in the image analysis software (HOROS). The imaging resident then completed the 30 normal series and evaluated the 10 cases with moderately enlarged medial retropharyngeal lymph nodes.
The two evaluators performed the contour delineation independently by manually setting ROIs, starting with the first slide in which the medial retropharyngeal lymph node became visible and continuing until the last slide with a visible part of the respective lymph node. The ROIs were set around the right and left lymph nodes. If there were concerns about precise contouring in a specific dataset, the evaluators would sit together and find a consensus. Once the annotations were completed, the subset of images containing the lymph nodes and ROIs were manually separated from the complete contrast‐enhanced series. This preprocessing step was performed to reduce the number of input images for the convolutional neural network.
Preprocessing also included the export of the collected ROIs to text files in comma separated values format (CSV), a procedure supported using standard tools of the viewer software. Those CSV files contained the ground truth in the form of pixel coordinates, which represent the edges of a polygon surrounding the lymph node. To train a convolutional neural network, a “mask” encoding this information on a pixel‐to‐pixel basis was needed for each image. Hence, the CSV files were processed to give 0/1 valued image arrays, with the value 1 indicating that the associated pixel lies inside the ROI, and 0 otherwise. As typical convolutional neural networks expect the pixel value within the image to be within 0 to 1, the DICOM images were preprocessed to extract the image data, and a soft‐tissue window‐like transformation was applied to scale each pixel value to that range. The aforementioned computational preprocessing as well as the model training were conducted by a data scientist (V.S.) with more than 20 years of experience.
2.4. Data partition
The training of convolutional neural networks usually requires an ample number of labeled images. As described above, the procedure to get to those is quite time‐consuming and involves well‐trained radiologists, which of course can only devote a limited amount of time to these tasks. It is hence of paramount importance to use the available image set as efficiently as possible. A standard method to better exploit a given set of ground truth data is to expand the image set by generating more images in addition to the given images. For example, take an image and slightly rotate or shift it. This has the additional benefit that the neural network will not be biased toward a fixed rotation axis while at the same time enlarging the dataset. Following these thoughts, the images of each patient were grouped into 10 consecutive slices (with respect to the z‐dimension) to retain some spatial information in that dimension. Next, for each of such a batch of images, two angles between −20° and 20° were chosen at random, and the same corresponding rotation was applied to all images in the same batch. With the original batch kept, the dataset was thus enlarged by a factor of three. At the end of this procedure, the dataset consisted of batches of 10 consecutive images. The collection of these batches was then split at random into three nonoverlapping sets with a split ratio of 80:10:10. The procedure implied that no batch of images was in more than one set. However, an original batch and one of its rotated versions might well end up in different sets. The same statement thus also amounts to the images of a patient—they might end up in different randomly chosen groups. Due to hardware constraints, the number of pixels in each image finally needed to be reduced. Downsampling the images by max‐pooling with a factor of three (taking the largest value in each three‐by‐three‐pixel square) was chosen, which is an averaging technique often used when training convolutional neural networks.
2.5. Model structure
A special type of convolutional neural network, the so‐called U‐Net architecture, was used. 1 Its name stems from the fact that its shape can be roughly pictured as an “U,” with several convolutions followed by averaging operations reducing the images in size more and more (the downward path of an “U”) while at the same time extracting more and more features. This is followed by many reverse convolutions (also called transposed convolutions in mathematical terms), which again enlarge the image increasingly, thereby combining the features into masks of increasing size (the upward path of the “U”). The exact structure can be examined using the source code, which is publicly available, as discussed in more detail later.
2.6. Model training
The training of such a convolutional neural network architecture usually requires two main steps. First, the optimal way to extract the features and combine them into masks needs to be found—this corresponds to finding the best set of convolutions. For this, a standard algorithm called Adam was used. 23 This algorithm finds in each step the convolutions that currently perform worse compared to others and updates them accordingly. Technically speaking, the cross entropy was used as a loss function, and a minibatch size of 10 images per batch was chosen. Second, the hyperparameters of such an architecture need to be adjusted, for example, how “fast” it is trained (technically called the learning rate). More precisely, the specific type of model architecture requires optimizing the dropout rate of the convolutional layers, which roughly corresponds to the fact that the network is forced to “forget” some of its learning from time to time during training. It is obvious that the same datasets should not be used for both steps, as otherwise they might influence each other. More importantly, to see the effects when changing, e.g., the learning rate, the convolutions need to be trained first with a particular choice for these parameters.
Technically, the procedure of adjusting these additional variables is called “hyperparameter optimization.” A new technique for this task was invented by a team at Google Brains and is called “population‐based training” (PBT). 24 In rough terms, it switches in an intelligent way between adjusting the convolutions and the hyperparameters such as the learning rate. The training procedure involved 2000 steps, where in each of those, a batch of roughly 100 images was used to update the convolutions followed by eventual updates of the hyperparameters based on the validation dataset.
2.7. Evaluation of network performance
For such a setup to work out, a quick judgment of whether a given configuration of convolutions and learning rates works well or not is essential. Technically speaking, such a quantity is called a “validation metric”; a single number tells how the network is performing. A well‐established one for segmentation tasks is the “intersection‐overuunion” (IoU), which measures how well the predicted mask corresponds to the true mask by computing the ratio of their intersection in relation to their union (see Figure 1). This also explains the threefold split into the training, validation, and test datasets: for updating the convolutions, the training dataset is used while the hyperparameters are adjusted in regard to the validation dataset—the test dataset is not used at all within training and only used to judge the final result. Throughout this study, the same metric for assessing results was used, which implies that the performance statements are all with respect to the IoU metric.
FIGURE 1.
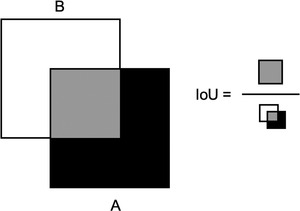
Intersection over UnionGray box represents the area of intersection, while the white and black boxes show the areas of union
2.8. Implementation and hardware resources
In principle, all steps described above can now be considered standards in the artificial intelligence community. However, their application to a particular project can often be time‐consuming by themselves and is also known to be error prone. To simplify future research ideas of the veterinary community as well as by themselves, the authors decided to bundle the technical steps into an open‐source Python package, which is now available on GitHub (https://github.com/volkherscholz/dicomml) and free to use for everybody. It expects as an input only the original DICOM images as well as the associated CSV files with the edge coordinates of the polygons surrounding the regions of interest. The authors hope that by utilizing this package, their colleagues will be enabled to perform their own studies regarding segmentation tasks.
Training a convolutional neural network architecture requires dedicated hardware resources involving specialized graphical processing units (GPUs). The authors are thankful to the University of Zurich Science Cloud for providing them with a virtual machine with 8 CPU cores, 32 GB of memory, and a Tesla T4 GPU attached. In utilizing these hardware resources, standard deployment tools (also known as containerization) known from cloud environments were applied.
3. RESULTS
3.1. Data
The examinations were performed with a 16‐slice CT scanner (Brilliance 16, Philips Medical Systems, Best, The Netherlands) with the dogs under general anesthesia and placed in sternal recumbency. The scanning parameters were set at 120 kVp, 280 mA, 1.0 s rotation time, 0.683 pitch, and a slice thickness of 1.5 mm with an increment of 0.75 mm. The field of view was adjusted to cover the head and the neck and collimated to the individual size of each patient. Iodinated contrast medium (2 ml/kg iv; Accupaque™ 350; GE Healthcare, Glattbrugg, Switzerland) was administered using a programmable injector (Accutron CT‐ D Medtron, Injector, SMD Medical Trade GmbH, Salenstein, Switzerland) through a peripheral venous catheter. The contrast‐enhanced images were acquired after a delay of 90 s. Analysis of the dataset revealed that the patients included in this study were adult, pure and mixed breed dogs with all types of skull conformation.
3.2. Model performance
The training procedure took approximately 12 h on the hardware described above. The best model achieved an average intersection over union of 36% ± 20% on the test dataset (median 39%, 25 percentiles at 22%, 75 percentiles at 51%).
Several examples to illustrate the high variation in performance of the resulting deep convolutional neural network are provided. Figure 2 shows images of a patient with asymmetry of the medial retropharyngeal lymph nodes for which the neural network performed the segmentation of the contours of the lymph nodes similar to their original appearance in the CT images. In comparison, Figure 3 shows images of another dog of a different breed and therefore different head conformations in which the neural network segmented the pixels containing the medial retropharyngeal lymph nodes but also included the immediate adjacent carotid artery as well as the medial contour of the right mandibular salivary gland on the right side. Performance can even vary within a patient, as illustrated in Figure 4, which shows consecutive images of the same dog, as shown in Figure 3. U‐Net performed more precise segmentation of the pixels compatible with the medial retropharyngeal lymph nodes from the surrounding structures at this more caudal location in the retropharyngeal area. Figure 5 shows the original contrast‐enhanced CT image as well as the input and corresponding output images created by U‐Net of a third, middle aged dog in which both medial retropharyngeal lymph nodes were of small size. The output image shows a mismatch of the contours between the original image and the pixels identified by the neural network as being the medial retropharyngeal lymph nodes.
FIGURE 2.
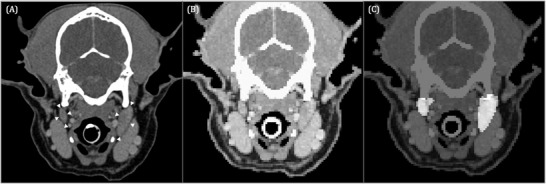
Transverse contrast‐enhanced CT image of a 9‐year‐old Shetland Sheepdog acquired in sternal recumbency (window width = 450, level = 140, 120 kVp, 280 mA) (A) at the level of the paracondylar processes showing the medial retropharyngeal lymph nodes (white arrowheads). Input image for U‐Net (B) and corresponding output image (C). The pixels identified by U‐Net as medial retropharyngeal lymph nodes are highlighted in image (C). Left in the images is right in the dog
FIGURE 3.
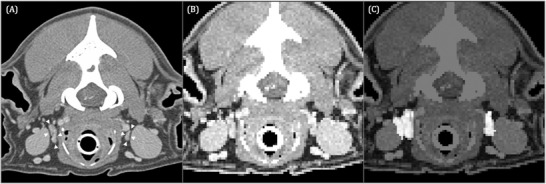
Transverse contrast‐enhanced CT image acquired in sternal recumbency (window width = 450, level = 140, 120 kVp, 280 mA) at the level of the atlanto‐occipital transition of a 12‐year‐old Golden Retriever (A) showing the medial retropharyngeal lymph nodes (white arrowheads). Input image for U‐Net (B) and corresponding output image (C). The pixels identified by U‐Net as medial retropharyngeal lymph nodes are highlighted in image (C). Image (C) shows inaccurate segmentation of the right medial retropharyngeal lymph node as the adjacent carotid artery, and the medial contour of the right mandibular salivary gland is highlighted as well
FIGURE 4.
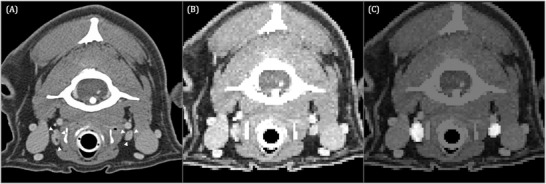
Transverse contrast‐enhanced CT image acquired in sternal recumbency (window width = 450, level = 140, 120 kVp, 280 mA) (A) at the level of the caudal atlas of the same dog shown in Figure 3 depicting the medial retropharyngeal lymph nodes (white arrowheads). Input image for U‐Net (B) and corresponding output image (C). The pixels identified by U‐Net as medial retropharyngeal lymph nodes are highlighted in image (C)
FIGURE 5.
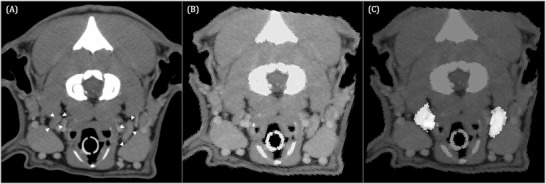
Transverse contrast‐enhanced CT image acquired in sternal recumbency (window width = 450, level = 140, 120 kVp, 280 mA) of a 5‐year‐old Boxer (A) at the level of the atlanto‐occipital transition showing the medial retropharyngeal lymph nodes (white arrowheads). Input image for U‐Net (B) and corresponding output image (C). Image (C) shows the mismatch of the contours between the original image and the pixels identified by U‐Net as being the medial retropharyngeal lymph nodes
4. DISCUSSION
The presented model reached a mean IoU of 36%, which is generally considered suboptimal performance given that an IoU score of 50% achieved by an image segmentation model for its prediction performance on the testing dataset is considered good performance. 25 In the authors` opinion, it still yielded acceptable results considering the circumstances of being one of the first models trained for automated image segmentation in veterinary medicine. The authors refer in that context to their example shown in Figure 3, which clearly shows a dissatisfactory performance of the model as contouring of the lateral margin of the right medial retropharyngeal lymph node was imprecise in the output image. However, as the remaining aspects of the lymph node were more exactly contoured by the network, the pixels highlighted in the output image as corresponding to the lymph node would still provide a first impression of the lymph node`s size and would help with visual organ delineation in a clinical setup.
This study was conducted with CT examinations of the head and neck of dogs, as this imaging modality provides the best insight into pathologic processes in these body areas and serves as the main imaging modality for the conception of RT plans. 26 , 27 , 28 There were several reasons for the selection of the medial retropharyngeal lymph nodes as topics for this pilot study.
This group of lymph nodes builds the largest lymph center of the head and neck 29 and as such acts as sentinel lymph nodes in neoplasms in these body regions. 30 , 31 Their size, shape, and appearance are important features in terms of cancer staging as well as assessment of the effectiveness of the selected radiation or chemotherapy treatment. 32 Their anatomy with their inherent relatively poor contrast to their surrounding tissues on CT slices as well as their variable appearance and boundaries on successive slices make the medial retropharyngeal lymph nodes a representative basic model for future tumor segmentation.
Contrast‐enhanced CT scans were chosen since the presence of contrast improves medial retropharyngeal lymph node visualization compared to noncontrast‐enhanced CT scans and because of the use of contrast‐enhanced scans for the conception of radiotherapy plans in oncologic patients. 33 With the far‐reaching goal of employing deep convolutional neural networks for this task entirely, it appeared logical to test whether such algorithms can successfully delineate the medial retropharyngeal lymph nodes.
Within veterinary medicine, the present study is one of the first to apply deep convolutional neural networks for image segmentation tasks as opposed to classification tasks. As described already in the introduction, one of the main working fields for automated image segmentation algorithms can very well be based within the field of veterinary radiation oncology. The emerging benefits of deep convolutional neural networks for automatic delineation and segmentation of either the OAR or the neoplastic masses, respectively, would probably help to overcome the known limitations of observer‐related variations and facilitate the tedious work of manual delineation of these structures and therefore optimize the workflow to conceive radiotherapy treatment (RT) plans. 2 , 26 , 34 In addition, in the future, AI‐based tools could also help with registration and segmentation of neoplastic masses and metastases, compare these to normal images and help with facilitating the labor‐intensive work of manually measuring and plotting detected size changes against the time and normal anatomy in follow‐up examinations. 34
Several reasons could potentially contribute to the lower performance of the network. First, the images had to be downsampled by a factor of three due to hardware constraints, which led to a much lower resolution. The lower resolution and relatively small number of pixels showing the medial retropharyngeal lymph nodes result in high sensitivity of the model to misidentification of the pixels resembling lymph node parenchyma. More precisely, the misclassification of even one or a few pixels to be within or not within the ROI can result in a degraded IoU if the total pixel count in the entire ROI is small. In contrast, a higher resolution image and thus an ROI with more pixels in total is less sensitive with regard to a few incorrectly predicted pixels. The main limiting factor here was the availability of GPUs. With the hardware configuration used, the hyperparameter optimization was only possible if the image size was restricted to 128 × 128 pixels per image. It is technically possible to combine several GPUs to push these boundaries, but this was not possible in the present setup due to the limited availability of such devices. Second, the dataset was based on only forty patients, and data augmentation techniques can only mitigate this bottleneck to some degree. Third, inherent to veterinary medicine, the head sizes of patients differ due to variation in breed, which constitutes a major challenge for neural networks. In particular, the variable relation of the medial retropharyngeal lymph nodes to their surrounding structures, such as the medial extending common carotid artery and branches, the dorsal adjacent bones and muscles and the lateral located mandibular salivary gland, is assumed to have a significant impact on their correct delineation. Nevertheless, the trained network achieved reasonable predictions on most images and was able to detect major clinical changes.
This study could well suffer from multiple causes of biases, the effects of which were considered and mitigated as much as possible by the authors. Due to the limited dataset to begin with, a selection bias cannot be ruled out, especially as all studies were conducted using the same imaging equipment at the same department. To counteract this initial bias, the authors took a random subset from the initial query result. However, with only 40 patients in total, it is well possible that good randomization was not achieved, especially as the distribution of normal to diseased patients was skewed (30 normal patients versus 10 diseased patients). To reduce the influence of the small sample size, the authors chose to split each patient's images into groups of 10 images each, which were then randomly rotated, and each such batch of images was then assigned by chance to one of the three datasets (training, validation, test). Another source of bias is due to hardware constraints – because of limited resources, the batch size per step of stochastic gradient descent was rather small and thus did not constitute a good random subset, which is also known to lead to suboptimal training results. In addition, and independent of the exact implementation, the fact that only two veterinary radiologists were annotating the images is almost certainly a possible source of rater bias. The authors tried to mitigate this fact by comparing their annotations and making sure that they are comparable, but of course this does not completely rule out a rater bias. The small number of patients did not allow a meaningful statistical analysis of inter‐ or intrarater agreement, a limitation that the authors hope to address in future studies. In summary, most possible causes of bias can be traced back to either the small sample size or the hardware constraints, and both such limitations should and hopefully will be lifted as the use of deep learning advances within veterinary medicine.
Future research steps, such as the extension of the hardware resources as well as the initial datasets, are required to improve the accuracy of the presented deep convolutional neural network. Of course, much further work is still needed before one can hope to use convolutional neural networks in clinical conditions for the automated conception of radiotherapy plans. In summary, this study constitutes a complementary step in that direction by illustrating the possibility of using a deep convolutional neural network for the segmentation of lymph nodes in canine patients and is in line with various other promising research projects describing the use of techniques from artificial intelligence in veterinary medicine.
5. LIST OF AUTHOR CONTRIBUTION
5.1. Category 1
(a) Conception and Design: Schmid, Lautenschlaeger, Scholz, Kircher
(b) Aquisition of Data: Schmid, Lautenschlaeger
(c) Analysis and Interpretation of Data: Schmid, Lautenschlaeger, Scholz
5.2. Category 2
(a) Drafting the Article: Schmid, Lautenschlaeger, Scholz
(b) Revising Article for Intellectual Content: Schmid, Lautenschlaeger, Scholz, Kircher
5.3. Category 3
(a) Final Approval of the Completed Article: Schmid, Lautenschlaeger, Scholz, Kircher
5.4. Category 4
(a) Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: Schmid, Lautenschlaeger, Scholz, Kircher
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
PREVIOUS PRESENTATION OR PUBLICATION DISCLOSURE
A version of the abstract of this study was presented at the EAVDI‐BID Spring 2022 Pre‐BSAVA Meeting.
REPORTING GUIDELINE DISCLOSURE
The “Checklist for Artificial Intelligence in Medical Imaging” (CLAIM Guideline) was used to draft this manuscript.
ACKNOWLEDGMENT
Open access funding provided by Universitat Zurich.
Schmid D, Scholz VB, Kircher PR, Lautenschlaeger IE. Employing deep convolutional neural networks for segmenting the medial retropharyngeal lymph nodes in CT studies of dogs. Vet Radiol Ultrasound. 2022;63:763–770. 10.1111/vru.13132
REFERENCES
- 1. Ronneberger O, Fischer P, Brox T. U‐Net: convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science. Springer; 2015; [Google Scholar]
- 2. Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multiorgan segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys. 2018;45(10):4558‐4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brønden LB, Eriksen T, Kristensen AT. Oral malignant melanomas and other head and neck neoplasms in Danish dogs–data from the Danish Veterinary Cancer Registry. Acta Vet Scand. 2009;51(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohn LA. Canine nasal disease: an update. Vet Clin North Am Small Anim Pract. 2020;50(2):359‐374. [DOI] [PubMed] [Google Scholar]
- 5. Bergman PJ. Canine oral melanoma. Clin Tech Small Anim Pract. 2007;22(2):55‐60. [DOI] [PubMed] [Google Scholar]
- 6. Henry CJ, Brewer WG Jr, Tyler JW, et al. Survival in dogs with nasal adenocarcinoma: 64 cases (1981‐1995). J Vet Intern Med. 1998;12(6):436‐439. [DOI] [PubMed] [Google Scholar]
- 7. Poirier VJ, Koh ESY, Darko J, Fleck A, Pinard C, Vail DM. Patterns of local residual disease and local failure after intensity modulated/image guided radiation therapy for sinonasal tumors in dogs. J Vet Intern Med. 2021;35(2):1062‐1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cancedda S, Sabattini S, Bettini G, et al. Combination of radiation therapy and firocoxib for the treatment of canine nasal carcinoma. Vet Radiol Ultrasound. 2015;56(3):335‐343. [DOI] [PubMed] [Google Scholar]
- 9. van Dijk LV, Van den Bosch L, Aljabar P, et al. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring. Radiother Oncol. 2020;142:115‐123. [DOI] [PubMed] [Google Scholar]
- 10. Christensen NI, Forrest LJ, White PJ, Henzler M, Turek MM. Single institution variability in intensity modulated radiation target delineation for canine nasal neoplasia. Vet Radiol Ultrasound. 2016;57(6):639‐645. [DOI] [PubMed] [Google Scholar]
- 11. Zhu W, Huang Y, Zeng L, et al. AnatomyNet: deep learning for fast and fully automated whole‐volume segmentation of head and neck anatomy. Med Phys. 2019;46(2):576‐589. [DOI] [PubMed] [Google Scholar]
- 12. Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44(12):6377‐6389. [DOI] [PubMed] [Google Scholar]
- 13. Unkelbach J, Bortfeld T, Cardenas CE, et al. The role of computational methods for automating and improving clinical target volume definition. Radiother Oncol. 2020;153:15‐25. [DOI] [PubMed] [Google Scholar]
- 14. Lustberg T, van Soest J, Gooding M, et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126(2):312‐317. [DOI] [PubMed] [Google Scholar]
- 15. Banzato T, Cherubini GB, Atzori M, Zotti A. Development of a deep convolutional neural network to predict grading of canine meningiomas from magnetic resonance images. Vet J. 2018;235:90‐92. [DOI] [PubMed] [Google Scholar]
- 16. Banzato T, Bernardini M, Cherubini GB, Zotti A. A methodological approach for deep learning to distinguish between meningiomas and gliomas on canine MR‐images. BMC Vet Res. 2018;14(1):317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoon Y, Hwang T, Lee H. Prediction of radiographic abnormalities by the use of bag‐of‐features and convolutional neural networks. Vet J. 2018;237:43‐48. [DOI] [PubMed] [Google Scholar]
- 18. Boissady E, de La Comble A, Zhu X, Hespel AM. Artificial intelligence evaluating primary thoracic lesions has an overall lower error rate compared to veterinarians or veterinarians in conjunction with the artificial intelligence. Vet Radiol Ultrasound. 2020;61(6):619‐627. [DOI] [PubMed] [Google Scholar]
- 19. Burti S, Longhin Osti V, Zotti A, Banzato T. Use of deep learning to detect cardiomegaly on thoracic radiographs in dogs. Vet J. 2020;262:105505. [DOI] [PubMed] [Google Scholar]
- 20. McEvoy FJ, Proschowsky HF, Müller AV, et al. Deep transfer learning can be used for the detection of hip joints in pelvis radiographs and the classification of their hip dysplasia status. Vet Radiol Ultrasound. 2021;62(4):387‐393. [DOI] [PubMed] [Google Scholar]
- 21. Park J, Choi B, Ko J, et al. Deep‐learning‐based automatic segmentation of head and neck organs for radiation therapy in dogs. Front Vet Sci. 2021;8:721612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kneissl S, Probst A. Comparison of computed tomographic images of normal cranial and upper cervical lymph nodes with corresponding E12 plastinated‐embedded sections in the dog. The Veterinary Journal. 2007;174:435‐438. [DOI] [PubMed] [Google Scholar]
- 23. Kingma DP, Ba Lei, Adam J,: A method for stochastic optimization. Paper presented as Conference paper at 3rd International Conference on Learning Representations, ICLR 2015; San Diego, CA, USA, May 7–9 [Google Scholar]
- 24. Jaderberg M, Dalibard V, Osindero S, et al. Population based training of neural networks. 2017; https://arxiv.org/abs/1711.09846
- 25. Balas VE, Hassanien AE, Chakrabarti S, Mandal L, Proceedings of International Conference on Computational Intelligence, Data Science and Cloud Computing: IEM‐ICDC 2020 (Lecture Notes on Data Engineering and Communications Technologies 62). Singapore: Springer; 2021; pp 192‐193. [Google Scholar]
- 26. Rattan R, Kataria T, Banerjee S, et al. Artificial intelligence in oncology, its scope, and future prospects with specific reference to radiation oncology. BJR Open. 2019;1(1):20180031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murakami K, Rancilio NJ, Plantenga JP, Moore GE, Heng HG, Lim CK. Interobserver reliability of computed tomographic contouring of canine tonsils in radiation therapy treatment planning. Vet Radiol Ultrasound. 2018;59(3):357‐364. [DOI] [PubMed] [Google Scholar]
- 28. Forrest LJ. Computed Tomography Imaging in Oncology. Vet Clin North Am Small Anim Pract. 2016;46(3):499‐513. [DOI] [PubMed] [Google Scholar]
- 29. Bezuidenhout AJ. The Lymphatic System. In: Evans HE, De Lahunta A., eds. Miller's Anatomy of the Dog. 4th ed. Saunders, an imprint of Elsevier Inc.; 2013;543‐544. [Google Scholar]
- 30. Thierry F, Longo M, Pecceu E, Zani DD, Schwarz T. Computed tomographic appearance of canine tonsillar neoplasia: 14 cases. Vet Radiol Ultrasound. 2018;59(1):54‐63. [DOI] [PubMed] [Google Scholar]
- 31. Herring ES, Smith MM, Robertson JL. Lymph node staging of oral and maxillofacial neoplasms in 31 dogs and cats. J Vet Dent. 2002;19(3):122‐126. [DOI] [PubMed] [Google Scholar]
- 32. Barbu A, Suehling M, Xu X, Liu D, Zhou SK, Comaniciu D. Automatic detection and segmentation of lymph nodes from CT data. IEEE Trans Med Imaging. 2012;31(2):240‐250. [DOI] [PubMed] [Google Scholar]
- 33. Gieger TL, Nolan MW. Treatment outcomes and target delineation utilizing CT and MRI in 13 dogs treated with a uniform stereotactic radiation therapy protocol (16 Gy single fraction) for pituitary masses: (2014‐2017). Vet Comp Oncol. 2021;19(1):17‐24. [DOI] [PubMed] [Google Scholar]
- 34. Langlotz CP. Will Artificial Intelligence Replace Radiologists?. Radiol Artif Intell. 2019;1(3):e190058. [DOI] [PMC free article] [PubMed] [Google Scholar]


