Abstract
Background
Responses to the systemic treatments commonly used to treat psoriasis vary. Biomarkers that accurately predict effectiveness and safety would enable targeted treatment selection, improved patient outcomes and more cost‐effective healthcare.
Objectives
To perform a scoping review to identify and catalogue candidate biomarkers of systemic treatment response in psoriasis for the translational research community.
Methods
A systematic search of CENTRAL, Embase, LILACS and MEDLINE was performed for relevant articles published between 1990 and December 2021. Eligibility criteria were studies involving patients with psoriasis (any age, n ≥ 50) reporting biomarkers associated with systemic treatment response. The main outcomes were any measure of systemic treatment efficacy or safety. Data were extracted by one reviewer and checked by a second; studies meeting minimal quality criteria (use of methods to control for confounding) were formally assessed for bias. Candidate biomarkers were identified by an expert multistakeholder group using a majority voting consensus exercise and mapped to relevant cellular and molecular pathways.
Results
Of 71 included studies (67 studying effectiveness outcomes and eight safety outcomes; four studied both), most reported genomic or proteomic biomarkers associated with response to biologics (48 studies). Methodological or reporting limitations frequently compromised the interpretation of findings, including inadequate control for key covariates, lack of adjustment for multiple testing, and selective outcome reporting. We identified candidate biomarkers of efficacy to tumour necrosis factor inhibitors [variation in CARD14, CDKAL1, IL1B, IL12B and IL17RA loci, and lipopolysaccharide‐induced phosphorylation of nuclear factor (NF)‐κB in type 2 dendritic cells] and ustekinumab (HLA‐C*06:02 and variation in an IL1B locus). None were supported by sufficient evidence for clinical use without further validation studies. Candidate biomarkers were found to be involved in the immune cellular crosstalk implicated in psoriasis pathogenesis, most notably antigen presentation, T helper (Th)17 cell differentiation, positive regulation of NF‐κB, and Th17 cell activation.
Conclusions
This comprehensive catalogue provides a key resource for researchers and reveals a diverse range of biomarker types and outcomes in the included studies. The candidate biomarkers identified require further evaluation in methodologically robust studies to establish potential clinical utility. Future studies should aim to address the common methodological limitations highlighted in this review to expedite discovery and validation of biomarkers for clinical use.
What is already known about this topic?
Responses to the systemic treatments commonly used to treat psoriasis vary.
Biomarkers that accurately predict effectiveness and safety would enable targeted treatment selection, improved patient outcomes and more cost‐effective healthcare.
What does this study add?
This review provides a comprehensive catalogue of investigated biomarkers of systemic treatment response in psoriasis.
A diverse range of biomarker types and outcomes was found in the included studies, serving as a key resource for the translational research community.
Responses to the systemic treatments commonly used to treat psoriasis vary. Biomarkers that accurately predict effectiveness and safety would enable targeted treatment selection, improved patient outcomes and more cost‐effective healthcare. This review provides a comprehensive catalogue of investigated biomarkers of systemic treatment response in psoriasis.
Linked Comment: A.D. Ormerod. Br J Dermatol 2022; 187:458–459.
Psoriasis is a common chronic inflammatory disease, estimated to affect at least 60 million individuals globally, 1 , 2 and causes major impact on quality of life. Disease severity, particularly with respect to body surface area involvement, often dictates the therapeutic approach. Topical agents are generally used for localized disease, and phototherapy or systemic agents for extensive disease and/or where there is significant involvement of high‐need sites or associated psoriatic arthritis. The advent of (now increasingly) powerful biologic therapies means that moderate‐to‐severe disease can be very effectively controlled in many. 3 However, variation in response, loss of benefit over time, toxicity, practical issues and high drug costs are all important barriers to effective management of the population as a whole. 4
Pre‐emptive identification of individuals with a higher likelihood of a safe and effective response to any selected treatment would enable targeted therapeutic selection, improved patient outcomes and more cost‐effective healthcare. Biomarkers are critical to enabling this ‘personalized medicine’ agenda (not just in psoriasis), and have been defined (broadly) by the Food and Drug Administration and National Institutes of Health as characteristics that are measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions. 5
Across medicine, including psoriasis, advances in omics technologies and bioinformatic approaches have unravelled biomarkers from different molecular levels that have been shown to associate with clinically relevant outcomes in several disease settings. 6 These technologies have driven a radical paradigm shift from analysis of single biomarkers to high‐throughput screens profiling assays in heterogeneous datasets. Efforts in psoriasis have been underpinned by major investment and collaboration. 7 , 8 In this context, collating up‐to‐date, accessible information on the status of biomarker discovery and validation is essential to avoid research waste and redundancy, and, crucially, expedite translation of the biomarker discovery pipeline into clinical practice.
The overall aim of this review is therefore to scope, collate and catalogue research investigating biomarkers of systemic treatment response in psoriasis. The specific aims are to (i) identify and catalogue studies relating to biomarkers of systemic treatment response in psoriasis as defined by efficacy and/or safety outcomes, (ii) select and functionally map biomarkers for which there is some evidence for potential predictive value, and (iii) evaluate study quality and highlight limitations to inform future biomarker research.
Materials and methods
This scoping review was performed by a multistakeholder group drawn from a large multidisciplinary European consortium with academic and industry partners (Biomarkers in Atopic Dermatitis and Psoriasis, www.biomap‐eu.imi) and the International Psoriasis Council (www.psoriasiscouncil.org). We included clinical‐academic dermatologists (ten), a patient representative, scientists with genetic (two), immunology (four) and bioinformatics expertise (four), systematic reviewers (two), and one information specialist. 8
Identification and cataloguing of studies of systemic treatment response biomarkers (stage 1)
Literature searches
A single strategy (Appendix S1, section 7; see Supporting Information) was used to search for both studies of biomarkers of disease progression (reported separately; see Ramessur et al.) 9 and biomarkers of treatment response. Electronic searches were performed by K.W. in CENTRAL, Embase, LILACS and MEDLINE on 7 December 2021 for studies in the English language, published between 1990 (chosen as this heralded the Human Genome Project start date) and December 2021.
Study selection
Criteria for study inclusion were established prior to study selection (Table 1). Titles and abstracts were single screened by one reviewer, with an independent second screening where requested (e.g. for abstracts where there was uncertainty regarding eligibility) (M.C., R.R., I.A.B., J.S., Marta Vergnano, S.H., S.R.). To assess the accuracy of our screening approach, every 10th excluded abstract was independently checked (500 in total) by a second screener (R.R.). From this list, full texts were screened by one reviewer, with decisions (inclusion or exclusion) checked by a second; any disagreements were resolved by consensus or through discussion with a senior member of the team (M.C., D.M., R.R.).
Table 1.
Eligibility criteria for the scoping review
| Review component | Criteria |
|---|---|
| Population | People with psoriasis, with or without psoriatic arthritis, already taking, or commencing, a systemic treatment for psoriasis were eligible. Systemic treatments include methotrexate, ciclosporin, acitretin, dimethylfumarate, etanercept, adalimumab, infliximab, ustekinumab, secukinumab, ixekizumab, brodalumab, guselkumab, certolizumab pegol, tildrakizumab, risankizumab, apremilast, tofacitinib, ruxolitinib, baricitinib or peficitinib, at any dose |
| Interventions | Genomic, epigenomic, transcriptomic, proteomic, cellular, microbiomic and metabolomic biomarkers were included |
| Physiological or radiographic biomarkers were excluded | |
| Comparators | Studies could be of one or more biomarkers |
| Outcomes |
Treatment response outcomes
Adverse event outcomes Adverse events leading to treatment withdrawal, serious adverse events, serious infection resulting in hospitalization, intravenous antibiotics or death, immune‐mediated disease (including demyelination disorders, inflammatory bowel disease, autoimmune hepatitis and cutaneous immune‐mediated disease), liver disease (advanced fibrosis/cirrhosis), major adverse cardiac events, bone marrow suppression |
| Study designs | Reviews and studies including fewer than 50 participants (excluding healthy control participants or other participants without psoriasis) were excluded |
| Studies were included providing the duration of systemic treatment was at least the period recommended to evaluate treatment response in summary of product characteristics documents. In included studies, the populations had to have < 50% of participants with psoriatic arthritis |
Data extraction and cataloguing
A minimal dataset (design, population characteristics, biomarkers, outcome measures and basic result details) was defined by the multistakeholder group following review of pilot data extraction from a pilot sample of studies. Data were extracted (R.R., M.C.) then cross‐checked by another researcher, and discrepancies were resolved by discussion (M.C., D.M., R.R.). For each biomarker type (genomic, transcriptomic, proteomic, metabolomic, cellular and mixed), study details were presented in structured tables subdivided by biomarker function (M.C., D.M.) using an informal classification. Studies meeting minimal study design quality criteria (studies with methods to control for confounding) underwent detailed review (stage 2).
Subset of studies undergoing additional data extraction and quality assessment of studies (stage 2)
Additional data were extracted on psoriasis clinical subtype, treatment history, study design and detailed results (including size and variance‐of‐effect estimates) (M.C., D.M., R.R.).
Quality assessment data were extracted by one researcher and checked by another (M.C., D.M., R.R.) with reference to domains within BIOCROSS 10 and QUIPS 11 – quality assessment tools specifically designed for evaluation of biomarker and prognostic studies – to quality assess studies in stage 2. Studies that adjusted for both sex and age of disease onset (or age) 12 (considered by the group to be the two most important covariates to control for to avoid a high risk of confounder bias) were considered to be at ‘low or moderate risk of bias’; all other studies were classified as ‘high risk of bias’. Other potential prognostic factors adjusted or controlled for in individual studies are detailed in the summary tables in Appendix S1 (section 3).
Other study quality assessment criteria evaluated included levels of attrition (losses to follow‐up) and adequacy of imputation of missing data, evidence of selective outcome reporting and adjustment for multiple statistical testing. 13 Further details of the quality assessment strategy are described in Appendix S1 (section 4).
Selection of candidate biomarkers for cellular and molecular pathway mapping (stage 3)
Given the breadth and heterogeneity of the studies reviewed in stage 2, we then selected biomarkers for cellular and molecular pathway mapping to aid interpretation of the findings and to direct future research (candidate biomarkers) based on consensus majority of the multistakeholder group (see Appendix S1, section 5 for details).
A biomarker‐based ‘disease map’ was built to represent mechanistic and associative links of the candidate biomarkers to psoriasis pathogenesis (methodology detailed in section 6 of Appendix S1) and significantly enriched biological processes were highlighted.
Results
Overview of all included studies (stage 1)
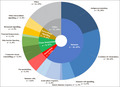
Following title and abstract screening, the full texts of 145 studies were sought, of which 71 met the review eligibility criteria (Figure 1; and see Appendix S1 for included and excluded studies). On checking every 10th excluded abstract, none were considered incorrectly excluded, adding validity to the accuracy of the chosen screening approach. The study designs included cohort studies (86%), registry studies (6%) and randomized controlled studies (6%). Investigated biomarkers covered a broad range of biological functions (Figure 2), although the majority related to immune processes.
Figure 1.
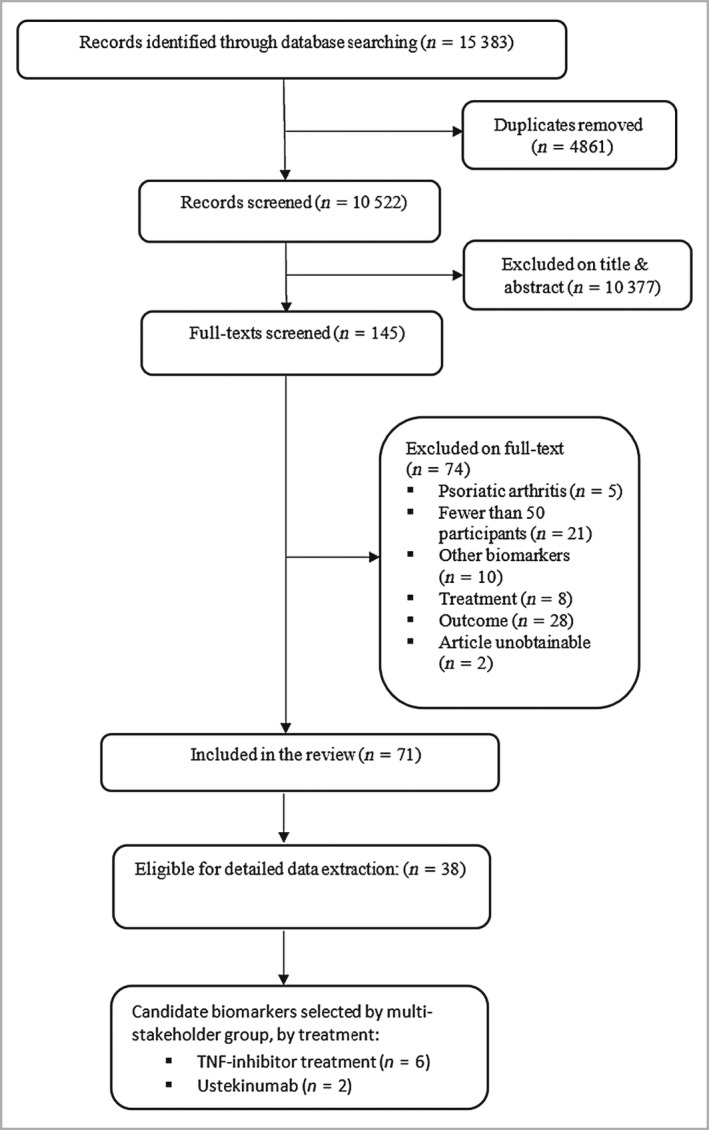
PRISMA flowchart showing the number of studies identified and eligible for inclusion. TNF, tumour necrosis factor.
Figure 2.
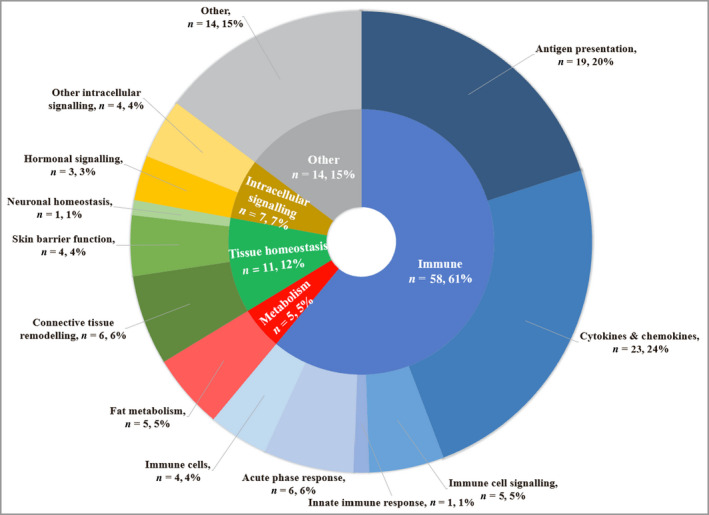
Primary functions of biomarkers in all included studies. Categories of biomarker function were devised using an informal classification, designed to capture the breadth of biomarker function in the included studies. Outer segments represent the number of biomarker studies examining biomarkers with a given primary function (n) and also grouped into broader functional categories (inner segment). Studies examining multiple biomarkers that have more than one function or single biomarkers with multiple key functions may be represented in more than one segment of the ring chart.
Consistent with the explosion of effort around biomarker discovery over the last decade, nearly all studies were published after 2010 (64 studies). As anticipated, the evidence base is largely dominated by studies of genomic (52%) and proteomic (21%) biomarkers (Table 2). In general, genomic biomarker studies evaluated more biomarkers per study than the proteomic biomarker studies, although the numbers of patients recruited were similar. No studies examining microbiomic biomarkers met the eligibility criteria. A full description of the study characteristics, categorized by biomarker type, is reported in Appendix S1 (section 2).
Table 2.
Summary characteristics of the included studies, overall and by type of biomarker
| Characteristic | Type of biomarker evaluated | All biomarkers | |||||
|---|---|---|---|---|---|---|---|
| Genomic | Transcriptomic | Proteomic | Metabolomic | Cellular | Mixed | ||
| Included studies (% of total) | 37 (52%) | 4 (6%) | 15 (21%) | 2 (3%) | 3 (4%) | 10 (14%) | 71 |
| Mean, median biomarkers per study | 30, 4 | 12, 9 | 13, 3 | 3, 3 | 3, 3 | 9, 9 | 21, 3 |
| Mean, median patients with psoriasis | 200, 130 | 142, 129 | 218, 128 | 85, 85 | 116, 95 | 150, 142 | 187, 137 |
| No. of studies evaluated further a (% of studies in biomarker category) | 24 (65%) | 2 (50%) | 7 (47%) | 0 | 0 | 5 (50%) | 38 (54%) |
| Candidate biomarkers (% of total) | 6 (86%) | 0 | 0 | 0 | 1 (14%) | 0 | 7 |
Studies that were eligible for detailed data extraction and quality assessment.
Across all of the included studies, 17 different systemic treatments were evaluated, the most common being tumour necrosis factor (TNF) inhibitors (etanercept 34 studies, adalimumab 31, infliximab 24), ustekinumab (27 studies) and methotrexate (11 studies); 36 studies evaluated more than one treatment. Conversely, few studies examined interleukin (IL)‐17 inhibitors (secukinumab, eight studies; ixekizumab, three), IL‐23 inhibitors (risankizumab, one study) or small‐molecule inhibitors (apremilast, two studies; tofacitinib, two; baricitinib, one).
Characteristics of studies that underwent detailed data extraction and quality assessment (stage 2)
Thirty‐eight studies fulfilled the criteria for further evaluation and quality assessment (Appendix S1, section 3), which revealed important similarities in cohort characteristics between studies. These studies had a narrow mean age range (42–50 years), mean Psoriasis Area and Severity Index (PASI) (15–23) and mean duration of psoriasis (16–24 years). Most studies were conducted in Europe or North America (68%), although the ethnicity of study participants was infrequently reported. Most studies did not report participants’ psoriasis subtype; where reported all included studies investigated plaque psoriasis. Details on past treatment use and the proportion of cohorts with psoriatic arthritis were generally not well reported, or not reported at all.
All studies – except one of methotrexate only 14 – evaluated responses to a biologic therapy: usually one or more of adalimumab, etanercept, infliximab and ustekinumab. The biomarker examined in the highest number of studies was HLA‐C*06:02 (14 studies).
Most studies evaluated treatment efficacy outcomes, reporting changes in PASI. Studies most frequently used dichotomous treatment response outcomes: usually one or more of ≥ 50% improvement in PASI (PASI 50), PASI 75 and PASI 90. Only two studies reported on associations with adverse events. 15 , 16 Other outcomes included drug survival and loss of response to treatment.
Quality assessment (stage 2)
Quality assessment of studies revealed at least one type of bias in nearly all studies (Appendix S1, sections 3 and 4). All studies that underwent further evaluation adjusted for confounding via their methods of analysis. Some studies also controlled for certain confounders by recruitment methods (e.g. controlling for ethnicity by recruiting a cohort of white patients and controlling for previous exposure to a biologic therapy by recruiting a biologic‐naive cohort). Both prespecified key covariates were controlled for in 10 of the 38 studies, so the possibility of the results being affected by bias arising from confounding could not be ruled out for most studies.
The included studies had several additional methodological limitations. Many analyses were limited by the lack of adjustment for multiple hypothesis testing; of the 21 studies that evaluated more than one biomarker, only five reported using such adjustments. Another source of bias was imputation methods used for dealing with data for patients lost to follow‐up. Additionally, there was evidence suggesting bias arising from selective outcome (or result) reporting in several studies. Overall, the quality assessment findings indicated that the results of all of the included studies should be interpreted with some caution: all studies except three were judged to be at high risk of bias.
Candidate biomarker associations (stage 3)
Seven different biomarkers (eight biomarker–outcome associations) were selected as candidate biomarkers for systemic treatment efficacy, based on the evidence available (Table 3): five genomic biomarkers for TNF inhibitor treatment (CARD14, CDKAL1, IL1B, IL12B and IL17RA loci), one cellular biomarker for adalimumab treatment (lipopolysaccharide‐induced phosphorylation of nuclear factor‐κB in type 2 dendritic cells) and two genomic biomarkers for ustekinumab treatment (HLA‐C*06:02 and variation in an IL1B locus).
Table 3.
Summary details of studies examining prioritized candidate biomarkers of treatment response
| Study and no. of patients | Biomarker and biomarker type | Systemic therapy | Number of review‐prespecified key covariates adjusted for | Outcome and timepoint | Results |
|---|---|---|---|---|---|
| HLA‐C*06:02 | |||||
| Burlando 2020 31 | HLA‐Cw6. Genomic | Adalimumab, etanercept, infliximab, golimumab, certolizumab pegol, ixekizumab, ustekinumab, secukinumab | Two: age/disease duration, sex | PASI 90 at weeks 16 and 48 | No significant difference between HLA‐Cw6 positive and negative groups (OR 1·53, 95% CI 0·22–30·7) at week 16 or 48 |
| n = 101 | |||||
| Chiu 2014 32 | HLA‐Cw6. Genomic | Ustekinumab | One: sex | PASI 50, PASI 75, PASI 90. Week 16 | No significant association for PASI 75 at week 18. Further result details not reported |
| n = 66 | |||||
| Costanzo 2018 33 | HLA‐Cw6. Genomic | Secukinumab | No review‐specified key covariates controlled for | PASI 90, change in PASI, adverse events. Week 16 | No significant difference for PASI 90 at week 16 (OR 0·75, 95% CI 0·44–1·28; P = 0·29) or for PASI change. Treatment‐emergent adverse events: no significant difference (P = 0·30) |
| n = 434 | |||||
| Dand 2019 20 | HLA‐C*06:02. Genomic | Adalimumab, ustekinumab | One: age at onset | PASI 75, PASI 90, PASI 100. 6 months | HLA‐C*06:02 associated with a better response to ustekinumab (OR 1·72, P = 0·02) and with a poorer response to adalimumab (OR 0·54, P < 0·001) at 6 months |
| n = 1326 | |||||
| Indhumathi 2017 14 | HLA‐Cw6. Genomic | Methotrexate | Two: sex, age at disease onset | PASI 50, PASI 75. Timepoint unclear | Significant association for PASI 75, OR 2·74 (95% CI 1·39–5·43, P = 0·0004) but timepoint for evaluation unclear |
| n = 189 | |||||
| Svedbom 2020 34 | HLA‐C*06:02. Genomic | Ustekinumab | No review‐specified key covariates controlled for | Drug survival, maintenance PASI. 2 years | No significant association (P = 0.11). No significant difference for PASI during maintenance treatment |
| n = 167 | |||||
| Talamonti 2017 21 | HLA‐C*06. Genomic | Ustekinumab | Two: sex, age at disease onset | PASI 50, PASI 75, PASI 90. Week 12 | Significant association for PASI 75 at 12 weeks, OR 3·28 (95% CI 1·92–5·59, P < 0·0001). Significant results also at weeks 28, 40 and 52, and for PASI 90 and PASI 100 from week 12 to week 52 |
| n = 255 | |||||
| Talamonti 2013 35 | HLA‐Cw6. Genomic | Ustekinumab | Two: sex, age at disease onset | PASI 50, PASI 75. Week 12 | Significant association for PASI 75 at 12 weeks, OR 13·4 (95% CI 1·6–12·6, P < 0·008). Also significant for PASI 75 at 28 weeks: OR 4·0 (95% CI 1·1–8·8, P = 0·016) |
| n = 51 | |||||
| van den Reek 2017 36 | HLA‐C*06. Genomic | Etanercept, adalimumab, ustekinumab | No review‐specified key covariates controlled for | PASI 75, change in PASI. 3 months | Unadjusted analysis: no significant associations for all three biologics (P > 0·3) for either outcome |
| n = 234 | |||||
| Zorlu 2022 37 | HLA‐Cw6. Genomic | Adalimumab, etanercept, infliximab, ustekinumab | One: age at onset | Drug survival (PASI < 50) | HLA‐Cw6 negativity significantly associated with treatment failure (HR 8·95, 95% CI 1·8–44·5) |
| n = 180 | |||||
| IL12B | |||||
| Indhumathi 2017 14 | IL12B. Genomic | Methotrexate | Two: sex, age at disease onset | PASI 50, PASI 75. Timepoint unclear | No significant association (P‐value NRa for adjusted analysis) |
| n = 189 | |||||
| Ovejero‐Benito 2018 38 | IL12B rs2546890. Genomic | Adalimumab, infliximab | No review‐specified key covariates controlled for | PASI 75. 3 months | Significant association, OR 0·12 (95% CI 0·01–0·95, P = 0·04) |
| n = 95 | |||||
| Prieto‐Perez 2017 39 | IL12B rs2546890. Genomic | Ustekinumab | No review‐specified key covariates controlled for | PASI 75. 4 months | No significant association (P = 0·35) |
| n = 69 | |||||
| Prieto‐Perez 2018 40 | IL12B rs2546890. Genomic | Etanercept, adalimumab, infliximab | No review‐specified key covariates controlled for | PASI 75. 3 months | Significant association, OR 3·22 (95% CI 1·23–8·40, P = 0·017) for TNF antagonist treatment (three treatments analysed in one grouping) |
| n = 144 | |||||
| van den Reek 2017 36 | IL12B rs3213094. Genomic | Etanercept, adalimumab, ustekinumab | No review‐specified key covariates controlled for | PASI 75, change in PASI. 3 months | Unadjusted: no significant association. PASI change, 3 months: significant association for ustekinumab, n = 66, P = 0·02 |
| n = 234 | |||||
| IL17RA | |||||
| Batalla 2018 41 | IL17RA rs4819554 and rs879577. Genomic | Adalimumab, etanercept, infliximab | No review‐specified key covariates controlled for | PASI 50, PASI 75. 12 weeks | Significant association for rs4819554 allele A (P = 0·01) for PASI 75 at 12 and 24 weeks. No significant association for rs879577 |
| n = 238 | |||||
| IL1B | |||||
| Loft 2018 42 | IL1B rs1143623, rs1143627. Genomic | Ustekinumab, adalimumab, infliximab, etanercept | One: sex | PASI change, drug survival. 3 months | Significant associations for PASI change with response to both ustekinumab and TNF antagonists for both SNPs. No significant associations for drug survival |
| n = 478 | |||||
| CARD14 | |||||
| Coto‐Segura 2016 43 | Five CARD14 SNPs. Genomic | Adalimumab, etanercept, infliximab | No review‐specified key covariates controlled for | PASI 75. Week 24 | Significant associations for rs11652075 (OR 3·71, 95% CI 1·30–10·5, P = 0·01) but not for the other SNPs |
| n = 116 | |||||
| CDKAL1 | |||||
| Coto‐Segura 2015 44 | Four CDKAL1 SNPs. Genomic | Adalimumab, etanercept, infliximab | Two: sex, age at disease onset | PASI 75. Week 24 | Significant associations for rs6908425 (OR 3·14, 95% CI 1·40–7·05) but not for the other SNPs |
| n = 116 | |||||
| Prieto‐Perez 2018 40 | CDKAL1 rs6908425. Genomic | Etanercept, adalimumab, infliximab | No review‐specified key covariates controlled for | PASI 75. 6 months | Significant association (OR 0·14, 95% CI 0·03–0·66, P = 0·01) |
| n = 144 | |||||
| LPS‐induced phosphorylation of NF‐κB in type 2 dendritic cells | |||||
| Andres‐Ejarque 2021 22 | LPS‐induced phosphorylation of NF‐κB in type 2 dendritic cells. Cellular | Adalimumab | Two: age, sex | PASI 75 at week 12 | Baseline NF‐κB translocation in dendritic cells correlated with lack of PASI 75 response (P = 0·01) |
| n = 67 | |||||
CI, confidence interval; FU, follow‐up; HR, hazard ratio; LOCF, last observation carried forward; LPS, lipopolysaccharide; NF, nuclear factor; OR, odds ratio; PASI, Psoriasis Area and Severity Index; PASI 75; ≥ 75% improvement in PASI; SNP, single‐nucleotide polymorphism; TNF, tumour necrosis factor. aNR, not reported: only studies that underwent detailed data extraction and quality assessment are included in the table. For further study details see Appendix S1, section 3.
All selected candidate biomarkers were at a single molecular level, although IL1B was also found to associate with etanercept efficacy at the genomic and proteomic levels. 17 Allele frequencies of genomic biomarkers were rarely reported, as were justifications for thresholds of significance used for nongenomic biomarkers, often limiting the interpretation of findings.
Pathway mapping of candidate biomarkers (stage 3)
Most candidate biomarkers were found to be involved in signalling pathways implicated in psoriasis pathogenesis, 18 including antigen processing and presentation (HLA‐C*06:02), T helper 17 cell differentiation (IL1B) and immune response (IL12B), and regulation of nuclear factor‐κB activity (CARD14, IL17RA) (see the interactive map of psoriasis biomarkers on the BIOMAP website). 19 The most enriched pathways among candidate biomarkers of response to TNF inhibitor treatment were cytokine‐mediated signalling pathways and granulocyte–macrophage colony‐stimulating factor production (section 6 and the associated figure in Appendix S1).
Discussion
This scoping review has identified a comprehensive catalogue of studies reporting data on a diverse range of biomarker types associated with outcomes to systemic psoriasis therapies. Most of these are related to biologics (TNF antagonists and ustekinumab) and methotrexate. These studies have focused on short‐term efficacy, with only one study addressing loss of response (secondary failure) in psoriasis, and very few addressing toxicity. Of the biomarkers reviewed in detail by the stakeholder group, seven (six genomic and one cellular) were selected as candidates for future research, mapping to immune pathways strongly implicated in disease pathogenesis and/or drug mechanisms, consistent with the principle that the ideal biomarker is on the causal pathway of interest.
Notably, HLA‐C*06:02, the primary susceptibility allele in psoriasis and the most extensively studied biomarker, showed a consistent association with ustekinumab response, and potential utility as a stratification tool to select those more likely to respond to ustekinumab (positive status) compared with TNF antagonists. 20 , 21 Of the remaining biomarkers selected, two (CDKAL1 and IL12B loci) also showed consistency of effect in two independent studies, positively associating with efficacy of TNF antagonists. No biomarkers were identified as being suitable for clinical use, reflecting the acknowledged need for further validation and performance testing.
The fact that most studies investigated first‐generation biologics (TNF inhibitors and ustekinumab) and methotrexate illustrates an important challenge for biomarker research: that is, the timeline for biomarker discovery and validation can outstrip the pace of change in therapeutics. Integration of co‐diagnostics into drug development programmes is one solution. Developing biomarkers with utility across drug classes and across diseases (for example TNF antagonists across immune‐mediated diseases) also enhances the value of the biomarker development pipeline, although this may miss disease‐ or drug‐specific mechanistic differences.
Genomic or proteomic biomarkers appear to dominate the research landscape, and the assessment of multiple biomarkers simultaneously reflects advances in high‐throughput biological assays and their application to larger‐scale cohorts. In this context, genomic biomarkers have an advantage given the clear causal direction between genomic biomarker and outcome. The identified candidate biomarkers of treatment efficacy mapped to immune pathways known to underpin psoriasis pathogenesis indicate the largely ‘hypothesis‐driven’ approach to date, where biomarkers are selected for study based on established knowledge of the role of a gene or molecule in disease pathogenesis. Very recently, studies have been performed using a less directed, ‘hypothesis‐free’ approach offering potential to uncover new mechanistic insights into drug response. This is exemplified in the study by Andres‐Ejarque et al., which identified enhanced nuclear factor‐κB signalling in type 2 dendritic cells as a biomarker of TNF antagonist nonresponse. 22 Genome‐wide association studies, similarly ‘hypothesis’ free, have not to date revealed strongly significant associations, due to the requirement of large sample sizes to offset the necessity for multiple‐testing adjustment.
The modest effect sizes observed in the included studies mean that the clinical utility of many of the reported biomarkers is likely to be limited. Combining multiple biomarkers of small effect size with established clinical ‘biomarkers’ (for example high body mass index and presence of psoriatic arthritis) 23 , 24 and indicators of drug exposure into risk prediction tools may better reflect the complexity of drug response, and hence the ability to accurately predict treatment response. 25 , 26
This review highlights key methodological and reporting limitations that had the potential to compromise the interpretation of findings in the included studies. Lack of measuring and adjusting for key covariates was a common limitation. Even studies that controlled for key covariates were at additional risk of bias due to one or more of: absence of consideration of the impact of missing data, lack of adjustment for multiple testing, and selective outcome reporting. Future studies should also consider possible confounding by the presence of other biomarkers to help identify independent associations with outcomes.
Few studies adequately imputed data from patients who discontinued treatment early, due to either lack of efficacy or adverse events. For a biomarker that is genuinely predictive of treatment efficacy, a difference in treatment discontinuation rates might be expected (between the biomarker present vs. absent groups). Therefore, the omission of such imputed data could underestimate associations between biomarker and treatment efficacy, and lead to bias.
Only one study reported biomarker results for patients taking placebo. 27 Data from patients receiving placebo or standard care treatments, or healthy volunteers (e.g. for mechanistic or functional biomarkers detected at baseline) 22 can be very informative in classifying and validating biomarker mechanisms. Such data are necessary to classify a biomarker as being predictive of a treatment response (only biomarker‐positive patients under the test treatment are likely to benefit) rather than being a prognostic biomarker (biomarker‐positive patients have better outcomes for both test treatment and placebo). Different randomized study designs have been proposed to address this issue. The most pragmatic approach may be to use data from regulatory efficacy trials, if available. Additionally, prospective randomized study designs specifically address the issue of validating a biomarker as being predictive. These include the ‘biomarker by treatment interaction’ and ‘biomarker strategy’ designs, among several others. 28 , 29
Heterogeneity of the outcome definition across studies also made the comparative assessment difficult for biomarker–outcome associations. There was variation in the timepoint for assessment of outcomes and in the dichotomous treatment response outcomes used. In future studies, analyses should be performed using both continuous and categorical data wherever possible, ideally using predefined categories and evaluation timepoints, to help reduce the risk of selective result reporting.
The main strength of the review is its breadth of scope – we believe this review to be the most comprehensive evaluation of biomarkers of systemic treatment response in psoriasis to date. The bibliographic database searches were extensive, allowing identification of nearly all relevant studies. However, there is a potential for the search strategy to have missed studies, such as trial reports, where keywords may not have been included in the abstract. This may bias towards some negative findings being missed. Eligibility criteria were designed to be as inclusive as was practicable, given that it was anticipated that a large number of studies would be included.
By excluding studies of fewer than 50 patients we may have missed important, early discovery studies, particularly those deploying high‐resolution information platforms. Other limitations were the exclusion of papers not reported in English and the single screening of some titles and abstracts, although we consider that our strategies for mitigation mean the risk of missing studies is low.
In conclusion, the wide extent of research on biomarkers of systemic treatment response revealed in this scoping review reflects the key unmet clinical need in psoriasis. The candidate biomarkers and pitfalls in reporting identified in this review can help focus future research efforts to expedite the validation of biomarkers for clinical use.
Synergized efforts, through interdisciplinary collaborations such as the BIOMAP project, 8 offer great opportunity for efficient and effective biomarker research, and also drive incorporation of biomarker discovery into drug development programmes. This has the potential to bring clinically useful biomarkers into psoriasis therapeutics, and enable rationalized early stratification of patients undergoing systemic treatment. 30
Author contributions
Mark Corbett: Data curation (lead); formal analysis (lead); methodology (lead); writing – original draft (lead); writing – review and editing (equal). Ravi Ramessur: Conceptualization (lead); data curation (equal); formal analysis (equal); methodology (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). David Marshall: Data curation (equal); formal analysis (supporting); methodology (equal). Marcio Acencio: Formal analysis (equal); visualization (equal); writing – review and editing (equal). Marek Ostaszewski: Formal analysis (equal); visualization (equal); writing – review and editing (equal). Ines Barbosa: Data curation (equal); formal analysis (equal); methodology (equal); project administration (equal); writing – review and editing (supporting). Nick Dand: Formal analysis (equal); methodology (supporting); writing – review and editing (equal). Paola Di Meglio: Formal analysis (supporting); visualization (equal); writing – review and editing (equal). Salma Haddad: Data curation (equal); formal analysis (equal); writing – review and editing (supporting). Andreas Hvirgel Moesgaard Jensen: Conceptualization (equal); methodology (supporting); writing – review and editing (supporting). Witte Koopmann: Funding acquisition (equal); writing – review and editing (supporting). Satveer Mahil: Conceptualization (supporting); formal analysis (equal); methodology (equal); writing – review and editing (equal). Seher Rahmatulla: Data curation (equal); formal analysis (supporting); writing – review and editing (supporting). Joe Rastrick: Funding acquisition (equal); writing – review and editing (supporting). Jake Saklatvala: Data curation (equal); formal analysis (equal); writing – review and editing (supporting). Stephan Weidinger: Conceptualization (supporting); funding acquisition (lead); project administration (supporting); writing – review and editing (supporting). Kath Wright: Data curation (equal); methodology (equal). Kilian Eyerich: Conceptualization (supporting); formal analysis (equal); methodology (supporting); writing – review and editing (supporting). Matladi Ndlovu: Conceptualization (equal); formal analysis (equal); methodology (supporting); writing – review and editing (supporting). Jonathan N W N Barker: Conceptualization (equal); formal analysis (equal); methodology (supporting); writing – review and editing (equal). Curdin Conrad: Formal analysis (equal); methodology (supporting); visualization (supporting); writing – review and editing (equal). Lone Skov: Conceptualization (supporting); data curation (supporting); formal analysis (equal); methodology (equal); visualization (supporting); writing – review and editing (equal). Catherine H. Smith: Conceptualization (lead); data curation (supporting); formal analysis (equal); funding acquisition (lead); methodology (equal); project administration (lead); supervision (lead); visualization (supporting); writing – review and editing (lead).
Funding sources
This scoping review was supported and funded by the International Psoriasis Council (IPC) and the BIOMAP (Biomarkers in Atopic Dermatitis and Psoriasis) consortium. This study was also supported by the Psoriasis Association, UK. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement number 821511 (BIOMAP). The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. The research also received funding from the National Institute for Health and Care Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. S.K.M. is funded by a Medical Research Council Clinical Academic Research Partnership award (MR/T02383X/1). N.D. is funded by Health Data Research UK (MR/S003126/1).
Conflicts of interest
P.D.M. has received research grants from UCB and consultancy or speaker honoraria from Novartis, UCB and Janssen. S.K.M. has received departmental funding from AbbVie, Celgene, Eli Lilly, Janssen‐Cilag, Novartis, Sanofi and UCB. K.E. has received honoraria and/or research grants from AbbVie, Almirall, Boehringer Ingelheim, Bristol Myers Squibb, Janssen, LEO, Lilly, Novartis, Pfizer and UCB. J.N.B. has received honoraria and/or research grants from AbbVie, Almirall, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Janssen, LEO, Lilly, Novartis, Samsung and Sun Pharma. C.C. has received honoraria and/or research grants from AbbVie, Actelion, Almirall, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Eli Lilly, Janssen, LEO Pharma, MSD, Novartis, Pfizer, Samsung and UCB. L.S. has received honoraria and/or research grants from AbbVie, Almirall, Bristol Myers Squibb, Celgene, Sanofi, UCB, Janssen, LEO Pharma, Lilly and Novartis. C.H.S. reports grants from a Medical Research Council‐funded stratified medicine consortium with multiple industry partners, grants from an Innovative Medicines Initiative (Horizon 2020)‐funded European consortium with multiple industry partners, and others from AbbVie, Novartis, Pfizer, Sanofi, Boehringer Ingelheim and SOBI, outside the submitted work; she is also chair of UK guidelines on biologic therapy in psoriasis. The remaining authors declare that they have no relevant conflicts of interest.
Data availability
The data are available in the Supporting Information.
Ethics statement
Ethics approval is not applicable to this study.
Supporting information
Appendix S1 Preliminary work and study design, overview of results, summary of quality assessment, candidate biomarker selection methodology, molecular and cellular pathway mapping, literature searches, and list of excluded studies.
Acknowledgments
The authors would like to thank Dr Marta Vergnano for her contribution to title and abstract screening. The interactive map of psoriasis biomarkers is hosted by ELIXIR Luxembourg.
References
- 1. Griffiths CEM, Armstrong AW, Gudjonsson JE et al. Psoriasis. Lancet 2021; 397:1301–15. [DOI] [PubMed] [Google Scholar]
- 2. Parisi R, Iskandar IYK, Kontopantelis E et al. National, regional, and worldwide epidemiology of psoriasis: systematic analysis and modelling study. BMJ 2020; 369:m1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Warren RB, Gooderham M, Burge R et al. Comparison of cumulative clinical benefits of biologics for the treatment of psoriasis over 16 weeks: results from a network meta‐analysis. J Am Acad Dermatol 2020; 82:1138–49. [DOI] [PubMed] [Google Scholar]
- 4. Puig L. The role of biologics in the treatment of moderate‐to‐severe plaque psoriasis. G Ital Dermatol Venereol 2017; 152:28–35. [DOI] [PubMed] [Google Scholar]
- 5. FDA–NIH Biomarker Working Group . BEST (Biomarkers, EndpointS, and other Tools) Resource . Silver Spring, MD and Bethesda, MD: U.S. Food and Drug Administration (FDA) and the National Institutes of Health (NIH), 2016. [PubMed]
- 6. Aydin B, Arga KY, Karadag AS. Omics‐driven biomarkers of psoriasis: recent insights, current challenges, and future prospects. Clin Cosmet Investig Dermatol 2020; 13:611–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yiu ZZN, Barker JNWN, Barnes MR et al. Meeting report: Psoriasis Stratification to Optimize Relevant Therapy Showcase. J Invest Dermatol 2021; 141:1872–8. [DOI] [PubMed] [Google Scholar]
- 8. Broderick C, Christian N, Apfelbacher C et al. The BIOMarkers in Atopic Dermatitis and Psoriasis (BIOMAP) glossary: developing a lingua franca to facilitate data harmonization and cross‐cohort analyses. Br J Dermatol 2021; 185:1066–9. [DOI] [PubMed] [Google Scholar]
- 9. Ramessur R, Corbett M, Marshall D et al. Biomarkers of disease progression in people with psoriasis: a scoping review. Br J Dermatol 2022; 187:481–93. 10.1111/bjd.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wirsching J, Grassmann S, Eichelmann F et al. Development and reliability assessment of a new quality appraisal tool for cross‐sectional studies using biomarker data (BIOCROSS). BMC Med Res Methodol 2018; 18:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayden JA, van der Windt DA, Cartwright JL et al. Assessing bias in studies of prognostic factors. Ann Intern Med 2013; 158:280–6. [DOI] [PubMed] [Google Scholar]
- 12. Naldi L. Risk factors for psoriasis. Curr Dermatol Rep 2013; 2:58–65. [Google Scholar]
- 13. Ensor JE. Biomarker validation: common data analysis concerns. Oncologist 2014; 19:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Indhumathi S, Rajappa M, Chandrashekar L et al. Pharmacogenetic markers to predict the clinical response to methotrexate in south Indian Tamil patients with psoriasis. Eur J Clin Pharmacol 2017; 73:965–71. [DOI] [PubMed] [Google Scholar]
- 15. Chiu HY, Chiu YM, Chang Liao NF et al. Predictors of hepatitis B and C virus reactivation in patients with psoriasis treated with biological agents: a 9‐year multicenter cohort study. J Am Acad Dermatol 2021; 85:337–44. [DOI] [PubMed] [Google Scholar]
- 16. Hoffmann JH, Knoop C, Enk AH et al. Baseline anti‐dsDNA concentrations and previous treatments predict response to adalimumab and etanercept: a retrospective investigation of 146 psoriasis patients. J Dermatol Sci 2014; 76:180–5. [DOI] [PubMed] [Google Scholar]
- 17. Liu Y, Qin G, Meng Z et al. IL‐1β, IL‐17A and combined phototherapy predicts higher while previous systemic biologic treatment predicts lower treatment response to etanercept in psoriasis patients. Inflammopharmacology 2019; 27:57–66. [DOI] [PubMed] [Google Scholar]
- 18. Mahil SK, Capon F, Barker JN. Update on psoriasis immunopathogenesis and targeted immunotherapy. Semin Immunopathol 2016; 38:11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. ELIXIR Luxembourg. Psoriasis biomarkers. Available at: https://imi‐biomap.elixir‐luxembourg.org/minerva/index.xhtml?id=psobiomarkers_map (last accessed 26 May 2022).
- 20. Dand N, Duckworth M, Baudry D et al. HLA‐C*06:02 genotype is a predictive biomarker of biologic treatment response in psoriasis. J Allergy Clin Immunol 2019; 143:2120–30. [DOI] [PubMed] [Google Scholar]
- 21. Talamonti M, Galluzzo M, van den Reek JM et al. Role of the HLA‐C*06 allele in clinical response to ustekinumab: evidence from real life in a large cohort of European patients. Br J Dermatol 2017; 177:489–96. [DOI] [PubMed] [Google Scholar]
- 22. Andres‐Ejarque R, Ale HB, Grys K et al. Enhanced NF‐κB signaling in type‐2 dendritic cells at baseline predicts non‐response to adalimumab in psoriasis. Nat Commun 2021; 12:4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wade R, Sharif‐Hurst S, Dias S. Patient characteristics as effect modifiers for psoriasis biologic treatment response: an assessment using network meta‐analysis subgroups. Syst Rev 2020; 9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Warren RB, Marsden A, Tomenson B et al. Identifying demographic, social and clinical predictors of biologic therapy effectiveness in psoriasis: a multicentre longitudinal cohort study. Br J Dermatol 2019; 180:1069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Savvateeva E, Smoldovskaya O, Feyzkhanova G et al. Multiple biomarker approach for the diagnosis and therapy of rheumatoid arthritis. Crit Rev Clin Lab Sci 2021; 58:17–28. [DOI] [PubMed] [Google Scholar]
- 26. Curtis JR, Xie F, Crowson CS et al. Derivation and internal validation of a multi‐biomarker‐based cardiovascular disease risk prediction score for rheumatoid arthritis patients. Arthritis Res Ther 2020; 22:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Correa da Rosa J, Kim J, Tian S et al. Shrinking the psoriasis assessment gap: early gene‐expression profiling accurately predicts response to long‐term treatment. J Invest Dermatol 2017; 137:305–12. [DOI] [PubMed] [Google Scholar]
- 28. Gosho M, Nagashima K, Sato Y. Study designs and statistical analyses for biomarker research. Sensors (Basel) 2012; 12:8966–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J Clin Oncol 2009; 27:4027–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muñoz‐Aceituno E, Martos‐Cabrera L, Ovejero‐Benito MC et al. Pharmacogenetics update on biologic therapy in psoriasis. Medicina (Kaunas) 2020; 56:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burlando M, Russo R, Clapasson A et al. The HLA‐Cw6 dilemma: is it really an outcome predictor in psoriasis patients under biologic therapy? A monocentric retrospective analysis. J Clin Med 2020; 9:3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chiu HY, Wang TS, Chan CC et al. Human leucocyte antigen‐Cw6 as a predictor for clinical response to ustekinumab, an interleukin‐12/23 blocker, in Chinese patients with psoriasis: a retrospective analysis. Br J Dermatol 2014; 171:1181–8. [DOI] [PubMed] [Google Scholar]
- 33. Costanzo A, Bianchi L, Flori ML et al. Secukinumab shows high efficacy irrespective of HLA‐Cw6 status in patients with moderate‐to‐severe plaque‐type psoriasis: SUPREME study. Br J Dermatol 2018; 179:1072–80. [DOI] [PubMed] [Google Scholar]
- 34. Svedbom A, Nikamo P, Stahle M. Interaction between smoking and HLA‐C*06:02 on the response to ustekinumab in psoriasis. J nvest Dermatol 2020; 140:1653–6. [DOI] [PubMed] [Google Scholar]
- 35. Talamonti M, Botti E, Galluzzo M et al. Pharmacogenetics of psoriasis: HLA‐Cw6 but not LCE3B/3C deletion nor TNFAIP3 polymorphism predisposes to clinical response to interleukin 12/23 blocker ustekinumab. Br J Dermatol 2013; 169:458–63. [DOI] [PubMed] [Google Scholar]
- 36. van den Reek J, Coenen MJH, van de L’Isle Arias M et al. Polymorphisms in CD84, IL12B and TNFAIP3 are associated with response to biologics in patients with psoriasis. Br J Dermatol 2017; 176:1288–96. [DOI] [PubMed] [Google Scholar]
- 37. Zorlu O, Bülbül Başkan E, Yazici S et al. Predictors of drug survival of biologic therapies in psoriasis patients. J Dermatolog Treat 2022; 33:437–42. [DOI] [PubMed] [Google Scholar]
- 38. Ovejero‐Benito MC, Prieto‐Perez R, Llamas‐Velasco M et al. Polymorphisms associated with adalimumab and infliximab response in moderate‐to‐severe plaque psoriasis. Pharmacogenomics 2018; 19:7–16. [DOI] [PubMed] [Google Scholar]
- 39. Prieto‐Perez R, Llamas‐Velasco M, Cabaleiro T et al. Pharmacogenetics of ustekinumab in patients with moderate‐to‐severe plaque psoriasis. Pharmacogenomics 2017; 18:157–64. [DOI] [PubMed] [Google Scholar]
- 40. Prieto‐Perez R, Solano‐Lopez G, Cabaleiro T et al. New polymorphisms associated with response to anti‐TNF drugs in patients with moderate‐to‐severe plaque psoriasis. Pharmacogenomics J 2018; 18:70–5. [DOI] [PubMed] [Google Scholar]
- 41. Batalla A, Coto E, Gomez J et al. IL17RA gene variants and anti‐TNF response among psoriasis patients. Pharmacogenomics J 2018; 18:76–80. [DOI] [PubMed] [Google Scholar]
- 42. Loft ND, Skov L, Iversen L et al. Associations between functional polymorphisms and response to biological treatment in Danish patients with psoriasis. Pharmacogenomics J 2018; 18:494–500. [DOI] [PubMed] [Google Scholar]
- 43. Coto‐Segura P, Gonzalez‐Fernandez D, Batalla A et al. Common and rare CARD14 gene variants affect the antitumour necrosis factor response among patients with psoriasis. Br J Dermatol 2016; 175:134–41. [DOI] [PubMed] [Google Scholar]
- 44. Coto‐Segura P, Batalla A, Gonzalez‐Fernandez D et al. CDKAL1 gene variants affect the anti‐TNF response among psoriasis patients. Int Immunopharmacol 2015; 29:947–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Preliminary work and study design, overview of results, summary of quality assessment, candidate biomarker selection methodology, molecular and cellular pathway mapping, literature searches, and list of excluded studies.
Data Availability Statement
The data are available in the Supporting Information.


