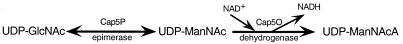Abstract
The Staphylococcus aureus serotype 5 capsular polysaccharide (CP5) has a repeating unit composed of (→4)-3-O-acetyl-β-d-ManNAcA-(1→4)-α-l-FucNAc (1→3)-β-d-FucNAc-(1→)n. Sixteen chromosomal genes (cap5A through cap5P) are involved in the synthesis of CP5. We recently demonstrated that Cap5P, a 2-epimerase, catalyzes the conversion of UDP–N-acetyl glucosamine (UDP-GlcNAc) to UDP–N-acetylmannosamine (UDP-ManNAc). In this study, we show that UDP-ManNAc is oxidized to UDP–N-acetylmannosaminuronic acid (UDP-ManNAcA) by a UDP-ManNAc dehydrogenase encoded by S. aureus cap5O. We expressed Cap5O in Escherichia coli and purified the recombinant protein. The UDP-ManNAc dehydrogenase activity of purified Cap5O was assessed by incubating Cap5P and UDP-GlcNAc (to produce UDP-ManNAc), together with Cap5O, NAD+, and a reducing agent. Enzymatic activity was quantitated indirectly by measuring the increase in absorbance at 340 nm resulting from NADH formation. The product of the reaction was confirmed as UDP-ManNAcA by gas chromatography-mass spectroscopy. A cap5O mutation, created by deletion of 727 bp in the 5′ end of the gene, was introduced by allelic replacement into S. aureus Reynolds, rendering it CP5 negative. Mice inoculated intravenously or subcutaneously with the wild-type strain Reynolds had greater numbers of S. aureus recovered from their kidneys (P = 0.019) or their subcutaneous abscesses (P = 0.0018), respectively, than did animals inoculated with the cap5O mutant. The results of this study indicate that S. aureus cap5O is essential for capsule production and that capsule promotes staphylococcal virulence in mouse models of abscess formation.
Staphylococcus aureus is responsible for a variety of infectious diseases ranging from cutaneous infections, such as abscesses, boils, and wound infections, to life-threatening infections such as bacteremia and endocarditis (23). Because S. aureus produces many adhesins, exoenzymes, and exotoxins, the pathogenesis of staphylococcal infections is multifactorial. In addition, the presence of an extracellular polysaccharide capsule promotes staphylococcal virulence in certain animal models of infection (25, 31). Most clinical isolates produce serotype 5 or 8 polysaccharide capsules (CP5 or CP8) (2, 9). These two types of capsule have a similar trisaccharide repeating unit that differs only in the linkages between the sugars and the sites of O acetylation: CP5, (→4)-3-O-acetyl-β-d-ManNAcA-(1→4)-α-l-FucNAc-(1→3)-β-d-FucNAc-(1→)n; CP8, (→3)-4-O-acetyl-β-d-ManNAcA-(1→3)-α-l-FucNAc-(1→3)-β-d-FucNAc-(1→)n.
Sixteen chromosomal genes (cap5A through cap5P) are involved in the synthesis of CP5 (29). We recently demonstrated that Cap5P is a 2-epimerase that catalyzes the conversion of UDP–N-acetylglucosamine (UDP-GlcNAc) to UDP–N-acetylmannosamine (UDP-ManNAc) (14). Adjacent to cap5P in the capsule gene region is cap5O. The putative amino acid sequence of Cap5O shows homology to Escherichia coli RffD, a UDP-ManNAc dehydrogenase involved in the biosynthesis of the enterobacterial common antigen (24). We showed that S. aureus cap5O functionally complemented an rffD mutation in E. coli and restored expression of the enterobacterial common antigen (13). We report here the in vitro enzymatic activity of Cap5O, which catalyzes the oxidation of UDP-ManNAc to UDP–N-acetylmannosaminuronic acid (UDP-ManNAcA), the putative donor of ManNAcA residues in CP5.
The deletion of a bacterial gene critical to capsule biosynthesis should yield an unencapsulated phenotype. However, we recently reported that a cap5P mutation did not affect S. aureus capsule expression. Capsule production by the cap5P mutant was explained by the presence on the S. aureus chromosome of a second gene (mnaA) that encodes a UDP-GlcNAc 2-epimerase (14). In this study, we showed that a cap5O mutant of S. aureus is unencapsulated and less virulent for mice compared with the parental strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. Luria-Bertani medium was used for growth of E. coli. S. aureus strains were grown in tryptic soy broth or on Columbia agar (Difco Laboratories, Detroit, Mich.) plates supplemented with 2% NaCl. The culture medium contained chloramphenicol (Cm) at 10 μg/ml, erythromycin (Em) at 5 μg/ml, or kanamycin (Km) at 25 μg/ml when required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| E. coli BL21(DE3) | F−ompT lon hsdSB (rB−mB−) DE3 | Novagen, Inc. |
| S. aureus | ||
| Reynolds | CP5 positive | 9 |
| JLO22 | 727-bp deletion in cap5O gene of Reynolds, CP5 negative | This study |
| RN4220 | Capsule negative, restriction negative | 27 |
| Plasmids | ||
| pET-24a+ | E. coli expression vector (Kmr) | Novagen, Inc. |
| pJCL24 | 9.1-kb EcoRI fragment from S. aureus Reynolds (cap5H to cap5P) in pLI50 | 21 |
| pKBK4 | 1.3-kb PCR amplicon carrying cap5O in pUC19 | 13 |
| pKBK5 | 9.1-kb EcoRI fragment (cap5H to cap5P) from pJCL24 in pUC19 | This study |
| pKBK9 | 4.1-kb AvaI-EcoRI fragment (cap5M to cap5P) from pKBK5 in pUC19 | This study |
| pKBK9-2 | 727-bp HpaI deletion (Δcap5O) of pKBK9 | This study |
| pKBK22 | 3.4-kb BamHI-EcoRI fragment (cap5M to cap5P; Δcap5O) from pKBK9-2 in pTS1 | This study |
| pKBK24 | 2.4-kb PCR amplicon carrying cap5O and cap5 flanking sequences in pLI50 | This study |
| pLI50 | Shuttle vector (Apr Cmr) | 16 |
| pNB1 | 1.3-kb XbaI-EcoRI fragment (cap5O) from pKBK4 in pET-28a+ | This study |
| pTS1 | Temperature-sensitive shuttle vector (Apr Cmr) | 26 |
| pUC19 | E. coli cloning vector (Apr) | New England Biolabs, Inc. |
Chemicals.
Reagents used in enzyme purification and activity assays were obtained from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise noted. Ultrapure reagents used for methanolysis, reduction, and derivatization of sugars were obtained from J. T. Baker, Inc. (Phillipsburg, N.J.); Sigma; or ICN Biomedicals, Inc. (Aurora, Ohio). Alditol acetate derivatives of GlcNAc and ManNAc (Aldrich Chemical Co., Milwaukee, Wis.) were used as standard sugars in gas chromatography-mass spectroscopy (GC-MS) analysis. Restriction endonucleases and other DNA modification enzymes were obtained from Life Technologies, Inc. (Gaithersburg, Md.), or New England Biolabs, Inc. (Beverly, Mass.).
Subcloning of cap5O into the pET-28a+ expression vector.
The 1.3-kb XbaI-EcoRI fragment from pKBK4 comprises a PCR amplicon containing the cap5O gene and upstream Shine-Dalgarno sequence; this fragment was ligated into pET-28a+ (Novagen, Inc., Madison, Wis.). In the resultant plasmid, pNB1, the 3′ end of the cap5O gene was fused in frame to the His tag sequence of the vector. PCR fidelity was confirmed by DNA sequencing.
Construction of S. aureus cap5O mutant.
The 9.1-kb EcoRI fragment (cap5H to cap5P) from pJCL24 was subcloned into pUC19 to create pKBK5. Digestion of pKBK5 with AvaI released a 5.1-kb fragment containing cap5H through cap5L. The digested plasmid was treated with Klenow fragment and deoxynucleoside triphosphates and ligated to create pKBK9 which carries a 4.1-kb AvaI-EcoRI insert (cap5M to cap5P). A 727-bp HpaI fragment was deleted from pKBK9 to create pKBK9-2. The deletion, encompassing nucleotide positions 9 to 734 of the 1,260-bp cap5O gene, resulted in a +1 frameshift. pKBK9-2 was digested with EcoRI and BamHI, and the 3.4-kb fragment (cap5O deletion and flanking cap5 sequence) was ligated into pTS1 to create plasmid pKBK22.
pKBK22 was electrotransformed (17) into S. aureus RN4220 and then transduced with phage 80α (10) into S. aureus type 5 strain Reynolds, in both instances selecting for Cmr colonies at 30°C. The mutation was introduced into the chromosome by allelic exchange. In brief, plasmid integrants in the chromosome were selected by plating of cells on tryptic soy agar containing Cm (5 μg/ml) at 42°C. Single colonies were then passaged three times at 30°C without antibiotic selection. Cms colonies were screened for CP5 production by colony immunoblot as previously described (20) with the use of CP5-specific polyclonal rabbit antiserum. DNA from CP5-negative mutant JLO22 was analyzed by PCR and Southern blot to confirm the presence of the mutation in cap5O. Lack of CP5 production by the cap5O mutant JLO22 was confirmed by immunodiffusion of capsular extracts with CP5-specific rabbit antiserum.
Purification of Cap5O.
A 100-ml culture of E. coli BL21(DE3) carrying pNB1 was grown at 37°C for 3 h on a platform shaker at 180 rpm. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the culture was incubated for another 3 h at 30°C. All subsequent purification steps were performed at 4°C. The culture was pelleted at 5,000 × g, and the bacterial cells were resuspended in 20 mM Tris-HCl buffer (pH 7.9) containing 5 mM imidazole. E. coli cells were lysed by three to four cycles through a French pressure cell (Aminco, Urbana, III.) at 800 lb/in2. The lysate was centrifuged at 39,000 × g for 20 min, and the supernatant was loaded onto a Ni2+ affinity column (Novagen). The column was washed sequentially with 5 and 60 mM imidazole in 20 mM Tris-HCl buffer, and the protein was eluted with 1 M imidazole in 20 mM Tris-HCl buffer. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing Cap5O were pooled and dialyzed against 50 mM Tris-HCl buffer (pH 8.5). The protein content of the purified enzyme was determined by the Bradford method (6) (Bio-Rad Laboratories, Hercules, Calif.) with bovine gamma globulin as the standard. Ammonium sulfate was added to the purified protein to a final concentration of 250 mM, and the enzyme solution was stored in aliquots at −70°C.
Dehydrogenase assay.
The activity of the purified enzyme was monitored by measuring the increase in absorption at 340 nm resulting from NADH formation. The assay mixture contained 0.5 mM dithiothreitol, 0.5 mM UDP-GlcNAc, 1.5 mM NAD+, 1.5 to 3 μg of purified Cap5P (14), and ∼70 μg of Cap5O in 0.2 ml of 50 mM Tris-HCl buffer (pH 8.5). All the reagents except for NAD+ were mixed and placed in a dry bath at 37°C. NAD+ was then added, and the absorbance at 340 nm was read at 0, 10, 20, 30, and 60 min in a Beckman UV-visible spectrophotometer. NADH formation was expressed in nanomoles, determined according to the Lambert-Beer law with an extinction coefficient for NADH of 6,220 M−1.
The effect of EDTA (2, 5, and 10 mM), magnesium (2 mM MgCl2 or MgSO4) and monovalent ions [150 mM KCl, NH4Cl, or (NH4)2SO4] on enzyme activity was determined. These reagents were included in the assay mixture prior to the addition of NAD+.
Identification of UDP-ManNAcA by GC-MS.
Dehydrogenase reaction mixtures were treated with 1 ml of 95% ethanol at 70°C for 10 min to precipitate proteins. UDP-amino sugars in the supernatant were converted to methyl glycosides by methanolysis with 0.5 ml of 1 M methanol containing 35 μl of acetyl chloride for 24 h at 85°C. The methyl glycosides were reduced overnight at room temperature with 3 mg of sodium borodeuteride in 0.3 ml of 95% ethanol. This step reduces the carboxylic acid of ManNAcA to an alcohol group and labels carbonyl carbon-6 with two deuterium atoms. Methyl glycosides were hydrolyzed with 0.5 M trifluoroacetic acid for 12 h at 100°C to remove the UDP moieties. Free sugars were reduced with 3 mg of sodium borodeuteride in 0.3 ml of 1 M ammonium hydroxide. This step converts the sugars into the corresponding alditols and labels the anomeric carbon-1 with one deuterium atom. The alditols were then acetylated with 0.1 ml of acetic anhydride and 0.1 ml of pyridine for 20 min at 100°C. Samples were washed with distilled water, partitioned with 0.5 ml of ethyl acetate, and analyzed by GC-MS (Saturn 2000; Varian, Palo Alto, Calif.) on a DB-17 column (30-m-by-0.25-mm inner diameter by 0.25-μm df). Electron impact ionization was used in all MS methods.
Mouse infection studies.
Male ICR and Swiss-Webster mice (6 to 7 weeks old) were obtained from Harlan Sprague-Dawley, Inc. (Indianapolis, Ind.). The mice were housed (up to four animals per cage) in a modified barrier facility under viral antibody-free conditions. Food and water were provided to the mice ad libitum. Animals were handled according to Brigham and Women's Hospital and Harvard Medical School institutional guidelines.
Staphylococci were cultivated on Columbia salt agar plates at 37°C for 24 h, and bacterial colonies were suspended in phosphate-buffered saline (0.01 M phosphate, 0.15 M NaCl; pH 7.2 to 7.4). The optical density of the bacterial suspensions was measured, and the samples were diluted to yield the appropriate numbers of CFU per milliliter.
In the renal abscess model (18), groups of four to six mice were injected in the tail vein with 4 × 106 or 4 × 105 CFU of S. aureus in a 0.2-ml inoculum. The number of CFU per milliliter of inoculum was verified by plate counts. Five days after bacterial challenge, mice were euthanized, and the kidneys were excised, weighed, and homogenized in 1 ml of tryptic soy broth. Serial dilutions of the homogenates were plated in duplicate on tryptic soy agar plates, and the results were expressed as the log CFU of S. aureus/gram of tissue. The lower limit of detection by culture was 1.1 log CFU/g of tissue. Two separate experiments were performed with each mouse strain.
In the subcutaneous model (7), bacterial suspensions were mixed with equal volumes of dextran beads (Cytodex-1 microcarriers; Sigma) prepared according to the manufacturer's instructions. Groups of three mice were injected subcutaneously in each flank with 0.2 ml of S. aureus suspensions ranging from 107 to 101 CFU. The numbers of CFU per milliliter of inoculum were verified by plate counts. Four days after bacterial challenge, mice were euthanized, and the abscesses were excised and homogenized in 1 ml of tryptic soy broth. Serial dilutions of the homogenates were plated in duplicate on tryptic soy agar plates, and the results were expressed as the log CFU of S. aureus/abscess. The lower limit of detection was 1.0 log CFU/abscess. In two separate experiments, Swiss-Webster mice were coinfected with a bacterial suspension containing equal numbers of the parental strain Reynolds and the cap5O mutant JLO22 mixed with dextran beads. The number of CP5-positive versus CP5-negative colonies recovered in each abscess was assessed by a colony immunoblot method (20).
Statistical analysis.
The results of quantitative renal cultures at each dose were compared with the Welch test, a modification of the unpaired Student's t test for comparing Gaussian populations with unequal standard deviations. A semiparametric weighted least-squares method (32) was used to compare the results of quantitative bacterial cultures performed over a range of doses in the subcutaneous abscess model. Data from coinfection experiments were analyzed by the unpaired Student's t test.
RESULTS
Purification and properties of Cap5O.
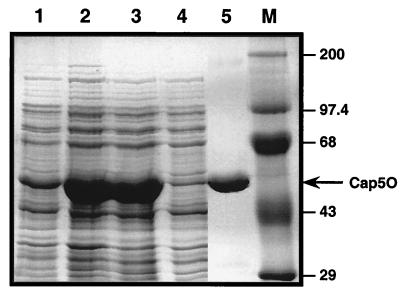
Cap5O was overexpressed from pNB1 in E. coli BL21(DE3). As shown in Fig. 1, most of the protein was recovered from the 39,000 × g supernatant, an indication that the protein was in a soluble form. The hydropathy plot of the deduced amino acid sequence of the protein confirmed its hydrophilic nature (data not shown). The histidine-tagged Cap5O, purified over a Ni2+ affinity column, showed a single band by SDS-PAGE, with a mass of ∼45.9 kDa (Fig. 1). This result is in agreement with the predicted mass of 45.6 kDa deduced from the nucleotide sequence of the cap5O gene. The isoelectric point predicted from the amino acid sequence of Cap5O was 4.8. N-terminal protein sequencing of purified Cap5O yielded the sequence MKLTVVGLGY, which confirmed the translational start site predicted by the nucleotide sequence (29). Purified Cap5O, at a concentration of ∼3.5 mg/ml, was stable for at least nine months when stored at −70°C in 50 mM Tris-HCl–250 mM ammonium sulfate (pH 8.5).
FIG. 1.
SDS-PAGE analysis of S. aureus Cap5O expression and purification in E. coli BL21(DE3). Lane 1, cell lysate from uninduced cells of E. coli; lane 2, cell lysate from IPTG-induced cells; lane 3, supernatant resulting from centrifugation at 39,000 × g of cell lysate; lane 4, column effluent reflecting unbound proteins; lane 5, purified Cap5O; lane M, molecular mass markers (expressed as kilodaltons).
Enzymatic function of Cap5O.
Because the substrate of Cap5O (UDP-ManNAc) is not commercially available, we produced UDP-ManNAc by incubating purified S. aureus Cap5P with its substrate UDP-GlcNAc as previously described (14). Purified Cap5O and the cofactor NAD+ were added, and the mixture was incubated at 37°C. As an indirect determination of Cap5O activity, we measured the increase in the absorbance at 340 nm resulting from NADH formation. A reaction buffer with a basic pH (ranging from 8 to 9) and a reducing agent (either dithiothreitol or β-mercaptoethanol) were both required for maximal production of NADH (data not shown). Furthermore, no enzymatic activity was detected if NAD+, Cap5O, Cap5P, or the substrate UDP-GlcNAc was omitted from the assay mixture. No NADH formation was observed if GlcNAc or UDP–N-acetylgalactosamine (UDP-GalNAc) was substituted for UDP-GlcNAc as the substrate in the assay (data not shown). The reaction mixture lacking Cap5P was used as a negative control sample for most experiments.
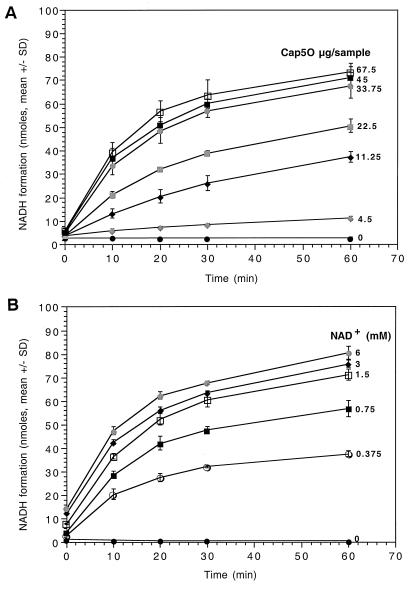
To determine the influence of Cap5O concentration on NADH production, we added increasing amounts of purified Cap5O to the enzyme reaction mixture containing 1.5 mM NAD+. As shown in Fig. 2A, NADH formation increased in a linear fashion with increasing amounts of Cap5O up to 33.75 μg. Similarly, when we added various concentrations of NAD+ (0 to 6 mM) to a reaction mixture containing 70 μg of Cap5O, the amount of NADH increased proportionally up to 1.5 mM NAD+ (Fig. 2B). We used 70 μg of Cap5O and 1.5 mM NAD+ in our standard assay. Under the standard conditions, the rate of NADH formation was 50 μmol/min/μg of Cap5O.
FIG. 2.
Time course of NADH formation with different amounts of Cap5O added (A) or with different concentrations of NAD+ added (B). The results are from three separate experiments. The error bars indicate the standard deviations.
Cap5O activity was unaffected by the addition of 2, 5, or 10 mM EDTA to the reaction mixture, indicating that exchangeable divalent cations were not required for enzyme activity (data not shown). Similarly, the addition of Mg2+ had no significant effect on the kinetics of the assay. The addition of 150 mM ammonium sulfate or ammonium chloride to the assay mixture produced no effect, whereas potassium chloride at the same concentration caused a modest inhibition (∼30%) of enzyme activity.
Identification of UDP-ManNAcA.
According to our proposed pathway for UDP-ManNAcA biosynthesis, UDP-GlcNAc is epimerized by Cap5P to UDP-ManNAc, which is then oxidized by Cap5O to UDP-ManNAcA. In the reaction products, all three sugars were expected to be present.
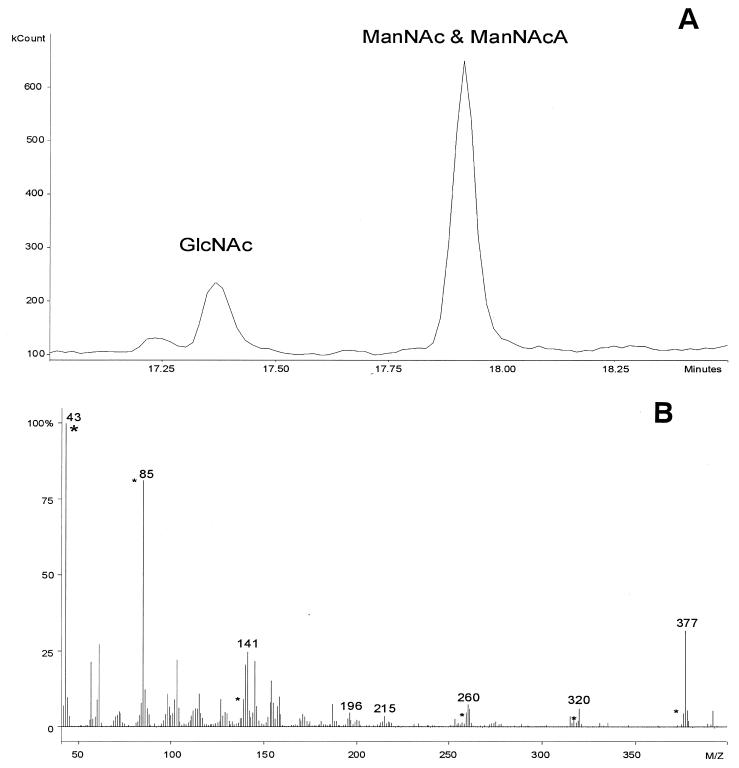
The products of the enzyme assay mixtures were reduced with sodium borodeuteride, hydrolyzed, and converted to alditol acetate derivatives. Analysis of gas chromatograms revealed two peaks with retention times of approximately 17.4 and 17.9 min (Fig. 3A), values corresponding to those for alditol acetate derivatives of authentic GlcNAc and ManNAc standards, respectively. Chromatograms from negative control samples with no Cap5P contained only the 17.4-min peak; MS analysis confirmed this peak to represent GlcNAc (data not shown). The peak at 17.9 min includes the derivatives of ManNAc and reduced ManNAcA. During the first reduction step with sodium borodeuteride, the carbon-6 of ManNAcA, but not ManNAc, is labeled with two deuterium atoms. Consequently, ManNAcA is two atomic mass units heavier than ManNAc. Thus, it was possible to distinguish the two enzymatic products accurately by MS analysis. The mass spectrum of the 17.9-min GC peak closely resembles that of a ManNAcA standard (peaks at (43, 85, 141, 196, 215, 260, 320, and 377) m/z; Fig. 3B). However, superimposed on the spectrum are peaks associated with ManNAc (peaks (43, 85, 139, 258, 318, and 375) m/z). By integration of the mass associated with the products of the reaction, the ratio of GlcNAc to ManNAc to ManNAcA was determined to be 1:3:22.
FIG. 3.
GC-MS analysis of hydrolyzed and derivatized enzyme assay mixtures. (A) Sample chromatograms showed two peaks at 17.4 and 17.9 min: retention times corresponding to those for derivatives of authentic GlcNAc and ManNAc, respectively. ManNAcA eluted together with ManNAc since the ManNAcA carboxyl group was reduced to an alcohol by sodium borodeuteride. (b) MS analysis at 17.9 min was shown to contain peaks for both ManNAc (marked with an asterisk) and ManNAcA (numbered peaks).
Essential role of the cap5O gene in CP5 expression.
A cap5O mutation was created by deletion of 727 bp in the 5′ end of the cap5O gene and subcloning the mutated fragment in a temperature-sensitive shuttle vector. The cap5O deletion was introduced by allelic exchange into the chromosome of S. aureus serotype 5 strain Reynolds, yielding mutant JLO22. To confirm the deletion in the chromosomal copy of cap5O in the mutant, genomic DNA from Reynolds and JLO22 was digested with HindIII, electrophoresed in an agarose gel, and analyzed by Southern blotting. Labeled pKBK4 (cap5O gene in pUC19) hybridized to 6.2- and 1.4-kb DNA bands from Reynolds and a single 6.9-kb DNA band from JLO22. The band sizes reflect the deletion of the 727-bp HpaI fragment (including an internal HindIII site) from the cap5O gene in the mutant JLO22. Mutant JLO22 was negative for CP5 production as determined by immunodiffusion and colony immunoblots with CP5-specific antiserum. Its growth rate in vitro was identical to that of the parental strain (data not shown).
Effect of cap5O deletion on staphylococcal virulence.
Two different strains of outbred mice were challenged intravenously with either S. aureus Reynolds or JLO22 to compare their virulence in the renal abscess model of infection. Swiss-Webster mice challenged with 4 × 105 CFU strain Reynolds had significantly (P = 0.019) higher numbers of CFU recovered per gram of kidney than mice challenged with mutant JLO22 (Table 2). However, this difference in infectivity could be overcome by increasing the inoculum to 4 × 106 CFU/mouse, in which case both groups of animals had similar numbers of staphylococci recovered from the kidney (data not shown). Significant differences in virulence were not seen at either inoculum when similar experiments were performed with ICR mice (Table 2).
TABLE 2.
Results of quantitative kidney cultures from Swiss-Webster mice inoculated intravenously with 4 × 105 CFU S. aureus
| Mouse strain (n) | Log CFU/g of kidney (mean ± SEM)
|
Pa | |
|---|---|---|---|
| Reynolds | JLO22 | ||
| ICR (9) | 3.55 ± 0.87 | 2.13 ± 0.33 | 0.1582 |
| Swiss-Webster (10–11) | 4.33 ± 0.60 | 2.58 ± 0.25 | 0.0190 |
Welch test, a modification of the unpaired Student's t test.
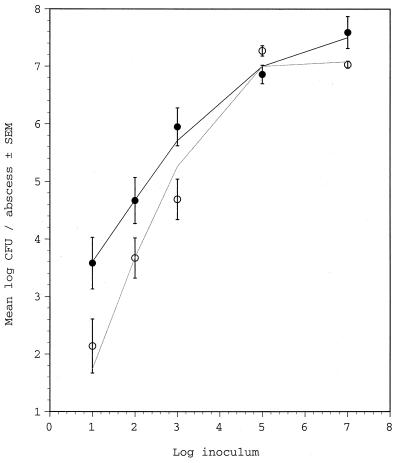
We challenged Swiss-Webster mice subcutaneously with S. aureus inocula ranging from 107 to 101 CFU. As shown in Fig. 4, animals challenged with the large inocula (107 or 105 CFU) showed similar bacterial densities (∼107 CFU/abscess) independent of the challenge strain. However, mice inoculated with 103, 102, or 101 CFU of the cap5O mutant JLO22 had significantly fewer CFU per abscess than mice inoculated with the wild-type strain Reynolds (P = 0.0018; Fig. 4). Significant differences between the strains were not observed when ICR mice were challenged with 107, 105, or 103 CFU S. aureus (data not shown).
FIG. 4.
Results of quantitative abscess cultures from groups of three to seven Swiss-Webster mice challenged subcutaneously with S. aureus Reynolds (solid symbols) or mutant JLO22 (open symbols). Curve fitting according to the statistical analysis for comparison of Reynolds and JLO22 groups is shown.
We challenged an additional group of 10 mice with a mixed inoculum containing equal numbers of strain Reynolds and JLO22 (either 103 or 102 total CFU). As shown in Table 3, between 74 and 81% of the bacteria recovered from the abscesses on day 4 were capsule positive. These data suggest that encapsulation promotes bacterial growth and/or survival within the abscess and confirm the results depicted in Fig. 4.
TABLE 3.
Results of coinfection experiments in which Swiss-Webster mice were challenged subcutaneously with equal numbers of the strains Reynolds and JLO22
| Log inoculum (CFU/mouse) | n | Log CFU/abscess (mean ± SEM) | Amt (%) of each strain recovered after 4 daysa (mean ± SEM)
|
Pb | |
|---|---|---|---|---|---|
| Reynolds | JLO22 | ||||
| 2.0 | 5 | 4.8 ± 1.1 | 81 ± 8 | 19 ± 8 | 0.0007 |
| 3.0 | 5 | 6.5 ± 0.1 | 74 ± 6 | 26 ± 6 | 0.0003 |
CP5-positive and CP5-negative colonies were scored by the colony immunoblot method (20).
Unpaired Student's t test.
DISCUSSION
DNA sequence analysis of the S. aureus cap5 and cap8 genes revealed that 16 genes [cap5(8)A through cap5(8)P], clustered on the bacterial chromosome, are involved in capsule biosynthesis (29). This information allowed us to compare the predicted amino acid sequences of cap5 and cap8 with sequences in the public databases and to assign putative functions to most of the genes (19, 29). The cap5 and cap8 gene clusters are almost identical in their flanking sequences (capA through capG and capL through capP), but they differ in the central serotype-specific gene region (capH through capK) (29). The function of only a few of the biosynthetic genes has been proven. We showed that the gene product of cap5H, one of the CP5-specific genes, O acetylates the third carbon on the ManNAcA residues of CP5 (5). In addition, we showed that the purified product of the cap5P gene is a UDP-GlcNAc 2-epimerase that catalyzes the conversion of UDP-GlcNAc to UDP-ManNAc (14). In this study, we characterized the S. aureus cap5O gene product as a UDP-ManNAc dehydrogenase.
The enzymatic activity of Cap5O was quantitated indirectly by mixing Cap5O with Cap5P and UDP-GlcNAc and measuring NADH production. The oxidation product was confirmed to be a UDP-ManNAcA by reduction of the reaction products with sodium borodeuteride and analysis of the derivatized products by GC-MS. Thus, we propose that the synthesis of UDP-ManNAcA in S. aureus occurs as presented in Fig. 5.
FIG. 5.
Synthesis of UDP-ManNacA in S. aureus.
GC-MS analysis of some of the coupled reaction mixtures containing both Cap5P and Cap5O revealed that all of the UDP-GlcNAc substrate was converted into UDP-ManNAc and UDP-ManNAcA. This is in contrast to the reversible Cap5P-mediated UDP-GlcNAc 2-epimerase reaction in which only ∼10% of the substrate was converted to UDP-ManNAc (14). It is likely that coupling the 2-epimerase reaction to the dehydrogenase reaction depletes the intermediate product UDP-ManNAc. As a result, the reversible 2-epimerase reaction is driven toward the formation of more UDP-ManNAc, the substrate for Cap5O.
The biochemical properties of S. aureus Cap5O are similar to those described for other microbial dehydrogenases. For example, both S. aureus Cap5O and an E. coli UDP-ManNAc dehydrogenase require a basic pH and a reducing agent to be present for maximal activity (12). Like other dehydrogenases with specificity for UDP-ManNAc (11, 22) or UDP-glucose (3), S. aureus Cap5O was unaffected by EDTA or divalent cations. Moreover, S. aureus UDP-ManNAc dehydrogenase contains the N-terminal NAD-binding domain (GXGXXG) typical of other dehydrogenases (33) requiring NAD+ as a cofactor.
The deletion of cap5O in S. aureus Reynolds yielded a CP5-negative mutant. Similarly, Sau et al. (30) showed that the serotype 8 strain Becker with a chemically induced mutation in cap8O was negative for CP8 production. Recombinant plasmids containing intact cap8O complemented the function of the mutated cap8O gene in trans. Similarly, we showed that introduction of the wild-type cap5O gene on a plasmid restored CP5 expression to a cap5O mutant of strain Newman (28).
The role of the S. aureus capsule in the pathogenesis of staphylococcal infections has been examined in a number of test systems. Serotype 5 and 8 strains of S. aureus were shown to resist opsonophagocytic killing by human polymorphonuclear leukocytes (8, 31). In addition, CP5 enhanced virulence in mouse models of lethality (31), bacteremia (31), and septic arthritis (25) and promoted long-term nasal colonization by S. aureus in mice (15). In contrast, both CP5 and CP8 attenuated staphylococcal virulence in a rat model of catheter-induced endocarditis (4). In 1991, we challenged inbred C57BL/6J mice intravenously with ∼5 × 106 CFU of the wild-type strain Reynolds, a capsule-deficient mutant created by transposon mutagenesis, or a chemically induced capsule-negative mutant (1). Because all of the mice developed renal abscesses and the numbers of bacteria recovered from the kidneys were similar, we concluded that CP5 did not influence renal abscess formation. The staphylococci in that study were harvested from logarithmic-phase broth cultures, in which little capsule is expressed (28, 31). In this study, we reexamined the role of capsule in renal abscess formation by challenging two different strains of mice with S. aureus cultivated under conditions known to optimize capsule expression (31). At a challenge inoculum of 4 × 105 CFU, significantly greater numbers of the parental strain Reynolds were recovered from the kidneys of Swiss-Webster mice compared with those challenged with the CP5-negative mutant JLO22. This effect on virulence was modest, however, since no differences between the two groups of animals were observed at a 10-fold-greater inoculum. Similar experiments carried out in ICR mice revealed no differences in virulence at either challenge dose. Mice are highly resistant to S. aureus infection and, in this model, an inoculum >105 CFU is essential for infectivity.
The subcutaneous abscess model proved to be a more sensitive model of infection since inocula as low as 10 CFU could provoke an infection by the wild-type S. aureus strain. Significantly fewer organisms were recovered from the subcutaneous abscesses of Swiss-Webster mice challenged with ≤103 CFU mutant JLO22 compared with the wild-type strain. Moreover, in coinfection experiments, 75 to 80% of the organisms recovered after 4 days were CP5 positive. This result is in agreement with our findings that the encapsulated wild-type strain is more virulent than the acapsular mutant in this model. However, no differences in virulence were observed when ICR mice were challenged subcutaneously with 107, 105, or 103 CFU. Taken together, these results suggest that the role of the capsule in the pathogenesis of staphylococcal infections is dependent not only on the bacterial growth conditions and inoculum size but also on the genetic background of the host.
The pathogenesis of the S. aureus subcutaneous infection model is clearly different from that of the renal abscess model. To induce subcutaneous abscesses, the bacteria are injected directly into the subcutaneous tissue in the presence of a foreign body (cytodex beads). It is likely that encapsulated staphylococci avoid uptake by phagocytes recruited to the site of infection, and thus capsule-positive S. aureus have a survival and growth advantage over staphylococci lacking a capsule. In contrast, a bolus dose of staphylococci is delivered intravenously to mice to provoke renal abscesses. The liver and spleen clear the majority of organisms, and only a small number of blood-borne organisms seed the kidney. We did not observe a significant difference in the number of parental or mutant S. aureus cells recovered from the kidney 24 h after bacterial challenge (unpublished observations). However, an S. aureus cap5O mutant lacking CP5 expression showed greater adherence to endothelial cells in vitro compared with the parental strain (28). It is likely that capsule expression augments staphylococcal survival within the kidney by enhancing its resistance to phagocytic uptake and killing.
In conclusion, we have purified the S. aureus cap5O gene product and demonstrated that it has UDP-ManNAc dehydrogenase activity. Cap5O is essential for CP5 expression, and a mutant lacking this enzyme is less virulent in two Swiss-Webster mouse models of abscess formation. Our findings are consistent with the observation that antibodies that neutralize the S. aureus capsule show some protection against S. aureus infections.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI29040 (to J. C. Lee) and AI09981 (to K. B. Kiser) from the National Institute of Allergy and Infectious Diseases.
We thank Jessica S. Lam and Adam J. Reitz, Jr., for their technical assistance and Vincent J. Carey, Jr., for his help on statistical analysis of the data.
REFERENCES
- 1.Albus A, Arbeit R D, Lee J C. Virulence of Staphylococcus aureus mutants altered in type 5 capsule production. Infect Immun. 1991;59:1008–1014. doi: 10.1128/iai.59.3.1008-1014.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeit R D, Karakawa W W, Vann W F, Robbins J B. Predominance of two newly described capsular polysaccharide types among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis. 1984;2:85–91. doi: 10.1016/0732-8893(84)90002-6. [DOI] [PubMed] [Google Scholar]
- 3.Arrecubieta C, Garcia E, Lopez R. Demonstration of UDP-glucose dehydrogenase activity in cell extracts of Escherichia coli expressing the pneumococcal cap3A gene required for the synthesis of type 3 capsular polysaccharide. J Bacteriol. 1996;178:2971–2974. doi: 10.1128/jb.178.10.2971-2974.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baddour L M, Lowrance C, Albus A, Lowrance J H, Anderson S K, Lee J C. Staphylococcus aureus microcapsule expression attenuates bacterial virulence in a rat model of experimental endocarditis. J Infect Dis. 1992;165:749–753. doi: 10.1093/infdis/165.4.749. [DOI] [PubMed] [Google Scholar]
- 5.Bhasin N, Albus A, Michon F, Livolsi P J, Park J-S, Lee J C. Identification of a gene essential for O-acetylation of the Staphylococcus aureus type 5 capsular polysaccharide. Mol Microbiol. 1998;27:9–21. doi: 10.1046/j.1365-2958.1998.00646.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Ford C W, Hamel J C, Stapert D, Yancey R J. Establishment of an experimental model of a Staphylococcus aureus abscess in mice by use of dextran and gelatin microcarriers. J Med Microbiol. 1989;28:259–266. doi: 10.1099/00222615-28-4-259. [DOI] [PubMed] [Google Scholar]
- 8.Karakawa W W, Sutton A, Schneerson R, Karpas A, Vann W F. Capsular antibodies induce type-specific phagocytosis of capsulated Staphylococcus aureus by human polymorphonuclear leukocytes. Infect Immun. 1988;56:1090–1095. doi: 10.1128/iai.56.5.1090-1095.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karakawa W W, Vann W F. Capsular polysaccharides of Staphylococcus aureus. Semin Infect Dis. 1982;4:285–293. [Google Scholar]
- 10.Kasatiya S S, Baldwin J N. Nature of the determinant of tetracycline resistance in Staphylococcus aureus. Can J Microbiol. 1967;13:1079–1086. doi: 10.1139/m67-144. [DOI] [PubMed] [Google Scholar]
- 11.Kawamura T, Ichihara N, Sugiyama S, Yokota H, Ishimoto N, Ito E. Biosynthesis of UDP-N-acetyl-D-glucosaminuronic acid and UDP–N-acetyl-d-mannosaminuronic acid in Micrococcus luteus. J Biochem. 1985;98:105–116. doi: 10.1093/oxfordjournals.jbchem.a135248. [DOI] [PubMed] [Google Scholar]
- 12.Kawamura T, Ishimoto N, Ito E. Enzymatic synthesis of uridine diphosphate N-acetyl-d-mannosaminouronic acid. J Biol Chem. 1979;254:8457–8465. [PubMed] [Google Scholar]
- 13.Kiser K, Lee J. Staphylococcus aureus cap5O and cap5P genes functionally complement mutations affecting enterobacterial common antigen biosynthesis in Escherichia coli. J Bacteriol. 1998;180:403–406. doi: 10.1128/jb.180.2.403-406.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiser K B, Bhasin N, Deng L, Lee J C. Staphylococcus aureus cap5P encodes a UDP–N-acetylglucosamine 2-epimerase with functional redundancy. J Bacteriol. 1999;181:4818–4824. doi: 10.1128/jb.181.16.4818-4824.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiser K B, Cantey-Kiser J M, Lee J C. Development and characterization of a Staphylococcus aureus nasal colonization model in mice. Infect Immun. 1999;67:5001–5006. doi: 10.1128/iai.67.10.5001-5006.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C Y, Buranen S L, Ye Z-H. Construction of single-copy integration vectors for Staphylococcus aureus. Gene. 1991;103:101–105. doi: 10.1016/0378-1119(91)90399-v. [DOI] [PubMed] [Google Scholar]
- 17.Lee J C. Electrotransformation of staphylococci. Methods Mol Biol. 1995;47:209–216. doi: 10.1385/0-89603-310-4:209. [DOI] [PubMed] [Google Scholar]
- 18.Lee J C, Betley M J, Hopkins C A, Perez N E, Pier G B. Virulence studies, in mice, of transposon-induced mutants of Staphylococcus aureus differing in capsule size. J Infect Dis. 1987;156:741–750. doi: 10.1093/infdis/156.5.741. [DOI] [PubMed] [Google Scholar]
- 19.Lee J C, Lee C Y. Capsular polysaccharides of Staphylococcus aureus. In: Goldberg J B, editor. Genetics of bacterial polysaccharides. Boca Raton, Fla: CRC Press, Inc.; 1999. pp. 185–205. [Google Scholar]
- 20.Lee J C, Liu M J, Parsonnet J, Arbeit R D. Expression of type 8 capsular polysaccharide and production of toxic shock syndrome toxin-1 are associated among vaginal isolates of Staphylococcus aureus. J Clin Microbiol. 1990;28:2612–2615. doi: 10.1128/jcm.28.12.2612-2615.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J C, Xu S L, Albus A, Livolsi P J. Genetic analysis of type 5 capsular polysaccharide expression by Staphylococcus aureus. J Bacteriol. 1994;176:4883–4889. doi: 10.1128/jb.176.16.4883-4889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lew H C, Nikaido H, Makela P H. Biosynthesis of uridine diphosphate N-acetylmannosaminuronic acid in rff mutants of Salmonella typhimurium. J Bacteriol. 1978;136:227–233. doi: 10.1128/jb.136.1.227-233.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 24.Meier-Dieter U, Starman R, Barr K, Mayer H, Rick P D. Biosynthesis of enterobacterial common antigen in Escherichia coli. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- 25.Nilsson I-M, Lee J C, Bremell T, Ryden C, Tarkowski A. The role of staphylococcal polysaccharide microcapsule expression in septicemia and septic arthritis. Infect Immun. 1997;65:4216–4221. doi: 10.1128/iai.65.10.4216-4221.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Connell C, Pattee P A, Foster T J. Sequence and mapping of the aroA gene of Staphylococcus aureus 8325-4. J Gen Microbiol. 1993;139:1449–1460. doi: 10.1099/00221287-139-7-1449. [DOI] [PubMed] [Google Scholar]
- 27.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohlmann-Dietze P, Ulrich M, Kiser K B, Doring G, Lee J C, Fournier J M, Botzenhart K, Wolz C. Adherence of Staphylococcus aureus to endothelial cells: influence of the capsular polysaccharide, the global regulator agr, and the bacterial growth phase. Infect Immun. 2000;68:4865–4871. doi: 10.1128/iai.68.9.4865-4871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 30.Sau S, Sun J, Lee C Y. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol. 1997;179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakker M, Park J-S, Carey V, Lee J C. Staphylococcus aureus serotype 5 capsular polysaccharide is antiphagocytic and enhances bacterial virulence in a murine bacteremia model. Infect Immun. 1998;66:5183–5189. doi: 10.1128/iai.66.11.5183-5189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisberg S. Applied linear regression. Wiley series in probability and mathematical statistics. xii. New York, N.Y: Wiley, Inc.; 1980. [Google Scholar]
- 33.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]