Summary
Embryonic stem cells (ESCs) are self-renewing and pluripotent. In recent years, factors that control pluripotency, mostly nuclear, have been identified. To identify non-nuclear regulators of ESCs, we screened an endogenously labeled fluorescent fusion-protein library in mouse ESCs. One of the more compelling hits was the cell-cycle-associated protein 1 (CAPRIN1). CAPRIN1 knockout had little effect in ESCs, but it significantly altered differentiation and gene expression programs. Using RIP-seq and SLAM-seq, we found that CAPRIN1 associates with, and promotes the degradation of, thousands of RNA transcripts. CAPRIN1 interactome identified XRN2 as the likely ribonuclease. Upon early ESC differentiation, XRN2 is located in the nucleus and colocalizes with CAPRIN1 in small RNA granules in a CAPRIN1-dependent manner. We propose that CAPRIN1 regulates an RNA degradation pathway operating during early ESC differentiation, thus eliminating undesired spuriously transcribed transcripts in ESCs.
Keywords: embryonic stem cells, pluripotency, differentiation, RNA degradation, RNA stability, stress granules, XRN2, CAPRIN1
Graphical abstract

Highlights
-
•
CAPRIN1 is highly expressed in murine ESCs and is required for ESC differentiation
-
•
CAPRIN1 is involved in RNA degradation in ESCs
-
•
CAPRIN1 acts through the exoribonuclease XRN2
-
•
CAPRIN1 is required for the cytoplasmic translocation of XRN2 from the nucleus
Viegas et al. explore the role of cell-cycle-associated protein 1 (CAPRIN1) in early murine-differentiated ESCs, and they show that CAPRIN1 promotes the degradation of thousands of RNA transcripts and that it interacts with exoribonuclease XRN2. The authors define a mechanism whereby CAPRIN1 (via XRN2) causes RNA degradation to eliminate undesired RNA transcripts during differentiation.
Introduction
Embryonic stem cells (ESCs), derived from the inner cell mass of the blastocyst, have the ability to self-renew and to differentiate into all cells of the organism. ESCs are regulated by master core transcription factors, including OCT4, SOX2, and NANOG.1 These factors have a broad spectrum of target genes in ESCs, and their effect is directed toward maintaining self-renewal and pluripotency. The mechanisms that control pluripotency and early ESC differentiation involve several levels of regulation including chomatin structure, transcription, and post-transcriptional regulation including RNA stability, translation, and post-translational modifications.2 In recent years, an increasing number of factors controlling pluripotency and differentiation of ESCs have been identified, most of which are nuclear (e.g., transcription factors) or chromatin related.3 However, it is becoming increasingly clear, based on a growing number of post-transcriptional regulators of pluripotency,4 that RNA metabolism and RNA binding proteins (RBPs) play key roles in ESCs and early differentiation. RBPs are actively involved in all stages of RNA metabolism, e.g., RNA processing, including capping, splicing, and polyadenylation; RNA editing; transport; stability; storage; export; degradation; and translational regulation.5
Although many pluripotency-related RBPs await discovery, several important factors have been identified in recent years. Notable examples include LIN28, which controls ESC identify and function by repressing the let-7 microRNA;6,7 SON, FOXP2, and MBNL proteins, which facilitate splicing and alternative splicing events in human ESCs;8,9,10,11,12 METTL3/4, mediating m6A RNA modification and stability;13,14,15 FIP1 and Nudt21, regulating alternative polyadenylation;16,17 the THO complex, controlling RNA export;18 CNOT proteins, mediating deadenylation of pluripotency-related targets;19 DDX6, modulating P-body homeostasis;20,21 and DAZL and DAP5, which play key roles in ESC regulation of translation.22,23 One interesting recent study demonstrated a role for the exosome complex in targeting several coding and non-coding RNA (ncRNA) transcripts in ESCs, including LINE1 elements, miRNAs, and long ncRNAs,24 likely originating from the promiscuous nature of transcription in ESCs.25
To identify regulators of ESCs, we screened an endogenously labeled fluorescent fusion-protein library in mouse ESCs, which we previously generated.26,27 Focusing on cytoplasmic RBPs, which show differential localization and/or expression during early ESC differentiation, we identified cell-cycle-associated protein 1 (CAPRIN1). Also known as RNG105, CAPRIN1 is a cytosolic RBP that is highly expressed in the brain and in actively proliferating cells.28 CAPRIN1 is part of a highly conserved protein family, and it binds RNA through an RNA granule (RG)-rich region as well as several arginine-glycine-rich (RGG) box motifs at the C terminus of the protein.29 CAPRIN1 is localized in RNA granules (RGs) / stress granules (SGs),30,31 and this localization is regulated by CAPRIN1’s phase separation properties though tyrosine phosphorylation, thereby tuning deadenylation and translation rates.32
By combining genetic, proteomic, and imaging approaches, we uncover an RNA degradation pathway, operating in early ESC differentiation, mediated by CAPRIN1 and its interaction within cytoplasmic non-SG foci with the otherwise nuclear ribonuclease XRN2.
Results
CAPRIN1 is downregulated during ESC differentiation and is localized in RGs
We previously generated an endogenously labeled fluorescent fusion-protein library in mouse ESCs.27 To identify proteins which are downregulated during ESC differentiation, we previously screened the library for fluorescence changes upon retinoic acid (RA) treatment and previously identified the cytoplasmic protein CAPRIN1, which showed reduced fluorescence upon early ESC differentiation27 (Figures 1A and 1B). We verified proper CAPRIN1-YFP localization using immunofluorescence (IF) with CAPRIN1 antibodies in WT R1 ESCs (Figure 1C) and expression of both endogenous CAPRIN1 and CAPRIN1-YFP by western blots in the CAPRIN1-YFP ESCs (Figure S1A). We validated its downregulation following RA treatment using both western blots (Figure 1D, shown for RA days 3 and 5) and RT-qPCR (Figure 1E, shown for RA day 3), as well as during embryoid bodies (EBs) formation (Figure 1F). Analyzing available ChIP-seq profiles for pluripotency factors,33 we identified binding sites for NANOG, OCT4, and SOX2 within the first intron of the Caprin1 gene (Figure S1B), supporting CAPRIN1’s high expression in ESCs. Interestingly, monitoring CAPRIN1-YFP localization dynamics during ESC self-renewal using live imaging, we observed an abrupt transition from a relative diffuse localization with a few small cytoplasmic granules during interphase (Figure 1G, left) to an almost complete aggregation in large cytoplasmic granules immediately after mitosis34 (Figure 1G, right; Video S1). CAPRIN1 was previously shown to localize to SGs in somatic cells.31,35 We therefore wished to test whether the same holds true for ESCs. To this end, we stressed the cells using sodium arsenite (NaAsO2, ARS, 200 μM) known to cause oxidative DNA damage.36 Time-lapse microscopy of our CAPRIN1-YFP ESCs revealed the formation of SGs within 30 m after the admistration of ARS (Figure 1H; Video S2) or the proteasome inhibitor MG132 (5 μM; Video S3). To validate that the ARS-induced CAPRIN1-YFP aggregates are indeed SGs, we subjected our CAPRIN1-YFP ESCs to stress and performed IF with known SG members, including DDX3X, G3BP2, and TIA1. In untreated ESCs, CAPRIN1 was diffusely localized throughout the cytoplasm (Figure S1C, top) but following exposure to ARS, CAPRIN1 was co-localized with all SG markers (Figures S1C bottom and S1D). This demonstrated that CAPRIN1 is a SG component also in ESCs. However, staining the non-stressed ESCs with SG markers (e.g., TIA1, Figure 1I) demonstrated that the CAPRIN1 foci which appear immediately after mitosis in non-stressed cells are not SGs. Taken together, these results demonstrate that in ESCs, CAPRIN1 is highly abundant and translocates into SGs after stress but that it also forms non-SG cytoplasmic foci in non-stressed conditions immediately after mitosis.
Figure 1.
CAPRIN1 changes expression and localization during early ESC differentiation
(A and B) Endogenously tagged CAPRIN1-YFP in undifferentiated mouse ESCs (A) and after 4 days of retinoic acid-induced differentiation (B). Scale bars, 10 μm.
(C) CAPRIN1-YFP (green) colocalizes with CAPRIN1 antibodies (red). Right panel: merge. Scale bars, 10 μm.
(D) Western blot for OCT4 (top), CAPRIN1 (middle), and GAPDH (bottom) in undifferentiated ESCs (left) and after 3-day (middle) or 5-day (right) retinoic acid treatment. The results from one of the three independent experiments.
(E) Relative expression (using real-time RT-PCR) of Caprin1 mRNA in undifferentiated (left) and RA-induced (right) ESCs, n = 3.
(F) Same as (E) in undifferentiated (left) and 3-day-old (middle) and 5-day-old embryoid bodies (right), n = 3.
(G) CAPRIN1-YFP localization in interphase (left), mitosis (middle), and immediately after mitosis (right). Scale bars, 5 μm.
(H) CAPRIN1-YFP ESCs (green, left) stained with anti-CAPRIN1 antibodies (middle, red) following 1 h treatment with sodium arsenite (200 μM, 2 h). Scale bars, 5 μm.
(I) Immunofluorescence of non-stressed CAPRIN1-YFP ESCs (left, green) using TIA1 antibodies (middle, red). Right: merge. Arrows point to the CAPRIN1 cytoplasmic aggregates, which show no TIA1 staining. Scale bars, 5 μm.
CAPRIN1 is dispensable in ESCs but important for early ESC differentiation
Next, to test whether CAPRIN1 plays a role in either ESC maintenance or differentiation, we used CRISPR-Cas9 to generate Caprin1-knockout (KO) ESCs. The depletion of CAPRIN1 was confirmed by western blots in undifferentiated ESCs (Figure 2A, top) and RA-induced cells (Figure 2A, bottom), by RT-qPCR (Figure 2B) and by DNA sequencing (not shown). Interestingly, of the two KO clones, both of which do not produce CAPRIN1 protein (Figure 2A), in RA-induced cells, only KO2 showed reduced RNA levels, whereas KO4 expresses Caprin1 RNA at similar levels to WT (Figure S2A, RNA sequencing [RNA-seq]; Figure S2B, RT-qPCR). The Caprin1-KO cells showed very mild effects in undifferentiated ESCs and remained as round alkaline phosphatase (AP)-positive colonies (Figure S2C), expressing normal levels of pluripotency factors (Figure 2C), although they displayed slower growth rates (Figure 2D). In contrast, when induced to differentiate into EBs, the Caprin1-KO clones displayed aberrant expression of differentiation markers including pluripotency, mesodermal, ectodermal, and endodermal genes (Figure 2E; Figures S2D–S2G). Elevated levels of NESTIN (Figure S2H) and reduced GATA4 (Figure S2I) in the Caprin1-KO cells was confirmed by IF in 4-day RA-induced cells. We also subjected the cells to directed differentiation into either beating cardiomyocytes, representing a mesodermal lineage, or neuronal progenitor cells (NPCs), representing ectodermal differentiation. In both cases, the Caprin1-KO ESCs failed to differentiate properly and showed significantly reduced numbers of beating foci (Figure 2F) and NPCs (Figure S2J). To ensure that these phenotypes were not, at least partly, due to potential off-target effects, we repeated the differentiation experiments following expression of WT Caprin1 cDNA in the KO cells. In all cases, the introduction of a WT copy of Caprin1 rescued the aberrant phenotypes (Figure 2F; Figure S2J).
Figure 2.
Aberrant differentiation of Caprin1-KO ESCs
(A) Western blot for CAPRIN1 in WT and two Caprin1-KO clones (KO2; KO4) ESCs (top) and RA-induced ESCs for 4 days (bottom). The results from one of the three independent experiments.
(B) Relative expression (RT-PCR) of Caprin1 mRNA in the WT (blue) and KO4 (gray) ESCs, n = 3.
(C) Relative expression (RT-PCR) of Nanog, Oct4, Sox2, and Klf4 in WT (blue) and KO4 (gray) ESCs. (None of the changes are statistically significant, U test), n = 3.
(D) Proliferation rates in WT (blue) and KO4 (red) ESCs. Shown are averages of four independent experiments. Error bars indicate standard deviations. Changes are significant (p < 0.005, U test).
(E) Relative expression (RT-PCR) of Oct4, Nodal, Nestin, and Sox17 in WT (blue) and KO4 (gray) ESCs after 4 days of RA-induced differentiation, n = 3.
(F) Number of beating cardiomyocytes differentiated from WT (left), KO4 (middle), and KO-Caprin1-add-back clones (right).
(G and H) Volcano plot of RNA-seq comparing WT and Caprin1-KO cells in undifferentiated ESCs (G) and in 4-day RA-induced cells (H).
(I) Volcano plots of RNA-seq results of three independent experiments comparing either WT versus Caprin1-KO ESCs (left) or WT versus Xrn2-KD ESCs (right).
(J) Venn diagram of the differentially expressed genes between WT versus Caprin1-KO ESCs and WT versus Xrn2-KD ESCs.
(K) GO analysis of the downregulated genes shared between the teratomas formed by the Caprin1-KO ESCs and the Xrn2-KD ESCs. Brain-related genes are dramatically enriched.
Next, we analyzed RNA-seq data from undifferentiated and RA-treated (4 days) WT and KO ESCs. Although undifferentiated ESCs had very few differentially expressed genes (DEGs) (Figure 2G), in agreement with their mild phenotypes, the RA-treated cells showed a much larger number of DEGs (Figure 2H). Gene ontology (GO) analyses suggested that the most prominent categories altered in the RA-induced Caprin1-KO cells are related to proliferation, developmental processes, and RNA metabolism (Table S1A). The very few DEGs in the undifferentiated Caprin1-KO ESC did not yield any GO enriched categories. To complement these experiments, we also performed teratoma assays by injecting WT or Caprin1-KO ESCs in severe combined immunodeficient (SCID) mice and analyzed RNA-seq of the formed teratomas 24 days later (Figure S2K). Overall, we observed a profound failure to activate neuronal genes (Figures 2I–2K), in line with CAPRIN1’s prominent expression in neurons37 and its role in long-term memory formation.38
Taken together, our results thus far suggest that CAPRIN1 is important for ESC differentiation and seems to be involved in RNA-related processes in early ESC differentiation and for neuronal commitment at later stages of development.
CAPRIN1 interacting transcripts detect a functional shift during ESC differentiation
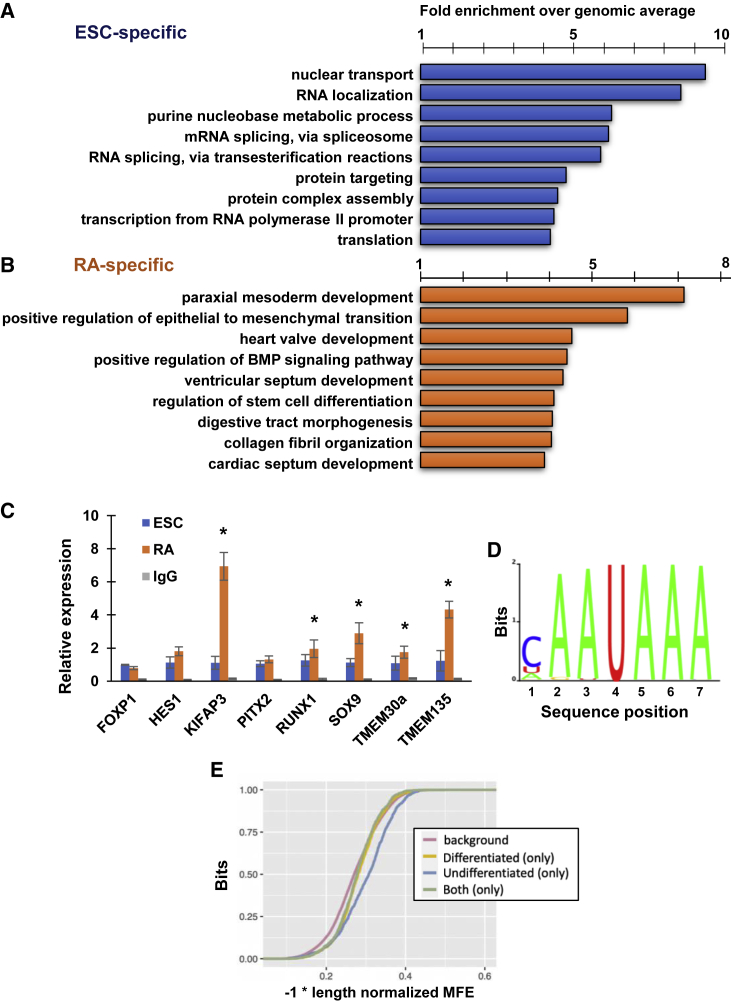
We next sought to identify the subset of RNAs associated with CAPRIN1 in both undifferentiated and RA-induced ESCs. We affinity-purified lysates from ESCs and RA-differentiated cells using CAPRIN1-specific antibodies, extracted total RNA, and subjected it to RNA-seq analysis (RNA immunoprecipitation sequencing - RIP-seq) (Figure S3A). Interestingly, we found that CAPRIN1 interacts with a significantly larger number of RNA transcripts in RA-induced cells, despite its lower levels of expression in these cells (Figure S3B). We identified 1,178 transcripts that associated, directly or indirectly, with CAPRIN1 in undifferentiated ESCs, and 2,116 transcripts in the RA-differentiated cells (Figure S3B), compared with IgG controls and Caprin1-KO cells (in two independent experiments) (Table S1B). GO analysis (geneontology.org) of CAPRIN1-bound RNAs that are ESC specific were related to nuclear transport, RNA localization, RNA splicing, protein targeting, transcription, and translation (Figure 3A). Interestingly, GO analysis of CAPRIN1-bound RNAs specific to RA-differentiated cells showed no enrichment in pathways related to RNA processing and, instead, highlighted pathways related to development, epithelial-mesenchymal transition, signaling, differentiation, and morphogenesis (Figure 3B), suggesting a shift from RNA and translation-related transcripts to transcripts related to differentiation and development. Comparing the list of interacting RNAs with the list of upregulated or downregulated RNAs in the Caprin1-KO cells, we observed that several developmental transcripts (e.g., Sox9), which directly interact with CAPRIN1, are elevated in the Caprin1-KO cells. We confirmed selected transcripts (Kifap3, Pitx2, Runx1, Sox9, Tmem30a, and Tmem135) by RIP-RT-PCR (Figure 3C). FoxP1 and Hes1, both of which showed no differential enrichment, served as controls. Reassuringly, five of the six transcripts showed higher levels in the RA-treated cells, validating our RIP-seq results.
Figure 3.
CAPRIN1 interacts with developmental transcripts in early differentiating ESCs
(A) Gene ontology (GO) enrichment analysis of CAPRIN1-interacting RNA transcripts specific for undifferentiated ESCs, identified by RIP-seq. Categories are sorted according to fold enrichment over genomic average. All categories are significant (FDR < 0.005).
(B) Same as (A) for RA-induced ESCs.
(C) Validation of RIP-seq by RIP-RT-PCR for selected transcripts in undifferentiated (blue) and RA-induced (orange) ESCs. IgG (gray) was used as control. (∗p < 0.05, U test), n = 3.
(D) Motif enrichment analysis for CAPRIN1. All RIP-seq data were analyzed for enriched motifs. The motif was identified specifically in undifferentiated ESCs, suggesting a mechanistic switch for CAPRIN1 interaction with RNAs during differentiation.
(E) Secondary structure enrichment analyses of CAPRIN1-bound RNAs in undifferentiated ESCs (blue); RA-induced ESCs (yellow), both (green) and a background dataset (red). Only undifferentiated ESCs are highly enriched for CAPRIN1-interacting RNAs with secondary structure (p < 2 × 10−16).
Finally, we searched for potential motifs or secondary structures through which CAPRIN1 might recognize and bind the interacting RNAs. Analyzing the RIP-seq results, we found that 3′ UTR sequences of genes that were pulled-down only in the undifferentiated ESCs were enriched for a characteristic sequence motif cAAUAAA (Figure 3D, p < 10−10, relative to 3′ UTRs of other genes expressed at the 0 h RNA-seq samples), resembling the canonical polyadenylation signal. Reassuringly, a similar signal was also previously identified in PAR-CLIP analysis of CAPRIN1 binding.39 No enrichments were found in 5′ UTRs or coding regions of transcripts in this set. Moreover, sequences of genes that were pulled-down only in differentiated ESCs or in both differentiated and undifferentiated ESCs were also not enriched for any specific sequence motif, suggesting that CAPRIN1-mediated mRNA degradation upon ESC differentiation might work via a mechanism that is distinct than the standard CAPRIN1 binding. We also tested the predicted structural preferences in 3′ UTR sequences of genes that were associated with CAPRIN1 binding. Indeed, 3′ UTRs of genes that were pulled-down only in undifferentiated ESCs were predicted to be more structured on average (lower length-normalized Minimal Free Energy prediction by RNA-fold), with 53% highly structured transcripts (length-normalized MFE < −0.3) in this group compared with only 34% in the background group (Figure 3E, p < 10−16). This analysis could suggest that CAPRIN1-mediated mRNA degradation in undifferentiated ESCs might also involve binding by, e.g., G3BP1 and UPF1 to highly structured mRNAs, which promotes their degradation.40
The observation that CAPRIN1 interacts with selected transcripts, which are upregulated in the Caprin1-KO RA-treated cells, raised the possibility that CAPRIN1 might be involved in degrading these transcripts during early ESC differentiation. To test this idea, we focused on Sox9 RNA, which is differentially expressed, and on FoxP1 RNA, as control. We treated our cells with the RNAPII inhibitor α-amanitin to inhibit transcription and collected RNA every 2 h for a total of 8 h and followed the RNA levels of Sox9 and FoxP1 in both WT and Caprin1-KO cells. Although FoxP1 RNA showed similar degradation rates (i.e., stability) in WT and KO cells (Figures S3C and S3E), Sox9 RNA was significantly more stable in the Caprin1-KO cells (Figures S3D and S3E). These data support the notion that CAPRIN1 might be involved in RNA degradation of developmental transcripts.
To test this idea genome-wide, we first performed thiol-linked alkylation for metabolic sequencing (SLAM-seq), a metabolic RNA-seq method that detects 4-thiouridine (4-SU) incorporation at single-nucleotide resolution.41 We treated both WT and Caprin1-KO ESCs, in either ESCs or RA differentiation conditions, with 4-SU for 24 h and chased with unlabeled uridine for the indicated time points (Figure S4A). Total RNA was then subjected to alkylation and RNA-seq. The RNA half-life was determined by calculating the normalized T > C conversion rate between time points. Cumulative fraction analysis of our SLAM-seq data showed a highly significant increase in RNA half-life in the Caprin1-KO cells, in both undifferentiated ESCs (Figure 4A) but most profoundly in RA-treated cells (Figure 4B) (in both cases p << 10−50, Kolmogorov-Smirnov test), demonstrating that RNA stability is globally elevated when CAPRIN1 is depleted and suggesting that CAPRIN1 promotes RNA degradation. Specifically, using a 1.5-fold cutoff, 2,468 and 2,729 transcripts displayed increased stability in undifferentiated and RA-induced Caprin1-KO ESCs, respectively, compared with merely 282 and 266 transcripts in ESCs and RA-treated cells, respectively (Figures 4C and 4D; Table S2A). Interestingly, GO analysis showed no significant enrichment in any particular biological categories, suggesting a widespread mechanism degrading spurious transcription associated with early differentiation.
Figure 4.
CAPRIN1 is involved in global RNA stability
(A and B) Cumulative fraction analysis of SLAM-seq in WT (cyan) and Caprin1-KO (red) undifferentiated ESCs (A) and in RA-induced ESCs (B). p << 10−10, n = 3.
(C and D) RNA half-life in WT (y axis) versus Caprin1-KO (x axis) in undifferentiated ESCs (C) and RA-induced ESCs (D).
(E–G) RNA decay rates (1/h; log2) for 3,132 fitted genes in cells treated with α-amanitin for 8 h. (E) wild-type (x axis) versus Caprin1-KO2 (y axis), (F) wild-type (x axis) versus Caprin1-KO4 (y axis), and (G) Caprin1-KO2 (x axis) versus Caprin1-KO4 (y axis). Color scale indicates the density of data (yellow = high, blue = low).
(H–J) RNA expression levels (TPM; log2) before α-amanitin treatment (0 h) for 3,132 fitted genes in cells treated with α-amanitin for 8 h. (H) Wild-type (x axis) versus Caprin1-KO2 (y axis), (I) wild-type (x axis) versus Caprin1-KO4 (y axis), and (J) Caprin1-KO2 (x axis) versus Caprin1-KO4 (y axis). Color scale indicates the density of data (yellow, high; blue, low).
To complement these experiments, we next performed degradation assays using α-amanitin in 4-day RA-induced ESCs. We concentrated on early differentiating cells (RA-treated) because our Caprin1-KO cells, as well as our RIP-seq and SLAM-seq experiments, all suggested a more prominent role for CAPRIN1 in early ESC differentiation. To this end, we induced WT and Caprin1-KO ESCs with RA for 4 days, added α-amanitin in the final 8 h of the experiment, and collected cells before α-amanitin addition, as well as 2, 4, and 8 h thereafter. All samples were then subjected to RNA-seq. In order to properly normalize our results, we added an additional normalization step and normalized standard TPM values in these experiment by a set 356 control transcripts (Figures S4B–S4G; Table S2B) that showed little to no degradation in WT cells. We then fitted an exponential decay model to the normalized data and extracted decay rates for each transcript. Comparison of decay rates in WT and two Caprin1-KO clones shows a global increase in RNA stability in the Caprin1-KO cells (Figures 4E–4G; Table S2C), confirming the involvement of CAPRIN1 in degrading RNA transcripts during early ESC differentiation. RNA-seq at time 0 confirmed no experimental bias (Figures 4H–4J).
We then compared the list of the CAPRIN1 RNA targets identified though our RIP-seq experiments (Figure S3A) with the differentially stabilized transcripts between WT and Caprin1-KO cells, identified through our α-amanitin degradation experiments. Reassuringly, when comparing CAPRIN1 binding targets with non-targets, we identified a stronger fold-reduction in degradation rates of CAPRIN1 binding targets in both undifferentiated ESCs (p < 10−6) and in RA-induced cells, where this trend was significantly stronger (p < 10−11) (Figure S4K). Taken together, our results identify a global role for CAPRIN1 in mediating RNA degradation during early ESC differentiation. Although the SLAM-seq experiments are using ribo-depleted RNA and are designed to pick nascent transcripts, in the α-amanitin experiments, we analyzed poly(A)+ RNA, and hence, we can conclude that most of the degraded transcript are likely mature processed RNAs.
CAPRIN1 acts via XRN2
So far, our results uncover an RNA degradation pathway during early ESC differentiation, mediated by CAPRIN1. Since CAPRIN1 is not, in itself, a ribonuclease nor has any known ribonuclease activity, we next sought CAPRIN1’s interaction partners in search of the mechanism by which CAPRIN1 mediates RNA degradation. Using CAPRIN1-specific antibodies, we pulled-down CAPRIN1 in ESCs and in RA-treated cells before and after stress (ARS, 200 μM, 2 h) and used LC-MS/MS analysis to identify the CAPRIN1-interacting proteins (Figures S5A–S5C). Using this approach, we identified 219 and 268 proteins that were reliably and reproducibly enriched in CAPRIN1-IP samples compared with controls in ESCs and in RA-induced cells, respectively (Table S3). A consensus interaction map of CAPRIN1’s binding partners in SGs identified several groups, including proteins involved in RNA methylation (m6A), DNA/RNA helicases, RBPs, and a group of nuclear proteins (Figure 5A). Our identified proteins significantly overlapped with CAPRIN1-interactome in HeLa cells, reported previously by Lu et al.42 GO analysis of CAPRIN1-interacting partners in RA-induced cells show that the most enriched biological processes were related to translation, RNA splicing, and RNA processing/metabolism, all of which, as expected, are related to RNA maintenance (Figure 5B). Similar GO terms emerged from CAPRIN1-interactomes in undifferentiated ESCs (Figure S5D). However, of the 392 interacting proteins (reproducible and specific), 130 (40%) were selectively identified in ESCs and 173 (34%) were selective for RA-induced cells (Figure 5C). Interestingly, although proteins related to the m6A machinery, including YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2 were ESC specific, 11 hnRNP proteins were selectively identified in RA-induced cells (compared with zero in ESCs) and 42 ribosomal proteins were selectively found in RA-induced cells (compared with three in ESCs and three which were common to both). These unique interactions could reflect the more prominent role observed for CAPRIN1 in early differentiating cells. Importantly, seeking potential ribonucleases in our CAPRIN1-interaction MS data, we identified a nuclear 5′-3′ exoribonuclease, XRN2, which has been shown to participate in transcription termination.43 Although XRN2 is considered to be a nuclear protein, it was discovered in a recent study of the proteome and compositional diversity of SGs, in both stressed HeLa cells as well as in untreated iPSC-derived motor neurons and in ALS samples.44 XRN2 was further identified in SGs in a more recent comprehensive study of SG proteome in U2OS cells,45 validating the presence of XRN2 in SG. Supporting these observations, in our MS data, CAPRIN1-XRN2 interaction was significantly enriched in stressed cells, suggesting that this interaction might be SG specific. To experimentally validate this interaction, we performed coIP experiments with CAPRIN1 and XRN2 using cellular fractionations (nuclear and cytoplasmic) in non-stressed and stressed conditions. In agreement with our MS data, we observed a weak interaction between XRN2 and CAPRIN1, which intensified upon stress (Figure 5D). Taken together, these data suggest that CAPRIN1 interacts with the exoribonuclease XRN2.
Figure 5.
CAPRIN1 interacts with XRN2
(A) LC-MS/MS of CAPRIN1-interactome.
(B) GO analysis of CAPRIN1-interactions in RA-induced ESCs.
(C) Venn diagram of CAPRIN1-interacting partners in ESCs (blue) and RA-induced ESCs (pink).
(D) CoIP of CAPRIN1 and XRN2. Shown are cytoplasmic (Cyto) and nuclear (Nuc) fractions of non-stressed (-ARS) and arsenite-treated (ARS, 200 μM, 2 h) ESCs pulled-down using CAPRIN1 antibodies (top) and blotted using XRN2 antibodies (second row from top). α-Tubulin (α-tub, third row) and histone H3 (H3, fourth row) were used to validate enrichment of cytoplasmic and nuclear fractions, respectively. Heavy- and light-chain IgGs are shown below. The results from one of the three independent experiments.
To confirm this interaction, we performed IF staining for XRN2 and CAPRIN1 in stressed and non-stressed conditions. As expected, in non-stressed cells, XRN2 was predominantly localized to the nucleus (Figure 6A, top). However, in stressed cells (ARS, 200 μM), a fraction of XRN2 is relocalized to CAPRIN1-positive SGs (Figure 6A, bottom). Although XRN2 is predominantly nuclear in ESCs, in RA-induced cells (non-stressed), we often observed a small fraction of XRN2 colocalizing with CAPRIN1 SGs (Figure 6B, arrows). These results validate the co-localization of XRN2 and CAPRIN1 within cytoplasmic foci in both stressed (SGs) and early differentiating cells (non-SGs), albeit to a lesser extent.
Figure 6.
CAPRIN1 is required for XRN2 relocalization into SGs
(A) Immunofluorescence (IF) for XRN2 (red) in non-stressed ESCs (top) and in sodium arsenite-treated ESCs (bottom). Blue: DAPI; green: CAPRIN1-YFP; right: merge. Scale bars, 10 μm.
(B) Same as (A) in non-stressed RA-induced ESCs. XRN2 can be found in small cytoplasmic foci (top panels) or dispersed in the cytoplasm (bottom panels).
(C) CAPRIN1 is required for XRN2 localization in SGs. Shown are IF images for XRN2 (red) in stressed (ARS-treated) WT (top) and stressed Caprin1-KO ESCs (bottom).
(D) CAPRIN1 is not entirely essential for the formation of SGs. Shown are IF images of XRN2 (red, top) and G3BP1 (green, bottom) in stressed (ARS-treated) Caprin1-KO ESCs. SGs are formed, although to a lesser extent, but XRN2 remains nuclear.
(E) Quantification of SG formation in Caprin1-KO ESCs. G3BP1-positive SGs were quantified in WT (blue) and Caprin1-KO (gray) ESCs following arsenite treatment. ∗p < 0.015, t test, n = 90 (WT) and n = 94 (Caprin1-KO).
Next, to establish whether XRN2 is indeed involved in CAPRIN1’s mediated degradation of developmental transcripts during early ESC differentiation, we first attempted to KO XRN2 in ESCs. Despite repeated attempts, we were unable to obtain homozygous KO clones, suggesting that XRN2 is essential for cell viability. Supporting this, whole-genome CRISPR-Cas9 screens in both human46 and mouse47 ESCs identified XRN2 as an essential gene. We therefore used lentiviral shRNAs to generate several stable knockdown (KD) clones of Xrn2 in ESCs (Figures S6A–S6C). We used non-targeting scrambled shRNAs to generate control cell lines. We selected two independent KD (shX3 and shX5, Figure S6A), with varying levels of XRN2 (Figures S6B and S6C). Xrn2-KD clones phenocopied the slower proliferation (Figure S6D) and the aberrant differentiation (Figures S6E and S6F) of the Caprin1-KO cells. Knockdown of Xrn2 did not prevent the formation of CAPRIN1 SGs following stress (Figure S6G, green) nor XRN2’s nuclear export (Figure S6G, red). We also tested whether XRN2 foci can form following depletion of another SG marker, namely DDX3X. We generated CRISPR-mediated Ddx3x-KO ESC clones (Figure S6H) and stained for XRN2 in non-stressed and stressed (ARS-treated) conditions. Although XRN2 was less conspicuous in these SGs, DDX3X depletion did not completely prevent XRN2’s SG relocalization following ARS treatment (Figure S6I) or in early differentiation (Figure S6J). To compare Caprin1-KO ESCs and Xrn2-KD ESCs more systematically and in in vivo setting, we injected these cells under the skin of SCID mice and performed RNA-seq on the formed teratomas 24 days later (Figures 2I and S2K). Interestingly, comparing the DEGs shared between the Caprin1-KO versus the Xrn2-KD ESCs (Figure 2J), we observed a striking enrichment in brain-related genes (Figure 2K). These results demonstrate that the precautious expression of neuronal markers in the Caprin1-KO ESCs (Figures 2E and S2F; Figure S2H) results in aberrant neuronal differentiation in later stages of differentiation, in line with CAPRIN1’s prominent expression in neurons37 and suggested role in long-term memory.38
Finally, we wondered how CAPRIN1 mediates its XRN2-dependent degradation regulation. Since the expression level of XRN2 is not dramatically different in Caprin1-KO cells, we tested whether CAPRIN1 is required for XRN2’s localization into SGs. To address this, we stressed WT and Caprin1-KO ESCs and stained for XRN2. Remarkably, although, as expected, in WT Caprin1+/+ ESCs, XRN2 was conspicuously present in SGs (Figure 6C, top), in the absence of CAPRIN1, XRN2 did not relocalize into SGs, and remained exclusively nuclear (Figure 6C, bottom). To test whether the failure of XRN2 to relocalize into SGs in stressed Caprin1-KO is due to suppressed formation of SGs in the absence of CAPRIN1, we repeated the experiment and co-stained the cells with the SG marker G3BP1. Following stress, SGs still formed in the Caprin1-KO cells, although to a lesser extent (Figures 6D and 6E), suggesting that CAPRIN1 is not essential, although it may play a role, in SG formation. Once again, however, the formed SGs were completely devoid of XRN2 (Figure 6D). Taken together, these results demonstrate that CAPRIN1 is directly involved in XRN2’s relocalization into SGs but is not completely essential for their formation.
Our combined results highlight CAPRIN1-XRN2 interaction as a novel differentiation-associated RNA degradation (DARD) process, by which undesired RNA transcripts are elimated during early ESC differentiation.
Discussion
Stem cell differentiation is controlled by many layers of regulation from transcription and post-transcription, RNA processing and export to translation, post-translational modifications, signaling, and more.48 In this study, we uncovered an RNA degradation pathway operating during the early stages of ESC differentiation, mediated by CAPRIN1 and XRN2. Although CAPRIN1 is largely dispensable in undifferentiated ESCs, it is required for ESC differentiation. Supporting a relatively minor role for CAPRIN1 in ESCs is the conspicuous absence of proteins related to cell cycle and proliferation from the MS interactome results. Although this may well be related to detection limits of our experiments, our data suggest that in ESCs, CAPRIN1 does not play a major role, if at all, in cell cycle progression. This idea is further supported by the very minor changes observed in gene expression in undifferentiated Caprin1-KO ESCs. By contrast, Caprin1-KO ESCs, as well as Xrn2-KD ESCs, show skewed spontaneous differentiation into EBs and teratomas, aberrant directed differentiation and showed major changes in gene expression in early differentiation. This suggests that the high expression of CAPRIN1 in ESCs is important not so much for the undifferentiated state, but for the initial stages of differentiation, a situation remiscent of a poised state. Indeed, in ESCs, CAPRIN1 binds RNA transcripts associated with RNA processing, suggesting a generic role, but switches to binding developmental transcripts during early differentiation. The fact that these thousands of transcripts do not report any enrichment in any specific GO term suggests that they consist of multiple different transcripts from different pathways, namely all those that are not required for the particular differentiation trajectory. Since serum-grown ESCs promiscuously express many genes and ncRNAs at low levels across the genome,25,49 this degradation pathway may be important to rid of excess RNA transcripts that are promiscuously transcribed. Interestingly, our motif search analysis identified a highly abundant motif recognized by CAPRIN1 in undifferentiated, but not in differentiated, ESCs. This demonstrates a shift in the mode of CAPRIN1 target recognition during early differentiation. In addition, it also appears that secondary structures play an important role in CAPRIN1 association with RNA at least in undifferentiated ESCs, suggesting an additional layer of regulation, acting possibly by masking this sequence recognition motif. Another plausible option is regulation by m6A, as our MS data identified most of the machinery required for this RNA modification.
Although many studies have explored different mechanisms of post-transcriptional regulation during development, mRNA decay remains a complex mechanism and is not well understood. (De)stabilization of transcripts represents an important post-transcriptional mechanism during early differentiation to ensure an appropriate transition to specific lineage formation.50 In our study, we employed several genome-wide approaches combined with time-course analysis to measure the dynamics of mRNA degradation rates during the early ESC differentiation in WT and Caprin1-KO cells. Our SLAM-seq and α-amanitin RNA-decay assays, both identified a global role for CAPRIN1 in mediating RNA degradation. Although both assays are designed to provide information on transcript degradation rates, SLAM-seq highlights recently transcribed transcripts, whereas α-amanitin enriches for the more mature transcripts, thus exposing different sets of transcripts. The fact that both sets showed the same global trend strengthens a global role for CAPRIN1 in mediating RNA degradation during early differentiation.
RNA degradation can take place in different compartments, both nuclear and cytosolic, at different stages of the life of the RNA transcript and can be mediated by different enzymes including endonucleases, 5′→3′ exonucleases, and 3′→5′ exonucleases.51 The CAPRIN1-mediated RNA degradation pathways uncovered here is likely executed by the 5′→3′ exonuclease XRN2, which was the only ribonuclease identified in our LC-MS/MS data of CAPRIN1-interacting proteins. It is likely that other ribonucleases interact with CAPRIN1/XRN2 but our MS conditions did not allow their detection. Since, in addition to its relocalization into SGs after stress, CAPRIN1 also resides in cytoplasmic granules in non-stressed cells, it is likely that the pathway reported here is not related to SGs. In fact, our genome-wide assays (α-amanitin; SLAM-seq) did not explore stress conditions and merely compared Caprin1-KO with WT cells and hence strongly support a general role not related, especially not exclusively, to SGs. Although considered a nuclear protein, XRN2 was already identified in non-biased screens of SG-residing proteins, placing it, at least in some conditions outside the nucleus.44,45 Interestingly, CAPRIN1 is required for XRN2 retention in SGs. In the absence of CAPRIN1, XRN2 remains nuclear even after stress, and thousands of RNA transcripts become stabilized. Importantly, although predominantly nuclear, XRN2 translocates to cytoplasmic granules also in non-stressed cells, mostly, albeit not exclusively, in early differentiating cells, where it resides together with CAPRIN1. This once again strongly supports a general, non-SG-related, role for CAPRIN1 in RNA metabolism during differentiation. How is binding specificity achieved? Our sequence motif analysis identified a general sequence motif highly resembling the polyadenylation signal in ESCs, but not in RA-induced cells, suggesting other modes of regulation and potentially explaining the relatively high number of transcripts affected by Caprin1 depletion.
Apart from XRN2, CAPRIN1’s interactome, as expected, consisted primarily of SG proteins, such as G3BP1/2, eukaryotic initiation factors, DNA-RNA helicases etc., in both ESCs and RA-induced cells. Several of CAPRIN1’s partners were already shown to be involved in pluripotency, differentiation and/or development. Examples include G3BP1, which was shown to be involved in neuronal development,52 eIF4E, a SG-residing regulator of translation initiation, which was shown to dramatically influence the efficiency of reprogramming somatic cells to iPSCs,53 and the ubiquitin ligase Trim25, one of 68 highly expressed RBPs in ESCs.54 Some of these interacting proteins may well play a role in RNA degradation, although other than XRN2 itself, none of the CAPRIN1 interacting partners has any known ribonuclease activity, leaving XRN2 as the primary candidate for exerting RNA degradation.
Hence, although the functional executor, XRN2, may have been identified, many questions remain. How is specificity achieved? What are the triggering signals? Is CAPRIN1-mediated RNA degradation specific to particular cell cycle stages, as its localization suggests? These and many other questions remain to be answered in the coming years.
Limitations of the study
In this study, we find that CAPRIN1 globally affects RNA stabilization in early-differentiated ESC. We show CAPRIN1-XRN2 interaction as a mechanism by which RNAs are degraded. However, we cannot rule out the possibility that CAPRIN1 might recruit other factors which participate in RNA degradation. We also found that XRN2 exits the nucleus in a CAPRIN1-dependent fashion, but we do not know how CAPRIN1 regulates XRN2 localization. Finally, we note that our study was done in mouse ESCs, and although both CAPRIN1 and XRN2 are conserved throughout evolution, it would be interesting to test the relevance in human cells. Very recently though, during the final revision of the current manuscript, two papers implicating CAPRIN1 in human disease were published, showing that CAPRIN1 haploinsufficiency causes a neurodevelopmental disorder55 and that a specific CAPRIN1 mutation is associated with early-onset ataxia.56 These studies strongly suggest that our findings are highly relevant also for humans and human cells.
STAR★Methods
Key resources table
| Reagent or resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| CAPRIN1 Polyclonal antibody | Proteintech | Cat#15112-1-AP; RRID: AB_2070016 |
| Purified G3BP antibody | BD Biosciences | Cat#611126; RRID: AB_398437 |
| TIA-1 (C-20) antibody | Santa Cruz Biotechnology | Cat#sc-1751; RRID: AB_2201433 |
| GATA4 antibody | Santa Cruz Biotechnology | Cat#sc-1237; RRID: AB_2108747 |
| NESTIN (C-termal) antibody | Sigma-Aldrich | Cat#N5413; RRID: AB_1841032 |
| XRN2 Polyclonal antibody | Abcam | Cat#ab72181; RRID: AB_2241927 |
| OCT3/4 (C-10) antibody | Santa Cruz Biotechnology | Cat#sc-5279; RRID: AB_628051 |
| Alpha-Tubulin antibody loading control | Abcam | Cat#ab4074; RRID: AB_2288001 |
| Histone H3 antibody (mAb) | Active Motif | Cat#39763; RRID: AB_2650522 |
| GAPDH antibody | Abcam | Cat#ab8245; RRID: AB_2107448 |
| Alexa Fluor 488 rabbit, mouse and goat | Thermo Fisher Scientific | Cat#A-21206; RRID: AB_2535792, Cat#A-21202; RRID: AB_141607, Cat#A11055; RRID: AB_2534102 |
| Alexa Fluor 568 rabbit, mouse and goat | Thermo Fisher Scientific | Cat#A-10042; RRID: AB_2534017, Cat#A-21124; RRID: AB_2535766, Cat# A-11057; RRID: AB_2534104 |
| HP mouse and rabbit | Jackson ImmunoResearch Labs | Cat#115-036-062; RRID: AB_2307346 Cat#111-035-144; RRID: AB_2307391 |
| Bacterial and virus strains | ||
| pLKO.1-puro shRNA | Addgene | Cat#8453 |
| pSpCas9(BB)-2A-GFP (PX458) | Addgene | Cat#48138 |
| Chemicals, peptides, and recombinant proteins | ||
| CHIR99021 | Axon Medchem | Cat#2425 |
| PD0325901 | Axon Medchem | Cat#1408 |
| Deposited data | ||
| Raw and analyzed data | This paper | GSE179002 |
| Experimental models: Cell lines | ||
| mESC R1-Caprin1YFP | Harikumar et al.27 | N/A |
| mESC R1-CAPRIN1 Knockout | This paper | N/A |
| mESC R1-ShRNA XRN2 knockdown | This paper | N/A |
| mESC R1-DDX3X Knockout | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Male NOD.SCID IL-2 Rg mice | Envigo | https://www.envigo.com/ |
| Oligonucleotides | ||
| Xrn2-F TGAAGATGAAATGATGGTTGCA | This paper | N/A |
| Xrn2-R CCTCCTTGAACGCTGCTGAT | This paper | N/A |
| Caprin1-F CAGCAGAATTTCAAGCGAGGC | This paper | N/A |
| Caprin1-R TTCATTTGCGGCATCCCTCT | This paper | N/A |
| Gapdh-F GTGTTCCTACCCCCAATGTGT | This paper | N/A |
| Gapdh-R ATTGTCATACCAGGAAATGAGCTT | This paper | N/A |
| Cfl1-F TGGCAGAGAAACTAGGTGGC | This paper | N/A |
| Cfl1-R CAGCCTGCAACACCCAAG | This paper | N/A |
| Lrp2-F TGCGCTTGTGACCCAGAATAT | This paper | N/A |
| Lrp2-R TGATTTTGTCACGACTGGCTACA | This paper | N/A |
| Slo2a1-F CCGCTCGGTCTTCAACAACA | This paper | N/A |
| Slo2a1-R CGATAGTGGTGAGGCTGCTC | This paper | N/A |
| Metrn-F TTGAACTGCACGAGGACCAA | This paper | N/A |
| Metrn-R CAAAATCACTGGTGCATGCAG | This paper | N/A |
| Snai-F CGAACCCACACATTGCCTTG | This paper | N/A |
| Snai-R GTGAGGGCAAGAGAAAGGCTT | This paper | N/A |
| Fbxo2-F AACTGCCCGGAGACAATGG | This paper | N/A |
| Fbxo2-R TGTCCAGTAGCTCCTCCCAG | This paper | N/A |
| Nanog-F AGGGTCTGCTACTGAGATGCT | This paper | N/A |
| Nanog-R CAACACCTGGTTTTTCTGCCACCG | This paper | N/A |
| Oct4-F AGCCGACAACAATGAGAACC | This paper | N/A |
| Oct4-R TGATTGGCGATGTGAGTGAT | This paper | N/A |
| Sox2-F TCCCCCCTTTTATTTTCCGTAG | This paper | N/A |
| Sox2-R CCTGATTCCAATAACAGAGCCG | This paper | N/A |
| Klf4-F CTATGCAGGCTGTGGCAAAACC | This paper | N/A |
| Klf4-R TTGCGGTAGTGCCTGGTCAGTT | This paper | N/A |
| Eomes-F TGCAAGAGAAAGCGCCTGTCTC | This paper | N/A |
| Eomes-R CAATCCAGCACCTTGAACGACC | This paper | N/A |
| Nodal-F GGGGGAGGAGTTTCATCCTA | This paper | N/A |
| Nodal-R ATGCTCAGTGGCTTGGTCTT | This paper | N/A |
| Col1a1-F CACCCTCAAGAGCCTGAGTC | This paper | N/A |
| Col1a1-R AGACGGCTGAGTAGGGAACA | This paper | N/A |
| Fabp7-F AACTGTAAGTCTGTGGTTCGGT | This paper | N/A |
| Fabp7-R CACGACCATCTTGCCATCCT | This paper | N/A |
| Nestin-F TCAGATCGCTCAGATCCTGGA | This paper | N/A |
| Nestin-R GGTGTCTGCAAGCGAGAGTTCT | This paper | N/A |
| Pax6-F GTTGTGTGAGTAAAATTCTGGGC | This paper | N/A |
| Pax6-R GAGTCGCCACTCTTGGCTTA | This paper | N/A |
| Sox17-F ACAGGAAAACCTCAGCATGTCACCT | This paper | N/A |
| Sox17-R CGCTTCCCCCGACCCTTGG | This paper | N/A |
| Foxa2-F AGGCCGCCCCGGGACTTAAC | This paper | N/A |
| Foxa2-R CCCTCGGGCTCCGCGTAGTA | This paper | N/A |
| Lamc1-F CCGTGGTACATACAGCGAGA | This paper | N/A |
| Lamc1-R GCACACACGGGCTATAAGGT | This paper | N/A |
| Gata4-F AAAACGGAAGCCCAAGAACCT | This paper | N/A |
| Gata4-R TGCTAGTGGCATTGCTGGAGT | This paper | N/A |
| Software and algorithms | ||
| Fiji (ImageJ) | ImageJ | https://imagej.nih.gov/ij/ |
| R Studio version 4.1.2 | R | https://www.r-project.org/ |
| IGV | IGV | https://igv.org |
| Other | ||
| Clarity™ Western ECL Substrate | Bio-Rad | Cat#1705060 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| RNeasy Mi Kit | Qiagen | Cat#74106 |
| Bolt™ 4 to 12%, Bis-Tris, 1.0 mm, Mi Protein Gels | Thermo Fisher Scientific | Cat#Nw04122box |
| TransIt-LT1 Transfection reagent | Mirus | Cat#MIR2300 |
| Paraformaldehyde 16% solution | Electron Microscopy Sciences | Cat#15710 |
| PureLink HiPure Plasmid DNA purification | Thermo Fisher Scientific | Cat#K210004 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Eran Meshorer (eran.meshorer@mail.huji.ac.il).
Materials availability
The cell lines (R1CAPRIN1-KO, R1ShScramble, R1ShXRN2 and R1DDX3X-KO cell lines) are available from the lead contact upon request.
Experimental model and subject details
Animal ethics
For teratoma assay, male NOD.SCID IL-2 Rg immunodeficient mice (Strain BALB/c) were used. Mice were housed in standard conditions (12h light/dark cycle) with food and water available ad libitum. All experimental procedures were approved by the ethics committee of the Hebrew University of Jerusalem, Israel (ethics number of the study NS-21-16536-4).
ESCs culture
Mouse R1 ESCs were cultured using standard procedures in DMEM medium containing 10% FBS, 1000U of leukemia inhibitory factor [LIF], 2 mM L-glutame, 1% nonessential amo acids, 50 units/ml penicillin, 50 μg/ml streptomycin and 0.1 mM β-mercaptoethanol with irradiated MEF feeders (GlobalStem) on gelatin-coated plates. For the 2i medium, two small-molecule inhibitors PD0325901 (1 mM, Axon Medchem) and CHIR99021 (3 mM, Axon Medchem), were added to the ESC media. Rescue experiments were performed by transient transfections of a plasmid containing Caprin1 cDNA.Cell cultures were maintained in a humidified atmosphere (5% CO2 at 37°C).
Method details
ESC 2i and RA-induced differentiation
For RA-induced differentiation, ESCs were grown on gelatin-coated dishes for up to 4 days in ESC medium without LIF and with 1 μM RA. MEF removal was achieved by passaging ESCs twice (approx. 25 min each) to gelatin-coated plates.
Immunofluorescence
Cells were grown on glass coverslips in 24 well-plates, fixed in 4% paraformaldehyde (15 m, room temp), washed (X3) in PBS (5 min, RT), permeabilized (0.5% Triton X-100, 5 m, RT) and incubated with the primary antibodies (1 h, RT or 4°C, overnight). Cells were washed (X3) in PBS (5 min, RT), incubated with secondary antibodies (1 h RT), washed again and finally mounted with DAPI medium in microscope slides.
Microscopy and imaging
We used the Revolution spinning disk (CSUX, Yokogawa) imaging system (Andor, UK) equipped with solid-state lasers (405, 488, 561 & 640 nm) mounted on a fully automated Olympus IX81 microscope. For time-lapse imaging we used the system equipped with an automated stage and an environmental chamber (LIS) controlling humidity, CO2, and temperature for live imaging. For imaging fixed samples and IF we used an Olympus IX71 microscope equipped with an Olympus color DP71 camera.
Western blotting
Cells were collected and lysed in RIPA buffer for 30 min on ice. The lysate was centrifuged for 10 minute and the supernatant collected. Protein was quantified with BCA Protein Assay Kit (Pierce). Protein was loaded on a Bolt 4-12 % Bis-Tris mini gel (Thermo Fisher Scientific), transferred to PVDF membrane and processed for western blotting. Briefly, PVDF membrane was blocked with 5% BSA for 45 min followed by incubation with primary antibody (2h RT). The PVDF membrane was washed in PBST (X3, 5 min each) followed by incubation in secondary antibodies anti-Horseradish Peroxidase for 45 min. The membrane was washed again in PBST (X3), and the western blot signal was visualized by addition of ECL onto membrane. Images were captured in BioRad image analyser using chemiluminescence.
Generation of Caprin1 and Ddx3x knock-out mouse ESCs
Guide RNAs targeting Exon 2 of Caprin1 and Exons 1 and 5 of Ddx3x were designed using the CRISPR design tool. Best target guides were selected. The target sequence for Caprin1: 5’-TCTCGGTCTAAAGATGCCCT-3’; for Ddx3x: 5’-AGTGGAAAATGCGCTCGGGC-3’ and 5’-ACTCCCACCAAGTGAACGAT-3’. This guide was cloned into mammalian expression vector PX458 (Addgene). Mouse R1 ESCs were transfected, and GFP positive cells were sorted after 48 h post-transfection. The sorted cells were clonally expanded and knock-out of Caprin1 was verified by DNA sequencing, western blotting and RT-qPCR.
RNA isolation, reverse transcription, and quantitative real-time PCR
Total RNA from ESCs and RA differentiated cells was prepared as described in the RNeasy Mi Kit (Qiagen, supplemented with RNase-free DNase set). Approximately 1000 ng total RNA was used for reverse transcription using High Capacity cDNA RT kit (Applied Biosystems) following the manufacturer’s instructions with a mix of random hexamers and poly(dT) primers. Quantitative real-time PCR was performed in a BioRad sequence detection system with diluted cDNA as template. Power SYBR Green PCR Master Mix (Applied Biosystems) was used for real-time PCR. Primers used for this experiment are listed on STAR Methods.
Genomic DNA isolation
Total genomic DNA was isolated using Sigma genomic DNA isolation kit following manufacturer’s instructions. Approximately 200 ng DNA was used for PCR to screen knock-out clones.
mRNA degradation assay
Caprin1 KO and R1 cells were cultured for 4 days in ESC medium without LIF and with 1 μM RA in the medium. On the fourth day the cells were exposed to 2 μg/ml of α-amanitin (Sigma Aldrich) and samples were harvested at different time points (0, 2, 4, 8 h). RNA was extracted and purified (Qiagen). Indexed RNA-Seq libraries were prepared from 500 ng total RNA using the KAPA Stranded mRNA-Seq Kit (Kapa Biosystems, Inc.) according to the manufacturer’s protocol. Libraries were size-selected with SPRI beads and quantified by QuBIT (Life Technologies). Average size of 300 bp was determined by TapeStation (Agilent Technologies, USA). Libraries are then sequenced using an Illuma Nextseq500 sequencer. Raw data quality control was done using FASTQC (Version 0.11.9). Reads were aligned to reference genome (GRCm38/mm10, Dec. 2011) with STAR (Version 2.4.0.1). Read normalization and differential gene expression analysis with multiple testing corrections were conducted using the R Bioconductor DESeq2 package in R Studio.
TPM values were fitted to an exponential decay model (X(t) = X0∗exp(-bt); b = degradation rate (1/h)), and transcripts with minimal degradation rate (b<0.05, corresponding to a half-life of >13.86 h) in WT cells were selected as a control set for normalization, resulting in a control set of 356 genes (Figure S4). All samples were subsequently normalized based on this control set, to a temporally constant average expression level of this control set. These normalized TPM values were fitted again to an exponential decay model, and the resulting degradation rates for each transcript were used for subsequent analysis.
Metabolic labeling and SLAM-Seq assay
Caprin1-KO and R1 cells were cultured with/without LIF and in the presence or absence of RA for 4 days. Cells were 4sU-metabolic labeled for 24 h, followed by a chase with unlabeled uridine for 0, 0.5, 1, 3, 6, 12, and 24 h. Samples were then subjected to RNA extraction (Qiagen) and thiol modification; the 4sU-containing transcripts were converted to C after reverse transcription and library construction. The mutations in each time point were integrated to calculate the half-life of each transcript.
Immunoprecipitation
Whole procedure of IP was performed in an ice bath. Cells were washed (X2) by re-suspending the cell pellet with 0.5 ml of ice-cold PBS buffer, centrifuged for 5 min at 2000 rpm, and the supernatant was discarded. 1 ml of pre-cooled cell lysis buffer was added to the cell pellet. Cells were incubated for 30 min on ice, vortexed for 30 sec every 5 min for 30 m. Cell lysates were centrifuged for 30 min at 20,000 g at 4oC. Supernatant was collected and used for IP (2 mg lysate was used for each IP reaction). Background was pre-cleared by incubating the supernatant with 5 μl protein A beads (Magna ChIP™ Protein A Magnetic Beads, 16-661) for 30 min. The reaction mix was placed on the magnet till supernatant became clear. Supernatants were collected. CAPRIN1 antibody (2 μg) was added along with 20 μl protein A beads to supernatants and incubated for 4 h. Eppendorfs were placed on the magnet till supernatants became clear. Supernatants were removed and discarded. The bead was washed 3 times with cell lysate buffer. 1) For mass-spec analysis the beads were boiled in 50 ul SDS-gel loading buffer (1X) for 5 min at 95⁰C and the supernatant was transferred to new tubes. The samples were processed for LC-MS/MS; 2). For RNA isolation from IP, the beads were mixed with RLT buffer and samples were processed according to RNA-isolation kit (Qiagen) following manufacturer’s instruction.
Teratoma assay
For teratoma generation, one million ESCs (Caprin1KOs, Xrn2KD or WT) were harvested and resuspended in ESC culture media with Matrigel™ (BD Biosciences). The cells were injected subcutaneously into the dorsal flank of NOD-SCID mice. 24 days after injection, mice were killed and teratomas were surgically dissected. RNA was extracted from the teratomas and sequenced. All animal experiments were conducted in accordance with the Hebrew University’s animal committee and approved by the ethics committee.
Mass spectrometry of proteins
The protein complex pull-down samples were analyzed as we previously described.57 Briefly, the proteins were resolved with SDS-PAGE. Proteins in gel pore were subject to in-gel trypsin digestion. The tryptic peptides were then analyzed using a Q-Exactive LC-MS/MS system (Thermo Fisher Scientific, MA, USA). MS/MS spectra were processed and converted to Mascot generic file (mgf) format that was searched using a Uniprot mouse protein sequence database for proteins identification and label-free quantitation using exponentially modified protein abundance index (emPAI).
Lentivirus production
Knockdown of XRN2 was performed using a stable expression of a short hairpin RNA (shRNA) targeting XRN2. sgRNA oligos were designed, annealed and cloned into the pLKO1-PURO vector. HEK293T cells were co-transfected with vector plasmid and packaging plasmids (pMD2.G and psPAX2) using the TransIT Transfection Reagent (Mirus). After 48 h, viral supernatants were collected and filtered though stericup-HV PVDF 0.45-μm filter. Lentiviral vectors expressing either scrambled pLKO1-PURO or mutant (ShXRN2) were transfected into ESCs with 8 μg/mL polybrene (Sigma). The selection process was initiated after 48 h of transfection, with 1 μg/ml puromycin to obtain stable clones. After 5 days of transfection, the surviving and stably proliferating clones were selected and expanded. Protein and DNA were extracted for further validation. shXRN2 target sequence: 5'-GTTGATGCCAGTAAACCTAAT-3'. Hairpin Sequence:
5'-CCGGGTTGATGCCAGTAAACCTAATCTCGAGATTAGGTTTACTGGCATCAACTTTTTG-3'
Library construction and RNA-sequencing
cDNA libraries for each sample were generated from total RNA using the Quantseq 3′ mRNA-Seq Library Prep kit for Illumina (Lexogen) following the manufacturer’s instructions. Libraries were size-selected with SPRI beads and quantified by QuBIT (Life Technologies). Average size of ∼250 bp was determined by TapeStation (Agilent Technologies, USA). Libraries are then sequenced using an Illumina Nextseq500 sequencer.
RNA and RIP sequencing analysis
Raw RNA/RIP-seq reads for each condition were mapped to the reference mouse genome using TopHat program. After running TopHat, the resulting alignment files were used to run Cufflinks that generates a transcriptome assembly file for each input condition. Further, these assembled files were combined using the Cuffmerge program. This merged assembly was the final file used to calculate gene and transcript expression in each sample. The reads and the merged assembly provided to Cuffdiff that calculates differential expression levels as well as tests the statistical significance.
Motif analysis
Mouse 3’UTR sequences were downloaded from ensembl biomart GRCm39, version 105.58 Sequences were analyzed by using R 4.1.0, with seqinr, dplyr, stringr and data.table packages. Sequences were divided into 3 groups: (1) genes expressed only in the undifferentiated ESCs, (2) genes expressed only in the differentiated ESCs and (3) genes expressed in both. Sequences were filtered so only the longest transcript for each gene was kept (to avoid biases in analysis by including multiple isoforms of the same gene). Streme version 5.4.159 with default parameters was used to find RNA motifs ranging between 6-30 bp. Background group included all genes expressed at 0 h. RNAfold program from the viennarna package version 2.5.060 with default parameters was used to calculate the minimum free energy of 3′UTR sequences. Results were analyzed using R 4.1.0, with the ggplot2 and patchwork packages.
Quantification and statistical analysis
The figures were assembled and statistical tests were performed using Microsoft Excel or R. The P-value was calculated by two-tailed paired Student’s t-test or U-test. Significance was defined as P < 0.05 or as indicated in each respective figure legend. All data are presented as mean Standard Error.
Acknowledgments
We thank the three anonymous peer reviewers for their constructive and helpful comments, Eran Hornstein (Weizmann Institute) for critical comments and suggestions, members of our lab for help with experiments, and the Life Sciences Institute Genomics and Imaging Facilities for help with high-throughput sequencing and microscopy. This work was supported by the National Natural Science Foundation of China (U20A2015 to M.A.E. and Youth Fund Project 32201214 to Y.L.), the Israel Science Foundation (ISF1140/2017 to E.M.) and the European Union’s Horizon Europe Research and Innovation Programme under the EIC Pathfinder-Open grant agreement #101099654 (RT-SuperES). E.M. is the incumbent of the Arthur Gutterman Professor Chair for Stem Cell Research. J.O.V. is supported by the European Union’s Horizon-2020 Marie Sk1odowska Curie EpiSyStem ITN Network (765966).
Author contributions
J.O.V. and G.K.A. designed and performed most of the experiments and wrote the paper. T.P. and S.P. helped with the experiments. J.E.P. and S.K.S. performed MS. D.K. helped with supervision, planning, and writing. M.R. performed bioinformatics and statistical analyses. Y.L. and M.E. performed SLAM-seq experiments. E.M. designed the experiments, supervised the project, and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published: December 9, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2022.11.014.
Supplemental information
(A) Differentially expressed genes – RNA-Seq, related to Figures 2G and 2H.
(B) RNA transcripts associated with CAPRIN1 – RIP-Seq, related to Figures 3 and S3B
(A) Differentially stabilized nascent RNA transcripts-SLAM-seq, related to Figures 4C and 4D.
(B) Stable transcripts used for normalization of α-amanitin degradation experiment, related to Figures S4B–S4G.
(C) Degradation rate of transcripts in WT and Caprin1-KO clones, normalized list, related to Figures 4E–4G.
Data and code availability
The RNA-seq and RIP-seq data have been deposited at Gene Expression Onmibus (GEO) accession number: GSE179002. SLAM-seq data: BioProject Accession number PRJNA734745.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 2.Wright J.E., Ciosk R. RNA-based regulation of pluripotency. Trends Genet. 2013;29:99–107. doi: 10.1016/j.tig.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Schlesinger S., Meshorer E. Open chomatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev. Cell. 2019;48:135–150. doi: 10.1016/j.devcel.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q., Hu G. Post-transcriptional regulation of the pluripotent state. Curr. Opin. Genet. Dev. 2017;46:15–23. doi: 10.1016/j.gde.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keene J.D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J., Ratanasirintrawoot S., Chandrasekaran S., Wu Z., Ficarro S.B., Yu C., Ross C.A., Cacchiarelli D., Xia Q., Seligson M., et al. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016;19:66–80. doi: 10.1016/j.stem.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Aaronson Y., Meshorer E. Stem cells: Regulation by alternative splicing. Nature. 2013;498:176–177. doi: 10.1038/nature12253. [DOI] [PubMed] [Google Scholar]
- 9.Han H., Irimia M., Ross P.J., Sung H.K., Alipanahi B., David L., Golipour A., Gabut M., Michael I.P., Nachman E.N., et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogrammg. Nature. 2013;498:241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livyatan I., Meshorer E. SON sheds light on RNA splicing and pluripotency. Nat. Cell Biol. 2013;15:1139–1140. doi: 10.1038/ncb2851. [DOI] [PubMed] [Google Scholar]
- 11.Lu X., Göke J., Sachs F., Jacques P.É., Liang H., Feng B., Bourque G., Bubulya P.A., Ng H.H. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat. Cell Biol. 2013;15:1141–1152. doi: 10.1038/ncb2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo G.W., Coufal N.G., Liang T.Y., Peng G.E., Fu X.D., Gage F.H. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K., et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geula S., Moshitch-Moshkovitz S., Domissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S., et al. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brumbaugh J., Di Stefano B., Wang X., Borkent M., Forouzmand E., Clowers K.J., Ji F., Schwarz B.A., Kalocsay M., Elledge S.J., et al. Nudt21 controls cell fate by connecting alternative polyadenylation to chomatin signaling. Cell. 2018;172:106–120.e21. doi: 10.1016/j.cell.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lackford B., Yao C., Charles G.M., Weng L., Zheng X., Choi E.A., Xie X., Wan J., Xing Y., Freudenberg J.M., et al. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33:878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Miao Y.L., Zheng X., Lackford B., Zhou B., Han L., Yao C., Ward J.M., Burkholder A., Lipchina I., et al. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogrammg. Cell Stem Cell. 2013;13:676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng X., Yang P., Lackford B., Bennett B.D., Wang L., Li H., Wang Y., Miao Y., Foley J.F., Fargo D.C., et al. CNOT3-dependent mRNA deadenylation safeguards the pluripotent state. Stem Cell Rep. 2016;7:897–910. doi: 10.1016/j.stemcr.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Stefano B., Luo E.-C., Haggerty C., Aigner S., Charlton J., Brumbaugh J., Ji F., Rabano Jiménez I., Clowers K.J., Huebner A.J., et al. The RNA helicase DDX6 controls cellular plasticity by modulating P-body homeostasis. Cell Stem Cell. 2019;25:622–638.e13. doi: 10.1016/j.stem.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viegas J.O., Meshorer E. The princess and the P: pluripotent stem cells and P-bodies. Cell Stem Cell. 2019;25:589–591. doi: 10.1016/j.stem.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Welling M., Chen H.-H., Muñoz J., Musheev M.U., Kester L., Junker J.P., Mischerikow N., Arbab M., Kuijk E., Silberstein L., et al. DAZL regulates Tet1 translation in murine embryonic stem cells. EMBO Rep. 2015;16:791–802. doi: 10.15252/embr.201540538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoffe Y., David M., Kalaora R., Povodovski L., Friedlander G., Feldmesser E., Ainbinder E., Saada A., Bialik S., Kimchi A. Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev. 2016;30:1991–2004. doi: 10.1101/gad.285239.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belair C., Sim S., Kim K.-Y., Tanaka Y., Park I.-H., Wolin S.L. The RNA exosome nuclease complex regulates human embryonic stem cell differentiation. J. Cell Biol. 2019;218:2564–2582. doi: 10.1083/jcb.201811148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D.G., Buetow K.H., et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edupuganti R.R., Geiger S., Lindeboom R.G.H., Shi H., Hsu P.J., Lu Z., Wang S.Y., Baltissen M.P.A., Jansen P.W.T.C., Rossa M., et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017;24:870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harikumar A., Edupuganti R.R., Sorek M., Azad G.K., Markoulaki S., Sehnalová P., Legartová S., Bártová E., Farkash-Amar S., Jaenisch R., et al. An endogenously tagged fluorescent fusion protein library in mouse embryonic stem cells. Stem Cell Rep. 2017;9:1304–1314. doi: 10.1016/j.stemcr.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B., David M.D., Schader J.W. Absence of Caprin-1 results in defects in cellular proliferation. J. Immunol. 2005;175:4274–4282. doi: 10.4049/jimmunol.175.7.4274. [DOI] [PubMed] [Google Scholar]
- 29.Shiina N., Shinkura K., Tokunaga M. A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 2005;25:4420–4434. doi: 10.1523/JNEUROSCI.0382-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., Mcinerney G.M., Ivanov P., et al. G3BP – Caprin1 – USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solomon S., Xu Y., Wang B., David M.D., Schubert P., Kennedy D., Schader J.W. Distinct Structural Features ofCaprin-1 Mediate Its Interaction with G3BP-1 and Its Induction of Phosphorylation of Eukaryotic Translation InitiationFactor 2α, Entry to Cytoplasmic Stress Granules, and Selective Interaction with a Subset of mRNAs. Mol. Cell. Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T.H., Tsang B., Vernon R.M., Sonenberg N., Kay L.E., Forman-Kay J.D. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science. 2019;365:825–829. doi: 10.1126/science.aax4240. [DOI] [PubMed] [Google Scholar]
- 33.Livyatan I., Aaronson Y., Gokhman D., Ashkenazi R., Meshorer E. BindDB: an integrated database and webtool platform for “reverse-ChIP” epigenomic analysis. Cell Stem Cell. 2015;17:647–648. doi: 10.1016/J.STEM.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Rai A.K., Chen J.-X., Selbach M., Pelkmans L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature. 2018;559:211–216. doi: 10.1038/s41586-018-0279-8. [DOI] [PubMed] [Google Scholar]
- 35.Kolobova E., Efimov A., Kaverina I., Rishi A.K., Schader J.W., Ham A.J., Larocca M.C., Goldenring J.R. Microtubule-dependent association of AKAP350A and CCAR1 with RNA stress granules. Exp. Cell Res. 2009;315:542–555. doi: 10.1016/j.yexcr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kessel M., Liu S.X., Xu A., Santella R., Hei T.K. Arsenic induces oxidative DNA damage in mammalian cells. Mol. Cell. Biochem. 2002;234–235:301–308. doi: 10.1023/A:1015927406142. [DOI] [PubMed] [Google Scholar]
- 37.Shiina N., Tokunaga M. RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J. Biol. Chem. 2010;285:24260–24269. doi: 10.1074/jbc.M110.108944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama K., Ohashi R., Shinoda Y., Yamazaki M., Abe M., Fujikawa A., Shigenobu S., Futatsugi A., Noda M., Mikoshiba K., et al. RNG105/caprin1, an RNA granule protein for dendritic mRNA localization, is essential for long-term memory formation. eLife. 2017;6:e29677. doi: 10.7554/eLife.29677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedersdorf M.B., Keene J.D. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014;15:R2. doi: 10.1186/gb-2014-15-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer J.W., Busa V.F., Shao Y., Leung A.K.L. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell. 2020;78:70–84.e6. doi: 10.1016/j.molcel.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzog V.A., Reichholf B., Neumann T., Rescheneder P., Bhat P., Burkard T.R., Wlotzka W., Von Haeseler A., Zuber J., Ameres S.L. Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods. 2017;14:1198–1204. doi: 10.1038/nmeth.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vu L., Ghosh A., Tran C., Tebung W.A., Sidibé H., Garcia-Mansfield K., David-Dirgo V., Sharma R., Pirrotte P., Bowser R., et al. Defining the Caprin-1 interactome in unstressed and stressed conditions. J. Proteome Res. 2021;20:3165–3178. doi: 10.1021/acs.jproteome.1c00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morales J.C., Richard P., Patidar P.L., Motea E.A., Dang T.T., Manley J.L., Boothman D.A. XRN2 links transcription termation to DNA damage and replication stress. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006107. e1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.C., Krach F., Yang D., Sen A., et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marmor-Kollet H., Siany A., Kedersha N., Knafo N., Rivkin N., Danino Y.M., Moens T.G., Olender T., Sheban D., Cohen N., et al. Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol. Cell. 2020;80:876–891.e6. doi: 10.1016/j.molcel.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yilmaz A., Peretz M., Aharony A., Sagi I., Benvenisty N. Defining essential genes for human pluripotent stem cells by CRISPR-Cas9 screening in haploid cells. Nat. Cell Biol. 2018;20:610–619. doi: 10.1038/s41556-018-0088-1. [DOI] [PubMed] [Google Scholar]
- 47.Shohat S., Shifman S. Genes essential for embryonic stem cells are associated with neurodevelopmental disorders. Genome Res. 2019;29:1910–1918. doi: 10.1101/gr.250019.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M., Belmonte J.C.I. Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet. 2017;18:180–191. doi: 10.1038/nrg.2016.156. [DOI] [PubMed] [Google Scholar]
- 49.Efroni S., Melcer S., Nissim-Rafinia M., Meshorer E. Stem cells do play with dice: A statistical physics view of transcription. Cell Cycle. 2009;8:43–48. doi: 10.4161/cc.8.1.7216. [DOI] [PubMed] [Google Scholar]
- 50.Neff A.T., Lee J.Y., Wilusz J., Tian B., Wilusz C.J. Global analysis reveals multiple pathways for unique regulation of mRNA decay in induced pluripotent stem cells. Genome Res. 2012;22:1457–1467. doi: 10.1101/gr.134312.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weskamp K., Barmada S.J. RNA degradation in neurodegenerative disease. Adv. Neurobiol. 2018;20:103–142. doi: 10.1007/978-3-319-89689-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zekri L., Chebli K., Tourrière H., Nielsen F.C., Hansen T.V.O., Rami A., Tazi J. Control of fetal growth and neonatal survival by the rasgap-associated endoribonuclease G3BP. Mol. Cell. Biol. 2005;25:8703–8716. doi: 10.1128/MCB.25.19.8703-8716.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tahmasebi S., Alain T., Rajasekhar V.K., Zhang J.P., Prager-Khoutorsky M., Khoutorsky A., Dogan Y., Gkogkas C.G., Petroulakis E., Sylvestre A., et al. Multifaceted regulation of somatic cell reprogrammg by mRNA translational control. Cell Stem Cell. 2014;14:606–616. doi: 10.1016/j.stem.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Kwon S.C., Yi H., Eichelbaum K., Föh S., Fischer B., You K.T., Castello A., Krijgsveld J., Hentze M.W., Kim V.N. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- 55.Pavinato L., Delle Vedove A., Carli D., Ferrero M., Carestiato S., Howe J.L., et al. CAPRIN1 haploinsufficiency causes a neurodevelopmental disorder with language impairment, ADHD and ASD. Brain. 2022:awac278. doi: 10.1093/brain/awac278. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delle Vedove A., Natarajan J., Zanni G., Eckenweiler M., Muiños-Bühl A., Storbeck M., Guillén Boixet J., Barresi S., Pizzi S., Hölker I., et al. CAPRIN1P512L causes aberrant protein aggregation and associates with early-onset ataxia. Cell. Mol. Life Sci. 2022;79:526. doi: 10.1007/s00018-022-04544-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alajem A., Biran A., Harikumar A., Sailaja B.S., Aaronson Y., Livyatan I., Nissim-Rafinia M., Sommer A.G., Mostoslavsky G., Gerbasi V.R., et al. Differential association of chomatin proteins identifies BAF60a/SMARCD1 as a regulator of embryonic stem cell differentiation. Cell Rep. 2015;10:2019–2031. doi: 10.1016/j.celrep.2015.02.064. [DOI] [PubMed] [Google Scholar]
- 58.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–D891. doi: 10.1093/NAR/GKAA942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey T.L. STREME: accurate and versatile sequence motif discovery. Bioinform. Oxf. Engl. 2021;37:2834–2840. doi: 10.1093/BIOINFORMATICS/BTAB203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Differentially expressed genes – RNA-Seq, related to Figures 2G and 2H.
(B) RNA transcripts associated with CAPRIN1 – RIP-Seq, related to Figures 3 and S3B
(A) Differentially stabilized nascent RNA transcripts-SLAM-seq, related to Figures 4C and 4D.
(B) Stable transcripts used for normalization of α-amanitin degradation experiment, related to Figures S4B–S4G.
(C) Degradation rate of transcripts in WT and Caprin1-KO clones, normalized list, related to Figures 4E–4G.
Data Availability Statement
The RNA-seq and RIP-seq data have been deposited at Gene Expression Onmibus (GEO) accession number: GSE179002. SLAM-seq data: BioProject Accession number PRJNA734745.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.