Summary
How the vascular and neural compartment cooperate to achieve such a complex and highly specialized structure as the central nervous system is still unclear. Here, we reveal a crosstalk between motor neurons (MNs) and endothelial cells (ECs), necessary for the coordinated development of MNs. By analyzing cell-to-cell interaction profiles of the mouse developing spinal cord, we uncovered semaphorin 3C (Sema3C) and PlexinD1 as a communication axis between MNs and ECs. Using cell-specific knockout mice and in vitro assays, we demonstrate that removal of Sema3C in MNs, or its receptor PlexinD1 in ECs, results in premature and aberrant vascularization of MN columns. Those vascular defects impair MN axon exit from the spinal cord. Impaired PlexinD1 signaling in ECs also causes MN maturation defects at later stages. This study highlights the importance of a timely and spatially controlled communication between MNs and ECs for proper spinal cord development.
Keywords: neuro-vascular communication, spinal cord, motor neuron, endothelial cell, Sema3C, PlexinD1, CNS vascularization, neurodevelopment, blood vessel
Highlights
-
•
Motor neurons (MNs) communicate with endothelial cells (ECs) of growing blood vessels
-
•
MN-EC crosstalk via Sema3C-PlexinD1 prevents vessel ingrowth into MN columns
-
•
Premature and aberrant MN column vascularization leads to impaired spinal MN axon exit
-
•
EC-specific PlexinD1 knockout embryos also display altered MN maturation
Vieira et al. describe a crosstalk between spinal motor neurons and endothelial cells of growing blood vessels during development. They show that motor neuron-derived Sema3C signals to PlexinD1/Nrp1 expressed in endothelial cells. This signaling pathway is required for proper spinal cord vascularization and motor neuron development.
Introduction
The development of the central nervous system (CNS) requires the precise integration of many different cell types into the brain and spinal cord, among them cells of the growing vasculature and developing neurons.1,2,3 At the same time as neural progenitor cells proliferate, differentiate, and neurons migrate and build neural networks, blood vessels (BVs) invade the CNS, grow, branch, and assemble into a functional vascular network.3 In the developing spinal cord, vascularization occurs in a highly temporal and spatial stereotypical manner. Spinal cord vascularization starts at embryonic day (E) 8.5–9.5 by the formation of the peri-neural vascular plexus (PNVP).4,5,6 Subsequently at around E10, from the PNVP, angiogenic sprouts invade the spinal cord ventrally between the floor plate and motor neuron (MN) columns. However, they strictly avoid entering into those areas during a developmental time window (from E10.5 until E12.5), despite the presence of pro-angiogenic molecules such as vascular endothelial growth factor (Vegf).6,7,8,9 Previous studies demonstrated that neural-derived signals regulate spinal cord vascularization,7,10,11,12,13,14,15 while BVs in turn influence neural cell development.16,17,18 Concurrently with spinal cord vascularization, the neural compartment also develops and reorganizes following a characteristic program.3,19,20,21 In particular, MNs arise from the MN progenitor domain at around E9.0 and migrate toward the ventro-lateral part of the spinal cord.22 There, MNs segregate and subcluster in different columns of MN subtypes.23 While segregating, MNs start projecting their axons out of the spinal cord to innervate different targets.24,25,26 At later embryonic stages, MNs receive inputs from afferent sensory neurons and interneurons,27,28,29 while they themselves also start establishing synapses with the targeted muscles by forming neuromuscular junctions.30
Initially discovered as axon guidance cues,31,32 the class 3 semaphorin (Sema3) family, including Sema3A to Sema3G, comprises secreted proteins that also regulate vascular processes.33,34 Class 3 semaphorins bind to Plexin receptors and different co-receptors, with neuropilin 1 (Nrp1) and neuropilin 2 (Nrp2) required for the signaling of all Sema3 family members except Sema3E.35 Sema3 binding to Nrp1 is dispensable for early brain vascularization, as is Sema3A.36 Among the Sema3 proteins, Sema3E is uniquely known to regulate angiogenesis during development by signaling through PlexinD1, including in the developing mouse retina, spinal cord, and heart.37,38,39 PlexinD1 is dynamically expressed in neurons and endothelial cells (ECs), and its expression in ECs has been implicated in intersomitic vessel formation and sprouting39,40,41,42 and CNS angiogenesis.37,38 The role of other members of the Sema3 family in developmental angiogenesis remains less characterized. In particular, a role for Sema3C is described in the inhibition of pathological angiogenesis,43 without knowledge of potential roles in spinal cord vascularization.
Here, we describe a molecular communication between MNs and ECs required to block premature vessel ingression into the MN columns. Using a single-cell sequencing dataset from the developing spinal cord,44 we identified Sema3C-PlexinD1 signaling as a mediator pathway between MNs and ECs. Cell-type-specific deletion of each of the molecules caused premature MN column vascularization. We show that keeping MNs avascular at these developmental stages is important to prevent subsequent MN developmental defects.
Results
The developing ventral spinal cord interactome implicates the semaphorin class 3 family in MN-EC communication
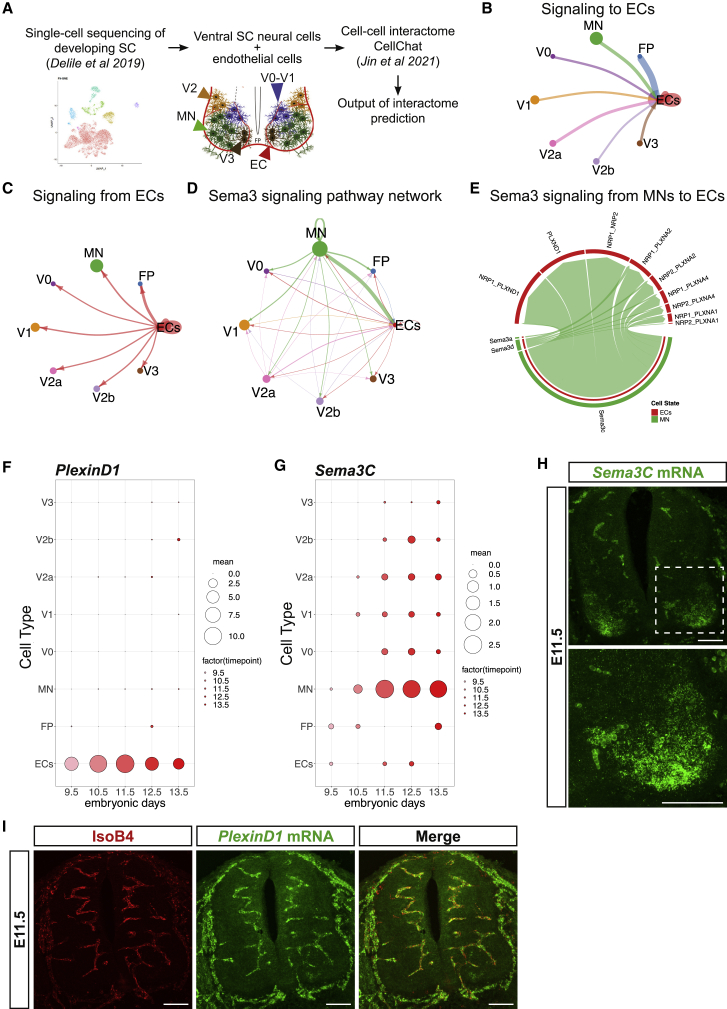
To understand the molecular mechanisms for vessel sprouting from the PNVP into the spinal cord at specific locations and branching, we re-analyzed a previously published single-cell RNA sequencing (scRNA-seq) dataset of the developing spinal cord (cervical to thoracic)44 to identify potential molecular interactions between ventral postmitotic neurons, floor plate (FP) cells, and ECs (Figure 1A). From this dataset we extracted the sequencing data from FP cells; all MNs; V0, V1, V2, and V3 interneurons; and the blood cell cluster that also contained the ECs (Figure 1A). To identify ECs within the blood cell cluster, we filtered the cluster using the canonical EC markers Pecam1, Cdh5, and Kdr.45,46,47 As we were interested in early spinal cord vascularization, when MN columns remain avascular (E9.5 to E11.5), the sequencing data from E9.5 to E11.5 were analyzed to identify potential cell-cell communication pathways between these cell types (Figure 1A). For this, we used the CellChat tool,48 which computationally analyses scRNA-seq data based on a database of ligand-receptor interactions to identify potential cell-to-cell molecular interactions. The prediction showed that all the different cell types were theoretically able to communicate with each other (Figure S1A; Table S1). This predicted communication was bidirectional, as neural cells could signal to ECs and vice versa (Figures 1B and 1C). We next identified the top five predominant signaling pathways appearing between neural cells and ECs of the ventral spinal cord. Those were the Notch, Epha, Laminin, Jam, and Sema3 signaling pathways (Figures 1D and SS1B–S1F). Remarkably, while four of those pathways (Notch, Epha, Laminin, and Jam) seemed to be equally present in the potential communication between ECs and all the other neural cell types analyzed, the Sema3 pathway appeared to be particularly strong between MNs and ECs (Figure 1D). Closer analysis of the identity of the ligands and receptors involved in the Sema3 pathway showed that the strongest predicted interaction was between MN-secreted Sema3C and EC-derived PlexinD1 (with or without Nrp1) (Figure 1E).
Figure 1.
MN-derived Sema3C is predicted to interact with EC-specific PlexinD1 in the embryonic developing spinal cord (SC)
(A) Schematic representation of the bioinformatic pipeline used to obtain the predicted cell-cell interactome.
(B and C) Graphs showing the predicted signaling from ventral neural cells to ECs (B) and from ECs to the different populations of ventral neural cells (C).
(D) Graph showing the interactions between the different cell types using the Sema3 signaling pathway network.
(E) Representation of the Sema3 signaling from MNs to ECs, showing the Sema3 family ligands expressed by MNs and their respective receptors expressed in ECs.
(F and G) Predicted expression of PlexinD1 (F) and Sema3C (G) in the different cell types between E9.5 abd E13.5, using previously published single-cell RNA-seq data.44
(H and I) Representative images of RNAscope Multiplex Fluorescent Assay using Sema3C probe (H) and PlexinD1 probe combined with staining for blood vessels (BVs) (IsoB4+) (I) at E11.5. Autofluorescence of erythrocytes is seen in the images, as in negative control (see Figure S2E). Inset shows higher magnification of MN columns. Scale bars, 100 μm.
As Sema3C-Nrp1-PlexinD1 signaling repels ECs during pathological angiogenesis,43 we asked whether Sema3C-Nrp1-PlexinD1 signaling could play a role in repelling ECs from entering into the MN columns, thereby maintaining them avascular for a defined developmental time window between E10.5 and E12.5 and hence contributing to the precise spatiotemporal cellular organization of the spinal cord. We first analyzed the temporal and spatial expression pattern of PlexinD1, Nrp1, and Sema3C in the developing mouse spinal cord using the same single-cell sequencing dataset.44 PlexinD1 mRNA was exclusively expressed in ECs from E9.5 to E13.5 (Figures 1F and 1I) at thoracic-brachial levels (Figure S1I), with its peak of expression occurring at E11.5 and then progressively decreasing (Figures S1G and S1H). Consistent with a previous study,7 Nrp1 was also expressed in ECs during spinal cord development (Figures S1J–S1M). Its expression peaked between E10.5 and E11.5 (Figures S1K and S1L). Sema3C expression appeared mainly in MNs (Figures 1G, 1H, and S2A) and, similarly to PlexinD1 and Nrp1, its expression peaked at E11.5 (Figures S2B and S2C). Sema3C was detected in all MNs, with particularly higher levels in the medial motor column (MMC) located at the brachial/lumbar level (Figure S2D); this was confirmed by RNAscope for Sema3C in combination with different MN markers (Figures S2E–S2G).
Global and EC-specific PlexinD1 loss causes premature MN column vascularization
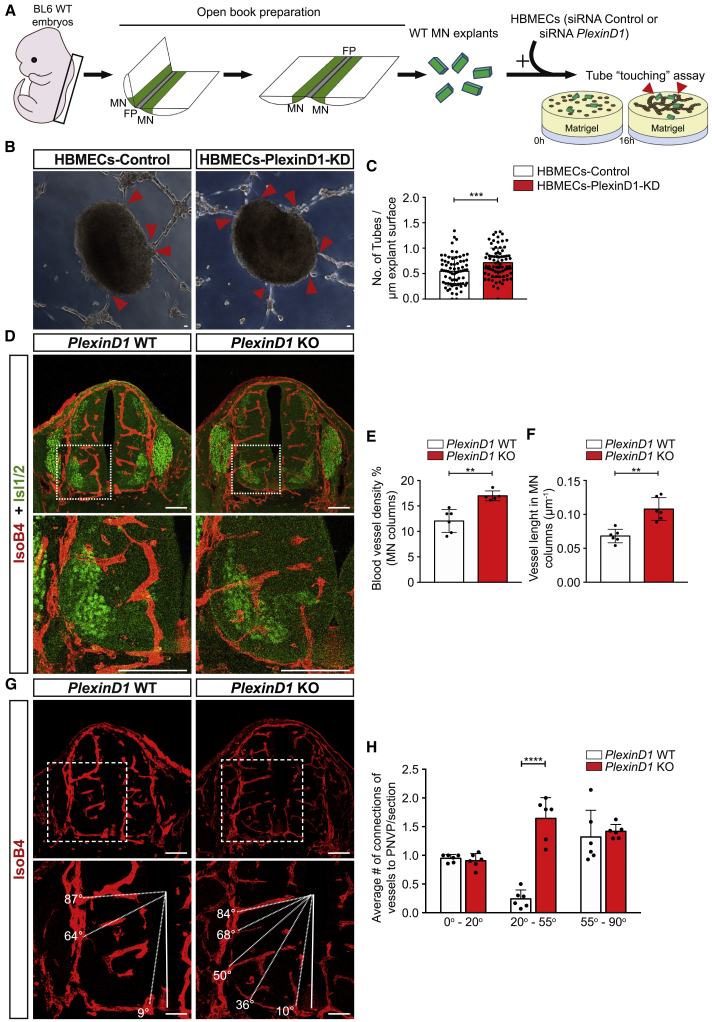
To evaluate whether PlexinD1 and Nrp1 in ECs are required for preventing sprouting BVs from entering into MN columns in mouse embryos, we performed an in vitro assay, which we had previously termed 3D “tube-touching” assay7 (Figure 2A). This assay consists of co-culturing MN explants from embryonic spinal cords with CNS ECs on a matrigel layer. In these conditions, ECs assemble into tubes and can either approach and touch the explant (attractive behavior) or not touch and be repelled from the explant (repulsive behavior).7 For this, MN explants were isolated from E11.5 mouse spinal cord (Figure 2A) and co-cultured with human brain microvascular ECs (HBMECs) previously transfected either with a control small interfering RNA (siRNA) or with siRNA to knockdown PlexinD1 or Nrp1 expression (Figures S3A and S3C). Quantification of the number of HBMEC tubes in contact with MN explants 16 h after starting the co-cultures showed that a higher number of PlexinD1 knockdown HBMEC tubes were attached to the explants compared to control HBMEC tubes (Figures 2B and 2C). No differences in explant size were observed between the conditions analyzed (Figure S3B). This indicates that MN explants exert a repulsive effect on ECs and that this depends on PlexinD1. An increased number of contacts between HBMECs-Nrp1-KD and MN explants, compared with HBMECs-Control, was also observed (Figures S3D–S3F). Together with the expression data and previous functional studies in ECs,43 these findings suggest that Nrp1 is also part of the PlexinD1 signaling complex in ECs.
Figure 2.
PlexinD1 regulates vascular patterning around MN columns
(A) Schematic representation of the steps of the in vitro “tube-touching” assay. BL6 WT mouse embryos were dissected at E11.5, and the isolated spinal cords (SCs) were flattened via open book preparation. MN explants were microdissected. WT MN explants were co-cultured in matrigel with HBMECs transfected with either siRNA Ctrl (HBMECs-Control) or siRNA PlexinD1 (HBMECs-PlexinD1-KD). Red arrowheads indicate contacts made between MN explants and EC tubes.
(B) Representative images of the tube-touching assay showing HBMEC tubes touching MN explants. Red arrowheads indicate contacts between HBMEC tubes and explants. Scale bars, 100 μm.
(C) Quantification of the number of either HBMECs-Control or HBMECs-PlexinD1-KD tubes touching MN explants, normalized to the explant perimeter (n = 72 explants siRNA Ctrl, n = 77 explants siRNA PlexinD1, from 2 independent experiments [each experiment consisting of 2 independent litters], parametric distribution, two-tailed unpaired Student’s t test). ∗∗∗p < 0.001.
(D) Representative images of SC sections at thoracic level stained for BVs (IsoB4+) and MNs (Isl1/2+) showing increased vessel ingression into the MNs columns of PlexinD1 KO embryos compared with WT littermates (E11.5). Insets showing higher magnifications of MN columns. Scale bars, 100 μm.
(E and F) Quantification of BV density (E) and BV length (F) in MN columns of WT and PlexinD1 KO embryos at E11.5. n = 6 WT, n = 5 PlexinD1 KO for BV density; n = 6 WT, n = 6 PlexinD1 KO for BV length, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗∗p < 0.01.
(G) Representative images of BVs (IsoB4+) ingression analysis in the ventral half of the SC at thoracic level in WT and PlexinD1 KO embryos (E11.5). Dashed lines indicate the angle of ingression into the spinal cord of the PNVP-derived sprouts. Insets showing higher magnifications of MN columns. Scale bars, 100 μm.
(H) Quantification of the angles of BV ingression into the ventral half of the SC (0°–90°). n = 6 WT and n = 6 PlexinD1 KO at E11.5, from two independent litters; two-way ANOVA with Sidak’s multiple comparisons test.∗∗∗∗p < 0.0001.
All data shown as mean ± SD.
To investigate whether repulsive PlexinD1 signaling in ECs might prevent premature ingression of BVs into MN columns in vivo, we assessed MN column vascularization at brachial and thoracic levels in the spinal cord of wild-type (WT) and PlexinD1 full knockout (KO) embryos (Figure S4A) at E11.5, a developmental stage when MN columns are avascular.7 The spinal cord of PlexinD1 KO embryos presented both significantly increased vascularization and increased vessel length in MN columns compared to PlexinD1 WT, with vessel sprouts crossing the central core of MN columns (Figures 2D–2F). Reflecting this increase in MN vascular density, vessel density in the total spinal cord was also increased (Figure S4B). This increased MN vascularization in PlexinD1 KO embryos was present at all the different spinal cord levels. We analyzed BV patterning with the ventral spinal cord by studying the ingression angle of BVs from the PNVP into the ventral spinal cord (from 0° to 90°, with the FP being the reference point for 0°).6,7 While in WT embryos, BVs avoided entering into the spinal cord through MN columns (located between 20° and 55°), in PlexinD1 KO embryos, BVs ingressed the spinal cord in between MN columns (Figures 2G and 2H), further indicating that PlexinD1 expression in ECs is necessary to restrain vessels from invading MNs columns from the PNVP.
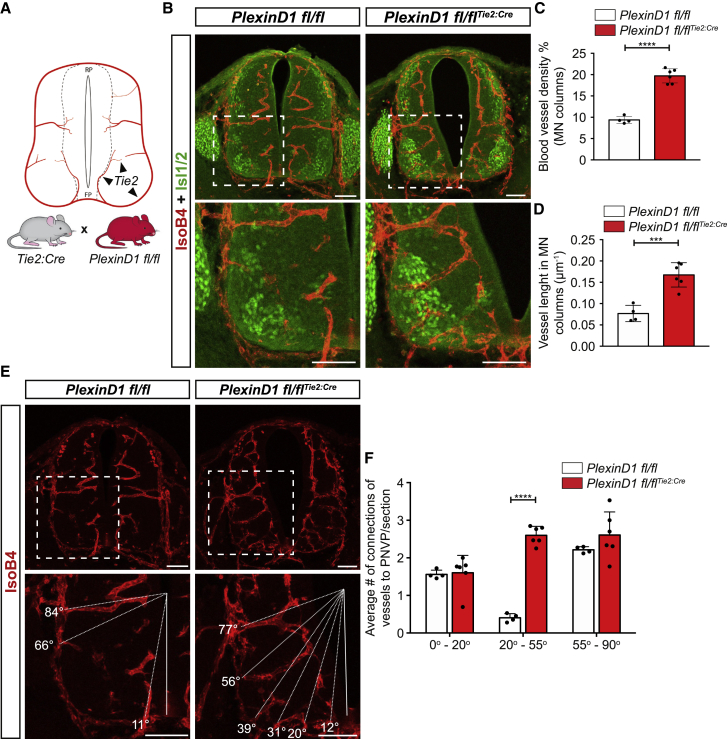
PlexinD1 is expressed at late embryonic stages (E15.5 onward) in dorsal root ganglia (DRG) sensory neurons, in which it is required to control sensory-MN connections.49,50 Although we could not detect PlexinD1 expression in any other spinal cord cell type apart from ECs at the earlier developmental time points (Figures 1I and S1I), we sought to confirm a cell-autonomous role of PlexinD1 in ECs by analyzing MN vascularization in conditional EC-specific PlexinD1 KO mouse embryos. These mice were generated by crossing PlexinD1 floxed mice (PlexinD1 fl/fl)51 with the EC-specific Tie2:Cre driver line52 and are from hereon termed PlexinD1 fl/flTie2:Cre (Figures 3A and S4C). As seen in global PlexinD1 KO embryos, we observed a significant increase of BVs ingressing into the MN columns at E11.5 at brachial and thoracic levels (Figures 3B–3D), indicating that specific deletion of PlexinD1 in ECs is sufficient to cause the premature ingression of BVs into MN columns. Reflecting this difference in MN vascular density, vessel density in the total spinal cord was also increased (Figure S4D). Similar to global PlexinD1 KO embryos, this phenotype in PlexinD1 fl/flTie2:Cre embryos was visible at all spinal cord levels. Analysis of the ingression pattern emphasized that a lack of PlexinD1 in ECs leads to more BVs entering into the spinal cord through MN columns (Figures 3E and 3F). Altogether, these results demonstrate that PlexinD1 in ECs is required during spinal cord development for preventing premature vascularization of MN columns.
Figure 3.
EC-specific loss of PlexinD1 causes premature MN column vascularization
(A) Scheme showing Tie2 expression in ECs of blood vessels to illustrate the use of Tie2-Cre to generate EC-specific PlexinD1 KO mice (PlexinD1 fl/flTie2:Cre) by crossing Tie2:Cre mice with PlexinD1 fl/fl mice.
(B) Representative images of spinal cord sections at thoracic level stained for BVs (IsoB4+) and MNs (Isl1/2+) showing increased vessel ingression into the MNs columns of PlexinD1 fl/flTie2:Cre embryo compared with control littermates at E11.5. Insets showing higher magnifications of MN columns. Scale bars, 100 μm.
(C and D) Quantification of BV density (C) and BV length (D) in MN columns of PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E11.5. n = 4 PlexinD1 fl/fl, n = 6 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗∗∗∗p < 0.0001 for (C) and ∗∗∗p < 0.001 for (D).
(E) Representative images of BV (IsoB4+) ingression analysis in the ventral half of the spinal cord at thoracic level in PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E11.5. Dashed lines indicate the ingression angle of the PNVP-derived sprouts into the spinal cord (SC). Insets showing higher magnifications of MN columns. Scale bars, 100 μm.
(F) Quantification of BV ingression angles into the ventral half of the SC (0°–90°). n = 4 PlexinD1 fl/fl, n = 6 PlexinD1 fl/flTie2:Cre at E11.5, from two independent litters; two-way ANOVA with Sidak’s multiple comparisons test.
All data shown as mean ± SD.
MN-specific Sema3C loss causes premature MN vascularization
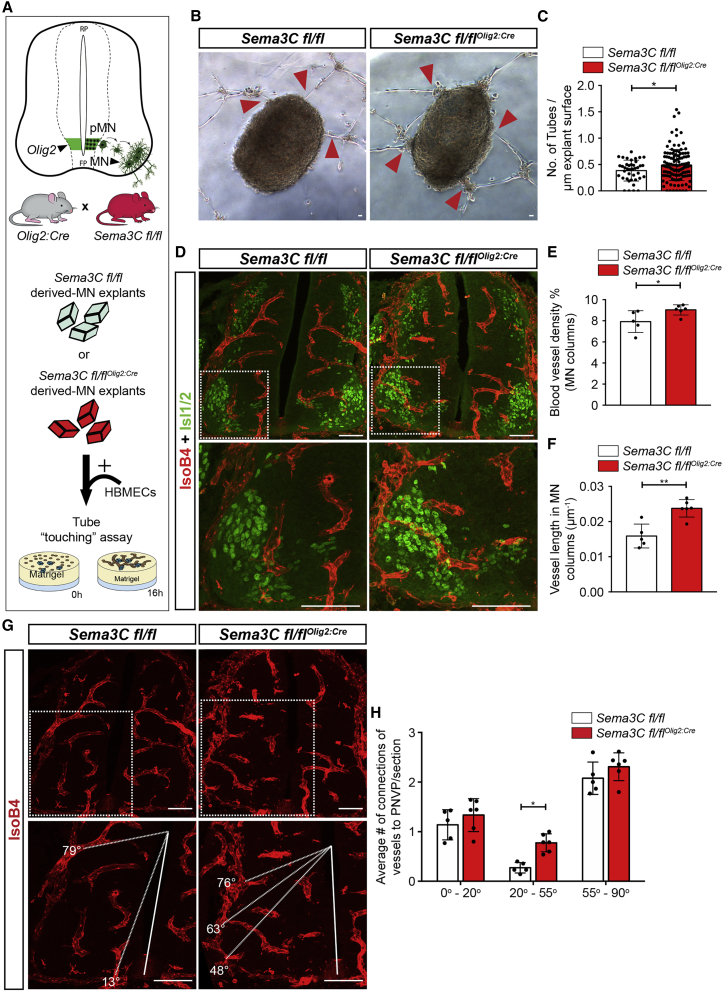
The analysis of predicted interactions between MNs and ECs (Figure 1E) and the validation of Sema3C expression in MNs in the developing spinal cord (Figures 1H and S2G) pointed to MN-derived Sema3C as the potential inducer of repulsive PlexinD1 signaling in spinal cord ECs. To explore the role of MN-derived Sema3C in MN column vascularization, we generated a MN-specific Sema3C conditional knockout mouse by crossing Sema3C floxed mice (Sema3C fl/fl)53 with the Olig2:Cre driver line54 that targets the MN progenitor domain (we term those mice from hereon Sema3C fl/flOlig2:Cre). The absence of Sema3C mRNA in spinal cord MNs at E11.5 indicated successful Cre-mediated Sema3C targeting (Figure S4E).
To study whether Sema3C acts as a repulsive cue for EC tubes, we co-cultured HBMECs with MN explants from control Sema3C fl/fl mouse embryos or Sema3C fl/flOlig2:Cre in the tube-touching assay (Figure 4A). Removal of Sema3C from MNs increased the number of tubes contacting MN explants (Figures 4B and 4C). The explant size was similar for both genotypes (Figure S4F). To investigate whether repulsive Sema3C signaling from MNs prevents premature ingression of BVs into the MN columns during development in vivo, we analyzed MN vascularization of Sema3C fl/flOlig2:Cre E11.5 spinal cords at brachial and thoracic levels. Evaluation of BV density and vessel length in MN columns of E11.5 Sema3C fl/flOlig2:Cre spinal cords showed premature vascularization of MN columns (Figures 4D–4F). Analysis of vessel patterning revealed that, in Sema3C fl/flOlig2:Cre, vessel sprouts ingressed into the spinal cord at angles where MN columns are positioned, while this was not the case in control littermates (Figures 4G and 4H). Vessel density in the total spinal cord was not altered (Figure S4G). These findings support the hypothesis that Sema3C acts as an inhibitory cue for EC ingression into MN columns via PlexinD1 signaling.
Figure 4.
Absence of Sema3C in MNs leads to premature MN column vascularization
(A) Top: scheme showing Olig2 expression in the motor neuron progenitor domain (pMN) to illustrate the use of Olig2-Cre to delete Sema3C in MNs (Sema3C fl/flOlig2:Cre) by crossing Olig2:Cre with Sema3C fl/fl mice. Bottom: schematic representation of the in vitro “tube-touching” assay co-culturing MN explants from Sema3C fl/fl and Sema3C fl/flOlig2:Cre embryos (E11.5) in matrigel with HBMECs.
(B) Representative images of the tube-touching assay showing HBMEC tubes touching MN explants. Red arrowheads pointing at connections between HBMEC tubes and explants. Scale bars, 100 μm.
(C) Quantification of the number of HBMEC tubes touching MN explants from Sema3C fl/fl or Sema3C fl/flOlig2:Cre embryos, normalized to the explant perimeter (n = 40 explants Sema3C fl/fl, n = 125 explants Sema3C fl/flOlig2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test). ∗p < 0.05.
(D) Representative images of spinal cord sections at brachial level stained for BVs (IsoB4+) and MNs (Isl1/2+) demonstrating increased vessel ingression into the MNs columns of Sema3C fl/flOlig2:Cre embryos compared with control littermates at E11.5. Insets showing higher magnifications of MN columns. Scale bars, 100 μm.
(E and F) Quantification of BV density (E) and BV length (F) in MN columns of Sema3C fl/fl or Sema3C fl/flOlig2:Cre at E11.5. n = 5 Sema3C fl/fl, n = 6 Sema3C fl/flOlig2:Cre from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗p < 0.05 for (E) and ∗∗p < 0.01 for (F).
(G) Representative images of BV (IsoB4+) ingression analysis in the ventral half of the spinal cord at brachial level in Sema3C fl/fl or Sema3C fl/flOlig2:Cre embryos at E11.5. Dashed lines indicate the angle of ingression into the spinal cord of the PNVP-derived sprouts. Insets showing higher magnifications of MN columns. Scale bars, 100 μm.
(H) Quantification of BV ingression angles into the ventral half of the spinal cord (E11.5) (0°–90°). n = 5 Sema3C fl/fl, n = 6 Sema3C fl/flOlig2:Cre, from two independent litters; two-way ANOVA with Sidak’s multiple comparisons test. ∗p < 0.05.
All data shown as mean ± SD
Sema3E is expressed in brachial MNs but not required to prevent premature MN vascularization
Besides Sema3C, Sema3E is also a ligand for PlexinD1,51,55 and Sema3E-PlexinD1 signaling controls BV patterning in developing intersomitic vessels and retina in the mouse and intersegmental vessels in zebrafish by inducing a repulsion in ECs upon PlexinD1 signaling.38,39,40,42 Prompted by this described role of Sema3E-PlexinD1 signaling, and despite the fact that our bioinformatic analysis, which grouped all MN pools together (see Figure 1), did not show Sema3E as a potential MN-derived ligand for EC-PlexinD1, we decided to explore further a potential role for Sema3E in preventing premature MN vascularization. Notably, Sema3E expression along the spinal cord at E12.5 and E13.5 is restricted to lateral motor column (LMC) MNs.56 Similarly, our expression analysis of Sema3E at E11.5 and E12.5 revealed that Sema3E is expressed in MNs at brachial levels (and also in the FP at all spinal cord levels), but not in thoracic regions (Figure S5A and S5B).
To investigate whether Sema3E provides an additional ligand for EC-PlexinD1 to prevent MN vascularization specifically at brachial levels of the developing spinal cord, we analyzed WT and Sema3E full KO littermate embryos at E11.5 (Figure S5C). Quantification of BV density in the MN columns did not reveal any significant difference between Sema3E WT and Sema3E KO embryos at the brachial level (Figure S5E), or when analyzing MN vascularization at brachial and thoracic levels together (Figures S5D and S5F). Similarly, total spinal cord vascularization was not altered (Figure S5G). Altogether, our findings suggest that Sema3E is dispensable for preventing premature MN column vascularization despite being expressed in MNs at brachial levels.
Impaired Sema3C-PlexinD1 signaling results in early MN axon patterning and late MN maturation defects
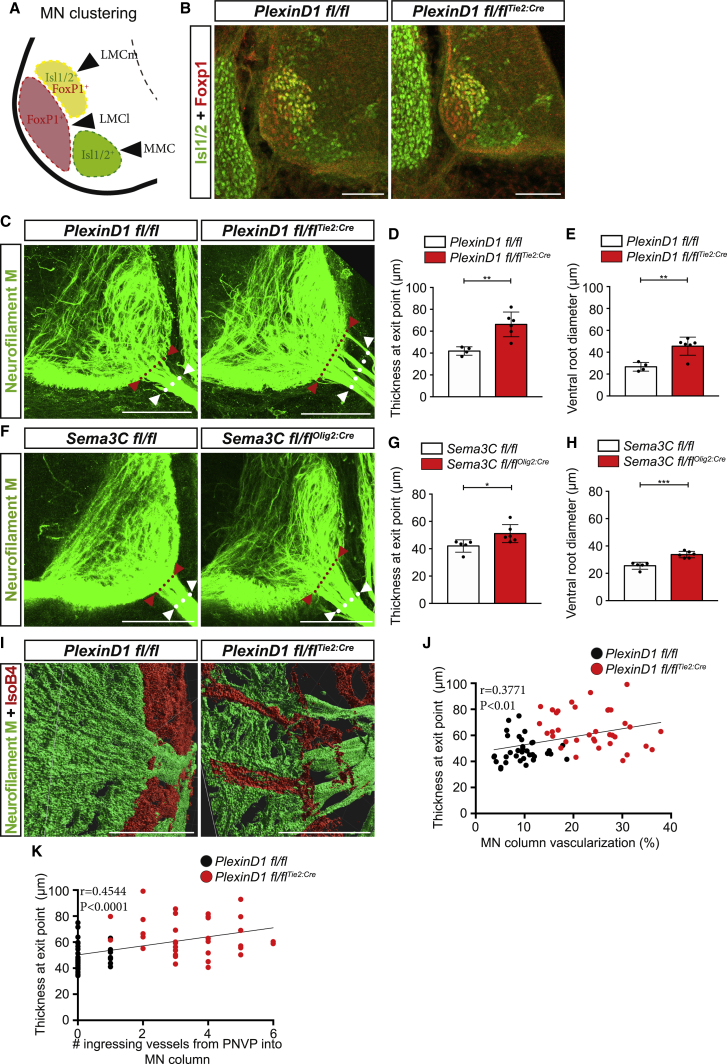
Why do MN columns remain avascular during a developmental time window? Different crucial phases of MN development occur within the developmental time window when MN columns remain avascular, including MN clustering and MN axon projection from the spinal cord into the periphery.24,25,54,57 We reasoned that premature invasion of BVs into MN columns could interfere with those early MN developmental processes. As such, to ensure their development, MNs would have developed mechanisms in place to repel BVs from invading. To address this hypothesis, we evaluated several different aspects of early MN development in PlexinD1 KO, PlexinD1 fl/flTie2:Cre, and Sema3C fl/flOlig2:cre, which have premature vascularization of MN columns. First, we assessed the MN area. PlexinD1 KO and PlexinD1 fl/flTie2:Cre embryos showed no significant differences in the MN column area when compared to their respective control littermates (Figures S6A and S6B), while Sema3C fl/flOlig2Cre embryos had only a slightly decreased MN area compared with control littermates (Figure S6C). Second, we analyzed the columnar positioning of MNs (medial LMCm, lateral LMCl, and MMC) but found no differences in PlexinD1 fl/flTie2:Cre compared with PlexinD1 fl/fl littermates (Figures 5A, 5B, and S6D), suggesting that MN cell body sorting is not affected. Third, we examined MN axon behavior during their exit from the spinal cord into the periphery at MN exit points (MEPs), where MN axons also fasciculate to form the ventral root. Analysis of the distance from the midline to the most ventral exit point showed no difference between genotypes, indicating that MNs are able to find the correct MEP (Figures S6E–S6I). However, analysis of the thickness of the axonal bundle at the MEP showed that MN axons take up a larger area as they exit the spinal cord in both PlexinD1 fl/flTie2:Cre and Sema3C fl/flOlig2Cre mouse lines compared to MN axons in control embryos (Figures 5C, 5D, 5F, and 5G). Consistently, analysis of the ventral root diameter also revealed that ventral roots of PlexinD1 fl/flTie2:Cre and Sema3C fl/flOlig2Cre mouse embryos were significantly expanded when compared to control littermates (Figures 5E and 5H). To further understand this aberrant exit of MN axons, we explored whether there could be a correlation between the shape of the axon bundles and ectopic vessel sprouting vessels within the MNs (Figures 5I and S6J). Co-labeling of axons and vessels and detailed imaging analysis showed that MN axon exit point thickness increased proportionally with vessel density (Figure 5J) and with the number of ingressing vessels from PNVP into MN columns (Figure 5K). These findings are consistent with the notion that ectopic and premature vessels in the MN columns present a physical obstacle for axonal projections from the CNS to the periphery, thereby forcing axons to navigate around them and thus causing axon spreading.
Figure 5.
Premature ingression of BVs impairs MN axons patterning
(A) Schematic representation of MN clustering and positioning at brachial level of the spinal cord at E11.5 embryos.
(B) Immunostaining showing the different MN columns at brachial level in PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E11.5. MN clustering into the medial division of the lateral motor column (LMCm, Isl1/2+ and FoxP1+), lateral division of the lateral motor column (LMCl, FoxP1+), and the medial motor column (MMC, Isl1/2+) is not affected in PlexinD1 fl/flTie2:Cre embryos. Scale bars, 100 μm.
(C) Representative images of 300-μm-thick sections stained for neurofilament M showing MN axons exiting the spinal cord at brachial level (red arrowheads and red dotted line indicate MN axon fascicle thickness at the motor neuron exit points [MEPs]; white arrowheads and dotted line indicate the ventral root diameter) in PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E11.5. Scale bars, 100 μm.
(D) Quantification of the MN axon fascicle thickness at the MEP in PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E11.5. n = 4 PlexinD1 fl/fl, n = 6 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗∗p < 0.01.
(E) Quantification of ventral root diameter at E11.5 in PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos. n = 4 PlexinD1 fl/fl, n = 6 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗∗p < 0.01.
(F) Representative images of 300-μm-thick sections stained for neurofilament M showing MN axons exiting the spinal cord at brachial level (red arrowheads and red dotted line indicate the MN axon fascicle thickness at the MEP, and white dotted line indicates the ventral root diameter) in Sema3C fl/fl and Sema3C fl/flOlig2:Cre embryos at E11.5. Scale bars, 100 μm.
(G) Quantification of the MN axon fascicle thickness at the MEP in Sema3C fl/fl and Sema3C fl/flOlig2:Cre embryos at E11.5. n = 5 Sema3C fl/fl, n = 6 Sema3C fl/flOlig2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗p < 0.05.
(H) Quantification of ventral root diameter Sema3C fl/fl and Sema3C fl/flOlig2:Cre embryos at E11.5. n = 5 Sema3C fl/fl, n = 6 Sema3C fl/flOlig2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗∗∗p < 0.001.
(I) Three dimensional representation of the MN axons (neurofilament M) and ingressing vessels (IsoB4) in MN columns of PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E11.5. Scale bar, 50 μm.
(J and K) Correlation between MN axon thickness at exit point at E11.5 with MN column vascularization (J) and with the number of ingressing vessels (K). n = 35 MN columns/MEPs from 3 embryos PlexinD1 fl/fl, n = 36 MN columns/MEPs from 3 embryos PlexinD1 fl/flTie2:Cre.
All data shown as mean ± SD
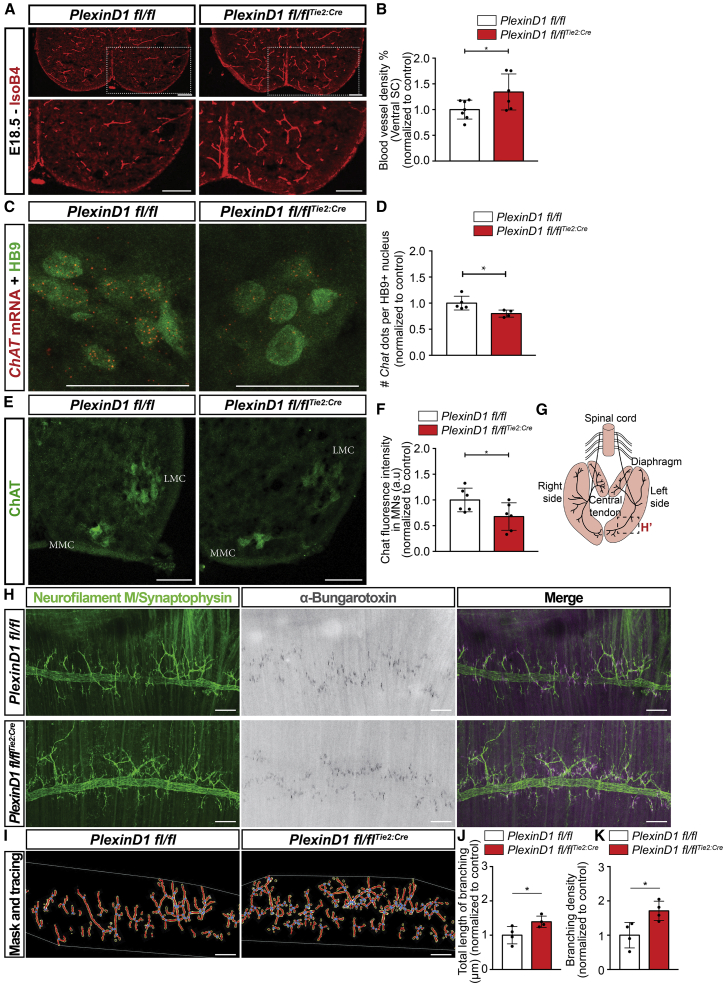
Next, we asked whether late MN development is also affected in the absence of PlexinD1 signaling in ECs. We focused on the PlexinD1 fl/flTie2:Cre mouse line and limited our analysis to E18.5 because the embryos die after birth due to cardiovascular defects.51 At this late embryonic stage, PlexinD1 fl/flTie2:Cre embryos still presented increased vascular density in the ventral spinal cord compared to control littermates (Figures 6A and 6B). Quantification of MN numbers, average soma size, and column area showed no difference between PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos (Figures S7A–S7H). We studied terminal differentiation and maturation profile of MNs by analyzing the expression pattern of a panel of genes identified as terminal differentiation markers for MNs (Nrg1, Sema5a, Mcam, Pappa, Glra2),58,59 neuronal activity (Fos), and genes belonging to the cholinergic pathway (Slc18a3, Slc5a7, ChAT, Slc10a4).60 Quantification of RNAscope signals of these genes in individual MNs showed that the transcription profile of Mcam, Glra2, Nrg1, Slc18a3, and ChAT was altered in PlexinD1 fl/flTie2:Cre compared with PlexinD1 fl/fl (Figures 6C, 6D, S8A–S8F, and S9A–S9E). We further analyzed at the protein level the expression of choline acetyltransferase (ChAT), the rate-limiting enzyme producing the neurotransmitter acetylcholine, and thus a marker of cholinergic function.61 Similar to the identified gene expression changes, quantification of ChAT expression in MNs showed a significant reduction in ChAT protein levels in PlexinD1 fl/flTie2:Cre embryos compared to WT littermates (Figures 6E and 6F), suggesting impaired cholinergic neurotransmission of MNs in PlexinD1 fl/flTie2:Cre embryos.
Figure 6.
PlexinD1 fl/flTie2:Cre embryos present increased spinal cord vascularization and altered MN maturation defects at E18.5
(A) Immunostaining for BVs (IsoB4+) in E18.5 spinal cords from PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos. Insets show higher magnification of one side of the ventral spinal cord. Scale bars, 100 μm.
(B) Quantification of BV density in the ventral spinal cord of PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos, normalized to control littermates at E18.5. n = 7 PlexinD1 fl/fl, n = 6 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗p < 0.05.
(C and D) Representative images of RNAscope for Chat co-stained with HB9 (C) and respective quantification of the number of Chat dots per HB9+ nucleus (D) (normalized to control littermates). Scale bars, 25 μm. n = 5 PlexinD1 fl/fl, n = 4 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗p < 0.05.
(E) Representative image of ChAT immunostaining in MNs from the medial motor column (MMC) and lateral motor column (LMC) of PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E18.5. Scale bars, 100 μm.
(F) Quantification of ChAT fluorescence intensity in PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E18.5. Data normalized to control. n = 6 PlexinD1 fl/fl, n = 6 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗p < 0.05.
(G) Scheme representing the innervation of diaphragm by the phrenic nerve. Inset: location where pictures shown in (H) were taken.
(H and I) Representative images (H) of the diaphragm of PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos at E18.5 stained to show the phrenic nerve and its branching (Neurofilament M + Synapthophysin), AchR+ clusters (α-bungaratoxin); (I) representation of the output result of image analysis for branching points and length (using the neurofilament + synapthophysin staining). Scale bars, 100 μm.
(J) Quantification of total length of phrenic nerve branching in the diaphragm muscle at E18.5. Data normalized to control. n = 4 PlexinD1 fl/fl, n = 4 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗p < 0.05.
(K) Quantification of branching density of phrenic nerve at E18.5 in PlexinD1 fl/fl and PlexinD1 fl/flTie2:Cre embryos. Data normalized to control littermates. n = 4 PlexinD1 fl/fl, n = 4 PlexinD1 fl/flTie2:Cre, from two independent litters; parametric distribution, two-tailed unpaired Student’s t test. ∗p < 0.05.
All data shown as mean ± SD.
Tracing of sensory axons arising from the DRG and innervating MNs (via DiI) and focusing on the general organization of sensory proprioceptive fibers showed no differences in the general arborization pattern, in the angle displayed by these most prominent bundles, or in their overall targeting of motor columns (Figures S7I–S7L). Analysis of the density of afferent inputs to MNs (by immunostaining for the vesicular acetylcholine transporter [VACht; cholinergic input] and vesicular GABA transporter [Vgat; inhibitory synapses]) revealed no differences between PlexinD1 fl/flTie2:Cre and control littermates (Figures S7M–S7Q). Together, these findings suggest that premature vascularization of MN columns does not alter afferent inputs but rather reduces the capacity of MNs for neurotransmission.
Finally, we also analyzed targeting and innervation of MN axons in the periphery, in particular intercostal muscles and the diaphragm. While targeting was not affected—axons reached their final muscle targets (Figures 6G, 6H, and S8G)—a closer analysis of nerve endings in the diaphragm revealed increased branching complexity of the phrenic nerve in PlexinD1 fl/flTie2:Cre embryos compared with PlexinD1 fl/fl littermates (Figures 6H–6K). However, the number of AChR+ clusters present in the muscle were not altered, suggesting that the additional branches might be immature or not functional (Figure S8H). These findings suggest that the fine-tuning of the phrenic nerve branching is altered after removing PlexinD1 from ECs.
Discussion
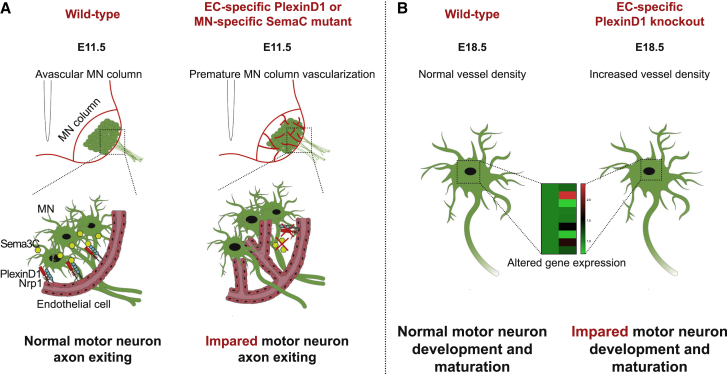
CNS development requires the integration of numerous molecular and cellular processes in a temporally and spatially coordinated manner. Among them, CNS vascularization coincides with neural cell proliferation, differentiation, migration, and network integration of many different neural cells. In both the developing brain and spinal cord, specific vascular patterns can be observed, whereby sprouting vessels avoid specific regions for particular periods of time and grow along stereotypical paths.6,7,8,18,62,63 Remarkably, MN columns in the developing embryonic spinal cord remain avascular between E9.5 and E12 despite expressing high levels of Vegf.7,9 Accounting for such an avascular period, it was shown that MNs express the soluble Vegf receptor sFlt1 in MNs, which titrates the existing Vegf, thus making vessels less responsive to this attractive factor.7 Here, we report a previously unknown molecular mechanism, whereby MNs express Sema3C to signal to EC-PlexinD1 in order to control the timing of BV ingression into the ventrolateral areas of the spinal cord where they are positioned (Figure 7). We propose that this mechanism allows MNs to undergo their own developmental processes without obstruction by BVs.
Figure 7.
Proposed model for MN-EC communication required for proper spinal cord vascularization and MN development
(A) At E11.5 , MN columns of WT embryos are avascular. MN-secreted Sema3C signals to PlexinD1/NRP1 in ECs, controlling their ingression into MN columns, thereby allowing proper MN development and maturation. However, in EC-specific PlexinD1 or MN-specific Sema3C mutants, this signaling is interrupted and BVs ingress into MN columns prematurely. MN development is impaired showing defects in axon organization at their exit into to the periphery at E11.5. (B) At E18.5 , MNs of embryos where PlexinD1 signaling in ECs is absent show dysregulated expression of terminal differentiation markers and functional factors.
BVs sprout from specific locations of the PNVP and begin to enter into the developing spinal cord where their paths are also stereotypic. Our in silico results, using a scRNA-seq dataset from embryonic mouse spinal cord,44 show that all the different neural cell types have the potential to signal to ECs, supporting a concept for neural-driven control of spinal cord vascularization beyond the initial process of Vegf secretion. This also implies a remarkable presence of different signaling pathways between particular cell types and ECs. Here, we validated one of those predicted crosstalk pathways (Sema3C-PlexinD1) by demonstrating that MN-specific Sema3C signals to PlexinD1 in ECs and that the removal of either one of the components of this signaling pair, in their cell type of origin, leads to premature ingression of BVs into MN columns (Figure 7). Interestingly, the expression of both genes peaks at E11.5 and is reduced at E12.5, coinciding with the developmental passage from avascular MN columns to the stage when MN columns are normally first invaded by vessels. The ability of Sema3C-PlexinD1 signaling to inhibit angiogenesis was demonstrated in pathological conditions.43 Our study demonstrates that this signaling pair is also important for BV patterning in a physiological developmental process.
A role for Sema3E has been described in BV development, such as in the initial formation of the dorsal aorta,64 during intersomitic vessel patterning in the mouse39 and zebrafish,42 and during BV growth in the mouse retina.37,38 In chick embryos, overexpression of Sema3E in the developing spinal cord via in ovo electroporation causes vessel repulsion.63 By analyzing Sema3E KO mouse embryos, we found that Sema3E expression is restricted to brachial levels, but it is not required at that spinal cord level for embryonic mouse spinal cord vascularization or for keeping MN columns avascular. Instead, we identified Sema3C as a molecular cue that is expressed in MNs along the entire spinal cord and that by signaling to PlexinD1/NRP1 in ECs (also expressed in all ECs along the spinal cord) restricts vessel ingression into MN columns.
Notably, deletion of Sema3C from MNs (Sema3C fl/flOlig2:Cre) resulted in a milder MN vascularization phenotype when compared to PlexinD1 fl/flTie2:Cre embryos. This may be explained by two different, but non-exclusive, mechanisms. First, other Sema3 members expressed in MNs and able to bind PlexinD156 might be compensating when Sema3C is absent, such as Sema3E. Second, PlexinD1 in ECs might also be activated by mechanical forces, independent of its ligands,65 and act as a mechanosensor to activate intracellular signaling pathways leading to cytoskeleton rearrangement of ECs and directed vessel growth. In pathological angiogenesis, Sema3C-PlexinD1/Nrp1 signaling in ECs has been described to crosstalk with the VEGF pathway in order to regulate vessel growth.43 Whether the same interaction occurs in this developmental setting requires further investigation.
Why do MNs prevent BV ingression for such a specific period of time? Our data suggest that this fine-tuned timely ingression of vessels into the MN columns is essential to ensure their appropriate development. Indeed, we observed that the premature vascularization of MN columns impairs proper MN axon exit (Figure 7). Thus, instead of projecting out of the spinal cord as a single bundle of axons, MN axons in PlexinD1 fl/flTie2:Cre mutants exit into the periphery segregated in more fascicles, leading to expanded ventral roots. Exactly in between the locations where those fascicles project out of the spinal cord, we identified vessel sprouts that had invaded those areas prematurely. This finding suggests that the presence of BVs acts as an obstacle for growing MN axons. Additionally, MN axons might have a tropism for BVs so that in the case of premature vascularization, MN axons associate with those vessels and use their surface as a track for further growing. MN axon navigation from the spinal cord toward their peripheral targets was described to be regulated by suppression of attractive signals within the spinal cord and by attraction from the MEP and peripheral targets.26,66 Our findings add another layer of regulation, whereby MN-derived Sema3C acts as a repulsive signal to prevent the invasion of BVs until axons have projected to the periphery in an organized manner.
At the end of embryonic development, MNs in PlexinD1 fl/flTie2:Cre show an altered pattern of expression of genes linked to terminal differentiation and functionality, among them reduced levels of ChAT, suggesting that MN functionality is impaired. As we did not observe differences in afferent inputs to MNs, decreased ChAT expression in PlexinD1 mutant embryos might be a consequence of aberrant EC-derived signals or of the targeting of MN axons in the periphery.67 PlexinD1 fl/flTie2:Cre also presents increased terminal branching of the phrenic nerve. Notably, reduced expression of ChAT and Nrg1 in MNs has been linked to abnormally increased nerve branching,68,69 suggesting that the observed altered expression of those genes in PlexinD1 fl/flTie2:Cre MNs might contribute to the hyperbranched phenotype. Whether those MN maturation defects and increased nerve branching are due to altered motor column vascularization and/or the effect of aberrant BVs on MN axon trajectories in the periphery would require further investigation. While it would be interesting to understand the impact of those phenotypes in postnatal or adult mice, PlexinD1 fl/flTie2:Cre embryos die at birth due to severe cardiac defects,51 preventing us from performing those studies. Overall, these data suggest that, in the absence of PlexinD1 signaling in ECs, MN connectivity and function are impaired.
These results could provide a new understanding of different pathological conditions with altered MN axon targeting and function as well as open potential new paths for translational research. For example, in CNS lesions, neuronal death, and the consequent failure to regenerate, precise neuronal networks may result at least in part from a chaotic and not guided vascularization of the injured tissue. In fact, the fibrotic scar is a major impediment to regenerating axons.70,71,72 Although ultimately required to support the surrounding environment, an early increased vascularization may not allow a time window for neurons to regenerate, extend their axons properly, and reconnect.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Isl1/2 | DSHB | Cat# 39.4D5; RRID:AB_2314683 |
| Rabbit anti-Foxp1 | Abcam | Cat# ab16645; RRID:AB_732428 |
| Goat anti-ChAT | Sigma | Cat# ab144P; RRID: AB_2079751 |
| Guinea-pig anti-VACht | Synaptic systems | Cat# 139105; RRID: AB_10893979 |
| Rabbit anti-VACht | Synaptic systems | Cat# 139103; RRID: AB_887864 |
| Mouse anti-VGAT | Synaptic systems | Cat# 131011; RRID:AB_2619818 |
| Rabbit anti-HB9 | Thaler et al. 199973 | N/A |
| Mouse anti-neurofilament M (RMO 270) | Thermo Scientific | Cat# 13-0700; RRID:AB_86551 |
| Rabbit anti-neurofilament M | Biolegend | Cat# 841001; RRID:AB_2565457 |
| Donkey anti-goat Alexa 647 | Jackson Immunoresearch | Cat# 705-605-147; RRID:AB_2340437 |
| Donkey anti-guinea pig Alexa 488 | Jackson Immunoresearch | Cat# 706-546-148; RRID:AB_2340473 |
| Donkey anti-guinea pig Alexa 594 | Jackson Immunoresearch | Cat# 706-586-148; RRID:AB_2340475 |
| Donkey anti-mouse Alexa 488 | Jackson Immunoresearch | Cat# 715-545-150; RRID:AB_2340846 |
| Donkey anti-rabbit Alexa 488 | Invitrogen | Cat# A21206; RRID:AB_2535792 |
| Donkey anti-rabbit Alexa 568 | Invitrogen | Cat# A10042; RRID:AB_2534017 |
| Isolectin GS-IB4 Alexa Fluor 568 | Invitrogen | Cat# I21412 |
| Isolectin GS-IB4 Alexa Fluor 647 | Invitrogen | Cat# I32450; RRID:SCR_014365 |
| α-Bungarotoxin Alexa 594 Conjugated | Thermofisher | Cat# B13423 |
| Alkaline phosphatase-coupled anti-DIG | Roche Diagnostics | Cat# 11093274910 |
| Chemicals, peptides, and recombinant proteins | ||
| Oligofectamine | Thermofisher | Cat# 12252011 |
| Corning Matrigel Matrix | Corning | Cat# 356234 |
| Nitroblue tetrazolium/5-bromo-3-chloro-3 indlyl phosphate | Promega | Cat# S3771 |
| Critical commercial assays | ||
| RNeasy Mini Kit | Qiagen | Cat# 74104 |
| DNase I | Thermo Scientific | Cat# EN0521 |
| Super-Script IV Vilo | Thermo Scientific | Cat# 11756-050 |
| Fast SYBR Green Master Mix | Thermo Scientific | Cat# 00408995 |
| Endopan 3 complete medium | PAN-Biotech | Cat# P04-0010K |
| Experimental models: Cell lines | ||
| Human brain microvascular endothelial cells (HBMECs) | Cell Systems | Cat# ACBRI 376 |
| Experimental models: Organisms/strains | ||
| PlexinD1 full KO | Gitler et al. 200441 | N/A |
| Tie2:Cre | Kisanuki et al. 200152 | JAX:008,863 |
| PlexinD1 fl/fl | Zhang et al. 200951 | JAX:018,319 |
| Sema3E full KO | Gu et al. 200539 | N/A |
| Olig2:Cre | Dessaud et al. 200754 | N/A |
| Sema3C fl/fl | Plein et al. 201553 | N/A |
| PlexinD1 fl/flTie2:Cre | This paper | N/A |
| Sema3C fl/flOlig2:Cre | This paper | N/A |
| C57BL/6N | Janvier Labs | N/A |
| Oligonucleotides | ||
| qRT-PCR Human PlexinD1 forward primer 5′-AACATCTCCAGCCAGAGCAG | This paper | N/A |
| qRT-PCR Human PlexinD1 reverse primer 5′-CCAGGAAGACCGCTGTGTAG | This paper | N/A |
| qRT-PCR Human Nrp1 forward primer 5′-CGCTCCCGCCTGAACTACCCT | This paper | N/A |
| qRT-PCR Human Nrp1 reverse primer 5′-TGAGGTGCGGGTGGAAGTGCC | This paper | N/A |
| qRT-PCR Human Gapdh forward primer 5′-TGCCGTCTAGAAAAACCTGC | This paper | N/A |
| qRT-PCR Human Gapdh reverse primer 5′-ACCCTGTTGCTGTAGCCAAA | This paper | N/A |
| ISH probe Mouse Sema3C forward primer 5′-AGGTCAGAGGACCAGGTATTCA | This paper | N/A |
| ISH probe Mouse Sema3C reverse primer 5′-GAGTGTTGTCCTTGGATTGTCA | This paper | N/A |
| ISH probe Mouse PlexinD1 forward primer 5′-GTACCAACTGTCGAGTGCCA | This paper | N/A |
| ISH probe Mouse PlexinD1 reverse primer 5′-TTCTCGAAGCGGTGGTCTTC | This paper | N/A |
| ISH probe Mouse Cre forward primer 5′-ACCAGGTTCGTTCACTCATGG | This paper | N/A |
| ISH probe Mouse Cre reverse primer 5′-AGGCTAAGTGCCTTCTCTACAC | This paper | N/A |
| ISH probe Mouse Sema3E forward primer 5′-CCACACGATCTACACCCGAG | This paper | N/A |
| ISH probe Mouse Sema3E reverse primer 5′-CACAGCAGAGGCTGATCCAA | This paper | N/A |
| siRNA Universal control | Sigma | Cat# SIC001 |
| siRNA against human PlexinD1 | Sigma | siRNA ID: SASI_Hs01_00194034 |
| siRNA against human Nrp1 | Sigma | siRNA ID: SASI_Hs02_00307190 |
| Software and algorithms | ||
| GraphPad Prism7 | GraphPad Software, Inc | RRID:SCR_002798 |
| Fiji | https://fiji.sc/ | RRID:SCR_002285 |
| Adobe Photoshop | Adobe Systems Inc | RRID:SCR_014199 |
| Adobe Illustrator | Adobe Systems Inc | RRID:SCR_010279 |
| Adobe InDesign | Adobe Systems Inc | RRID:SCR_021799 |
| R | https://www.r-project.org | RRID:SCR_001905 |
| Imaris | Oxford Instruments | RRID:SCR_007370 |
| Other | ||
| Endopan 3 complete medium | PAN-Biotech | Cat# P04-0010K |
| RNA scope probe for mouse Sema3c | ACD Bio-Techne | Cat# 441441-C3 |
| RNA scope probe for mouse PlexinD1 | ACD Bio-Techne | Cat# 405931 |
| RNA scope probe for mouse Nrp1 | ACD Bio-Techne | Cat# 471621 |
| RNA scope probe for mouse Slc18a3 | ACD Bio-Techne | Cat# 448771 |
| RNA scope probe for mouse Slc5a7 | ACD Bio-Techne | Cat# 439941 |
| RNA scope probe for mouse Chat | ACD Bio-Techne | Cat# 408731 |
| RNA scope probe for mouse Nrg1 | ACD Bio-Techne | Cat# 418181 |
| RNA scope probe for mouse Sema5a | ACD Bio-Techne | Cat# 508091 |
| RNA scope probe for mouse Glra2 | ACD Bio-Techne | Cat# 510301 |
| RNA scope probe for mouse Slc10a4 | ACD Bio-Techne | Cat# 544771 |
| RNA scope probe for mouse Pappa | ACD Bio-Techne | Cat# 443921 |
| RNA scope probe for mouse Mcam | ACD Bio-Techne | Cat# 406321 |
| RNA scope probe for mouse Fos | ACD Bio-Techne | Cat# 316921 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Carmen Ruiz de Almodovar (carmen.ruizdealmodovar@ukbonn.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All experimental protocols, handling, use and care of mice were conducted in accordance with the local authorities and animal welfare officers (Germany: Regierungspräsidium Karlsruhe, Germany; animal projects T38/19, I19/13, I21/02. UK: a project license to C.R. from the UK Home Office that was reviewed the UCL Institute of Ophthalmology Animal Welfare and Review Body (AWERB)).
Mouse strains
PlexinD1 full KO,41 Tie2:Cre,52 PlexinD1 fl/fl,51 Sema3E full KO,39 Olig2:Cre54 and Sema3C fl/fl53 transgenic mice were previously described. To specifically delete PlexinD1 from ECs, PlexinD1 fl/fl were crossed with PlexinD1fl/+Tie2:Cre, the generated mice were termed PlexinD1 fl/flTie2:Cre. To specifically delete Sema3C in MNs, Sema3C fl/fl mice were crossed with Sema3C fl/+ Olig2:Cre mice, the generated mice were termed Sema3C fl/flOlig2:Cre. To define the gestational stages, mice were mated and the morning of vaginal plug formation was considered as E0.5. C57Bl/6N mice were purchased from Janvier Labs and Charles River Laboratories.
Method details
Cell culture
Human brain microvascular endothelial cells (HBMECs) (ACBRI 376, Cell Systems) were cultured in well plates coated with 0.1% gelatin in Endopan 3 complete medium for ECs (P04-0010K, PAN-Biotech) supplemented with 10% FBS, 100 U ml−1 of penicillin and 100 μg mL−1 of streptomycin (15140122, ThermoFisher) in a 5% CO2 humidified incubator at 37°C. HBMECs were used from passages 6 to 10 for the experiments.
Tissue processing
Mouse embryos were collected at different embryonic stages (E9.5 to E12.5 and E18.5) and fixed in 4% formaldehyde/PBS (DEPC-treated) at 4°C overnight. Afterward, mouse embryos were transferred to 30% sucrose in PBS (DEPC-treated) at 4°C until sinking and subsequently embedded in optimal cutting temperature (OCT) compound and stored at −20°C or −80°C until the day of sectioning. Tissue blocks were cut between 20 and 40 μm thickness using a cryostat (MICROM HM560) and collected on Super-Frost Plus slides (Menzel-Glaeser, Braunschweig, Germany). For thick sections, after fixation embryos were kept in PBS until sectioning. On the day of sectioning, mouse embryos were embedded in 5% low-melting agarose and cut on a vibratome (VT1200S, Leica) to obtain 300 μm thick slices.
qRT-PCR
RNA from mouse tissue or human brain microvascular endothelial cells (HBMECs) (ACBRI 376, Cell Systems) was extracted using the RNeasy Mini Kit (74104, Qiagen). RNA was treated with DNase I (EN0521, Thermo Scientific) and afterward reverse transcribed into cDNA using Super-Script IV Vilo (11756-050, Thermo Scientific) following the manufacturer’s protocol. mRNA expression levels were assessed by qRT-PCR using Fast SYBR Green Master Mix (00,408,995, Thermo Scientific), relative to the expression level of the housekeeping gene Gapdh. All qRT-PCR primers used in this work are described in the key resources table methods.
In situ hybridization
For detection of target mRNA expression and localization, ISH was performed using the following protocol: the hybridization step was performed by incubating frozen sections with digoxigenin (DIG)-labeled antisense riboprobes at 68°C overnight. DIG-Riboprobes were detected with alkaline phosphatase-coupled anti-DIG antibody (1:500 or 1:1500, Roche Diagnostics, Mannheim, Germany). The alkaline phosphatase reaction was performed with nitroblue tetrazolium/5-bromo-3-chloro-3-indolyl phosphate (NBT-BCIP, Promega) as a chromogenic substrate, which formed a violet precipitate after few hours. Total incubation times varied between 1 and 16 h, depending on the specific probe used. Sense probes were used in parallel as negative controls, which did not generate any specific signal. Sections were imaged under a Zeiss Axiovert 200 fluorescence microscope equipped with an AxioCam MRc camera or confocal microscope Zeiss LSM800.
Immunohistochemistry
Frozen sections were washed once with PBS, permeabilized and blocked in 2% normal donkey serum in 0.3% Triton X-100/PBS for 1 h and incubated with primary antibodies in blocking buffer at 4°C overnight. The primary antibodies used were the following: Isl1/2 (39.4D5, 1:100, DSHB), Foxp1 (ab16645, 1:300, Abcam), ChAT (Ab144P, 1:200, Sigma), guinea-pig VAChT (139105, 1:200, Synaptic systems), rabbit VAChT (139103, 1:200, Synaptic systems), VGAT (131011, 1:100, Synaptic systems) and rabbit anti-HB9 #6055 (1:8000)73). After washing, all the conjugated secondary antibodies were used at 1:500 dilution and incubated at room temperature (RT) for 2 h. The following secondary antibodies were used: donkey α-goat Alexa 647 (705-605-147, Jackson ImmunoResearch), donkey α-guinea pig Alexa 488 (706-546-148, Jackson ImmunoResearch), donkey α-guinea pig Alexa 594 (706-586-148, Jackson ImmunoResearch), donkey α-mouse Alexa 488 (715-545-150, Jackson ImmunoResearch), donkey α-rabbit Alexa 488 (A21206, Invitrogen) and donkey α-rabbit Alexa 568 (A10042, Invitrogen). Blood vessels were labeled using Isolectin GS-IB4 Alexa Fluor 568 or 647 conjugate (I21412 and I32450, 1:250, Invitrogen) during incubation with secondary antibodies. 300 μm vibratome sections were blocked in 5% normal donkey serum and 1% Triton X-100 in PBS at 4°C overnight. Following blocking, sections were incubated for 72 h with primary antibodies against neurofilament M (RMO 270, 13-0700, 1:300, Thermo Scientific and 841001, 1:300, Biolegend) diluted in blocking buffer. After three washes for 30 min, sections were incubated with secondary antibodies donkey α-mouse Alexa 488 (715-545-150, 1:500, Jackson ImmunoResearch), donkey α-rabbit Alexa 488 (A21206, Invitrogen) and Isolectin GS-IB4 Alexa Fluor 568 or 647 conjugate (I21412 and I32450, 1:250, Invitrogen) in blocking buffer at 4°C for 24 h. Finally, sections were washed three times for 30 min and mounted. Images were acquired on a confocal microscope (Zeiss LSM800) with x20/0.8 Plan-APOCHROMAT and on a Nikon AR1 confocal microscope with x20/0.75 Plan-Fluor Objective. Image processing was performed using NIH ImageJ software or IMARIS software (version 9.5.1) for the three-dimensional reconstructions.
Muscle innervation immunohistochemistry
Muscles were dissected under a stereomicroscope, washed in PBS and blocked in 10% normal donkey serum in 0.3% Triton X-100/PBS at 4°C overnight. Afterward, tissue was incubated with primary antibody against neurofilament M (841001, 1:300, Biolegend) diluted in blocking buffer at 4°C for 72 h. After washing, the conjugated secondary antibody donkey α-rabbit Alexa 488 (1:500, A21206, Invitrogen), α-Bungarotoxin Alexa 594 conjugated – to label AchR (1:50, B13423, Thermofisher) - and Isolectin GS-IB4 Alexa Fluor 647 conjugate (I32450, 1:250, Invitrogen) were used and incubated at 4°C overnight. Finally, tissues were washed, mounted and imaged. Images were acquired on a confocal microscope (Zeiss LSM800) with x20/0.8 Plan-APOCHROMAT and 40x/1.30C Plan-APOCHROMAT.
Due to the high background in the neurofilament 488 channel, Adobe Photoshop 2022 (Adobe Systems, Inc., San Jose, CA) was used to paint the background black. The black background was then selected using the magic wand tool (non-contiguously, tolerance of 30) and the images were saved without any compression. Images were then inserted into Angiotool software74 to then process for junction density and total length. For the quantification of the number of AchR+ clusters, images were segmented using the Trainable Weka Segmentation function from ImageJ software and clusters with more than 4 pixels were counted.
DiI tracing
E18 embryos were sacrificed, washed in PBS and submerged in 4% paraformaldehyde at 4°C overnight. 100 μL drops of Neuro-DiI (60016, Biotium)-ethanol solutions were applied to a slide glass and let evaporate to form a thin layer of Neuro-DiI crystals. Tungsten needles (10130-20, FST) were used to scratch-collect Neuro-DiI crystals and insert them into DRGs to label the dorsal root. The corresponding ventral root was severed to avoid leaked labeling into ventral motor neurons columns. Neuro-DiI-injected tissue was incubated in 4% paraformaldehyde at 37°C for seven days. Next, spinal cords were cryopreserved in PBS-sucrose 30% at 4°C overnight, rinsed and cryosectioned at a thickness of 50 μm. Confocal tilescan images were acquired as z-stacks with a Plan-Apochromat 20X/0.95 N.A. autocorrect air objective (20×/0.8 N.A. at 1X magnification) using a CellDiscoverer 7 (automated boxed LSM900 confocal microscope; all from Zeiss).
RNAscope multiplex fluorescent assay
Sema3c (Mm-Sema3c-C3, 441441-C3), PlexinD1 (Mm-Plxnd1, 405931), Nrp1 (Mm-Nrp1-C3, 471621), Slc18a3 (Mm-Slc18a3, 448771), Slc5a7 (Mm-Slc5a7-C3, 439941), ChAT (Mm-Chat-C2, 408731), Nrg1 (Mm-Nrg1, 418181), Sema5a (Mm-Sema5a-C3, 508091), Glra2 (Mm-Glra2, 510301), Slc10a4 (Mm-Slc10a4, 544771), Pappa (Mm-Pappa, 443921), Mcam (Mm-Mcam, 406321) and Fos (Mm-Fos-C2, 316921) RNAscope probes (all from Advanced Cell Diagnostics) were used for the detection of their respective mRNA presence in embryonic spinal cord sections. RNAscope was performed on fixed-frozen 20 μm tissue sections following the manufacturer’s instructions (Advanced Cell Diagnostics). For the co-staining with MN markers HB9, Isl1/2 or Foxp1, the sections were blocked and stained with primary antibodies followed by fixation with 4% PFA and then proceeded with direct Protease III treatment, RNAscope probe binding and processing following the manufacturer’s instructions. Blood vessels were visualized using Isolectin GS-IB4 Alexa Fluor 568 or 647 conjugate (I21412 and I32450, 1:250, Invitrogen) at RT for 2 h and, together with secondary antibodies, were incubated after the RNAscope processing. As negative control a RNAscope probe detecting dapb (bacterial gene) was used, and as positive control we used the 3-Plex-positive control RNAscope probe targeting Polr2a, Ppib and Ubc (both provided by the manufacturer). Images were acquired on a confocal microscope (Zeiss LSM800) with x20/0.8 Plan-APOCHROMAT and 40x/1.30C Plan-APOCHROMAT. Quantification of the different mRNA transcripts in HB9+ nuclei were counted manually.
siRNA transfection
siRNA Universal control (SIC001, Sigma), siRNA against human PlexinD1 (siRNA ID: SASI_Hs01_00194034, Sigma) and siRNA against human Nrp1 (siRNA ID: SASI_Hs02_00307190, Sigma) were used to transfect HBMECs. Transfection of HBMECs (12 × 104 cells) in 6-well plates (657-160, Greiner Cell Star) with siRNA (final concentration of 200 nM) was performed with Oligofectamine according to the transfection protocol (12252011, ThermoFisher). Briefly, HBMECs were transfected in Opti-MEM reduced serum medium (51985034, ThermoFisher) for 4 h. Afterward, cells were washed and cultured in Endopan 3 complete medium for 24 h and further used in the tube-touching assay or for RNA extraction to verify successful PlexinD1 or Nrp1 knockdown.
Spinal cord MN explant dissection
Dissection of MN explants from embryonic spinal cords was performed as previously described.7 Briefly, spinal cords from E11.5 embryos were dissected on L15 medium (L5520, Sigma) containing 5% horse serum, 50 U ml−1 of penicillin and 50 μg mL−1 of streptomycin via open-book preparation.75 Spinal cords were flattened and MN columns were isolated and cut in explants with approximately 1 mm of size under a stereomicroscope. Due to the technical procedure followed to dissect and prepare these explants, neural progenitors can also be present in the generated explants. To reduce the impact of it, in the experiments using explants from WT embryos (co-cultured with HBMECs-PlexinD1-KD or Ctrl; and HBMECs-Nrp1-KD and Ctrl), explants deriving from the same embryo/dissection have been used in both the KD and Ctrl conditions.
Tube-touching assay
Tube-touching assays were performed in μ-Slide Angiogenesis wells (81506, ibidi GmbH) using 10 μL of Corning Matrigel matrix (356234, Corning) per well and allowed to polymerize at 37°C for 30 min 50 μL of HBMECs suspension (in HBMECs culture medium without VEGF, FGF-2 and FBS (= starving medium)) containing 1x104 cells and MN explants was added per well (3–4 explants per well). Cells together with the explants were incubated in a humidified chamber at 37°C, 5% CO2 for 16-18 h. Images were acquired using a microscope Zeiss Axiovert 200 M with 5x/0,16 EC Plan-NEOFLUAR Objective. The tubes touching the explant were counted manually and quantification was done with investigators blinded to the experimental condition.
Quantification of blood vessel density and ingression angles
Spinal cord vascularization analysis was performed at the brachial and thoracic levels for all lines except for Sema3E WT and KO embryos, in which only the brachial level was considered (as Sema3E is only expressed at brachial level). Total spinal cord vascularization was calculated as the area covered by IsoB4 staining per spinal cord area (excluding floor plate and ventricle). MN column vascularization was determined as the percentage of the area covered by IsoB4 staining in the MN column area (occupied by Isl1/2+ cells). Quantifications were performed using the NIH ImageJ software. Blood vessel ingression was analyzed as previously described,6 whereby the floor plate was taken as reference with the value of 0°. Using the Angle Tool of NIH ImageJ software, the angle of BVs ingressing from the PNVP into the spinal cord was measured. Quantification of the vessel length was performed using the software “Angiotool.”74
Analysis of MEP position, quantification of thickness of MN axon bundle at MEP and measure of ventral root diameter
The position of MEPs was measured by quantifying the circumferential distance from the ventral midline to the most ventral bundle at exit points using neurofilament M staining. Similarly, the circumferential distance from the ventral midline to the average position of all of the exiting motor axon bundles was quantified using neurofilament M staining. To normalize for embryo size, the distance was divided by the embryo height (the distance between the dorsal-ventral midlines) as previously described.76 Thickness of MN axon bundles at MEP was quantified by measuring the distance between the most ventral and most dorsal axon bundle (labeled with neurofilament M) leaving the spinal cord. Ventral root diameter was quantified in 300 μm vibratome sections using NIH ImageJ software. Maximum intensity projections were created from confocal z-stacks containing the entire ventral root. The diameter of the ventral root was measured at a distance of 30 μm from the MEP.
Analysis of MN number, soma size, and synaptic input density
At E18.5, MNs were detected by staining for HB9. To determine the total number of MNs, HB9+ cells located in the ventral spinal cord were manually counted with ImageJ. Quantification of soma size was performed by measuring the area of HB9+/ChAT+ cells with ImageJ. For counting the total area occupied by the input synapsis (density), the VAChT + and VGAT+ staining area overlapping the MNs was measured and normalized for the respective MN soma area.
In silico analysis
Previously published single-cell RNA sequencing data from the embryonic spinal cord44 were downloaded from ArrayExpress (accession number E-MTAB-7320) and analyzed using R (version 4.0.2; http://www.R-project.org/). Firstly, selection of the ventral neural cells and ECs was performed: ECs were extracted from the cluster identified as “Blood” by identifying the cells expressing the commonly accepted EC markers Cdh5, Pecam1 and Kdr, while the ventral neural cells were already identified. To predict the cell-cell communication between ECs and ventral neural cells, we used the recently developed R toolkit Cellchat.48 The dataset was analyzed using the pipeline published by Jin et al. 202148 (github.com/sqjin/CellChat) with minor changes: instead of using the default method for the computation of the communication probability, we selected the method type “truncatedMean” with “trim = 0.07”, as the previously described Vegf-Kdr interaction in developing motor neurons7 was not predicted. The output results from the CellChat analysis are available in Table S1. For the plot of Sema3C, PlexinD1 and Nrp1 expression (Figures 1F, 1G, S1J, and S2C) we used the original pipeline of Delile et al.44 with a minor change: no threshold in the minimum number of cells expressing the genes was applied.
Quantification and statistical analysis
All results are plotted as the mean for each group, and all include error bars that represent means ± SD. For each experiment at least two independent experiments (or litters) were analyzed. Statistical significances between two groups were calculated using parametric two tailed unpaired Student’s t test. One-way ANOVA followed by Sidak’s multiple comparisons was used to determine the statistical significances of three or more groups. Outlier detection has been performed using the Outlier calculator (GraphPad Prism) with an Alpha = 0.05 significance level. Statistically significant results are indicated in the figures using ∗p < 0.05, ∗∗p < 0.01 ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001. Sample sizes are provided in the figure legends.
Analyses were performed blinded to the experimental conditions. The statistical analysis was performed using GraphPad Prism (version 7.0)
Acknowledgments
We thank Ruiz de Almodóvar’s lab for constructive discussions. We thank veterinarians and animal care takers at Heidelberg University and at UCL Institute of Ophthalmology for their support. We thank S. Offermanns (MPI for hearth lung research, Bad Nauenheim, Germany) for his support during the revision of the study. This project was supported by DFG grants from SFB1366 (“Vascular control of organ function”, project number 394046768-SFB 1366 to C.R.d.A. and A.F.) and SFB1158 (“From nociception to chronic pain: structure-function properties of neural pathways and their reorganization” to C.R.d.A.); the state of Baden-Württemberg Foundation special program “Angioformatics single cell platform” (to C.R.d.A.); and British Heart Foundation fellowships to E.I. (FS/19/29/34367) and A.N.A. (FS/14/67/31294). C.R.d.A. is supported by European Research Council consolidator grant (ref. 864875). D.B. was supported by European Research Council Starting Grant 335590 and Giovanni Armenise-Harvard Foundation Career Development Award. C.R. was supported by a Wellcome Investigator award (ref. 205099/Z/16/Z). F.B. is a member of the Nordrhein-Westfalen (NRW) network iBehave. C.R.d.A. and F.B. are members of the excellence cluster ImmunoSensation2. F.B. is a member of the DFG Collaborative Research Centers (SFBs) 1089 and 1158 and the DFG priority programs (SPP) 2395 and is a recipient of the Roger de Spoelberch Prize. This work was supported by the Open Access Publication Fund of the University of Bonn.
Author contributions
J.R.V. and C.R.d.A. designed the study and wrote the paper. J.R.V. performed the experimental work and most of the data collection and analysis. B.S., I.P., P.H., G.S., H.A. E.I., A.N.A., A.M., L.G., E.D., R.B., and S.D. performed experiments. J.R.V. and G.S. performed the bioinformatic analysis. J.R.V., B.S., S.D., I.P., P.H., G.S., H.A., A.F., D.B., F.B., C.R., and C.R.d.A. provided reagents/samples and participated in data discussion and interpretation. All authors provided input during the manuscript preparation. C.R.d.A. and C.R. acquired the funding and supervised the study.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of science.
Published: December 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.neuron.2022.12.005.
Supplemental information
Data and code availability
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the appropriated methods section.
-
•
This paper does not report original code. The modifications to the original codes are available upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Tam S.J., Watts R.J. Connecting vascular and nervous system development: angiogenesis and the blood-brain barrier. Annu. Rev. Neurosci. 2010;33:379–408. doi: 10.1146/annurev-neuro-060909-152829. [DOI] [PubMed] [Google Scholar]
- 2.Walchli T., Wacker A., Frei K., Regli L., Schwab M., Hoerstrup S., Gerhardt H., Engelhardt B. Wiring the vascular network with neural cues: A CNS perspective. Neuron. 2015;87:271–296. doi: 10.1016/j.neuron.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 3.Paredes I., Himmels P., Ruiz de Almodovar C. Neurovascular communication during CNS development. Dev. Cell. 2018;45:10–32. doi: 10.1016/j.devcel.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 4.Kurz H., Gartner T., Eggli P.S., Christ B. First blood vessels in the avian neural tube are formed by a combination of dorsal angioblast immigration and ventral sprouting of endothelial cells. Dev. Biol. 1996;173:133–147. doi: 10.1006/dbio.1996.0012. [DOI] [PubMed] [Google Scholar]
- 5.Cox C.M., Poole T.J. Angioblast differentiation is influenced by the local environment: FGF-2 induces angioblasts and patterns vessel formation in the quail embryo. Dev. Dyn. 2000;218:371–382. doi: 10.1002/(SICI)1097-0177(200006)218:2<371::AID-DVDY10>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.James J.M., Gewolb C., Bautch V.L. Neurovascular development uses VEGF-A signaling to regulate blood vessel ingression into the neural tube. Development. 2009;136:833–841. doi: 10.1242/dev.028845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Himmels P., Paredes I., Adler H., Karakatsani A., Luck R., Marti H.H., Ermakova O., Rempel E., Stoeckli E.T., Ruiz de Almodovar C. Motor neurons control blood vessel patterning in the developing spinal cord. Nat. Commun. 2017;8 doi: 10.1038/ncomms14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakao T., Ishizawa A., Ogawa R. Observations of vascularization in the spinal cord of mouse embryos, with special reference to development of boundary membranes and perivascular spaces. Anat. Rec. 1988;221:663–677. doi: 10.1002/ar.1092210212. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz de Almodovar C., Fabre P., Knevels E., Coulon C., Segura I., Haddick P., Aerts L., Delattin N., Strasser G., Oh W.J., et al. VEGF mediates commissural axon chemoattraction through its receptor Flk1. Neuron. 2011;70:966–978. doi: 10.1016/j.neuron.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wild R., Klems A., Takamiya M., Hayashi Y., Strahle U., Ando K., Mochizuki N., van Impel A., Schulte-Merker S., Krueger J., et al. Neuronal sFlt1 and Vegfaa determine venous sprouting and spinal cord vascularization. Nat. Commun. 2017;8 doi: 10.1038/ncomms13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuoka R.L., Marass M., Avdesh A., Helker C.S., Maischein H.M., Grosse A.S., Kaur H., Lawson N.D., Herzog W., Stainier D.Y. Radial glia regulate vascular patterning around the developing spinal cord. Elife. 2016;5:e20253. doi: 10.7554/eLife.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieira J.R., Shah B., Ruiz de Almodovar C. Cellular and molecular mechanisms of spinal cord vascularization. Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.599897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenman J.M., Rajagopal J., Carroll T.J., Ishibashi M., McMahon J., McMahon A.P. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322:1247–1250. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 14.Daneman R., Agalliu D., Zhou L., Kuhnert F., Kuo C.J., Barres B.A. Wnt/β-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:641–646. doi: 10.1073/pnas.0805165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhnert F., Mancuso M.R., Shamloo A., Wang H.T., Choksi V., Florek M., Su H., Fruttiger M., Young W.L., Heilshorn S.C., Kuo C.J. Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science. 2010;330:985–989. doi: 10.1126/science.1196554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai H.H., Niu J., Munji R., Davalos D., Chang J., Zhang H., Tien A.C., Kuo C.J., Chan J.R., Daneman R., Fancy S.P.J. Oligodendrocyte precursors migrate along vasculature in the developing nervous system. Science. 2016;351:379–384. doi: 10.1126/science.aad3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paredes I., Vieira J.R., Shah B., Ramunno C.F., Dyckow J., Adler H., Richter M., Schermann G., Giannakouri E., Schirmer L., et al. Oligodendrocyte precursor cell specification is regulated by bidirectional neural progenitor-endothelial cell crosstalk. Nat. Neurosci. 2021;24:478–488. doi: 10.1038/s41593-020-00788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tata M., Wall I., Joyce A., Vieira J.M., Kessaris N., Ruhrberg C. Regulation of embryonic neurogenesis by germinal zone vasculature. Proc. Natl. Acad. Sci. USA. 2016;113:13414–13419. doi: 10.1073/pnas.1613113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francius C., Harris A., Rucchin V., Hendricks T.J., Stam F.J., Barber M., Kurek D., Grosveld F.G., Pierani A., Goulding M., Clotman F. Identification of multiple subsets of ventral interneurons and differential distribution along the rostrocaudal axis of the developing spinal cord. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laumonnerie C., Tong Y.G., Alstermark H., Wilson S.I. Commissural axonal corridors instruct neuronal migration in the mouse spinal cord. Nat. Commun. 2015;6:7028. doi: 10.1038/ncomms8028. [DOI] [PubMed] [Google Scholar]
- 21.Briscoe J., Small S. Morphogen rules: Design principles of gradient-mediated embryo patterning. Development. 2015;142:3996–4009. doi: 10.1242/dev.129452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jessell T.M. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 23.Price S.R., Briscoe J. The generation and diversification of spinal motor neurons: Signals and responses. Mech. Dev. 2004;121:1103–1115. doi: 10.1016/j.mod.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 24.Lieberam I., Agalliu D., Nagasawa T., Ericson J., Jessell T.M. A Cxcl12-CXCR4 chemokine signaling pathway defines the initial trajectory of mammalian motor axons. Neuron. 2005;47:667–679. doi: 10.1016/j.neuron.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Bonanomi D., Pfaff S.L. Motor axon pathfinding. Cold Spring Harb. Perspect. Biol. 2010;2:a001735. doi: 10.1101/cshperspect.a001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonanomi D., Valenza F., Chivatakarn O., Sternfeld M.J., Driscoll S.P., Aslanian A., Lettieri K., Gullo M., Badaloni A., Lewcock J.W., et al. p190RhoGAP Filters Competing Signals to Resolve Axon Guidance Conflicts. Neuron. 2019;102:602–620.e9. doi: 10.1016/j.neuron.2019.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladle D.R., Pecho-Vrieseling E., Arber S. Assembly of motor circuits in the spinal cord: Driven to function by genetic and experience-dependent mechanisms. Neuron. 2007;56:270–283. doi: 10.1016/j.neuron.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Levine A.J., Lewallen K.A., Pfaff S.L. Spatial organization of cortical and spinal neurons controlling motor behavior. Curr. Opin. Neurobiol. 2012;22:812–821. doi: 10.1016/j.conb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osseward P.J., 2nd, Pfaff S.L. Cell type and circuit modules in the spinal cord. Curr. Opin. Neurobiol. 2019;56:175–184. doi: 10.1016/j.conb.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H., Xiong W.C., Mei L. To build a synapse: Signaling pathways in neuromuscular junction assembly. Development. 2010;137:1017–1033. doi: 10.1242/dev.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolodkin A.L., Matthes D.J., O'Connor T.P., Patel N.H., Admon A., Bentley D., Goodman C.S. Fasciclin IV: Ssequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron. 1992;9:831–845. doi: 10.1016/0896-6273(92)90237-8. [DOI] [PubMed] [Google Scholar]
- 32.Luo Y., Raible D., Raper J.A. Collapsin: A protein in brain that induces the collapse and paralysis of neuronalgrowth cones. Cell. 1993;75:217–227. doi: 10.1016/0092-8674(93)80064-l. [DOI] [PubMed] [Google Scholar]
- 33.Sakurai A., Doci C., Gutkind J.S. Semaphorin signaling in angiogenesis, lymphangiogenesis and cancer. Cell Res. 2012;22:23–32. doi: 10.1038/cr.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao B., Liu S., Tan X., Lu P., Wang D., Xu H. Class-3 semaphorins: Potent multifunctional modulators for angiogenesis-associated diseases. Biomed. Pharmacother. 2021;137 doi: 10.1016/j.biopha.2021.111329. [DOI] [PubMed] [Google Scholar]
- 35.Alto L.T., Terman J.R. Semaphorins and their signaling mechanisms. Methods Mol. Biol. 2017;1493:1–25. doi: 10.1007/978-1-4939-6448-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vieira J.M., Schwarz Q., Ruhrberg C. Selective requirements for NRP1 ligands during neurovascular patterning. Development. 2007;134:1833–1843. doi: 10.1242/dev.002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukushima Y., Okada M., Kataoka H., Hirashima M., Yoshida Y., Mann F., Gomi F., Nishida K., Nishikawa S.I., Uemura A. Sema3E-PlexinD1 signaling selectively suppresses disoriented angiogenesis in ischemic retinopathy in mice. J. Clin. Invest. 2011;121:1974–1985. doi: 10.1172/JCI44900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J., Oh W.J., Gaiano N., Yoshida Y., Gu C. Semaphorin 3E-Plexin-D1 signaling regulates VEGF function in developmental angiogenesis via a feedback mechanism. Genes Dev. 2011;25:1399–1411. doi: 10.1101/gad.2042011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu C., Yoshida Y., Livet J., Reimert D.V., Mann F., Merte J., Henderson C.E., Jessell T.M., Kolodkin A.L., Ginty D.D. Semaphorin 3E and plexin-D1 control vascular pattern independently of neuropilins. Science. 2005;307:265–268. doi: 10.1126/science.1105416. [DOI] [PubMed] [Google Scholar]
- 40.Torres-Vazquez J., Gitler A.D., Fraser S.D., Berk J.D., Van N.P., Fishman M.C., Childs S., Epstein J.A., Weinstein B.M. Semaphorin-plexin signaling guides patterning of the developing vasculature. Dev. Cell. 2004;7:117–123. doi: 10.1016/j.devcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Gitler A.D., Lu M.M., Epstein J.A. PlexinD1 and semaphorin signaling are required in endothelial cells for cardiovascular development. Dev. Cell. 2004;7:107–116. doi: 10.1016/j.devcel.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 42.Zygmunt T., Gay C., Blondelle J., Singh M., Flaherty K., Means P., Herwig L., Krudewig A., Belting H.G., Affolter M., et al. Semaphorin-PlexinD1 signaling limits angiogenic potential via the VEGF decoy receptor sFlt1. Dev. Cell. 2011;21:301–314. doi: 10.1016/j.devcel.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang W., Hu J., Uemura A., Tetzlaff F., Augustin H.G., Fischer A. Semaphorin-3C signals through Neuropilin-1 and PlexinD1 receptors to inhibit pathological angiogenesis. EMBO Mol. Med. 2015;7:1267–1284. doi: 10.15252/emmm.201404922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delile J., Rayon T., Melchionda M., Edwards A., Briscoe J., Sagner A. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development. 2019;146 doi: 10.1242/dev.173807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalucka J., de Rooij L.P., Goveia J., Rohlenova K., Dumas S.J., Meta E., Conchinha N.V., Taverna F., Teuwen L.A., Veys K., et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 46.Khan S., Taverna F., Rohlenova K., Treps L., Geldhof V., de Rooij L., Sokol L., Pircher A., Conradi L.C., Kalucka J., et al. EndoDB: A database of endothelial cell transcriptomics data. Nucleic Acids Res. 2019;47:D736–D744. doi: 10.1093/nar/gky997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohlenova K., Goveia J., Garcia-Caballero M., Subramanian A., Kalucka J., Treps L., Falkenberg K.D., de Rooij L.P., Zheng Y., Lin L., et al. Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metab. 2020;31:862–877.e14. doi: 10.1016/j.cmet.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pecho-Vrieseling E., Sigrist M., Yoshida Y., Jessell T.M., Arber S. Specificity of sensory-motor connections encoded by Sema3e-Plxnd1 recognition. Nature. 2009;459:842–846. doi: 10.1038/nature08000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukuhara K., Imai F., Ladle D., Katayama K.i., Leslie J., Arber S., Jessell T., Yoshida Y. Specificity of monosynaptic sensory-motor connections imposed by repellent Sema3E-PlexinD1 signaling. Cell Rep. 2013;5:748–758. doi: 10.1016/j.celrep.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Singh M.K., Degenhardt K.R., Lu M.M., Bennett J., Yoshida Y., Epstein J.A. Tie2Cre-mediated inactivation of plexinD1 results in congenital heart, vascular and skeletal defects. Dev. Biol. 2009;325:82–93. doi: 10.1016/j.ydbio.2008.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kisanuki Y.Y., Hammer R.E., Miyazaki J.i., Williams S., Richardson J.A., Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 53.Plein A., Calmont A., Fantin A., Denti L., Anderson N.A., Scambler P.J., Ruhrberg C. Neural crest-derived SEMA3C activates endothelial NRP1 for cardiac outflow tract septation. J. Clin. Invest. 2015;125:2661–2676. doi: 10.1172/JCI79668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dessaud E., Yang L.L., Hill K., Cox B., Ulloa F., Ribeiro A., Mynett A., Novitch B.G., Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 55.Gay C.M., Zygmunt T., Torres-Vazquez J. Diverse functions for the semaphorin receptor PlexinD1 in development and disease. Dev. Biol. 2011;349:1–19. doi: 10.1016/j.ydbio.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen S., Funkelstein L., Livet J., Rougon G., Henderson C.E., Castellani V., Mann F. A semaphorin code defines subpopulations of spinal motor neurons during mouse development. Eur. J. Neurosci. 2005;21:1767–1776. doi: 10.1111/j.1460-9568.2005.04021.x. [DOI] [PubMed] [Google Scholar]
- 57.Huber A.B., Kania A., Tran T.S., Gu C., De Marco Garcia N., Lieberam I., Johnson D., Jessell T.M., Ginty D.D., Kolodkin A.L. Distinct roles for secreted semaphorin signaling in spinal motor axon guidance. Neuron. 2005;48:949–964. doi: 10.1016/j.neuron.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Catela C., Chen Y., Weng Y., Wen K., Kratsios P. Control of spinal motor neuron terminal differentiation through sustained Hoxc8 gene activity. Elife. 2022;11:e70766. doi: 10.7554/eLife.70766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel T., Hammelman J., Closser M., Gifford D.K., Wichterle H. General and cell-type-specific aspects of the motor neuron maturation transcriptional program. bioRxiv. 2021 doi: 10.1101/2021.03.05.434185. [DOI] [Google Scholar]
- 60.Halder N., Lal G. Cholinergic system and its therapeutic importance in inflammation and autoimmunity. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.660342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blusztajn J.K., Wurtman R.J. Choline and cholinergic neurons. Science. 1983;221:614–620. doi: 10.1126/science.6867732. [DOI] [PubMed] [Google Scholar]
- 62.Vasudevan A., Long J.E., Crandall J.E., Rubenstein J.L.R., Bhide P.G. Compartment-specific transcription factors orchestrate angiogenesis gradients in the embryonic brain. Nat. Neurosci. 2008;11:429–439. doi: 10.1038/nn2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi T., Takase Y., Yoshino T., Saito D., Tadokoro R., Takahashi Y. Angiogenesis in the developing spinal cord: Blood vessel exclusion from neural progenitor region is mediated by VEGF and its antagonists. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meadows S.M., Fletcher P.J., Moran C., Xu K., Neufeld G., Chauvet S., Mann F., Krieg P.A., Cleaver O. Integration of repulsive guidance cues generates avascular zones that shape mammalian blood vessels. Circ. Res. 2012;110:34–46. doi: 10.1161/CIRCRESAHA.111.249847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mehta V., Pang K.L., Rozbesky D., Nather K., Keen A., Lachowski D., Kong Y., Karia D., Ameismeier M., Huang J., et al. The guidance receptor plexin D1 is a mechanosensor in endothelial cells. Nature. 2020;578:290–295. doi: 10.1038/s41586-020-1979-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suter T.A.C.S., Jaworski A. Cell migration and axon guidance at the border between central and peripheral nervous system. Science. 2019;365:eaaw8231. doi: 10.1126/science.aaw8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martins L.F., Brambilla I., Motta A., de Pretis S., Bhat G.P., Badaloni A., Malpighi C., Amin N.D., Imai F., Almeida R.D., et al. Motor neurons use push-pull signals to direct vascular remodeling critical for their connectivity. Neuron. 2022;110 doi: 10.1016/j.neuron.2022.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolpowitz D., Mason T.B., Dietrich P., Mendelsohn M., Talmage D.A., Role L.W. Cysteine-rich domain isoforms of the neuregulin-1 gene are required for maintenance of peripheral synapses. Neuron. 2000;25:79–91. doi: 10.1016/s0896-6273(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 69.Brandon E.P., Lin W., D'Amour K.A., Pizzo D.P., Dominguez B., Sugiura Y., Thode S., Ko C.P., Thal L.J., Gage F.H., Lee K.F. Aberrant patterning of neuromuscular synapses in choline acetyltransferase-deficient mice. J. Neurosci. 2003;23:539–549. doi: 10.1523/jneurosci.23-02-00539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hellal F., Hurtado A., Ruschel J., Flynn K.C., Laskowski C.J., Umlauf M., Kapitein L.C., Strikis D., Lemmon V., Bixby J., et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science. 2011;331:928–931. doi: 10.1126/science.1201148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruschel J., Bradke F. Systemic administration of epothilone D improves functional recovery of walking after rat spinal cord contusion injury. Exp. Neurol. 2018;306:243–249. doi: 10.1016/j.expneurol.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 72.Ruschel J., Hellal F., Flynn K.C., Dupraz S., Elliott D.A., Tedeschi A., Bates M., Sliwinski C., Brook G., Dobrindt K., et al. Systemic administration of epothilone B promotes axon regeneration after spinal cord injury. Science. 2015;348:347–352. doi: 10.1126/science.aaa2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thaler J., Harrison K., Sharma K., Lettieri K., Kehrl J., Pfaff S.L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron. 1999;23:675–687. doi: 10.1016/s0896-6273(01)80027-1. [DOI] [PubMed] [Google Scholar]
- 74.Zudaire E., Gambardella L., Kurcz C., Vermeren S. A computational tool for quantitative analysis of vascular networks. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Langlois S.D., Morin S., Yam P.T., Charron F. Dissection and culture of commissural neurons from embryonic spinal cord. J. Vis. Exp. 2010 doi: 10.3791/1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim M., Fontelonga T.M., Lee C.H., Barnum S.J., Mastick G.S. Motor axons are guided to exit points in the spinal cord by Slit and Netrin signals. Dev. Biol. 2017;432:178–191. doi: 10.1016/j.ydbio.2017.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the appropriated methods section.
-
•
This paper does not report original code. The modifications to the original codes are available upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.