Abstract
The misfolding and pathological aggregation of the α-synuclein forming insoluble amyloid deposit is associated with Parkinson’s disease, the second most common neurodegenerative disease in the world population. Characterizing the self-assembly mechanism of α-synuclein is critical for discovering treatments against synucleinopathies. The intrinsically disordered property, high degrees of freedom, and macroscopic timescales of conformational conversion make its characterization extremely challenging in vitro and in silico. Here, we systematically investigated the dynamics of monomer misfolding and dimerization of the full-length α-synuclein using atomistic discrete molecular dynamic simulations. Our results suggested that both α-synuclein monomers and dimers mainly adopted unstructured formations with partial helices around N-terminus (residues 8–32) and various β-sheets spanned the residues 35–56 (N-terminal tail) and residues 61–95 (NAC region). C-terminus mostly assumed unstructured formation wrapping around the lateral surface and elongation edge of the β-sheet core formed by N-terminal tail and NAC regions. Dimerization enhanced the β-sheets formations along with a decrease in unstructured content. The inter-peptide β-sheets were mainly formed by the N-terminal tail and NACore (residues 68–78) regions, suggesting that these two regions played critical roles in the amyloid aggregation of α-synuclein. Interactions of the C-terminus with the N-terminal tail and NAC region were significantly suppressed in the α-synuclein dimer, indicating that interaction of the C-terminus with the N-terminal tail and NAC regions could prevent α-synuclein aggregation. These results on the structural ensembles and early aggregation dynamics of α-synuclein will help understanding the nucleation and fibrillization of α-synuclein.
Graphical Abstract
Introduction
The misfolding and pathological accumulation of α-synuclein forming insoluble amyloid deposits in the central nervous system is associated with a series of neurodegenerative diseases, including Parkinson’s disease (PD), dementia with Lewy bodies (DLB), and multiple-system atrophy (MSA)1–4. The α-synuclein is widely abundant in the presynaptic terminals of neurons5. In addition, α-synuclein is known as a natively unfolded protein or intrinsically disordered protein (IDP) due to the lack of a stable native structure6. Similar to the other amyloid degenerative diseases-related peptides (e.g., Aβ, amylin, and Tau)7–9, the aggregation kinetics of α-synuclein followed a sigmoidal pattern with three distinct phases, where the initial lag phase corresponding to the nucleation of monomers into oligomers and proto-fibrils followed by rapid elongation of proto-fibrils before reaching the saturation of mature fibrils10–12. The amyloid aggregation of α-synuclein is very sensitive to solution conditions (e.g., temperature, pH, and salt concentration)13–15. Despite the atomic polymorphous fibrillar structures of α-synuclein structure, featured of cross-β-sheet motifs, have been determined by cryo-electron microscopy (cryo-EM)16, nuclear magnetic resonance (NMR)17, 18, micro-electron diffraction (micro-ED)19, as well as quenched hydrogen/deuterium (H/D) exchange20 experiments. Mounting experimental evidence suggested that the soluble low-molecular-weight oligomers formed during the early nucleation stages of α-synuclein are much more cytotoxic than the mature fibrils21, 22. However, the molecular mechanism of α-synuclein fibrillization is not fully understood, due to the oligomers of α-synuclein formed during the early stages being usually heterogeneous and unstable23. Therefore, the characterization of the monomeric and oligomeric conformations of α-synuclein is critical for both understanding their pathological amyloid aggregations mechanism and the future therapeutic strategies development against PD.
The full-length α-synuclein is composed of 140 amino acids which could be divided into three regions at the level of amino acid sequence24, 25: a positively charged amphiphilic helical N-terminus (residues 1–60), a highly hydrophobic and amyloidogenic central non-amyloid-β component (NAC, residues 61–95) region, and a strongly acidic and negatively charged unstructured C-terminus. There are seven 11-residue imperfect helical repeats with a conserved motif (i.e., KTKEGV) from the first two regions (residue 1–87), which play a crucial role in the α-synuclein interaction with lipid-membrane5. The NAC region is responsible for the pathological aggregation of α-synuclein1, 5, 11, 26. For example, the fibrillization of α-synuclein is significantly inhibited when some residues from NAC are deleted27. Prior experimental and computational studies demonstrated that the segments (i.e., residues 68–78, also known as NACore) from NAC region of α-synuclein could spontaneously self-assemble into well-ordered cross-β fibrillar structures19, 28. The unstructured C-terminal region has been implicated in regulating the nuclear localization and interactions of α-synuclein with proteins, metals, and small molecules11, 26, 29, 30. Monomers of α-synuclein are dramatically disordered and lack stable conformations, regardless of the neuronal cells and buffer conditions31. Monomeric of α-synuclein is also known populated with dynamically collapsed conformations with helix-rich or β-sheet-rich formations, which may assist the aggregation of α-synuclein13, 32, 33. For example, the conformations of monomeric α-synuclein are more compact under physiological cell conditions than the buffer31. The α-synuclein monomers are more extended at the neutral environment than the acidic conditions, stable dimer conformations only exist in the natural conditions34. Previous experimental study suggested that the aggregation and neurotoxicity of α-synuclein was initiated by dimerization35. The presence of a partially folded intermediate, mediated by a decrease in pH or an increase in temperature, is strongly correlated with the enhanced formation of a-synuclein fibrils13. However, the conformational features of α-synuclein in the monomers and oligomers are still elusive; the relationship between the dynamical conformations in the monomeric state and aggregation propensity also remains to be established.
Here, we systemically studied the monomer and dimer (i.e., the smallest aggregates at neutral pH conditions34) structures of the full-length α-synuclein using all-atom discrete MD (DMD) simulations. DMD is an efficient and predictive MD algorithm widely used to study protein folding, misfolding, and aggregation36–38. For each molecular system, 100 independent DMD simulations starting from the different initial states were performed with the duration time of each DMD trajectory up to 800 ns in the monomer and 1000 ns in the dimer. Our results demonstrated that both α-synuclein monomers and dimers predominantly adopted unstructured formations with partial local helices and β-sheets. The partial helices in α-synuclein monomers and dimers were mainly formed by the residues 8–32 of the N-terminus, consistent with prior X-ray and NMR measurements39–41. Diverse β-sheets mostly spanned around the N-terminal tail and NAC region, in which the N-terminal tail (residues 35–56) adopted a typical β-hairpin motif and incorporated with the NAC region forming various β-sheets. One similar β-hairpin motif formed by the N-terminal tail was also reported in prior experimental literature42. Similar β-sheet regions were also present in various prior biophysical and biochemical approaches that determined amyloid fibril and unfolded structures of α-synuclein20, 43–51. C-terminus in α-synuclein monomer and dimer were very dynamic, mostly assumed unstructured formations, consistent with prior NMR measurements20, 50, 52, 53. The C-terminus preferred dynamically wrapping around the lateral surface and elongation edge of the β-sheet core formed by the N-terminal tail and NAC regions. Interactions between C-terminus and N-terminal tail and between C-terminus and NAC region were significantly suppressed in the α-synuclein dimer, indicating that the dynamic wrapping and capping of the C-terminus around the β-sheet core may prevent amyloid aggregation of α-synuclein. Overall, our simulation results uncovered the conformation and self-assembly dynamics of the full-length α-synuclein, which will help better understanding the pathological aggregation of α-synuclein.
Results and discussion
The conformational dynamics of α-synuclein monomers.
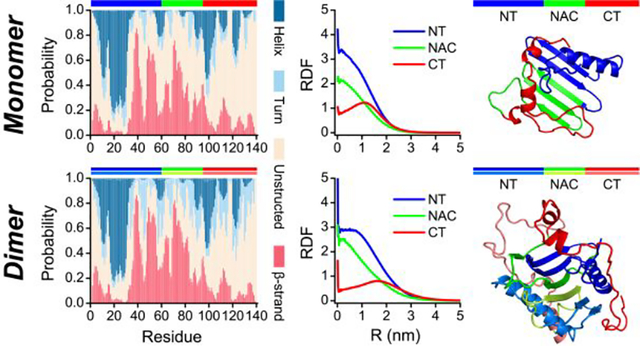
The amino acid sequence and initial structure of α-synuclein used in our DMD simulation are shown in Figure 1. The solution structure of the α-synuclein monomer is still elusive. Thus, we chose the micelle-bound α-synuclein monomer54 (PDB: 1XQ8) as the initial structure, which has been extensively employed as the starting structure in numerous computational studies for the conformational investigation of α-synuclein55–57. The equilibrium assessment by examining the time evolution of the several structural parameters (including radius gyration (Rg), number of hydrogen bonds, and secondary structure contents) of the representative trajectory, randomly selected from 100 independent trajectories, suggesting the monomeric simulation reached the steady states in the last 400 ns (Figure S1a). Not many changes in the time-evolution of ensemble-averaged conformational parameters (e.g., Rg, number of hydrogen bonds, and secondary structure contents) over 100 independent simulations during the last 400 ns indicated that all the simulation systems were reasonably converged (Figure S1b). The conformational dynamic analysis revealed that the helix-rich extended α-synuclein monomer readily collapsed into a compact structure with more residue-pairwise contacts and fewer hydrogen bonds within 100 ns (Figures S1a&2). Time evolution of each residue’s secondary structure revealed that residues from N-terminus still mainly adopted helical formations, but most residues from the NAC region converted into β-sheet structures. The negatively charged C-terminal region was still populated with unstructured formations (random coil and bend structures) wrapping around the core collapsed by the N-terminal tail and NAC regions (snapshots in Figures S1a&2). Transient partial helix and β-sheet were also observed around the C-terminal region, but these structured formations were easily converted into unstructured formations. Another recent site-specific structural dynamics of α-synuclein study also showed that structured formations (e.g., dynamic helix and β-sheet) around N-terminus and NAC region were more abundant than C-terminus due to the C-terminus mostly remaining unstructured formations41, 58. Monomers of α-synuclein collapsing into both helix and β-sheet abundant formations were consistent with numerous prior experimental studies13, 32, 33. The monomeric α-synuclein was dramatically disordered under buffer conditions and neuronal cell conditions31. The de novo protein structure determination based on the incorporation of short-distance crosslinking data according to experimental proteomics measurements as constraints in DMD simulations revealed that the conformational ensemble of α-synuclein was rather compact globular conformations57. Using inter-dye distance distributions from bulk time-resolved Förster resonance energy transfer as restraints in DMD simulations, Chen et al. found some globular conformations of α-synuclein were extremely stable with a lifetime of more than milliseconds56. The dynamically collapsed conformations with both helix-rich and β-sheet-rich structures observed in our DMD simulations were consistent with prior experimental measurements13, 31–33.
Figure 1. Amino acid sequence and initial structure of α-synuclein.
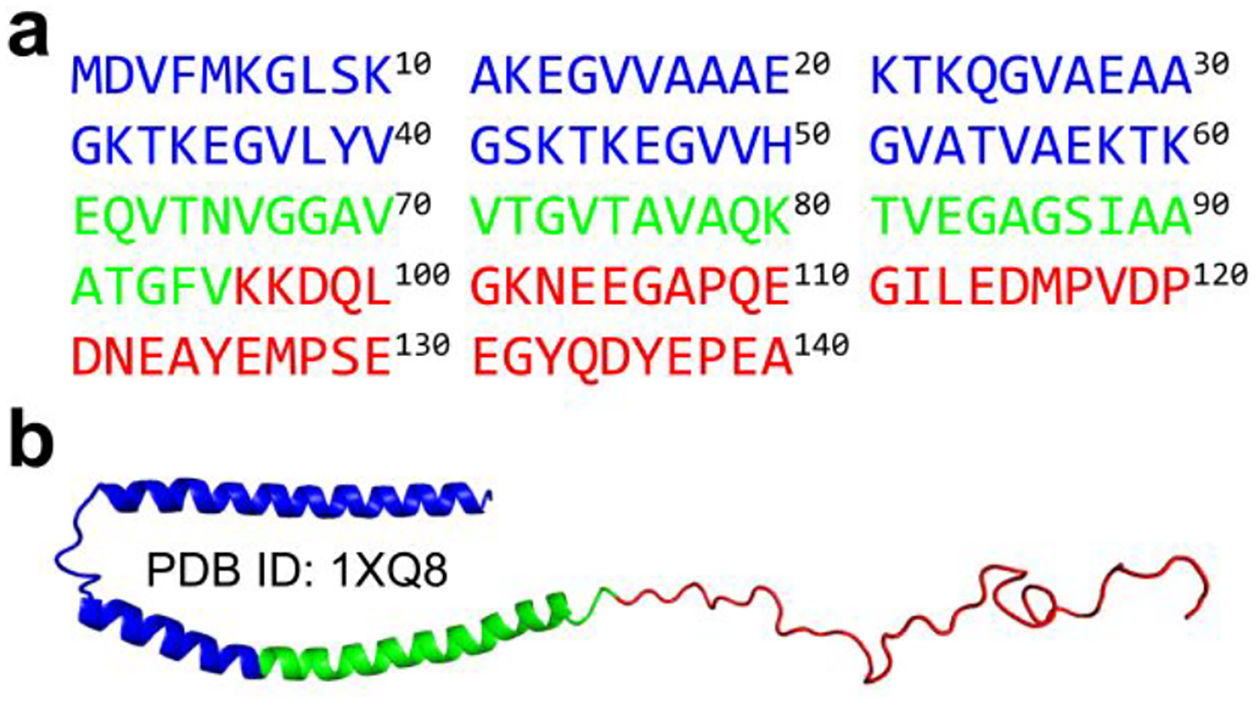
The amino acid sequence a) and initial structure b) of full-length α-synuclein used in our DMD simulation. The N-terminus, NAC region, and C-terminus are colored by blue, green, and red, respectively.
The dimerization and conformation dynamics analysis of α-synuclein.
Similar to the monomeric simulations, the α-synuclein peptides also spontaneously collapsed into compact structures populated with both helix and β-sheet in two-peptide simulation by examining the time evolution of radius gyration, the number of contacts and hydrogen bonds, and secondary structure of each residue (Figure S2). The dimerization of α-synuclein monitored by the time evolution of intermolecular hydrogen bonds and contacts featured a significant heterogeneity with diverse lifetimes in dimeric states among the simulations (Figure S2&3). The increase of intermolecular contacts accompanied by the rise of inter-peptide main-chain hydrogen bonds indicating dimerization may render inter-peptide β-sheets, which was confirmed by the representative snapshots of dimer structures (Figure S2&3). Dimers of α-synuclein were mostly dynamic with limited duration time (length of consecutive red pixel regions along the time axes in Figure 4a&b) during the course of simulations. Although dimers with a certain number of intermolecular contacts and hydrogen bonds persisted until the end of the simulation were also observed (Figure 3&4&S2). Although dimers α-synuclein featured lower potential energy than two isolated monomers on average, a large population of isolated monomers could adopt conformations with much lower potential energy than most dimeric structures (Figure S3). Therefore, some α-synuclein dimers were unstable and readily dissociated into monomers due to the broad potential energy overlapping between isolated monomeric and dimeric states in two-peptide simulation. The probability distribution and potential mean force as a function of the number of intermolecular contacts suggested that dimer conformation became favorable as the number of intermolecular contact up to 24 or more (Figure 4a). Thus, the conformations with the number of intermolecular contacts of more than 24 were used for the structural analysis of α-synuclein dimers. The first-passage time of two isolated α-synuclein nucleated into a dimer with a number of inter-molecular contacts larger than 24 and the average lifetime of the corresponding dimer in each independent DMD simulation featured a high heterogeneity among the simulations (Figure S4), indicating multiple long-timescale independent simulations were necessary for investigating the dimerization dynamics of α-synuclein to get enough conformational sampling. Secondary structure of each residue and representative snapshots along simulations suggested that residues from N-terminus, NAC region, and C-terminus preferred to adopt helix, β-sheet, and unstructured formations, respectively, in both monomer and dimer states (Figure S2&3). The β-sheet formations around the NAC region were more populated in the dimeric state than the monomeric states, indicating dimerization may promote the β-sheet structures of NAC segments (Figure 2&3).
Figure 4. Dimerization dynamics analysis of α-synuclein.
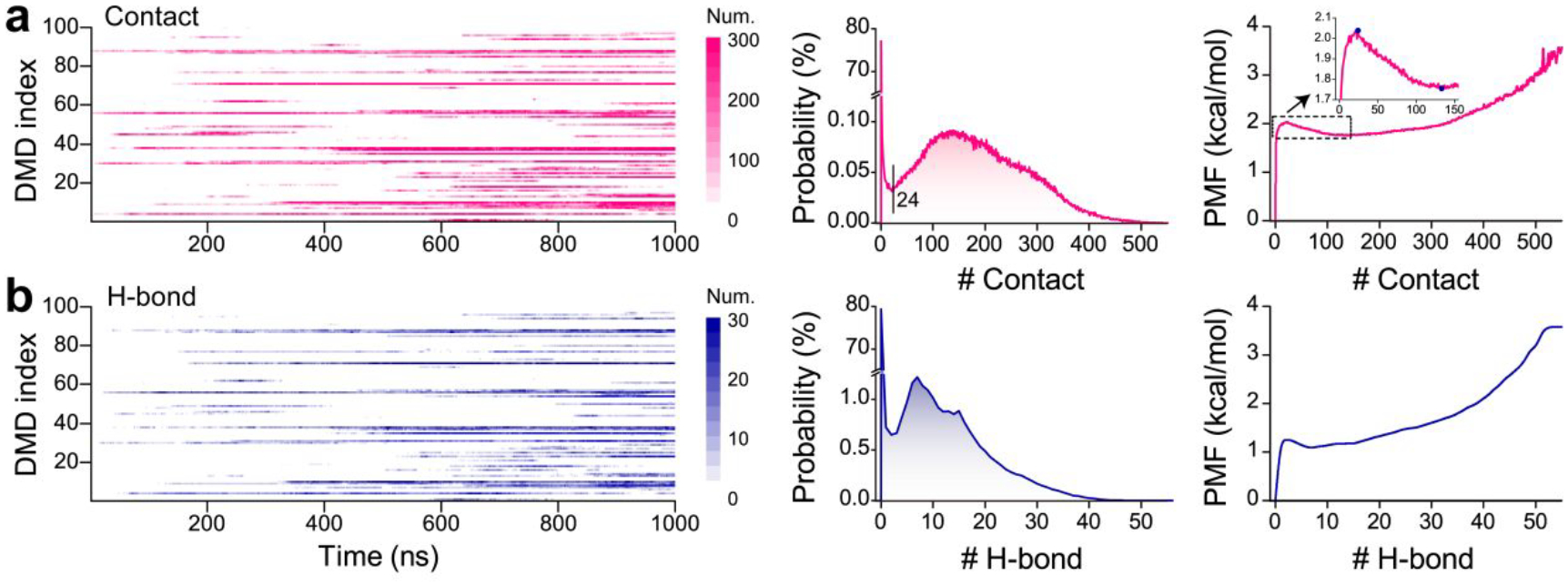
The time evolution and probability distribution of intermolecular contacts a) and hydrogen bonds b) in each trajectory. All the 1000 ns simulation data of each independent trajectory is used for the probability distribution calculation.
Figure 3. The conformational dynamics of α-synuclein dimerization.
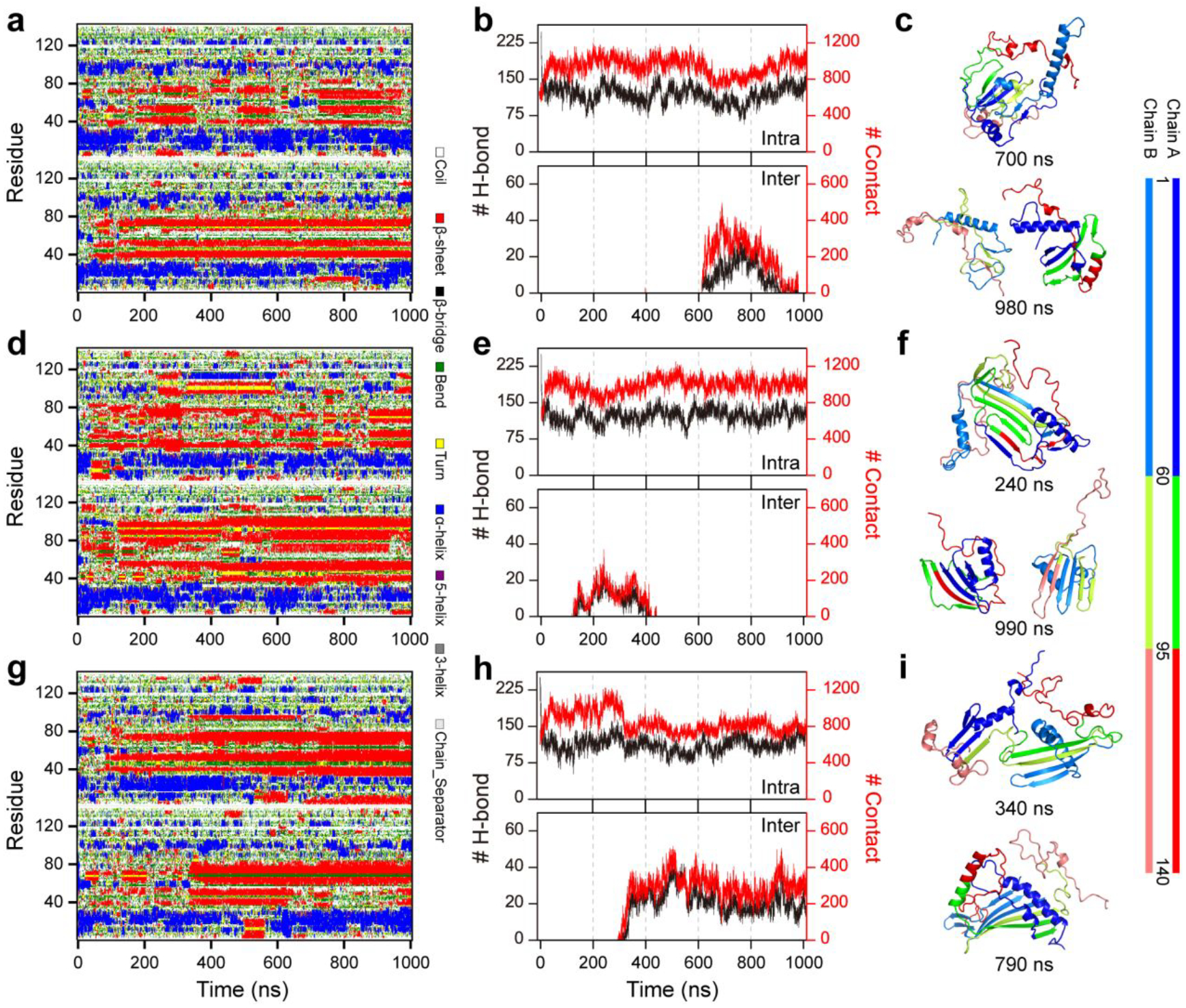
The dimerization dynamics are monitored by the time evolution of the secondary structure of each residue (a, d, &g) and the number of contacts and main-chain hydrogen bonds (b, e, & h). The representative snapshots along the simulation time are also present (c, f, &i). For clarity, the N-terminus, NAC region, and C-terminus are colored by blue, green, and red, respectively. Three trajectories were randomly selected from 100 independent DMD dimerization simulations.
Figure 2. The conformational dynamics of α-synuclein monomer.
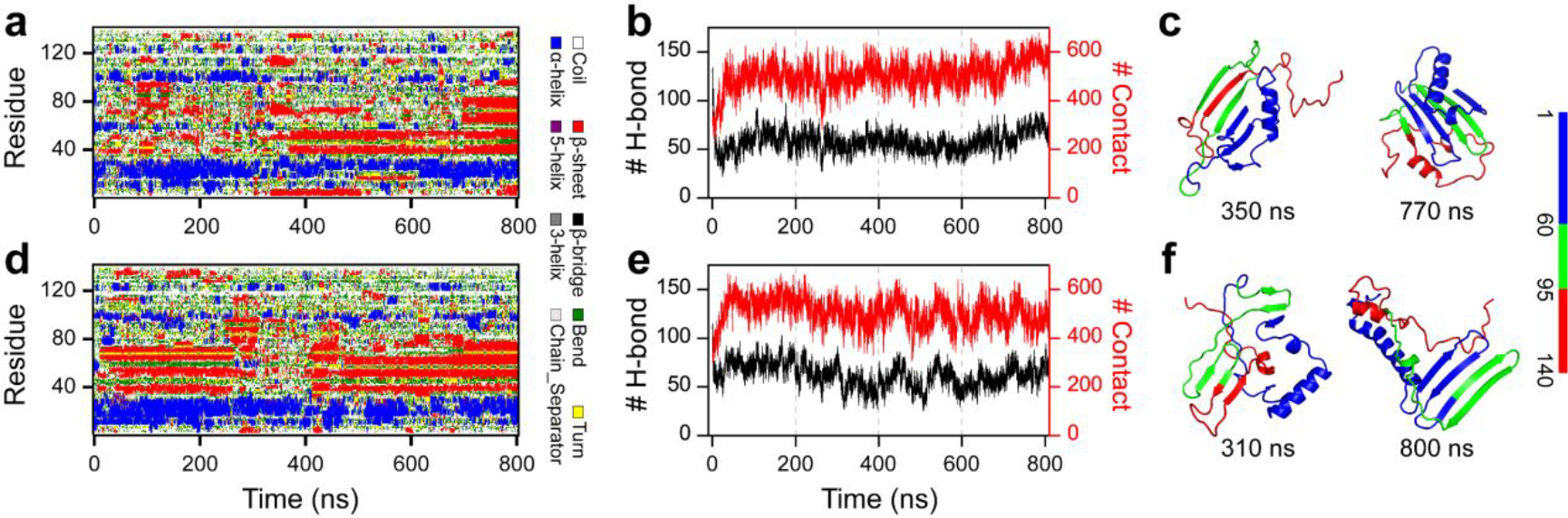
The conformational dynamics of the α-synuclein monomer are monitored by the time evolution of the secondary structure of each residue (a&d), the number of contacts and main-chain hydrogen bonds (b&e) and representative snapshots (c&f). For clarity, the N-terminus, NAC region, and C-terminus are colored blue, green, and red, respectively. Two trajectories are randomly selected from 100 independent DMD simulations.
Secondary structure analysis of the monomeric and dimeric α-synuclein.
We further analyzed the secondary structure of α-synuclein monomer and dimer. The unstructured formations (i.e., random coil and bend) were the dominant species in both α-synuclein monomer and dimer with a probability of ~49.0% and ~38.9% (Figure 5a), respectively. The propensity of each residue to adopt random coil and bend structures showed that the unstructured formations mainly spanned the C-terminal region (Figure 5b), which agreed with prior NMR measurements40, 41. Significant conformational flexibility of residues 110–140 was also observed in prior solid-state and solution-state NMR, Electron Paramagnetic Resonance, and limited proteolysis assays20, 50, 52, 53. Compared to the α-synuclein monomer, the unstructured propensities of residues from the tail of the N-terminus and NACore were suppressed in the dimer. The average helical formations were very similar in α-synuclein monomer (~18.6%) and dimer (~18.8%). The helices were mainly formed by residues 8–32 from N-terminal regions (Figure 5c). Prior crystal structure of α-synuclein segment with residues ranging 1–72 carried by maltose-binding protein also had two helical regions around residues 1–13 and 20–34, which was very similar to our simulation results39. Multi-dimensional heteronuclear NMR spectroscopy measurement also showed the helical structures were mainly presented around residues 6–37 in the N-terminus of the α-synuclein40, 41. The helix-rich conformations around the above N-terminal region were also observed in the all-atom MD55 and DMD simulations56, 57. The solid-state and solution NMR spectroscopy found these N-terminal residues adopted a well-defined α-helical secondary structure to target and anchor α-synuclein to the membrane59. Partial helices with a weaker average probability than the N-terminal region were also observed in C-terminus (e.g., around residue 90–130). The α-synuclein dimers featured more turn structures than monomers (Figure 5d). Dimeric α-synuclein had more β-sheet formations (~26.4%) than the monomeric α-synuclein (~22.8%). Residue with a strong β-sheet propensity was mostly located in the tail of the N-terminus and NAC regions. The stability of polymorphic α-synuclein dimers derived from the experimentally determined fibrils by standard MD simulation also featured similar β-sheet regions29. Interestingly, the β-sheet regions in the various biophysical and biochemical approaches determined amyloid fibril and unfolded structures of α-synuclein were also present around residues L38–K43, V48–N65, V70–Q79, and G86–K9720, 43–51 (Figure 5e). The conformational cluster analysis (Figure S5) further confirmed that both monomers and dimers of α-synuclein mainly adopted unstructured formations; partial helix and β-sheet formation were also present around N-terminal and NAC regions, respectively.
Figure 5. Secondary structure analysis of α-synuclein monomer and dimer.
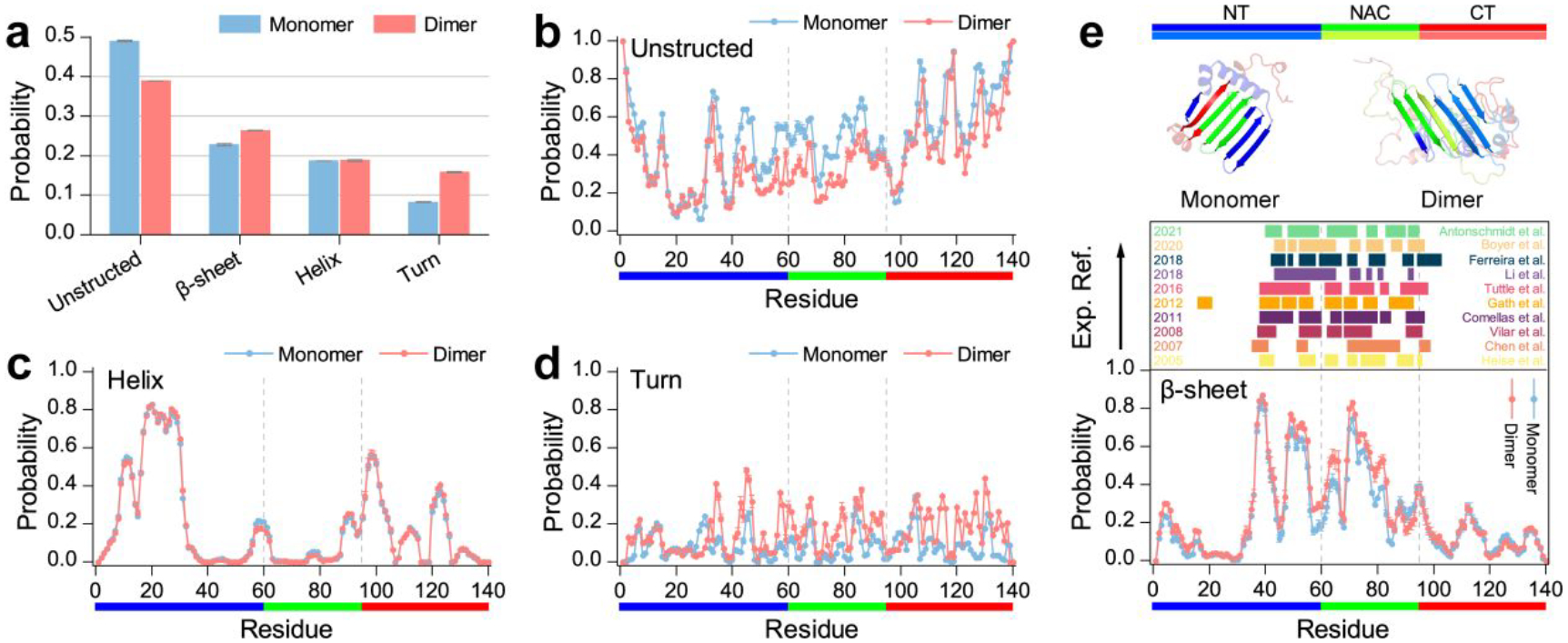
a) The averaged probability of each secondary structure of α-synuclein monomer and dimer. The averaged propensity of each residue adopted unstructured structure b), helix c), turn d), and β-sheet e) formations in α-synuclein monomer and dimer. The β-sheet propensity profile (lower panel) is compared with the regions of the sequence found experimentally to form the β-sheet core region (upper panel) of the oligomer and fibrils (color bars) according to the references indicated in the inset. All the last 400 ns simulation trajectories of the α-synuclein monomer DMD simulation are used for the conformational analysis. Dimers of α-synuclein with the number of inter-molecular contact up to 24 or more are selected for the secondary structure analysis. The error bar of each propensity is estimated by computing the mean difference between the first and last 50 independent DMD simulations.
Intra-peptide residue-pairwise contact frequency analysis of α-synuclein monomers.
To characterize the dominant inter-residue interactions in α-synuclein monomers, we calculated the main-chain and side-chain residue-pairwise contact frequency maps (Figure 6a). The high intra-chain contact frequencies along the diagonal around residues 8–32 (greater than 0.40) suggested a strong helical tendency in this region (snapshot 1 in Figure 6b), consistent with the secondary structure propensity analysis (as discussed in Figure 5c) and prior experimental measurements39–41. The tail of the N-terminus featured a typical β-hairpin contact pattern perpendicular to the diagonal with the β-strand spanned residues 35–44 and 47–56 (snapshot 2 in Figure 6b). An extended β-hairpin structure formed by residues 37–54 with the β-strands around residues 38–43 and 48–53 stabilized by β-wrap proteins AS69 was also reported by an experimental study42. These two β-strands regions (i.e., residues 35–44 and 47–56) incorporated with the NAC region forming various both parallel and anti-parallel β-sheets were also observed (corresponding contact patterns and snapshots 3~5 shown in Figure 6a&b). Representative β-sheet motifs (e.g., residues 37–44 vs 70–77, residues 52–56 vs 62–66, and residues 47–56 vs 69–78) revealed that these β-sheets were mainly stabilized by the interactions among hydrophobic residues from the tail of N-terminus and NAC region. The interactions between the N-terminal tail and NAC region forming diverse β-sheets were also supported by an extensive set of intramolecular paramagnetic relaxation enhancement (PRE) data from measurement32. The familial PD mutations around the N-terminal tail region (including E46K2, H50Q60, G51D61, A53T62, and A53E63) could affect the pathogenic aggregation of α-synuclein49, 64, 65, indicating the residues from the tail of N-terminus may participate in forming β-sheet dominated fibrillar formations with NAC region. For example, the fibrillization of α-synuclein was inhibited when the β-hairpin structure around residues 37–54 was wrapped by AS6942, due to the interactions between residues 37–54 and NAC region were prevented. The rigid core of α-synuclein oligomers incorporated residues 30–60 and the NAC region was also reported in the prior experiment66. Residues from the NAC region, which were necessary for the fibrillization of α-synuclein27, 67, had a strong propensity to form various β-hairpins (e.g., residues 61–67 vs 70–76, residues 70–78 vs 81–89, residues 70–82 and 86–98) driven by hydrophobic interactions. Such β-hairpin motifs formed by residues from the NAC region were also supported by NMR measurements32. The β-hairpin-rich formations within the NAC segment were also observed in the conformational ensemble of α-synuclein monomers determined by short-distance crosslinking constraint-guided DMD simulations56, 57. A β-hairpin formation formed by the tail of the NAC region and the head of the C-terminus (residues 93–100 vs 108–115) driven by the interaction between residues V95 and L113 was observed (illustrated as snapshot 10 in Figure 6). Residues from C-terminus mostly adopted unstructured formations along with partial dynamic structures (Figure 2&3). Transient helical patterns were very short, with the residue length 6~11 (snapshots 9 and 11 illustrated in Figure 6). Dynamic short helices within the C-terminal region were consistent with the NMR Cα chemical shift40. In addition, we also observed long-range unstructured contact patterns between N- and C-terminus (snapshot 12 illustrated in Figure 6).
Figure 6. Contact frequency analysis of α-synuclein monomer.
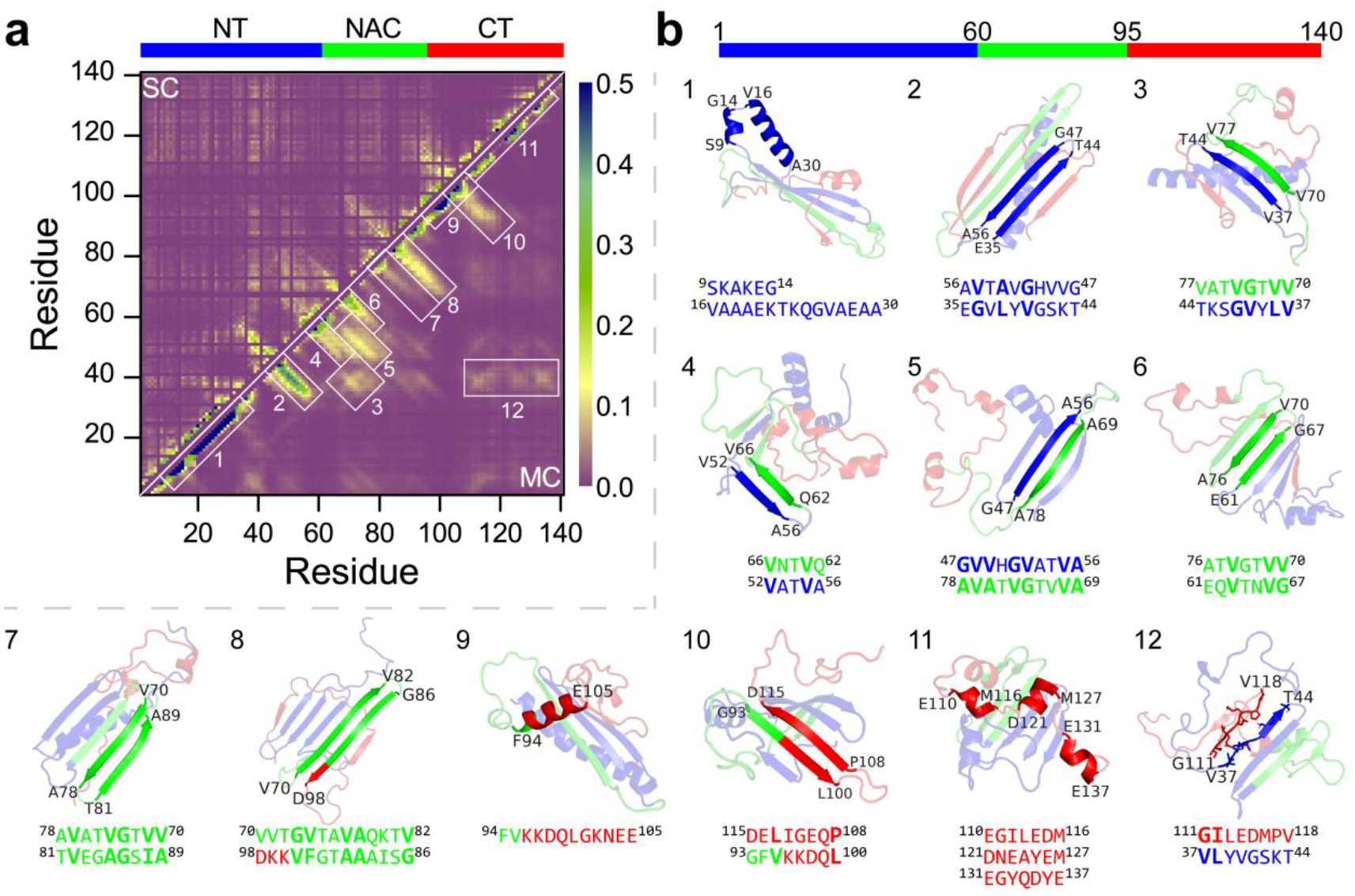
The residue-pairwise contact frequency map of α-synuclein monomer is computed both between main-chain atoms (MC-MC) and between side-chain atoms (SC-SC) based on the last 400 ns trajectories of 100 independent DMD simulations after reaching steady state a). The representative structured motifs with high contact frequency patterns, mostly corresponding to the helices or β-sheets, labelled as 1–12 in the contact frequency map, are also presented b).
The side-chain residue-pairwise contact frequency suggested residues from the C-terminus could dynamically interact with both N-terminus and NAC regions but didn’t induce obviously β-sheet structured contact patterns (Figure 6a). The time evolution of the contact surface areas of the NAC region with N- and C-terminus and the representative snapshot suggested partial N-terminus (residues 35–56) preferred to incorporate with the NAC region forming β-sheet formations with relative stable contact surface areas (Figure S6a&b). Residues 1–35 populated with helical formation featured weakly side-chain contact frequency with the NAC region and C-terminus (Figure 6a). C-terminal could dynamically wrap around the NAC and N-terminal tail regions (Figure S6a&b) with unstructured formations. The dramatically faint β-sheet patterns formed by the C-terminus participated with the NAC region or N-terminal tail (Figure 6a), as well as the weak β-sheet propensity of the C-terminus (Figure 5e), suggested that C-terminus could also dynamically adopt β-sheet formations capped to the β-sheet elongation edges (Figure S6a&b). Contact surface area probability distribution showed that the interface between the C-terminus and the NAC region was smaller than the contact area of the N-terminus with the NAC region or C-terminus (Figure S6c). The radius distribution function of each Cα atom showed that most residues from the N-terminal and NAC regions were buried inside, whereas residues from the C-terminus preferred to expose outside (Figure S6d). Prior studies suggested that hydrophobic interactions between the C-terminal tail and the NAC region may lead to globular formations31, 57, 68. The α-synuclein with the C-terminus truncated was more prone to aggregate than the wild-type of α-synuclein69. Enhancing the exposure of the N-terminus and the beginning of the NAC region of α-synuclein upon disruption of the contacts with the C-terminus through calcium binds could accelerate fibrilization of α-synuclein70. Overall, these studies indicated the dynamic interaction between the C-terminus with NAC and N-terminal regions might modulate the aggregation of α-synuclein.
Residues from the NACore and N-terminal tail played critical roles in the dimerization of α-synuclein.
Residue-pairwise contact of the dimeric α-synuclein was also analyzed to investigate the aggregation mechanism. Both the helix and β-sheet contact patterns and the corresponding structures observed in the monomeric α-synuclein (Figure 6) were also present in the α-synuclein dimers (Figures 7a&S4). For example, the N-terminal tail and NAC region were still populated with various β-sheet formations (snapshots 2–7 in Figure S7b). The difference of each residue-pairwise intra-molecular contact frequency of α-synuclein between the dimer and monomer was also calculated by subtracting each residue-pairwise contact in the monomer from the corresponding value in the dimer (Figure 7b). Compared to α-synuclein monomers, C-terminus interacted with N-terminal and NAC regions were suppressed, and the β-sheet formations around N-terminus were slightly enhanced (Figure 5e and snapshots 12 in Figure S7b). For example, formations of residue 100–140 capping to the N-terminal tail and NAC regions featured with the β-sheet pattern were decreased in α-synuclein dimers (Figure 7b). The intermolecular residue-pairwise main-chain contact showed that inter-peptide β-sheets were mainly formed among NAC regions, and NAC segments incorporated with N-terminal tail β-sheets were also observed (Figure 7c). The specific interactions between monomers within the polymorphic fibril-like α-synuclein dimers by all-atom MD simulations also showed that both N-terminal and the NACore domains play a role in the dimerization of all polymorphic alpha-synuclein dimers29. Representative residue-pairwise contact motifs with obvious β-sheet structured patterns were mostly formed by NACore (residues 68–78) participated with residues from the N-terminal tail and NAC region forming various β-sheets (e.g., snapshots 1, 2, 3, 5, 6, 8, and 9 in Figure 7d). In addition, the N-terminal tail (e.g., residues 35–56) involved β-sheets conformations were also abundant (e.g., snapshots 5, 6, 7, 8, 9, 10, 11, and 12 in Figure 7d). Overall, both NACore and N-terminal tail segments played curial roles in forming intermolecular β-sheet dimeric conformations of α-synuclein. The NAC and N-terminal tail regions displayed a less interacted tendency with C-terminus in the α-synuclein dimer than the monomer, indicating the presence of the C-terminus around the NAC and N-terminal tail region may hinder α-synuclein aggregation. In addition, residues from the N-terminal tail (i.e., residues 35–56) and NACore (i.e., residues 68–78) region had a smaller number of contacts with the C-terminus in dimer than monomer (Figure S8). Prior experimental studies have also shown that preventing the C-terminus from interacting with N-terminus and NAC regions could accelerate the aggregations of α-synuclein69, 70.
Figure 7. The residue-pairwise contact frequency of dimeric α-synuclein.
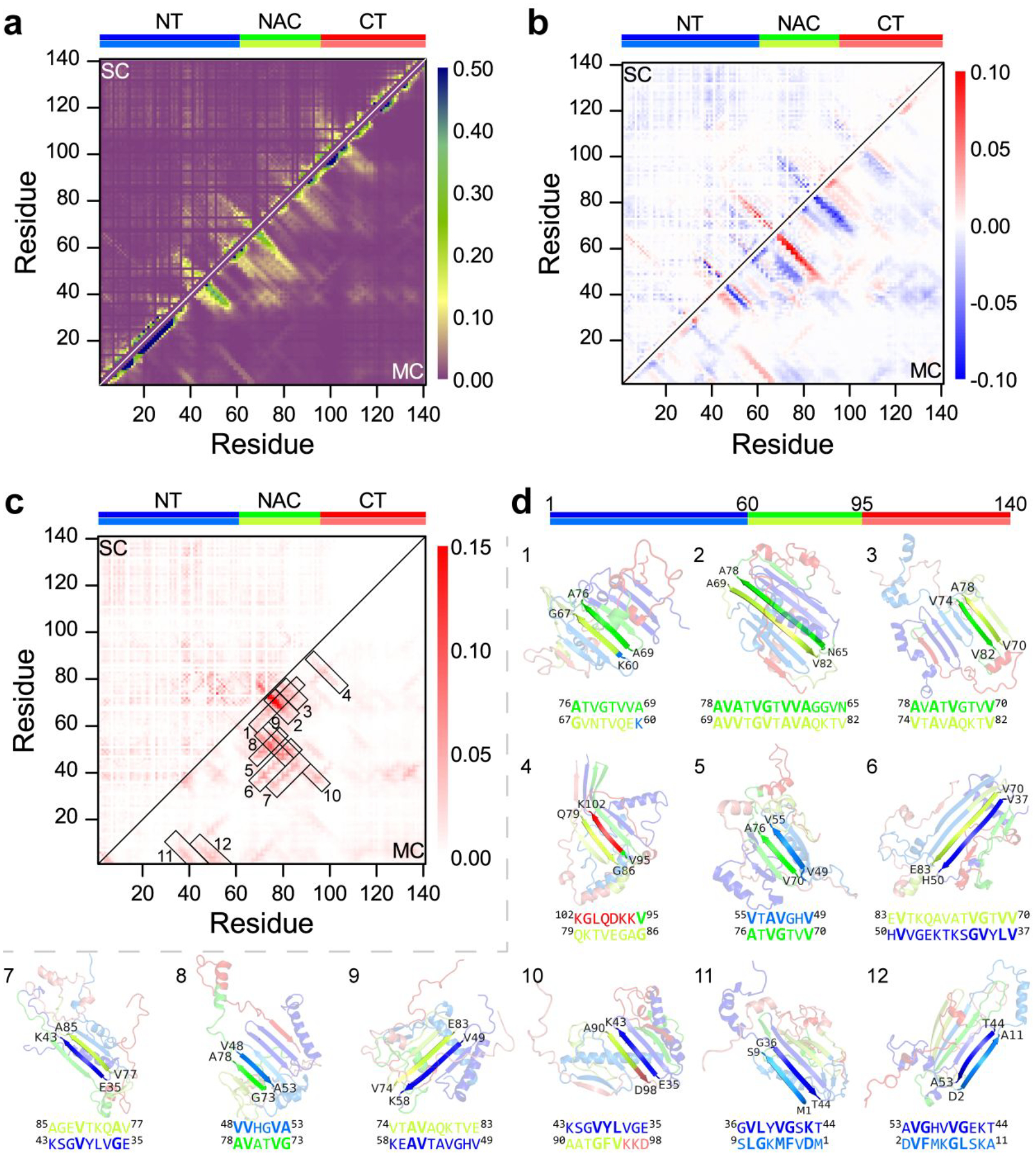
a) The intra-chain residue-pairwise contact frequency of α-synuclein in the dimer. b) The difference of each residue-pairwise intra-molecular contact frequency of α-synuclein in the dimeric and monomeric state by subtracting each residue-pairwise contact in monomer from the corresponding value in the dimer. c) The intermolecular residue-pairwise contact frequency in the α-synuclein dimer. The representative β-sheet structures with high contact frequency patterns labelled as 1–12 in the contact frequency map are also presented d).
Conformational free energy landscape analysis.
To better understand the conformational features of α-synuclein monomers and dimers, we calculated the potential of mean force (PMF; also known as the effective free-energy landscape) as a function of β-sheet and α-helix contents (Figure. 8a&c). There was only one flat and broad energy basin with the helix and β-sheet contents of 10%~30% and 10%~40%, indicating conformations of α-synuclein monomers and dimers were very diverse. The typical snapshots randomly selected for the basin center of each system (snapshots 3–10 in Figure 8a and snapshots 2–7 in Figure 8c) further validated the diversity of α-synuclein monomers and dimers. Low β-sheet (<0.1) or low helical (<0.1) content formations were unfavourable with high free energy (snapshots 1, 2 and 12 in Figure 8a, and snapshots 1 and 8 in Figure 8c), suggested that the intrinsically disordered protein α-synuclein assumed with partial local helices and β-sheets. Secondary structure (Figure 5) and residue-pairwise contact frequency (Figure 6&7) analyses, as well as the representative snapshots in Figure 8, suggested that local helical formation mostly formed around N-terminus (residues 8–32). Diverse β-sheets were mostly formed by residues from the N-terminal tail (residues 35–56) and NAC region. The C-terminal residues predominantly adopted unstructured formations wrapping around the local β-sheet core (representative snapshots in Figure 8). The radius distribution function of each Cα atom from each region revealed that residues from the NAC region were mostly buried inside, while the C-terminal residues were exposed outside (Figure 8b&d).
Figure 8. The conformational free energy landscape of α-synuclein monomers and dimers.
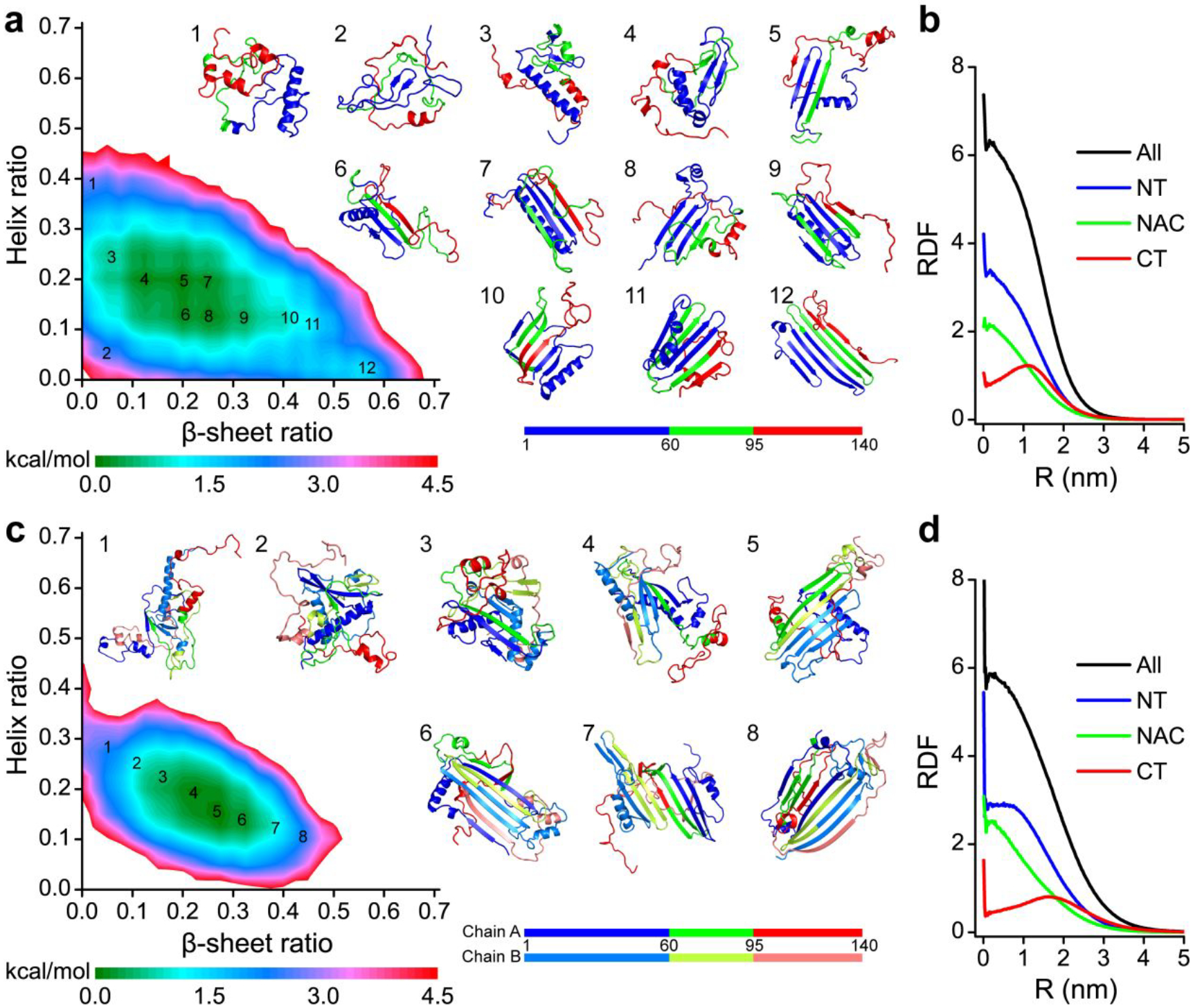
The potential mean force as a function of β-sheet and helix contents of α-synuclein monomers a) and dimers c). Representative structures labelled in the PMFs are also shown. The radius distribution function of Cα atom from N-terminus, NAC region, and C-terminus in α-synuclein monomers b) and dimers d).
Conclusions
We systematically investigated the conformational dynamics of monomer folding/misfolding and dimerization for full-length α-synuclein with long timescale all-atom DMD simulations, accumulatively 80.0 μs for monomers and 100.0 μs for dimers. Our results demonstrated that both α-synuclein monomers and dimers predominantly adopted unstructured formations with partial local structured formations around N-terminal and NAC regions. Helical formations in α-synuclein monomers and dimers are mainly formed by the residues 8–32 of the N-terminus, consistent with prior X-ray and NMR measurements39–41 and computational simulations55–57. Diverse β-sheets mostly spanned around the N-terminal tail and NAC region, in which residues 35–56 from the N-terminal tail adopted a typical β-hairpin motif and incorporated with the NAC region forming various β-sheets. An experimental study also reported a similar extended β-hairpin structure formed by residues 37–54 stabilized by β-wrap proteins AS6942. In addition, the residues 68–78 of NACore displayed significant β-sheet tendency, agreed with prior experimental and computational studies19, 28, 29. C-terminus mostly assumed unstructured formation wrapping around the β-sheet core formed by the N-terminal tail and the NAC region. Compared to the α-synuclein monomers, the intermolecular interaction among N-terminal tail and NACore forming inter-peptide β-sheet enhanced the average β-sheet content in α-synuclein dimers. Residue-pairwise contact frequency suggested that the C-terminus dynamic capping with the N-terminal tail and NAC region hindered the dimerization of α-synuclein, indicating that preventing the C-terminus from interacting with N-terminus and NAC regions could accelerate the aggregations of α-synuclein agreed with prior experimental results69, 70. Overall, our study provides a full picture of the monomer conformations and dimerization dynamics of α-synuclein, which will help better understanding the pathology of α-synuclein aggregation.
Materials and Methods.
Simulation systems.
The amino acid sequence and initial structure of α-synuclein used in our simulation is taken from protein data bank (PDB: 1XQ8) determined by solution NMR spectroscopy measurement in the micelle-bound form (Figure. 1), which is composed of two helical regions formed by residues V3-V37 and K45-T92 and an extended hydrophilic C-terminus54. According to the previous studies, both α-synuclein monomer and dimer existed in the natural conditions, and dimerization accelerated the formation of neurotoxic aggregates and amyloid fibrils in vitro34, 35. To understand the aggregation of mechanism of α-synuclein, we systematically performed both one- and two-peptide simulations using all-atom DMD. For each molecular system, 100 independent trajectories were obtained starting with different initial state. In the two-peptide system, two isolated α-synuclein monomers were randomly placed in the simulation box with the intermolecular distances at least 1.5 nm. The durations of each DMD trajectory in one- and two-peptide systems were 0.8 μs and 1.0 μs, respectively.
Discrete molecular dynamics (DMD) simulations.
All the simulations were performed using the all-atom DMD algorithm71 with the Medusa force field, which has been benchmarked for the accurate prediction of protein stability change upon mutation and protein–ligand binding affinity72, 73. DMD is a unique type of MD algorithm, where the continuous potential functions in the standard MD simulations were replaced by discrete step functions. A comprehensive description of the atomistic DMD algorithm can be found in prior studies36, 71, 74. Similar to the standard MD, both bonded interactions (including covalent bonds, bond angles, and dihedrals) and non-bonded interactions (including van der Waals, hydrogen bond, and electrostatic terms) were considered in DMD37. To reduce the computational cost, the implicit solvent model of effective energy function (EEF1) for proteins in the solution proposed by Lazaridis and Karplus was used to model solvation effects75. The solvation free energy of a protein molecule is a sum of group contributions, which are determined from values for small model compounds75. The accuracy of Medusa force field with EEF1 implicit solvent model has been well benchmarked in protein folding and aggrgation37, 72, 76. With significantly enhanced sampling efficiency and rapid computational speed, DMD simulations have been widely used in studying protein folding, amyloid aggregation, and small molecule/nanoparticle–protein interactions by both our group7, 8, 77, 78 and others38, 79–81. The units of mass, time, length, and energy used in our DMD simulations with an implicit water model are 1 Da, ~50 fs, 1 Å, and 1 kcal/mol, respectively. The DMD program is available via Molecules in Action, LLC (http://www.moleculesinaction.com).
Analysis methods.
The Secondary structure analysis was performed using the dictionary of secondary structure of protein (DSSP) method82. A hydrogen bond was considered to be formed if the distance between the backbone N and O atoms less than 3.5 Å and the angle of NH···O was more than 120°76. A residue pairwise contact was defined if they had at least one heavy atom contact within the cutoff distance of 0.65 nm. The two-dimensional potential of mean force, PMF, was constructed using – kbTlnP(x, y), where kb is Boltzmann constant, T denotes the temperature of 300 K, and P(x, y) corresponds to the probability of selected reaction ordinates, x and y. The radial distribution function g(r) of atom in each system corresponding to the complex center was calculated by the following equation g(r) = Nr,r+dr/(4πr2dr), where Nr,r+dr is the number of atoms within distances of r and r+dr away from the center of the complex28. Cluster analysis was performed using the Daura algorithm83–85 and the Ca root-mean-square deviation cutoff of 0.80 nm and 1.0 nm for monomer and dimer of α-synuclein structured regions (residue 1–103).
Supplementary Material
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China under the Grant No. 11904189 (Sun), K.C.Wong Magna Fund in Ningbo University, China (Sun), US National Science Foundation CBET-1553945 (Ding), and US National Institutes of Health R35GM119691 (Ding). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSFC, NIH and NSF.
Footnotes
Supporting Information
The Supporting Information is available free of charge at:
Equilibrium assessment of the monomeric α-synuclein simulation; the dimerization and conformation dynamics analysis of α-synuclein; dimerization free energy landscape analysis; the dimerization time analysis of α-synuclein; representative structures of α-synuclein monomer and dimer; contact surface area analysis of α-synuclein monomer; intra-molecular residue-pairwise contact frequency of α-synuclein dimer; the interaction of C-terminus with N-terminus and NAC region analysis (PDF).
Declaration of Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- (1).Goedert M; Jakes R; Spillantini MG The Synucleinopathies: Twenty Years On. J Parkinsons Dis 2017, 7 (s1), S51–S69. DOI: 10.3233/JPD-179005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zarranz JJ; Alegre J; Gomez-Esteban JC; Lezcano E; Ros R; Ampuero I; Vidal L; Hoenicka J; Rodriguez O; Atares B; Llorens V; Gomez Tortosa E; del Ser T; Munoz DG; de Yebenes JG The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol 2004, 55 (2), 164–173. DOI: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- (3).Ugalde CL; Lawson VA; Finkelstein DI; Hill AF The role of lipids in alpha-synuclein misfolding and neurotoxicity. J Biol Chem 2019, 294 (23), 9016–9028. DOI: 10.1074/jbc.REV119.007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Shahnawaz M; Mukherjee A; Pritzkow S; Mendez N; Rabadia P; Liu X; Hu B; Schmeichel A; Singer W; Wu G; Tsai AL; Shirani H; Nilsson KPR; Low PA; Soto C Discriminating alpha-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 2020, 578 (7794), 273–277. DOI: 10.1038/s41586-020-1984-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yasuda T; Nakata Y; Mochizuki H alpha-Synuclein and neuronal cell death. Mol Neurobiol 2013, 47 (2), 466–483. DOI: 10.1007/s12035-012-8327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Cattani J; Subramaniam V; Drescher M Room-temperature in-cell EPR spectroscopy: alpha-Synuclein disease variants remain intrinsically disordered in the cell. Phys Chem Chem Phys 2017, 19 (28), 18147–18151. DOI: 10.1039/c7cp03432f. [DOI] [PubMed] [Google Scholar]
- (7).Sun Y; Kakinen A; Wan X; Moriarty N; Hunt CPJ; Li Y; Andrikopoulos N; Nandakumar A; Davis TP; Parish CL; Song Y; Ke PC; Ding F Spontaneous Formation of beta-sheet Nano-barrels during the Early Aggregation of Alzheimer’s Amyloid Beta. Nano Today 2021, 38. DOI: 10.1016/j.nantod.2021.101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sun Y; Kakinen A; Xing Y; Pilkington EH; Davis TP; Ke PC; Ding F Nucleation of beta-rich oligomers and beta-barrels in the early aggregation of human islet amyloid polypeptide. Biochim Biophys Acta Mol Basis Dis 2019, 1865 (2), 434–444. DOI: 10.1016/j.bbadis.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Faridi A; Sun Y; Okazaki Y; Peng G; Gao J; Kakinen A; Faridi P; Zhao M; Javed I; Purcell AW; Davis TP; Lin S; Oda R; Ding F; Ke PC Mitigating Human IAPP Amyloidogenesis In Vivo with Chiral Silica Nanoribbons. Small 2018, 14 (47), e1802825. DOI: 10.1002/smll.201802825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Biswas S; Bhadra A; Lakhera S; Soni M; Panuganti V; Jain S; Roy I Molecular crowding accelerates aggregation of alpha-synuclein by altering its folding pathway. Eur Biophys J 2021, 50 (1), 59–67. DOI: 10.1007/s00249-020-01486-1. [DOI] [PubMed] [Google Scholar]
- (11).Kumari P; Ghosh D; Vanas A; Fleischmann Y; Wiegand T; Jeschke G; Riek R; Eichmann C Structural insights into alpha-synuclein monomer-fibril interactions. Proc Natl Acad Sci U S A 2021, 118 (10). DOI: 10.1073/pnas.2012171118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Gaspar R; Meisl G; Buell AK; Young L; Kaminski CF; Knowles TPJ; Sparr E; Linse S Secondary nucleation of monomers on fibril surface dominates alpha-synuclein aggregation and provides autocatalytic amyloid amplification. Q Rev Biophys 2017, 50, e6. DOI: 10.1017/S0033583516000172. [DOI] [PubMed] [Google Scholar]
- (13).Uversky VN; Li J; Fink AL Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem 2001, 276 (14), 10737–10744. DOI: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- (14).Munishkina LA; Cooper EM; Uversky VN; Fink AL The effect of macromolecular crowding on protein aggregation and amyloid fibril formation. J Mol Recognit 2004, 17 (5), 456–464. DOI: 10.1002/jmr.699. [DOI] [PubMed] [Google Scholar]
- (15).Buell AK; Galvagnion C; Gaspar R; Sparr E; Vendruscolo M; Knowles TP; Linse S; Dobson CM Solution conditions determine the relative importance of nucleation and growth processes in alpha-synuclein aggregation. Proc Natl Acad Sci U S A 2014, 111 (21), 7671–7676. DOI: 10.1073/pnas.1315346111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Li Y; Zhao C; Luo F; Liu Z; Gui X; Luo Z; Zhang X; Li D; Liu C; Li X Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy. Cell Res 2018, 28 (9), 897–903. DOI: 10.1038/s41422-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hwang S; Fricke P; Zinke M; Giller K; Wall JS; Riedel D; Becker S; Lange A Comparison of the 3D structures of mouse and human alpha-synuclein fibrils by solid-state NMR and STEM. J Struct Biol 2019, 206 (1), 43–48. DOI: 10.1016/j.jsb.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lv G; Kumar A; Huang Y; Eliezer D A Protofilament-Protofilament Interface in the Structure of Mouse alpha-Synuclein Fibrils. Biophys J 2018, 114 (12), 2811–2819. DOI: 10.1016/j.bpj.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rodriguez JA; Ivanova MI; Sawaya MR; Cascio D; Reyes FE; Shi D; Sangwan S; Guenther EL; Johnson LM; Zhang M; Jiang L; Arbing MA; Nannenga BL; Hattne J; Whitelegge J; Brewster AS; Messerschmidt M; Boutet S; Sauter NK; Gonen T; Eisenberg DS Structure of the toxic core of alpha-synuclein from invisible crystals. Nature 2015, 525 (7570), 486–490. DOI: 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Vilar M; Chou HT; Luhrs T; Maji SK; Riek-Loher D; Verel R; Manning G; Stahlberg H; Riek R The fold of alpha-synuclein fibrils. Proc Natl Acad Sci U S A 2008, 105 (25), 8637–8642. DOI: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kayed R; Dettmer U; Lesne SE Soluble endogenous oligomeric alpha-synuclein species in neurodegenerative diseases: Expression, spreading, and cross-talk. J Parkinsons Dis 2020, 10 (3), 791–818. DOI: 10.3233/JPD-201965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Cascella R; Chen SW; Bigi A; Camino JD; Xu CK; Dobson CM; Chiti F; Cremades N; Cecchi C The release of toxic oligomers from alpha-synuclein fibrils induces dysfunction in neuronal cells. Nat Commun 2021, 12 (1), 1814. DOI: 10.1038/s41467-021-21937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Nguyen PH; Ramamoorthy A; Sahoo BR; Zheng J; Faller P; Straub JE; Dominguez L; Shea JE; Dokholyan NV; De Simone A; Ma B; Nussinov R; Najafi S; Ngo ST; Loquet A; Chiricotto M; Ganguly P; McCarty J; Li MS; Hall C; Wang Y; Miller Y; Melchionna S; Habenstein B; Timr S; Chen J; Hnath B; Strodel B; Kayed R; Lesne S; Wei G; Sterpone F; Doig AJ; Derreumaux P Amyloid Oligomers: A Joint Experimental/Computational Perspective on Alzheimer’s Disease, Parkinson’s Disease, Type II Diabetes, and Amyotrophic Lateral Sclerosis. Chem Rev 2021, 121 (4), 2545–2647. DOI: 10.1021/acs.chemrev.0c01122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Guerrero-Ferreira R; Kovacik L; Ni D; Stahlberg H New insights on the structure of alpha-synuclein fibrils using cryo-electron microscopy. Curr Opin Neurobiol 2020, 61, 89–95. DOI: 10.1016/j.conb.2020.01.014. [DOI] [PubMed] [Google Scholar]
- (25).Evans SR; West C; Klein-Seetharaman J Similarity of the non-amyloid-beta component and C-terminal tail of monomeric and tetrameric alpha-synuclein with 14-3-3 sigma. Comput Struct Biotechnol J 2021, 19, 5348–5359. DOI: 10.1016/j.csbj.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Emamzadeh FN Alpha-synuclein structure, functions, and interactions. J Res Med Sci 2016, 21, 29. DOI: 10.4103/1735-1995.181989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Giasson BI; Murray IV; Trojanowski JQ; Lee VM A hydrophobic stretch of 12 amino acid residues in the middle of alpha-synuclein is essential for filament assembly. J Biol Chem 2001, 276 (4), 2380–2386. DOI: 10.1074/jbc.M008919200. [DOI] [PubMed] [Google Scholar]
- (28).Sun Y; Kakinen A; Zhang C; Yang Y; Faridi A; Davis TP; Cao W; Ke PC; Ding F Amphiphilic surface chemistry of fullerenols is necessary for inhibiting the amyloid aggregation of alpha-synuclein NACore. Nanoscale 2019, 11 (24), 11933–11945. DOI: 10.1039/c9nr02407g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lan-Mark S; Miller Y Insights into the Interactions that Trigger the Primary Nucleation of Polymorphic alpha-Synuclein Dimers. ACS Chem Neurosci 2022, 13 (3), 370–378. DOI: 10.1021/acschemneuro.1c00754. [DOI] [PubMed] [Google Scholar]
- (30).Carboni E; Lingor P Insights on the interaction of alpha-synuclein and metals in the pathophysiology of Parkinson’s disease. Metallomics 2015, 7 (3), 395–404. DOI: 10.1039/c4mt00339j. [DOI] [PubMed] [Google Scholar]
- (31).Theillet FX; Binolfi A; Bekei B; Martorana A; Rose HM; Stuiver M; Verzini S; Lorenz D; van Rossum M; Goldfarb D; Selenko P Structural disorder of monomeric alpha-synuclein persists in mammalian cells. Nature 2016, 530 (7588), 45–50. DOI: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- (32).Esteban-Martin S; Silvestre-Ryan J; Bertoncini CW; Salvatella X Identification of fibril-like tertiary contacts in soluble monomeric alpha-synuclein. Biophys J 2013, 105 (5), 1192–1198. DOI: 10.1016/j.bpj.2013.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Bartels T; Choi JG; Selkoe DJ alpha-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 2011, 477 (7362), 107–110. DOI: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bernstein SL; Liu D; Wyttenbach T; Bowers MT; Lee JC; Gray HB; Winkler JR Alpha-synuclein: stable compact and extended monomeric structures and pH dependence of dimer formation. J Am Soc Mass Spectrom 2004, 15 (10), 1435–1443. DOI: 10.1016/j.jasms.2004.08.003. [DOI] [PubMed] [Google Scholar]
- (35).Roostaee A; Beaudoin S; Staskevicius A; Roucou X Aggregation and neurotoxicity of recombinant alpha-synuclein aggregates initiated by dimerization. Mol Neurodegener 2013, 8, 5. DOI: 10.1186/1750-1326-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Urbanc B; Borreguero JM; Cruz L; Stanley HE Ab initio discrete molecular dynamics approach to protein folding and aggregation. Methods Enzymol 2006, 412, 314–338. DOI: 10.1016/S0076-6879(06)12019-4. [DOI] [PubMed] [Google Scholar]
- (37).Ding F; Tsao D; Nie H; Dokholyan NV Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure 2008, 16 (7), 1010–1018. DOI: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Proctor EA; Dokholyan NV Applications of Discrete Molecular Dynamics in biology and medicine. Curr Opin Struct Biol 2016, 37, 9–13. DOI: 10.1016/j.sbi.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Zhao M; Cascio D; Sawaya MR; Eisenberg D Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation. Protein Sci 2011, 20 (6), 996–1004. DOI: 10.1002/pro.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Eliezer D; Kutluay E; Bussell R Jr.; Browne G Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol 2001, 307 (4), 1061–1073. DOI: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- (41).Sahay S; Ghosh D; Dwivedi S; Anoop A; Mohite GM; Kombrabail M; Krishnamoorthy G; Maji SK Familial Parkinson disease-associated mutations alter the site-specific microenvironment and dynamics of alpha-synuclein. J Biol Chem 2015, 290 (12), 7804–7822. DOI: 10.1074/jbc.M114.598607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Mirecka EA; Shaykhalishahi H; Gauhar A; Akgul S; Lecher J; Willbold D; Stoldt M; Hoyer W Sequestration of a beta-hairpin for control of alpha-synuclein aggregation. Angew Chem Int Ed Engl 2014, 53 (16), 4227–4230. DOI: 10.1002/anie.201309001. [DOI] [PubMed] [Google Scholar]
- (43).Antonschmidt L; Dervisoglu R; Sant V; Tekwani Movellan K; Mey I; Riedel D; Steinem C; Becker S; Andreas LB; Griesinger C Insights into the molecular mechanism of amyloid filament formation: Segmental folding of alpha-synuclein on lipid membranes. Sci Adv 2021, 7 (20). DOI: 10.1126/sciadv.abg2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Boyer DR; Li B; Sun C; Fan W; Zhou K; Hughes MP; Sawaya MR; Jiang L; Eisenberg DS The alpha-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc Natl Acad Sci U S A 2020, 117 (7), 3592–3602. DOI: 10.1073/pnas.1917914117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Guerrero-Ferreira R; Taylor NM; Mona D; Ringler P; Lauer ME; Riek R; Britschgi M; Stahlberg H Cryo-EM structure of alpha-synuclein fibrils. Elife 2018, 7. DOI: 10.7554/eLife.36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Li B; Ge P; Murray KA; Sheth P; Zhang M; Nair G; Sawaya MR; Shin WS; Boyer DR; Ye S; Eisenberg DS; Zhou ZH; Jiang L Cryo-EM of full-length alpha-synuclein reveals fibril polymorphs with a common structural kernel. Nat Commun 2018, 9 (1), 3609. DOI: 10.1038/s41467-018-05971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Tuttle MD; Comellas G; Nieuwkoop AJ; Covell DJ; Berthold DA; Kloepper KD; Courtney JM; Kim JK; Barclay AM; Kendall A; Wan W; Stubbs G; Schwieters CD; Lee VM; George JM; Rienstra CM Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein. Nat Struct Mol Biol 2016, 23 (5), 409–415. DOI: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gath J; Habenstein B; Bousset L; Melki R; Meier BH; Bockmann A Solid-state NMR sequential assignments of alpha-synuclein. Biomol NMR Assign 2012, 6 (1), 51–55. DOI: 10.1007/s12104-011-9324-3. [DOI] [PubMed] [Google Scholar]
- (49).Comellas G; Lemkau LR; Nieuwkoop AJ; Kloepper KD; Ladror DT; Ebisu R; Woods WS; Lipton AS; George JM; Rienstra CM Structured regions of alpha-synuclein fibrils include the early-onset Parkinson’s disease mutation sites. J Mol Biol 2011, 411 (4), 881–895. DOI: 10.1016/j.jmb.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chen M; Margittai M; Chen J; Langen R Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J Biol Chem 2007, 282 (34), 24970–24979. DOI: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- (51).Heise H; Hoyer W; Becker S; Andronesi OC; Riedel D; Baldus M Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci U S A 2005, 102 (44), 15871–15876. DOI: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Miake H; Mizusawa H; Iwatsubo T; Hasegawa M Biochemical characterization of the core structure of alpha-synuclein filaments. J Biol Chem 2002, 277 (21), 19213–19219. DOI: 10.1074/jbc.M110551200. [DOI] [PubMed] [Google Scholar]
- (53).Der-Sarkissian A; Jao CC; Chen J; Langen R Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J Biol Chem 2003, 278 (39), 37530–37535. DOI: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- (54).Ulmer TS; Bax A; Cole NB; Nussbaum RL Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem 2005, 280 (10), 9595–9603. DOI: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- (55).Bhattacharya S; Xu L; Thompson D Molecular Simulations Reveal Terminal Group Mediated Stabilization of Helical Conformers in Both Amyloid-beta42 and alpha-Synuclein. ACS Chem Neurosci 2019, 10 (6), 2830–2842. DOI: 10.1021/acschemneuro.9b00053. [DOI] [PubMed] [Google Scholar]
- (56).Chen J; Zaer S; Drori P; Zamel J; Joron K; Kalisman N; Lerner E; Dokholyan NV The structural heterogeneity of alpha-synuclein is governed by several distinct subpopulations with interconversion times slower than milliseconds. Structure 2021, 29 (9), 1048–1064 e1046. DOI: 10.1016/j.str.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Brodie NI; Popov KI; Petrotchenko EV; Dokholyan NV; Borchers CH Conformational ensemble of native alpha-synuclein in solution as determined by short-distance crosslinking constraint-guided discrete molecular dynamics simulations. PLoS Comput Biol 2019, 15 (3), e1006859. DOI: 10.1371/journal.pcbi.1006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Mehra S; Sahay S; Maji SK alpha-Synuclein misfolding and aggregation: Implications in Parkinson’s disease pathogenesis. Biochim Biophys Acta Proteins Proteom 2019, 1867 (10), 890–908. DOI: 10.1016/j.bbapap.2019.03.001. [DOI] [PubMed] [Google Scholar]
- (59).Fusco G; De Simone A; Gopinath T; Vostrikov V; Vendruscolo M; Dobson CM; Veglia G Direct observation of the three regions in alpha-synuclein that determine its membrane-bound behaviour. Nat Commun 2014, 5, 3827. DOI: 10.1038/ncomms4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Appel-Cresswell S; Vilarino-Guell C; Encarnacion M; Sherman H; Yu I; Shah B; Weir D; Thompson C; Szu-Tu C; Trinh J; Aasly JO; Rajput A; Rajput AH; Jon Stoessl A; Farrer MJ Alpha-synuclein p.H50Q, a novel pathogenic mutation for Parkinson’s disease. Mov Disord 2013, 28 (6), 811–813. DOI: 10.1002/mds.25421. [DOI] [PubMed] [Google Scholar]
- (61).Kiely AP; Asi YT; Kara E; Limousin P; Ling H; Lewis P; Proukakis C; Quinn N; Lees AJ; Hardy J; Revesz T; Houlden H; Holton JL alpha-Synucleinopathy associated with G51D SNCA mutation: a link between Parkinson’s disease and multiple system atrophy? Acta Neuropathol 2013, 125 (5), 753–769. DOI: 10.1007/s00401-013-1096-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Polymeropoulos MH; Lavedan C; Leroy E; Ide SE; Dehejia A; Dutra A; Pike B; Root H; Rubenstein J; Boyer R; Stenroos ES; Chandrasekharappa S; Athanassiadou A; Papapetropoulos T; Johnson WG; Lazzarini AM; Duvoisin RC; Di Iorio G; Golbe LI; Nussbaum RL Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276 (5321), 2045–2047. DOI: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- (63).Pasanen P; Myllykangas L; Siitonen M; Raunio A; Kaakkola S; Lyytinen J; Tienari PJ; Poyhonen M; Paetau A Novel alpha-synuclein mutation A53E associated with atypical multiple system atrophy and Parkinson’s disease-type pathology. Neurobiol Aging 2014, 35 (9), 2180 e2181–2185. DOI: 10.1016/j.neurobiolaging.2014.03.024. [DOI] [PubMed] [Google Scholar]
- (64).Xu L; Nussinov R; Ma B Coupling of the non-amyloid-component (NAC) domain and the KTK(E/Q)GV repeats stabilize the alpha-synuclein fibrils. Eur J Med Chem 2016, 121, 841–850. DOI: 10.1016/j.ejmech.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Bhattacharyya D; Kumar R; Mehra S; Ghosh A; Maji SK; Bhunia A Multitude NMR studies of alpha-synuclein familial mutants: probing their differential aggregation propensities. Chem Commun (Camb) 2018, 54 (29), 3605–3608. DOI: 10.1039/C7CC09597J. [DOI] [PubMed] [Google Scholar]
- (66).Paslawski W; Mysling S; Thomsen K; Jorgensen TJ; Otzen DE Co-existence of two different alpha-synuclein oligomers with different core structures determined by hydrogen/deuterium exchange mass spectrometry. Angew Chem Int Ed Engl 2014, 53 (29), 7560–7563. DOI: 10.1002/anie.201400491. [DOI] [PubMed] [Google Scholar]
- (67).Holec SAM; Liu SL; Woerman AL Consequences of variability in alpha-synuclein fibril structure on strain biology. Acta Neuropathol 2022, 143 (3), 311–330. DOI: 10.1007/s00401-022-02403-w. [DOI] [PubMed] [Google Scholar]
- (68).McClendon S; Rospigliosi CC; Eliezer D Charge neutralization and collapse of the C-terminal tail of alpha-synuclein at low pH. Protein Sci 2009, 18 (7), 1531–1540. DOI: 10.1002/pro.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Huang S; Mo X; Wang J; Ye X; Yu H; Liu Y alpha-Synuclein phase separation and amyloid aggregation are modulated by C-terminal truncations. FEBS Lett 2022, 596 (11), 1388–1400. DOI: 10.1002/1873-3468.14361. [DOI] [PubMed] [Google Scholar]
- (70).Stephens AD; Zacharopoulou M; Moons R; Fusco G; Seetaloo N; Chiki A; Woodhams PJ; Mela I; Lashuel HA; Phillips JJ; De Simone A; Sobott F; Schierle GSK Extent of N-terminus exposure of monomeric alpha-synuclein determines its aggregation propensity. Nat Commun 2020, 11 (1), 2820. DOI: 10.1038/s41467-020-16564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Peng S; Ding F; Urbanc B; Buldyrev SV; Cruz L; Stanley HE; Dokholyan NV Discrete molecular dynamics simulations of peptide aggregation. Phys Rev E Stat Nonlin Soft Matter Phys 2004, 69 (4 Pt 1), 041908. DOI: 10.1103/PhysRevE.69.041908. [DOI] [PubMed] [Google Scholar]
- (72).Yin S; Ding F; Dokholyan NV Eris: an automated estimator of protein stability. Nat Methods 2007, 4 (6), 466–467. DOI: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- (73).Yin S; Biedermannova L; Vondrasek J; Dokholyan NV MedusaScore: an accurate force field-based scoring function for virtual drug screening. J Chem Inf Model 2008, 48 (8), 1656–1662. DOI: 10.1021/ci8001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Sun Y; Ding F alphaB-Crystallin Chaperone Inhibits Abeta Aggregation by Capping the beta-Sheet-Rich Oligomers and Fibrils. J Phys Chem B 2020, 124 (45), 10138–10146. DOI: 10.1021/acs.jpcb.0c07256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Lazaridis T; Karplus M Effective energy functions for protein structure prediction. Curr Opin Struct Biol 2000, 10 (2), 139–145. DOI: 10.1016/s0959-440x(00)00063-4. [DOI] [PubMed] [Google Scholar]
- (76).Sun Y; Ding F Thermo- and pH-responsive fibrillization of squid suckerin A1H1 peptide. Nanoscale 2020, 12 (11), 6307–6317. DOI: 10.1039/c9nr09271d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Zhang Y; Liu Y; Zhao W; Sun Y Hydroxylated single-walled carbon nanotube inhibits beta2m21–31 fibrillization and disrupts pre-formed proto-fibrils. Int J Biol Macromol 2021, 193 (Pt A), 1–7. DOI: 10.1016/j.ijbiomac.2021.10.103. [DOI] [PubMed] [Google Scholar]
- (78).Tang H; Sun Y; Ding F Hydrophobic/Hydrophilic Ratio of Amphiphilic Helix Mimetics Determines the Effects on Islet Amyloid Polypeptide Aggregation. J Chem Inf Model 2022, 62 (7), 1760–1770. DOI: 10.1021/acs.jcim.1c01566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Emperador A; Orozco M Discrete Molecular Dynamics Approach to the Study of Disordered and Aggregating Proteins. J Chem Theory Comput 2017, 13 (3), 1454–1461. DOI: 10.1021/acs.jctc.6b01153. [DOI] [PubMed] [Google Scholar]
- (80).Serpa JJ; Popov KI; Petrotchenko EV; Dokholyan NV; Borchers CH Structure of prion beta-oligomers as determined by short-distance crosslinking constraint-guided discrete molecular dynamics simulations. Proteomics 2021, 21 (21–22), e2000298. DOI: 10.1002/pmic.202000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Bunce SJ; Wang Y; Stewart KL; Ashcroft AE; Radford SE; Hall CK; Wilson AJ Molecular insights into the surface-catalyzed secondary nucleation of amyloid-beta40 (Abeta40) by the peptide fragment Abeta16–22. Sci Adv 2019, 5 (6), eaav8216. DOI: 10.1126/sciadv.aav8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Kabsch W; Sander C Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22 (12), 2577–2637. DOI: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- (83).Daura X; Gademann K; Jaun B; Seebach D; van Gunsteren WF; Mark AE Peptide folding: When simulation meets experiment. Angew Chem Int Edit 1999, 38 (1–2), 236–240. DOI: Doi . [DOI] [Google Scholar]
- (84).Koukos PI; Glykos NM Grcarma: A fully automated task-oriented interface for the analysis of molecular dynamics trajectories. J Comput Chem 2013, 34 (26), 2310–2312. DOI: 10.1002/jcc.23381. [DOI] [PubMed] [Google Scholar]
- (85).Glykos NM Software news and updates. Carma: a molecular dynamics analysis program. J Comput Chem 2006, 27 (14), 1765–1768. DOI: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


