Abstract
Background
Identification of those at risk of more severe psoriasis and/or associated morbidities offers opportunity for early intervention, reduced disease burden and more cost‐effective healthcare. Prognostic biomarkers of disease progression have thus been the focus of intense research, but none are part of routine practice.
Objectives
To identify and catalogue candidate biomarkers of disease progression in psoriasis for the translational research community.
Methods
A systematic search of CENTRAL, Embase, LILACS and MEDLINE was performed for relevant articles published between 1990 and December 2021. Eligibility criteria were studies involving patients with psoriasis (any age, n ≥ 50) reporting biomarkers associated with disease progression. The main outcomes were any measure of skin severity or any prespecified psoriasis comorbidity. Data were extracted by one reviewer and checked by a second; studies meeting minimal quality criteria (longitudinal design and/or use of methods to control for confounding) were formally assessed for bias. Candidate biomarkers were identified by an expert multistakeholder group using a majority voting consensus exercise, and mapped to relevant cellular and molecular pathways.
Results
Of 181 included studies, most investigated genomic or proteomic biomarkers associated with disease severity (n = 145) or psoriatic arthritis (n = 30). Methodological and reporting limitations compromised interpretation of findings, most notably a lack of longitudinal studies, and inadequate control for key prognostic factors. The following candidate biomarkers with future potential utility were identified for predicting disease severity: LCE3D, interleukin (IL)23R, IL23A, NFKBIL1 loci, HLA‐C*06:02 (genomic), IL‐17A, IgG aHDL, GlycA, I‐FABP and kallikrein 8 (proteomic), tyramine (metabolomic); psoriatic arthritis: HLA‐C*06:02, HLA‐B*27, HLA‐B*38, HLA‐B*08, and variation at the IL23R and IL13 loci (genomic); IL‐17A, CXCL10, Mac‐2 binding protein, integrin b5, matrix metalloproteinase‐3 and macrophage‐colony stimulating factor (proteomic) and tyramine and mucic acid (metabolomic); and type 2 diabetes mellitus: variation in IL12B and IL23R loci (genomic). No biomarkers were supported by sufficient evidence for clinical use without further validation.
Conclusions
This review provides a comprehensive catalogue of investigated biomarkers of disease progression in psoriasis. Future studies must address the common methodological limitations identified herein to expedite discovery and validation of biomarkers for clinical use.
What is already known about this topic?
The current treatment paradigm in psoriasis is reactive.
There is a need to develop effective risk‐stratified management approaches that can proactively attenuate the substantial burden of disease.
Prognostic biomarkers of disease progression have therefore been the focus of intense research.
What does this study add?
This review is the first to scope, collate and catalogue research investigating biomarkers of disease progression in psoriasis.
The review identifies potentially promising candidate biomarkers for further investigation and highlights common important limitations that should be considered when designing and conducting future studies in this area.
This review is the first to scope, collate, and catalogue research investigating biomarkers of disease progression in psoriasis. The review identifies potentially promising candidate biomarkers for further investigation and highlights common important limitations that should be considered when designing and conducting future studies in this area.
Plain language summary available online
Psoriasis is a common chronic inflammatory disease estimated to affect at least 60 million people globally. 1 , 2 It is multifactorial in origin, caused by interplay between a strong genetic component, the immune system and environmental risk factors. 3 Skin involvement varies in extent, severity and course, and is associated with major negative impact on quality of life and social and psychological wellbeing. The associated increased risk of comorbidities including psoriatic arthritis (PsA), cardiometabolic syndrome and depression, 4 adds to this burden especially in those with severe disease.
Effective treatment of psoriasis is now possible with the advent of multiple targeted biologics. The current treatment paradigm is reactive but there are compelling reasons to adopt a more proactive, risk‐stratified approach. Early intervention with effective treatment for psoriasis reduces cumulative impact and, by reducing the inflammatory burden, may also reduce or prevent cardiometabolic disease. Equally importantly, predicting those at risk of comorbidities may facilitate more targeted implementation of primary preventative measures and/or expedite diagnosis, especially where early treatment can prevent irreversible damage and disability as is the case for PsA. 5
A biomarker is a defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions. 6 Their use is well established in many disciplines including genetics, infection, cancer, cardiovascular disease and immune‐mediated diseases such as rheumatoid arthritis. 7 , 8 In contrast, there are no biomarkers that are part of routine clinical care to identify those at risk of disease progression in psoriasis, despite the clear clinical need, and sustained and intense research effort. Improvements in high‐throughput biological assays and computational technologies have driven an increase in biomarker discovery with consequent need to develop effective strategies for selection, validation and implementation (http://www.psort.org.uk). This explosion of effort also mandates synergized efforts to collate high‐quality evidence to avoid future research waste.
The overall aim of this review is therefore to scope, collate and catalogue the research investigating biomarkers of potential clinical utility in psoriasis. The first of these reviews focuses on biomarkers of disease progression and a second review will focus on treatment response. The specific aims of this review are to (i) identify and catalogue studies relating to biomarkers of disease progression in psoriasis as defined by disease severity, and/or development of comorbidities, (ii) select and functionally map any biomarkers for which there is evidence for prognostic value and (iii) evaluate study quality and highlight limitations to inform future biomarker research.
Materials and methods
This scoping review was performed by a multistakeholder group drawn from a large multidisciplinary European consortium with academic and industry partners, Biomarkers in Atopic Dermatitis and Psoriasis (https://www.biomap‐imi.eu), and the International Psoriasis Council (www.psoriasiscouncil.org). We included clinical‐academic dermatologists (10), a patient representative, immunology/genetic scientists with expertise in genetics (two), immunology (four) and bioinformatics (four), systematic reviewers (two), and an information specialist. Preliminary work was performed to inform the study design and is detailed in Section 1 of Appendix S1 (see Supporting Information).
Identification and cataloguing of studies of disease progression biomarkers (stage 1)
Literature searches
A single strategy (Appendix S1, Section 7) was used to search for both studies of biomarkers of progression and biomarkers of treatment response (reported separately). Electronic searches were performed in Cochrane Central Register of Controlled Trials (CENTRAL), Embase, LILACS and MEDLINE on 7 December 2021 for studies in the English language, published between 1990 (chosen because this heralded Human Genome Project start date) and December 2021 (K.W.).
Study selection
Criteria for study inclusion were established prior to study selection (Table 1). While a longitudinal study design is required to evaluate the prognostic potential of any given biomarker, we chose to also include studies with a cross‐sectional design given our overarching aim to encompass the breadth of biomarker research. Titles and abstracts were single‐screened by one reviewer, with an independent second screening where requested (e.g. where there was uncertainty regarding eligibility) (M.C., R.R., I.A.B., J.S., M.V., S.H., S.R.). To assess the accuracy of our screening approach, every tenth excluded abstract was independently checked (500 in total) by a second screener (R.R.). From this list, full texts were screened by one reviewer, with decisions (inclusion/exclusion) checked by a second; any disagreements were resolved by consensus or through discussion with a senior member of the team (M.C., D.M.).
Table 1.
Eligibility criteria for the scoping review
| Review component | Criteria |
|---|---|
| Population | People with psoriasis, regardless of past or current treatments were included |
| Interventions | Genomic, epigenomic, transcriptomic, proteomic, cellular, microbiomic and metabolomic biomarkers were included |
| Physiological or radiographic biomarkers were excluded | |
| Comparators | Studies could be of one or more biomarkers |
| Outcomes | Eligible psoriasis severity outcomes: |
| • Mild: as defined by objectively validated scoring measures (e.g. PGA clear/almost clear/mild, BSA < 2%, PASI < 2) or health care utilization (e.g. primary care management only, use of topical treatment only) | |
| • Severe: as defined by objectively validated scoring measures (e.g. PGA moderate/severe, BSA > 10%, PASI > 10), erythroderma, development of phenotypes with major impact (e.g. face, scalp, genitalia, palms, soles) or healthcare utilization (e.g. referral to secondary care, hospital admissions, use of systemic treatment) | |
| • Severity outcomes defined only as mild, moderate or severe, or reported only using continuous data were also eligible | |
| Eligible comorbidity outcomes: | |
| • Immune‐mediated inflammatory disease: rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, coeliac disease | |
| • Cardiometabolic syndrome: metabolic syndrome or its components (obesity, hyperlipidaemia, hypertension, diabetes mellitus), cardiovascular disease (ischaemic heart disease, stroke), liver disease (liver fibrosis, liver cirrhosis). Results relating to components of cardiometabolic syndrome which were reported on a continuous scale without further classification were also eligible | |
| • PsA: axial/ spondyloarthropathy, oligoarticular, polyarticular, arthritis mutilans, dactylitis, enthesitis | |
| • Psychiatric outcomes: anxiety, depression | |
| • Death | |
| Study designs | Reviews and studies including fewer than 50 participants (excluding any healthy controls or other participants who did not have psoriasis) were excluded |
| All other study designs were eligible providing they compared outcomes of patients with the biomarker with those of patients without the biomarker, or compared outcomes associated with different biomarker levels. Cross‐sectional and case–control studies were eligible for inclusion, regardless of the temporality of the possible association between biomarker and outcome |
PGA, Physician’s Global Assessment; BSA, body surface area; PASI, Psoriasis Area and Severity Index; PsA, psoriatic arthritis.
Data extraction and cataloguing
A minimal dataset (design, population characteristics, biomarkers, outcome measures and basic result details) was defined by the multistakeholder group following review of pilot data extraction from a sample of studies. Data were extracted (R.R., M.C.) then cross‐checked by another researcher, and discrepancies were resolved by discussion (M.C., D.M., R.R.). For each biomarker type (genomic, transcriptomic, proteomic, metabolomic, microbiomic, cellular and mixed), study details were presented in structured tables subdivided by biomarker function (M.C., D.M.) using an informal classification. Studies meeting minimal study design quality criteria (longitudinal studies, cross‐sectional studies with methods to control for confounding) underwent detailed review (stage 2).
Subset of studies undergoing additional data extraction and quality assessment of studies (stage 2)
Additional data were extracted on psoriasis clinical subtype, treatment history, study design and detailed results (including size and variance of effect estimates) (M.C., D.M., R.R.).
Quality assessment data were extracted by one researcher and checked by another (M.C., D.M., R.R.) with reference to domains within BIOCROSS 9 and QUIPS, quality assessment tools specifically designed for evaluation of biomarker/prognostic studies, to quality assess studies in stage 2. 10 Studies that adjusted for both sex and age of disease onset (or age) 11 (considered by the group to be the two most important prognostic factors to control for in order to avoid a high risk of confounder bias) were considered to be at ‘low or moderate risk of bias’; all other studies were classified as ‘high risk of bias’. Other potential prognostic factors adjusted or controlled for in individual studies were detailed in the summary tables in Appendix S1 (Section 3).
Other study quality assessment criteria evaluated included levels of attrition (losses to follow‐up) and adequacy of imputation of missing data in longitudinal studies, adequacy of outcome measurement/assessment, evidence of selective outcome reporting and adjustment for multiple statistical testing. 12 Further details on quality assessment strategy are described in Appendix S1 (Section 4).
Selection of candidate biomarkers for cellular and molecular pathway mapping (stage 3)
Given the breadth and heterogeneity of studies reviewed in stage 2, we then selected biomarkers for cellular and molecular pathway mapping to aid interpretation of findings and to direct future research (candidate biomarkers) based on consensus majority of the multistakeholder group (see Appendix S1, Section 5 for details).
A biomarker‐based ‘disease map’ was built to represent mechanistic and associative links of the candidate biomarkers to psoriasis pathogenesis (methodology detailed in Section 6 of Appendix S1) and significantly enriched biological processes were highlighted.
Results
Overview of all included studies (stage 1)
Following title and abstract screening, the full texts of 246 studies were sought, of which 181 studies met the review eligibility criteria (Figure 1; see Appendix S1 for included/excluded studies). On checking every tenth excluded abstract (n = 500), none were considered incorrectly excluded, adding validity to the accuracy of the chosen screening approach. Of the 181 studies included, nine had a longitudinal design, 170 were case–control or cross‐sectional and two were meta‐analyses. No biomarkers were found to be evaluated in formal trials for the purpose of clinical use.
Figure 1.
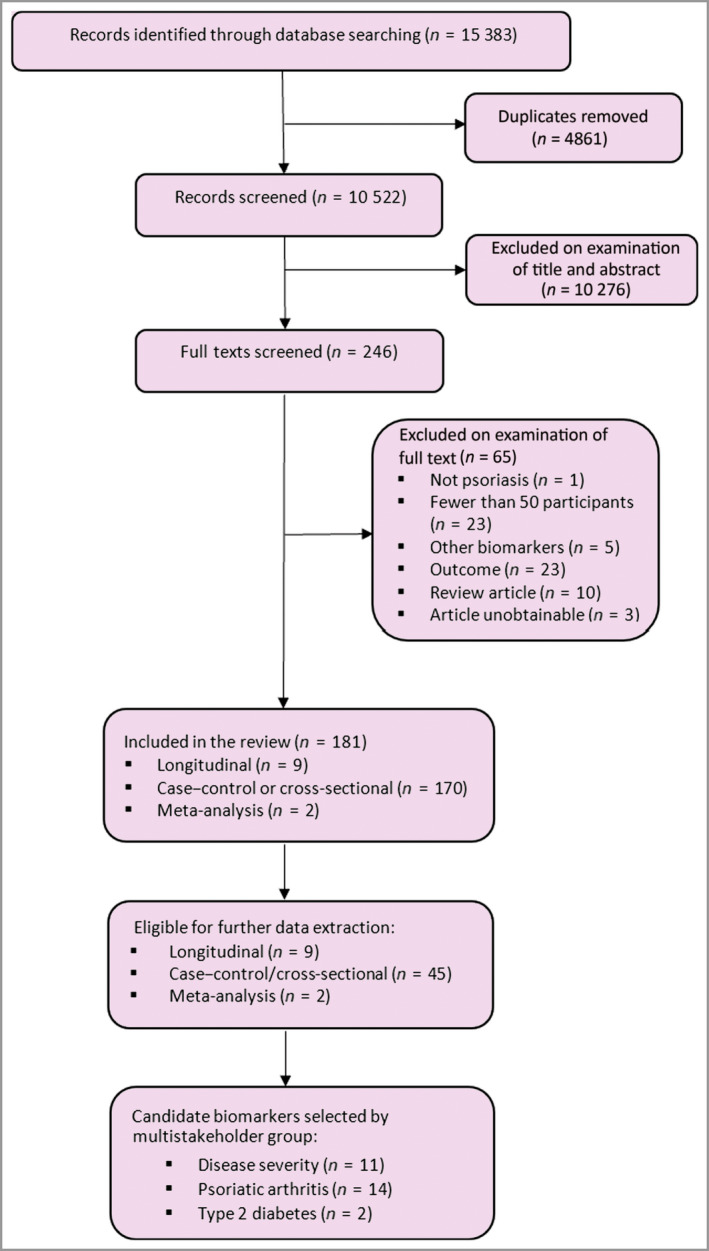
PRISMA flowchart showing the number of studies identified and eligible for inclusion. [Colour figure can be viewed at wileyonlinelibrary.com]
Most studies were published in the last decade (only 17 studies were published before 2010). The evidence‐base was dominated by studies of proteomic biomarkers (49% of the total), although – per study – the genomic biomarker studies evaluated more biomarkers and recruited more patients than the proteomic biomarker studies (Table 2). Only one study examining microbiomic biomarkers met the review’s eligibility criteria. 13 A full description of study characteristics, categorized by biomarker type, is reported in Appendix S1 (Section 2).
Table 2.
Summary characteristics of studies, overall and by type of biomarker
| Type of biomarker evaluated | Totals, all biomarkers | |||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | Genomic | Transcriptomic | Proteomic | Metabolomic | Microbiomic | Cellular | Mixed | |
| No. of included studies (% of total) | 41 (23%) | 7 (4%) | 88 (49%) | 8 (4%) | 1 (1%) | 7 (4%) | 29 (16%) | 181 |
| Mean/median biomarkers per study | 20/3 | 16/1 | 8/2 | 53/1 | N/A | 3/2 | 9/5 | 13/2 |
| Mean/median no. of psoriasis patients | 1305/398 | 76/73 | 235/74 | 89/88 | N/A | 161/96 | 130/72 | 371/92 |
| No. of studies evaluated further a (% of studies in category) | 19 (46%) | 2 (29%) | 28 (31%) | 3 (38%) | 0 (0%) | 3 (43%) | 6 (21%) | 61 (34%) |
| Candidate biomarkers (% of total) | 10 (45%) | 0 | 10 (45%) | 2 (9%) | 0 | 0 | 0 | 22 |
Studies which were eligible for further data extraction and quality assessment.
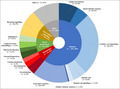
For any given biomarker, supportive evidence of association with outcomes of interest was not seen across multiple data types (e.g. genome, transcriptome, proteome). C‐reactive protein (CRP) was evaluated in 23 studies, one or more interleukins were evaluated in 19 studies, tumour necrosis factor in 18 studies, and HLA‐C*06:02 in 17 studies. Biomarkers subject to investigation covered a broad range of biological functions (Figure 2), although the majority related to immune processes.
Figure 2.
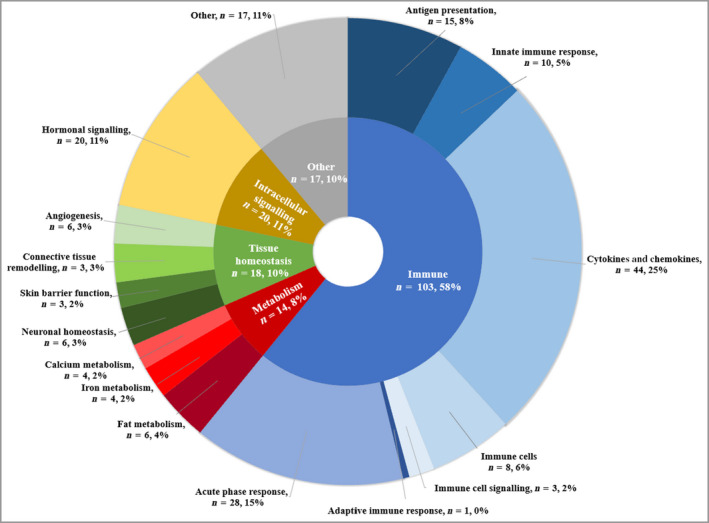
Primary functions of biomarkers in all included studies. Categories of biomarker function were devised using an informal classification, designed to capture the breadth of biomarker function in included studies. Segments represent the number of biomarker studies examining biomarkers with a given primary function (n). Studies examining multiple biomarkers which have more than one function or single biomarkers with multiple functions may be represented in more than one segment of the ring chart. [Colour figure can be viewed at wileyonlinelibrary.com]
Characteristics of studies that underwent detailed data extraction and quality assessment (stage 2)
Overall, 61 studies fulfilled the criteria for further evaluation and quality assessment (Appendix S1, Section 3), but only nine had a longitudinal design (the ideal study design). A total of 11 studies had sample sizes of more than 1000, two of which were genome‐wide meta‐analyses of several studies investigating genetic biomarkers predictive of the presence of PsA. 14 , 15 More genomic biomarker studies were eligible for detailed data extraction than proteomic biomarker studies (i.e. they used methods to control for confounding more often).
Across all biomarker categories, data on key patient cohort characteristics were often not reported. Where reported, the mean ages of the psoriasis cohorts were mostly in the range of 40–60 years. Additionally, 31 studies (51%) did not report on duration of psoriasis. Mean Psoriasis Area and Severity Index (PASI) varied from < 5 to > 30. In studies where ethnicity details were reported, most studies involved participants of white ethnicity.
Most studies reported disease severity and/or PsA (54 studies) as outcomes, with few studies presenting results for the other eligible comorbidity outcomes. Eight studies reported comorbidity outcomes related to at least one of the components of metabolic syndrome. As expected, there was variation across studies in the way outcomes were reported. For disease severity outcomes, the proteomic biomarker studies tended to report results based only on continuous data (59%) and often reported correlation coefficients, when compared with the genomic studies, which tended to report results based on categorical outcome measures (continuous results data alone were reported in 30% of studies).
There was a high degree of variability in approach used for disease classification. Across studies where categorical disease severity data were reported, different severity thresholds were used, e.g. ‘mild’ psoriasis may be classed as PASI < 5 in one study but as PASI < 10 in another. For PsA outcomes, only four studies reported using methods to reduce the possibility that some patients with psoriasis may have had undeveloped PsA (which might bias the result towards no biomarker association). 16 , 17 , 18 , 19 Furthermore, details on past or current treatment use were generally not well reported.
Quality assessment (stage 2)
Quality assessment of studies revealed at least one type of bias for every study (Appendix S1, Sections 3 and 4). Overall, 55 studies (93%) adjusted for prognostic factors by their methods of analysis, three studies (5%) matched patients at the design stage, and three studies (5%) used both methods to control for prognostic factors. Only 14 studies adjusted for both key prognostic factors, so the possibility of bias arising from confounding cannot be ruled out in most studies. For studies examining multiple biomarkers, analyses were also frequently limited by the lack of adjustment of results for multiple hypothesis testing. Overall, the quality assessment findings indicated that the results of all included studies should be interpreted with caution.
Candidate biomarker associations (stage 3)
Biomarker‐outcome associations examined in multiple studies often showed conflicting results, with studies frequently addressing study quality issues in a different manner and to a varying extent. A consensus was reached on 22 different candidate biomarkers (27 biomarker‐outcome associations) for psoriasis progression based on the evidence available (Table 3).
Table 3.
Summary details of studies examining candidate biomarkers of psoriasis progression
| Study/number of psoriasis patients | Biomarker type/study design | Biomarker(s) examined | Number of key prognostic factors adjusted for | Outcome(s)/key results |
|---|---|---|---|---|
| HLA‐C*06:02 | ||||
| Douroudis et al. 29 | Genomic/case–control | 1: HLA‐C*06:02 | 2: Age, sex | PsA and hypertension: hypertension (OR 0·73, 95% CI 0·61–0·88) and PsA (OR 0·68, 95% CI 0·58–0·80) were significantly associated with HLA‐C*06:02‐negative status in fully adjusted model |
| N = 9286 | ||||
| Eder et al. 29 | Genomic/case–control | 10: HLA alleles | 1: Age of onset | PsA: HLA‐C*06 was less frequent in the PsA group than in the PsC group (OR 0·58, P < 0·001). This result remained significant in the early‐onset psoriasis subgroup but not the late‐onset subgroup |
| N = 1047 | ||||
| Gudjonsson et al. 27 | Genomic/cross‐sectional | 1: HLA‐Cw*0602 | 1: Age of onset | Severity: significant association between Cw*0602 positive status and disease severity (P < 0·001). Late onset in patients who were Cw*0602‐positive was associated with less severe psoriasis compared with those with early onset (P = 0·0028, r = −0·13) |
| N = 1019 | ||||
| Gudjonsson et al. 25 | Genomic/cross‐sectional | 1: HLA‐Cw6 allele | No review‐specified key prognostic factors controlled for | Severity: mean disease severity score was higher in the Cw6+ group (1·67 vs. 1·45, P = 0·005). The difference between the groups increased when excluding patients with PsA (1·62 vs. 1·27, P < 0·001). No results reported for severity scoring based only on the extent of the skin lesions. Mean age at onset was 17·2 years in the Cw6+ group compared with 24·5 years in the Cw6− group |
| N = 369 | PsA: no significant difference between groups, P = 0·135 | |||
| Julia et al. 26 | Genomic/ Case–control | 32: loci with previous genome‐wide evidence of association with psoriasis | 1: Age of onset | Severity: HLA‐C‐positive status associated with severe disease. OR 1·29 (95% CI 1·11–1·51, P = 0·033 after correction for multiple testing) |
| N = 2005 | PsA: HLA‐C‐positive status was significantly associated with PsC in comparison with PsA (P = 1·69 × 10−6) | |||
| Suomela et al. 20 | Genomic/cross‐sectional | 2: HLA‐Cw6, HCR (CCHCR1) | 2: Sex, age of onset | Severity: statistically significant lower PASI in patients who were Cw6+ compared with Cw6− (P < 0·007) but mean PASI were not reported |
| N = 379 | ||||
| HLA‐B*27, HLA‐B3*8 & HLA‐B*08 | ||||
| Eder et al. 28 | Genomic/case–control | 10: HLA alleles | 1: Age of onset | PsA: Multivariate regression analysis results. Three HLA alleles more frequent in PsA than in psoriasis: HLA‐B*27 (OR 5·17, P < 0·001), HLA‐B*08 (OR 1·61, P = 0·009) and HLA‐B*38 (OR 1·65, P = 0·026) |
| N = 1047 | ||||
| IL12B, IL23A & IL23R | ||||
| Eiris et al. 31 | Genomic/cross‐sectional | 5: IL12B, IL23R and IL23A polymorphisms | No review‐specified key prognostic factors controlled for | Severity: IL23R rs11209026 GG vs. AG + AA. GG associated with severity, OR 2·11 (95% CI 1·13–3·95), P = 0·02 |
| N = 405 | PsA: IL23R rs11209026 GG vs. AG + AA. OR 2·77 (95% CI 1·15–6·68) | |||
| Type 2 diabetes: significant associations with IL12B rs6887695‐CC (OR 2·90, 95% CI 1·09–7·69, P = 0·03), IL12B rs3212227‐CC (OR 5·90, 95% CI 1·35–25·73, P = 0·035) and IL23R rs2201841‐GG (OR 2·69, 95% CI 1·09–6·66, P = 0·027). BMI reported as confounding the rs6887695 result. No other significant diabetes associations | ||||
| Nikamo et al. 32 | Genomic/case–control | 20: Genes involved in the IL‐23 and NF‐κB signalling pathway | 2: Sex, age of onset | Severity: after multiple testing correction – common IL‐23R (rs7530511) allele associated with disease severity – OR 0·71 (95% CI 0·57 to 0·88, P = 0·002). IL‐23A (rs2066808) [OR 0·53 (95% CI 0·37 to 0·76, P < 0·001)] and IL‐23R (rs2201841) [OR 1·26 (95% CI 1·07 to 1·48, P = 0·006)] |
| N = 1411 | ||||
| LCE3D | ||||
| Julia et al. 26 | Genomic/case–control | 32: Loci with previous genome‐wide evidence of association with psoriasis | 1: Age of onset | Severity: having two copies of the LCE3D risk allele was associated with moderate‐to‐severe disease, OR1·38 (95% CI 1·19–1·59, Pc = 0·0005) |
| N = 2005 | ||||
| NFKBIL1 | ||||
| Nikamo et al. 32 | Genomic/case–control | 20: Genes involved in the IL‐23 and NF‐κB signalling pathway | 2: Sex, age of onset | Severity: minor NFKBIL1 allele associated with severe disease: OR 1·39 (95% CI 1·12–1·72, P = 0·002) |
| N = 1411 | ||||
| IL13 | ||||
| Eder et al. 40 | Genomic/case–control | 3: IL13 polymorphisms | No review‐specified key prognostic factors controlled for | PsA: the combination of non‐smoking and rs1800925*CC was associated with increased risk of PsA (OR 2·04, 95% CI 1·37–3·03, P < 0·001) |
| N = 897 | ||||
| IgG aHDL | ||||
| Paiva‐Lopes et al. 41 | Proteomic/cross‐sectional | 4: IgG aHDL, aApoA‐I, aApoE, and aPON1 antibodies | 2: Sex, age of onset | Severity: in the multivariate regression IgG aHDL was significantly associated with PASI; β coefficient 1·02 (P < 0·001). Patients with PASI > 10 had higher mean levels of IgG aHDL (P = 0·010). Comorbidities such as hypertension and diabetes mellitus were not associated with antibodies (P‐values between 0·23 and 1·00) |
| N = 67 | ||||
| GlycA | ||||
| Joshi et al. 42 | Proteomic/cross‐sectional | 1: GlycA | 1: Sex | Severity: in multivariate regression analyses, adjusting for all variables, GlycA correlated with psoriasis severity assessed by BSA. Cohort 1: β coefficient 0·21 (P = 0·01); Cohort 2: β coefficient 0·40 (P < 0·001). No adjusted results for categorical severity |
| N = 273 (cohort 1: n = 122; cohort 2: n = 151) | ||||
| I‐FABP | ||||
| Sikora et al. 43 | Proteomic/case–control | 1: Intestinal fatty acid binding protein (I‐FABP) | 1: Sex | Severity: after adjustment for all covariates I‐FABP was significantly associated with moderate‐to‐severe disease: OR 3·47 (95% CI 1·20–10·07, P < 0·05) for each 100 pg mL−1 increase |
| N = 80 | ||||
| Kallikrein 8 | ||||
| Eissa et al. 44 | Proteomic/case–control | 7: Kallikreins | 1: Sex | Severity: in the multivariate regression KLK8 correlated positively with PASI (r = 0·43, P = 0·001) |
| N = 152 | ||||
| CXCL10 | ||||
| Abji et al. 18 | Proteomic/longitudinal | 2: CXCL10, CRP | No review‐specified key prognostic factors controlled for | PsA: an increase of 100 pg mL−1 in baseline CXCL10 resulted in a 30% increase in the odds of progression to PsA (OR 1·3, 95% CI 1·1–1·5, P = 0·004). CXCL10 significantly higher in converters at baseline (median 927 pg mL−1) than after PsA diagnosis (median 492 pg mL−1), P < 0·001 |
| N = 91 | ||||
| Abji et al. 17 | Proteomic/longitudinal | 1: CXCL10 | No review‐specified key prognostic factors controlled for | PsA: significant decline in CXCL10 levels over time prior to PsA development in converters (to PsA, P < 0·001) but not in nonconverters. There was significant evidence of a difference in the trend of CXCL10 levels between converters and nonconverters (P = 0·02) at preconversion |
| N = 81 | ||||
| Mac‐2 binding protein and integrin b5 | ||||
| Cretu et al. 21 | Proteomic/case–control | 6: CD5‐like protein, myeloperoxidase, CRP, Mac‐2‐binding protein (M2BP), integrin b5 (ITGβ5), matrix MMP‐3 | 2: Sex, age of onset | PsA: multivariate regression suggested that two of the three biomarkers were associated with PsA, compared with psoriasis without PsA. ORs relate to a twofold protein level increase: ITGβ5: OR 3·82 (95% CI 2·18–6·70), M2BP: OR 32·32 (95% CI 4·90–213·3). Additionally, ITGβ5 and M2BP levels were significantly correlated with each other, r = 0·24 |
| N = 200 | ||||
| MMP‐3 and M‐CSF | ||||
| Jadon et al. 19 | Proteomic/cross‐sectional | 4: Osteoprotegerin, dickkopf‐1; macrophage‐colony stimulating factor (M‐CSF); matrix metalloproteinase‐3 (MMP3) | 2: Sex, age of onset | PsA: MMP‐3 concentrations were significantly higher in patients with PsA compared with no‐PsA [adjusted OR 1·02 per ng mL−1 increase; 95% CI 1·01–1·03 (P = 0·001)] |
| N = 444 | Patients with PsA had significantly lower M‐CSF concentrations than no‐PsA [adjusted OR 0·44 per ng mL−1 increase 95% CI 0·24–0·82 (P = 0·01)] | |||
| Chandran et al. 45 | Mixed/cross‐sectional | 12: IL‐12, IL‐12p40, IL‐17, TNFSF14, MMP‐3, RANK ligand, osteoprotegerin (OPG), cartilage oligomeric matrix protein, C‐propeptide of type II collagen (CPII), collagen fragment neoepitopes Col2‐3/4 (C2C), Col2‐3/4short and CRP | 2: Sex, age of onset | PsA: multivariate model results indicated that increased levels of MMP‐3 (OR 1·28, 95% CI 1·02–1·60; P = 0·03) were independently associated with PsA when compared with patients with psoriasis only |
| N = 52 | ||||
| Tyramine and mucic acid | ||||
| Kishikawa et al. 46 | Metabolomic/cross‐sectional | 417 metabolites | 2: Sex, age | Severity: significant correlations between PASI scores and levels of tyramine and mucic acid (P = 0·0019 and P = 0·014, respectively) |
| N = 92 | PsA: significantly increased tyramine levels in patients with PsA compared with PsC (P = 1·4 × 10−5) | |||
N, number of patients with psoriasis; PsA, psoriatic arthritis; PsC, cutaneous psoriasis; BMI, body mass index; OR, odds ratio; CI, confidence interval; IL, interleukin; CRP, C‐reactive protein; NF‐κB, nuclear factor kappa B. Further study details and full quality assessment details are provided in Appendix S1 (see Supporting Information).
Five genomic biomarkers (LCE3D, IL23R, IL23A, NFKBIL1 loci and HLA‐C*06:02) and five proteomic biomarkers [interleukin (IL)‐17A, IgG aHDL, GlycA, I‐FABP and kallikrein 8] and one metabolomic biomarker (tyramine) demonstrated potential as candidate biomarkers of psoriasis severity. HLA‐C*06:02‐positive status was found to be associated with disease severity in three studies, although one study found an association for HLA‐C*06:02‐negative status. 20
Six genomic biomarkers (HLA‐C*06:02, HLA‐B*27, HLA‐B*38, HLA‐B*08, and variation at the IL23R and IL13 loci), six proteomic biomarkers [IL‐17A, CXCL10, Mac‐2 binding protein, integrin b5, matrix metalloproteinase (MMP)‐3 and macrophage‐colony stimulating factor (M‐CSF)] and two metabolomic biomarkers (tyramine and mucic acid) were selected as candidate biomarkers of PsA in psoriasis. These were all examined in cross‐sectional or case–control study designs, except for CXCL10. 18 Variation in IL12B and IL23R loci were considered candidate biomarkers of type 2 diabetes in psoriasis.
Thresholds of significance used for proteomic biomarkers were rarely justified in included studies. Several proteomic biomarker studies reported large beta coefficients (e.g. the association between Mac‐2 binding protein and integrin b5 with PsA observed by Cretu et al.), 21 which were difficult to contextualize in terms of potential clinical utility owing to insufficient information on the expected variation in biomarker levels in individuals with psoriasis. Furthermore, dichotomous interpretations of test results risk overlooking potentially important information across the range of biomarker levels.
Pathway mapping of candidate biomarkers (stage 3)
Most of the candidate biomarkers were found to be involved in signalling pathways implicated in psoriasis pathogenesis, 22 notably antigen processing presentation (HLA‐C*06:02, HLA‐B27, HLA‐B38, HLA‐B08, integrin b5) and leucocyte recruitment (CXCL10, M‐CSF) and activation (IL13, IL23R, IL23A, Mac‐2 binding protein) (see interactive map of psoriasis biomarkers: https://imi‐biomap.elixir‐luxembourg.org/minerva/index.xhtml?id=psobiomarkers_map).
Discussion
This scoping review provides a comprehensive catalogue of biomarker studies investigating disease progression in psoriasis. From the diverse range of biomarker types and outcomes examined in the included studies, we identify candidate biomarkers (10 genomic, 10 proteomic and two metabolomic) but note that none have sufficient evidence for clinical use without further validation. We detail methodological and reporting limitations to avoid in future studies and so expedite biomarker discovery and validation.
Several genomic biomarker studies investigated HLA‐C*06:02, which is perhaps not surprising given this is the major genetic determinant of disease susceptibility. 23 Most confirmed established associations with disease severity 20 , 24 , 25 , 26 , 27 and lack of PsA. 25 , 26 , 28 , 29 However, Eder et al. 28 and others 30 have highlighted the difficulty in verifying these associations owing to potential ascertainment bias that arises from the strong relationship between HLA‐C*06:02 and psoriasis age of onset. Furthermore, extensive linkage disequilibrium between class I HLA genes makes it challenging to distinguish the effects of other PsA‐associated HLA biomarkers (e.g. HLA‐B*27, HLA‐B*38 and HLA‐B*08) that are potentially independent of HLA‐C*06:02. The predictive ability of HLA‐B and HLA‐C alleles should ideally be confirmed in appropriately powered prospective cohort studies.
Genes encoding members of the IL‐23‐mediated signalling pathway, a key pathway for psoriasis pathogenesis, were found to be candidate genomic biomarkers of disease severity (IL23A, IL23R), PsA (IL23R) and type 2 diabetes (IL12B, IL23R). The two studies investigating these biomarkers examined different polymorphisms at each locus. 31 , 32 These polymorphisms were frequently found to be in linkage disequilibrium with each other, therefore there was a consequent variation in the direction effect reported for each association. Polymorphisms near HLA‐C, IL23R and LCE3A loci were also found to be associated with PsA in the two large genome‐wide meta‐analyses included in the review, 15 , 33 and in independent cross‐sectional/case–control studies. 20 , 25 , 26 , 27 , 28 , 31 , 32
A broad definition of severity was used in this review, including both biomarkers that may be reflective of current disease activity and those that may be predictive of future severity. The former require validation in longitudinal studies to establish potential predictive clinical utility. Of the candidate proteomic biomarkers, only CXCL10 was examined in a longitudinal study design. 17 , 18 It is therefore unclear whether these biomarkers reflect current disease activity and PsA, rather than being predictive of future disease progression. Nevertheless, we note that several biomarkers involving leucocyte recruitment and cytokine‐mediated signalling were associated with both severe psoriasis and PsA.
Most biomarkers explored have putative roles in psoriasis pathogenesis or are established markers of relevant underlying disease processes such as systemic inflammation (e.g. CRP) or skin barrier function (e.g. kallikreins) indicating a bias towards a candidate approach to biomarker discovery. Few studies employed ‘hypothesis‐free’ approaches, with the notable exceptions of two genome‐wide meta‐analyses of studies investigating biomarkers associated with the presence of PsA in psoriasis, 14 , 15 , 30 or explored the utility of including multiple biomarkers to enhance precision. Numerous studies identified biomarkers of nonspecific systemic inflammation (e.g. acute phase reactants such as CRP), which can be elevated in several inflammatory processes, 34 and therefore may lack specificity to common psoriasis comorbidities. In addition, while several studies did suggest potential value of markers of connective tissue remodelling (e.g. MMP3 and M‐CSF), understanding whether these are specific to PsA will be important, particularly given the burden of other conditions associated with psoriasis that may also increase levels, e.g. osteoarthritis, 34 inflammatory bowel disease 35 and cardiovascular disease. 36
The review highlights several important methodological and reporting limitations that should be considered when designing and conducting future biomarker studies. The investigation of biomarker associations with psoriasis progression outcomes was often not the primary objective of included studies. This meant that information on key patient cohort characteristics (including participants’ ethnicity, age, age of onset of psoriasis and psoriasis subtype) were often not reported. This limits the degree to which findings from these studies can be extrapolated to the population seen in clinical practice and hampers comparative assessment between studies.
The importance of identifying, measuring and adjusting for key prognostic factors of disease progression outcomes was also highlighted in this review. Fewer than one‐third of the case–control and cross‐sectional studies adjusted for confounding. Future studies should also consider possible confounding by the presence of other biomarkers to help identify independent associations with outcomes.
For disease severity outcomes, studies frequently reported a single recording of disease severity. This made it difficult to determine whether candidate biomarkers associate with chronic disease severity. Future studies should assess and report the disease severity comprehensively by using more than one measure and record previous treatments as a proxy measure for chronic severity. Analyses should also be performed using both continuous and categorical data wherever possible to allay concerns about the possibility of selective outcome reporting. Categories used should be clearly defined and easy to justify clinically. Future studies that evaluate comorbidities as outcomes should be clear on how comorbidity status is classified or diagnosed, particularly for comorbidities that may have a subclinical phase (e.g. PsA and type 2 diabetes).
The main strength of the review is its breadth of scope – eligibility criteria were designed to be as inclusive as was practicable, given that it was anticipated that a large number of studies would be included. We chose to include cross‐sectional and case–control studies, of which there were many, to capture the research landscape. However, the ideal study is longitudinal in design, and very few of these studies were included in our review. Such studies are essential to test the value/utility of biomarkers in predicting future outcomes and therefore in helping the identification of objective indications for early intervention. Furthermore, applying a study size cutoff in this review of ≥ 50 participants – irrespective of biomarker type – may have potentially overlooked informative well‐conducted studies of epigenetic and transcriptomic biomarkers, where obtaining larger sample sizes is less feasible. Other limitations were the exclusion of papers not reported in English and the single‐screening of some titles and abstracts. These approaches were taken because of the need to balance timeliness of results with methodological rigour.
Although we have identified a putative list of candidates for further research, this does not mean that those biomarkers not on the list should be excluded from further research. This is especially true of more recent studies, where the initial, and often promising ‘biomarker discovery’ study reports only findings from multiple analytes in a small number of individuals and lacks the necessary validation in larger cohorts. 37
To optimize this next step, we identify a need for longitudinal studies where carefully selected analytes are tested in many individuals, with accurate reporting of baseline disease stage. There is also increasing acknowledgment that a single variable is rarely likely to be a good predictor of disease progression, and therefore algorithms and risk prediction models that combine the predictive power of multiple biomarkers are likely to be required to develop clinically useful tools. 38 , 39 The inclusion of clinical characteristics can also improve translational models of inflammatory disease, 40 thereby maximizing their future clinical utility as stratification tools.
The extent of research on biomarkers of disease progression revealed in this scoping review illustrates the unmet clinical need in psoriasis. Common important methodological limitations highlighted in this review should be carefully considered in future studies. Synergized efforts, through interdisciplinary collaborations such as the BIOMAP project, will be crucial in facilitating cross‐consortium agreements on terminology and harmonization of outcomes of interest.
Author contributions
Ravi Ramessur: Conceptualization (equal); data curation (lead); formal analysis (equal); methodology (supporting); writing – original draft (lead); writing – review and editing (equal). Mark Corbett: Data curation (lead); formal analysis (equal); methodology (equal); visualization (equal); writing – original draft (equal); writing – review and editing (equal). David Marshall: Data curation (equal); formal analysis (equal); methodology (equal); writing – review and editing (supporting). Marcio Acencio: Formal analysis (supporting); investigation (equal); visualization (equal); writing – review and editing (equal). Ines Barbosa: Data curation (equal); formal analysis (equal); methodology (supporting); writing – review and editing (supporting). Nick Dand: Formal analysis (equal); investigation (equal); methodology (supporting); writing – review and editing (equal). Paola Di Meglio: Formal analysis (supporting); visualization (equal); writing – review and editing (equal). Salma Haddad: Data curation (equal); formal analysis (supporting); writing – review and editing (supporting). Andreas Hvirgel Moesgaard Jensen: Conceptualization (supporting); methodology (supporting); writing – review and editing (supporting). Witte Koopmann: Funding acquisition (supporting); investigation (supporting); writing – review and editing (equal). Satveer Mahil: Conceptualization (supporting); formal analysis (equal); methodology (equal); writing – review and editing (equal). Marek Ostaszewski: Formal analysis (supporting); investigation (supporting); visualization (equal); writing – review and editing (equal). Seher Rahmatulla: Data curation (equal); formal analysis (supporting); writing – review and editing (supporting). Joe Rastrick: Conceptualization (supporting); funding acquisition (supporting); writing – review and editing (supporting). Jake Saklatvala: Data curation (equal); formal analysis (equal); methodology (supporting); writing – review and editing (supporting). Stephan Weidinger: Funding acquisition (lead); supervision (supporting); writing – review and editing (supporting). Kath Wright: Data curation (equal); formal analysis (supporting); methodology (equal). Kilian Eyerich: Conceptualization (equal); formal analysis (equal); supervision (supporting). Matladi Ndlovu: Data curation (equal); formal analysis (equal); methodology (supporting); writing – review and editing (supporting). Jonathan N W N Barker: Conceptualization (equal); formal analysis (supporting); methodology (equal); writing – review and editing (equal). Lone Skov: Conceptualization (supporting); formal analysis (equal); methodology (equal); visualization (supporting); writing – review and editing (equal). Curdin Conrad: Conceptualization (equal); formal analysis (equal); methodology (equal); visualization (supporting); writing – review and editing (equal). Catherine H. Smith: Conceptualization (lead); data curation (supporting); formal analysis (equal); funding acquisition (lead); methodology (lead); project administration (lead); supervision (lead); visualization (equal); writing – original draft (equal); writing – review and editing (equal).
Funding sources
This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 821511 (BIOMAP). The JU receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. The research also received funding from the International Psoriasis Council, and support by the National Institute for Health and Care Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care. S.K.M. is funded by a Medical Research Council Clinical Academic Research Partnership award (MR/T02383X/1). N.D. is funded by Health Data Research UK (MR/S003126/1).
Conflicts of interest
S.K.M. has received departmental funding from AbbVie, Celgene, Eli Lilly, Janssen‐Cilag, Novartis, Sanofi and UCB. P.D.M. has received research grants from UCB and consultancy/speaker honoraria from Novartis, UCB and Janssen. K.E. has received honoraria and/or research grants from AbbVie, Almirall, Boehringer Ingelheim, Bristol Myers Squibb, Janssen, LEO Pharma, Lilly, Novartis, Pfizer and UCB. C.C. has received honoraria and/or research grants from AbbVie, Actelion, Almirall, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Janssen, LEO Pharma, Eli Lilly, MSD, Novartis, Pfizer, Samsung and UCB. J.N.B. has received honoraria and/or research grants from AbbVie, Almirall, Amgen, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Janssen, LEO Pharma, Lilly, Novartis, Samsung and Sun Pharma. L.S. has received honoraria and/or research grants from AbbVie, Almirall, Bristol Myers Squibb, Celgene, Sanofi, UCB, Janssen, LEO Pharma, Lilly and Novartis. C.H.S. reports grants from an MRC‐funded stratified medicine consortium with multiple industry partners, grants from IMI (Horizon 2020)‐funded European consortium with multiple industry partners, and others from AbbVie, Novartis, Pfizer, Sanofi, Boehringer Ingelheim and SOBI, outside the submitted work; and is Chair of UK guidelines on biologic therapy in psoriasis.
Data availability
The data that support the findings of this study will be publicly available following publication.
Supporting information
Appendix S1 Preliminary work and study design, overview of results, summary of quality assessment, candidate biomarker selection methodology, molecular and cellular pathway mapping, literature searches, list of excluded studies.
Acknowledgments
This scoping review was supported and funded by the IPC and the BIOMAP consortium. This study was also supported by the Psoriasis Association, UK. The authors would like to thank Marta Vergnano for her contribution in title and abstract screening. The interactive map of psoriasis biomarkers is hosted by ELIXIR Luxembourg. Open access funding was enabled and organized by ProjektDEAL.
Plain language summary available online
References
- 1. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker JNW. Psoriasis. Lancet 2021; 397:1301–15. [DOI] [PubMed] [Google Scholar]
- 2. Parisi R, Iskandar IYK, Kontopantelis E et al. National, regional, and worldwide epidemiology of psoriasis: systematic analysis and modelling study. BMJ 2020; 369:m1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dand N, Mahil SK, Capon F et al. Psoriasis and genetics. Acta Derm Venereol 2020; 100: adv00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davidovici BB, Sattar N, Prinz JC et al. Psoriasis and systemic inflammatory diseases: potential mechanistic links between skin disease and co‐morbid conditions. J Invest Dermatol 2010; 130:1785–96. [DOI] [PubMed] [Google Scholar]
- 5. Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA 2018; 320:1360–72. [DOI] [PubMed] [Google Scholar]
- 6. FDA‐NIH Biomarker Working Group . BEST (Biomarkers, EndpointS, and other Tools) Resource. Bethesda, MD: National Institutes of Health, 2016. [Google Scholar]
- 7. Jog NR, James JA. Biomarkers in connective tissue diseases. J Allergy Clin Immunol 2017; 140:1473–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS 2010; 5:463–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wirsching J, Grassmann S, Eichelmann F et al. Development and reliability assessment of a new quality appraisal tool for cross‐sectional studies using biomarker data (BIOCROSS). BMC Med Res Methodol 2018; 18:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayden JA, van der Windt DA, Cartwright JL et al. Assessing bias in studies of prognostic factors. Ann Intern Med 2013; 158:280–6. [DOI] [PubMed] [Google Scholar]
- 11. Naldi L. Risk Factors for Psoriasis. Current Dermatology Reports 2013; 2:58–65. [Google Scholar]
- 12. Ensor JE. Biomarker validation: common data analysis concerns. Oncologist 2014; 19:886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dei‐Cas I, Giliberto F, Luce L et al. Metagenomic analysis of gut microbiota in non‐treated plaque psoriasis patients stratified by disease severity: development of a new Psoriasis‐Microbiome Index. Sci Rep 2020; 10:12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patrick MT, Stuart PE, Raja K et al. Genetic signature to provide robust risk assessment of psoriatic arthritis development in psoriasis patients. Nat Commun 2018; 9:4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stuart PE, Nair RP, Tsoi LC et al. Genome‐wide association analysis of psoriatic arthritis and cutaneous psoriasis reveals differences in their genetic architecture. Am J Hum Genet 2015; 97:816–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loft ND, Skov L, Rasmussen MK et al. Genetic polymorphisms associated with psoriasis and development of psoriatic arthritis in patients with psoriasis. PLOS ONE 2018; 13:e0192010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abji F, Lee KA, Pollock RA et al. Declining levels of serum chemokine (C‐X‐C motif) ligand 10 over time are associated with new onset of psoriatic arthritis in patients with psoriasis: a new biomarker? Br J Dermatol 2020; 183:920–7. [DOI] [PubMed] [Google Scholar]
- 18. Abji F, Pollock RA, Liang K et al. Brief report: CXCL10 is a possible biomarker for the development of psoriatic arthritis among patients with psoriasis. Arthritis Rheumatol 2016; 68:2911–16. [DOI] [PubMed] [Google Scholar]
- 19. Jadon DR, Sengupta R, Nightingale A et al. Serum bone‐turnover biomarkers are associated with the occurrence of peripheral and axial arthritis in psoriatic disease: a prospective cross‐sectional comparative study. Arthritis Res Ther 2017; 19:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suomela S, Kainu K, Onkamo P et al. Clinical associations of the risk alleles of HLA‐Cw6 and CCHCR1*WWCC in psoriasis. Acta Derm Venereol 2007; 87:127–34. [DOI] [PubMed] [Google Scholar]
- 21. Cretu D, Gao L, Liang K et al. Differentiating psoriatic arthritis from psoriasis without psoriatic arthritis using novel serum biomarkers. Arthritis Care Res (Hoboken) 2018; 70:454–61. [DOI] [PubMed] [Google Scholar]
- 22. Mahil SK, Capon F, Barker JN. Update on psoriasis immunopathogenesis and targeted immunotherapy. Semin Immunopathol 2016; 38:11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng BJ, Sun LD, Soltani‐Arabshahi R et al. Multiple loci within the major histocompatibility complex confer risk of psoriasis. PLOS Genet 2009; 5:e1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gudjonsson JE, Karason A, Antonsdottir A et al. Psoriasis patients who are homozygous for the HLA‐Cw 0602 allele have a 2.5‐fold increased risk of developing psoriasis compared with Cw6 heterozygotes. Br J Dermatol 2003; 148:233–5. [DOI] [PubMed] [Google Scholar]
- 25. Gudjonsson JE, Karason A, Antonsdottir AA et al. HLA‐Cw6‐positive and HLA‐Cw6‐negative patients with psoriasis vulgaris have distinct clinical features. J Invest Dermatol 2002; 118:362–5. [DOI] [PubMed] [Google Scholar]
- 26. Julia A, Tortosa R, Hernanz JM et al. Risk variants for psoriasis vulgaris in a large case‐control collection and association with clinical subphenotypes. Hum Mol Genet 2012; 21:4549–57. [DOI] [PubMed] [Google Scholar]
- 27. Gudjonsson JE, Karason A, Runarsdottir EH et al. Distinct clinical differences between HLA‐Cw 0602 positive and negative psoriasis patients‐‐an analysis of 1019 HLA‐C‐ and HLA‐B‐typed patients. J Invest Dermatol 2006; 126:740–5. [DOI] [PubMed] [Google Scholar]
- 28. Eder L, Chandran V, Pellet F et al. Human leucocyte antigen risk alleles for psoriatic arthritis among patients with psoriasis. Ann Rheum Dis 2012; 71:50–5. [DOI] [PubMed] [Google Scholar]
- 29. Douroudis K, Ramessur R, Barbosa IA et al. Differences in clinical features and comorbid burden between HLA‐C*06:02 carrier groups in >9,000 people with psoriasis. J Invest Dermatol 2022; 142:1617–28.e10. [DOI] [PubMed] [Google Scholar]
- 30. Bowes J, Ashcroft J, Dand N et al. Cross‐phenotype association mapping of the MHC identifies genetic variants that differentiate psoriatic arthritis from psoriasis. Ann Rheum Dis 2017; 76:1774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eiris N, Gonzalez‐Lara L, Santos‐Juanes J et al. Genetic variation at IL12B, IL23R and IL23A is associated with psoriasis severity, psoriatic arthritis and type 2 diabetes mellitus. J Dermatol Sci 2014; 75:167–72. [DOI] [PubMed] [Google Scholar]
- 32. Nikamo P, Lysell J, Stahle M. Association with genetic variants in the IL‐23 and NF‐κB pathways discriminates between mild and severe psoriasis skin disease. J Invest Dermatol 2015; 135:1969–76. [DOI] [PubMed] [Google Scholar]
- 33. Ansar W, Ghosh S. C‐reactive protein and the biology of disease. Immunol Res 2013; 56:131–42. [DOI] [PubMed] [Google Scholar]
- 34. Takei I, Takagi M, Ida H et al. High macrophage‐colony stimulating factor levels in synovial fluid of loose artificial hip joints. J Rheumatol 2000; 27:894–9. [PubMed] [Google Scholar]
- 35. O’Sullivan S, Gilmer JF, Medina C. Matrix metalloproteinases in inflammatory bowel disease: an update. Mediators Inflamm 2015; 2015:964131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu P, Sun M, Sader S. Matrix metalloproteinases in cardiovascular disease. Can J Cardiol 2006; 22 (Suppl. B):25b–30b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kraus VB. Biomarkers as drug development tools: discovery, validation, qualification and use. Nat Rev Rheumatol 2018; 14:354–62. [DOI] [PubMed] [Google Scholar]
- 38. Savvateeva E, Smoldovskaya O, Feyzkhanova G, Rubina A. Multiple biomarker approach for the diagnosis and therapy of rheumatoid arthritis. Crit Rev Clin Lab Sci 2021; 58:17–28. [DOI] [PubMed] [Google Scholar]
- 39. Curtis JR, Xie F, Crowson CS et al. Derivation and internal validation of a multi‐biomarker‐based cardiovascular disease risk prediction score for rheumatoid arthritis patients. Arthritis Res Ther 2020; 22:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eder L, Chandran V, Pellett F et al. IL13 gene polymorphism is a marker for psoriatic arthritis among psoriasis patients. Ann Rheum Dis 2011; 70:1594–8. [DOI] [PubMed] [Google Scholar]
- 41. Paiva‐Lopes MJ, Batuca JR, Gouveia S et al. Antibodies towards high‐density lipoprotein components in patients with psoriasis. Arch Dermatol Res 2020; 312:93–102. [DOI] [PubMed] [Google Scholar]
- 42. Joshi AA, Lerman JB, Aberra TM et al. GlycA is a novel biomarker of inflammation and subclinical cardiovascular disease in psoriasis. Circ Res 2016; 119:1242–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sikora M, Stec A, Chrabaszcz M et al. Intestinal fatty acid binding protein, a biomarker of intestinal barrier, is associated with severity of psoriasis. J Clin Med 2019; 8:1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eissa A, Cretu D, Soosaipillai A et al. Serum kallikrein‐8 correlates with skin activity, but not psoriatic arthritis, in patients with psoriatic disease. Clin Chem Lab Med 2013; 51:317–25. [DOI] [PubMed] [Google Scholar]
- 45. Chandran V, Cook RJ, Edwin J et al. Soluble biomarkers differentiate patients with psoriatic arthritis from those with psoriasis without arthritis. Rheumatology 2010; 49:1399–405. [DOI] [PubMed] [Google Scholar]
- 46. Kishikawa T, Arase N, Tsuji S et al. Large‐scale plasma‐metabolome analysis identifies potential biomarkers of psoriasis and its clinical subtypes. J Dermatol Sci 2021; 102:78–84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Preliminary work and study design, overview of results, summary of quality assessment, candidate biomarker selection methodology, molecular and cellular pathway mapping, literature searches, list of excluded studies.
Data Availability Statement
The data that support the findings of this study will be publicly available following publication.


