Summary
Crassulacean acid metabolism (CAM) photosynthesis has evolved repeatedly across the plant tree of life, however our understanding of the genetic convergence across independent origins remains hampered by the lack of comparative studies. Here, we explore gene expression profiles in eight species from the Agavoideae (Asparagaceae) encompassing three independent origins of CAM.
Using comparative physiology and transcriptomics, we examined the variable modes of CAM in this subfamily and the changes in gene expression across time of day and between well watered and drought‐stressed treatments. We further assessed gene expression and the molecular evolution of genes encoding phosphoenolpyruvate carboxylase (PPC), an enzyme required for primary carbon fixation in CAM.
Most time‐of‐day expression profiles are largely conserved across all eight species and suggest that large perturbations to the central clock are not required for CAM evolution. By contrast, transcriptional response to drought is highly lineage specific. Yucca and Beschorneria have CAM‐like expression of PPC2, a copy of PPC that has never been shown to be recruited for CAM in angiosperms.
Together the physiological and transcriptomic comparison of closely related C3 and CAM species reveals similar gene expression profiles, with the notable exception of differential recruitment of carboxylase enzymes for CAM function.
Keywords: Agavoideae, carboxylation, Crassulacean acid metabolism photosynthesis, comparative transcriptomics, Yucca
Introduction
The repeated origin of phenotypes across the tree of life has long fascinated biologists, particularly in cases in which such phenotypes are assembled convergently, that is, using the same genetic building blocks. Documented examples of convergent evolution in which the same genetic mechanisms are involved include the repeated origins of betalain pigmentation in the Caryophyllales (Sheehan et al., 2020), the origins of caffeine biosynthesis in eudicots (Denoeud et al., 2014), and the multiple transitions to red flowers in Ipomoea (Streisfeld & Rausher, 2009), among others. In all these cases, careful analysis of the genetic components underlying the repeated phenotypic evolution was driven by recruitment or loss of function of orthologous genes. Such convergence in the genetic mechanism suggests that the evolutionary path toward these phenotypes is relatively narrow, meaning the phenotype can only be obtained through a small set of very important molecular changes.
Such shared molecular mechanisms of repeated phenotypic evolution are especially surprising when observed across larger clades. For example, across all flowering plants, the large number of independent origins (c. 100) of both C4 and Crassulacean acid metabolism (CAM) photosynthesis imply relatively straightforward genetic and evolutionary paths from the ancestral C3 photosynthetic pathway (Edwards, 2019; Heyduk et al., 2019a). The overall photosynthetic metabolic pathway in C4 and CAM species is largely conserved; CO2 is converted to a four carbon acid by phosphoenolpyruvate carboxylase (PPC) and either moved to adjoining cells (C4) or stored in the vacuole overnight (CAM). The four carbon acids are then decarboxylated, resulting in high concentrations of CO2 in the cells in which Rubisco is active. Although some aspects of these photosynthetic pathways can vary among independent lineages, such as decarboxylation pathways in C4 lineages (Christin et al., 2009; Bräutigam et al., 2014), the same homologue of some genes has been repeatedly recruited for carbon concentration. In three independently derived C4 grass lineages, five out of seven photosynthetic genes examined had the same gene copy (orthologues) recruited, despite the presence of alternative copies (paralogues) of each gene (Christin et al., 2013). In Cleome gynandra (Cleomaceae) and Zea mays (Poaceae), transcription factors that induce expression of C4 photosynthetic genes in the required cell‐specific manner were orthologous, despite > 140 million years (Myr) of evolution separating the two lineages (Aubry et al., 2014).
While C4 is known for the unique Kranz anatomy that allows the carbon concentrating mechanism to function efficiently, CAM instead relies on the temporal separation of CO2 assimilation and conversion of CO2 into sugars. The diurnal cycle of primary CO2 fixation and photosynthesis in CAM plants is thought to require a close integration with the circadian clock, although how that is explicitly accomplished remains unknown. Studies have shown that only a handful of core clock genes differ in their expression between C3 and CAM species (Yang et al., 2017; Yin et al., 2018), although many of these studies have relied on comparisons of distantly related species, confounding changes attributable to evolutionary distance with those that underlie the evolution of CAM photosynthesis. While the core clock seems largely similar in CAM and C3 species, 24‐h expression profiles for genes involved in carboxylation, decarboxylation, sugar metabolism, and stomatal movement have been shown to differ between C3 and CAM species (Ceusters et al., 2014; Ming et al., 2015; Abraham et al., 2016; Heyduk et al., 2018a; Wai et al., 2019), suggesting a regulatory link between clock genes and genes contributing to CAM function.
A hallmark of CAM is the evening expression of phosphoenolpyruvate carboxylase (PPC) genes, which produce the enzyme required for the initial fixation of atmospheric CO2 into an organic acid in both C4 and CAM plants. Unlike Rubisco, which has affinities for both CO2 and O2, PPC has only carboxylase function, which it uses to convert bicarbonate and phosphoenolpyruvate (PEP) into oxaloacetate (OAA). The carboxylating function of PPC is used by all plants to supplement intermediate metabolites into the tricarboxylic acid (TCA) cycle, and therefore PPC genes are present in all plant lineages in multiple copies. Phosphoenolpyruvate carboxylase enzymes used by the CAM pathway are active in the evening and night, whereas TCA‐related PPC enzymes are likely to have constitutive expression across the diel cycle, with perhaps higher activity during the day. Transcriptomic investigations of CAM species have shown that expression of the PPC genes involved in CAM is induced to much higher levels at dusk and overnight (Ming et al., 2015; Brilhaus et al., 2016; Yang et al., 2017; Heyduk et al., 2018a, 2019b): expression levels of CAM PPCs can be 100–1000× higher than PPC homologues contributing to housekeeping functions.
There are two main families of PPC genes in flowering plants: PPC1, which is typically present in 2–6 copies in most lineages (Deng et al., 2016), and PPC2, which shares homology with a PPC gene copy found in bacteria, and is typically found in single or low copy in plant genomes. PPC1 forms a homotetramer, whereas PPC2 requires the formation of a hetero‐octamer with PPC1 to function (O’Leary et al., 2009). PPC1 is used in the TCA cycle in plants, and in all published cases within angiosperms a PPC1 gene copy is recruited for CAM (and C4) function. New work in the lycophyte Isoetes taiwanensis has shown novel recruitment of PPC2 into the CAM pathway, rather than PPC1 (Wickell et al., 2021). PPC2 has been shown to be involved in pollen maturation, fatty acid production in seeds, and possibly root development and salt sensing (Gennidakis et al., 2007; Igawa et al., 2010; Wang et al., 2012), although overall consensus on PPC2 function in plants remains elusive.
To understand both the evolution of CAM, as well as the recruitment of PPC homologues in independent origins of CAM, we built upon existing physiological and transcriptomic data in the Agavoideae (Asparagaceae) by investigating additional species. Crassulacean acid metabolism has evolved three times independently in the Agavoideae (Fig. 1): once in Agave sensu lato (Agave s.l., includes the genera Agave, Manfreda, and Polianthes), once in Yucca, and once in Hesperaloe (Heyduk et al., 2016b). Previous research compared gene expression and physiology in closely related C3 and CAM Yucca species (Heyduk et al., 2019b) and, separately, in species that ranged from weak CAM (low amounts of nocturnal CO2 uptake) to strong CAM in Agave s.l. (Heyduk et al., 2018b). Gene expression profiles for key CAM genes in the C3 Yucca species studies showed CAM‐like expression, especially when drought stressed, suggesting that perhaps Yucca or even the Agavoideae as a whole was primed for the evolution of CAM due to gene regulatory networks and expression patterns that existed in a C3 ancestor. Here we conducted additional RNA sequencing in two species of Hesperaloe (CAM) and one species of Hosta (C3) to assess (1) how gene expression varies in timing of expression and in response to drought stress across Agavoideae and (2) to what extent have the three independent origins of CAM in the Agavoideae been involved in recruitment of the same carboxylating enzyme gene homologues.
Fig. 1.
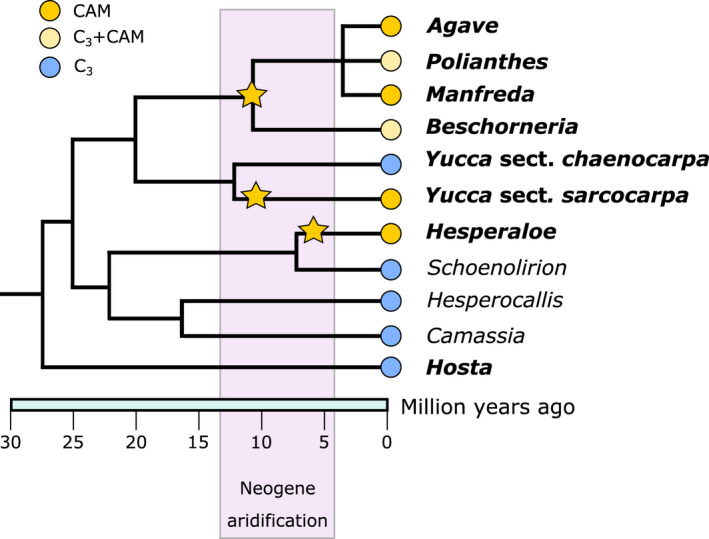
Simplified phylogeny of the Agavoideae, with estimated topology and mean divergence times from McKain et al. (2016), and aridification timing based on Eronen et al. (2012). Bolded taxa names are the species/genera included in this study. Tips are labelled according to photosynthetic pathway as described by previous work (Heyduk et al., 2016b, 2018b, 2019b), and yellow stars indicate hypothesised origins of Crassulacean acid metabolism photosynthesis.
Materials and Methods
Plant growth and physiological sampling
Plants of Hesperaloe parviflora (accession: PARL 436) and Hesperaloe nocturna (accession: PARL 435) were grown from seed acquired in 2014 from the USDA Germplasm Resources Information Network (GRIN). Hesperaloe plants were kept in the University of Georgia (UGA) Plant Biology glasshouses with once weekly watering. Hosta plants were purchased for New Hampshire Hostas (https://www.nhhostas.com/) in January 2018 and kept on a misting bench at the same glasshouses until experimentation began in March 2018. Replicates of each species (n = 4, 4 and 6 for H. parviflora, H. nocturna, and H. venusta, respectively) were placed into a walk‐in Conviron growth chamber, with day length set to 12 h (lights on at 7:00 h), day : night temperatures 30 : 17°C, humidity at 30%, and maximum PAR (c. 400 μmol m−2 s−1 at plant level).
Plants were acclimated in the growth chamber for 4 d before sampling and watered to saturation daily. On day 1, plants were sampled every 2 h, beginning at 1 h after the lights were turned on, for gas exchange using a Li‐Cor 6400XT. Due to the small size of the plants, only two replicates of Hosta had Li‐Cor measurements taken; one replicate of Hesperaloe nocturna was not measured due to an ant infestation in the pot. After day 1, water was withheld for 5 d in all plants with the exception of Hosta, which were all removed from the experiment at this point. On day 7, all the remaining plants' water status had dropped to 8% soil water content, and plants were measured again for gas exchange. After day 7, plants were re‐watered and one more day of gas exchange sampling was conducted on day 9. Triplicate leaf tissue samples per plant were collected for titratable acidity measurements 2 h before lights turned on (pre‐dawn sample) and 2 h before lights turned off (pre‐dusk sample) on days 1 and 7. Samples for leaf titrations were immediately flash frozen and stored at −80°C.
Leaf acid titrations were conducted as in Heyduk et al. (2018b); briefly, frozen leaf discs were quickly weighed and placed into 60 ml of 20% EtOH. Samples were boiled until the volume was reduced to half, then 30 ml of diH2O was added. Samples were reduced to half again and a final volume of 30 ml of diH2O was added. Samples were allowed to cool then titrated to pH 7.0 using 0.002 M NaOH. Total micromoles H+ per gram of frozen mass was calculated as (ml NaOH × 0.002 M) g−1. Pre‐dusk values were subtracted from pre‐dawn values to determine the change, or ΔH+, per replicate. All statistical analyses were conducted in R v.3.5.0 (R Core Team, 2021). Physiology data for Agave, Polianthes, Beschorneria, and Yucca species were taken from previously published data (Heyduk et al., 2016a, 2018b); for comparison, we include soil moisture data as measured from the three independent studies during the drought stress (Supporting Information Table S1).
RNA sequencing and assembly
Tissue for RNA sequencing was collected every 4 h from each of the three species, from four replicate plants per species. For H. nocturna and H. parviflora, samples were collected from both well watered and drought‐stressed plants (days 1 and 7). For H. venusta, only well watered samples were collected. Tissue was flash frozen in N2, then stored at −80°C. RNA was isolated using a Qiagen RNeasy Plant Kit, purified with Ambion Turbo DNase, and quantified by a NanoDrop spectrophotometer and Agilent Bioanalyzer v.2100 (Santa Clara, CA, USA). RNA‐sequencing libraries were constructed with a KAPA Stranded RNA‐seq kit at half reaction volume and barcoded separately using dual barcodes (Glenn et al., 2019). Library concentrations were measured by quantitative PCR, pooled in sets of 28–29 libraries, and sequenced with PE 75 bp reads on an Illumina NextSeq system at the Georgia Genomics and Bioinformatics Core at the University of Georgia. Raw reads from sequencing Hesperaloe and Hosta species are available on the NCBI Sequence Read Archive (SRA), under BioProject PRJNA755802.
Raw reads were processed with Trimmomatic v.0.36 (Bolger et al., 2014) and paired reads were assembled de novo for each of the three species (H. parviflora, H. nocturna, and Hosta venusta) in Trinity v.2.5.1 (Grabherr et al., 2011). Reads were initially mapped to the entire Trinity‐assembled transcriptome for each species with Bowtie v.2.0 (Langmead & Salzberg, 2012). Trinity ‘isoforms’ that had < 2 transcripts mapped per million (TPM) abundance, or constituted < 20% of total component expression, were removed. Transcriptome assemblies of sister species, including Agave bracteosa, Polianthes tuberosa, and Beschorneria yuccoides (Heyduk et al., 2018b) had already been filtered by the same thresholds as above. All six filtered assemblies had open reading frames (ORFs) predicted by Transdecoder v.2.1 (Grabherr et al., 2011) using both ‘LongOrfs’ and ‘Predict’ functions and keeping only the best scoring ORF per transcript.
To sort the predicted Transdecoder sequences into gene families, we generated orthogroups circumscribed from nine reference genomes downloaded from the Phytozome portal (Goodstein et al., 2012), with a particular focus on monocots. Translated primary transcript sequences were downloaded for Acorus americanus v.1.1 (DOE‐JGI, http://phytozome‐next.jgi.doe.gov/), Arabidopsis thaliana v.Araport11 (Cheng et al., 2017), Asparagus officinalis v.1.1 (Harkess et al., 2017), Ananas comosus v.3 (Ming et al., 2015), Amborella trichopoda v.1 (Amborella Genome Project, 2013), Brachypodium distachyon v.3.1 (International Brachypodium Initiative, 2010), Dioscorea alata v.2.1 (Bredeson et al., 2022), Musa acuminata v.1 (D’Hont et al., 2012), Oryza sativa v.7 (Ouyang et al., 2007), Sorghum bicolor v.3.1.1 (McCormick et al., 2018), and Setaria italica v.2.2 (Bennetzen et al., 2012) from Phytozome. Translated coding sequences from these genomes were clustered using OrthoFinder v.2.2.7 (Emms & Kelly, 2019). In addition to the above published genomes, preliminary draft genome annotations (primary translated transcripts) for Yucca aloifolia and Yucca filamentosa were secondarily added to the orthogroup analyses using the ‐b flag of OrthoFinder (pre‐publication permission from JGI was obtained for Yucca annotation use). Finally, transdecoder translated coding sequences for the Agavoideae species were then added to the orthogroup circumscription again using the ‐b flag.
Expression analysis
Reads were remapped using kallisto (Bray et al., 2016) onto the filtered transcriptomes (iso_pct > 20, TPM > 2, transdecoder best scoring ORF) for the de novo assemblies of Hesperaloe and Hosta. For the two Yucca genomes, existing RNA‐seq reads (from Heyduk et al., 2019b) were mapped onto the annotated primary transcripts using Kallisto. Because previously published expression analysis of Agave, Beschorneria, and Polianthes was done on transcriptomes filtered the same way (iso_pct > 20, TPM > 2) (Heyduk et al., 2018b), expression data in the form of read counts and TPM values for genes were used as previously published. Count and TPM matrices for all taxa analysed here, as well as orthogroup annotations, are available on github (www.github.com/kheyduk/AgavoideaeComparative).
Read counts for the two Hesperaloe species, Hosta, and the two Yucca species were imported into R for initial outlier filtering in edgeR (Robinson et al., 2010) and subsequent time‐structured expression analysis in maSigPro (Conesa et al., 2006; Nueda et al., 2014). The latter program fits read count data to regressions, taking into account treatments (well watered and drought stress), and asks whether a polynomial regression of degree n (chosen to be 5, or one less the number of timepoints) is a better fit to each gene than a straight line. Genes with expression patterns across time can be best explained by a polynomial regression are from this point forwards referred to as ‘time structured’. While read counts are required for the maSigPro analysis, all comparative expression plots presented use TPM normalised expression. Time‐structured expression information for Agave, Beschorneria, and Polianthes was taken from previously published data (Heyduk et al., 2018b).
Circadian gene expression
Previous studies in CAM have shown that a few circadian regulators get re‐wired in the evolution of CAM (Moseley et al., 2018; Wai et al., 2019). To determine whether these patterns held in more closely related C3 and CAM species, the expression of circadian clock genes was compared between members of the Agavoideae. From the list of genes that had significantly time‐structured expression from maSigPro for each species, we assessed gene family presence/absence data from the OrthoFinder gene circumscriptions. Shared gene family presence in the time‐structured expression was assessed using the UpSetR package (Conway et al., 2017) in R 4.0.4 (R Core Team, 2021). A curated list of Arabidopsis thaliana circadian genes was used to examine the extent to which time‐structured expression of circadian genes was shared across all eight species. Finally, for circadian‐annotated genes, we used jtk_cycle (Hughes et al., 2010) and Lomb‐Scargle (Glynn et al., 2005) methods implemented in metaCycle (Wu et al., 2016) to obtain period, lag and amplitude for genes with a period expression pattern. Cycling patterns for Agave and Beschorneria were excluded from further analysis, as their resolution (number of replicates and time points) was lower than other species due to dropped libraries (Heyduk et al., 2018b). We then used ANOVA to assess whether there were differences in average phase across OrthoFinder gene families between CAM and C3 species, as well as between Hosta and the other Agavoideae species. P‐values were corrected for multiple testing using the Benjamini–Hochberg correction.
PPC evolution
The PPC1 and PPC2 gene families were identified in the OrthoFinder‐circumscribed orthogroups by searching for annotated Arabidopsis PPC1 and PPC2 copies. Both orthogroups were manually inspected for completeness by checking if all known genes from sequenced and annotated genomes were properly sorted into those two orthogroups. Only sequences that were at least 50% of the length of the longest sequence (based on coding sequence) were retained and aligned. Because de novo transcriptomes often contain allelic variation assembled as separate contigs, we developed a threshold to collapse highly similar sequences within a species. Using gene annotations from the two Yucca species, we calculated pairwise sequence similarities within each species by gene combination (PPC1 or PPC2). The highest similarity within a gene was used as a cutoff; it represented the highest sequence similarity that existed within a species across separate gene copies. Because the Yucca annotation is based on genomic sequence, we felt confident that separate assembled genes represented loci, rather than alleles. The highest within‐species similarity in Yucca for PPC1 was 96.23% and 99.71% for PPC2. Sequences from the de novo transcriptomes were then collapsed within a species if they were more similar than these percentages; instead of using ambiguity codes, we used the longest sequence as the representative for the collapsed sequences. To get in‐frame coding sequence alignment, each orthogroup protein and CDS output from transdecoder was used to align the coding sequences using Pal2nal (Suyama et al., 2006). Phylogenetic trees for PPC1 and PPC2 were estimated on the in‐frame coding sequence alignments using IQTree v.2.0 Nguyen et al., 2015; Minh et al., 2020) and 1000 rapid bootstrap replicates, using built‐in ModelFinder to determine the best substitution model. The resulting tree for each gene family, along with the in‐frame coding sequence alignment, were used to estimate shifts in molecular evolution using codeml in Paml (Yang, 2007). Specifically, we tested branch, sites and branch (clade)–sites models. We compared branch models to a null M0 model with a single ⍵ value, the M2a sites model (positive selection) to the null M1a (nearly neutral; Wong et al., 2004), the branch–sites model A to the null (fixed_omega = 1, omega = 1), and the clade C model to M2a_rel. For branch, branch–sites and clade models, we labelled the two PPC1 Agavoideae lineages and estimated ⍵ separately; for PPC2, we labelled the single Agavoideae stem branch. Due to low phylogenetic resolution within the Agavoideae, specific tests for independent CAM origins within the subfamily were not feasible. Fasta files of multispecies alignments and Newick gene trees are available at www.github.com/kheyduk/AgavoideaeCAM.
Results
CAM in the Agavoideae
Gas exchange and leaf titratable acidity amounts implicated CAM in both Hesperaloe species and C3 photosynthesis in Hosta venusta (Fig. 2; Tables S2, S3). Although we did not sample Hosta venusta under drought‐stressed conditions, its thin leaf morphology (Heyduk et al., 2016b) and shady, mesic habitat suggests it is very unlikely to use any mode of CAM photosynthesis. Most of the CAM species in the Agavoideae examined still rely at least partially on daytime CO2 fixation by Rubisco (Fig. 2). Furthermore, Yucca, Agave and Manfreda all appear to downregulate CAM under drought stress, as seen in both their gas exchange patterns and titratable acidity levels under drought relative to well watered status (Fig. 2). Hesperaloe parviflora had slightly higher CO2 uptake at night than did H. nocturna, although both had appreciable levels of acid accumulation and, unlike Yucca and Agave sensu lato species, had a slight upregulation of CAM under drought stress. Finally, as previously described (Heyduk et al., 2018b), Polianthes tuberosa and Beschorneria yuccoides are C3 + CAM, and both are able to facultatively use CAM under drought stress.
Fig. 2.
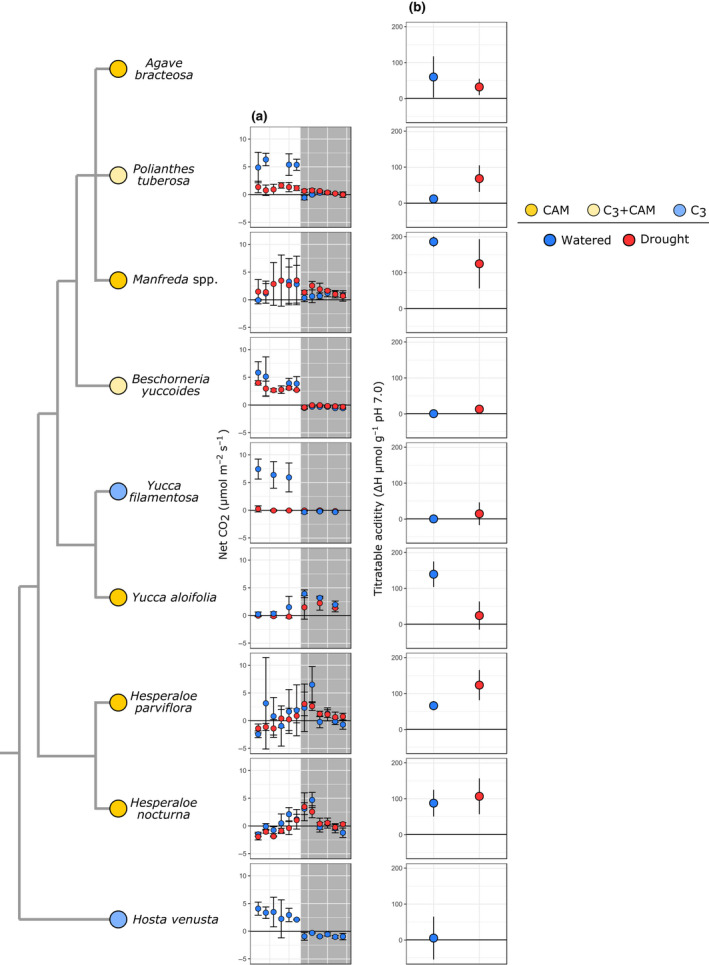
Photosynthetic physiology of species in the Agavoideae. Species relationships are represented by the cladogram to the left; A (net photosynthesis, or the net flux of CO2 per second per area), with shaded areas representing night time points (a); and the daily change in titratable leaf acidity (H μmol g−1 in the early morning – late afternoon) (b); are shown per species, with means and standard errors calculated from replicates of each species. Colours next to species names indicate Crassulacean acid metabolism (CAM) (bright yellow), C3 + CAM (pale yellow), and C3 (blue). Data for all species except Hesperaloe and Hosta comes from previous work (Heyduk et al., 2018b, 2019b).
Cross‐Agavoideae comparisons
The number of transcripts that showed significant time‐structured expression varied across species, with the fewest in the C3 species Hosta venusta (n = 5576) and the highest in the CAM species Agave bracteosa (n = 28 856). Although Hosta was only assessed under well watered conditions, drought conditions are unlikely to have increased the total number of time‐structured transcripts: across species, the transcripts that were both time structured and differentially expressed under drought represent a small proportion of the total time‐structured transcripts (with the notable exception of Agave; Fig. 3a). All species that used C3 photosynthesis or exhibit weak CAM had fewer transcripts that had a significant change in diurnal expression, with the exception of Polianthes tuberosa, which uses CAM facultatively more so than does Beschorneria yuccoides (Fig. 2). Many gene families (923) were time structured in all eight species (Table S4); 731 additional gene families were time structured in all species with the exception of Hosta (Fig. 3b; Table S5). This latter set included some canonical CAM genes, including both PPC1 and PPC2, as well as phosphoenolpyruvate carboxylase kinase (PPCK), a kinase dedicated to the phosphorylation of PPC and thought to be required for efficient CAM (Taybi et al., 2000), auxin‐related response genes, and some genes related to light reactions (e.g. photosystem II reaction centre protein D).
Fig. 3.
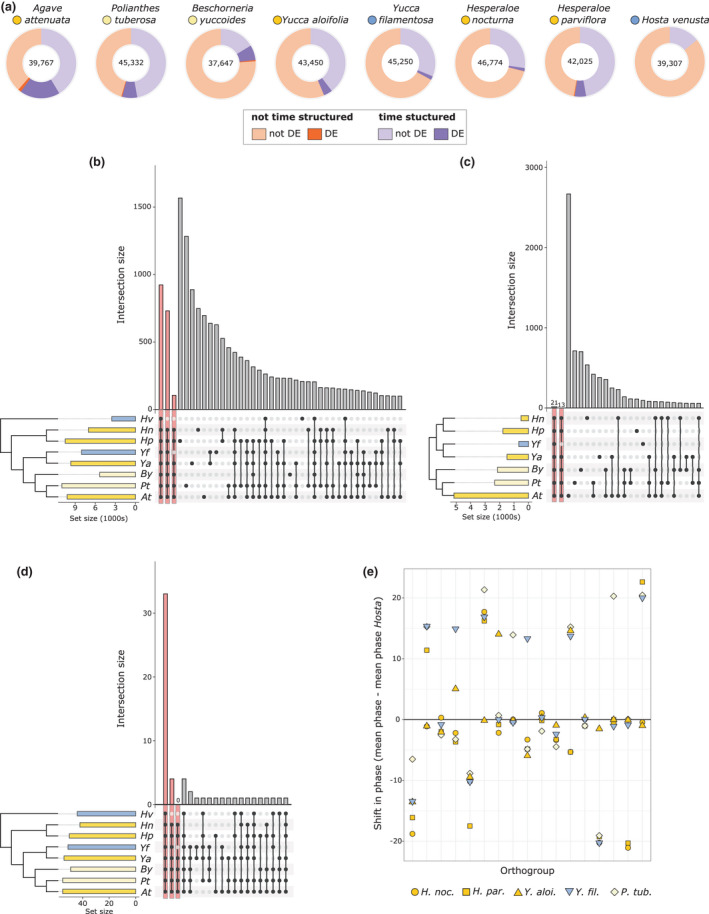
(a) Total number of transcripts assessed in each species (centre), with a proportion of the transcripts showing significant time‐structured expression (purple shades). A subset of both time‐structured and not time‐structured transcripts (orange shades) were differentially expressed (DE) under drought conditions (darker shades). (b) upSet plot showing overlap in gene families (orthogroups from OrthoFinder) that were time structured across the eight species; bars on left of the species names indicate total number of gene families per species, colours indicate Crassulacean acid metabolism (CAM) (bright yellow), C3 + CAM (pale yellow), and C3 (blue). The same colour scheme is used for panels (b–e). The first three bars highlighted in pink indicate orthogroups shared across all species, across all species except Hosta, and across all CAM species. (c) Comparison of gene families that had differential expression under drought stress across seven of the eight species (Hosta venusta was not droughted). (d) Comparison of gene families with core circadian clock annotations that had time‐structured expression across all eight species. (e) Shift in mean phase relative to H. venusta (mean phase of species – mean phase in H. venusta) in 17 gene families that had significantly different (P < 0.01) cycling across species as indicated by metaCycle.
The number of genes responsive to drought stress was far lower than the total number with time‐structured expression, and the majority of drought‐responsive genes had time‐structured expression in at least one condition (watered or drought) (Fig. 3a,c). Agave had the largest number of differentially expressed genes under drought (c. 20%, Fig. 3a), while H. nocturna had the fewest (c. 2%). In both Yucca species, all drought‐responsive genes were time structured in their expression. Examination of shared gene families of drought‐responsive genes across the species showed that many gene families were unique to a particular species (Fig. 3c), suggesting that the drought response in the Agavoideae is variable and lineage specific.
Of the gene families with circadian clock annotations, over half (33/58) had significant time‐structured expression in all eight species (Fig. 3d). Comparisons of phase (timing of peak expression) across the eight species resulted in few differences in phase between species. In a comparison of CAM species (excluding Agave and Beschorneria due to low replicates/resolution; please refer to methods) vs C3 species, only four gene families had a significant shift in phase: Pseudo‐response regulator 9 (PPR9), Alfin‐like (AFL), telomere binding protein (TRFL), and a gene of unknown function (no Arabidopsis homologue, and Blast hits are uncharacterised proteins) (Table S6). The comparison of phase changes between Hosta and the remainder of the Agavoideae species produced only a single gene family that had a shift in average timing of expression: TRFL, the same gene family found to be different between CAM and C3 species. In general, expression patterns across species were highly similar; of the 265 gene families that were (1) common in all eight species and (2) significant cyclers as assessed by metaCycle, only 17 had a shift in phase when testing for species as an explanatory factor (P < 0.01) (Table S7). To understand the evolution of phase, the mean phase per species of each of these 17 gene families was calculated, and from it we subtracted the mean phase of the gene family in Hosta (Fig. 3e). In the majority of these 17 gene families, the mean phase shift was low or was an instance in which one species had a large phase shift different from the remaining species (Fig. 3e), but none had a concerted C3‐to‐CAM shift. In general, timing of expression was similar across all eight species in the majority of gene families.
PPC expression
Gene tree reconstruction of sequences placed in PPC1 and PPC2 gene families by OrthoFinder was largely consistent with previous analyses (Fig. 4) (Deng et al., 2016; Heyduk et al., 2019a). The PPC1 tree shows a duplication event within monocot evolutionary history, after the divergence of the Dioscorales (represented by Dioscorea alata) from the lineage leading to the last common ancestor of Asparagales and Poales (although D. alata appears to have a lineage‐specific duplication). The monocot duplication event is independent from a similar duplication event in ancestral eudicots (Christin et al., 2014; Silvera et al., 2014). The placement of the Acorus americanus gene in the PPC2 phylogeny as sister to all other sampled angiosperm homologues except Amborella is likely to be a result of the lack of other eudicots taxa in the analyses, or possibly eudicot‐like mutations in the A. americanus PPC2 gene. Regardless, the remainder of the gene tree is concordant with species relationships.
Fig. 4.
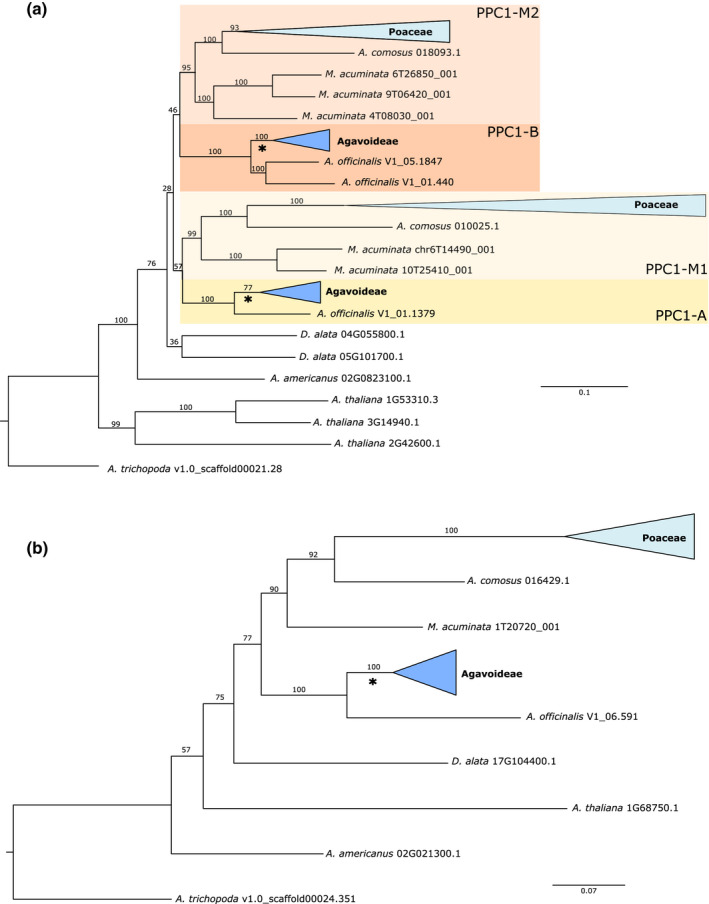
Gene trees estimated with IQTree for PPC1 (a) and PPC2 (b). Members of Poaceae and Agavoideae are collapsed for readability. All rapid bootstrap values are reported. Branches used for branch, branch × sites and clade model tests in codeml are subtended by an asterisk.
Although both major clades of PPC1 were expressed in the Agavoideae, the overall expression levels of PPC1‐B transcripts were much higher than for PPC1‐A, particularly in CAM species (Fig. 5). PPC1‐B expression also increased under drought notably in Polianthes tuberosa, a species known to engage in facultative CAM upon drought stress (Fig. 2). PPC2 transcripts were highly expressed in both Yucca aloifolia and Beschorneria yuccoides, strong CAM and facultative‐CAM species, respectively (Fig. 6). Expression of PPC2 increased with drought in Beschorneria, consistent with increased CAM activity under drought conditions (Fig. 2). Three gene copies of PPC2 were identified in the Yucca aloifolia genome, and all three had characteristic CAM‐like expression, with a peak before the onset of the dark period. Notably, PPC2 is also expressed in a CAM‐like pattern, albeit at lower levels, in the C3 Yucca filamentosa (Fig. 6). This finding is consistent with previous RNA‐seq analyses of Yucca (Heyduk et al., 2019b). Hesperaloe nocturna gene expression is not shown in Fig. 5 because the lengths of PPC transcripts were too short, and therefore were filtered out from our gene tree estimation and subsequent expression analyses.
Fig. 5.
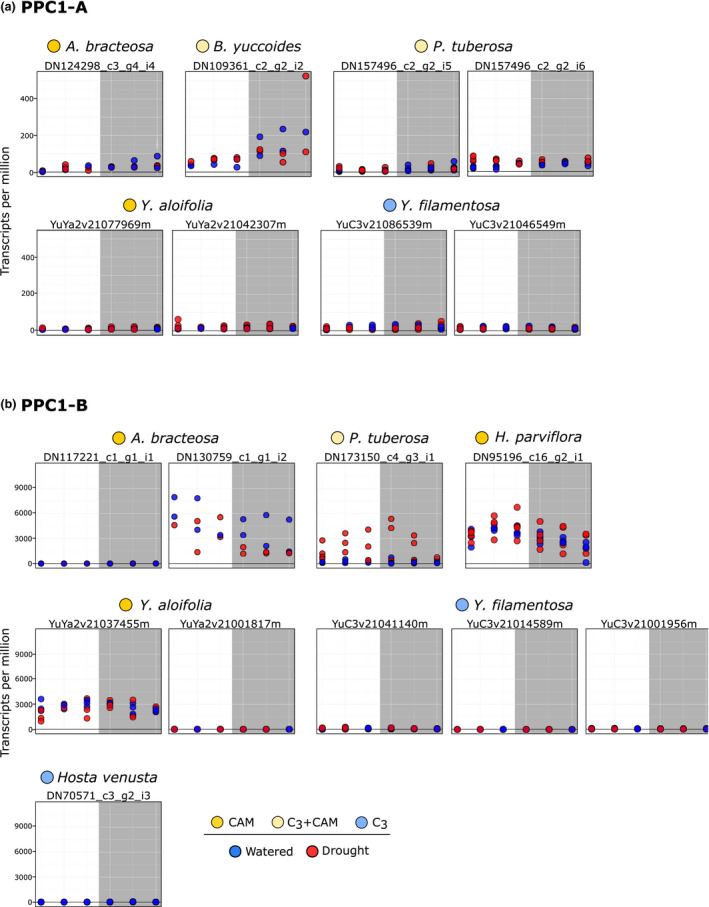
Expression (transcripts per million) of PPC1 transcripts from the core Agavoideae species, shown separately for the two clades (a) and (b). Dots represent individual samples, with blue, well watered and red, drought stressed. Colours next to species names indicate Crassulacean acid metabolism (CAM) (bright yellow), C3 + CAM (pale yellow), and C3 (blue). Grey boxes indicates night time points. Transcripts are only shown here if they passed length and percentage identity filtering. Hosta venusta did not have drought‐stressed samples taken for RNA sequencing.
Fig. 6.
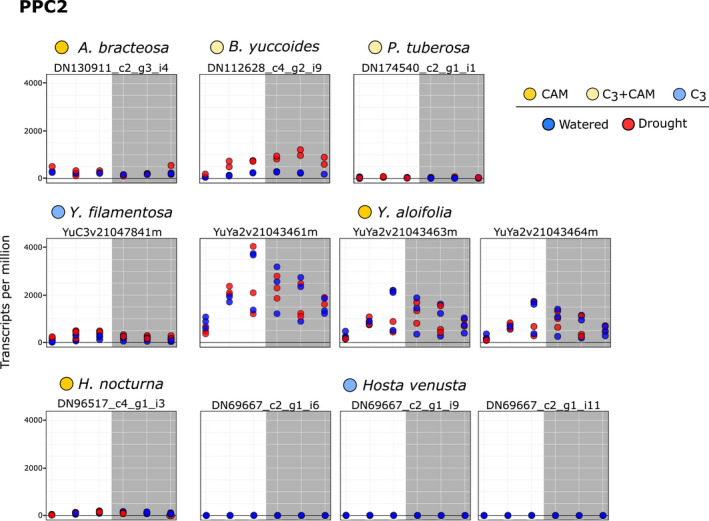
Expression (transcripts per million) of PPC2 transcripts from the core Agavoideae species. Dots represent individual samples, with blue, well watered and red, drought stressed. Colours next to species names indicate Crassulacean acid metabolism (CAM; bright yellow), C3 + CAM (pale yellow), and C3 (blue). Grey boxes indicates night time points. Transcripts are only shown here if they passed length and percentage identity filtering.
Molecular evolution of PPC genes
Assessment of changes in the strength and mode of selection assessed by the branch model revealed a significant shift in ω for PPC1‐A, but not PPC1‐B or PPC2. PPC1‐A had a reduced ω relative to the background rate, suggesting increased purifying selection consistent with this gene's role in housekeeping pathways (Fig. 4; Tables 1, 2). The sites model tests for positive selection were not significant for either PPC1 or PPC2 in the Agavoideae. However, PPC1‐B had significant positive selection on some sites in the Agavoideae genes (Table 1), and Bayesian Empirical Bayes analysis revealed only one site under positive selection with a posterior probability > 95%: a transition from an alanine to an asparagine at position 591. PPC1‐B also exhibited shifts on constraint in the clade‐sites test, with the Agavoideae having a third class of sites with weaker purifying selection compared with the background rate (0.44 on the foreground, 0.21 on the background, proportion of sites = 0.27). PPC2 similarly only had a significant rejection of the sites null model in favour of the alternative clade model, with a third class of sites that had an elevated ω relative to background (0.52 on foreground, 0.18 on background, proportion of sites = 0.36) (Table 2). Together these results suggest that specific amino acid residues in Agavoideae PPC1‐B and PPC2 genes may be evolving under relaxed or positive selection.
Table 1.
Results from tests of selection on PPC1.
| Branch | Model | ⍵ 1 | lnL | np 2 | Significance 3 |
|---|---|---|---|---|---|
| na | M0 | ⍵ = 0.077 | −52767.838 | 110 | na |
| PPC1‐A | Branch | ⍵b = 0.078, ⍵f = 0.013 | −52762.554 | 111 | LR = 10.57, P = 0.001 |
| PPC1‐B | Branch | ⍵b = 0.077, ⍵f = 0.065 | −52767.705 | 111 | LR = 0.265, P = 0.608 |
| na | Sites, M1a (nearly neutral) | ⍵1 = 0.059 (p1 = 0.92), ⍵2 = 1 (p2 = 0.08) | −52185.176 | 111 | na |
| na | Sites, M2a (positive selection) | ⍵1 = 0.059 (p1 = 0.92), ⍵2 = 1 (p2 = 0.05), ⍵3 = 1 (p3 = 0.03) | −52185.176 | 113 | LR = 0 |
| PPC1‐A | MA null | 0[⍵b = 0.059, ⍵f = 0.059, p = 0.92], 1[⍵b = 1, ⍵f = 1, p = 0.08], 2a[⍵b = 0.059, ⍵f = 1, p = 0], 2b[⍵b = 1, ⍵f = 1, p = 0] | −52185.176 | 112 | na |
| PPC1‐A | MA | 0[⍵b = 0.059, ⍵f = 0.059, p = 0.92], 1[⍵b = 1, ⍵f = 1, p = 0.08], 2a[⍵b = 0.059, ⍵f = 39.8, p = 0.002], 2b[⍵b = 1, ⍵f = 1, p = 0] | −52185.176 | 113 | LR = 0 |
| PPC1‐B | MA null | 0[⍵b = 0.059, ⍵f = 0.059, p = 0.92], 1[⍵b = 1, ⍵f = 1, p = 0.08], 2a[⍵b = 0.059, ⍵f = 1, p = 0], 2b[⍵b = 1, ⍵f = 1, p = 0] | −52185.176 | 112 | na |
| PPC1‐B | MA | 0[⍵b = 0.059, ⍵f = 0.059, p = 0.92], 1[⍵b = 1, ⍵f = 1, p = 0.08], 2a[⍵b = 0.059, ⍵f = 39.8, p = 0.002], 2b[⍵b = 1, ⍵f = 39.8, p = 0.0002] | −52182.449 | 113 | LR = 5.45, P = 0.019 |
| na | M2a_rel (null for clade‐sites C tests) | ⍵1 = 0.02 (p1 = 0.72), ⍵2 = 1 (p2 = 0.02), ⍵3 = 0.22 (p3 = 0.26) | −51414.294 | 113 | na |
| PPC1‐A | Clade‐sites C | 0[⍵b = 0.02, ⍵f = 0.02, p = 0.72], 1[⍵b = 1, ⍵f = 1, p = 0.016], 2[⍵b = 0.23, ⍵f = 0.27, p = 0.26] | −51413.361 | 114 | LR = 1.87, P = 0.17 |
| PPC1‐B | Clade‐sites C | 0[⍵b = 0.02, ⍵f = 0.02, p = 0.71], 1[⍵b = 1, ⍵f = 1, p = 0.02], 2[⍵b = 0.21, ⍵f = 0.44, p = 0.27] | −51392.506 | 114 | LR = 43.58, P < 0.001 |
Values reported for background, foreground, in which foreground is the branch of interest in the Agavoideae; for sites models, three classes of omegas and proportion of sites (p) in each class are reported; for branch × sites models, foreground and background values for omega are reported plus a proportion of sites in each site class (0, 1, 2a and 2b).
Number of parameters.
Based on a likelihood ratio test which is χ2 distributed. na, null models that were not tested against any other null.
Table 2.
Results from tests of selection on PPC2.
| Model | ⍵ 1 | lnL | np 2 | Significance 3 |
|---|---|---|---|---|
| M0 | ⍵ = 0.126 | −27536.821 | 44 | na |
| Branch | ⍵b = 0.126,⍵f = 0.115 | −27536.753 | 45 | LR = 0.136, P = 0.71 |
| Sites, M1a (nearly neutral) | ⍵1 = 0.07 (p1 = 0.82), ⍵2 = 1 (p2 = 0.18) | −26777.829 | 45 | na |
| sites, M2a (positive selection) | ⍵1 = 0.07 (p1 = 0.82), ⍵2 = 1 (p2 = 0.09), ⍵3 = 1 (p3 = 0.09) | −26777.829 | 47 | LR = 0 |
| MA null | 0[⍵b = 0.065, ⍵f = 0.065, p = 0.82], 1[⍵b = 1, ⍵f = 1, p = 0.18], 2a[⍵b = 0.07, ⍵f = 1, p = 0], 2b[⍵b = 1, ⍵f = 1, p = 0] | −26777.829 | 46 | na |
| MA | 0[⍵b = 0.065, ⍵f = 0.065, p = 0.82], 1[⍵b = 1, ⍵f = 1, p = 0.18], 2a[⍵b = 0.07, ⍵f = 1, p = 0], 2b[⍵b = 1, ⍵f = 1, p = 0] | −26777.829 | 47 | LR = 0 |
| M2a_rel (null for clade‐sites C tests) | ⍵1 = 0.02 (p1 = 0.59), ⍵2 = 1 (p2 = 0.09), ⍵3 = 0.23 (p3 = 0.33) | −26537.435 | 47 | na |
| Clade‐sites C | 0[⍵b = 0.01, ⍵f = 0.01, p = 0.55], 1[⍵b = 1, ⍵f = 1, p = 0.09], 2[⍵b = 0.18, ⍵f = 0.52, p = 0.36] | −26508.092 | 48 | LR = 58.68, P < 0.001 |
Values reported for background, foreground, in which foreground is the branch of interest in the Agavoideae; for sites models, three classes of omegas and proportion of sites in each class are reported; for branch × sites models, foreground and background values for omega are reported plus proportion of sites in each site class (0, 1, 2a and 2b).
Number of parameters.
Based on a likelihood ratio test that is χ2 distributed. na, null models that were not tested against any other null.
Discussion
Evolution of CAM in the Agavoideae
Previous work estimated three independent origins of CAM in the Agavoideae: one in the genus Hesperaloe, one in the genus Yucca, and one in Agave s.l. (Heyduk et al., 2016b). However, this initial estimation was based on carbon isotope values, which cannot separate C3 + CAM from C3 in the majority of cases (Winter et al., 2015). Detailed physiological measurements under both well watered and drought‐stressed conditions revealed that Polianthes had the ability to upregulate CAM under drought stress, although maintains low‐level CAM even under well watered conditions. Beschorneria has a very slight CAM increase at night, indicated by a small shift in titratable acidities and a decrease in nighttime respiration (Heyduk et al., 2018b). Yucca species are divided, in that nearly half are expected to use C3 and the other half are likely to be CAM; these inferences are based on carbon isotopes, and warrant more detailed physiological assessment beyond the two species included in this study and a handful of others (Smith et al., 1983; Heyduk et al., 2016a). The presence of CAM was confirmed in Hesperaloe, with both species in this study exhibiting strong CAM (Fig. 2). Hosta showed no evidence of CAM in our study, although it was not drought stressed. Based on gene expression patterns detected here, as well as anatomical traits and carbon isotope values (Heyduk et al., 2016b), we do not expect Hosta to be able to upregulate CAM under drought stress. Together these separate physiological assessments across the Agavoideae confirmed the presence of CAM in Hesperaloe, Yucca and Agave s.l. and furthered our understanding of intermediate CAM species (e.g. Polianthes and Beschorneria).
Conservation and novelty in gene expression
Across diverse plant species, roughly 20–60% of transcripts show some time‐of‐day differential expression (Covington et al., 2008; Hayes et al., 2010; Filichkin et al., 2011; Lai et al., 2020), however Arabidopsis has up to 89% of transcripts cycling under at least one experimental time course condition (Michael et al., 2008). In Sedum album, which has the ability to facultatively upregulate CAM, there is a slight increase in the number of cycling transcripts when plants use CAM compared with C3 (41% vs 35%, respectively) (Wai et al., 2019). The number of time‐structured transcripts in Agavoideae species varied, with Hosta having the fewest transcripts that were time structured, and Agave having the greatest number. The number of time‐structured transcripts did not correlate with the presence of CAM; for example, Y. filamentosa and Y. aloifolia had similar numbers of time‐structured transcripts, despite differences in photosynthetic pathway. Similarly, Agave and Polianthes both had many transcripts with diel variation, despite Polianthes being only weakly, facultatively CAM (Fig. 3). Beschorneria, which is sister to Polianthes and Agave, showed the smallest number of time‐structured transcripts, although it is also the weakest CAM species measured across these species.
Very few gene families had time‐structured expression across all CAM species (n = 105 in all CAM, n = 126 in strong CAM). Two key genes related to CAM – PPC2 (although, notably, not PPC1) and PPCK – were time structured in all species except Hosta (i.e. including the C3 Y. filamentosa). PPCK in particular has been shown to have direct and reciprocal clock connections; knockdowns of PPCK in Kalanchoë fedtschenkoi had significantly reduced CAM and the lack of circadian oscillation in PPCK perturbed oscillation patterns of core clock genes (Boxall et al., 2017). Knockdown of PPC1 in Kalanchoë laxiflora also resulted in changes to the oscillation patterns and amplitude of clock genes, although notably a different set of core clock genes was affected by PPC1 knockdowns relative to PPCK (Boxall et al., 2020). The integration of the circadian clock and CAM pathway genes is clearly important for CAM physiology, although the presence and cycling of these genes does not, alone, lead to CAM ability; Y. filamentosa has cycling of these gene families (e.g. PPC2), but expression levels are either too low or transcripts are affected by other post‐translational modifications to render them insufficient for CAM (Heyduk et al., 2019b). Moreover, we found a lack of shared gene families with shifts to time‐structured expression across CAM species in the Agavoideae, which suggests three hypotheses: (1) the repeated evolution of CAM has involved lineage‐specific changes to the molecular networks rather than parallelisms, (2) gene re‐wiring happened in the ancestor of the Agavoideae and facilitated the repeated evolution of CAM, or (3) the overall scope of re‐wiring of gene expression into the clock is limited for CAM.
Assessing the 24‐h time‐structured variation of gene expression in CAM and C3 lineages has confirmed the important role of clock integration with CAM metabolic genes, but generally has not revealed any master regulator of CAM. Most studies highlight the conservation of circadian clock components and the timing of their expression, regardless of the photosynthetic pathway (Moseley et al., 2018; Wai & VanBuren, 2018; Yin et al., 2018; Wai et al., 2019). Researchers have focused on the few aspects of the clock that are different between C3 and CAM comparisons. For example, PRR9 in Opuntia (CAM) was shown to have a change in phase compared with the Arabidopsis orthologue (Mallona et al., 2011), comparisons between Kalanchoë and Arabidopsis showed phase shifts in some evening elements, including ELF3/4 and LUX (Moseley et al., 2018), and Agave had shifted expression of RVE, a clock output gene, relative to Arabidopsis (Yin et al., 2018). In the Agavoideae, the majority of circadian gene families had shared phase of expression across all eight species. Of those gene families that had a significant species effect in the phase of expression, few had extreme phase shifts or showed consistent C3 vs CAM differences. Our findings, together with those of other studies assessing core circadian regulators in CAM lineages, pointed to an overall conservation of the circadian clock, even in plants with a strong CAM physiology (Boxall et al., 2020). However, it is worth noting that the majority of comparative transcriptomics studies in CAM, including this one, assessed temporal variation in expression over a single day : night period, making it difficult to pinpoint which genes were responsible for clock inputs into the CAM pathway, and which were downstream targets. Many studies still rely on distant outgroups for comparison (typically Arabidopsis), and therefore continue to confound changes associated with CAM to those that have arisen simply due to evolutionary divergence. Future work on the nature of gene expression and evolution in CAM species should endeavour to use free‐running conditions to better assess the roles of the circadian clock in CAM species, and should carefully select comparison species to minimise evolutionary distance. Regardless, it seems unlikely that large perturbations to the circadian clock are required for the evolution of CAM from a C3 ancestor; instead, changes to promoter sequences and regulatory regions of genes contributing to CAM may have a larger role to play in altering the timing and magnitude of their expression.
Unlike the relatively conserved number and type of genes that exhibited time‐structured variation in gene expression, the response to drought was highly lineage specific. A large proportion of gene families was uniquely differentially expressed in a singular species. Although the present study includes comparisons across three separate experiments (Heyduk et al., 2018b, 2019b), even species drought stressed in the same experiment showed vastly different responses to drought. While plant water status was monitored in all experiments by the proxy of soil moisture content (Table S1), even the same soil moisture content can variably affect plants of different ages, genotypes and species. Agave's strong differential regulation to drought is surprising given its constitutive CAM physiology that is thought to buffer against the effects of drought stress. Indeed, the majority of CAM species studied here was affected by drought: Y. aloifolia, A. bracteosa, and Manfreda sp. all exhibited decreases in titratable leaf acidity and, in Yucca and Manfreda, drops in nocturnal CO2 assimilation. The effects of drought stress on CAM physiology are vastly understudied, although work has been done in facultative‐CAM species (Cushman et al., 2008; Wai et al., 2019; Heyduk et al., 2021). Both the physiological and gene expression data presented here suggested that full CAM species are not immune to the effects of drought, and indeed exhibit strong physiological and transcriptional responses. Finally, for all species studied here, the majority of drought‐responsive genes were also time structured; in other words, constitutively expressed genes were infrequently affected by drought stress.
Gene recruitment for CAM photosynthesis
In almost all published instances of C4 or CAM evolution (please refer to Wickell et al., 2021 for a notable exception), the PPC gene copy that was recruited was from a gene family known as the ‘plant’ PPCs, or PPC1. PPC1 is used by all plants for the replenishment of intermediates in the TCA cycle, and a singular copy typically gets re‐wired for C4 or CAM (Heyduk et al., 2019a). The clear CAM‐like expression of PPC2 in Yucca aloifolia and, to a lesser extent, Beschorneria yuccoides, suggests that both of these species have recruited PPC2 as an alternate carboxylating enzyme for CAM. PPC1 is still expressed in both of these species, and supports previous work that suggests PPC2 that forms a hetero‐octamer with PPC1 (O’Leary et al., 2011), although this remains to be tested in the Agavoideae. Although we cannot say for certain PPC2 protein is produced, it seems unlikely that the transcripts would be expressed so highly (> 1000 TPM) in Yucca aloifolia with no functional consequence. Moreover, expression of PPC2 in Yucca aloifolia peaks right before the onset of the night period, consistent with expression patterns of PPCs in other Agavoideae in this study, as well as expression profiles of PPC in other lineages (Ming et al., 2015; Abraham et al., 2016; Yang et al., 2017; Heyduk et al., 2018a; Wai et al., 2019). The overall carboxylase activity of PPC2 in the Agavoideae remains to be studied, but could lend further clues to how this atypical gene copy was recruited into the CAM pathway.
Unlike C4 PPCs, in which convergent amino acid substitutions seem key to the recruitment of PPC1 gene copies into the C4 pathway (Christin et al., 2007; Rosnow et al., 2014; Goolsby et al., 2018), evidence for convergent evolution at the molecular level in CAM is lacking. A comparison of PPC sequences between Kalanchoë and Phalaenopsis did reveal a shared amino acid change from R/H/K to D and was shown to significantly increase the activity of PPC (Yang et al., 2017). However, this amino acid substitution is not ubiquitous in CAM species; it is absent from Ananas (Yang et al., 2017) and all members of the Agavoideae examined here (please refer to the github repository for Fasta files), suggesting that either the shared mutation is due to homoplasy, or may be convergent but not essential for CAM. Our results further suggest that overall PPC genes are conserved, even when they are being recruited into the CAM pathway (Tables 1 and 2). In general, the lability of CAM as a phenotype, as well as the wide diversity of lineages in which it evolves, seems to allow variable pathways to organise the genetic requirements, including which major copy of the main carboxylating enzyme, PPC, is recruited. Increasing the number of CAM lineages studied physiologically and genomically will allow us to determine whether novel mechanisms of evolving CAM – such as the recruitment of PPC2 in the Agavoideae – are indeed rare, or more common across green plants.
By comparing RNA‐seq data across closely related species that span multiple origins of CAM, we have shown that most gene families have diurnal variation in gene expression, regardless of photosynthetic status. In particular, core circadian clock genes are similarly expressed across all the species examined here. By contrast, drought response was highly lineage specific, and suggests that lineages have fine tuned or independently evolved their drought response gene networks. While historically CAM in the Agavoideae has been thought to be the result of three independent origins, we cannot rule out a single origin of CAM with subsequent reversals to C3. However, reversals to C3 from CAM appear to be rare in angiosperms, and the recruitment of PPC2 for CAM function in Yucca (and to a lesser extent in Beschorneria) supports the inference of independent origins of CAM in the Agavoideae and furthers the idea that the evolutionary routes to CAM are markedly variable.
Author contributions
KH conducted physiology experiments, sampled for RNA, prepared sequencing libraries and analysed data; EVM conducted molecular evolution analyses; JL‐M helped with framing and data interpretation; all three authors contributed to writing and editing the manuscript.
Supporting information
Table S1 Soil moisture values for species included here under well watered and drought conditions.
Table S2 Raw Li‐Cor data for Hesperaloe parviflora, Hesperaloe nocturna and Hosta venusta.
Table S3 Titratable acidity measurements for Hesperaloe parviflora, Hesperaloe nocturna and Hosta venusta.
Table S4 Gene families with time‐structured expression across all eight species.
Table S5 Gene families with time‐structured expression in all species but Hosta venusta.
Table S6 Analysis of circadian‐annotated genes in metaCycle, using all species except Agave and Beschorneria, which had too few replicates and time points after data cleaning.
Table S7 Abbreviated ANOVA results of testing effect of species on phase of gene expression.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
The authors would like to acknowledge the UGA Plant Biology glasshouse staff for the help in maintaining plants, the Georgia Advanced Computing Resources Center and the University of Hawaiʻi Information Technology Services – Cyberinfrastructure for computational support, and Amanda L. Cummings and Richard Field for assistance with glasshouse and lab work. This work was supported by funding from NSF (DEB1442199 to JL‐M) and a Yale Institute for Biospheric Studies Donnelley Fellowship (to KH). We thank the Department of Energy Joint Genome Institute and collaborators for access to pre‐publication data for Acorus americanus v.1.1, Yucca aloifolia and Yucca filamentosa.
Data availability
Raw sequencing data generated in this study are available on the NCBI SRA, under BioProject PRJNA755802. Processed RNA‐seq counts and TPM files, as well as Fasta files for PPC1 and PPC2 molecular evolution analyses, are available at https://github.com/kheyduk/AgavoideaeComparative.
References
- Abraham PE, Yin H, Borland AM, Weighill D, Lim SD, De Paoli HC, Engle N, Jones PC, Agh R, Weston DJ et al. 2016. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nature Plants 2: 16178. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project . 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Aubry S, Kelly S, Kümpers BMC, Smith‐Unna RD, Hibberd JM. 2014. Deep evolutionary comparison of gene expression identifies parallel recruitment of trans‐factors in two independent origins of C4 photosynthesis. PLoS Genetics 10: e1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J et al. 2012. Reference genome sequence of the model plant Setaria . Nature Biotechnology 30: 555–561. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxall SF, Dever LV, Kneřová J, Gould PD, Hartwell J. 2017. Phosphorylation of phosphoenolpyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate crassulacean acid metabolism species Kalanchoë fedtschenkoi . Plant Cell 29: 2519–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxall SF, Kadu N, Dever LV, Kneřová J, Waller JL, Gould PJD, Hartwell J. 2020. Kalanchoë PPC1 is essential for Crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes. Plant Cell 32: 1136–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Schliesky S, Külahoglu C, Osborne CP, Weber APM. 2014. Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD‐ME, NADP‐ME, and PEP‐CK C4 species. Journal of Experimental Botany 65: 3579–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near‐optimal probabilistic RNA‐seq quantification. Nature Biotechnology 34: 525–527. [DOI] [PubMed] [Google Scholar]
- Bredeson JV, Lyons JB, Oniyinde IO, Okereke NR, Kolade O, Nnabue I, Nwadili CO, Hřibová E, Parker M, Nwogha J et al. 2022. Chromosome evolution and the genetic basis of agronomically important traits in greater yam. Nature Communications 13: 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhaus D, Bräutigam A, Mettler‐Altmann T, Winter K, Weber APM. 2016. Reversible burst of transcriptional changes during induction of crassulacean acid metabolism in Talinum triangulare . Plant Physiology 170: 102–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceusters J, Borland AM, Taybi T, Frans M, Godts C, De Proft MP. 2014. Light quality modulates metabolic synchronization over the diel phases of Crassulacean acid metabolism. Journal of Experimental Botany 65: 3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C‐Y, Krishnakumar V, Chan AP, Thibaud‐Nissen F, Schobel S, Town CD. 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. The Plant Journal 89: 789–804. [DOI] [PubMed] [Google Scholar]
- Christin P‐A, Arakaki M, Osborne CP, Bräutigam A, Sage RF, Hibberd JM, Kelly S, Covshoff S, Wong GK‐S, Hancock L et al. 2014. Shared origins of a key enzyme during the evolution of C4 and CAM metabolism. Journal of Experimental Botany 65: 3609–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P‐A, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP. 2013. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biology and Evolution 5: 2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P‐A, Salamin N, Savolainen V, Duvall MR, Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Current Biology 17: 1241–1247. [DOI] [PubMed] [Google Scholar]
- Christin P‐A, Samaritani E, Petitpierre B, Salamin N, Besnard G. 2009. Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biology and Evolution 1: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Nueda MJ, Ferrer A, Talón M. 2006. maSigPro: a method to identify significantly differential expression profiles in time‐course microarray experiments. Bioinformatics 22: 1096–1102. [DOI] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33: 2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. 2008. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biology 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Tillett RL, Wood JA, Branco JM, Schlauch KA. 2008. Large‐scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and Crassulacean acid metabolism (CAM). Journal of Experimental Botany 59: 1875–1894. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Denoeud F, Aury J‐M, Baurens F‐C, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M et al. 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488: 213–217. [DOI] [PubMed] [Google Scholar]
- Deng H, Zhang L‐S, Zhang G‐Q, Zheng B‐Q, Liu Z‐J, Wang Y. 2016. Evolutionary history of PEPC genes in green plants: implications for the evolution of CAM in orchids. Molecular Phylogenetics and Evolution 94: 559–564. [DOI] [PubMed] [Google Scholar]
- Denoeud F, Carretero‐Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G et al. 2014. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345: 1181–1184. [DOI] [PubMed] [Google Scholar]
- Edwards EJ. 2019. Evolutionary trajectories, accessibility, and other metaphors: the case of C4 and CAM photosynthesis. The New Phytologist 223: 1752–1755. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eronen JT, Fortelius M, Micheels A, Portmann FT, Puolamäki K, Janis CM. 2012. Neogene aridification of the Northern Hemisphere. Geology 40: 823–826. [Google Scholar]
- Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, Michael TP, Chory J, Kay SA, Mockler TC. 2011. Global profiling of rice and poplar transcriptomes highlights key conserved circadian‐controlled pathways and cis‐regulatory modules. PLoS ONE 6: e16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennidakis S, Rao S, Greenham K, Uhrig RG. 2007. Bacterial‐ and plant‐type phosphoenolpyruvate carboxylase polypeptides interact in the hetero‐oligomeric class‐2 PEPC complex of developing castor oil seeds. The Plant Journal 52: 839–849. [DOI] [PubMed] [Google Scholar]
- Glenn TC, Nilsen RA, Kieran TJ, Sanders JG, Bayona‐Vásquez NJ, Finger JW, Pierson TW, Bentley KE, Hoffberg SL, Louha S et al. 2019. Adapterama I: universal stubs and primers for 384 unique dual‐indexed or 147,456 combinatorially‐indexed Illumina libraries (iTru & iNext). PeerJ 7: e7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Chen J, Mushegian AR. 2005. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb–Scargle periodograms. Bioinformatics 22: 310–316. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goolsby EW, Moore AJ, Hancock LP, De Vos JM, Edwards EJ. 2018. Molecular evolution of key metabolic genes during transitions to C4 and CAM photosynthesis. American Journal of Botany 105: 602–613. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al. 2011. Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nature Biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkess A, Zhou J, Xu C, Bowers JE, Van der Hulst R, Ayyampalayam S, Mercati F, Riccardi P, McKain MR, Kakrana A et al. 2017. The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nature Communications 8: 1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes KR, Beatty M, Meng X, Simmons CR, Habben JE, Danilevskaya ON. 2010. Maize global transcriptomics reveals pervasive leaf diurnal rhythms but rhythms in developing ears are largely limited to the core oscillator. PLoS ONE 5: e12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, Burrell N, Lalani F, Leebens‐Mack J. 2016a. Gas exchange and leaf anatomy of a C3‐CAM hybrid, Yucca gloriosa (Asparagaceae). Journal of Experimental Botany 67: 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, Hwang M, Albert V, Silvera K, Lan T, Farr K, Chang T‐H, Chan M‐T, Winter K, Leebens‐Mack J. 2018a. Altered gene regulatory networks are associated with the transition from C3 to Crassulacean acid metabolism in Erycina (Oncidiinae: Orchidaceae). Frontiers in Plant Science 9: 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, McKain MR, Lalani F, Leebens‐Mack J. 2016b. Evolution of CAM anatomy predates the origins of Crassulacean acid metabolism in the Agavoideae (Asparagaceae). Molecular Phylogenetics and Evolution 105: 102–113. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Moreno‐Villena JJ, Gilman IS, Christin P‐A, Edwards EJ. 2019a. The genetics of convergent evolution: insights from plant photosynthesis. Nature Reviews Genetics 20: 485–493. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Ray JN, Ayyampalayam S, Leebens‐Mack J. 2018b. Shifts in gene expression profiles are associated with weak and strong Crassulacean acid metabolism. American Journal of Botany 105: 587–601. [DOI] [PubMed] [Google Scholar]
- Heyduk K, Ray JN, Ayyampalayam S, Moledina N, Borland A, Harding SA, Tsai C‐J, Leebens‐Mack J. 2019b. Shared expression of Crassulacean acid metabolism (CAM) genes predates the origin of CAM in the genus Yucca . Journal of Experimental Botany 70: 6597–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, Ray JN, Leebens‐Mack J. 2021. Leaf anatomy is not correlated to CAM function in a C3+CAM hybrid species, Yucca gloriosa . Annals of Botany 4: 437–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. 2010. JTK CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome‐scale data sets. Journal of Biological Rhythms 25: 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Fujiwara M, Tanaka I, Fukao Y, Yanagawa Y. 2010. Characterization of bacterial‐type phosphoenolpyruvate carboxylase expressed in male gametophyte of higher plants. BMC Plant Biology 10: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Brachypodium Initiative . 2010. Genome sequencing and analysis of the model grass Brachypodium distachyon . Nature 463: 763–768. [DOI] [PubMed] [Google Scholar]
- Lai X, Bendix C, Yan L, Zhang Y, Schnable JC, Harmon FG. 2020. Interspecific analysis of diurnal gene regulation in panicoid grasses identifies known and novel regulatory motifs. BMC Genomics 21: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped‐read alignment with Bowtie 2. Nature Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallona I, Egea‐Cortines M, Weiss J. 2011. Conserved and divergent rhythms of Crassulacean acid metabolism‐related and core clock gene expression in the cactus Opuntia ficus‐indica . Plant Physiology 156: 1978–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick RF, Truong SK, Sreedasyam A, Jenkins J, Shu S, Sims D, Kennedy M, Amirebrahimi M, Weers BD, McKinley B et al. 2018. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. The Plant Journal 93: 338–354. [DOI] [PubMed] [Google Scholar]
- McKain MR, McNeal JR, Kellar PR, Eguiarte LE, Pires JC, Leebens‐Mack J. 2016. Timing of rapid diversification and convergent origins of active pollination within Agavoideae (Asparagaceae). American Journal of Botany 103: 1717–1729. [DOI] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM et al. 2008. Network discovery pipeline elucidates conserved time‐of‐day‐specific cis‐regulatory modules. PLoS Genetics 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R, VanBuren R, Wai CM, Tang H, Schatz MC, Bowers JE, Lyons E, Wang M‐L, Chen J, Biggers E et al. 2015. The pineapple genome and the evolution of CAM photosynthesis. Nature Genetics 47: 1435–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ‐Tree 2: new models and efficient methods for phylogenetic inference in the genomic era. Molecular Biology and Evolution 37: 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley RC, Mewalal R, Motta F, Tuskan GA, Haase S, Yang X. 2018. Conservation and diversification of circadian rhythmicity between a model Crassulacean acid metabolism plant Kalanchoë fedtschenkoi and a model C3 photosynthesis plant Arabidopsis thaliana . Frontiers in Plant Science 9: 1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L‐T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ‐Tree: a fast and effective stochastic algorithm for estimating maximum‐likelihood phylogenies. Molecular Biology and Evolution 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nueda MJ, Tarazona S, Conesa A. 2014. Next maSigPro: updating maSigPro bioconductor package for RNA‐seq time series. Bioinformatics 30: 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary B, Park J, Plaxton WC. 2011. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post‐translational controls of non‐photosynthetic PEPCs. Biochemical Journal 436: 15–34. [DOI] [PubMed] [Google Scholar]
- O’Leary B, Rao SK, Kim J, Plaxton WC. 2009. Bacterial‐type phosphoenolpyruvate carboxylase (PEPC) functions as a catalytic and regulatory subunit of the novel class‐2 PEPC complex of vascular plants. The Journal of Biological Chemistry 284: 24797–24805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud‐Nissen F, Malek RL, Lee Y, Zheng L et al. 2007. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Research 35: D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2021. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnow JJ, Edwards GE, Roalson EH. 2014. Positive selection of Kranz and non‐Kranz C4 phosphoenolpyruvate carboxylase amino acids in Suaedoideae (Chenopodiaceae). Journal of Experimental Botany 65: 3595–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan H, Feng T, Walker‐Hale N, Lopez‐Nieves S, Pucker B, Guo R, Yim WC, Badgami R, Timoneda A, Zhao L et al. 2020. Evolution of l‐DOPA 4,5‐dioxygenase activity allows for recurrent specialisation to betalain pigmentation in Caryophyllales. The New Phytologist 227: 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera K, Winter K, Rodriguez BL, Albion RL, Cushman JC. 2014. Multiple isoforms of phosphoenolpyruvate carboxylase in the Orchidaceae (subtribe Oncidiinae): implications for the evolution of Crassulacean acid metabolism. Journal of Experimental Botany 65: 3623–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Hartsock TL, Nobel PS. 1983. Ecophysiology of Yucca brevifolia, an arborescent monocot of the Mojave desert. Oecologia 60: 10–17. [DOI] [PubMed] [Google Scholar]
- Streisfeld MA, Rausher MD. 2009. Genetic changes contributing to the parallel evolution of red floral pigmentation among Ipomoea species. The New Phytologist 183: 751–763. [DOI] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. Pal2nal: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC. 2000. A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in crassulacean acid metabolism‐induced leaves of the common ice plant. Plant Physiology 123: 1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai CM, VanBuren R. 2018. Circadian regulation of pineapple CAM photosynthesis. In: Ming R, ed. Genetics and genomics of pineapple. Cham, Switzerland: Springer International Publishing, 247–258. [Google Scholar]
- Wai CM, Weise SE, Ozersky P, Mockler TC, Michael TP, VanBuren R. 2019. Time of day and network reprogramming during drought induced CAM photosynthesis in Sedum album . PLoS Genetics 15: e1008209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liu R, Wu G, Lang C, Chen J, Shi C. 2012. Specific downregulation of the bacterial‐type PEPC gene by artificial microRNA improves salt tolerance in Arabidopsis . Plant Molecular Biology Reporter 30: 1080–1087. [Google Scholar]
- Wickell D, Kuo L‐Y, Yang HP, Ashok AD, Irisarri I, Dadras A, de Vries S, de Vries J, Huang Y‐M, Li A et al. 2021. Underwater CAM photosynthesis elucidated by Isoetes genome. Nature Communications 12: 6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Holtum JAM, Smith JAC. 2015. Crassulacean acid metabolism: a continuous or discrete trait? The New Phytologist 208: 73–78. [DOI] [PubMed] [Google Scholar]
- Wong WSW, Yang Z, Goldman N, Nielsen R. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. 2016. MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics 32: 3351–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hu R, Yin H, Jenkins J, Shu S, Tang H, Liu D, Weighill DA, Cheol Yim W, Ha J et al. 2017. The Kalanchoë genome provides insights into convergent evolution and building blocks of Crassulacean acid metabolism. Nature Communications 8: 1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. Paml 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yin H, Guo H‐B, Weston DJ, Borland AM, Ranjan P, Abraham PE, Jawdy SS, Wachira J, Tuskan GA, Tschaplinski TJ et al. 2018. Diel rewiring and positive selection of ancient plant proteins enabled evolution of CAM photosynthesis in Agave . BMC Genomics 19: 588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Soil moisture values for species included here under well watered and drought conditions.
Table S2 Raw Li‐Cor data for Hesperaloe parviflora, Hesperaloe nocturna and Hosta venusta.
Table S3 Titratable acidity measurements for Hesperaloe parviflora, Hesperaloe nocturna and Hosta venusta.
Table S4 Gene families with time‐structured expression across all eight species.
Table S5 Gene families with time‐structured expression in all species but Hosta venusta.
Table S6 Analysis of circadian‐annotated genes in metaCycle, using all species except Agave and Beschorneria, which had too few replicates and time points after data cleaning.
Table S7 Abbreviated ANOVA results of testing effect of species on phase of gene expression.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Raw sequencing data generated in this study are available on the NCBI SRA, under BioProject PRJNA755802. Processed RNA‐seq counts and TPM files, as well as Fasta files for PPC1 and PPC2 molecular evolution analyses, are available at https://github.com/kheyduk/AgavoideaeComparative.


