Abstract
In 2019, a new coronavirus was identified that has caused significant morbidity and mortality worldwide. Like all RNA viruses, severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) evolves over time through random mutation resulting in genetic variations in the population. Although the currently approved coronavirus disease 2019 vaccines can be given to those over 5 years of age and older in most countries, strikingly, the number of people diagnosed positive for SARS-Cov-2 is still increasing. Therefore, to prevent and control this epidemic, early diagnosis of infected individuals is of great importance. The current detection of SARS-Cov-2 coronavirus variants are mainly based on reverse transcription-polymerase chain reaction. Although the sensitivity of reverse transcription-polymerase chain reaction is high, it has some disadvantages, for example, multiple temperature changes, long detection time, complicated operation, expensive instruments, and the need for professional personnel, which brings considerable inconvenience to the early diagnosis of this virus. This review comprehensively summarizes the development and application of various current detection technologies for novel coronaviruses, including isothermal amplification, CRISPR-Cas detection, serological detection, biosensor, ensemble, and microfluidic technology, along with next-generation sequencing. Those findings offer us a great potential to replace or combine with reverse transcription-polymerase chain reaction detection to achieve the purpose of allowing predictive diagnostics and targeted prevention of SARS-Cov-2 in the future.
Key Points
| Emerging variants of severe acute respiratory syndrome coronavirus 2 are still spreading rapidly in our community, resulting in serious health issues and a global economic crisis. |
| US Food and Drug Administration-approved nucleic acid-based molecular assays including reverse transcription-polymerase chain reaction and rapid antigen tests have been the dominant techniques for the detection of severe acute respiratory syndrome coronavirus 2, the drawbacks of these two methods hinder the current coronavirus disease 2019 testing paradigm. |
| Novel fast, sensitive, and accurate coronavirus detection technologies are urgently needed. |
| Recent advances in the development of molecular diagnostic technologies, including isothermal amplification, CRISPR-Cas, biosensors, sequencing, and microfluidic-based technology offer us great potential for the rapid and sensitive detection of coronavirus disease 2019. |
Introduction
In late 2019, several cases of pneumonia of unknown origin were reported in Wuhan, China. This pneumonia was later considered to be a respiratory viral infection pneumonia, and its causative pathogen was identified as severe acute respiratory syndrome coronavirus [1], which was named severe acute respiratory syndrome coronavirus 2 (SARS-COV-2, coronavirus disease 2019 [COVID-19]) by the World Health Organization. As of 5 June, 2022, the coronavirus had resulted in approximately 529 million infections and more than 6 million deaths worldwide [2].
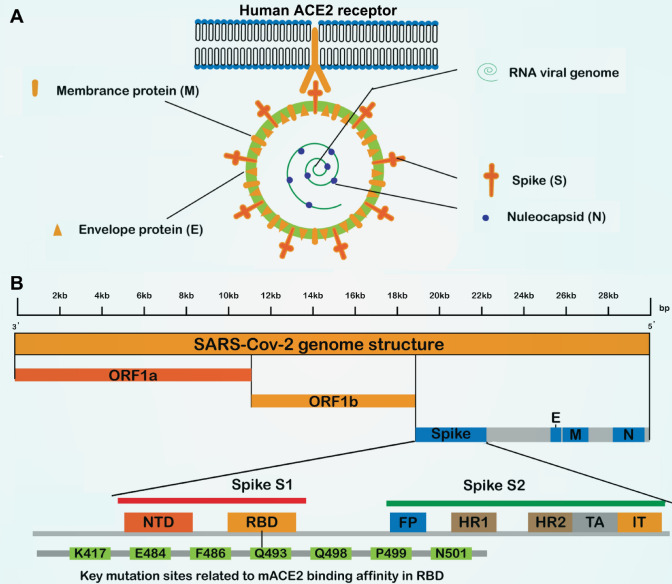
Severe acute respiratory syndrome coronavirus 2 and severe acute respiratory syndrome coronavirus belong to the β-COV of the Coronavirus family of the Reticuloviridae, a single-stranded RNA virus without segmentation. As shown in Fig. 1, the S protein is a key protein for coronavirus infection [3]. The affinity of SARS-Cov-2 to ACE2 is 10–20 times higher than that of severe acute respiratory syndrome coronavirus to ACE2, and the S protein is a key target for clinical therapy and diagnosis [4]. Although an authorized vaccine against the S protein has been developed, the viral surface S protein is frequently mutated, thus leading to vaccinated individuals still having a high probability of contracting this coronavirus [5], and the prevention and control of the epidemic are becoming increasingly challenging as the virus mutates.
Fig. 1.
Schematic of severe acute respiratory syndrome coronavirus 2 (SARS-Cov2) structure, detection method, and genome annotation of spike proteins. A SARS-Cov-2 virus structure includes surface proteins (S), envelope protein (E), membrane protein (M), nucleocapsid (N) protein, and RNA genome. B The structure of the SARS-Cov-2 genome includes two ORF1a, ORF1b, and four structural proteins: N protein, M protein, E protein, and S protein
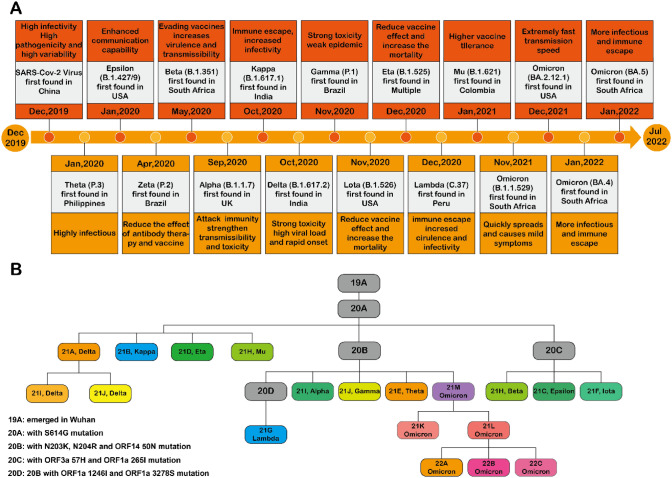
Currently, the eight variants of interest include Lambda (C.37), MU (B.1.621), Zeta (P.2), Eta (B.1.525), Thela (P.3), Lota (B.1.526), Kappa (B.1.617.1), and Epsilon (B.1.427, B.1.429). The five variants of concern including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (including BA.1, BA.2, BA.3, BA.4, BA.5, and descendent lineages) [B.1.1.529] and the two variants under monitoring released from the World Health Organization reduce the role of vaccines (Fig. 2), and the difficulty of disease prevention and treatment is also increasing. Therefore, there is a need to find a rapid, simple, and sensitive detection technique as an alternative to reverse transcription-polymerase chain reaction (RT-PCR) technology or combined RT-PCR. Early detection of the virus through accurate techniques is currently the most important aspect of outbreak-targeted prevention and prognostic capacity.
Fig. 2.
Schematic timeline of the emergence of coronavirus 2019 pandemic and severe acute respiratory syndrome coronavirus 2 (SARS-Cov2) variants. A Timeline of coronavirus 2019 pandemic from December 2019 to May 2022. B The emergence of SARS-Cov-2 virus variants in three main categories: variants of interest, variants of concern, and variants under monitoring. The eight variants of interest are Lambda (C.37), MU (B.1.621), Zeta (P.2), Eta (B.1.525), Thela (P.3), Lota (B.1.526), Kappa (B.1.617.1), and Epsilon (B.1.427,B.1.429); the five variants of concern are Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (includes BA.1, BA.2, BA.3, BA.4, BA.5, and descendent lineages) [B.1.1.529]; two variants are being monitored
Classical Coronavirus Detection Technology
RT-PCR Test
Rapid and accurate disease diagnosis is essential for the effective treatment and prevention of long-term sequelae [6, 7]. At present, more than 200 SARS-COV-2 assays have been approved by the US Food and Drug Administration, 19 representative test kits are shown in Table 1, and the method mostly used for SARS-COV-2 detection is RT-PCR (a diagram of the RT-PCR process is shown in Fig. 3A). This technique is the gold standard for SARS-CoV-2 detection as it allows direct detection of the presence of viral RNA and yields sensitive and reliable results in a high-throughput manner within a few hours [8, 9].
Table 1.
US Food and Drug Administration-approved test method for SARS-Cov-2
| Type | Source target | Result time | Performance | Number | Example |
|---|---|---|---|---|---|
| PCR | RNA | 45–120 min | LOD: 100–75,000 copies/mL | 156 | Xpert Xpress SARS-CoV-2 |
| ddPCR | RNA | 2–4 h | LOD: 625 copies/mL | 1 | Bio-RADSARS-CoV-2 ddPCR Test |
| NEAR | RNA | 5–15 min | LOD: 125 copies/mL | 1 | ID NOW COVID-19 test |
| RT-LAMP | RNA | N/A | LOD: 1000 copies/mL | 1 | AQ-TOP COVID-19 Detection Kit PLUS |
| OMEGA | RNA | 1 h | LOD: 4000 copies/mL | 1 | iAMP COVID-19 detection kit |
| SHERLOCK | RNA | 1 h | LOD: 900 copies/mL | 1 | CRISPR SARS-CoV-2 |
| DETECTR | RNA | N/A | LOD: 20,000 copies/mL | 2 | SARS-CoV-2 DETECTR Reagent Kit |
| NGS | RNA | 4–6 h | LOD: 250–1000 copies/mL | 5 | Explify Respiratory |
| uNMR | RNA | N/A | LOD: 2000 copies /mL | 1 | T2SARS-CoV-2 Panel |
| LFA | IgM/IgG Ag | 15–20 min |
Sen: 93.3–100% Spe: 94.4–100% |
22 | BinaxNOW COVID-19 Ag Card |
| ELISA | IgM/IgG Ag | 20 min |
Sen: 92.5–100% Spe: 97.5–100% |
12 | CareStart COVID-19 Antigen Test |
| CLIA | IgM/IgG | 30 min |
Sen: 85.4–100% Spe: 64.7–100% |
18 | AdviseDx SARS-CoV-2 IgM |
| EIA | IgM/IgG | 2 h |
Sen: 92.2% Spe: 99.6% |
2 | Platelia SARS-CoV-2 Total Ab test |
| MIA | IgM/IgG | N/A |
Sen: 88–100% Spe: 98.8–99.8% |
4 | xMAP SARS-CoV-2 Multi-Antigen IgG |
| ECLIA |
IgM/IgG IL6 |
20 min |
Sen: 84–100% Spe: 63–99.8% |
2 | Elecsys Anti-SARS-CoV-2 Cobas |
| ECS | IgG, cytokine | 2 h |
Sen: 91.4% Spe: 100% |
1 | ePlex SARS-CoV-2 test |
| PRI | Antigen | N/A | Spe: 97.7% | 1 | Maverick SARS-CoV-2 Multi-Antigen |
| Microarrays | IgM/IgG | 1.5 h | Spe: 99.8% | 1 | MosaiQ COVID-19 Antibody Magazine |
| IFM | Antigen | 3 h |
Sen: 97.6% Spe: 96.6% |
1 | LumiraDx SARS-CoV-2 Ag Test |
COVID-19 coronavirus disease 2019, DETECTR (DNA endonuclease-targeted CRISPR reporter, ddPCR droplet digital PCR, ELISA enzyme-linked immunoassay, h hours, min minutes, LFA lateral flow assay, N/A , NGS next-generation sequencing, PCR polymerase chain reaction, RT-LAMP reverse-transcription-loop mediated isothermal amplification, SARS-Cov-2 severe acute respiratory syndrome coronavirus 2, Sen sensitivity, SHERLOCK Specific High Sensitivity Enzymatic Reporter Unlocking, Spe specificity
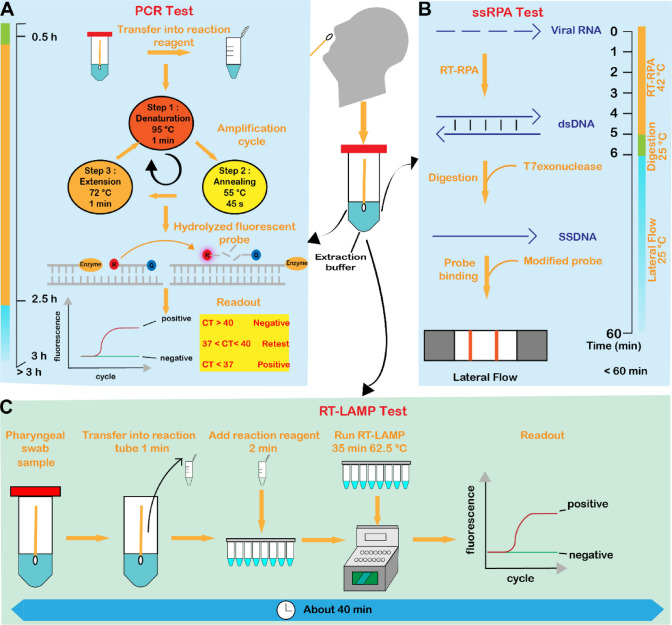
Fig. 3.
Nucleic acid-based severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) tests. A Basic steps of polymerase chain reaction (PCR) assay: (1) transfer the sample into the reaction reagent; (2) RNA amplification: denaturation, annealing, extension; (3) hydrolysis of the fluorophore; and (4) readout. The whole process lasted for more than 3 h. B Basic steps of recombinase polymerase amplification (RPA) detection: Step 1: The target RNA is reverse transcribed to form DNA, and then isothermally amplified. Step 2: The amplified product is transferred to a buffer containing T7 nucleic acid exonuclease and incubated for 1 min to digest the reverse strand of the double-stranded DNA (dsDNA) amplification product to form single-stranded DNA (SSDNA) amplicons homologous to the target RNA. Step 3: the SSDNA amplicons are bound to biotin and FAM-modified detection probes, immobilized on the detection line (biotin binding) and capture the rabbit anti-FAM-gold nanoparticles complexes, producing a positive result. The entire process takes less than 60 min. C Loop-mediated isothermal amplification (LAMP) assay steps (the entire process from adding samples and reagents to running reverse transcription (RT)-LAMP is completed in about 40 minutes [min]). CT cycle threshold, h hours, RT-RPA reverse transcription-recombinase polymerase amplification, s seconds, ssRPA single-strand Recombinase Polymerase Amplification
Currently, the SARS-CoV-2 RNA detection reagents certified by the Food and Drug Administration are based on the RT-PCR technology, which is being continuously updated, a good example of this is the Cepheid Xpert Xpress assay [10]. The speed of detection has been greatly improved (45 minutes to obtain results), and now this assay has a fairly high specificity and sensitivity. However, the disadvantages of RT-PCR are unavoidable. These disadvantages include: (1) stringent requirements for sample collection, as improper collection can result in false negatives; (2) high cost of instrumentation, accessories, and maintenance; (3) complexity of instrument operation; (4) the requirement for highly qualified technicians; and (5) unsuitability for rapid on-site testing and on-site analysis. Thus, we still need to develop a new detection technology that replaces or assists RT-PCR technology for SARS-COV-2 detection to meet the current requirements for virus detection in epidemic prevention and control.
With the continuous variation of SARS-Cov-2, most of the RT-PCR primers or probes designed for coronavirus detection inevitably led to a decrease in sensitivity or even a false-negative result. In a study of six SARS-Cov-2 variants (α, β, γ, δ, ο, and Fin-796H), detection using five detection reagents (DCan, Bio-Germ, EasyDiagnosis, LiveRiver, and Sansure) showed the β and δ variants adversely affected the sensitivity of the BioGerm and Sansure [10]. Therefore, RT-PCR detection technology needs to be constantly optimized to cope with the continuous mutating SARS-Cov-2. The Chung et al. group developed a multiplex RT-PCR by using specific primers or probes targeting nine mutations (ΔHV 69/70, K417T, K417N, L452R, E484K, E484Q, N501Y, P681H, and P681R) on the S gene, and they found that this method exhibited better sensitivity in 250 clinically positive samples than those of the commercial VirSNiP Covid-19 variant kit [11]. Another study showed a multiplex, quantitative, RT-PCR-based molecular beacon technology, the sensitivity and specificity of this technology can achieve 100% in 26 confirmed positive patients [12]. These studies suggested that the RT-PCR-based diagnostic method needs to be optimized as viral mutations continue.
Detection of Antibodies
For virus serological antibody detection, commonly used methods include the enzyme-linked immunoassay (ELISA), chemiluminescence immunoassay, immunofluorescence assay, and the colloidal gold immunochromatography assay. Among them, ELISA is the gold standard in serology, providing quantitative and extremely sensitive assays [13]. Antibody testing has been a complementary method to PCR methods for the detection of neo-coronaviruses and helps to screen suspected and asymptomatic patients.
Detection of antibodies to SARS-CoV-2-specific protein in serum plays a very important role in the detection of prior infection with the new coronavirus [14]. Zhang et al. developed a rapid and sensitive magnetofluidic immuno-PCR technique [15], which preloads a programmable magnetic arm in the analytical reagent to attract and transport magnetically captured specific antibodies, followed by the use of a microthermal immuno-PCR performed with a circulator and a fluorescence detector to detect specific antibodies. To validate the sensitivity of the method, the team analyzed 108 clinical serum samples and obtained a sensitivity of 93.8% (45/48) and a specificity of 98.3% (59/60). Magnetic fluid immuno-PCR overcomes lengthy workflows and bulky instrumentation and has a great potential for rapid and sensitive serological testing.
In another study, Li et al. developed a rapid and simple lateral flow immunoassay [16]. This research group used gold nanoparticles (Au Nps) and SARS-Cov-2 antigens in combination to form Au NP-Ag conjugates. The anti-human IgM antibody is immobilized on the M line (M = IgM) of the horizontal flow test strip and the anti-human IgG antibody is immobilized on the G line (G = IgG). On the C quality- control line (C = control), the anti-rabbit IgG antibody is immobilized. When the target serum contains IgM or IgG antibodies, it binds specifically to the Au NP-Ag conjugate via anti-human IgM or IgG antibodies. To verify the sensitivity and specificity of the method, the team collected 525 blood samples from patients with COVID-19. Of these, 397 were clinically confirmed cases. A total of 352 samples were positive (sensitivity of 88%), and 12 of 128 non-infected individuals tested positive (specificity of 90.63%). Because of the influence of time on the appearance of antibodies in the body (antibodies usually appear 1 week after infection), the research team tested 58 patients in Wuhan (8–33 days after infection). The test results showed that 94.83% of patients were positive for both IgM and IgG, 1.72% were positive for IgM, and 3.45% were positive for IgG, with an overall sensitivity of 100%. To simplify the methodology, the authors also tested finger peripheral blood from ten patients (seven positive patients and three negative patients). The results showed that four of the seven patients were double-antibody positive and three were IgM positive (IgM antibodies appeared earlier), while all three healthy individuals were negative for both antibodies.
These experimental results suggest that an AuNP-based antibody detection method may be advantageous. It has high sensitivity and specificity, a short detection time (results can be obtained in 15 min), and requires simple equipment, which meet the requirements of disease screening. This rapid test has enormous potential for rapid screening of SARS-CoV-2 infection, but there are limitations to the early detection of serum antibodies owing to the late appearance of antibodies in vivo. Therefore, the detection of serum antibodies is used more as an assessment of disease progression.
Detection of Antigen
Currently, SARS-COV-2 antigen detection is for N protein antigen sites. Compared with PCR methods, although the accuracy of antigen detection is lower, it is shorter, easier, cheaper to perform, and more suitable for point-of-care testing (POCT) detection. Therefore, most of the Food and Drug Administration-approved home self-testing assays are based on antigen detection. Early screening allows for rapid and cost-effective detection of COVID-19 in symptomatic individuals and contacts of confirmed cases, thus necessary action can be promptly taken. The nucleocapsid protein (NP) is the main structural protein expressed by SARS-Cov-2 and has been identified as an ideal target in the early stages of SARS-Cov-2 infection.
Diao et al. evaluated the use of SARS-Cov-2 NP as a target of viral antigens and established a rapid, simple, and accurate SARS-Cov-2 antigen detection test based on a fluorescent immunochromatographic method [17]. They used nitrocellulose membranes labeled with SAR-Cov-2 NP and antigen-specific antibodies to detect the NP antigen. When the sample contains the SARS-Cov-2 NP antigen, a double antibody sandwich is formed and a fluorescent signal is detected, whereas no fluorescent lines are formed when the target antigen is not present in the sample. A fluorescence reaction line is formed in the control region regardless of the presence or absence of the NP antigen as the assay control. The research team analyzed 253 participants, of whom two participants were excluded because of the invalid NP results. Simultaneously, diagnostic accuracy of NP antigen testing was measured by blindly performing RT-PCR as the reference standard, in which samples with a cycle threshold (CT) of RT-PCR of ≤40 were considered as SARS-CoV-2 positive. Of the 251 valid results, 201 (80.1%) had a CT value of <40, 46 had 37 < CT ≤ 40 (18.3%), 155 had a CT value of <37 (81.7%), and 50 had a CT value of >40 (19.9%), and the sensitivity, specificity, and concordance of the fluorescence immunochromatographic assay, using RT-PCR as the reference standard, were 75.6%, 100%, and 80.5%, respectively. These findings all indicate that the sensitivity of the study method is lower than that of the PCR assay, but it has a rapid and simple operation, making antigen detection a better method for home or POCT detection of SARS-Cov-2.
Traditional virus antigen detection methods are prone to false-negative results and low accuracy, which are detrimental to the clinical diagnosis and treatment of patients [18]. New research has produced a multi-antibody combination modified graphene transistor sensor [19], a detection device that targets different regions of the SARS-Cov-2 S protein by modifying S1 protein monoclonal, CR3022 monoclonal, and N3021 nano antibodies on the surface of graphene field-effect transistors. It achieves rapid detection of the SARS-Cov-2 antigen by a synergistic action. The research team evaluated 43 single-tube clinical samples and 17 10-in-1 mixed samples using the device. The results showed a 100% overlap with RT-PCR, demonstrating high accuracy, and the average detection time of this method was only 38.9 s. This assay technique solves the problem of a low precision of antigen detection, and its rapid and high-throughput advantages make it a valuable tool for the rapid diagnosis and screening of COVID-19; however, obviously, more testing with much higher numbers of patients will be needed before clinical application.
The use of antigen or antibody assays in virus detection is quite well established, but the poor specificity and the time lag before antibodies appear in vivo have meant that compared with PCR detection techniques, antigen and antibody assays are more often used as complementary assays for SARS-Cov-2 nucleic acid detection. However, this series of recent studies have demonstrated that combining traditional immunological methods with emerging nanomaterials (which act as a signal amplification), such as AuNPs [16], graphene [19], or an electrochemical detection platform [20], is a new direction of prognostic capacity for future antigen and antibody detection.
Novel Coronavirus Detection Technology
Isothermal Amplification Technology
Isothermal amplification technology is a general term for a biomolecular technique that amplifies specific DNA or RNA at a specific temperature. Among these techniques, loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) are the most widely used in viral nucleic acid detection [21]. Compared with conventional amplification methods, isothermal amplification of nucleic acids simplifies the equipment requirements. The reaction time is also greatly reduced, and the sensitivity is comparable to that of RT-PCR, which can better meet the need for a rapid and simple detection of COVID-19 nucleic acid (Fig. 3).
Polymerase Amplification Technology
Polymerase amplification (RPA) is a nucleic acid thermostable amplification technology developed by Armes and coworkers in 2006 [22] and it relies on three enzymes: recombinases capable of binding single-stranded nucleic acids, single-stranded DNA-binding proteins, and strand-substituted DNA polymerases. Recombinases and primers form complexes that search for homologous sequences on the template. After localization, a chain exchange reaction will be initiated to exponentially amplify the target region on the template. Currently, RPA technology has made great progress in virus detection. Back in 2012, Rohrman et al. reported a RPA-based HIV detection assay [23]. Since then, RPA has shown high sensitivity and specificity in the detection of H7N9 [24] and dengue virus [25].
Following the COVID-19 outbreak, Kim et al. reported an improved version of the reverse transcription (RT)-RPA test using the single-strand recombinase polymerase amplification method [26], which applies rapid amplification of double-stranded DNA, conversion to single-stranded DNA, and a sequence-specific hybridization-based readout by a lateral flow device (Fig. 3B). Single-strand recombinase polymerase amplification combines the speed of established RT-RPA with the sequence specificity of single-stranded DNA. The team analyzed a limited number of clinical samples, only 18, using single-strand recombinase polymerase amplification technology and compared them to RT-PCR, concluding 100% sensitivity and 100% specificity. In addition, single-strand recombinase polymerase amplification had a better reaction time and a lower detection limit (eight copies per 50-µL reaction in 8 minutes or four copies in 10 minutes for the naked eye) compared with RT-PCR.
In another study, Xia et al. reported the introduction of an improved one-pot RT-RPA by Gendx Biotechnology, called reverse transcription-enzymatic recombinase amplification [27]. This method features a fluorescence resonance energy transfer designed to detect the N and S genes of SARS-Cov-2, combined with WEPEAR (a whole-course encapsulated process for more essential adaptation from RNA), and they also designed two different probes, fluorescence resonance energy transfer and NFO (primer named by author). The fluorescence resonance energy transfer probe detects RNA genes by fluorescence enhancement, while the NFO affinity probe detects RNA using lateral flow strips, making the detection method simpler. The team demonstrated through five independent experiments that the method has a low detection limit of 1 copy and high accuracy (10 out of 11 samples showed positive results). More importantly, the assay can meet the requirements of home detection and POCT.
LAMP
Loop-mediated isothermal amplification technology is a nucleic acid amplification technology proposed by the Notomiet research team in 2000 [28]. This technique uses two to three sets of specially designed primers, including forward internal primers (FIP, F1C, and F2) and reverse inner primers (BIP, B1C, and B2), outer primers F3, B3, and bst (Bacillus stearothermophilus), and DNA polymerase with strand-substitution activity, so that the primers at both ends of the template are bound during the reaction. The circular single-stranded structure that appears in the loop ensures that the primers can bind smoothly to the template under isothermal conditions for an amplification reaction [29].
In 2004, lhira et al. applied LAMP to the detection of herpes simplex virus in cerebrospinal fluid with a sensitivity of up to 25 copies/reaction [30]. Lee et al. applied it to the detection of MERS-CoV virus with a 100% agreement with the clinical diagnosis [31]. Curtis et al. applied LAMP technology to the detection of the HIV virus with a sensitivity of 10–100 copies/reaction, and the assay could be completed within 60 min [32]. These studies indicate that LAMP technology is quite mature in virus detection, and reverse transcription (RT)-LAMP technology has been developed and clinically validated worldwide for SARS-COV-2 nucleic acid detection (Table 2).
Table 2.
LAMP-based virus detection method
| Virus | Year | Sensitivity | Testing time | Technology | Ref |
|---|---|---|---|---|---|
| HIV | 2009 | 120 copies | 35 min | One-step RT-LAMP | [74] |
| 2008 | 10–100 copies | 1 h | RT-LAMP | [32] | |
|
Japanese encephalitis virus |
2006 | 1 PFU | 1 h | Real-time RT-LAMP | [75] |
| Chikungunya virus | 2007 | 20 copies | 1 h | One-Step, single-tube RT-LAMP | [76] |
| Human papillomavirus | 2007 | 1000 copies/tube | 59 min | LAMP | [77] |
| Dengue virus | 2013 | 10 copies | 1 h | Single-tube RT-LAMP | [78] |
| West Nile virus | 2004 | 0.1 PFU | 17 min | One-Step, single-tube RT-LAMP | [79] |
| Mumps virus | 2005 | 0.1 PFU | 1 h | RT-LAMP | [80] |
| H5N1 | 2007 | 0.01–0.1 PFU | 35 min | RT-LAMP | [81] |
| SARS | 2004 | 0.01 PFU | 11-60 min | One-Step, single-tube RT-LAMP | [82] |
| MERS | 2015 | 5–50 PFU | 30–50 min | Asymmetric five-primer RT-LAMP | [83] |
| 2018 | 15–20 copies | 30 min | Fluorescent RT-LAMP using Qprobes | [84] | |
| 2016 | 4 copies | 30–60 min | One-pot RT-LAMP | [31] | |
| SARS-CoV-2 | 2020 | 100 RNA molecules | 30 min | Two-color RT-LAMP | [35] |
| 2020 | 500 copies | 30 min | Single-step RT-LAMP | [34] |
h hour, RT-LAMP reverse-transcription-loop mediated isothermal amplification, MERS Middle East respiratory syndrome, min minutes, N/A , PFU , Ref reference, SARS-Cov-2 severe acute respiratory syndrome coronavirus 2
At the beginning of the SARS-Cov-2 outbreak, Kitagawa et al. used the purified and quantitative SARS-Cov-2 viral RNA provided by the National Institute of Infectious Diseases in Japan as a standard sample for molecular diagnosis and assessed the sensitivity of the RT-LAMP method analysis using serial ten-fold dilutions [33]. The results showed that the minimum amount of RNA detectable in 35 min was 1.0 × 10 copies/μL. They screened 76 patients, 30 were positive and 46 were negative, using conventional RT-quantitative PCR (qPCR) technology, while the RT-LAMP test identified 32 positives and 44 negatives. A 97.4% consistency was achieved using RT-PCR and RT-LAMP detection. Compared with RT-PCR, Kitagawa et al. found that RT-LAMP has the advantages of ease of use, a short detection time, and the ability to judge results directly by the naked eye under natural light, which is supportive of RT-LAMP as a point-of-care screening method for new coronaviruses [33]. Jiang et al. developed a LAMP-based assay in another study and screened 260 clinical samples, including 213 negative patients. The results from 47 positive patients showed a sensitivity of 500 copies/mL, and the method achieved 100% analytical specificity compared to a wide range of closely and distantly related viruses, fungi, and bacteria, and human DNA, as well as 99.5% specificity and 91.4% sensitivity compared to RT-PCR. The positive predictive value was 97.7%, and the negative predictive value was 98.1% [34].
Another group developed a primer test for the N gene of SARS-Cov-2, this was a two-color RT-LAMP assay for RNA [35] based on the detection of DNA using a pH indicator to react in a weakly buffered environment. With a positive reaction, the pH decreases, resulting in a color change from red to yellow (as shown in Fig. 3A), and the results are judged based on the color. To verify the feasibility of the method, the team tested 768 specimens with the method and compared the results with those of RT-PCR. The overall specificity of RT-LAMP was 99.7%. In these samples (i.e., positive samples), the sensitivity of RT-LAMP was 97.5% (Fig. 3C). The same research team developed a swab-to-RT-LAMP method based on the two-color RT-LAMP technique. This method does not require a prior RNA isolation step and retains excellent specificity (99.5%), but it is compatible with the RT-LAMP analysis. Compared to this RT-LAMP assay, its sensitivity is lower (86% at a CT <30), but the sensitivity of the thermal swab for RT-LAMP detection is higher. They also developed a multiplex LAMP sequencing protocol that uses a Tn5 transposase marker to rapidly identify thousands of positive RT-LAMP assays from next-generation sequencing runs, it can be used as a validation protocol for RT-LAMP reaction results (to identify false-positive samples) [35]. The data from this study suggest that dual-color RT-LAMP detection of SARS-Cov-2 is feasible. The RT-LAMP dual-color method is faster and more convenient than RT-PCR, but RT-LAMP is only suitable for individuals with a high viral load. For individuals with a low viral load and using a direct detection by pharyngeal swabs, the detection sensitivity of RT-LAMP is quite low [36].
CRISPR-Cas-Based Detection Technology
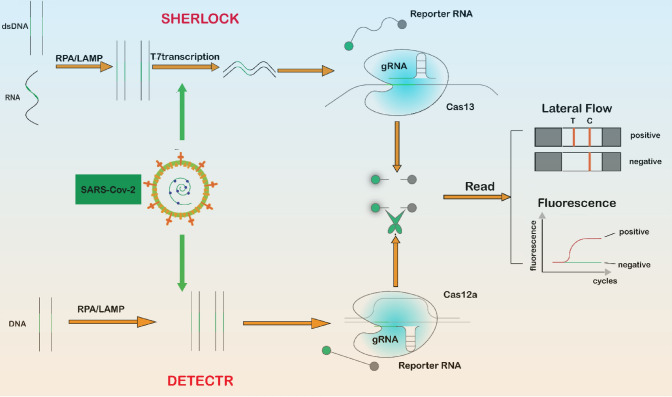
CRISPR-Cas gene editing technology specifically recognizes target gene sequences by guide RNA, so that Cas protease is activated to effectively cut the DNA double strand and cause DNA double-strand breaks, thus enabling gene knockout or knockin [37–39]. In 2017, Zhang’s team used CRISPR-Cas technology to detect Zika and dengue viruses, and named this new detection system SHERLOCK (Specific High Sensitivity Enzymatic Reporter Unlocking) [40]. This technique uses Cas13a after cutting the target RNA, it remains active and then proceeds to cut non-target RNA, a feature called collateral cleavage. In 2018, Doudna’s team found that the Cas12 enzyme can be activated after binding to the target sequence under the guidance of CRISPR RNA. The properties of other single-stranded DNA in the crazy cleavage system were introduced into the RPA amplification step and the RPA+Cas12a nucleic acid molecule was developed. The detection technique is named DETECTR (DNA endonuclease-targeted CRISPR reporter) [41]. After the SARS-Cov-2 outbreak, many research teams used CRISPR-Cas gene editing technology to detect SARS-Cov-2 RNA, showing high sensitivity and specificity (Fig. 4).
Fig. 4.
CRISPR-based severe acute respiratory syndrome coronavirus 2 detection. Schematic of two CRISPR-based detection methods: (1) SHERLOCK (Specific High Sensitivity Enzymatic Reporter Unlocking): DNA or RNA in the sample is isothermally amplified by recombinase polymerase amplification (RPA) or loop-mediated isothermal amplification (LAMP), the amplicon is converted into RNA by binding to T7 transcriptase, Cas13 is activated when it encounters the target RNA, and the RNA is cut with fluorescent molecules to release fluorescent signals. (2) DETECTR (DNA endonuclease-targeted CRISPR reporter): the target DNA is isothermally amplified by RPA or LAMP, and when Cas12a is activated when it encounters the target DNA and randomly cuts the RNA with fluorescent molecules, releasing a fluorescent signal. dsDNA double-stranded DNA
Cas13 Protein-Based Detection
Zhang’s team developed an improved SHERLOCK-based crown detection method, called STOP (SHERLOCK Testing in One Pot), at the beginning of the outbreak [42]. The conventional SHERLOCK method involves two separate reaction steps. There is liquid handling and opening of the tubes, which increases the potential for sample cross-contamination, whereas the STOP method does not require sample extraction. The amplification step and CRISPR-mediated detection step can be integrated at one temperature, and with only one liquid handling step and a simple visual reading similar to a pregnancy test, STOP detection of SARS-Cov-2 is divided into three steps: 1, 60 °C for 10 min to lyse patient samples containing the virus; 2, 60 °C for 1 h to detect using STOPCovid RNA Virus; and (3) 2 min to visually read the test results. Using this method, the team repeatedly tested specimens from 12 patients who were positive for novel coronavirus and five patients who were negative for novel coronavirus. Of these, 11 of the 12 patients tested positive on three repeat tests, and one tested positive twice. Among the negative patients, all five patients tested negative. The STOP method is not only highly sensitive and specific, but because of the simplified method of operation and equipment, it can be used by non-professionals at POCT, in many under-resourced situations or perhaps, even at home. Zhang’s team then further optimized the method to simplify the nucleic acid extraction step with a simple magnetic bead enrichment method, enabling a one-step CRISPR nucleic acid assay (STOPCovid-2) [42]. The team obtained a sensitivity of 93.1% and a specificity of 98.5% in a total of 402 clinical samples, and the time was greatly reduced, taking only 15–40 min to detect the results. The continuous improvement of the CRISPR assay for SARS-Cov-2 by Zhang et al.’s team in a short period of time shows the great potential for virus detection using CRISPR technology.
In August 2021, the Collins group at Massachusetts Institute of Technology developed a simple, saliva sample-based detection technique using SHERLOCK technology, minimal instrumented SHERLOCK (miSHERLOCK) [43]. The device first uses 10 mM of dithiothreitol and 5 mM of EGTA mixed with saliva samples and heated to 95 °C for 3 minutes, which effectively inactivates nucleases in saliva and lyses viral particles without inhibiting the performance of the downstream detection. The purified RNA was then transferred to the reaction chamber on the polyethersulfone membrane and the SHERLOCK reaction was initiated. Fluorescence readings were observed through a light transmittance 55 minutes later. The team performed the miSHERLOCK analysis on saliva samples from 27 patients with RT-PCR-positive COVID-19 and 21 healthy individuals. The results showed that miSHERLOCK had a sensitivity of 96% and a specificity of 95% for detecting the virus in saliva samples. In addition, RNA synthesized from the N50iy, (B.1.1.7, B.1.351, and P.1) and E484k mutations in SARS-Cov-2 variants were mixed with saliva samples for the miSHERLOCK analysis. Positive rates of more than 95% were obtained for limit of detection values of 49,000 copies/mL (N501y), 1100 copies/mL (Y144del), and 1200 copies/mL (E484k) [43]. The study demonstrated that the miSHERLOCK assay device can not only effectively detect SARS-Cov-2-positive samples, but also can efficiently diagnose SARS-Cov-2 variants through a simple workflow and flexible module design, making it a suitable assay technology for POCT.
Cas12 Protease-Based Detection
In 2020, Broughton et al. developed a CRISPR-Cas12-based lateral flow assay that is fast (less than 40 min), easy to implement, and accurate [44]. This method uses RT-LAMP to simultaneously reverse transcribe and isothermally amplify RNA extracted from the nasopharynx and detects the samples by the CRISPR-Cas12-based lateral flow assay. The team validated the assay with clinical samples from 36 patients with SARS-Cov-2 infection and 42 patients with other viral respiratory infections. Compared to RT-PCR, the results showed that the method had a 95% positive predicative concordance and 100% negative predictive consistency. The DETECTR assay has comparable accuracy to current conventional RT-PCR, and it is a fast and simple device (can be configured in a few days). It is easy to report (lateral flow strips) and is suitable for large-scale screening of novel coronavirus 2019 (nCoV-2019) in airports and clinics, as well as in locations that cannot be equipped with large-scale laboratories. In another study, Wang et al. integrated RT-LAMP and Cas12a lyase into a single reaction system and developed a visual assay called opvCRISPR [45]. Analysis of 50 clinical samples infected with SARS-Cov-2 yielded results that were 100% consistent with those of the US Centers for Disease Control and Prevention. Both the DETECTR assay and the opvCRISPR detection method showed that the combination of LAMP isothermal amplification technology and CRISPR detection technology has high sensitivity and specificity. It was also shown to be a rapid and simple method for SARS-Cov-2 detection. It can meet the current needs for new coronavirus detection.
Rapid Tandem Integrated Nucleic Acid Method
Doudna’s research team developed a new nucleic acid detection technique called Fast Integrated Nuclease Detection In Tandem (FIND-IT) [46]. In December 2020, her research team published version 1.0 of this technology by using Cas13 [47]. After cutting the RNA, non-specific cut single-stranded RNA with fluorescent molecules is converted to a fluorescent signal to determine the result, this method can reach a sensitivity of 100 copies/µL in 30 min. The published FIND-IT method incorporates the Csm6 enzyme, which allows direct RNA detection in a suitable form for predictive diagnosis as well as a wide range of other diagnostic or research applications. They fused Cas13 (LbuCas13) with Csm6, a dimeric RNA endonuclease from the type III CRISPR-Cas system, to enable signal amplification and further to improve RNA detection efficiency. LbuCas13 alone cannot detect nucleic acids at concentrations of 31–125 copies/µL within 20 minutes, but assays containing both LubCas13 and Csm6 can detect nucleic acid samples at concentrations of 31 copies/µL within 20 min. The accuracy of 125 copies/µL, 63 copies/µL, and 31 copies/µL was detected in 16, 30, and 60 minutes, respectively, reaching 95%. They also developed a detector that includes a microfluidic chip with a reaction chamber, a heating module that keeps the reaction at 37 °C, and a compact fluorescent imaging system, containing target RNA and LbuCas3-TtCsm6. The fluorescence signal was observed for 1 hour. The results showed that the fluorescence signal of the SARS-Cov-2 genome containing 400 copies/µL increased approximately 4.7 times over 1 hour, while the negative control without target RNA showed only a 1.7-fold increase in the fluorescence signal. This significant difference suggests that it is feasible to apply this response to microfluidics. The research team also tested clinical samples. In four positive and four negative samples, FIND-IT detected samples with CT values between 8 and 20 at 5 min with a CT value of 25, which was higher than the negative control. Samples with a CT value of 29 were higher than the negative control at 20 min, and samples with a CT value of 29 were also higher than the negative control at 40 min, while negative samples were detected below the negative control. Based on 296 patients in the IGI testing laboratory, 82% of the positive samples could be detected with FIND-IT [46]. These results suggest that fusion of LubCas13 and Csm6 ensures the sensitivity and specificity of the reaction. In this case, the reaction time was significantly shortened (the fastest result was 5 min), and the amplification step was omitted, which meets the current requirements for rapid and simple detection and therefore has a great market potential.
CRISPR-Cas9-based SHERLOCK and DETECTR technologies show great potential in virus detection; however, we noted that these two technologies need isothermal amplification before viral RNA detection. Nevertheless, FIND-IT technology can directly detect viral RNA without amplification, which makes CRISPR-Cas gene editing technology promising for targeted virus detection and diagnostic testing.
Biosensor Detection
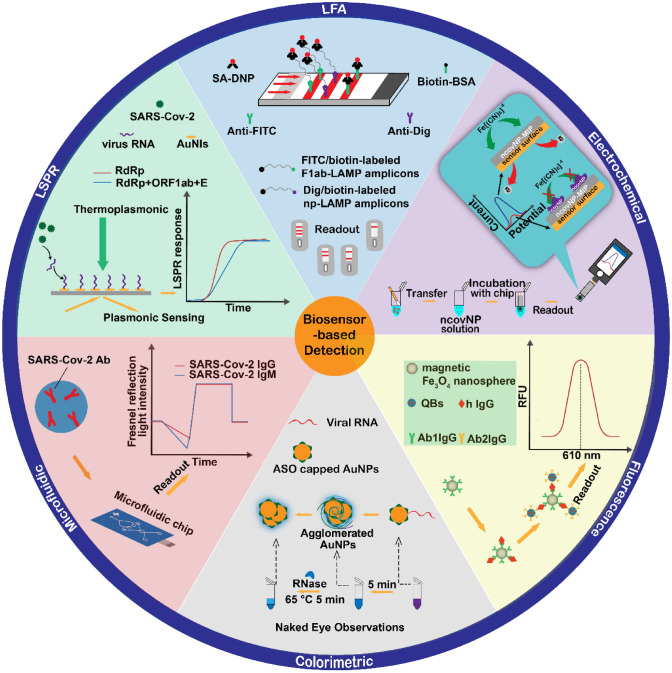
A biosensor is composed of a chemical or biological receptor and a sensor. The receptor interacts specifically with the target analyte and the sensor recognizes the process and converts it into a quantitative signal [48]. It can provide an inexpensive, sensitive, rapid, miniaturized, and portable platform compared with traditional laboratory assays [49]. Biosensors have also been widely used in human respiratory viruses (Table 3). In 2018, Dziąbowska al. detected the influenza virus by electrochemical biosensors [50]. In 2019, Layqah and Eissa detected MERS viruses by electrochemical biosensors [51]. After the COVID-19 outbreak, the SARS-Cov-2 biosensor detection has also made great progress (Fig. 5).
Table 3.
Application of biosensors in virus detection
| Virus | Type | Nanomaterials | Target | LOD | Result time (minutes) | Ref |
|---|---|---|---|---|---|---|
| MERS | Electrochemistry | Carbon electrodes modified with AU NPs | Spike protein S1 | 1.04 pg/mL | 20 | [51] |
| Colorimetric | Au NPs | E, ORF1a | 1 pmol/uL | 10 | [85] | |
| Colorimetric | Ag NPs | DNA | 1.53 nM | N/A | [86] | |
| HIV | Photonic | Photonic crystals | gp 120envelope |
104–108 copies/mL |
30 | [87] |
| Surface plasmon Resonance | Ag NPs | DNA | 195 pmol/L | N/A | [88] | |
| H1N1 | Colorimetric | Au immunostrip | Hemagglutinin | 2.27 PFU/mL | < 10 | [89] |
| Colorimetric |
Magnetic nanobeads Au NPs |
Hemagglutinin | 2.6 PFU/mL | N/A | [90] | |
| H5N1 | Fluorescence | Au NPs | RHA protein |
3.5 ng/mL (serum) |
30 | [91] |
| DENV | Surface plasmon Resonance | Graphene nanoparticles | E | 0.08 pm | 8 | [92] |
| SARS-Cov | Surface plasmon Resonance |
Gold-binding polypeptides |
Antibody | 200 ng/mL | 10 | [93] |
| SARS-Cov-2 | Colorimetric | Au NPs | N | 0.18 ng/uL | 10 | [94] |
| Colorimetric | Au NPs | RdRp | 0.5 ng | < 30 | [95] | |
| Lateral flow |
Au NPs Dye streptavidin-coated polymer nanoparticles |
IgM ORF1ab and N |
12–25 copies/uL |
15 < 2 |
[96] | |
| [56] | ||||||
| Fluorescence | QDs and MnFe3O4 nanospheres | IgG | 4 pg/ml | N/A | [53] | |
| Microfluidic | Au NPs | S | 0.5 pM | 30 | [97] | |
| Optomagnetic | Iron oxide NPs | RdRp | 0.4 fm | 100 | [98] | |
| Field-effect transistor | Graphene oxide nanosheets Au NPs |
S N |
24.2 copies/mL 6.9 copies/μL |
5 | [99] | |
| [100] |
Ag NPs Silver nanoparticles, Au NPs gold nanoparticles, DENV dengue virus, LOD limit of detection, MERS Middle East respiratory syndrome, N/A not applicable, QDs Quantum dots, Ref reference, SARS-Cov severe acute respiratory syndrome coronavirus, SARS-Cov-2 severe acute respiratory syndrome coronavirus 2, PFU plaque-forming unit
Fig. 5.
Biosensor applications for severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) detection, summarizing the recently reported biosensors for SARS-Cov-2 detection, including lateral flow biosensors, electrochemical biosensors, colorimetric biosensors, fluorescence biosensors, microfluidic biosensors, and localized surface plasmon resonance biosensors. Ab antibody, AuNPs gold nanoparticles, LAMP loop-mediated isothermal amplification, SA-DNP streptavidin-dinitrophenol, BSA bovine serum albumin, Dig digoxin, ASO antisense oligonucleotide, RdRp RNA-dependent RNA polymerase, AuNIs gold nanoislands, FITC fluorescein isothiocyanate
Optical Biosensors
Optical biosensors are the most widely used sensors for virus detection and include colorimetric, fluorescent, localized surface plasmon resonance (LSPR), and photonic detection techniques. Such sensors have the advantages of being fast, simple, inexpensive, label free, reusable, and highly sensitive. Ventura’s team developed a fast colorimetric-based biosensor using the unique optical properties and biocompatibility of Au NPs [52]. The biosensor consists of an Au NP colloidal solution of Au NPs modified by the corresponding antibody to the SARS-Cov2 protein. In the presence of an antigenic antibody, Au NP forms a layer of nanoparticles on the surface of the virus particles. The LSPR coupling between the nanoparticles results in the color of colloidal solution changing from red to blue. By comparing 94 RT-PCR samples (45 positive and 49 negative), the team found that the biosensor had a sensitivity of 96% and a specificity of 98%. The sensor can detect virus particles in less than 3 min, and the sensor targets the virus itself rather than the RNA, making it useful not only as a diagnostic tool, but it can also determine the extent of a patient’s infection. In another study, Guo et al. developed an optical sensor using a fluorescence method [53]. The team combined the characteristics of magnetic Fe3O4 nanospheres and quantum dots, which are excellent fluorescent materials, owing to their uniform diameter distribution, large surface area, good solubility in water, high dispersion, and strong magnetic properties. A magnet-based fluorescent-linked immunosorbent assay was developed for the detection of IgG antibodies in human serum. When IgG is present in the sample to be tested, it forms an immune complex with quantum dot nanobeads linked to 1 anti-Fe3O4 and 2 anti-Fe3O4, which releases a strong fluorescence signal after magnetic separation, and the concentration of antibody in the serum can be determined by the change in the fluorescence signal. The research team diluted 20 serum samples to 1 in 50,000 and compared them with the fluorescent-linked immunosorbent assay. The results showed no statistically significant difference between the fluorescent-linked immunosorbent assay and ELISA, demonstrating the reliability of the sensor. The sensor is more suitable for population screening because of its higher linear range, sensitivity, and simplicity to traditional ELISA methods.
Electrochemical Biosensor
An electrochemical biosensor is a type of electronic device biosensor, which includes field effect transistors, three-electrode potential sensors, and amperometric systems. It is characterized by miniaturization, low cost, and mass production. Recent studies have reported a molecularly imprinted polymer-based electrochemical biosensor that detects NCov NP by combining the chemical and thermal stability of molecularly imprinted polymers with a biosensor [54]. The key components of this sensor are a sensor chip and a thin film electrode. The interface is connected to a molecularly imprinted polymer that selectively binds NCov NP and is connected to the chip and a constant potential meter. Raziq et al. tested COVID-19 samples confirmed by RT-PCR by using differential pulse voltammetry [54]. The sensor responded linearly to NCov NP over a concentration range of 2.22–111 fM with a limit of detection of 15 fM and a limit of quantification of 50 fM. In addition, it did not react with other interfering proteins, which confirmed the ability of the sensor to detect NCov NP in complex biological media. Yousefi et al. developed a reagent-free electrochemical biosensor for rapid and sensitive detection of the SARS-CoV-2 [55]. The sensor combines a specific antibody with double-stranded DNA and the REDOX probe ferrocene to form a negatively charged connector. The negatively charged connector is adsorbed on the surface and the presence of virus particles affects the time the connector is adsorbed on the surface. Viral particles are detected by timing amperometry. The team was able to detect viral particles at concentrations as low as 4000 copies/mL in 10 minutes by analyzing a patient’s saliva sample. The sensor can be stored for up to 9 months, making it fast, sensitive, stable, inexpensive, and portable. However, the specificity of the method is affected to some extent by a cross-reaction between the sensor and the SARS-Cov-1 viral spike protein. The team believes that the SARS-Cov-1 virus is now extinct in humans and this cross-reactivity is tolerable in clinical trials [55].
LAMP-Based Lateral Flow Biosensor
Zhu et al. devised a method combining multiple RT-LAMP with a nanoparticle-based lateral flow biosensor [56], which uses two sets of primers to amplify both ORF1ab and N genes of SARS-Cov-2 in a single tube reaction. Using FITC (fluorescein)-/digoxin- and biotin-labeled primers, multiplex RT-LAMP produced a large amount of FITC-/digoxin- and biotin-attached duplex amplicons, which can be detected by a lateral flow biosensor through immunoreactions between the FITC/digoxin-labeled duplex and the anti-FITC/digoxin antibody, and a biotin/streptavidin interaction between the biotin-labeled duplex and the streptavidin-labeled polymerase nanoparticle. The accumulated nanoparticles can form a visible red-colored band to indicate detection of the ORF1ab and N gene without instrumentation. The multiplex RT-LAMP-lateral flow biosensor method showed 100% sensitivity (33 positive samples) using 129 respiratory samples (33 SARS-Cov-2 positive specimens) initially analyzed by RT-PCR by the research team. All samples were detected with 100% specificity (all 96 negative samples tested negative), and the entire experiment could be completed from sampling to the result in less than 1 hour. The assay is fast, and the advantage of simple equipment makes this technique a viable assay for initial virus diagnostics.
Dual-Function Plasma Photothermal Biosensor
Qiu et al. combined plasma with a photothermal effect and LSPR to develop a dual-function LSPR biosensor [57]. Localized surface plasmon resonance is a surface-conducted electron coherent oscillation driven by strong photons. It can be modulated when coupled to the surface of a plasmonic material. Because of the enhancement of the plasma field near the nanostructure, the LSPR sensing system shows a high response to local changes in sensitivity, including refractive index changes and molecular binding, and thus, LSPR is an ideal method for real-time and label-free detection of micro- and nano-scale analytes. In this method, AuNI chips use hybridization of nucleic acid sequences selected from SARS-Cov-2. When the plasmonic resonance frequency is irradiated, heat is generated on the chip. The localized photothermal effect heat not only increases the temperature of in-situ hybridization on the chip and enhances the hybridization response, but also accurately distinguishes similar gene sequences and avoids false positives. The high sensitivity of the dual-function LSPR biosensor for SARS-CoV-2 and low detection limit of 0.22 pm are advantages that support the detection of specific targets in multi-gene mixtures by the dual-function LSPR biosensor. This is a nucleic acid hybridization technique for detecting synthesized SARS-CoV-2 oligonucleotides, providing an alternative and promising potential for future clinical application.
Microfluidic Chip-Based Biosensor
Microfluidic technologies are platforms for many diagnostic tests, including RT-PCR, RT-LAMP, nested PCR, nucleic acid hybridization, ELISA, fluorescence-based analysis, rolling circle amplification, aptamers, sample preparation multiplex, porous silicon nanowire forest, silica sol-gel coating/adhesion, and CRISPR. They provide faster, cheaper, easier to use, and more sensitive platforms, thus microfluidic devices have great potential to become an alternative method for detecting viral RNA [58]. In recent years, microfluidic devices have been used to detect a variety of viruses, such as rotavirus, Ebola virus, and Zika virus [59–61] and good results have been achieved.
Zhou's team developed a microfluidic chip detection technique based on the specific binding reaction of an antigen and antibody [62]. Repeated exposure to the SARS-Cov-2 nucleocapsid antigen immobilized on a microfluidic chip resulted in a complete immunoconjugate within 1 min, thus enabling ultra-rapid detection of specific virus N protein antibodies in the patient’s serum. By using reciprocating-flowing ELISA, Zhou et al. found that the reciprocating-flowing ELISA chip strategy achieved a precise diagnosis with a 100% true positive and negative rate in 13 patients with suspected COVID-19. Moreover, the detection time was shortened to 5 min and the limit of quantification was as low as 4.14 pg/mL, achieving an ultra-fast detection of antibodies targeting the nucleoprotein antigens of SARS-CoV-2. Recently, Xu et al. constructed a novel all-fiber reflection microfluidic biosensor that combines an all-fiber optical systems, a microfluidic chip, and multi-mode fiber biological probes [63], it is based on the Fresnel reflection mechanism and immunoassay principles for the detection of SARS-COV-2 IgG and IgM. The team tested the sensor on 30-fold diluted serum samples and obtained limit of detection values of 24.6 ng/mL (IgG) and 13.5 ng/mL (IgM) in serum samples with a detection time of 7 min. Microfluidic biosensor technology is a virus detection method that is integrated into chip technology. The advantage over other biosensors is that microfluidics technology allows a flexible combination of a variety of biological and chemical experiments on a tiny platform.
Wearable Biosensors
Synthetic biology is an emerging discipline that studies engineering approaches to genetically engineered artificial biological systems and systems biology [64]. The development of synthetic biology has provided a new research direction for virus detection biosensors. Among them, freeze-dried cell-free synthetic biotechnology was developed for Ebola virus detection in 2014 [65]. After the SARS-CoV-2 outbreak, freeze-dried cell-free technology and CRISPR detection technology were used on masks to develop wearable and non-invasive SARS-CoV-2 detection at room temperature [66]. This wearable detection technique is different from traditional nasopharyngeal sampling or serum sampling. Breath sampling technology collects respiratory droplets for aerosol detection. The mask detection system consists of four modules: hydration reservoir, large surface area collection sample pad, wax-patterned µPAD, and a lateral flow assay strip. Capillaries transport the collected droplets and aerosols to the module containing the freeze-dried lysate to lyse the virus particles. The target sequence is amplified by RT-RPA, followed by the detection of amplicons with a CRISPR detection system, and finally visualization of the result determination with an integrated lateral flow assay. The method takes about 1.5 h from activation to the result, with a detection limit of 500 copies. This wearable biosensor based on freeze-dried cell-free technology is the first wearable personalized technology comparable to the sensitivity of traditional laboratory assays, but still has some limitations, such as the inability to detect in high humidity or underwater [66].
Other Methods of Detection
In addition to the methods discussed above, researchers in various countries have developed aptamer-based assays, droplet digital PCR (ddPCR)-based assays, and sequencing-based assays for coronavirus diagnostic testing.
Detection Methods Based on Aptamer
A nucleic acid aptamer is a segment of DNA that is an antibody-like molecule. After screening and enrichment by SELEX (systematic evolution of ligands by exponential enrichment) technology, it can recognize a specific target molecule. The advantages are based on its small molecular weight and low production cost compared with antibodies, and thus it is expected to replace antibodies for SARS-CoV-2 detection. In 2020, Song et al. used an aptamer selection strategy based on ACE2 competition and a machine screening procedure to create aptamers with high affinity for SARS-Cov-2 RBD, including Cov2-RBD-1C and Cov2-RBD-4C [67]. Woo et al. developed a ligation reaction by SplintR ligase followed by transcription by T7 RNA polymerase [68], and the resulting transcripts formed RNA aptamers. The aptamer binds to a fluorescent dye and produces fluorescence when the target RNA is present. This method is called SENSR, and it produces results in as little as 30 min and achieves a 95% positive predictive value and a 100% negative predictive value.
Detection Method Based on ddPCR
Droplet digital PCR is a third-generation PCR technique for the absolute quantitation of nucleic acid molecules. Prior to PCR amplification, the technique requires the processing of samples to micro-droplets, to allow for a “single molecule quantitative analysis” of the sample. Compared with traditional PCR, ddPCR has the advantages of high sensitivity, precision, repeatability, and resistance to inhibitors. It may be that ddPCR is superior to gold standard RT-PCR in the detection and predictive diagnosis of SARS-COV-2 [69].
A recent paper reported that they tested 74 clinical samples (36 feces samples, 36 sputum samples, and 2 throat swabs) from 43 recovering patients with COVID-19 with ddPCR and qPCR, 41 samples were positive and 33 were negative using ddPCR. The positive rate of ddPCR (55.41%) was significantly higher than that of qPCR (33.49%). To verify the accuracy of ddPCR in low viral load samples, the team performed a comparative test on 18 retained samples from nine discharged patients, in which qPCR showed negative results while ddPCR showed 12 positive results (only one patient showed negative results in both samples). This showed that ddPCR significantly improved the accuracy of diagnosis in low viral load samples and reduced the number of false-negative cases reported [70]. Mao's team developed a rapid, accurate, and quantitative SARS-Cov-2 detection system that combines PCR with ddPCR. The system cleverly utilizes a microfluidic chip that generates droplets with a diameter of 30 µm at high speed through a multi-stage bifurcation structure. Coupled with a micro-heating array for rapid PCR (controlled by a high-speed temperature-cooling system), the system can detect samples with target sequences down to 5 copies/µL in less than 5 minutes, with a significantly higher accuracy than RT-PCR at low concentrations [71].
Detection Methods Based on Sequencing Technology
The principle of using sequencing technology for the detection of neo-coronaviruses involves high-throughput sequencing of all nucleic acids in suspected samples or nucleic acids from specific targets, and the detection of all pathogenic microorganisms or their target sequences in the samples through bioinformatics analysis. Next-generation sequencing is the most advanced method in the personalization of medical services. Bhoyar et al. used this COVID-Seq method on 752 clinical samples in duplicate for a total of 1536 samples [72]. The 1536 specimens were analyzed within a sequencing time of 11 h and an analysis timeline of 6 hours, the COVIDseq test has comparable sensitivity, precision, and accuracy to RT-PCR technology showing that high-throughput gene sequencing techniques can guarantee sensitivity. Traditional gene sequencing techniques are more often used for the confirmation of patient specimens with high clinical suspicion, but the PCR tests may be negative for many reasons, including negligible viral load, scant analytic sensitivity, improper specimen types, poor or suboptimal specimen collection procedures, testing too early or too late after infection, or changeability in viral shedding. A recent study has shown an innovative assay that combines the LAMP amplification technique with a sequencing detection technology, called LAMP-Seq [73]. The LAMP-seq analysis of 676 clinical samples showed a sensitivity of 100% and a specificity of 99.7% at lower material costs, which suggested that it represents a predictive diagnostic strategy for frequent testing as a surveillance program in public health.
Perspective on POCT
With the continuous updating and development of testing technology, in vitro diagnostic technology has developed in two directions: first, testing units that are mainly laboratory oriented, which are developed in the direction of high precision and integration; second, testing units are mainly for families, small clinics, and pharmacies, which are being developed to be simple, easy to carry, fast, and convenient for personal health management. In this context, POCT technology, which is easy to carry, simple to operate, and fast, has rapidly developed. Point-of-care testing is divided into three types: (1) solid-phase POCT detection, based on antigen-antibody immunoassay detection; (2) liquid-phase POCT (small chemiluminescence technology and microfluidic chip technology); and (3) molecular POCT (nucleic acid molecular detection technology based on PCR detection technology). At present, POCT detection technology has been applied for many types of analysis, including blood glucose, blood gas analysis, myocardial markers, and infectious disease detection. With the global outbreak of COVID-19, many COVID-19 POCT detection products have also appeared on the market, such as the Accula SARS-Cov-2 Test. These kits include a disposable microfluidic cuvette and a high-precision reader (handheld or benchtop analyzer) for sample detection. These personalized assays can produce results in less than 30 minutes, significantly reducing the test time, but the deficiencies in the quality-control system of the POCT assay (lack of controlled tests) make it challenging to ensure the stability and accuracy of each batch compared with traditional assays. In addition, inadequate user understanding of the operation instructions can lead to inaccurate test results. In addition, it remains a challenge to use the large amount of data generated by POCT for an intelligent analysis to monitor the health status of the population and rationalize the allocation of medical resources. However, it is hoped that the big data generated by POCT can become a powerful support for public health and epidemiological surveillance. The current set of problems are gradually being solved by applying the latest detection technology to POCT. To improve the accuracy of detection, POCT will be the most important tool for SARS-Cov-2 or other epidemiological testing in the future.
Conclusions
The SARS-Cov-2 pandemic has brought more virus detection technologies to the forefront. At present, RT-PCR is still the most popular method for detecting SARS-Cov-2 and its variants. As a complementary method to RT-PCR, antigen detection is more acceptable for home-based detection. Serum antibody detection is also an important method to monitor the changes of antibodies in the middle and late stages of the infection. Novel methods, such as isothermal amplification technology, CRISPR technology, biosensors, and microfluidic chip technology is still under development, although these diagnostic techniques have the advantages of simplicity of operation, short detection time, inexpensive equipment, portability, availability of equipment, and high sensitivity and specificity comparable to RT-PCR. However, most of the above techniques have not been tested and validated on a large scale, or with various types of clinical samples. Further, the stability of reagents, the cost of equipment, and the automation of these technologies are still uncertain. Therefore, there is still a long way to go before the wide application of these technologies enters clinical testing, or to the home, small clinics, pharmacies, supermarkets, and airports for public infection control and to aid in the strategic management of the ongoing pandemic.
Declarations
Funding
No sources of funding were received for the preparation of this article.
Conflicts of interest/competing interests
The authors have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Code availability
Not applicable.
Author contributions
HW created the concepts and XH and HW performed the literature search, manuscript drafting, table preparation, and figure illustration. HW, JW, JG, and LX elaborated and consolidated the manuscript. All authors have read and approved the final version of the manuscript.
References
- 1.Zhou Y, Zhang L, Xie YH, Wu J. Advancements in detection of SARS-CoV-2 infection for confronting COVID-19 pandemics. Lab Invest. 2022;102(1):4–13. doi: 10.1038/s41374-021-00663-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borberg E, Granot E, Patolsky F. Ultrafast one-minute electronic detection of SARS-CoV-2 infection by 3CL(pro) enzymatic activity in untreated saliva samples. Nat Commun. 2022;13(1):6375. doi: 10.1038/s41467-022-34074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahimi A, Mirzazadeh A, Tavakolpour S. Genetics and genomics of SARS-CoV-2: a review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics. 2021;113:1221–1232. doi: 10.1016/j.ygeno.2020.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp D, Wang N, Corbett K, Goldsmith J, Hsieh C, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh H, Ahn H, Tripathi A. A closer look into FDA-EUA approved diagnostic techniques of Covid-19. ACS Infect Dis. 2021;7(10):2787–2800. doi: 10.1021/acsinfecdis.1c00268. [DOI] [PubMed] [Google Scholar]
- 6.Jarrom D, Elston L, Washington J, Prettyjohns M, Cann K, Myles S, et al. Effectiveness of tests to detect the presence of SARS-CoV-2 virus, and antibodies to SARS-CoV-2, to inform COVID-19 diagnosis: a rapid systematic review. BMJ Evid Based Med. 2022;27(1):33–45. doi: 10.1136/bmjebm-2020-111511. [DOI] [PubMed] [Google Scholar]
- 7.Kaminski MM, Abudayyeh OO, Gootenberg JS, Zhang F, Collins JJ. CRISPR-based diagnostics. Nat Biomed Eng. 2021;5(7):643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- 8.Reynard C, Allen JA, Shinkins B, Prestwich G, Goves J, Davies K, et al. COVID-19 rapid diagnostics: practice review. Emerg Med J. 2022;39(1):70–76. doi: 10.1136/emermed-2021-211814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Han Y, Yang J, Ma Y, Li J, Zhang R. Impact of SARS-CoV-2 variants on the analytical sensitivity of rRT-PCR assays. J Clin Microbiol. 2022;60(4):e0237421. doi: 10.1128/jcm.02374-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung HY, Jian MJ, Chang CK, Lin JC, Yeh KM, Chen CW, et al. Emergency SARS-CoV-2 variants of concern: novel multiplex real-time RT-PCR assay for rapid detection and surveillance. Microbiol Spectr. 2022;10(1):e0251321. doi: 10.1128/spectrum.02513-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dikdan RJ, Marras SAE, Field AP, Brownlee A, Cironi A, Hill DA, et al. Multiplex PCR assays for identifying all major severe acute respiratory syndrome coronavirus 2 variants. J Mol Diagn. 2022;24(4):309–319. doi: 10.1016/j.jmoldx.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amanat F, Stadlbauer D, Strohmeier S, Nguyen THO, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26(7):1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yong SEF, Anderson DE, Wei WE, Pang J, Chia WN, Tan CW, et al. Connecting clusters of COVID-19: an epidemiological and serological investigation. Lancet Infect Dis. 2020;20(7):809–815. doi: 10.1016/S1473-3099(20)30273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P, Chen L, Hu J, Trick AY, Chen FE, Hsieh K, et al. Magnetofluidic immuno-PCR for point-of-care COVID-19 serological testing. Biosens Bioelectron. 2022;195:113656. doi: 10.1016/j.bios.2021.113656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diao B, Wen K, Zhang J, Chen J, Han C, Chen Y, et al. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin Microbiol Infect. 2021;27(2):289.e1–4. doi: 10.1016/j.cmi.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peaper DR, Landry ML. Laboratory diagnosis of viral infection. Handb Clin Neurol. 2014;123:123–147. doi: 10.1016/B978-0-444-53488-0.00005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai C, Guo M, Wu Y, Cao BP, Wang X, Wu Y, et al. Ultraprecise antigen 10-in-1 pool testing by multiantibodies transistor assay. J Am Chem Soc. 2021;143(47):19794–19801. doi: 10.1021/jacs.1c08598. [DOI] [PubMed] [Google Scholar]
- 20.Peng R, Pan Y, Li Z, Qin Z, Rini JM, Liu X. SPEEDS: a portable serological testing platform for rapid electrochemical detection of SARS-CoV-2 antibodies. Biosens Bioelectron. 2022;197:113762. doi: 10.1016/j.bios.2021.113762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou Y, Mason MG, Botella JR. Evaluation and improvement of isothermal amplification methods for point-of-need plant disease diagnostics. PLoS ONE. 2020;15(6):e0235216. doi: 10.1371/journal.pone.0235216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohrman BA, Richards-Kortum RR. A paper and plastic device for performing recombinase polymerase amplification of HIV DNA. Lab Chip. 2012;12(17):3082–3088. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abd El Wahed A, Weidmann M, Hufert FT. Diagnostics-in-a-suitcase: development of a portable and rapid assay for the detection of the emerging avian influenza A (H7N9) virus. J Clin Virol. 2015;69:16–21. doi: 10.1016/j.jcv.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teoh BT, Sam SS, Tan KK, Johari J, Danlami MB, Hooi PS, et al. Detection of dengue viruses using reverse transcription-loop-mediated isothermal amplification. BMC Infect Dis. 2013;13:387. doi: 10.1186/1471-2334-13-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim Y, Yaseen AB, Kishi JY, Hong F, Saka SK, Sheng K, et al. Single-strand RPA for rapid and sensitive detection of SARS-CoV-2 RNA. medRxiv. 2020 doi: 10.1101/2020.08.17.20177006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia S, Chen X. Single-copy sensitive, field-deployable, and simultaneous dual-gene detection of SARS-CoV-2 RNA via modified RT-RPA. Cell Discov. 2020;6(1):37. doi: 10.1038/s41421-020-0175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gill P, Hadian AA. AS-LAMP: a new and alternative method for genotyping. Avicenna J Med Biotechnol. 2020;12(1):2–8. [PMC free article] [PubMed] [Google Scholar]
- 30.Ihira M, Yoshikawa T, Enomoto Y, Akimoto S, Ohashi M, Suga S, et al. Rapid diagnosis of human herpesvirus 6 infection by a novel DNA amplification method, loop-mediated isothermal amplification. J Clin Microbiol. 2004;42(1):140–145. doi: 10.1128/JCM.42.1.140-145.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SH, Baek YH, Kim YH, Choi YK, Song MS, Ahn JY. One-pot reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) for detecting MERS-CoV. Front Microbiol. 2016;7:2166. doi: 10.3389/fmicb.2016.02166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtis KA, Rudolph DL, Owen SM. Rapid detection of HIV-1 by reverse-transcription, loop-mediated isothermal amplification (RT-LAMP) J Virol Methods. 2008;151(2):264–270. doi: 10.1016/j.jviromet.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Kitagawa Y, Orihara Y, Kawamura R, Imai K, Sakai J, Tarumoto N, et al. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J Clin Virol. 2020;129:104446. doi: 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang M, Pan W, Arasthfer A, Fang W, Ling L, Fang H, et al. Development and validation of a rapid, single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) system potentially to be used for reliable and high-throughput screening of COVID-19. Front Cell Infect Microbiol. 2020;10:331. doi: 10.3389/fcimb.2020.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. 2020;12(556):eabc7075. doi: 10.1126/scitranslmed.abc7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba MM, Bitew M, Fokam J, Lelo EA, Ahidjo A, Asmamaw K, et al. Diagnostic performance of a colorimetric RT -LAMP for the identification of SARS-CoV-2: a multicenter prospective clinical evaluation in sub-Saharan Africa. EClinicalMedicine. 2021;40:101101. doi: 10.1016/j.eclinm.2021.101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ran F, Hsu P, Lin C, Gootenberg J, Konermann S, Trevino A, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geng J, Xia X, Teng L, Wang L, Chen L, Guo X, et al. Emerging landscape of cell-penetrating peptide-mediated nucleic acid delivery and their utility in imaging, gene-editing, and RNA-sequencing. J Control Release. 2022;341:166–183. doi: 10.1016/j.jconrel.2021.11.032. [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Zeng F, Zhang M, Huang F, Wang J, Guo J, et al. Emerging landscape of cell penetrating peptide in reprogramming and gene editing. J Control Release. 2016;226:124–137. doi: 10.1016/j.jconrel.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Gootenberg J, Abudayyeh O, Lee J, Essletzbichler P, Dy A, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, Ma E, Harrington L, Da Costa M, Tian X, Palefsky J, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joung J, Ladha A, Saito M, Kim NG, Woolley AE, Segel M, et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Puig H, Lee RA, Najjar D, Tan X, Soeknsen LR, Angenent-Mari NM, et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci Adv. 2021;7(32):eabh2944. doi: 10.1126/sciadv.abh2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang R, Qian C, Pang Y, Li M, Yang Y, Ma H, et al. opvCRISPR: one-pot visual RT-LAMP-CRISPR platform for SARS-Cov-2 detection. Biosens Bioelectron. 2021;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu TY, Knott GJ, Smock DCJ, Desmarais JJ, Son S, Bhuiya A, et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat Chem Biol. 2021;17(9):982–988. doi: 10.1038/s41589-021-00842-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fozouni P, Son S, Díaz de León Derby M, Knott GJ, Gray CN, D'Ambrosio MV, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184(2):323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozer T, Geiss BJ, Henry CS. Review-chemical and biological sensors for viral detection. J Electrochem Soc. 2020;167(3):037523. doi: 10.1149/2.0232003JES. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samson R, Navale G, Dharne M. Biosensors: frontiers in rapid detection of COVID-19. 3 Biotech. 2020;10(9):385. doi: 10.1007/s13205-020-02369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dziąbowska K, Czaczyk E, Nidzworski D. Detection methods of human and animal influenza virus: current trends. Biosensors (Basel). 2018;8(4):94. doi: 10.3390/bios8040094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Layqah LA, Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Mikrochim Acta. 2019;186(4):224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ventura BD, Cennamo M, Minopoli A, Campanile R, Censi SB, Terracciano D, et al. Colorimetric test for fast detection of SARS-CoV-2 in nasal and throat swabs. ACS Sens. 2020;5(10):3043–3048. doi: 10.1021/acssensors.0c01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo J, Wang Y, Niu S, Li H, Tian Y, Yu S, et al. Highly sensitive fluorescence-linked immunosorbent assay for the determination of human IgG in serum using quantum dot nanobeads and magnetic FeO nanospheres. ACS Omega. 2020;5(36):23229–23236. doi: 10.1021/acsomega.0c02987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raziq A, Kidakova A, Boroznjak R, Reut J, Öpik A, Syritski V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens Bioelectron. 2021;178:113029. doi: 10.1016/j.bios.2021.113029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yousefi H, Mahmud A, Chang D, Das J, Gomis S, Chen JB, et al. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J Am Chem Soc. 2021;143(4):1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- 56.Zhu X, Wang X, Han L, Chen T, Wang L, Li H, et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick G, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 58.Basiri A, Heidari A, Nadi MF, Fallahy MTP, Nezamabadi SS, Sedighi M, et al. Microfluidic devices for detection of RNA viruses. Rev Med Virol. 2021;31(1):1–11. doi: 10.1002/rmv.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang B, Kong J, Fang X. Bandage-like wearable flexible microfluidic recombinase polymerase amplification sensor for the rapid visual detection of nucleic acids. Talanta. 2019;204:685–692. doi: 10.1016/j.talanta.2019.06.031. [DOI] [PubMed] [Google Scholar]
- 60.Ye X, Xu J, Lu L, Li X, Fang X, Kong J. Equipment-free nucleic acid extraction and amplification on a simple paper disc for point-of-care diagnosis of rotavirus A. Anal Chim Acta. 2018;1018:78–85. doi: 10.1016/j.aca.2018.02.068. [DOI] [PubMed] [Google Scholar]
- 61.Fernández-Carballo BL, McBeth C, McGuiness I, Kalashnikov M, Baum C, Borrós S, et al. Continuous-flow, microfluidic, qRT-PCR system for RNA virus detection. Anal Bioanal Chem. 2018;410(1):33–43. doi: 10.1007/s00216-017-0689-8. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, Tan Y, Fu Q, Lin M, He J, He S, et al. Reciprocating-flowing on-a-chip enables ultra-fast immunobinding for multiplexed rapid ELISA detection of SARS-CoV-2 antibody. Biosens Bioelectron. 2021;176:112920. doi: 10.1016/j.bios.2020.112920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu W, Liu J, Song D, Li C, Zhu A, Long F. Rapid, label-free, and sensitive point-of-care testing of anti-SARS-CoV-2 IgM/IgG using all-fiber Fresnel reflection microfluidic biosensor. Mikrochim Acta. 2021;188(8):261. doi: 10.1007/s00604-021-04911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11(5):367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pardee K, Green A, Ferrante T, Cameron D, DaleyKeyser A, Yin P, et al. Paper-based synthetic gene networks. Cell. 2014;159(4):940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen PQ, Soenksen LR, Donghia NM, Angenent-Mari NM, de Puig H, Huang A, et al. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat Biotechnol. 2021;39(11):1366–1374. doi: 10.1038/s41587-021-00950-3. [DOI] [PubMed] [Google Scholar]
- 67.Song Y, Song J, Wei X, Huang M, Sun M, Zhu L, et al. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal Chem. 2020;92(14):9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 68.Woo CH, Jang S, Shin G, Jung GY, Lee JW. Sensitive fluorescence detection of SARS-CoV-2 RNA in clinical samples via one-pot isothermal ligation and transcription. Nat Biomed Eng. 2020;4(12):1168–1179. doi: 10.1038/s41551-020-00617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ishak A, AlRawashdeh MM, Esagian SM, Nikas IP. Diagnostic, prognostic, and therapeutic value of droplet digital PCR (ddPCR) in COVID-19 patients: a systematic review. J Clin Med. 2021;10(23):5712. doi: 10.3390/jcm10235712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu C, Shi Q, Peng M, Lu R, Li H, Cai Y, et al. Evaluation of droplet digital PCR for quantification of SARS-CoV-2 virus in discharged COVID-19 patients. Aging. 2020;12(21):20997–21003. doi: 10.18632/aging.104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin H, Wu Z, Shi N, Qi Y, Jian X, Zhou L, et al. Ultrafast multiplexed detection of SARS-CoV-2 RNA using a rapid droplet digital PCR system. Biosens Bioelectron. 2021;188:113282. doi: 10.1016/j.bios.2021.113282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhoyar R, Jain A, Sehgal P, Divakar M, Sharma D, Imran M, et al. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS ONE. 2021;16(2):e0247115. doi: 10.1371/journal.pone.0247115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ludwig KU, Schmithausen RM, Li D, Jacobs ML, Hollstein R, Blumenstock K, et al. LAMP-Seq enables sensitive, multiplexed COVID-19 diagnostics using molecular barcoding. Nat Biotechnol. 2021;39(12):1556–1562. doi: 10.1038/s41587-021-00966-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hosaka N, Ndembi N, Ishizaki A, Kageyama S, Numazaki K, Ichimura H. Rapid detection of human immunodeficiency virus type 1 group M by a reverse transcription-loop-mediated isothermal amplification assay. J Virol Methods. 2009;157(2):195–199. doi: 10.1016/j.jviromet.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toriniwa H, Komiya T. Rapid detection and quantification of Japanese encephalitis virus by real-time reverse transcription loop-mediated isothermal amplification. Microbiol Immunol. 2006;50(5):379–387. doi: 10.1111/j.1348-0421.2006.tb03804.x. [DOI] [PubMed] [Google Scholar]
- 76.Parida MM, Santhosh SR, Dash PK, Tripathi NK, Lakshmi V, Mamidi N, et al. Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol. 2007;45(2):351–357. doi: 10.1128/JCM.01734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hagiwara M, Sasaki H, Matsuo K, Honda M, Kawase M, Nakagawa H. Loop-mediated isothermal amplification method for detection of human papillomavirus type 6, 11, 16, and 18. J Med Virol. 2007;79(5):605–615. doi: 10.1002/jmv.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Teoh BT, Sam SS, Tan KK, Danlami MB, Shu MH, Johari J, et al. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J Clin Microbiol. 2015;53(3):830–837. doi: 10.1128/JCM.02648-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42(1):257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okafuji T, Yoshida N, Fujino M, Motegi Y, Ihara T, Ota Y, et al. Rapid diagnostic method for detection of mumps virus genome by loop-mediated isothermal amplification. J Clin Microbiol. 2005;43(4):1625–1631. doi: 10.1128/JCM.43.4.1625-1631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Imai M, Ninomiya A, Minekawa H, Notomi T, Ishizaki T, Van Tu P, et al. Rapid diagnosis of H5N1 avian influenza virus infection by newly developed influenza H5 hemagglutinin gene-specific loop-mediated isothermal amplification method. J Virol Methods. 2007;141(2):173–180. doi: 10.1016/j.jviromet.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 82.Hong TC, Mai QL, Cuong DV, Parida M, Minekawa H, Notomi T, et al. Development and evaluation of a novel loop-mediated isothermal amplification method for rapid detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2004;42(5):1956–1961. doi: 10.1128/JCM.42.5.1956-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhadra S, Jiang Y, Kumar M, Johnson R, Hensley L, Ellington A. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV) PLoS ONE. 2015;10(4):e0123126. doi: 10.1371/journal.pone.0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shirato K, Semba S, El-Kafrawy SA, Hassan AM, Tolah AM, Takayama I, et al. Development of fluorescent reverse transcription loop-mediated isothermal amplification (RT-LAMP) using quenching probes for the detection of the Middle East respiratory syndrome coronavirus. J Virol Methods. 2018;258:41–48. doi: 10.1016/j.jviromet.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim H, Park M, Hwang J, Kim JH, Chung DR, Lee KS, et al. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019;4(5):1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- 86.Teengam P, Siangproh W, Tuantranont A, Vilaivan T, Chailapakul O, Henry CS. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal Chem. 2017;89(10):5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shafiee H, Lidstone EA, Jahangir M, Inci F, Hanhauser E, Henrich TJ, et al. Nanostructured optical photonic crystal biosensor for HIV viral load measurement. Sci Rep. 2014;4:4116. doi: 10.1038/srep04116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xia L, Song J, Xu R, Liu D, Dong B, Xu L, et al. Zinc oxide inverse opal electrodes modified by glucose oxidase for electrochemical and photoelectrochemical biosensor. Biosens Bioelectron. 2014;59:350–357. doi: 10.1016/j.bios.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 89.Bhardwaj J, Sharma A, Jang J. Vertical flow-based paper immunosensor for rapid electrochemical and colorimetric detection of influenza virus using a different pore size sample pad. Biosens Bioelectron. 2019;126:36–43. doi: 10.1016/j.bios.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 90.Oh S, Kim J, Tran VT, Lee DK, Ahmed SR, Hong JC, et al. Magnetic nanozyme-linked immunosorbent assay for ultrasensitive influenza A virus detection. ACS Appl Mater Interfaces. 2018;10(15):12534–12543. doi: 10.1021/acsami.8b02735. [DOI] [PubMed] [Google Scholar]
- 91.Pang Y, Rong Z, Wang J, Xiao R, Wang S. A fluorescent aptasensor for H5N1 influenza virus detection based-on the core-shell nanoparticles metal-enhanced fluorescence (MEF) Biosens Bioelectron. 2015;66:527–532. doi: 10.1016/j.bios.2014.10.052. [DOI] [PubMed] [Google Scholar]
- 92.Omar NAS, Fen YW, Abdullah J, Mustapha Kamil Y, Daniyal W, Sadrolhosseini AR, et al. Sensitive detection of Dengue virus type 2 E-proteins signals using self-assembled monolayers/reduced graphene oxide-PAMAM dendrimer thin film-SPR optical sensor. Sci Rep. 2020;10(1):2374. doi: 10.1038/s41598-020-59388-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park T, Hyun M, Lee H, Lee S, Ko S. A self-assembled fusion protein-based surface plasmon resonance biosensor for rapid diagnosis of severe acute respiratory syndrome. Talanta. 2009;79(2):295–301. doi: 10.1016/j.talanta.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moitra P, Alafeef M, Dighe K, Frieman MB, Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14(6):7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kumar V, Mishra S, Sharma R, Agarwal J, Ghoshal U, Khanna T, et al. Development of RNA-based assay for rapid detection of SARS-CoV-2 in clinical samples. Intervirology. 2022;65(4):181–187. doi: 10.1159/000522337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang C, Wen T, Shi FJ, Zeng XY, Jiao YJ. Rapid Detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5(21):12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Funari R, Chu KY, Shen AQ. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tian B, Gao F, Fock J, Dufva M, Hansen MF. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 99.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 100.Alafeef M, Dighe K, Moitra P, Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14(12):17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]