Abstract
The expression of virulence determinants in Staphylococcus aureus is controlled by global regulatory loci (e.g., sar and agr). The sar locus is composed of three overlapping transcripts (sar P1, P3, and P2 transcripts from P1, P3, and P2 promoters, respectively), all encoding the 372-bp sarA gene. The level of SarA, the major regulatory protein, is partially controlled by the differential activation of sar promoters. We previously partially purified a ∼12 kDa protein with a DNA-specific column containing a sar P2 promoter fragment. In this study, the putative gene, designated sarR, was identified and found to encode a 13.6-kDa protein with homology to SarA. Transcriptional and immunoblot studies revealed the sarR gene to be expressed in other staphylococcal strains. Recombinant SarR protein bound sar P1, P2, and P3 promoter fragments in gel shift and footprinting assays. A sarR mutant expressed a higher level of P1 transcript than the parent, as confirmed by promoter green fluorescent protein fusion assays. As the P1 transcript is the predominant sar transcript, we confirmed that the sarR mutant expressed more SarA than the parental strain. We thus proposed that SarR is a regulatory protein that binds to the sar promoters to down-regulate P1 transcription and the ensuing SarA protein expression.
Staphylococcus aureus is a major human pathogen capable of causing a wide spectrum of infections ranging from superficial abscesses, pneumonia, endocarditis, to sepsis (4)). The ability of S. aureus to cause a multitude of human infections is probably attributable to an impressive array of extracellular and cell wall-associated virulence determinants that are coordinately expressed in this organism (35). The coordinate expression of many of these virulence determinants in S. aureus is regulated by global regulatory elements such as sar and agr (11, 23). These regulatory elements, in turn, control the transcription of a wide variety of unlinked genes, many of which have been implicated in pathogenesis.
The global regulatory locus agr encodes a two-component, quorum sensing system that is involved in the generation of two divergent transcripts, RNAII and RNAIII, from two distinct promoters, P2 and P3, respectively. RNAIII is the regulatory molecule of the agr response and hence responsible for the up-regulation of extracellular protein production and down-regulation of cell wall-associated protein synthesis during the postexponential phase (20, 34). The RNAII molecule, driven by the P2 promoter, encodes a four-gene operon, agrBDCA, with AgrC and AgrA corresponding to the sensor and activator proteins of a two-component regulatory system. Additionally, agrD, in concert with agrB, participates in the generation of an octapeptide with quorum sensing functions (21, 28). The autoinducing peptide would stimulate the transcription of the agr regulatory molecule RNAIII, which ultimately interacts with target genes to modulate transcription (34) and possibly translation (31).
In contrast to agr, the sar locus activates the synthesis of both extracellular (e.g., alpha- and beta-hemolysins) and cell wall proteins (e.g., fibronectin binding protein) in S. aureus (11). The sar locus is composed of three overlapping transcripts [sar P1 [0.56 kb], sar P3 [0.8 kb], and sar P2 [1.2 kb] transcripts), each with a common 3′ end but initiated from three distinct promoters (P1, P3, and P2 promoters). Due to their overlapping nature, each of these transcripts encodes the major 372-bp sarA gene, yielding the 14.5-kDa SarA protein (2). DNA footprinting studies revealed that the SarA protein binds to the promoters of several target genes (14), including agr, hla (alpha-hemolysin gene), spa (protein A gene), and fnbA (fibronectin binding protein A gene), thus implicating SarA to be a regulatory molecule that can modulate target gene transcription via both agr-dependent and agr-independent pathways (5, 14, 15). Presumably, with the agr-dependent pathway of target gene activation, the SarA protein binds to the agr promoter to stimulate RNAIII transcription; RNAIII, in turn, interacts with target genes (e.g., hla) to modulate transcription. With the SarA-dependent but agr-independent pathway, the SarA protein will interact directly with target gene (e.g., hla and spa) (14) promoters to control gene expression.
Deletion and promoter fusion analyses indicated that the regions upstream of the sar P2 promoter and between the P1 and P3 promoters may have a modulating role in SarA expression, possibly by controlling transcription from the sar P1 promoter, the predominant promoter within the sar locus (6, 27) (Fig. 1A). Using a DNA-specific column containing a 49-bp sequence upstream of the sar P2 promoter that shares homology with the region between the P1 and P3 promoters, we previously described the purification of a ∼12-kDa protein (27). In this study, we report the cloning and sequencing of the putative 345-bp gene, designated sarR, encoding this DNA binding protein (predicted molecular size of 13.6 kDa). SarR shares sequence homology with SarA. Purified recombinant SarR was found to bind to the sar promoters, as confirmed by gel shift and footprinting studies. Allelic replacement of the sarR gene with an ermC antibiotic marker disclosed that transcription from the sar P1 and the native P2-P3-P1 promoter, as determined by flow cytometry and fluorescence spectrophotometric assays, was increased in the sarR mutant compared with the parental strain. As the P1 transcript is the predominant sar transcript, we confirmed by immunoblotting that an increase in sar P1 transcription in the sarR mutant would lead to enhanced SarA protein expression. Based on the data presented here, we propose that SarR is a regulatory protein that binds to the sar promoter region to down-regulate sar P1 transcription and the ensuing SarA protein expression.
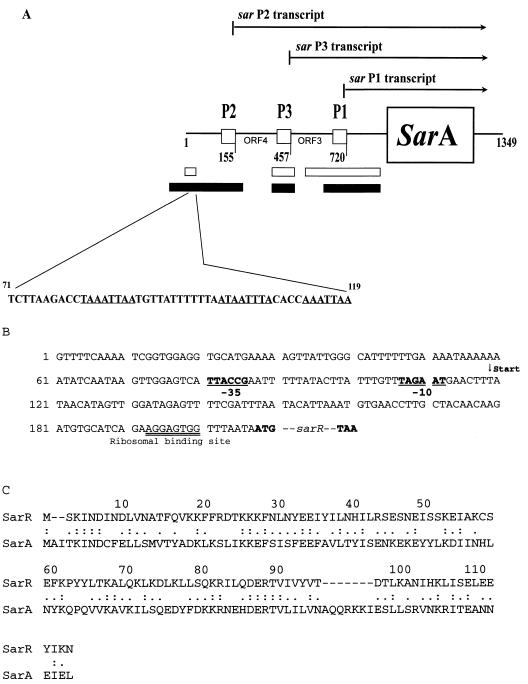
FIG. 1.
(A) Schematic of the sar promoters and transcripts analyzed in this study. Positions of the transcription start sites (146, 409, and 711 bp upstream of the translation start) for P1, P3, and P2 promoters are depicted according to published sequence (2). The P1, P3, and P2 transcripts have previously been designated a sarA, sarC, and sarB transcripts. The 49-bp sequence outlined was used to construct a DNA-specific column as described elsewhere (27). Relative positions of the sar promoter fragments used in gel shift and footprinting studies are indicated (filled boxes); the promoter fragments for the GFP transcriptional fusion assays are represented by empty boxes. (B) Promoter region of sarR. The transcription start site has been mapped by primer extension (data not shown) to position 119. The putative −10 and −35 promoter boxes are in bold and underlined. (C) Alignment of SarR with SarA. Colons represent identity; periods indicate conservative substitutions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Phage φ11 was used as the transducing phage for S. aureus strains. S. aureus strain RN4220, a restriction-deficient derivative of strain 8325-4 (32), was used as the initial recipient for the transformation of plasmid constructs by electroporation, following the protocol of Schenk and Laddaga (40).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Reference or source | Comments |
|---|---|---|
| S. aureus | ||
| RN4220 | 32 | Mutant of 8325–4 that accepts foreign DNA |
| RN6390 | 32 | Laboratory strain that maintains its hemolytic pattern when propagated on sheep erythrocyte agar (parental strain) |
| RN6911 | 34 | agr mutant of RN6390 with Δagr::tetM mutation |
| ALC488 | 9 | sar mutant with sarA::ermC mutation |
| ALC1713 | This study | sarR mutant of RN6390 with ΔsarR::ermC mutation |
| Cowan I | 17 | Laboratory strain |
| DB | 10 | Clinical blood isolate previously used in adhesion and endocarditis studies |
| Newman | 29 | Laboratory strain |
| S. epidermidis | Strain from collection at Utrecht University Hospital | |
| S. haemolyticus | Strain from collection at Utrecht University Hospital | |
| S. saprophyticus | Strain from collection at Utrecht University Hospital | |
| E. coli | ||
| XL-1 Blue | 26 | Host strain for cloning |
| DH5α | 26 | Host strain for cloning |
| Plasmids | ||
| pCR2.1 | Invitrogen | E. coli cloning vector for direct cloning of PCR products |
| pBluescript | Stratagene | E. coli cloning vector |
| pUC18 | 26 | E. coli cloning vector |
| pACYC177 | New England Biolabs | E. coli cloning vector |
| pCL52.1 | 24 | Temperature-sensitive E. coli-S. aureus shuttle vector |
| pET11b | Novagen | Expression vector for E. coli |
| pALC926 | This study | pUC18 containing a 49-bp fragment upstream of the P2 promoter of the sar locus |
| pALC1357 | This study | pET11b containing the 345-bp sarR gene at the NdeI/BamHI site |
| pALC1361 | This study | pACYC177 with a ∼4-kb ClaI fragment containing the sarR region of RN6390 |
| pALC1627 | This study | pBluescript with a 2.5-kb EcoRI/ClaI fragment containing the sarR gene subcloned from pALC1361 |
| pALC1687 | This study | pBluescript with a 290-bp deletion of the sarR gene in pALC1627 |
| pALC1696 | This study | pCL52.1 with a 290-bp sarR deletion replaced by the ermC gene at the EcoRV/SalI site |
S. aureus cells were grown at 37°C with aeration in CYGP or 03GL broth (32, 33) or tryptic soy broth supplemented with antibiotics when necessary. 03GL and NYE agar (40) containing antibiotics were routinely used for the selection of S. aureus transformants; Luria-Bertani medium was used for growing Escherichia coli. Antibiotics were used at the following concentrations: for S. aureus erythromycin at 5 μg/ml, tetracycline at 5 μg/ml, and chloramphenicol at 10 μg/ml; E. coli, ampicillin at 50 μg/ml, chloramphenicol at 30 μg/ml, erythromycin at 200 μg/ml, and spectinomycin at 75 μg/ml.
Cloning of the sarR gene and construction of the sarR mutant.
In a previous study (27), we partially purified the SarR protein by using a DNA-specific column containing a 49-bp DNA fragment (nucleotides [nt] 71 to 119) covalently linked to Sepharose (13). The first 14 residues in the amino terminus were determined by microsequencing at the core facility at our institution. A BLAST search of the S. aureus genome data bank at the Institute for Genomic Research (TIGR) revealed a partial open reading frame (ORF) of 47 amino acids. Using these data, we successfully amplified by PCR a 141-bp fragment with two degenerate oligonucleotides, 5′-ATG(T/A)(C/G)(A/T)AAAAT(T/C)AA(T/C)GATAT(T/C)AA(T/C)GATTTT-3′ and 5′-ATT(T/A)G/C(A/T)(T/C)TC(T/A)(G/C)(A/T)(A/T)C(G/T)(T/C)AA(A/G)AT(A/G)TG(A/G)TT(T/C)AA-3′. The PCR fragment was cloned into the vector pCR2.1 (Invitrogen). Southern hybridization of enzyme-restricted chromosomal DNA of the parental strain RN6390 with a radiolabeled 141-bp DNA probe revealed a single ∼4-kb ClaI-digested hybridizing fragment. To clone this fragment, ClaI-digested chromosomal DNA in the range of 3 to 5 kb was resolved in an agarose gel, excised, purified, and ligated to the ClaI site of pACYC177 in E. coli DH5α. Positive-reacting clones, all containing the ∼4-kb ClaI fragment, were identified. Sequencing of one of these clones revealed a 345-bp ORF with identity to the partial 47-amino-acid sequence of SarR as predicted from the S. aureus genome.
Deletion and insertion mutagenesis was performed with a Stratagene Quick Change kit to introduce a deletion and a mutation concomitantly into the sarR gene. In brief, the ∼4-kb ClaI DNA fragment containing the sarR gene in recombinant pACYC177 was cloned into pBluescript to serve as a template for mutagenesis. An oligonucleotide (5′-22GCATGAAAAAGA[T]TATC[T]GGGCATTT45-338GTGAGTCTAACGAT[A]ATCTCATCTAAA363-3′ [native nucleotides are indicated in brackets]), and its complement were used to construct a deletion and to introduce an exogenous EcoRV restriction site into the sarR gene (restriction site underlined: intact sarR gene from nt 208 to 555). After amplification with the recombinant pBluescript template, the PCR product was digested with DpnI to remove methylated template DNA (i.e., pBluescript with the native sarR gene) and transformed into XL1-Blue cells to select for ampicillin-resistant colonies. Successful deletion and mutation in the resultant clones were confirmed by restriction analysis with EcoRV and finally verified by automated DNA sequencing. The ermC gene was then ligated to the EcoRV site of the mutated construct. The fragment containing an ermC replacement of the sarR gene was cloned into the temperature-sensitive shuttle vector pCL52.1 (39), which was then transformed into RN4220 by electroporation (40) followed by transduction into RN6390 with phage φ11 as described elsewhere (11). Transductants were selected at 30°C on erythromycin- and tetracycline-containing plates.
S. aureus RN6390 harboring the recombinant pCL52.1 was grown overnight at 30°C in liquid medium in the presence of erythromycin, diluted 1:1,000 in fresh medium, and propagated at 42°C, a temperature nonpermissive for the replication of pCL52.1. This cycle was repeated four times, and the cells were replicate plated onto 03GL plates containing erythromycin and erythromycin plus tetracycline to select for tetracycline-sensitive but erythromycin-resistant colonies, representing mutants with double crossovers. The mutations were confirmed by Southern hybridization with sarR and ermC probes.
Southern blot hybridization.
Chromosomal DNA of assorted staphylococcal species was isolated from lysostaphin-treated cells as previously described (11), restriction digested, resolved in agarose gels, and transferred onto a Hybond N+ membrane (Amersham, Arlington Heights, III.). Hybridization was performed under high-stringency conditions with 32P-labeled DNA probes as described elsewhere (11). The blots were subsequently washed and autoradiographed.
Purification of proteins.
The intact 345-bp sarR gene was amplified by PCR using RN6390 chromosomal DNA as the template and primers containing flanking restriction sites (NdeI and BamHI) to facilitate cloning into expression vector pET11b (Novagen). The recombinant plasmid containing the sarR gene was transformed to E. coli BL21(DE3)pLysS. Enhanced expression of SarR was induced by adding IPTG (isopropyl-1-thio-β-d-galactopyranoside) to a 2-liter growing culture (37°C) at an optical density at 650 mm (OD650 of 0.7. After 4 h of additional growth, cells were harvested, resuspended in buffer (25 mM Tris-Cl, 1 mM EDTA) [pH 8.0], 100 mM NaCl, 10% sucrose, 1 mM dithiothreitol [DTT]), flash-frozen and thawed twice, and clarified by centrifugation at 4°C (45,000 rpm for 1 h). After precipitation with 80% ammonium sulfate, the pellets were dissolved in bufferA (10 mM Tris-Cl [pH 7.5], 1 mM EDTA, 100 mM NaCl, 10% glycerol, 1 mM DTT), dialyzed against buffer A, and applied to a Resource-Q column in an AKTA purifier (Pharmacia, Piscataway, N.J.). The flowthrough was reapplied to a Resource-S column and eluted with a NaCl gradient. The fractions were analyzed in a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel. Fractions containing the putative SarR protein were pooled, dialyzed against buffer A with 40% glycerol, and stored at −80°C. The authenticity of the SarR protein was confirmed by determining the N-terminal 15 residues with microsequencing. The concentration of the purified protein was determined with the Bio-Rad protein assay solution (Bio-Rad Laboratories, Richmond, Calif.), using bovine serum albumin as the standard.
Production of anti-SarR monoclonal antibodies.
Purified SarR protein was used to immunize two (BALB/c × SJL/J)F1 mice (100 μg of each) to obtain monoclonal antibodies as described elsewhere (22). The titers of the immune sera were determined by an enzyme-linked immunosorbent assay (ELISA) in which diluted sera were added to microtiter wells precoated with SarR (5 μg/ml) as described by Jones et al. (22). After splenic fusion, antibodies from limited dilutions were screened by an ELISA with immobilized SarR protein. Monoclonal antibodies were then purified from culture supernatants with a protein A-agarose column as described previously (22).
RNA isolation and Northern analysis.
Overnight cultures of S. aureus were diluted 1:50 in CYGP broth with appropriate antibiotics and grown to mid-log (OD650 = 0.7 with an 18 mm borosilicate glass tube), late log (OD650 = 1.1), and postexponential (OD650 = 1.7) phases. The cells were pelleted and processed with 1 ml of Trizol (Gibco-BRL, Gaithersburg, Md.) in combination with 0.1-mm-diameter sirconia-silica beads in a Fast Prep reciprocating shaker (Bio 101, San Diego, Calif.) as described elsewhere (8). Ten micrograms of total cellular RNA from each sample was electrophoresed through a 1.5% agarose–0.66 M formaldehyde gel in MOPS (morpholinepropanesulfonic acid) running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA [pH 7.2]). RNA was transferred onto Hybond N+ membranes (Amersham) under mild alkaline conditions by using a Turboblotter system (Schleicher Schuell, Keene, N.H.) as described by the manufacturer. RNA was fixed to the membrane by baking at 80°C for 1 h. For detection of specific transcripts, gel-purified DNA probes were radiolabeled with [32P] dCTP by using the random-primed method (Ready-To-Go labeling kit; Pharmacia) and hybridized under aqueous conditions at 65°C. The blots were subsequently washed and autoradiographed.
Promoter fusion analysis with the gfpuvr reporter gene.
To confirm the effect of the sarR mutation on sar promoter activities, we cloned sar promoter fragments (P1, P2, P3, and combined P2-P3-P1) (27) into shuttle vector pALC1484, which is a derivative of pSK236 containing the recombinant gfpuvrgene. Briefly, the gfpuvrgene was constructed by introducing a S65T mutation into gfpuv (Clontech, Palo Alto, Calif.), thereby facilitating a shift in the excitation maxima from 395 to 488 nm (19). The sar promoter fragments were then cloned into pALC1484, upstream of the gfpuvrreporter gene. After sequence confirmation, the recombinant pALC1484s were then electroporated into RN4220 and transduced into S. aureus strains RN6390 and its isogenic sarR mutant (11).
The activities of sar promoter fragments linked to the gfpuvrreporter gene in RN6390 and its isogenic sarR mutant were assayed by flow cytometry. Bacterial cell suspensions obtained at different parts of the growth cycle were analyzed by fluorescence-activated cell sorting (FACS) in a FACScan (Becton Dickinson, Franklin Lakes, N.I.). After filtering of bacterial samples through a 5-μm-pore-size filter to remove large aggregates, bacteria were detected by side scatter as described by Russo-Marie et al. (38). Fluorescence and side scatter data were collected with logarithmic amplifiers. The fluorescence data were reported in fluorescence units as specified by the FACScan.
To obtain more quantitative fluorescence data, each of the above gfpuvrreporter constructs was diluted 1:100 from overnight cultures into fresh CYGP medium and, beginning at the second hour, sampled hourly (200 μl) for 10 h to encompass the growth cycle from log to stationary phases. The samples were analyzed for total fluorescence and OD605 in microtiter wells in a multi purpose fluorescence spectrophotometer (FL600; BioTek Instruments, Winooski, Vt.). The fluorescence units and ODs were recorded as given by the instrument. The background was ∼200 to 300 fluorescence units, with variations of less than 100 units between duplicate samples.
Cell extract preparation and Western analysis.
Cell extracts from mid-log, late log, and early stationary phases (representing OD650 of 0.7, 1.1, and 1.7, respectively, in an 18-mm borosilicate tube) were prepared from RN6390, the isogenic sarR mutant, and other staphylococcal strains. Cells were grown in CYGP broth (50 ml) supplemented with the appropriate antibiotics. After pelleting, the cells were resuspended in 0.5 ml of TEG buffer (25 mM Tris-HCl, 5 mM EGTA [pH 8.0], and cell extracts were prepared from lysostaphin-treated cells as described by Mahmood and Khan (25).
Equivalent amounts of cellular proteins were separated in SDS–12% polyacrylamide gels and transferred onto nitrocellulose membranes as described elsewhere (41). The blots were incubated at room temperature (RT) with a 1:1,000 or 1:2,000 dilution of anti-SarR or anti-SarA monoclonal antibody for 3 h, followed by another hour of incubation with a 1:10,000 dilution of goat anti-mouse alkaline phosphatase conjugate (Jackson ImmunoResearch, West Grove, Pa.). Immunoreactive bands were detected as described by Blake et al. (3). SeaBlue-prestained protein standards (Novex, San Diego, Calif.) were used for molecular weight estimations.
Gel shift analysis and DNase I footprinting.
Gel shift assays were performed to determine the interaction of purified SarR with sar promoters. DNA fragments were end labeled with [γ-32P] ATP by using polynucleotide kinase. Labeled DNA fragments were incubated at RT for 20 min with the indicated amounts of purified protein in 25 μl of binding buffer (25 mM Tris-HCl) [pH 7.5], 0.1 mM EDTA, 75 mM NaCl, 1 mM DTT, 5% glycerol) containing 0.5 μg of calf thymus DNA. The reaction mixtures were analyzed by nondenaturing polyacrylamide gel electrophoresis as described elsewhere (14). The band shifts were detected by exposing dried gels to film.
Footprinting assays with linear DNA template and DNase I were performed using a modification of the method previously described (16). A 49-bp fragment upstream of the sar P2 promoter region (27) was cloned into the BamHI site of pUC18, yielding pALC926. A 109-bp EcoRI/HindIII fragment from pALC926 was gel purified and end labeled with γ-32P. PCR fragments containing sar P1 (nt 531 to 859 and 620 to 859) and P3 (nt 364 to 525) promoter regions were also used in footprinting reactions. To label these PCR products, only one of the primers was end labeled with γ-32P in the amplification reactions, yielding PCR products labeled at one end. For the assay, the binding reactions were carried out in a 100-μl reaction volume containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 2 mM DTT, 10 μg of Borine serum albumin, 0.4 μg of calf thymus DNA, template DNA, and various amounts of the SarR protein at RT for 30 min. DNase I (0.02 U; Boehringer Mannheim, Indianapolis, Ind.) was added and allowed to incubate for 1 min at RT. The reaction mixtures were then extracted with phenol-chloroform. DNA was ethanol precipitated, resuspended in loading buffer (98% formamide, 10 mM EDTA [pH 8.0], 0.025% [wt/vol] xylene cyanol FF, 0.025% [wt/vol] bromophenol blue), and analyzed on a 6% denaturing polyacrylamide sequencing gel. Positions of the protected regions were identified by comparing the footprint with the A+G sequencing ladder of the same fragment (26).
Nucleotide sequence accession number.
The sarR sequence has been assigned. GenBank accession no. AF207707.
RESULTS
In a previous study, we demonstrated that the sar promoters are differentially expressed during the growth cycle, with P1 and P2 promoters being most active during the exponential phase and the P3 promoter activated postexponentially (27). Because of the complexity in promoter activation and the ensuing expression of SarA, we previously hypothesized that the promoter region upstream of the sarA gene may serve as a binding site for one or more trans-acting factors (2, 27). Taking advantage of a P2 promoter sequence (2) that shares homology with a region upstream of the sar P1 promoter (Fig. 1A), we previously used a DNA-specific column, containing the 49-bp P2 promoter sequence, to partially purify a ∼12-kDa protein that bound to sar promoter fragments (27). To further characterize this protein and investigate its regulatory function, we report here the cloning and characterization of the sarR gene product, using biochemical, immunological, and genetic approaches.
Cloning and sequence analysis of the sarR gene encoding a 13.6-kDa protein.
To identify the gene encoding SarR, we blotted the ∼12-kDa protein onto a polyvinylidene difluoride membrane for N-terminal sequencing. The first 14 amino acids were X(K)IND(I)NDLVNA(S/T)F (X is an unknown residue; residues in parentheses are putative). In searching the data bank of the S. aureus genome (www.tiger.org), we obtained a partial ORF of 47-amino-acid sequence that corresponds to the N-terminal sequence of the ∼12-kDa protein. By using two degenerate oligonucleotides of 30 bases each, we amplified a 141-bp fragment to probe a chromosomal digest of S. aureus strain RN6390 (data not shown), thus allowing us to identify a ∼4-kb ClaI-hybridizing fragment. A plasmid DNA library containing ∼3- to 5-kb ClaI fragments constructed in pACYC177 (26) was then screened with the 141-bp PCR-generated probe. A positive clone (pALC1361) yielding a ∼4-kb insert at the ClaI site of pACYC177 vector was identified. In determining the sequence of the insert and comparing the insert sequence with that of the 141-bp probe, we were able to obtain the DNA sequence of the putative gene, which we called sarR (Fig. 1B). The predicted SarR protein contains 115 amino acids, with a predominance of charged residues (34%) and a predicted molecular size of 13,689 Da. The sarR gene has a putative Shine-Dalgarno sequence (AGGAGTGG) lying 7 bp upstream of the translation start, with typical initiation (ATG) and termination (TAA) condons. To ascertain the transcription start site and the putative promoter boxes, we mapped the 5′ end of the sarR transcript by primer extension, using an internal primer of the noncoding strand positioned near the N terminus of the sarR coding region (data not shown). The transcription initiation site is located 88 bp upstream of the translation start, thereby allowing us to identify the putative -10 and -35 promoter boxes as TAGAAT and TTACCG, respectively (Fig. 1B).
In searching the GenBank database for related proteins, we found that the entire SarR protein shares sequence similarity with SarA, with a high probability score of 1.8e−7 (Fig. 1C). There were also other SarR homologs in the S. aureus database (University of Oklahoma S. aureus genome database). Like SarA, the SarR protein has a deduced basic pI (9.23). The sequence similarity between SarR and SarA is 51%, with 28% identity (Fig. 1C). In limiting the homology to specific regions, we found that residues 52 to 75 of SarR share homology with residues 54 to 77 of SarA, which, in turn, have a limited but regional sequence similarity to the DNA binding domain of VirF (residues 175 to 198), a transcription regulator of virulence gene expression in Shigella flexneri (13, 18).
Overexpression of SarR and production of monoclonal antibodies.
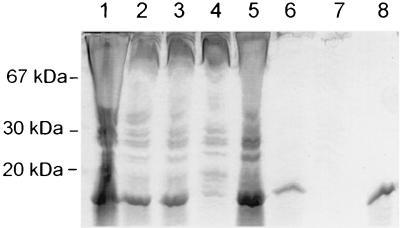
To obtain a large amount of SarR for our studies, we cloned the sarR gene into pET11b and over expressed the gene product under an IPTG-inducible promoter in E. coli BL21. The expression, purification, and purity of the SarR protein are shown in Fig. 2. The SarR protein was expressed primarily in the cytosolic fraction (Fig. 2, lane 2). After 80% ammonium sulfate precipitation (Fig. 2, lane 5), the redissolved proteins were dialyzed and applied to an anion-exchange column (Resource-Q; Pharmacia), only to be found in the flowthrough (Fig. 2, lane 6). The flowthrough was then applied to a cation-exchange column (Resource-S; Pharmacia) and eluted with a salt gradient (see Materials and Methods for details). Using this purification scheme, we were able to purify SarR to near homogeneity (Fig. 2, lane 8). The authenticity of SarR was confirmed by N-terminal sequencing. The purified SarR was then used to immunize mice for the production of anti-SarR monoclonal antibodies. Three monoclonal antibodies, designated 2A7, 2C7, and 5E4, were obtained. Despite the similarity between SarR and SarA, cross-reactive studies indicated that anti-SarR monoclonal antibodies reacted with SarR and not SarA on immunoblots (data not shown).
FIG. 2.
Purification of SarR from the pET11b expression vector. Equivalent volumes of protein fractions obtained during the purification process were applied to an SDS–12% polyacrylamide gel. Lane 1, whole-cell lysate of E. coli containing pALC1357 (pET11b with the sarR gene); lane 2, supernatant of the cell lysate after clarification by centrifugation; lane 3, supernatant before 40% ammonium sulfate precipitation; lane 4, pellet resulting from 40% ammonium sulfate precipitation; lane 5, pellet from 80% ammonium sulfate precipitation; lane 6, flowthrough of the redissolved 80% ammonium sulfate precipitant as applied to a MonoQ column (Pharmacia); lane 7, flowthrough from the MonoS column (Pharmacia); lane 8, NaCl elution from the MonoS column. N-terminal sequencing confirmed the identity of the purified SarR protein.
Evidence for the existence of sarR in other staphylococcal strains.
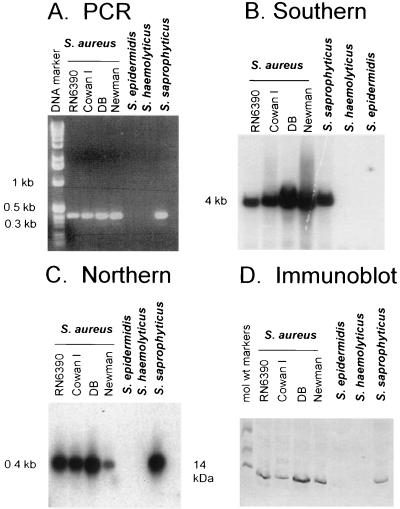
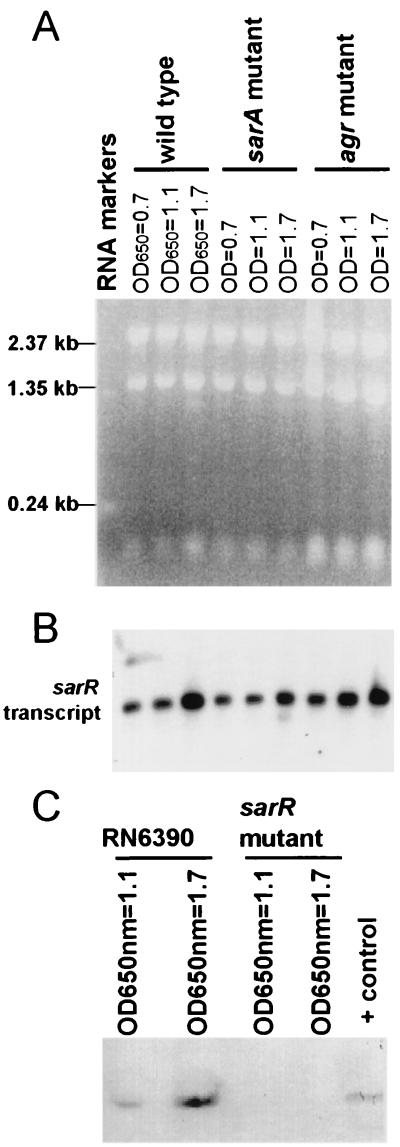
To examine whether the sarR gene is present in a variety of S. aureus strains and in other staphylococcal species, we analyzed S. aureus strains RN6390, Newman, Cowan I (17), and DB (12) and one clinical isolate each of S. epidermidis, S. haemolyticus, and S. saprophyticus (from Utrecht University Hospital, Utrecht, The Netherlands) by PCR, Southern, Northern, and immunoblot analyses (Fig. 3). PCR analysis of these strains revealed the presence of the sarR gene in all four S. aureus isolates as well as in S. saprophyticus. Surprisingly, the ∼380-bp fragment was missing in products of PCRs with chromosomal DNA templates of S. epidermidis and S. haemolyticus (Fig. 3A). Southern blot analysis also confirmed the results of the PCRs with a ∼4-kb ClaI fragment hybridizing to the sarR probe (Fig. 3B). Northern blot analyses were also performed to assess the expression of the sarR transcript in these strains. In accord with the PCR and Southern blot data, the sarR transcript was detected in all staphylococcal strains except S. epidermidis and S. haemolyticus (Fig. 3C). Additional Northern analysis with S. aureus strain RN6390 indicated that the transcription of sarR peaked during the postexponential phase (see below). Notably, the size of the sarR transcript was ∼0.4 kb, suggesting that the transcript is likely to be monocistronic. To correlate gene transcription with protein expression, we probed for the presence of SarR in these strains by immunoblotting. For this experiment, equivalent amounts of cell extracts of various strains (25 μg of protein each) harvested from stationary phase cultures were blotted onto nitrocellulose and probed with anti-SarR monoclonal antibody followed by goat anti-mouse alkaline phosphatase conjugate. As anticipated, a band corresponding to ∼14 kDa was detected in S. aureus strains RN6390, DB, Cowan I, and Newman and a clinical isolate of S. saprophyticus. As with the Northern blot data, SarR protein was not expressed in the S. epidermidis and S. haemolyticus isolates (Fig. 3D). Collectively, these results suggested that the sarR gene is actively transcribed in S. aureus and a clinical isolate of S. saprophyticus but not in S. epidermidis and S. haemolyticus. More importantly, the expression of SarR correlates with gene transcription. Nevertheless, our data did not eliminate the possibility that the sarR homologs in S. epidermidis and S. haemolyticus may be too divergent from the S. aureus counterpart and hence would not be detectable under high-stringency conditions.
FIG. 3.
PCR amplification of sarR-like genes in S. aureus strains RN6390, Cowan I, DB, and Newman, S. epidermidis, S. haemolyticus, and S. saprophyticus using primers 5′-208ATGAGTAAAATTAATGATATTAAT231-3′ and 5′-589TCGTTCAATGTTATTAAACG569-3′. (B) Southern blot of the above strains, restricted with ClaI and probed with a 345-bp sarR probe (nt 208 to 552). (C) Northern blot of total cellular RNA (10 μg each) of the above strains, probed with a sarR probe. (D) Cell lysates of the above strains, immunoblotted onto nitrocellulose and probed with anti-SarR monoclonal antibody 2A7 at a 1:2,000 dilution.
Binding of SarR to sar promoter fragments by gel shift and footprinting assays.
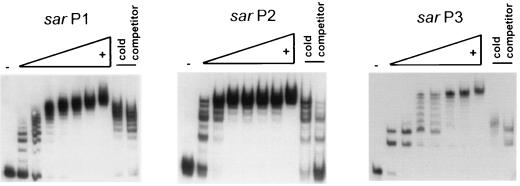
Recognizing SarR binds to the sar P2 promoter region (2, 27), we examined the interaction between SarR and various sar promoter fragments with gel shift and footprinting assays. Accordingly, purified recombinant SarR from E. coli was used in gel shift assays with assorted DNA fragments of the sar promoter region including P2 (nt 1 to 196 [2]) P3 (nt 364 to 525), and P1 (nt 531 to 859). As expected, the mobility of the labeled DNA fragments became more hindered with increasing concentrations of SarR in gel shift assays (Fig. 4). The unusual laddering pattern of the band shifts was observed with all three sar promoter fragments. One plausible explanation is that each of the sar promoter fragments may contain multiple binding sites; alternatively, the binding of SarR in multimeric form to a common site or multiple sites within each of the sar promoter fragment may be plausible. Detailed stoichiometric studies to ascertain these possibilities are in progress. A comparison of the relative binding of SarR and SarA to the sar P1 promoter is of interest. More specifically, the amount of SarA required to completely retard the mobility of 2 to 5 ng of radiolabeled sar P1 fragment was 10 times more than that of SarR, thus indicating the higher avidity of SarR than SarA for the sar P1 promoter fragment (data not shown).
FIG. 4.
Gel shift assay of end-labeled 32P fragment of sar P1 (nt 531 to 859) 2, P2 (nt 1 to 196), and P3 (nt 364 to 525) promoters. Increasing amounts (30, 60, 100, 150, 200, 250, and 300 ng) of purified SarR were applied to the reaction mixtures. In competition assays, 50- and 100-fold excesses of unlabeled DNA fragments were added.
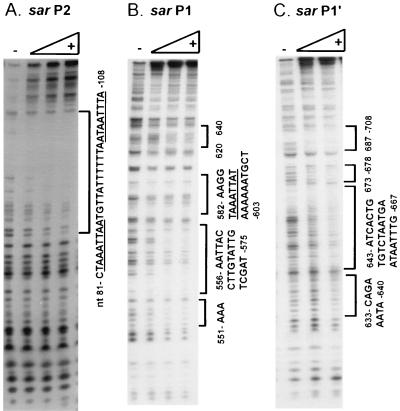
To determine the binding site of SarR and verify the specificity of binding to the sar promoter region, DNase I footprinting analysis was performed. To identify the SarR binding site, a 109-bp EcoRI-HindIII fragment derived from pUC18 containing the 49-bp sequence (27) was end labeled at the EcoRI or HindIII site separately and subjected to DNase I footprinting with various concentrations of SarR. Analysis of the footprint of the plus strand (EcoRI site end labeled) (Fig. 5A) disclosed the protected region (nt 81TAAATTAATGTTATTTTTTAATAATTTA108) (2) to be extremely AT rich (96%), thus implying specific binding of SarR to this region but not to the more GC-rich polylinker region of pUC18, even when higher concentrations of SarR were used in the assay. A similar protection site was also found for the minus strand (HindIII site end labeled) (data not shown). The SarR protected region was found to consist of a 7- to 8-base sequence (TAAATTAA, with the last base variable) conserved in both strands (e.g., 101ATAATTTA108 being the complement of TAAATTAA) and throughout the sar promoter region (27).
FIG. 5.
DNase I footprinting assays of SarR with end-labeled 32P sar P2 (49-bp fragment), P1 (nt 531 to 859), and P1′ (nt 620 to 859) promoter fragments. The sequence was deduced from G+A ladder reactions run in parallel by the standard method (26). The following amounts of SarR were applied to the sar P2 and P1 reactions: 30, 60, and 100 ng. With sar P1′, only lanes containing 30 and 60 ng of SarR protein are shown. The binding site of SarR on the sar P3 promoter (underlined) was also mapped: 373 TTACTAAATTAAAAAAATTA402 (footprinting data not shown) (2). The underlined nucleotides represent the 7- or 8-base sequence conserved in the binding site throughout the sar promoter region.
We also determined the binding of SarR to other sar promoter regions. In a previous study (27), we had shown that an inverted repeat region (nt 553 to 593) upstream of the sar P1 promoter may play a role in repressing sar P1 transcription. Recognizing that SarR binds to a large P1 fragment in gel shift assays (Fig. 4), we performed footprinting analysis with two different DNA fragments upstream of the sar P1 promoter (329 bp [nt 531 to 859] and 240 bp [nt 620 to 859] [2]). Using 32P-end-labeled sense strand, the SarR-protected region on the 329-bp sar P1 promoter fragment was found to comprise several regions including nt 551 to 553, 556 to 575, 582 to 603 (586TAAATTAT593), and 620 to 640 (Fig. 5B). In analyzing the smaller 240-bp P1 fragment, four additional protected regions, downstream of the above binding sites, were uncovered: nt 633 to 640, 643 to 667, 673 to 678, and 687 to 708 (Fig. 5C). Thus, the inverted repeat region (nt 553 to 593), which has previously been shown to play a putative role in repressing P1 transcription (27), is also part of the SarR binding sites. We also uncovered the SarR binding site on the sar P3 promoter: 373TTACTAAATTAAAAAAATTA402 (footprinting data not shown) (2). In comparing the broad binding sites protected by SarR, a common feature is their highly AT-rich nature. More remarkably, the 7- to 8-bp conserved sequence (TAAATTAA) was included within the SarR binding sites in each of the sar promoter fragments (P2, P1, and P3).
Expression of the sarR gene in RN6390 and its isogenic sar and agr mutants.
During the growth cycle, the major sar gene product such as SarA partially mediates its effect by binding to the agr promoter to influence RNAII and RNAIII transcription. To ascertain if the sarR gene is modulated by sar or agr (i.e., acting downstream of the sar or agr regulatory cascade), we assayed for sarR transcription in parental strain RN6390 and its isogenic agr and sar mutants. To ensure that comparable amounts of total cellular RNA were applied to each lane, we compared rRNA bands stained with ethidium bromide among the lanes (Fig. 6A). As displayed in Fig. 6B, the transcription of sarR in RN6390 could be detected in mid-log phase and was maximally expressed during the postexponential phase. Accounting for minor experimental variations, the observation that sarR transcription was not significantly altered in sar and agr mutants indicated that sar and agr did not regulate sarR as one would expect if these loci lie downstream of the sarR regulatory cascade. This notion was support by the finding (described below) that a mutation in sarR affects sar and agr transcriptions.
FIG. 6.
Expression of sarR in parental strain RN6390 and its sar (ALC488) and agr (RN6911) mutants. (A) Northern blots of sarR transcript in RN6390 and its isogenic sar and agr mutants. Ten micrograms of total cellular RNA was applied to each lane. The sarR probe was a 345-bp fragment (nt 208 to 552). OD650 of 0.7, 1.1, and 1.7 represent mid-log, late log, and early stationary phases, respectively, as predicted from the growth cycle. (B) Ethidium bromide stain of the above RNA gel prior to transfer to a hybridization membrane. (C) Expression of SarR on an immunoblot probed with anti-SarR antibody 2C7. Each lane contains 25 μg of cell extract of RN6390 grown to late log and early stationary phases. Cells at mid-log phase expressed little SarR; as expected, SarR was not detected in the sarR mutant ALC1713 (data not shown). The positive control lane contains 0.1 μg of purified SarR protein.
We also determined the expression of the SarR protein during the growth cycle by immunoblotting. Using anti-SarR monoclonal antibody 2C7 (1:1,000 dilution), we probed an immunoblot of cell extracts of RN6390 derived from cells grown to late-log (OD650 = 1.1) and postexponential (OD650 = 1.7) phases. Employing ∼25 μg of cellular proteins in each lane, we found that the expression of SarR corresponded quite well with the pattern of sarR transcription, with SarR expression detectable at late log phase and maximal during the postexponential phase (Fig. 6C).
Expression of sar in a sarR mutant.
Aware that the SarR protein likely modulates SarA expression by virtue of its binding to the sar promoter region, we proceeded to construct a sarR deletion mutant by replacing the sarR gene with an ermC gene in strain RN6390 (see Materials and Methods). Northern analysis confirmed that the transcription of sarR was disrupted in sarR mutant ALC1713 (data not shown). To analyze the effect of sarR on individual sar promoters, we cloned P2 (nt 1 to 180 plus 197 bp upstream), P3 (nt 364 to 525), P1 (nt 620 to 859), and combined (or native) P2-P3-P1 promoter (nt 1 to 859 plus 197 bp upstream) (2, 27) upstream of the gfpuvrreporter gene in shuttle plasmid pALC1484. Using flow cytometry to evaluate promoter activity, we found that the sar P1 and combined P2-P3-P1 promoters were more active in the sarR mutant than the parental control (mean fluorescence of 5.01 ± 0.29 [log scale] in RN6390 versus 5.84 ± 0.13 in the sarR mutant and 5.49 ± 0.21 in RN6390 versus 8.44 ± 0.24 in the mutant, for P1 and combined promoters, respectively, during the postexponential phase). However, the relative weakness of the sar P3 and P2 promoters compared with the P1 promoter (∼20- to 30-fold less than P1) (27), coupled with the relative stability of the green fluorescent protein (GFP) reporter, rendered flow cytometry less useful to record small variations in GFP expression during the growth cycle among 10,000 organisms gated for this experiment. Not surprisingly, we failed to detect differences in activation of the weaker sar P3 and P2 promoters between the parent and the isogenic sarR mutant by flow cytometry (27). More specifically, the level of P2 and P3 activation as detected by flow cytometry was only slightly above background levels (data not shown).
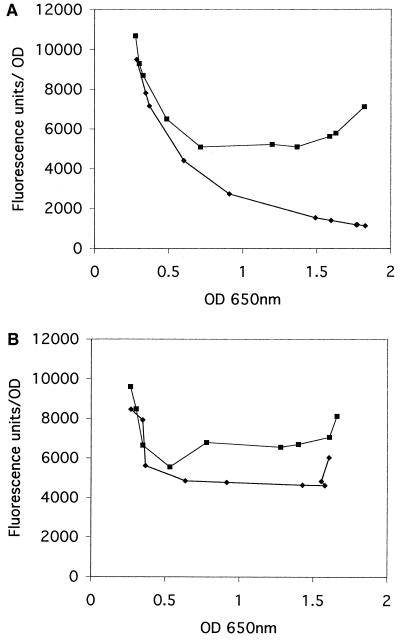
To obtain more quantitative fluorescence data for a larger number of bacterial cells, we used a multifunction fluorescence spectrophotometer in a microtiter format (FL600; BioTek Instruments) to measure the OD and total fluorescence of samples (200 μl) obtained serially during the growth cycle. To minimize the variation in fluorescence attributable to cell density, we plotted fluorescence per OD unit against OD over a 10-h period (extending from log to stationary phase). The data showed that the sar P1-GFP fusion activity in the sarR mutant was higher than that in the parental strain (RN6390) throughout the growth cycle (Fig. 7A). The early decline in fluorescence for both strains is likely attributable to the carryover of the GFP from the overnight inoculum. Following the decline, the highest level of GFP fusion in the mutant occurred late in the stationary phase (at∼7 to 10 h after initial culture dilution) (Fig. 7A), at a time when the sarR transcript level and the expression of SarR in the parent were highest (Fig. 6B and C). Curiously, the level of GFP fusion for the sar P1 promoter in the parental strain declined after the initial dilution, but was higher than the background (∼300 fluorescence units), during the growth cycle. This finding is likely attributable to a steady decrease in P1 promoter activation (per bacterial cell) in the parental strain as the cell cycle progressed (Fig. 7A). Additionally, a lack of contribution from upstream promoters (i.e., P3 and P2) to modulate P1 activity in this promoter fragment may conceivably play a role. Similar studies were also conducted for the combined or native sar P2-P3-P1 promoter linked to the gfpuvrreporter. In this instance, the combined promoter activity in the sarR mutant was also higher than that of the parent (Fig. 7B). As with the P1 promoter, the level of activity for the combined promoter decreased after initial culture dilution for both strains and then increased during the postexponential phase. Of interest, the increase in combined promoter activity with growth in the parental strain seemed to suggest that the sequence element upstream of P1 may have contributed to the overall increase in combined promoter activity during the postexponential phase. However, we were not able to demonstrate any differences in fluorescence for the P2 or P3 promoter GFP fusion between the sarR mutant and its isogenic parent. Notably, the fluorescence of the P2 and P3 promoters was only slightly above background; thus the fluorescent assays may not be sensitive enough to detect subtle differences in P2 and P3 promoter activities.
FIG. 7.
Promoter activation of sar P1 and combined P2-P3-P1 promoters fused to a gfpuvrreporter gene as evaluated in a fluorescence spectrophotometer (FL600; BioTek Instruments). (A) Recombinant shuttle plasmid pALC1484 containing the sar P1 promoter linked to gfpuvr(excitation maxima at 488 nm) was introduced into strain RN6390 (♦) and its isogenic sarR mutant ALC1713 (▪). A negative control (RN6390 containing pALC1484 with no promoter fragment) showed no significant background fluorescence (background = ∼300 fluorescence units [data not shown]). Cells were obtained hourly (200 μl of each in duplicate) during the growth cycle (from h 2 to 10 after an initial dilution of 1:100 in fresh medium) to obtain fluorescence and OD values in the same instrument. To minimize variations in fluorescence attributable to cell density, the data are presented as average of reported fluorescence per OD unit in triplicate samples plotted against the mean OD. The error bar was too small to be discerned (typically less than 100 fluorescence units). The experiment was repeated at least thrice; one representative experiment is shown. (B) Plot similar to that in panel A except that the combined sar P2-P3-P1 promoter fragment was used in place of the P1 promoter in the recombinant pALC1484 containing the gfpuvrreporter gene. In similar assays with the individual sar P2 and P3 promoters linked to the gfpuvrreporter in the isogenic pair, we detected no differences in GFPuvr expression between the parental strain and the sarR mutant. However, the level of fluorescence associated with individual P2 and P3 promoters was very low and only slightly above background levels.
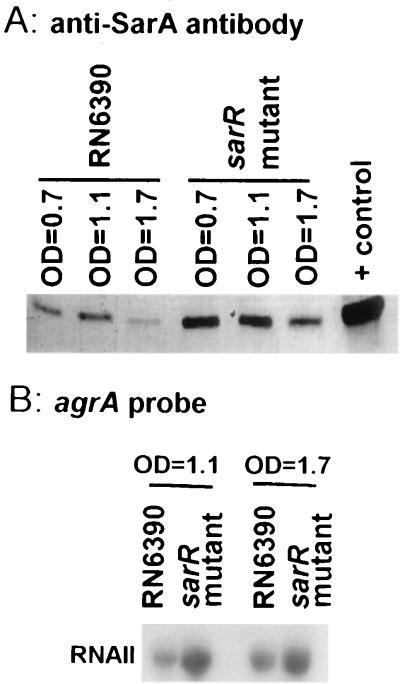
As the P1 transcript is the predominant sar transcript (27), we reasoned that an intact sarR gene may repress the expression of SarA, the major sar regulatory molecule. Accordingly, we obtained cell extracts of the isogenic sarR strains during various stages of the growth cycle. Using cell extracts (25 μg of protein each) of the sarR mutant obtained at different phases of the growth cycle, we probed an immunoblot with anti-SarA monoclonal antibody 1D1 (15). As shown in Fig. 8A, the sarR mutant expressed higher levels of SarA protein than the isogenic parent at ODs representing mid-log, late log, and stationary phases. Notably, in both the parental and mutant strains, SarA expression was maximal during the late log phase and tapered toward the stationary phase. Presumably, the reduction in SarA expression in the parental strain during the stationary phase may be partially explained by increased proteolytic activity and hence processing of SarA in stationary cells (27, 35). Additional immunoblot analyses with increased amounts of cell extracts (at 25 μg each) from mid-log, late log, and stationary phases also confirmed higher expression of SarA in the sarR mutant than in the parental strain (data not shown). Taken together, these data support our original hypothesis that SarR is likely a DNA binding protein that binds to the sar promoter to down-regulate SarA expression.
FIG. 8.
Effect of the sarR mutation on SarA and agr expression. (A) SarA expression during mid-log, late log, and early stationary phases; (B) agrA (RNAII) transcription. (A) Immunoblot of cell extracts (5 μg of protein each) of RN6390 and the sarR mutant (harvested at mid-log, late log, and stationary phases) probed with anti-SarA monoclonal antibody 1D1 at 1:2,000 dilution. The positive control lane contains 0.5 μg of purified SarA. Similar results were obtained with 25 μg of protein per lane. (B) Northern blot of the RNAII (agrA probe) transcript in RN6390 and the sarR mutant (10 μg of total RNA each). The agrA probe corresponds to nt 3830 to 4342 according to the published sequence 23.
In a prior study, we showed that the level of SarA correlates with the extent of agr activation (15). Based on our data on SarA expression in sarR mutant ALC1713, we predicted that transcription of RNAII of agr would be enhanced in a sarR mutant compared with the parental strain. Northern analysis of sarR mutant ALC1713 with an agrA (RNAII) probe confirmed this prediction (Fig. 8B). Collectively, these data supported the notion that SarR likely down-regulates SarA expression, probably by binding to the sar promoter to down-modulate sar P1 transcription, resulting in the modulation of target genes (e.g., agr) downstream of the sar regulatory cascade.
DISCUSSION
The sar locus is characterized by a 372-bp sarA ORF the expression of which is partially controlled by the sar triple-promoter system. Within the triple-promoter region are multiple repeats and potential peptide coding regions that can form a complex network of regulatory elements for sar promoter activation (2, 6). In prior studies, we demonstrated that the level of SarA, the major sar regulatory molecule (6, 16), correlates with the extent of agr activation (15). Because of the complexity of the sar promoter region, we speculated that a regulatory protein(s) likely binds to the region to modulate SarA expression. Deletion analysis has implicated the P3-P1 interpromoter region to play an essential role in down-regulating P1 transcription and hence SarA expression during the growth cycle (6, 27). Based on these studies, we speculate that the P3-P1 interpromoter region may serve as a binding site for a repressor protein (27). Curiously, the region upstream of the P1 promoter but downstream of the P3 promoter is homologous with another AT-rich region upstream of the P2 promoter (Fig. 1A). Accordingly, we previously partially purified from crude cell lysates a ∼12-kDa protein (SarR) with a DNA-specific column containing a sar P2 promoter fragment (27). In this study, we have significantly extended the characterization of the SarR protein by characterizing the gene, revealing its sar promoter binding activity, and analyzing its binding sites on the sar promoters. More importantly, a mutation in sarR led to increases in sar P1 (Fig. 7A) and combined P2-P3-P1 (native) (Fig. 7B) promoter activities. The result of such a mutation is an increase in SarA expression and subsequent activation of agr, a major sar target gene, thus justifying the premise that SarR is a DNA binding protein that down-regulates SarA expression.
Using the N-terminal sequence of the crudely purified SarR protein and the availability of a partial amino acid sequence from the S. aureus genome (TIGR), we were able to amplify part of the sarR gene with degenerate oligonucleotides and thus generate a probe to screen for a pACYC177 plasmid clone containing the intact sarR gene. The SarR protein was found to have a small molecular size (13.7 kDa), a deduced basic pI (9.8), and a predominance of charged amino acid (34%) all features consistent with a DNA binding protein. Gel shift and footprinting assays indeed confirmed the DNA binding activity of SarR to the sar promoter regions. Secondary structure analysis with the PHD program (37) revealed that SarR consists primarily of α-helices (75%), which implies a single-domain structure. A homology search revealed that SarA is homologous to the entire SarR protein, with 51% similarity between the two. Interestingly, the homology is relatively global (Fig. 1C). Whether SarA and SarR belong to a new family of regulatory proteins that do not possess the classic DNA binding motifs (e.g., helix-turn-helix and leucine zipper) is entirely speculative. Nevertheless, the region from residues 54 to 77 of SarA, which shows homology with the corresponding region of SarR, also has sequence similarity to the DNA binding domain (residues of 175 to 198) of VirF, a positive transcriptional regulator of virulence genes in Shigella flexneri (13, 18).
The binding site of SarR on the 49-bp fragment derived from the sar P2 promoter in pUC18 has been determined to be a 28-bp AT-rich sequence (nt 81 to 108) (2) situated 12 bp upstream of the P2 −35 promoter box (Fig. 5A). The footprinting reaction appeared to be specific because the adjacent GC-rich region from pUC18 was not protected by SarR. In a previous study, we speculated that a 7 to 8-bp sequence (TAATTAA), repeated eight times in both sense and nonsense strands throughout the sar promoter region, may serve as a binding site for a DNA binding protein. Despite a high AT content, this motif is found only once within a 6-kb sequence of agr. In examining the 28-bp SarR binding site on the P2 promoter, this motif was found twice (underlined in Fig. 5A). Likewise, the motif was also found in the SarR-protected sites of the sar P1 (underlined in Fig. 5B) and P3 (legend to Fig. 5) promoter regions. However, unlike that of the P2 promoter, this motif was found only once within the SarR binding site on the sar P1 promoter (nt 576 to 583). Remarkably, this 8-bp sequence was found within an inverted repeat region (nt 553 to 593), previously thought to be essential to the repression of P1 transcription (27). More importantly, SarR binds to this inverted repeat region within the sar P1 promoter. Using synthesized and end-labeled 8-bp fragment (TAAATTAA) and SarR in gel shift assays, we did not detect any binding of this sequence motif to SarR. We surmise that the fragment may be too small to preserve a binding site, since a 34-bp sequence (nt 567 to 600) (2) upstream of the P1 promoter but containing the 8-bp sequence did bind to SarR (reference 27 and unpublished data). The presence of multiple binding repeats would be consistent with the laddering pattern as observed in gel shift assays of SarR with the sar P1, P2, and P3 promoters (Fig. 4). However, given the abundance of shifted bands (Fig. 4) relative to binding sites on the sar P1, P2, and P3 promoters, SarR may bind to these sites in multimers as well. Given the observations that the SarA protein can exist in dimer form (36) and that SarR is homologous with SarA, it is also conceivable that heterodimers of SarA and SarR can bind to the sar promoters to elicit the binding pattern that we have repeatedly observed in gel shift assays of whole-cell extracts with sar promoter fragments (unpublished data).
The effect of SarR on sar-related transcription in S. aureus is complex. The complexity arises from the observation that multiple regulatory proteins likely bind to the sar promoter region. For instance, SigB, a stress-induced transcription factor, binds to the sar P3 promoter upstream of P1, resulting in activation of P3 (30) and an associated decrease in P1 expression (1, 7, 27). Likewise, SarA also binds to its own promoter (reference 27 and unpublished data). Finally, recent inquiry into the S. aureus genome (at TIGR) hints at the presence of other SarA homologs. Thus, SarR likely represents a series of proteins that bind to the sar promoter to modulate SarA expression in S. aureus. The multiplicity of regulatory proteins that likely bind to the sar promoter implies that the correlation of sarR expression to the activation of individual sar promoters, being more complex, is less likely to be linear. Despite these complexities, our studies clearly indicate that one of the major effects of SarR is to down-modulate sar P1 transcription from the strongest sar promoter (2, 27). This premise is supported by several findings: (i) an augmentation in sar P1 promoter activity by FACS; (ii) an enhancement in sar P1 activity toward the postexponential phase in the sarR mutant as documented by a more quantitative fluorescence-based assay; and (iii) a notable increase in sar P1 activity in the sarR mutant during the postexponential phase, at a time when SarR expression is normally maximal in the parental strain (Fig. 7A and 6C).
Although SarR has been shown to bind to the sar P3 and P2 promoter regions by gel shift and footprinting assays, the specific effect of this protein on the sar P3 and P2 promoters (i.e., increase or decrease in promoter activation during various growth phases) is less clear. In particular, we could not establish any significant differences in P3 promoter activity during the growth cycle between the isogenic pair, using both FACS and a more sensitive and quantitative fluorescence spectrophotometric assay. We reported previously that a decrease in P3 promoter activity, as found in a sigB mutant, was associated with a concomitant increase in sar P1 promoter activity and hence elevated SarA expression (7). It is thus tempting to speculate that SarR, with its maximal expression concurring with peaked SigB activity during the postexponential phase, may influence sar P3 promoter and hence down-regulate the P1 promoter downstream (Fig. 1A and 7A), possibly by virtue of a partial promoter occlusion as has been observed in E. coli (1).
The effect of SarR on the combined or native sar promoter driving SarA expression was also found to be down-regulatory, as confirmed by increased P2-P3-P1 fusion activity in the sarR mutant compared with the parent (Fig. 7B). Contrasting with the downward trend of the P1 sample in the parental strain (Fig. 7A), the activity of the combined or native sar promoter in the parental strain increased as the cells transitioned to stationary phase. As P1 is the predominant sar promoter, these data also implied that the sequence upstream of the P1 promoter (i.e., P3 and/or P2) in the parental strain probably plays an important role in up-regulating P1 promoter activity. However, the exact manner in which they contribute to the increase in P1 activity as the cell cycle progresses is not clearly defined. Thus, the main effect of the SarR protein is likely to down-modulate the combined sar P2-P3-P1 promoter activity, probably via an effect on the sar P1 promoter. The heightened increase in the combined promoter activity during the postexponential phase in the sarR mutant also hints at the effect of unopposed activator(s) in the absence of SarR. The candidate activators include SarA (27), and possibly other SarA homologs, acting either alone or in combination.
Thus, the major effect of SarR on binding to the sar promoter is a reduction in P1 and in the combined (P2-P3-P1) sar promoter activity, thus leading to a decrease in SarA protein expression, preferentially in the late log and stationary phases. This argument was bolstered by our immunoblot studies in which we demonstrated that a sarR mutant expressed a higher level of SarA than its parental counterpart in mid-log, late-log, and stationary-phase cells (Fig. 8A). Interestingly, maximal SarA expression in the isogenic pair was observed during the late log rather than stationary phase. With the parental strain, the decrease in SarA expression during the stationary phase may be paritally explained by maximal SarR expression. In the case of the sarR mutant, we speculated that proteolytic processing (27, 35) or other unknown factor(s) may serve to decrease SarA protein expression during the stationary phase.
Because of the structural complexity of the sar promoter region (2, 27), it seems logical to surmise that activator and repressor proteins likely bind to the sar promoter to modulate the expression of SarA, the major sar regulatory molecule. Recent transcriptional fusion studies suggested that SarA is auto regulatory (27), with SarA likely to be an activator for its own expression (27). In contrast, activation of SigB likely leads to a down-regulation in SarA expression (7). The combination of activator (SarA) and down-modulators (SigB and SarR) for SarA expression argues for the complex interaction between regulatory proteins and the sar promoter to modulate downstream genes.
ACKNOWLEDGMENTS
The contribution of the S. aureus genome database at TIGR and at the University of Oklahoma to this work is gratefully acknowledged. We thank Willem Van Wamel for the S. epidermidis, S. haemolyticus, and S. saprophyticus strains from Utrecht University Hospital, The Netherlands.
This work was supported in part by NIH grants AI30061 and AI37142. A. L. Cheung is a recipient of an AHA-Genentech Established Investigator Award from the American Heart Association.
REFERENCES
- 1.Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1981;29:939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- 2.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blake M S, Johnston K H, Russell-Jones G J, Gotschlich E C. A rapid sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 4.Boyce J M. Epidemiology and prevention of nosocomial infections. In: Crossley K B, Archer G L, editors. The staphlococci in human diseases. New York, N.Y: Churchill Livingstone; 1997. pp. 309–329. [Google Scholar]
- 4a.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A L, Bayer M G, Heinrichs J H. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol. 1997;179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung A L, Chien Y T, Bayer A S. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect Immun. 1999;67:1331–1337. doi: 10.1128/iai.67.3.1331-1337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A L, Eberhardt K, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 9.Cheung A L, Eberhardt K, Heinrichs J H. Infect. Immun. 2243–2249. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung A L, Fischetti V A. The role of fibrinogen in staphylococcal adherence to catheters in vitro. J Infect Dis. 1990;161:1177–1186. doi: 10.1093/infdis/161.6.1177. [DOI] [PubMed] [Google Scholar]
- 11.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung A L, Krishnan M, Jaffe E A, Fischetti V A. Fibrinogen acts as a bridging molecule in the adherence of Staphylococcus aureus to cultured human endothelial cells. J Clin Investig. 1991;87:2236–2245. doi: 10.1172/JCI115259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung A L, Projan S J. Cloning and sequencing of sarA: a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien C-T, Manna A C, Projan S J, Cheung A L. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar dependent gene regulation. J Biol Chem. 1999;274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 15.Chien Y, Manna A C, Cheung A L. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol Microbiol. 1998;30:991–1001. doi: 10.1046/j.1365-2958.1998.01126.x. [DOI] [PubMed] [Google Scholar]
- 16.Chien Y, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;237:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 17.Doran J E, Raynor R H. Fibronectin binding to protein A-containing staphylococci. Infect Immun. 1981;33:683–689. doi: 10.1128/iai.33.3.683-689.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale T L. Genetic basis of virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heim R, Cubitt A B, Tsien R Y. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 20.Janzon L, Arvidson S. The role of the δ-hemolysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji G, Beavis R, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 22.Jones K F, Manjula B N, Johnston K H, Hollingshead S K, Scott J R, Fischetti V A. Location of variable and conserved epitopes among the multiple serotypes of streptococcal M protein. J Exp Med. 1985;161:623–628. doi: 10.1084/jem.161.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornblum J, Kreiswirth B, Projan S J, Ross H, Novick R P. Agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 373–402. [Google Scholar]
- 24.Lee C Y. Cloning of genes affecting capsule expression in Staphylococcus aureus strain M. Mol Microbiol. 1992;6:1515–1522. doi: 10.1111/j.1365-2958.1992.tb00872.x. [DOI] [PubMed] [Google Scholar]
- 25.Mahmood R, Khan S A. Role of upstream sequences in the expression of the staphylococcal enterotoxin B. gene. J Biol Chem. 1990;265:4652–4656. [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Manna A C, Bayer M G, Cheung A L. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol. 1998;180:3828–3836. doi: 10.1128/jb.180.15.3828-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayville P, Ji G, Beavis R, Yang H, Goger M, Novick R P, Muir T W. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc Natl Acad Sci USA. 1999;96:1218–1223. doi: 10.1073/pnas.96.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDevitt D, Francois P, Vaudaux P, Foster T J. Cloning and sequencing of the clumping factor of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 30.Miyazaki E, Chen J M, Ko C, Bishai W R. The Staphylococcus aureus rsbW gene encodes an anti-sigma factor of SgiB. J Bacteriol. 1999;181:2846–2851. doi: 10.1128/jb.181.9.2846-2851.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–4577. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novick R P. The staphylococcus as a molecular genetic system. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH; 1990. pp. 1–40. [Google Scholar]
- 33.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 34.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3977. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Projan S J, Novick R P. The molecular basis of pathogenicity. In: Crossley K B, Archer G L, editors. The staphylococci in human diseases. New York, N.Y: Churchill Livingstone; 1997. pp. 55–81. [Google Scholar]
- 36.Rechtin T M, Gillaspy A F, Schumacher M A, Brennan R G, Smeltzer M S, Hurlburt B K. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol Microbiol. 1999;33:307–316. doi: 10.1046/j.1365-2958.1999.01474.x. [DOI] [PubMed] [Google Scholar]
- 37.Rost B, Sander C, Schneider R. PHS—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 38.Russo-Marie F, Roederer M, Sager B, Herzenberg L A, Kaiser D. β-Galactosidase activity in single differentiating bacterial cells. Proc Natl Acad Sci USA. 1998;90:8194–8198. doi: 10.1073/pnas.90.17.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sau S, Sun L, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule gene in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schenk S, Laddaga R A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 41.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]