Abstract
NK cells monitor altered molecular patterns in tumors and infected cells through an ample array of receptors. Two families of evolutionarily distant receptors have converged to enable human NK cells to sense levels of HLA class I ligands, frequently abnormal in altered cells. Whilst different forms of polymorphism are a hallmark of killer‐cell immunoglobulin‐like receptors and their classic HLA‐A, B, and C ligands, genetic diversity of killer‐cell lectin‐like receptors for the non‐classical HLA‐E (CD94/NKG2 heterodimers) is less conspicuous and has attracted less attention. A common pattern of diversification in both receptor families is evolution of pairs of inhibitory and activating homologs for a common ligand, the genes encoding activating receptors being more frequently affected by copy number variation (CNV). This is exemplified by the gene encoding the activating NKG2C subunit (KLRC2 or NKG2C), which marks an NK‐cell subpopulation that differentiates or expands in response to cytomegalovirus. We have studied NKG2C diversity in 240 South European individuals, using polymerase chain reaction and sequencing methods to assess both gene CNV and single‐nucleotide polymorphisms (SNPs) affecting its promoter, coding and 3′‐untranslated (3′UT) regions. Sequence analysis revealed eight common SNPs—one in the promoter, two in the coding sequence, and five in the 3′UT region. These SNPs associate strongly with each other, forming three conserved extended haplotypes (frequencies: 0.456, 0.221, and 0.117). Homo‐ and heterozygous combination of these, together with complete gene deletion (0.175) and additional haplotypes with frequencies lower than 0.015, generate a diversity of NKG2C genotypes of potential immunological importance.
Keywords: alleles, genetic polymorphism, HLA‐E, human genetics, natural killer cell lectin‐like receptors, NKG2C receptor
1. INTRODUCTION
Natural killer cells monitor altered molecular patterns in infected and tumor cells, including downregulation of MHC class I expression. The latter function is achieved by means of inhibitory receptors, belonging to different protein superfamilies that have converged in using common signaling motifs and pathways. Among them, human killer‐cell immunoglobulin‐like receptors (KIR) and mouse lectin‐like Ly49 molecules have independently evolved as families of polymorphic receptors, each family member recognizing subsets of MHC class I allotypes. 1 , 2 , 3 In contrast, CD94/NKG2 heterodimers recognize in each species a non‐classical MHC molecule of low polymorphism presenting the leader peptides encoded in other MHC class I loci. 4 , 5 , 6 , 7 In humans, their HLA‐E ligand can also present a similar peptide encoded by cytomegalovirus (CMV) protein UL40. 8 , 9 , 10 All these families of inhibitory receptors have evolved homologs with NK‐cell activating capacity. One of them, mouse Ly49H, enables NK cells to recognize the murine CMV‐encoded decoy MHC molecule m157, Ly49H genetic variation explaining resistance or susceptibility to the virus. 11 , 12 No human NK‐cell receptor has been demonstrated to play such a critical role in virus recognition or resistance. However, activating CD94/NKG2C heterodimers mark a human NK‐cell subpopulation that differentiates or expands in response to human CMV 10 , 13 , 14 ; and KIR2DS1 has been proposed to contribute to recognition of CMV‐infected cells through ill‐defined mechanisms. 15
KIR, like their murine Ly49 analogues, 16 exhibit extreme gene‐copy number variation and allelic polymorphism, only inferior to that of their MHC ligands. 3 , 17 , 18 In contrast, members of the CD94/NKG2 family are generally considered minimally polymorphic. However, human NKG2C, encoded in the NK complex on chromosome 12 by the KLRC2 gene (more commonly referred to as NKG2C i ), 19 , 20 is characterized by a complete gene deletion that affects in homo‐ or heterozygosis ca. one third of individuals in different world populations. 21 , 22 , 23 , 24 Furthermore, a few single nucleotide polymorphisms (SNPs) are seen in its coding and non‐coding regions (Figure 1), but they have rarely been investigated. 19 , 21 , 25 , 26 , 27 To better characterize NKG2C and its role in immunity, we have studied the distribution of its gene polymorphisms in healthy donors of South European origin.
FIGURE 1.
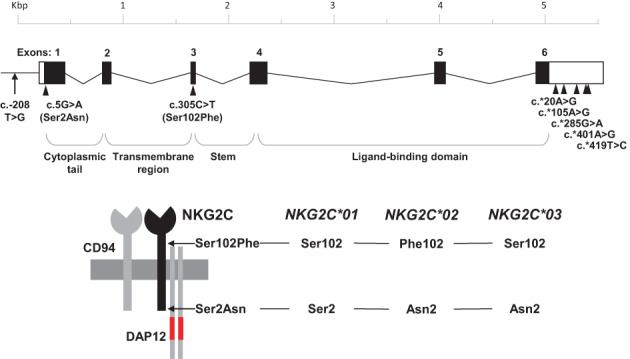
Schemes representing the NKG2C gene, its single‐nucleotide polymorphisms studied herein, and the CD94/NKG2C heterodimer coupled to the signaling molecule DAP12 (immunoreceptor tyrosine‐based activation motifs marked in red). Polymorphic amino acids defining alleles NKG2C*01, *02 and *03 are represented on the NKG2C molecule. Exons and introns are depicted approximately to scale.
2. MATERIALS AND METHODS
2.1. Subjects and samples
We studied a cohort of 240 unrelated healthy controls recruited in our centers in Madrid and Barcelona, and a collection of widely available cell lines, mainly ones established in International Histocompatibility Workshops. After informed consent, genomic DNA was isolated with standard methods from peripheral blood, mononuclear cell (PBMC) suspensions obtained by density gradient centrifugation, or cultured cell lines. Total cytoplasmic RNA was isolated from PBMCs with the RNeasy Plus kit (Qiagen GmbH, D‐40724), and cDNA was synthesized using a first‐strand synthesis kit (GE Healthcare‐Fisher Scientific).
2.2. Sequence analysis
The complete coding sequence (CDS) of NKG2C, and parts of its flanking 5′ and 3′ sequences were amplified in a single 5832‐bp fragment from 28 genomic DNA samples using long‐range polymerase chain reaction (PCR) with primers KLRC2F–383 and KLRR+485b (promoter and 3′‐untranslated region [UTR], respectively, Table S1, and Figure S1). Approximately 100 ng of DNA were amplified with Advantage 2 polymerase mix (BD‐Clontech, Palo Alto) in 20 μl of buffer containing 1 μM of each primer, under the following conditions: initial denaturation at 96°C for 2 min; 10 cycles of 20 s at 95°C, 30 s of primer annealing at 66°C and 7 min extension at 72°C; then 30 cycles of 95°C for 20s, 60°C for 30 s 72°C for 7 min; and a final extension at 72°C for 2 min. The resulting amplicons were separated by electrophoresis in 2% agarose gels, visualized after ethidium bromide staining, and sequenced with the Sanger method and internal primers (Table S1) that covered all six exons. Complete coding sequences were obtained from 19 donors, and partial ones spanning most exons, from an additional nine donors.
Additionally, we sequenced the polymorphic first and third exons of NKG2C in 75 donors, after amplifying them in a single 2871‐bp segment with the same forward primer and reverse KLRC2Ri4 + 197 (intron 4, Table S1, and Figure S1). PCRs were performed in 20 μl of Advantage 2 polymerase mix (BD‐Clontech, Palo Alto, CA, USA) containing 125 μM deoxyribonucleotide triphosphates, ~100 ng of DNA and 1 μM of each primer, in the following conditions: 2 min at 96°C; 10 cycles of 20 s at 95°C, 30 s at 64°C and 4 min at 72°C; 30 cycles of 20 s at 95°C, 30 s at 62°C and 4 min at 72°C; and 2 min at 72°C. Amplified products were verified by agarose gel electrophoresis, and exons 1 and 3 were sequenced as above. Sequences covering ~300 bp of the 5′‐UT and promoter regions upstream of exon 1 were obtained from 85 donors using the two aforementioned genomic fragments.
For 3′UTR analysis, an additional 587‐bp fragment was amplified in all 240 donors with KLRFg669 (exon 6) and KLRR+523 (3′UTR, Table S1, and Figure S1) in 20 μl of buffer (67 mM Tris–HCl, pH 8.8, 16 mM [NH4]2SO4, 2 mM MgCl2, 0.01% Tween‐20, 125 μM deoxyribonucleotide triphosphates) containing 0.6 U of BioTaq polymerase (Bioline), 0.5 μM of each primer and 30–300 ng of DNA. PCR conditions were 2 min at 96°C; 30 cycles of 10 s at 95°C, 20 s at 60°C and 20 s at 72°C; and 2 min at 72°C. Amplified products were verified by electrophoresis in 2% agarose gel and sequenced with primer KLRFg669.
Complementary DNA (cDNA) of NKG2C was amplified with a semi‐nested PCR. The first PCR contained primers targeting the 5′‐ and 3′UTR (KLRC2F‐26 and KLRC2R + 16, respectively, Table S1). For the second PCR, the reverse primer was substituted by KLRRa544 (exon 6, Table S1). Both PCRs were performed in 20 μl of BioTaq buffer containing 0.5 U of polymerase, 1 μM of each primer and 1 μl of cDNA (or product from the first PCR). PCR conditions for both reactions were 95°C for 2 min; 30 cycles at 95°C for 10 s, 55°C for 30 s, 72°C for 1 min, and 72°C for 1 min. The final PCR product, of 706 bp, spanned most of the CDS (exons 1–6).
Sanger sequencing was performed using BigDye cycle sequencing kits, after eliminating excess PCR primers and nucleotides with ExoSAP‐IT (Thermo Fisher Scientific). Sequencing reactions were purified with BigDye‐XTerminator (Thermo Fisher Scientific) and analyzed in an ABI Prism 3100‐Avant Genetic analyzer (Applied Biosystems) in the central facility of IDIPHISA.
2.3. PCR genotyping of NKG2C polymorphisms
NKG2C copy‐number variation was determined as in Moraru et al. 28 To assess the two common SNPs in the CDS (c.5G > A and c.305C > T), we developed a PCR‐SSP (sequence‐specific primers) assay comprising four reactions – two for detecting exon 1 polymorphisms (c.5G > A) and another two for exon 3 (c.305C > T) (Tables 1 and 1S). Each reaction contained ~100 ng of genomic DNA and 1 U of BioTaq polymerase in 15 μl of buffer including primers mixes for each SNP and an internal positive control (IPC) band (Table 1). PCR conditions were 2 min at 96°C, 30 cycles of 20 s at 95°C, 20 s at 61°C and 1 min at 72°C, then 4 min at 72°C, in ABI2720 thermocyclers (Applied Biosystems). Exon 1 and 3 products were ~400‐bp and 1123‐bp long, respectively (Figure 2).
TABLE 1.
Primers and conditions for polymerase chain reaction‐SSP of single‐nucleotide polymorphisms in the NKG2C coding and promoter regions.
| Coding sequence | Specific primers | IPC primers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Reaction | Specificity | Forward | Reverse | μM | Size (bp) | Forward | Reverse | μM | Size (bp) |
| 1 | c.5G | KLRC2F‐274b | KLRRg5 | 0.67 | 405 | FDRA360 | RDRA633 | 0.10 | 607 |
| 2 | c.5A | KLRC2F‐274b | KLRRa5b | 0.67 | 403 | FDRA360 | RDRA633 | 0.10 | 607 |
| 3 | c.305C | KLRC2Fi1‐186 | KLRRc305 | fwd 1.00, rev 0.33 | 1123 | COCHFi8‐86 | COCHR11 + 95 | 0.33 | 1987 |
| 4 | c.305T | KLRC2Fi1‐186 | KLRRt305 | 1.00 | 1123 | COCHFi8‐86 | COCHR11 + 95 | 0.33 | 1987 |
| Promoter region | Specific primers | IPC primers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Reaction | Specificity | Forward | Reverse | μM | Size (bp) | Forward | Reverse | μM | Size (bp) |
| 1 | c.‐208 T | KLRC2F‐358 | KLRRt‐208 | 0.67 | 200 | FDRA360 | RDRA633 | 0.13 | 607 |
| 2 | c.‐208G | KLRC2F‐358 | KLRRg‐208b | 0.67 | 195 | FDRA360 | RDRA633 | 0.13 | 607 |
Note: Primer sequences in Table S1.
Abbreviations: Fwd, forward; IPC, internal positive control; rev, reverse.
FIGURE 2.
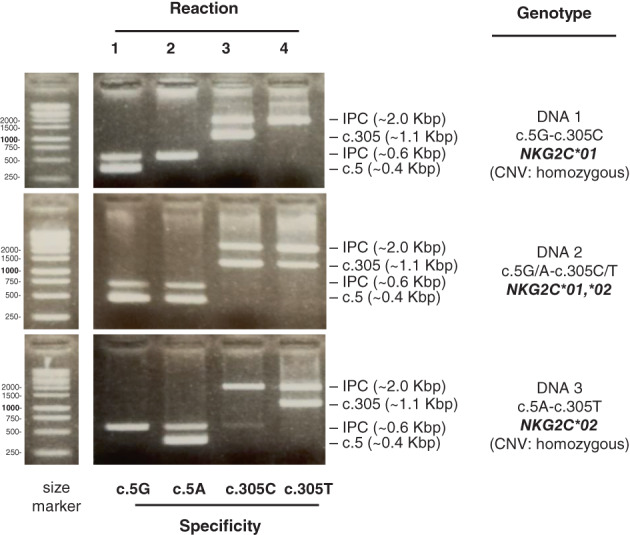
Polymerase chain reaction‐SSP genotyping of NKG2C coding polymorphisms. Three DNA samples with different known genotypes were assayed with the four SSP reactions for the single‐nucleotide polymorphisms of exons 1 (c.5G > A) and 3 (c.305C > T), as indicated. This assay does not inform of sample homo‐ or hemizygosity (CNV), which was determined as in Moraru et al. 28 Exact amplicon sizes are given in Table 1.
Another PCR‐SSP assay, designed for rapid genotyping of the g.‐208 T > G polymorphism, was carried out combining, in two separate reactions, an NKG2C‐promoter‐specific forward primer, KLRC2F‐358, and either of two reverse allele‐specific primers, KLRRt‐208 and KLRRg‐208b (Tables 1 and S1). Reaction mixes contained ~100 ng genomic DNA and 1 U of BioTaq polymerase in 15 μl of PCR buffer. Amplification conditions were 2 min at 96°C; 10 cycles of 20 s at 95°C, 20 s at 65°C and 30s at 72°C; 25 cycles of 20 s at 95°C, 20 s at 62°C and 30s at 72°C; then 4 min at 72°C. The method was validated on 42 sequenced individuals.
3. RESULTS
3.1. Sequence analysis of the NKG2C coding region
We explored NKG2C diversity in a South European population by first using a panel of 101 healthy unrelated donors to identify prevalent polymorphisms along with, possibly, new alleles, and their distribution. This was accomplished through DNA sequencing of the CDS using source templates including cDNA (12 donors), a genomic DNA segment covering the whole NKG2C gene (28 donors), and a partial ~2.9‐kbp genomic amplicon that contained its first and third exons, known to be polymorphic (75 donors). A subset of the donors was sequenced with more than one method with consistent results. These analyses of the NKG2C CDS revealed only two already described SNPs: c.5G > A (exon 1, Ser2Asn, rs28403159) and c.305C > T (exon 3, Ser102Phe, rs1141715) (Figure 1). As previously reported in other populations, 21 , 26 and further analyzed in the following section, the two SNPs are tightly linked to each other in Spaniards, defining two alleles differing at two amino acids, NKG2C*01 (Ser2‐Ser102) and NKG2C*02 (Asn2‐Phe102), whose distribution is described below. However, we found that such linkage is incomplete—asparagine 2 is rarely found with serine 102, defining allele NKG2C*03, as we have described elsewhere 29 ; whilst we observed no examples of the reciprocal combination (Ser2‐Phe102), which could define a fourth allele. Sequence analysis of cDNA synthesized from PBMCs RNA generally demonstrated no trace of alternative splicing, except for one donor in whom intron 1 retention was detectable in part of the transcripts (Figure S2).
3.2. A PCR‐SSP typing assay to study the distribution of the coding dimorphisms NKG2C c.5G > A and c.305C > T
To facilitate higher‐throughput study of NKG2C CDS polymorphism and its putative association with disease and immune responses, we designed a PCR‐SSP typing method that, in combination with our method for assessing CNV, 28 allowed us to detect all possible genotypes of the two SNPs in exons 1 and 3. The assay is based on four reactions, each of which is specific for one of the alternative bases of the two polymorphic sites (Table 1, Figure 2). As is a common quality measure in PCR‐SSP, each reaction includes an internal positive control that alerts of technical failure when neither a specific nor an IPC band is present. For optimization and validation of this technique, it was applied to 75 samples previously analyzed by direct sequencing. We also applied it to characterize the NKG2C c.5G > A and c.305C > T SNPs in a panel of worldwide available reference cell lines, which includes examples of all common genotypes (Table 2) and should help local adjustment and validation of the method in other centers.
TABLE 2.
NKG2C alleles of a panel of reference cell lines.
| Cell lines | NKG2C alleles |
|---|---|
| BM16, BM21, BOLETH, DEU, E4181324, IBW9, JESTHOM, KAS116, LUY, PAR, PITOUT, RML, SWEIG, NK3.3, NK92, YT, DU145 | *01,*01 |
| CB6B, DUCAF, HOM‐2, HOR, KT12, LE023, MZ070782, PLH, RSH, WDV, WT24, HEK‐293T | *01,del |
| AMALA, BH, COX, DBB, OLGA, PE117, T7527, WT8, YAR, NKL, HeLa | *01,*02 |
| SPO010, WT51, HMy‐2 C1R | *02,del |
| SK‐BR3 | del,del |
Finally, the PCR‐SSP method was further used to genotype the NKG2C CDS in our cohort of 240 donors, whose CNV for NKG2C had been previously determined. Direct sequencing and PCR‐SSP analyses of 186 unambiguous individuals (hemizygotes and homozygotes for one or both SNPs) confirmed the nearly complete association of c.5G and c.305C (allele NKG2C*01), on one hand, and c.5A and c.305T (allele NKG2C*02), on the other; with only two c.5A‐c.305C exceptions, corresponding to allele NKG2C*03.
The distribution of coding NKG2C polymorphisms in the whole sample, including allelic, carrier and genotypic frequencies, is shown in Figure 3 and Table S2. NKG2C*01 was the major allele, carried by 90.4% of the individuals; followed by NKG2C*02, found in 25.0% of the population. Two individuals, including the reference donor in whom the allele was originally reported, carried NKG2C*03 (c.5A and c.305C). In addition, as in earlier reports of Spaniards and other populations, 21 , 22 , 23 , 30 eight healthy donors (3.33%) lacked a NKG2C gene on both chromosomes. Eight of the nine possible CDS genotypes, considering CNV, were found in this sample; three genotypes (NKG2C*01,*01, NKG2C*01,del and NKG2C*01,*02) had frequencies ranging ~19%–47%, and, together, they accounted for more than 90% of the population. The frequencies of the two observed SNPs are similar to those observed previously in two East Asian populations 21 , 25 (Table S2).
FIGURE 3.
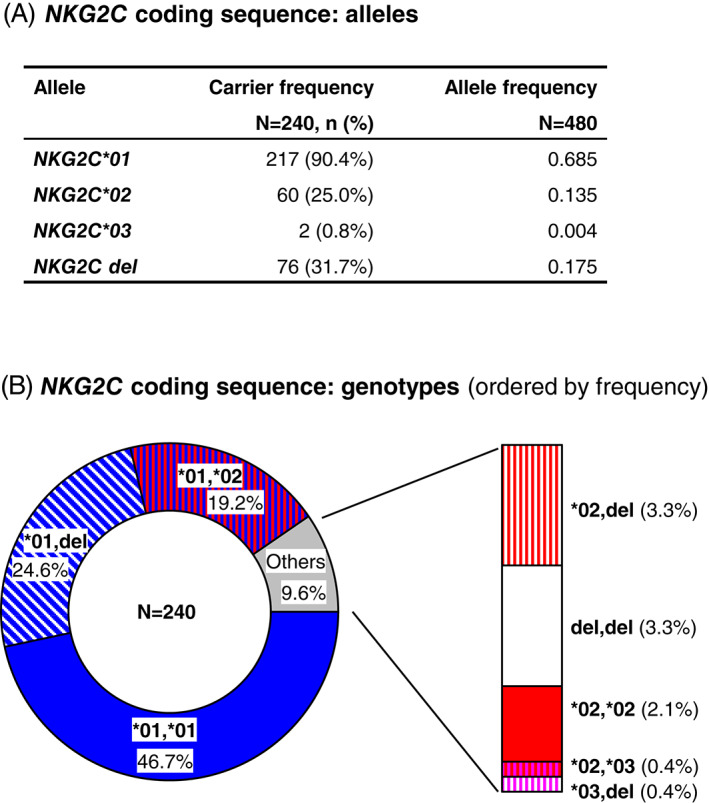
Distribution of NKG2C CDS polymorphisms: allele (A) and genotype (B) frequencies. Colors: blue, allele NKG2C*01; red, allele NKG2C*02; pink, NKG2C*03; white, gene deletion
3.3. Analysis of the NKG2C promoter region
Considering the known variability in NKG2C surface expression, 14 we extended our genetic study to non‐coding regions that might participate in regulation. Sequence analysis of ~300 bp 5′ of the start codon in 85 donors detected a single SNP, c.‐208 T > G. To facilitate the analysis of the complete sample, we set up an assay in which each allele is identified by a PCR‐SSP reaction (Figure 4). After validation in 42 sequenced DNAs, we completed the analysis of c.‐208 T > G in the whole cohort. Its frequencies, considering CNV (assessed as in Moraru et al.), 28 are shown in Table 3. As it will be analyzed in more detail in the last section, the minor allele c.‐208G was seen exclusively in presence of c.5A, being found in 51 of 60 donors carrying the NKG2C*02 allele defined by the latter SNP (56 of 65 chromosomes). In contrast, the major allele c.‐208 T was seen in association with all CDS allotypes: 100% of NKG2C*01 and *03 donors/alleles, and a minority of NKG2C*02 cases—9 of 60 donors (65 chromosomes) carrying this allele (global carrier frequency of c.‐208 T‐NKG2C*02, 3.75%).
FIGURE 4.
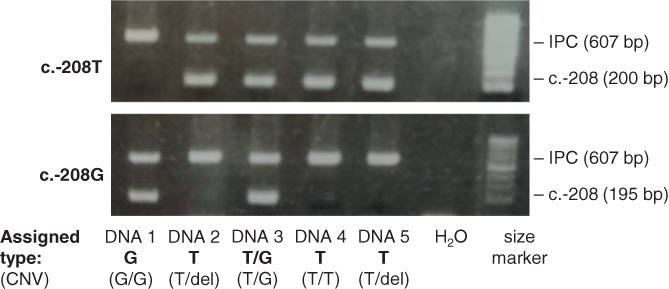
Polymerase chain reaction‐SSP genotyping of NKG2C promoter polymorphism. Five DNA samples with different genotypes were assayed with two SSP reactions: one (top) for c.‐208T, and one (bottom) for c.‐208 > G. This assay does not inform of sample homo‐or hemizygosity (CNV), which was determined as in Moraru et al. 28
TABLE 3.
Frequencies of NKG2C non‐coding polymorphism
| Frequencies | ||
|---|---|---|
| SNP | Major allele (carrier %) | Minor allele (carrier %) |
| Promoter region | ||
| c.‐208 T > G (rs76592132) | 0.708 (91.2%) | 0.117 (21.2%) |
| 3′‐untranslated region | ||
| c.*20A > G (rs2981593) | 0.469 (69.2%) | 0.356 (55.8%) |
| c.*105A > G (rs3003) | 0.696 (91.2%) | 0.129 (23.7%) |
| c.*285G > A (rs61917664) | 0.596 (81.2%) | 0.229 (37.9%) |
| c.*401A > G (rs2981594) | 0.469 (69.2%) | 0.356 (55.8%) |
| c.*419 T > C (rs2981595) | 0.696 (91.2%) | 0.129 (23.7%) |
Note: Allele frequencies sum 0.825; the remaining 0.175 corresponds to gene deletion, determined as in Moraru et al. 28
3.4. Analysis of the NKG2C 3′UTR
We also carried out an analysis of the NKG2C 3′UTR. By means of DNA sequencing in all 240 individuals, we found five SNPs: c.*20A > G, c.*105A > G, c.*285G > A, c.*401A > G, c.*419 T > C, whose frequencies are given in Table 3. No examples of the previously reported SNPs c.*8‐9GC > AA 21 were detected. Unambiguous phase analysis of 68 hemizygotes showed that, of all potential combinatorial associations, the five detected 3′UTR polymorphisms combine in, almost exclusively, three haplotypes: in order of frequency, ‘aagat’, ‘gaagt’, and ‘ggggc’ (Figure 5A). As we analyze further in the following section, both ‘aagat’ and ‘gaagt’ associate with NKG2C*01, whilst ‘ggggc’ associates with NKG2C*02. Those three haplotypes also explain virtually all 3′UTR genotypes seen in donors with a full NKG2C‐gene dose. Only two variants of these nearly fixed 3′UTR haplotypes were detected: a ‘gaggt’ motif found in one hemizygote (the variant nucleotide is highlighted), and a ‘ggagc’ variant seen in two NKG2C*01,*02 heterozygotes (genotype: ‘aagat,ggagc’). The frequencies of all five 3′UTR haplotypes and their genotypic combinations are represented in Figure 5.
FIGURE 5.
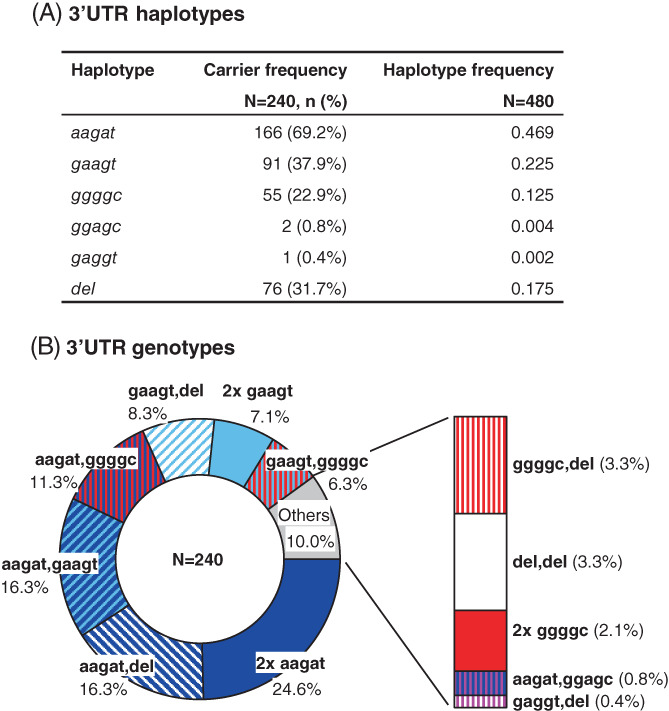
Distribution of NKG2C 3′untranslated region haplotype (A) and genotype (B) frequencies. Colors of common haplotypes: dark and light blue, aagat and gaagt, respectively (both associated with allele NKG2C*01); red, ggggc (associated with allele NKG2C*02); white, gene deletion
3.5. Analysis of extended NKG2C haplotypes
The presented data on the distribution of eight NKG2C polymorphisms (one in the promoter, two in the CDS, and five in the 3′UTR) revealed that, in at least certain gene regions, they tend to form conserved haplotypes, like those defining alleles NKG2C*01 and *02, and the three common 3′UTR haplotypes. We therefore examined whether haplotypes extending across the three regions could also be identified. To that end, we took advantage of the fact that phasing ambiguities could be ruled out in at least 143 individuals—68 lacking NKG2C on one chromosome, and 75 being homozygous at all eight sites. An additional 48 donors were homozygous for two of the three gene segments, being heterozygous only at the 3′UTR (42 individuals) or the CDS (six individuals), which, again, enabled us to phase their allotypes at all three regions. We thus defined unambiguously seven extended haplotypes (1a–d, 2a, 2d and 3 in Figure 6A), whose combinations explained the genotypes of all but three individuals of the whole cohort. Of those eight‐locus haplotypes, only three (1a, 1b and 2a) had frequencies greater than 0.1, and, together with the null haplotype, had a cumulative frequency of 0.969. These three dominant haplotypes were formed by: c.–208 T, allele NKG2C*01 in the CDS and either ‘aagat’ or ‘gaagt’ in the 3′UTR (haplotypes 1a and 1b, respectively); and by NKG2C*02 preceded by c.–208G and followed by the third 3′UTR motif, ‘ggggc’ (haplotype 2a).
FIGURE 6.
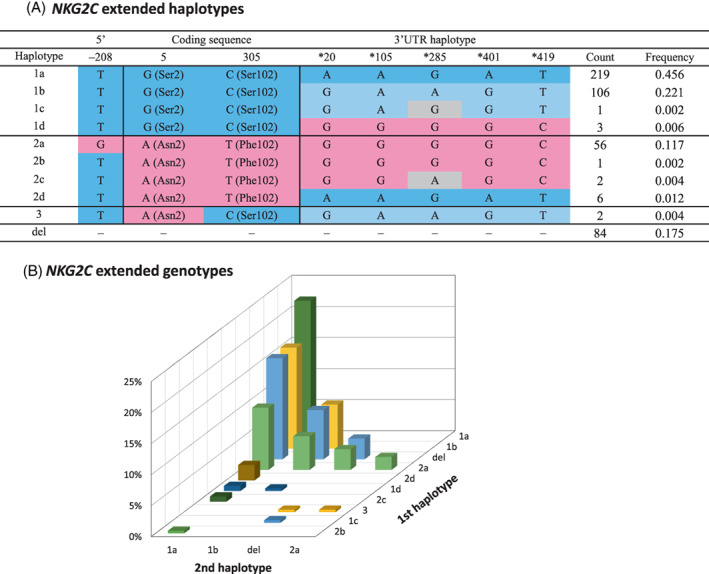
(A) NKG2C extended haplotypes and their frequencies (N = 480 haplotypes). Single‐nucleotide polymorphisms and 3′untranslated region (UTR) haplotypes commonly associated with allele NKG2C*01 are depicted in blue, whereas those typically seen with NKG2C*02 are marked in pink; variant nucleotides in 3′UTR haplotypes are shaded in gray. (B) Frequencies of extended NKG2C genotypes (promoter‐CDS‐3′UTR). Haplotypes are ordered according to their frequencies. For the sake of visual clarity, bar colors are not based on those of (A), and they depend only on the first haplotype. Numbers are provided in Table S3
As stated, only three individuals had genotypes not explained by the seven defined extended haplotypes. These remaining, unfitting genotypes were accounted for by two variants of haplotype 2a – haplotype 2b, which differed from it only at the promoter (c.–208 T, one donor); and haplotype 2c, having both c.–208 T and the ‘ggagc’ variant motif described in the previous section (two individuals). The NKC of Europeans thus harbors a minimum of nine NKG2C sequences. Random combinations of CNV and those allotypes in the population could theoretically produce 45 different genotypes, 18 of which were actually seen in the studied sample, with frequencies detailed in Figure 6B and Table S3.
4. DISCUSSION
The CD94/NKG2 family of lectin‐like NK‐cell receptors and their HLA‐E ligand appear well conserved by contrast with the extreme polymorphism of KIR and their classic HLA‐A, B, C ligands. However, both HLA‐E and its receptors display some degree of diversity. HLA‐E is generally represented by two common alleles, E*01:01 and E*01:03, with different expression levels attributed to one amino acid substitution in the peptide‐binding domain. 31 On the receptors side, a common ancestor locus underwent duplication and diversification to generate a family of genes that encode the different members of CD94/NKG2 heterodimers. As in other receptors, including KIR, one extreme example of divergent evolution in this family is a single ligand being recognized by pairs of receptors with opposed function – NK‐cell inhibition (CD94/NKG2A) versus activation (CD94/NKG2C). Other family members have less‐well characterized functions, and alternative splicing generates additional isoforms (NKG2A/B and NKG2E/H) which might confer an additional layer of functional diversification. 32 , 33 Finally, several coding and non‐coding sequence polymorphisms appear to be present in all the CD94 and NKG2 genes. 19 , 21 , 25 , 26 , 27 In sharp contrast with KIR, only NKG2C among lectin‐like receptors for HLA is commonly affected by gene copy‐number variation. Nonetheless, a common feature of both the KLRC‐ and the KIR‐gene complex is CNV affecting particularly the genes encoding activating receptors. 3 , 21 , 34
Here we have studied in detail the genotypic diversity of the NKG2C gene by analyzing the distribution of both CNV and a series of coding and non‐coding SNPs. We have verified that several previously reported polymorphisms of uncertain distribution are represented in a South European population. Furthermore, we have confirmed that coding polymorphisms define two conserved alleles (NKG2C*01 and *02), whilst the only detected hybrid of these (allele NKG2C*03) is represented at a low frequency. Our results also reveal the distribution of some SNPs in potential regulatory regions and demonstrate that these tend to be inherited with CDS polymorphisms in conserved haplotypes. Taking advantage of availability of a high number of hemizygous subjects, we have managed to establish unambiguously the composition of nine such extended haplotypes. In essence, the most prevalent allele in the coding region, NKG2C*01, is always seen with the same promoter allele, but combines with either of two conserved 3′UTR haplotypes; whilst the other common CDS allele, NKG2C*02, associates strongly with the alternative promoter allele and the third common 3′UTR motif; and all other combinations represent only ~3% of haplotypes. The homo‐ and heterozygous combinations of those haplotypes and NKG2C deletion generate a wide diversity of genotypes, the most common of which is seen in only 22% of individuals, revealing a noticeable level of population diversity. The fact that these polymorphisms are shared at detectable frequency by distant populations such as South Europeans and East Asians suggests existence of some evolutionary advantage in NKG2C diversity. The functional significance of NKG2C sequence polymorphism is unknown. Analyses of the sequence surrounding c.‐208 T > G with programs Consite 35 and sTRAP 36 , 37 , ii suggest that the SNP might modify its binding affinity for YY1 and other transcription factors (not shown). Furthermore, precomputed predictions on NKG2C cDNA in the RNA22 tool 38 (Computational Medicine Center at Thomas Jefferson University) indicate, with p < 0.01, that nucleotides 260–308 of its 3′UTR (including c.*285G > A) could be targets for multiple microRNAs. iii However, potential significance of these findings is uncertain, since no models have been proposed for regulation of NKG2C‐gene expression. In addition, the non‐conservative changes Ser2Asn (intracellular) and Ser102Phe (extracellular stem) might modulate receptor expression or function but, unfortunately, predictions are hindered by lack of crystal structures that include the affected regions of NKG2C (or its NKG2A homolog). 39 , 40 , 41 Further investigation is therefore warranted to ascertain whether and how genetic diversity modifies NKG2C function and, possibly, immune responses and health conditions conditioned by the receptor. We expect that the information presented herein should facilitate such studies.
AUTHOR CONTRIBUTIONS
Judit Asenjo and Manuela Moraru designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. Karima Al‐Akioui‐Sanz performed experiments. Aura Muntasell and Mireia Altadill contributed samples. Miguel López‐Botet designed the study and revised the manuscript. Carlos Vilches designed the study, directed research, and wrote the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Figure S1 Sequencing and genotyping strategy
Figure S2 Alternative splicing of NKG2C mRNA: partial retention of intron 1 in one donor is revealed using forward and reverse sequencing primers on the same RT‐PCR amplicon.
Table S1 PCR and sequencing primers.
Table S2. Distribution of SNPs in the NKG2C CDS in different populations.
Table S3. Genotypic combinations of extended NKG2C haplotypes (promoter, CDS and 3′UTR): absolute numbers and frequencies (% in parentheses). N = 240 individuals
ACKNOWLEDGMENTS
This article is dedicated to the memory of Benny P. Shum. We thank Dr. Elvira Ramil, from the DNA sequencing core facility of Instituto de Investigación Sanitaria Puerta de Hierro – Segovia de Arana, for continued support. This work was funded by grant PID2019‐110609RB‐C22/AEI/10.13039/501100011033 (AEI/FEDER, EU). MM and JA were hired by the latter grant and by GCB15152947MELE from the Asociación Española contra el Cáncer Foundation. Karima Al‐Akioui‐Sanz was supported sequentially by grant PEJ‐2017‐AI/BMD‐7377, with co‐financing by EU Youth Employment Initiative, European Social Fund (91.89%), and Consejería de Educación, Juventud y Deporte de la Comunidad de Madrid; and by grant SAF2016‐80363‐C2‐2‐R (AEI/FEDER, EU).
Asenjo J, Moraru M, Al‐Akioui‐Sanz K, et al. Diversity of NKG2C genotypes in a European population: Conserved and recombinant haplotypes in the coding, promoter, and 3′‐untranslated regions. HLA. 2022;100(5):469‐478. doi: 10.1111/tan.14734
Judit Asenjo and Manuela Moraru shared first authorship.
Funding information AEI/FEDER, Grant/Award Number: PID2019‐110609RBC22/AEI/10.13039/501100011033; Consejería de Educación, Juventud y Deporte, Comunidad de Madrid, Grant/Award Number: SAF2016‐80363‐C2‐ 2‐R; EU Youth Employment Initiative, European Social Fund, Grant/Award Number: PEJ‐2017‐AI/BMD‐7377; Fundación Científica Asociación Española Contra el Cáncer, Grant/Award Number: GCB15152947MELE; Consejería de Educación, Juventud y Deporte de la Comunidad de Madrid; European Social Fund
Endnotes
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Abi‐Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin‐like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201(8):1319‐1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson SK, Ortaldo JR, McVicar DW. The ever‐expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79‐89. [DOI] [PubMed] [Google Scholar]
- 3. Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217‐251. [DOI] [PubMed] [Google Scholar]
- 4. Braud VM, Allan DS, O'Callaghan CA, et al. HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795‐799. [DOI] [PubMed] [Google Scholar]
- 5. Lee N, Llano M, Carretero M, et al. HLA‐E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci USA. 1998;95(9):5199‐5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vance RE, Kraft JR, Altman JD, Jensen PE, Raulet DH. Mouse CD94/NKG2A is a natural killer cell receptor for the nonclassical major histocompatibility complex (MHC) class I molecule Qa‐1(b). J Exp Med. 1998;188(10):1841‐1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carretero M, Cantoni C, Bellón T, et al. The CD94 and NKG2‐a C‐type lectins covalently assemble to form a natural killer cell inhibitory receptor for HLA class I molecules. Eur J Immunol. 1997;27(2):563‐567. [DOI] [PubMed] [Google Scholar]
- 8. Tomasec P, Braud VM, Rickards C, et al. Surface expression of HLA‐E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287(5455):1031‐1033. [DOI] [PubMed] [Google Scholar]
- 9. Ulbrecht M, Martinozzi S, Grzeschik M, et al. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA‐E and prevents NK cell‐mediated lysis. J Immunol. 2000;164(10):5019‐5022. [DOI] [PubMed] [Google Scholar]
- 10. Hammer Q, Ruckert T, Borst EM, et al. Peptide‐specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat Immunol. 2018;19(5):453‐463. [DOI] [PubMed] [Google Scholar]
- 11. Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296(5571):1323‐1326. [DOI] [PubMed] [Google Scholar]
- 12. Smith HR, Heusel JW, Mehta IK, et al. Recognition of a virus‐encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA. 2002;99(13):8826‐8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gumá M, Angulo A, Vilches C, Gómez‐Lozano N, Malats N, López‐Botet M. Imprint of human cytomegalovirus infection on the NK ceH receptor repertoire. Blood. 2004;104(12):3664‐3671. [DOI] [PubMed] [Google Scholar]
- 14. López‐Botet M, Muntasell A, Vilches C. The CD94/NKG2C + NK‐cell subset on the edge of innate and adaptive immunity to human cytomegalovirus infection. Semin Immunol. 2014;26(2):145‐151. [DOI] [PubMed] [Google Scholar]
- 15. van der Ploeg K, Chang C, Ivarsson MA, Moffett A, Wills MR, Trowsdale J. Modulation of human leukocyte antigen‐C by human cytomegalovirus stimulates KIR2DS1 recognition by natural killer cells. Front Immunol. 2017;8:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rahim MM, Makrigiannis AP. Ly49 receptors: evolution, genetic diversity, and impact on immunity. Immunol Rev. 2015;267(1):137‐147. [DOI] [PubMed] [Google Scholar]
- 17. Wroblewski EE, Parham P, Guethlein LA. Two to tango: co‐evolution of hominid natural killer cell receptors and MHC. Front Immunol. 2019;10:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD—the Immuno polymorphism database. Nucleic Acids Res. 2013;41(D1):D1234‐D1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Houchins JP, Yabe T, McSherry C, Miyokawa N, Bach FH. Isolation and characterization of NK cell or NK/T cell‐specific cDNA clones. J Mol Cell Immunol. 1990;4(6):295‐304. [PubMed] [Google Scholar]
- 20. Renedo M, Arce I, Rodríguez A, et al. The human natural killer gene complex is located on chromosome 12p12‐p13. Immunogenetics. 1997;46(4):307‐311. [DOI] [PubMed] [Google Scholar]
- 21. Hikami K, Tsuchiya N, Yabe T, Tokunaga K. Variations of human killer cell lectin‐like receptors: common occurrence of NKG2‐C deletion in the general population. Genes Immun. 2003;4(2):160‐167. [DOI] [PubMed] [Google Scholar]
- 22. Miyashita R, Tsuchiya N, Hikami K, et al. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int Immunol. 2004;16(1):163‐168. [DOI] [PubMed] [Google Scholar]
- 23. Moraru M, Cisneros E, Gómez‐Lozano N, et al. Host genetic factors in susceptibility to herpes simplex type 1 virus infection: contribution of polymorphic genes at the interface of innate and adaptive immunity. J Immunol. 2012;188(9):4412‐4420. [DOI] [PubMed] [Google Scholar]
- 24. Muntasell A, López‐Montañés M, Vera A, et al. NKG2C zygosity influences CD94/NKG2C receptor function and the NK‐cell compartment redistribution in response to human cytomegalovirus. Eur J Immunol. 2013;43(12):3268‐3278. [DOI] [PubMed] [Google Scholar]
- 25. Seo J, Park JS, Nam JH, et al. Association of CD94/NKG2A, CD94/NKG2C, and its ligand HLA‐E polymorphisms with Behcet's disease. Tissue Antigens. 2007;70(4):307‐313. [DOI] [PubMed] [Google Scholar]
- 26. Shum BP, Flodin LR, Muir DG, et al. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168(1):240‐252. [DOI] [PubMed] [Google Scholar]
- 27. Cantoni C, Biassoni R, Pende D, et al. The activating form of CD94 receptor complex: CD94 covalently associates with the Kp39 protein that represents the product of the NKG2‐C gene. Eur J Immunol. 1998;28(1):327‐338. [DOI] [PubMed] [Google Scholar]
- 28. Moraru M, Cañizares M, Muntasell A, de Pablo R, López‐Botet M, Vilches C. Assessment of copy‐number variation in the NKG2C receptor gene in a single‐tube and characterization of a reference cell panel, using standard polymerase chain reaction. Tissue Antigens. 2012;80(2):184‐187. [DOI] [PubMed] [Google Scholar]
- 29. Asenjo J, Muntasell A, López‐Botet M, Moraru M, Vilches C. Complete genomic characterization of a new KLRC2 allele, NKG2C*03. HLA. 2021;98:259‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muntasell A, Pupuleku A, Cisneros E, et al. Relationship of NKG2C copy number with the distribution of distinct cytomegalovirus‐induced adaptive NK cell subsets. J Immunol. 2016;196(9):3818‐3827. [DOI] [PubMed] [Google Scholar]
- 31. Strong RK, Holmes MA, Li P, Braun L, Lee N, Geraghty DE. HLA‐E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. 2003;278(7):5082‐5090. [DOI] [PubMed] [Google Scholar]
- 32. Dukovska D, Fernández‐Soto D, Valés‐Gómez M, Reyburn HT. NKG2H‐expressing T cells negatively regulate immune responses. Front Immunol. 2018;9:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bellón T, Heredia AB, Llano M, et al. Triggering of effector functions on a CD8+ T cell clone upon the aggregation of an activatory CD94/kp39 heterodimer. J Immunol. 1999;162(7):3996‐4002. [PubMed] [Google Scholar]
- 34. Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7(6):753‐763. [DOI] [PubMed] [Google Scholar]
- 35. Sandelin A, Wasserman WW, Lenhard B. ConSite: web‐based prediction of regulatory elements using cross‐species comparison. Nucleic Acids Res. 2004;32:W249‐W252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thomas‐Chollier M, Hufton A, Heinig M, et al. Transcription factor binding predictions using TRAP for the analysis of ChIP‐seq data and regulatory SNPs. Nat Protoc. 2011;6(12):1860‐1869. [DOI] [PubMed] [Google Scholar]
- 37. Manke T, Heinig M, Vingron M. Quantifying the effect of sequence variation on regulatory interactions. Hum Mutat. 2010;31(4):477‐483. [DOI] [PubMed] [Google Scholar]
- 38. Miranda KC, Huynh T, Tay Y, et al. A pattern‐based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203‐1217. [DOI] [PubMed] [Google Scholar]
- 39. Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA‐E. Proc Natl Acad Sci USA. 2008;105(18):6696‐6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Petrie EJ, Clements CS, Lin J, et al. CD94‐NKG2A recognition of human leukocyte antigen (HLA)‐E bound to an HLA class I leader sequence. J Exp Med. 2008;205(3):725‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sullivan LC, Clements CS, Beddoe T, et al. The heterodimeric assembly of the CD94‐NKG2 receptor family and implications for human leukocyte antigen‐E recognition. Immunity. 2007;27(6):900‐911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Sequencing and genotyping strategy
Figure S2 Alternative splicing of NKG2C mRNA: partial retention of intron 1 in one donor is revealed using forward and reverse sequencing primers on the same RT‐PCR amplicon.
Table S1 PCR and sequencing primers.
Table S2. Distribution of SNPs in the NKG2C CDS in different populations.
Table S3. Genotypic combinations of extended NKG2C haplotypes (promoter, CDS and 3′UTR): absolute numbers and frequencies (% in parentheses). N = 240 individuals
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


