Abstract
The present study aimed to characterize and compare the skin and gut microbial communities of rohu at various post‐harvest stages of consumption using quantitative real‐time polymerase chain reaction and 16S rRNA‐based amplicon sequencing. Real‐time PCR amplification detected higher copy numbers for coliform bacteria—Escherichia coli, Salmonella enterica and Shigella spp. in the marketed fish—compared to fresh and frozen samples. The 16S rRNA data revealed higher alpha diversity measurements in the skin of fish from different retail markets of Dhaka city. Beta ordination revealed distinct clustering of bacterial OTUs for the skin and gut samples from three different groups. At the phylum level, Proteobacteria was most abundant in all groups except the Fusobacteria in the control fish gut. Although Aeromonas was found ubiquitous in all types of samples, diverse bacterial genera were identified in the marketed fish samples. Nonetheless, low species richness was observed for the frozen fish. Most of the differentially abundant bacteria in the skin samples of marketed fish are opportunistic human pathogens enriched at different stages of postharvest handling and processing. Therefore, considering the microbial contamination in the aquatic environment in Bangladesh, post‐harvest handling should be performed with proper methods and care to minimize bacterial transmission into fish.
Keywords: bioinformatics, fish, metagenomics, microbial contamination, post‐harvest
Significance and Impact of the Study: Rohu (Labeo rohita) is the most popular fish in Bangladesh. In this manuscript, we have characterized bacterial community and quantified coliforms from the skin and gut samples of rohu across the supply chains (live fresh, frozen and marketed) using TaqMan real‐time PCR and Illumina‐based 16S rRNA sequencing to identify microbial community differences in the post‐harvest stages. The qPCR and sequence data revealed transmission of opportunistic pathogens in the marketed fish samples from the environment. The results raised significant health concerns and advocated proper post‐harvest handling and processing of fish in the megacities of Bangladesh.

Introduction
Fish provides around 60% of the animal protein intake in Bangladesh and the fisheries sector continues to grow at a rate of around 8% per annum (Belton et al. 2011). Bangladesh is now the fourth‐largest producer of inland fishes after China, India and Myanmar (Bhowmick and Crumlish 2016). More than 17 million people are directly involved in the fisheries industry including farming, fishing, processing and trading (Shamsuzzaman et al. 2017). However, due to widespread environmental pollutions and poor hygienic fish handling practices, there is an increasing risk of microbial contamination of fish and fishery products, and foodborne illness to the public caused by poor handling and consumption of contaminated food products. The rearing environment and water used for the fish processing are identified as potential sources of microbial contamination in the farms (Siddique et al. 2021), natural habitats (Majumdar et al. 2014), wholesale markets (Hossain et al. 2018; Foysal et al. 2019) and processed fish (Sanjee and Karim 2016). Each year, about 30 million people are infected by cholera and diarrhoea, mostly through contaminated water and food (Van Egmond et al. 2007).
While the fish itself is a natural reservoir of several bacteria potentially pathogenic for humans such as Mycobacterium spp., Streptococcus iniae, Vibrio alginolyticus and V. vulnificus, other pathogens associated with foodborne illness include V. cholerae, Escherichia coli, Aeromonas spp., Salmonella spp., Listeria monocytogenes and Clostridium perfringens (Novotny et al. 2004). Notably, in our recent study, a diverse range of bacterial genera including Escherichia, Vibrio, Klebsiella, Acinetobacter, Enterobacter, Proteus, Streptococcus, Staphylococcus, Serratia, Aeromonas, Pseudomonas and Flavobacterium were detected from the gut and skin samples of marketed hilsa fish in Bangladesh (Foysal et al. 2019). More importantly, even in frozen fish and crustaceans' species, pathogenic bacteria were shown to present according to several studies performed in Dhaka, the capital city of Bangladesh, highlighting the severity of microbial contamination in local fish products (Rokibul et al. 2013; Samia et al. 2014; Noor et al. 2021). However, in all studies, samples were collected from a specific point, and thus the sources of pathogen transmission in various stages of post‐harvest have not been identified.
Among the fish cultured in Bangladesh, rohu (Labeo rohita) is most popularly consumed, accounting for 25% of total production (Foysal et al. 2020). Due to the high consumption rate of rohu in this nation (Belton et al. 2011; Foysal et al. 2020), plus previous contamination reports arising from unhygienic processing and inappropriate storage environments (Foysal et al. 2019), the high risk of foodborne diseases is a major concern. It is therefore important to characterize the microbiota of this fish in different stages of post‐harvest to identify the sources of contamination. Owing to the limitations of plate‐based morphological and biochemical characterization, culture‐independent high‐throughput sequencing (HTS) has been used to trace down the potential sources of microbial contamination in post‐harvest fishes and food products (Bledsoe et al. 2016; Antunes‐Rohling et al. 2019; Xing et al. 2021). In this study, we aimed to identify and compare the skin and gut microbiota of live, frozen and marketed rohu fish samples, with the aid of culture‐independent quantitative real‐time polymerase chain reaction (RT‐PCR) and 16S rRNA gene sequencing technology. The results would help to reveal and understand the endogenous and exogenous bacteria present in rohu, especially those that are important foodborne pathogens, and thus allowing the development of prevention strategies and better food safety policies to safeguard public health safety.
Results and discussion
Following sample pooling, nine pairs of fish gut and skin samples were acquired, with three pairs each prepared from fresh, marketed and frozen fish, respectively.
TaqMan real‐time PCR detection of coliform bacteria
The real‐time qPCR efficiency for E. coli, S. enterica and Shigella spp. was 91·2% with R 2 value of 0·999. In the skin, E. coli (P‐value <0·001), S. enterica (P‐value = 0·008) and Shigella spp. (P‐value = 0·002) had higher copy numbers in marketed samples, compared to fresh control and frozen fish where the differences were found even higher for E. coli (P‐value <0·001), S. enterica (P‐value = 0·002) and Shigella spp. (P‐value = 0·001). However, no significant difference in copy number was observed between fresh and frozen fish skin samples. In the gut however, only E. coli was detected from all three sources with higher copy numbers in the marketed samples than fresh (P‐value = 0·021) and frozen (P‐value = 0·012) samples. Shigella spp. was found only in all samples in low copy numbers. No S. enterica was detected in the gut samples from any sources (Table 1).
Table 1.
Escherichia coli and coliforms in live fresh, marketed and frozen fish with TaqMan assay
| Bacteria (cells per gram) | Skin | Gut | ||||
|---|---|---|---|---|---|---|
| Fresh | Marketed | Frozen | Fresh | Marketed | Frozen | |
| E. coli | b2·4 ± 0·6 × 102 | a1·08 ± 0·4 × 104 | b2·3 ± 0·5 × 102 | b1·6 ± 0·4 × 102 | a4·8 ± 0·5 × 102 | b1·5 ± 0·3 × 102 |
| S. enterica | b3·8 ± 0·2 × 101 | b6·7 ± 0·3 × 101 | b3·2 ± 0·3 × 101 | — | — | — |
| Shigella spp. | b6.1 ± 0·4 × 101 | a4·2 ± 0·5 × 102 | b5·9 ± 0·7 × 101 | b1·2 ± 0·2 × 101 | b1·2 ± 0·3 × 101 | b1·1 ± 0·2 × 101 |
Rows with different superscript letters indicate significantly different values between the skin and gut samples from different sources; fresh, marketed and frozen samples (n = 5). Skin and gut samples were compared separately.
Attributing to poor waste disposal and hygiene, widespread environmental pollution as a result of rapid urbanization and the lack of public awareness, transmission of opportunistic and pathogenic bacteria into fish from environmental sources are common in Bangladesh (Rahman et al. 2007; Foysal et al. 2019). Therefore, we aimed to quantify and compare the coliform bacteria in the gut and skin microbial compositions of live, market and frozen rohu, which is the most consumed fish in this nation. The qPCR results suggest skin contamination of marketed fish by coliforms. Similar to a recent study (Ava et al. 2020), the higher copy number of E. coli, Shigella and S. enterica may have a link to water use for cleaning and basket to hold fish in the market. The frozen fish in the supermarket usually undergone an extra step of hygienic cleaning and processing and that might remove some bacteria before freezing. Hence, good practice in post‐harvest processing can reduce the chance of coliform contamination in marketed fish.
Differences in rohu fish skin and gut microbiota between groups
After quality filtering and merging of overlapping paired‐end reads, 689 364 sequences were retained and clustered into 218 OTUs. Taxonomic classification of these OTUs subsequently revealed 12 phyla and 88 genera.
We first examined the fish skin microbiota of each group. Alpha diversity analysis revealed that the market fish skin samples had the highest bacterial richness and diversity, which were significantly greater than that in the control fresh and frozen groups (Fig. 1a). The marketed fish also had 16 unshared genera, not found in fresh and frozen fish (Fig. 1b). Beta diversity analysis based on the unweighted (R 2 = 0·8224, P‐value <0·001) and weighted (R 2 = 0·7223, P‐value <0·001) UniFrac distance metric further demonstrated clear separations among the fresh, market and frozen fish skin samples, indicating that there are distinct differences in microbial composition between groups (Fig. 1c,d).
Figure 1.
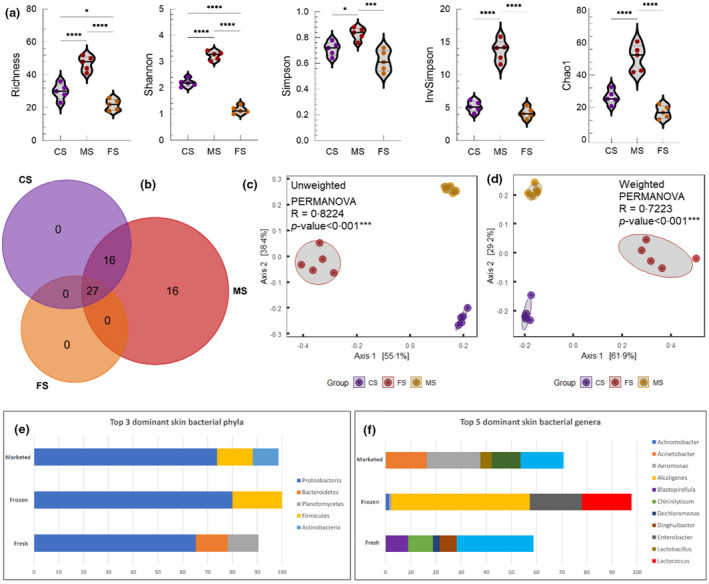
Diversity and composition of the skin microbiota in samples collected from fresh, frozen and marketed fish. (a) Alpha diversity in terms of richness, Shannon, Simpson, InvSimpson and Chao1. (b) The number of shared and unique genera in the skin of fresh, frozen and marketed fish. (c, d) Unweighted and weighted UniFrac distance metric. Predominant bacteria at phylum (e) and genus (f) level in the skin of fresh, frozen and marketed fish. Abbreviations: CS, control skin; FS, frozen skin; MS, marketed skin. [Colour figure can be viewed at wileyonlinelibrary.com]
We next analysed the relative abundances of bacterial phyla and genera in each group. At the phylum level, Proteobacteria was the most dominant bacterial phylum present in fresh, frozen and market fish skin samples, at 65·3, 80 and 73·8%, respectively (Fig. 1e). It was then followed by Bacteroidetes (12·9%) and Planctomycetes (12·4%) within the control group. Firmicutes was the second most common bacterial phylum present in both frozen and market groups, at 19·8 and 14·4%, respectively, and with Bacteroidetes (0·14%) being the third most abundant phylum in the former and Actinobacteria (10·4%) in the latter. At the genus level, the control samples were dominated by bacteria of environmental origin including Shewanella (30·5%), Chitinilyticum (9·9%), Blastopirellula (8·9%), Dinghuibacter (6·9%) and Dechloromonas (2·6%), while in the frozen samples Alcaligenes (55%), Enterobacter (20·7%) and Lactococcus (19·7%) made up at least 95% of the total bacterial population. Among the market fish skin samples, Aeromonas (21·2%), Shewanella (17·1%), Acinetobacter (16·4%), Pseudomonas (11·5%) and Lactobacillus (4·5%) constituted the top five most abundant bacterial genera (Fig. 1f).
Differential abundance analysis subsequently revealed 28 distinguishing bacterial genera (Table 2), among which 17 of them were significantly more abundant in the market group as compared to their counterparts in control fresh and frozen groups. These include Rothia, Acinetobacter, Aeromonas, Pseudomonas, Streptococcus, Staphylococcus, Enterococcus and Escherichia‐Shigella. The remaining 11 genera, comprising primarily environmental organisms, were detected with significantly higher abundances in the control group than that of the comparison groups. Nevertheless, Pseudomonas had significantly higher abundance in the skin samples of marketed fish (Fig. 3). It is also important to note that these distinguishing genera, if not undetected, were the least abundant in the frozen samples.
Table 2.
Distinguished genera in the gut and skin of control fresh, frozen and market fish
| Bacterial genera | Fresh | Frozen | Marketed | Adjusted P |
|---|---|---|---|---|
| Skin | ||||
| Dinghuibacter | 6·85 ± 0·28 | 0 | 0 | <0·001 |
| Rothia | 0 | 0·02 ± 0·03 | 3·38 ± 0·16 | <0·001 |
| Acinetobacter | 0 | 0·02 ± 0·02 | 16·39 ± 0·97 | <0·001 |
| Pseudomonas | 0·33 ± 0·08 | 0·27 ± 0·26 | 11·5 ± 0·61 | <0·001 |
| Psychrobacter | 0 | 0 | 3·31 ± 0·22 | <0·001 |
| Chitinilyticum | 9·94 ± 0·7 | 0 | 0 | <0·001 |
| Nitrospira | 0·29 ± 0·02 | 0 | 0 | <0·001 |
| Dechloromonas | 2·59 ± 0·26 | 0 | 0 | <0·001 |
| Brochothrix | 0 | 0 | 0·51 ± 0·06 | <0·001 |
| Leucobacter | 0 | 0 | 1·11 ± 0·13 | <0·001 |
| Aeromonas | 1·44 ± 0·18 | 0·87 ± 0·66 | 21·2 ± 2·36 | <0·001 |
| Vitreoscilla | 0 | 0 | 0·07 ± 0·01 | <0·001 |
| Reyranella | 1·32 ± 0·17 | 0 | 0 | <0·001 |
| Soonwooa | 0 | 0 | 0·06 ± 0·01 | <0·001 |
| Carnobacterium | 0 | 0 | 0·12 ± 0·02 | <0·001 |
| Roseomonas | 0·2 ± 0·03 | 0 | 0 | <0·001 |
| Flavobacterium | 1·07 ± 0·14 | 0 | 0·16 ± 0·03 | <0·001 |
| Shewanella | 30·49 ± 3·63 | 0 | 17·1 ± 1·46 | <0·001 |
| Enterococcus | 0 | 0 | 0·62 ± 0·09 | <0·001 |
| Aquicella | 0·23 ± 0·04 | 0 | 0 | 0·001 |
| Staphylococcus | 0 | 0·07 ± 0·13 | 1·31 ± 0·16 | 0·001 |
| Corynebacterium | 0 | 0·01 ± 0·02 | 0·95 ± 0·15 | 0·001 |
| Arthrobacter | 0 | 0 | 0·61 ± 0·11 | 0·003 |
| Paeniglutamicibacter | 0 | 0 | 4·04 ± 0·75 | 0·003 |
| Pirellula | 0·39 ± 0·07 | 0 | 0 | 0·003 |
| Halomonas | 1·01 ± 0·19 | 0 | 0 | 0·004 |
| Chryseobacterium | 1·74 ± 0·32 | 0 | 0·07 ± 0·07 | 0·004 |
| Escherichia–Shigella | 0 | 0·01 ± 0·01 | 0·1 ± 0·01 | 0·005 |
| Gut | ||||
| Brachybacterium | 0·53 ± 0·06 | 0 | 0 | <0·001 |
| Cetobacterium | 46·72 ± 2·97 | 0·27 ± 0·3 | 9·34 ± 4·92 | <0·001 |
| Plesiomonas | 0·93 ± 0·12 | 0·03 ± 0·03 | 0·05 ± 0·02 | <0·001 |
| Serratia | 0·89 ± 0·14 | 0 | 0 | 0·001 |
Each value displayed under the ‘Fresh’, ‘Frozen’ and ‘Marketed’ columns represents mean relative abundance with standard deviation (n = 5).
Figure 3.

Circular LEfSe cladogram representing the phylogenetic distribution of bacterial lineage in six different groups. The lineage with LDA scores of 2·0 or above is displayed here. The red, green, blue, purple, cyan and orange colours indicate CG, CS, FG, FS, MG and MS, respectively. The dot at the centre represents the OTUs at the phylum level while the outer circle of dots denotes OTUs at genus level. The order, family and genus that are significantly different between groups are given in the upper right corner with respective colour codes. Abbreviations: CS, control skin; FS, frozen skin; MS, marketed skin; CG, control gut; FG, frozen gut; MS, marketed gut. [Colour figure can be viewed at wileyonlinelibrary.com]
We also examined the rohu fish gut microbial composition in each group. For alpha diversity, apparent but non‐significant greater bacterial richness and diversity were observed in the control fresh group while Shannon and InviSimpson had significantly higher values for fresh samples compared to frozen fish (Fig. 2a). In all, 17 shared genera in three groups and one unique genus in the marketed fish gut was observed (Fig. 2b). Beta diversity analysis further revealed distinct and significant clustering of samples belonging to each group, both in terms of unweighted (R 2 = 0·3324, P‐value <0·002) and weighted (R 2 = 0·2224, P‐value <0·004) UniFrac distance metric (Fig. 2c,d).
Figure 2.

Diversity and composition of the gut microbiota in samples collected from fresh, frozen and marketed fish. (a) Alpha diversity in terms of richness, Shannon, Simpson, InvSimpson and Chao1. (b) The number of shared and unique genera in the gut of fresh, frozen and marketed fish. (c, d) Unweighted and weighted UniFrac distance metric. Predominant bacteria at phylum (e) and genus (f) level in the gut of fresh, frozen and marketed fish. Abbreviations: CG, control gut; FG, frozen gut; MS, marketed gut. [Colour figure can be viewed at wileyonlinelibrary.com]
In the control group, Proteobacteria and Fusobacteria were found predominantly, with roughly comparable abundances 52·1 and 46·7%, respectively (Fig. 2e). In contrast, the frozen group was contained almost entirely Proteobacteria, with a mean relative abundance of 99·4%, while the market group contained mostly Proteobacteria (85%), followed by 9·3 and 5·6% of Fusobacteria and Firmicutes, respectively. At the genus level, both Aeromonas (51·3%) and Enterobacter (48%), and Aeromonas (47·9%) and Cetobacterium (46·7%) were the two most predominant bacterial genera identified in frozen and control fresh groups, respectively (Fig. 2f). In the market group, the most predominant bacterial genus was Aeromonas, at 83·7% (Fig. 3), followed by Cetobacterium (9·3%) and Lactococcus (5·4%). Only four significant bacterial genera were identified (Table 2). Brachybacterium, Cetobacterium, Plesiomonas and Serratia were all significantly more abundant in the control fresh than that of the frozen and market ones.
In the present study, most of the bacteria identified from control fresh, marketed and frozen fish including species of Aeromonas, Cetobactrium and Shewanella, that are the normal flora of gut and skin of fish (Austin 2006; Egerton et al. 2018; Ramírez et al. 2018; Foysal et al. 2020). However, relative and differential abundance in marketed samples suggested rapid colonization of opportunistic bacteria and coliforms from environmental sources, mostly from unhygienic handling and water used for processing and preservation. Differentially abundant reads for genera Pseudomonas, Staphylococcus, Enterobacter and Corynebacterium with more accurate Bonferroni correction revealed the transmission of environmental bacteria and opportunistic pathogens into marketed fish at different stages of handling, transportation, processing and marketing. These results are consistent to previous studies that investigate microbial communities in marketed and processed fish in Bangladesh. Molecular‐based studies identified opportunistic pathogen including Aeromonas, Vibrio, Streptococcus, Staphylococcus and Serratia from the marketed hilsa (Tenualosa ilisha) fish for human consumption in Bangladesh (Hossain et al. 2018; Foysal et al. 2019). Another plate‐based study identified high prevalence of pathogenic bacteria including Pseudomonas, Salmonella, Shigella, Vibrio, Listeria and Staphylococcus from the marketed and processed sea fish samples in Dhaka city (Rokibul et al. 2013; Noor et al. 2021). However, compared to previous studies on sea and hilsa fish (Hossain et al. 2018; Foysal et al. 2019; Noor et al. 2021), we found low Listeria, Streptococcus, Salmonella, Shigella and Serratia abundance in marketed rohu fish. The sea fish and hilsa mostly caught at southern part of Bangladesh and thus the longer transportation time, frequent exposure to environmental contaminants (air, water, soil) and mixture with other fish samples in the local market during selling probably associated with more pathogen transmission in these fish species. In contrast, rohu fish in the present study were collected from Gazipur, a city next to Dhaka. Therefore, distance between fish harvested and selling point together with fish handling during transportation and marketing play a crucial role in faecal contamination in major cities of Bangladesh.
In the present study, though we could not able to detect some previously reported pathogen in marketed and processed fish, yet the number and abundance for some opportunistic pathogens in marketed fish were found significantly higher. Acinetibacter, Enterobacter, Aeromonas, Pseudomonas and Staphylococcus were detected at high copy numbers in the marketed fish. Aeromonas species are ubiquitous in the environment, especially in the aquatic systems and many species are reported to play role in fish diseases (Janda and Abbott 2010; Igbinosa et al. 2012) wherein Acinetibacter, Pseudomonas and Enterobacter accelerate fish spoilage through the production of histamines (Kim et al. 2001; Kuley et al. 2017). In Bangladesh, the fishes after caught are transported in live condition to local retail markets in big drums with water from different sources. The higher Aeromonas and Enterobacter abundance in the gut can be linked to transfer of bacteria in the gut from the transporting water. On the other hand, Staphylococcus that is mostly food‐borne pathogen and commonly found on animal skin where their percentage of abundance indicate the degree of spoilage (Gutiérrez et al. 2012; Bujjamma and Padmavathi 2015). Therefore, the presence of these bacteria can be correlated with poor fish quality in the city markets of Bangladesh.
The processed frozen fish in the present study found to contain more Proteobacteria and Firmicutes, and low abundance of opportunistic pathogens and coliforms. The Enterobater and Alcaligenes possesses Psychrophillic characteristics (Macaulay et al. 1963; Arulkumar et al. 2019) and lactic acid bacteria including Lactococcus and Lactobacillus can survive in ultra‐low temperature, identified in processed fish and meat (Kato et al. 2000; Matamoros et al. 2009). The higher concentration of the selected bacteria in frozen fish hence can be correlated to their growth capabilities at low temperature storage. Nevertheless, the lower diversity of bacteria in frozen fish primarily linked to faster processing, cared handling, use of chemicals and preservatives that slowed down and inhibit the growth of microbes. Therefore, transmission of pathogen in fish can be controlled with good post‐harvest handling and transportation practices.
In Bangladesh, fish are well cooked before consumption, so there is no direct risk associated with bacterial contamination. However, widespread transmission of opportunistic pathogens results in drug‐resistant isolates in the aquatic species that posing continuous threat to public health safety (Hossain et al. 2018; Foysal et al. 2019; Siddique et al. 2021). To overcome the problem and considering environmental safety, appropriate steps should be taken to keep fish out of possible transmission during processing, preservation and selling at the market. Some strong initiatives by the government in recent times have proven worthwhile against environmental pollutions as reflected by the results of the present study. To prevent further contamination in marketed fish, exposure could be minimized through application of advanced automated fish processing techniques (Komlatsky et al. 2019) or proper handling during processing, chilling, use of legislated antimicrobials like nitrites, sulphides and organic acids (Ghaly et al. 2010; Nagarajarao 2016).
Overall, the present study revealed the transmission of environmental and opportunistic bacteria into fish at post‐harvest handling in the capital city of Bangladesh. However, low volume of samples in HTS and qPCR are some of the limitations for this study. Therefore, future research is recommended on more varieties of fish from pond, river and ocean as well as from different cities to know the correlation between bacteria and factors that hasten microbial contamination. To tackle the impacts of widespread pollution on fisheries sectors, government should take appropriate awareness and training programmes for the farmers, workers associated with processing, transportation and preservation, and sellers to reduce the chance of microbial contamination into fish at different stages of post‐harvest.
Materials and methods
Animal ethics
The collection of samples and anesthetization of fish were carried out following the guidelines and recommendations of the Guidelines for the Use of Fishes in Research published by the American Fisheries Society (2014) since Bangladesh has no specific guidelines for the use of fish in research. The research was approved and strictly supervised by the Dean, Graduate Research Committee of the Department of Biotechnology, Bangabandhu Sheikh Mujibur Rahman Agriculture University, Gazipur, Bangladesh.
Sample collection and processing
For this study, three monoculture ponds for rohu farming in Salna, Gazipur district (24·0290°N, 90·3862°E), Dhaka, Bangladesh, were selected. The live fresh fish cultured for 8 months with an average weight of 831 ± 26 g were randomly selected from three different ponds, five from each pond. The freshly caught live fish was used as control to compare the bacterial diversity with marketed and frozen fish. Following the supply chain, 15 fish (814 ± 31 g) were randomly collected from Kawranbazar (23·7516°N, 90·3943°E), Mogbazar (23·7494°N, 90·4090°E) and Malibagh (23·7466°N, 90·4128°E) fish retail markets, five from each market between 24 and 48 h of harvest. At the same time, 15 frozen fish (832 ± 36 g) samples were collected from a retail supermarket located at Kawranbazar, Mogbazar and Malibagh. A total of 45, 30 dead and 15 freshly caught live fishes were collected from fish culture ponds, retail markets and supermarkets (Fig. S1). We have collected marketed and frozen samples after 48 h post‐harvest and processing as this time‐point is reported as an indicator for the growth of fish spoilage microbes (Golden and Arroyo‐Gallyoun 1997; Rezaabad et al. 2017; Tsironi and Taoukis 2017).
Collected fish samples were kept at 4°C (excluding the live fishes) and transported immediately (<1 h) to our laboratory at the Institute of Genetic Engineering and Biotechnology, Bangabandhu Sheikh Mujibur Rahman Agricultural University, Gazipur, Dhaka, Bangladesh, and stored at −80°C until processing. In all, 15 live fish were euthanized by dipping into 250 mg l−1 solution of MS‐222 (Sigma‐Aldrich, Steinheim, Germany), stored at −80°C for 15 min and dissected together with other dead fishes stored previously to collect intestine samples. From each fish, a skin sample was prepared by swabbing the scale surface and the fish body (after descaling) prior to resuspension in 50 μl phosphate‐buffered saline (PBS). Subsequently, skin samples prepared from the respective three fishes of the same pond, retail market and supermarket were homogenized and pooled together (n = 5). The gut sample was prepared by excising the whole gut, separation of interior, mid and distal gut, followed by rinsing with PBS and dissected into small pieces. Approximately 300 g of samples, 100 g from each part of the gut containing the mucosa and the gut content were then transferred into a 2 ml Eppendorf tube and homogenized using beads in a tissue lyser (Qiagen, Hilden, Germany). The gut samples of the respective three fishes for the same pond (n = 5), retail market (n = 5) and supermarket (n = 5) were pooled together for the DNA extraction.
DNA extraction
The bacterial DNA from the pooled samples was extracted using DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer's instructions. Extracted DNA was quantified using the NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA), diluted into 50 ng μl−1 final concentration with DEPC treated water and preserved at −20°C until further use.
TaqMan quantitative real‐time PCR for the detection of coliform bacteria
Standard plasmid and curve for E. coli, Salmonella enterica and Shigella spp. were constructed according to the methods described earlier (Liu et al. 2019). Briefly, purified fragments were cloned into pGEM®‐T Easy Vector System (Thermo Fisher Scientific), transformed into One Shot™ TOP10 Chemically Competent E. coli (Invitrogen, CA, USA). Plasmid was extracted using Plasmid Miniprep Kit (Qiagen). The purity and concentration of DNA were checked in NanoDrop 2000 cc (Thermo Fisher Scientific) and Qubit™ 3 Flurometer (Thermo Fisher Scientific). After calculation of plasmid DNA copy number using formula, 6·02 × 1023 × (ng μl−1 × 10−9)/ bp × 660, concentration of 100–109 cell per μl was achieved by serial dilution, and used as qPCR standards. Real‐time TaqMan PCR assay was performed using rfbE, hilA, ipaH pathogen‐specific virulence genes (Table S1) targeted E. coli, S. enterica and Shigella spp. Real‐time TaqMan assay was carried out in 25 μl final mixture containing 12·5 μl TaqMan Fast Advanced Master Mix (Thermo Fisher Scientific), 1 μl of each forward and reverse primer (10 μmol l−1), 1 μl probe (5 μmol l−1), 1 μl of genomic DNA and 8·5 μl of nuclease‐free water (Sigma‐Aldrich, Steinheim, Germany). The following qPCR conditions were used: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 55°C for 10 s and 72°C for 30 s. All runs included positive and negative controls (nuclease‐free water). Absolute copy number of unknown samples were determined based on corresponding standard (Rao et al. 2013).
Illumina MiSeq sequencing
PCR amplification of the V3–V4 region of the 16S rRNA gene was performed on each DNA sample using the primer sets as specified in Illumina's 16S metagenomic sequencing library preparation protocol. Except the primer set, the PCR mixture was prepared as described in the previous section for the detection of E. coli and coliforms. PCR parameters were initial denaturation at 95°C for 30 s, followed by 34 amplification cycles of 95°C for 10 s, 55°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 5 min. PCR products were examined using 1% agarose gel electrophoresis. Amplified PCR products were purified using Agencourt Ampure XP beads (Beckman Coulter, Brea, CA), followed by indexing PCR using the Nextera XT index kit (Illumina, San Diego, CA). Sequencing of the barcoded amplicons was performed on an Illumina MiSeq instrument using the 2 × 300 paired‐end v3 chemistry (600 cycles).
Bioinformatics
Sickle program (https://github.com/najoshi/sickle) was used for quality trimming of raw sequencing reads with the following parameters: ‐q 20 –l 200. Read quality was checked before and after trimming in FastQC pipeline (https://github.com/s‐andrews/FastQC). MeFiT program used for merging of overlapping pair‐end reads with default parameters (Parikh et al. 2016). Filtering of merged overlapping sequences, de novo greedy clustering into operational taxonomic units (OTUs) at 97% identity threshold level were performed using Micca version 1.7.2 (Albanese et al. 2015). Taxonomic classification of OTUs was performed using the micca classify command against SILVA database (release 1.32) (Quast et al. 2012).
Rarefaction was performed at the depth value of 7132. Alpha and beta diversity estimations were performed using the microbiomeSeq R package (https://github.com/umerijaz/microbiomeSeq). For alpha diversity, the species richness, Shannon, Simpson, InvSimpson and Chao1 diversity metrics were employed. For beta diversity analysis, principal coordinate analysis (PCoA) based on unweighted and weighted UniFrac distance metric was used. Nonparametric statistical analysis of the distance metric was performed using the permutational multivariate analysis of variance method.
Statistical analysis
All statistical analysis were performed in Rstudio (v4·1·2) (R Core Team 2013). Data were subjected to Shaprio–Wilk's and Levene's tests to assess the normal distribution and homogeneity of the variances. Distinguishing genera between groups was analysed using anova with Bonferroni correction. Differences with adjusted P‐values <0·01 were considered significant. The qPCR data were compared with one‐way anova with Tukey's HSD between groups for skin and marketed samples.
Calculations
Standard curve for the qPCR: y = mx + b, where m = slope and b = intercept.
Absolute quantification of DNA copies in unknown sample: , where n = C T .
Conflict of Interests
Authors declare no conflict of interest.
Authors’ Contribution
Conceptualization: Md Javed Foysal and Md Mahbubur Rahman. Sample collection, methodology and data curation: A.Q.M. Robiul Kawser, Md Javed Foysal, Hazrat Ali and Sulav Indra Paul. Formal analysis: Eng Guan Chua and Muhammad A.B. Siddik. Writing—first draft: A.Q.M. Robiul Kawser, Md Javed Foysal and Eng Guan Chua. Writing—reviewing and editing: Adnan Mannan, Md Mahbubur Rahman and Alfred Tay.
Supporting information
Figure S1. An outline of experimental design and sample collection procedures from rohu fish across the supply chain.
Table S1. Primers and probes used for TaqMan real‐time PCR.
Acknowledgements
The authors would like to acknowledge Helicobacter Research Laboratory, University of Western Australia and Harry Perkins Institute of Medical Research for providing sequencing and computational analysis supports. Open access publishing facilitated by Curtin University, as part of the Wiley ‐ Curtin University agreement via the Council of Australian University Librarians. [Correction added on 6 July 2022, after first online publication: CAUL funding statement has been added.]
The authors Kawser and Foysal contributed equally to this work.
Data Availability Statement
The raw sequence data in fastq format are currently available in National Centre for Biotechnology Information (NCBI) under the BioProject accession number PRJNA667752.
References
- Albanese, D. , Fontana, P. , De Filippo, C. , Cavalieri, D. and Donati, C. (2015) MICCA: a complete and accurate software for taxonomic profiling of metagenomic data. Sci Rep 5, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes‐Rohling, A. , Calero, S. , Halaihel, N. , Marquina, P. , Raso, J. , Calanche, J. , Beltrán, J.A. , Álvarez, I. et al. (2019) Characterization of the spoilage microbiota of hake fillets packaged under a modified atmosphere (MAP) rich in CO2 (50% CO2/50% N2) and stored at different temperatures. Foods 8, 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulkumar, A. , Paramasivam, S. , Rameshthangam, P. and Paramithiotis, S. (2019) Evaluation of psychrophilic, mesophilic, histamine forming bacteria and biogenic amine content in the muscle of mud spiny lobster, Panulirus polyphagus (HERBST, 1793) during ice storage. J Food Saf 39, e12582. [Google Scholar]
- Austin, B. (2006) The bacterial microflora of fish, revised. Scientific World J 6, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ava, A. , Faridullah, M. , Lithi, U. and Roy, V. (2020) Incidence of Salmonella and Escherichia coli in fish farms and markets in Dinajpur, Bangladesh. Bangladesh J Sci Ind Res 55, 65–72. [Google Scholar]
- Belton, B. , Karim, M. , Thilsted, S. , Collis, W. and Phillips, M. (2011) Review of aquaculture and fish consumption in Bangladesh. The WorldFish Center 53, 1–71. [Google Scholar]
- Bhowmick, B. and Crumlish, M. (2016) Aquaculture health management and biosecurity practises in South West of Bangladesh. Bangladesh J Vet Med 14, 263–269. [Google Scholar]
- Bledsoe, J.W. , Peterson, B.C. , Swanson, K.S. and Small, B.C. (2016) Ontogenetic characterization of the intestinal microbiota of channel catfish through 16S rRNA gene sequencing reveals insights on temporal shifts and the influence of environmental microbes. PLoS One 11, e0166379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujjamma, P. and Padmavathi, P. (2015) Prevalence of Staphylococcus aureus in fish samples of local domestic fish market. Intl J Curr Microbiol Appl Sci 4, 427–433. [Google Scholar]
- Egerton, S. , Culloty, S. , Whooley, J. , Stanton, C. and Ross, R.P. (2018) The gut microbiota of marine fish. Front Microbiol 9, 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foysal, M. , Momtaz, F. , Kawsar, A. , Rahman, M. , Gupta, S. and Tay, A. (2020) Next‐generation sequencing reveals significant variations in bacterial compositions across the gastrointestinal tracts of the Indian major carps, rohu (Labeo rohita), catla (Catla catla) and mrigal (Cirrhinus cirrhosis). Lett Appl Microbiol 70, 173–180. [DOI] [PubMed] [Google Scholar]
- Foysal, M.J. , Momtaz, F. , Robiul Kawser, A. , Chaklader, M.R. , Siddik, M.A. , Lamichhane, B. , Tay, A.C.Y. , Rahman, M.M. et al. (2019) Microbiome patterns reveal the transmission of pathogenic bacteria in hilsa fish (Tenualosa ilisha) marketed for human consumption in Bangladesh. J Appl Microbiol 126, 1879–1890. [DOI] [PubMed] [Google Scholar]
- Ghaly, A.E. , Dave, D. , Budge, S. and Brooks, M. (2010) Fish spoilage mechanisms and preservation techniques. Am J Appl Sci 7, 859. [Google Scholar]
- Golden, D.A. and Arroyo‐Gallyoun, L. (1997) Relationship of frozen‐food quality to microbial survival Quality in frozen food. Boston, MA: Springer. [Google Scholar]
- Gutiérrez, D. , Delgado, S. , Vázquez‐Sánchez, D. , Martínez, B. , Cabo, M.L. , Rodríguez, A. , Herrera, J.J. and García, P. (2012) Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Appl Env Microbiol 78, 8547–8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, Z.Z. , Farhana, I. , Tulsiani, S.M. , Begum, A. and Jensen, P.K. (2018) Transmission and toxigenic potential of Vibrio cholerae in hilsha fish (Tenualosa ilisha) for human consumption in Bangladesh. Front Microbiol 9, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbinosa, I.H. , Igumbor, E.U. , Aghdasi, F. , Tom, M. and Okoh, A.I. (2012) Emerging Aeromonas species infections and their significance in public health. Scientific World Journal 2012, 625023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J.M. and Abbott, S.L. (2010) The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23, 35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, Y. , Sakala, R. , Hayashidani, H. , Kiuchi, A. , Kaneuchi, C. and Ogawa, M. (2000) Lactobacillus algidus sp. nov., a psychrophilic lactic acid bacterium isolated from vacuum‐packaged refrigerated beef. Intl J Syst Evol Microbiol 50, 1143–1149. [DOI] [PubMed] [Google Scholar]
- Kim, S.‐H. , Field, K.G. , Chang, D.‐S. , Wei, C.‐I. and An, H. (2001) Identification of bacteria crucial to histamine accumulation in Pacific mackerel during storage. J Food Prot 64, 1556–1564. [DOI] [PubMed] [Google Scholar]
- Komlatsky, V. , Podoinitsyna, T. , Verkhoturov, V. and Kozub, Y. (2019) Automation technologies for fish processing and production of fish products. J Physics: Conference Series 1399, 044050. [Google Scholar]
- Kuley, E. , Durmus, M. , Balikci, E. , Ucar, Y. , Regenstein, J.M. and Özoğul, F. (2017) Fish spoilage bacterial growth and their biogenic amine accumulation: inhibitory effects of olive by‐products. Int J Food Prop 20, 1029–1043. [Google Scholar]
- Liu, Y. , Cao, Y. , Wang, T. , Dong, Q. , Li, J. and Niu, C. (2019) Detection of 12 common food‐borne bacterial pathogens by TaqMan real‐time PCR using a single set of reaction conditions. Front Microbiol 10, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay, D. , Hawirko, R. and James, N. (1963) Effect of pasteurization on survival of certain psychrophilic bacteria. Appl Microbiol 11, 90–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar, P.R. , Sarker, B.S. , Dey, S.C. , Saha, D. , Adhikary, R.K. and Mondal, S. (2014) Bacterial hazards especially pathogenic to human in consumable marine fishes of Noakhali Sadar, Bangladesh. Am J Food Tech 9, 257–265. [Google Scholar]
- Matamoros, S. , Leroi, F. , Cardinal, M. , Gigout, F. , Chadli, F.K. , Cornet, J. , Prevost, H. and Pilet, M. (2009) Psychrotrophic lactic acid bacteria used to improve the safety and quality of vacuum‐packaged cooked and peeled tropical shrimp and cold‐smoked salmon. J Food Prot 72, 365–374. [DOI] [PubMed] [Google Scholar]
- Nagarajarao, R.C. (2016) Recent advances in processing and packaging of fishery products: a review. Aquatic Procedia 7, 201–213. [Google Scholar]
- Noor, R. , Acharjee, M. , Ahmed, T. , Das, K.K. , Paul, L. , Munshi, S.K. , Urmi, N.J. , Rahman, F. et al. (2021) Microbiological study of major sea fish available in local markets of Dhaka city, Bangladesh. J Microbiol Biotechnol Food Sci 2021, 2420–2430. [Google Scholar]
- Novotny, L. , Dvorska, L. , Lorencova, A. , Beran, V. and Pavlik, I. (2004) Fish: a potential source of bacterial pathogens for human beings. Vet Med 49, 343. [Google Scholar]
- Parikh, H.I. , Koparde, V.N. , Bradley, S.P. , Buck, G.A. and Sheth, N.U. (2016) MeFiT: merging and filtering tool for illumina paired‐end reads for 16S rRNA amplicon sequencing. BMC Bioinformatics 17, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , Peplies, J. and Glöckner, F.O. (2012) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Res 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2013). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. http://www.R‐project.org/ [Google Scholar]
- Rahman, M. , Huys, G. , Rahman, M. , Albert, M.J. , KüHn, I. and MöLlby, R. (2007) Persistence, transmission, and virulence characteristics of Aeromonas strains in a duckweed aquaculture‐based hospital sewage water recycling plant in Bangladesh. Appl Env Microbiol 73, 1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, C. , Coronado, J. , Silva, A. and Romero, J. (2018) Cetobacterium is a major component of the microbiome of giant Amazonian fish (Arapaima gigas) in Ecuador. Animals 8, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, X. , Huang, X. , Zhou, Z. and Lin, X. (2013) An improvement of the 2ˆ (−delta delta CT) method for quantitative real‐time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 3, 71. [PMC free article] [PubMed] [Google Scholar]
- Rezaabad, M.K. , Khodanazary, A. and Hosseini, S.M. (2017) Effect of chitosan treatments and vacuum packaging on the shelf life of spangled emperor lethrinus nebulosus fillets stored in refrigerator. J Packag Technol Res 1, 157–164. [Google Scholar]
- Rokibul, M.H. , Mrityunjoy, A. , Eshita, D. , Kamal, K. , Tasnia, A. , Muhammad, A. , Kazi, K. and Rashed, N. (2013) Microbiological study of sea fish samples collected from local markets in Dhaka city. Intl Food Res J 20, 1491. [Google Scholar]
- Samia, S. , Galib, H. , Tanvir, A. , Basudeb, C. , Walliullah, M. , Tasnia, A. , Sakil, M.M. , Afsana, F. et al. (2014) Microbiological quality analysis of shrimps collected from local market around Dhaka city. Intl Food Res J 21, 33. [Google Scholar]
- Sanjee, S.A. and Karim, M. (2016) Microbiological quality assessment of frozen fish and fish processing materials from Bangladesh. Intl J Food Sci 2016, 8605689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsuzzaman, M.M. , Islam, M.M. , Tania, N.J. , Al‐Mamun, M.A. , Barman, P.P. and Xu, X. (2017) Fisheries resources of Bangladesh: present status and future direction. Aquac Fish 2, 145–156. [Google Scholar]
- Siddique, A.B. , Moniruzzaman, M. , Ali, S. , Dewan, M. , Islam, M.R. , Islam, M. , Amin, M.B. , Mondal, D. et al. (2021) Characterization of pathogenic Vibrio parahaemolyticus isolated from fish aquaculture of the southwest coastal area of Bangladesh. Front Microbiol 12, 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsironi, T.N. and Taoukis, P.S. (2017) Effect of storage temperature and osmotic pre‐treatment with alternative solutes on the shelf‐life of gilthead seabream (Sparus aurata) fillets. Aquac Fish 2, 39–47. [Google Scholar]
- Van Egmond, H.P. , Schothorst, R.C. and Jonker, M.A. (2007) Regulations relating to mycotoxins in food. Anal Bioanal Chem 389, 147–157. [DOI] [PubMed] [Google Scholar]
- Xing, J. , Xu, X. , Luo, X. , Zheng, R. , Mao, L. , Zhang, S. , Lu, J. and Shen, J. (2021) Characterization of microbial community in cold‐chain hairtail fish by high‐throughput sequencing technology. J Food Protect 84, 1080–1087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. An outline of experimental design and sample collection procedures from rohu fish across the supply chain.
Table S1. Primers and probes used for TaqMan real‐time PCR.
Data Availability Statement
The raw sequence data in fastq format are currently available in National Centre for Biotechnology Information (NCBI) under the BioProject accession number PRJNA667752.


