Abstract
G protein-coupled receptors (GPCRs) are the largest class of cell surface receptors in the genome and the most successful family of targets of FDA-approved drugs. New frontiers in GPCR drug discovery remain, however, as achieving receptor subtype selectivity and controlling off- and on-target side effects are not always possible with classic agonist and antagonist ligands. These challenges may be overcome by focusing development efforts on allosteric ligands that confer signaling bias. Biased allosteric modulators (BAMs) are an emerging class of GPCR ligands that engage less well-conserved regulatory motifs outside the orthosteric pocket and exert pathway-specific effects on receptor signaling. The unique ways that BAMs texturize receptor signaling present opportunities to fine-tune physiology and develop safer, more selective therapeutics. Here, we provide a conceptual framework for understanding the pharmacology of BAMs, explore their therapeutic potential and discuss strategies for their discovery.
Keywords: G protein-coupled receptor (GPCR), allosteric modulation, signaling bias, functional selectivity, β-arrestin, drug development
Capitalizing on bias and allosterism in the development of safer GPCR-targeted therapeutics
G protein-coupled receptors (GPCRs, see Glossary) are the largest superfamily of transmembrane proteins in the human genome, with over 800 members [1]. These receptors play essential roles in a wide array of fundamental physiological processes [2]. Historically, GPCRs have been the most successful class of drug targets [3]. Over 700 drugs, ~35% of all drugs approved by the Food and Drug Administration (FDA), target 134 unique GPCRs [3]. GPCRs also remain one of the most promising targets for future drug discovery [4, 5]. Unfortunately, over the last decade, the probability of a new drug reaching product launch has remained low. Greater than 90% of agents entering phase I trials fail to achieve FDA approval, predominantly because of lack of efficacy or safety concerns [6]. This and the economics of drug development - time to market averaging 7–12 years at a cost of $1.3 billion USD per drug [7] - have created an environment where pharmacologists are rethinking the properties that define a good developmental candidate.
GPCRs are pleiotropically coupled transducers that activate different families of closely related effector proteins, including G proteins (e.g., Gs, Gi/o, Gq, G12) and β-arrestins (e.g., β-arrestin1, β-arrestin2), to mediate distinct cellular and physiological effects. Biased signaling, ligand-directed signaling, and functional selectivity are equivalent terms that, applied to GPCRs, describe preferential receptor coupling to a subset of potential effectors, resulting in the activation of a subset of the signaling pathways in a receptor’s full repertoire. Studies of biased agonism form a comparatively new area of GPCR investigation that are transforming how GPCR signaling is conceptualized. It is now appreciated that biased ligands are discoverable through complementary functional assays designed to pharmacologically distinguish between different receptor signaling modes, and that the bias properties of lead compounds can be improved through rational drug design and optimization. Beyond their utility in basic research, the practical value of biased ligands is their potential to selectively stimulate therapeutically relevant signaling and avoid on-target side effects.
Balanced (i.e., unbiased) agonists or antagonists, activate or inhibit, respectively, receptor signaling pathways uniformly through the binding of the same receptor site engaged by the endogenous ligand (i.e., the orthosteric site). There are inherent disadvantages to this mechanism, with uniform activation of signaling pathways and disruption of endogenous ligand rhythms leading to the potential for on-target side effects and highly conserved orthosteric determinants making it difficult to achieve receptor subtype selectivity, leading to off-target side effects (Figure 1, Key Figure). Classical allosterism predicted 50 years ago that there are regulatory sites outside of the orthosteric pocket that should be able to regulate receptor signaling by controlling receptor conformation. An idea first applied to hemoglobin, allosterism also found favor in explaining how the conformational space of GPCRs can be constrained to affect signaling activity. Application of allosteric principles to GPCRs has led to an appreciation that motifs outside of the orthosteric binding pocket can affect orthosteric ligand binding and signaling efficacy. The identification of allosteric ligands that exert pathway-specific effects has given rise to new classes of biased allosteric modulators (BAMs) with intriguing properties.
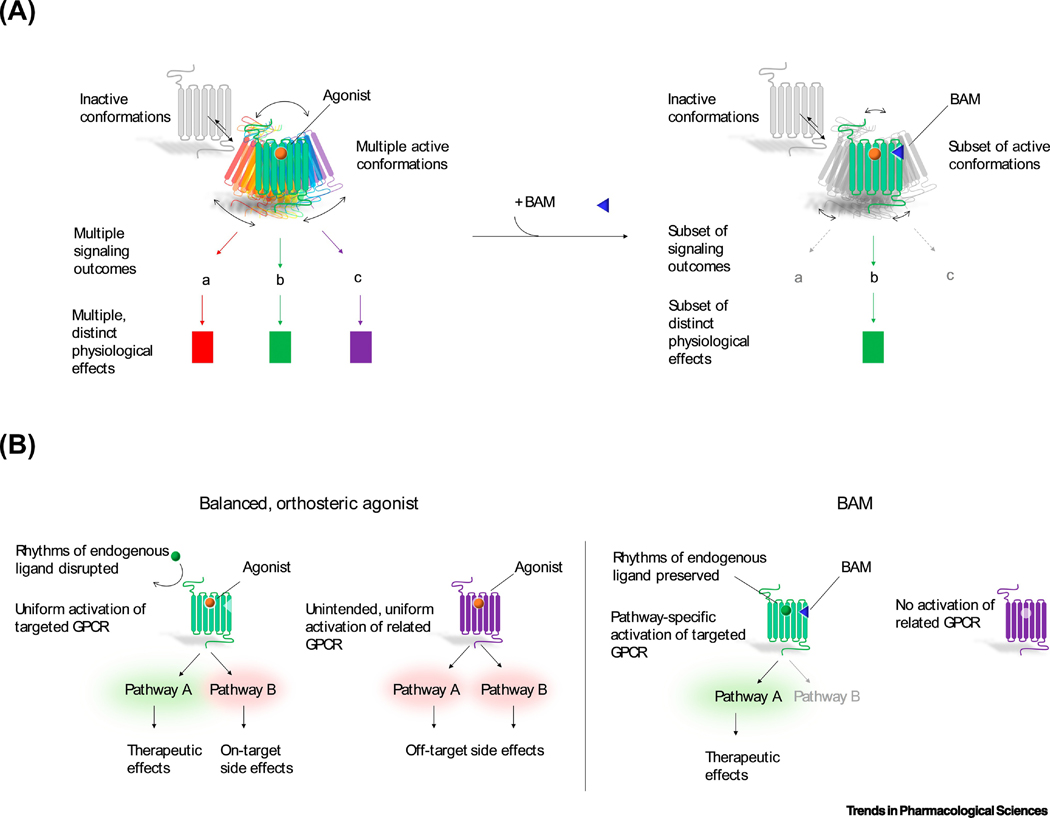
Figure 1, Key Figure. The molecular basis of biased allosteric modulation and its potential therapeutic advantages.
(A) Depiction of the molecular basis of biased allosteric modulation, based on the empirically supported conformational hypothesis of biased signaling. When a GPCR is activated by a balanced, orthosteric agonist (left), the receptor samples multiple active conformations, which permit receptor association with distinct effector proteins, including Gα family proteins (e.g., Gαs, Gαi/o, Gαq), G protein-receptor kinases and β-arrestins. These effectors give rise to multiple downstream signaling pathways (e.g., changes in cAMP levels, Ca2+ mobilization, ERK activation), which, in turn, lead to distinct cellular and physiological outcomes. Biased allosteric modulators (BAMs, right) confer more restricted pharmacological action. In the presence of a BAM, a GPCR samples only a subset of potential active conformations, leading to recruitment of only a subset of potential effectors. Consequently, signaling proceeds through a subset of potential pathways, producing a more limited repertoire of cellular and physiological effects. (B) Representation of the potential therapeutic advantages of BAMs. Balanced, orthosteric agonists (left) disrupt rhythms of endogenous ligands by blocking endogenous ligand binding and produce both on- and off-target side effects. By contrast, BAMs (right) do not prevent endogenous ligand binding, gain receptor subtype selectivity by binding to less well-conserved allosteric motifs and gain pathway selectivity by activating only a subset of signaling pathways.
BAMs provide unprecedented spatial, temporal, and signal pathway specificity, offering new strategies for designing better, more selective GPCR-targeted therapeutics (Figure 1). Resolving the mechanisms of action of BAMs and leveraging these mechanisms to obtain a therapeutic benefit, however, requires a conceptual framework for understanding their pharmacology and practical strategies for their discovery. In this review, we synthesize late-breaking reports to answer the questions: What are BAMs? Why seek them as therapeutics? And, how can drug discovery campaigns for biased and allosteric ligands be designed to maximize the probability of success? In addressing these questions, we provide an analytical framework for understanding biased allosteric modulation, highlight a growing body of work suggesting how it may be therapeutically beneficial, and discuss the use of cell-based functional screens to identify biased and allosteric GPCR ligands. As their potential value in creating safer therapies is realized, BAMs may well sustain a new era of GPCR drug discovery.
What are biased allosteric modulators?
BAMs are GPCR regulators that engage allosteric sites and change signaling outcomes in a non-uniform manner. Decoding their behavior at the receptor, cell, and system levels requires an understanding of both biased GPCR signaling and GPCR allosteric modulation.
Biased GPCR signaling
The ability of GPCRs to relay signals across the cell membrane depends on conformational rearrangements of their transmembrane helices, extracellular loops, and intracellular loops [8]. While signal transduction was initially attributed only to G proteins, with β-arrestins recognized for mediating desensitization, it is now understood that β-arrestins also act as independent GPCR signaling conduits [8]. β-arrestins serve as adaptors to scaffold and mediate signaling via well-recognized signaling proteins, including mitogen-activated protein kinases (MAPKs), serine/threonine kinase AKT, phosphoinositide 3-kinase (PI3K), the tyrosine kinase SRC, and nuclear factor-κB (NF-κB) [8].
Through differential effector coupling, GPCRs can signal through a large selection of pathways. Some GPCR agonists can activate a single signaling pathway or a subset of signaling pathways more effectively than they activate others, a phenomenon known as biased agonism. Different types of biased ligands have been identified, including those that preferentially promote receptor activation of G proteins over β-arrestins (i.e., G protein-biased agonists), β-arrestins over G proteins (i.e., β-arrestin-biased agonists), and specific G protein subtypes over others (e.g., Gi over Gs) [9, 10]. The degree of signaling bias can be quantified in relation to a reference ligand [11]. The physical basis of biased agonism may be the ability of the biased compound to stabilize an alternative receptor conformation while binding the orthosteric pocket in a distinctive manner [12, 13]. Because it supports the existence of multiple active receptor conformations, biased agonism is not consistent with a two-state model of receptor signaling [8, 14]. It is, however, proof-of-concept for pathway-specific efficacy.
A case for extending the search for biased ligands beyond the orthosteric pocket
Pathway-selective drugs will be especially useful for GPCRs for which one signaling arm controls therapeutic endpoints and the other side-effects [15]. Biased ligands are desirable drug candidates because of their pathway specificity, but it remains unclear how to identify which GPCRs are better discovery candidates and, for a GPCR of interest, how to achieve functional selectivity. Modern information theory formulated by Claude Shannon in 1940s, may provide a solution [16, 17]. Shannon divided communication into stages which include message encoding, transmission, and decoding. In analogy to thermodynamic entropy, he defined a concept of a bit entropy H to describe a property of messages that logarithmically increases with their uncertainty (i.e., number of interpretations). We can apply this idea to GPCR signaling. Like its thermodynamic counterpart, GPCR system information entropy H increases as the potential enabled outcomes increase. Receptor systems transducing messages that must avoid corruption should have low bit entropies, implying significant non-zero probabilities for only a limited number of conformations stabilized by agonist occupancy.
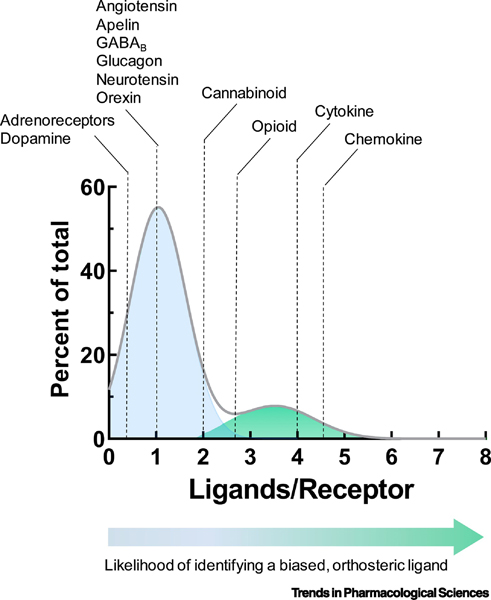
To estimate the degree of information inherent in non-odorant GPCR signaling (Figure 2 and Table 1) we determined the average number of messages (m), in our case ligands per receptor, for GPCR families with the hypothesis H = log2(1+m). Thus, the number of states of the system is 2H, with m=0 corresponding to a one-state system, m=1 to a two-state system, m=3 to a four-state system, and so on. The results can be fit to a sum of gaussians, which indicates that the receptor families fall into two groups (Figure 2), with the chemokine receptors forming the majority of the second peak of high-entropy receptors. The first group of GPCRs has a mean bit entropy of ~1, corresponding to a two-state system. Thus, these GPCRs are predicted to encode/decode only one outside message and most likely are examples of a two-state full-on/full-off model. They include, among others, adrenergic, dopaminergic, neurotensin, angiotensin and GABAB receptor families. For receptor families comprising this first peak, ligands binding only orthosteric determinants and conferring signaling bias would most probably be difficult to find. In this situation, engaging sites outside the orthosteric pocket may be required to produce a non-binary outcome.
Figure 2. Analysis of endogenous receptor-ligand interactions suggests the need to look beyond the orthosteric pocket to develop biased GPCR ligands.
An analysis of non-odorant, rhodopsin family GPCRs was completed to determine inherent informational entropy. For each receptor family, the number of endogenous ligands was divided by the number of receptor variants to yield a “Ligands/Receptor” value indicative of informational entropy. The percent of total receptor families with a given Ligands/Receptor value is shown. Results were fit to a sum of gaussians, indicating that receptors analyzed fall into two main categories with different mean entropies. Mean ± SD for peak 1 and peak 2 are 0.97 ± 0.55 and 3.93 ± 1.6 ligands/receptor, respectively.
Table 1.
Non-odorant human GPCRs and endogenous orthosteric ligands
| Receptor Family1 | Receptor Variants2 | # of Receptor Variant: | Endogenous Ligands2 | # of Ligands | Ligands per Receptor |
|---|---|---|---|---|---|
| 5-HT | 5-Hydroxytryptamine | 13 | 5-HT | 1 | 0.1 |
| Muscarinic | M1–5 | 5 | Acetylcholine | 1 | 0.2 |
| Dopamine | D1–5 | 5 | Dopamine | 1 | 0.2 |
| Lysophosphatidic Acid | LPA1–5 | 5 | LPA | 1 | 0.2 |
| Adrenoreceptors | β1–3/α1A,B,D/a2A–C | 9 | Adrenaline/noradrenaline | 2 | 0.2 |
| Histamine | H1–4 | 4 | Histamine | 1 | 0.3 |
| Proteinase activated | PAR1–4 | 4 | Thrombin | 1 | 0.3 |
| Nicotinic acid receptors | GPR109A,B / GPR81 | 3 | Niacin | 1 | 0.3 |
| Sphingosine 1 - phosphatate | S1P1–5 | 5 | S1P/SPC | 2 | 0.4 |
| Adenosine | Adenosine | 4 | Adenosine/inosine | 2 | 0.5 |
| Melanin Concentrating Hormone | MCH1–2 | 2 | MCH | 1 | 0.5 |
| Neuromedin U | NMU1–2 | 2 | Neuromedin U | 1 | 0.5 |
| Thyrotropin releasing hormone | TRH1–2 | 2 | TRH | 1 | 0.5 |
| Vasopressin and Oxytocin | V1a,b/V2/OT | 4 | Vasopressin/oxytocin | 2 | 0.5 |
| Somatostatin | Sst1–5 | 5 | Somatostatin-14/28/cortistatin | 3 | 0.6 |
| Glutamate, metabotropic | mGluR1–8 | 8 | Glutamate/asparate/LSOP/NNAG/CSA | 5 | 0.6 |
| P2Y | P2Y1/2/4/6/11-14 | 8 | ATP/ADP/UTP/UDP/UDP-glucose | 5 | 0.6 |
| Galanin | GAL1–3 | 3 | Galanin/galp | 2 | 0.7 |
| Prostanoid | DP1–2/EP1–4/// | 9 | Prostaglandin/prostacyclin/thromboxane/// | 6 | 0.7 |
| Melanocortin | MC1–5 | 5 | MSHa/b/g/ACTH | 4 | 0.8 |
| Neuropeptide Y | Y1/2/4–6 | 5 | NPY/YY/PYY/PP | 4 | 0.8 |
| Anaphylatoxin | C5a | 1 | C5a/C3a | 2 | 1.0 |
| Angiotensin | AT1–2 | 2 | Angiotensin II/III | 2 | 1.0 |
| Apelin | Apelin | 1 | Apelin variants | 1 | 1.0 |
| Bombesin | BB1–3 | 3 | GRP/NMB/GRP18-27 | 3 | 1.0 |
| GABAB | GABAB | 1 | GABA | 1 | 1.0 |
| Ghrelin | GHSR1a | 1 | Ghrelin | 1 | 1.0 |
| Glucagon | GIP/GHRH/Secretin | 3 | GIP/GHRH/Secretin | 6 | 1.0 |
| Gonadotropin-releasing hormone | GnRH | 2 | GnRH1–2 | 2 | 1.0 |
| G protein-coupled oestrogen | GPE | 1 | Oestrogen | 1 | 1.0 |
| Melatonin | MT1–2 | 2 | Melatoni n/acetyl-serotonin | 2 | 1.0 |
| Motolin | Motolin | 1 | Motolin | 1 | 1.0 |
| Neuropeptide | NPS | 1 | Neuropeptide S | 1 | 1.0 |
| Neuropeptides B and W | NPBW1–2 | 2 | Neuropeptide w and B | 2 | 1.0 |
| Neurotensin | NTSR1–2 | 2 | Neurotensin/neuromedin N | 2 | 1.0 |
| Orexin | OX1–2 | 2 | Orexin A and B | 2 | 1.0 |
| Platelet Activating factor | PAF-R | 1 | PAF | 1 | 1.0 |
| Prokineticin | PK1–2 | 2 | PROK1–2 | 2 | 1.0 |
| Urotensin II | UT | 1 | U-II | 1 | 1.0 |
| Glucagon | Glucagon/GLP-1/GLP-2 | 3 | Glucagon/GLP-1/GLP-2 | 3 | 1.0 |
| Leukotriene family | BLT1/2 RvE1/// | 8 | Leukotrienes/lipoxins/RvE1/// | 10 | 1.3 |
| Relaxin | RXFP1–4 | 4 | H1-3 relaxin/INSL3/5 | 5 | 1.3 |
| Glycoprotein hormone | LH/FSH/TSH | 3 | FSH/LH/CG/TSH | 4 | 1.3 |
| Chemokine | CX3CR1/XCR1 | 2 | CX3CL1/lymphotactin/// | 3 | 1.5 |
| Endothelin | ET1–2 | 2 | ET1–/3 | 3 | 1.5 |
| Tachykinin | NK13 | 3 | SP/NKA/NKB/neuropeptide K/neuropeptide g | 5 | 1.7 |
| Cannabinoid | CB1–2 | 2 | Anandamide/2-arachidonylglycerol/// | 4 | 2.0 |
| Cholecystokinin | CCK1–2 | 2 | CCK-4/CCK-8/CCK-33/gastrin | 4 | 2.0 |
| Corticotropin releasing factor | CRF1–2 | 2 | CRH/urocortin1-3 | 4 | 2.0 |
| Free fattty acid-short chain | FFA2–3 | 2 | C2–5 | 4 | 2.0 |
| GPR119 | GPR119 | 1 | OEA/PEA | 2 | 2.0 |
| KISS1 family neuropeptide | KISS1 NPFF/PRP/QRFP | 5 | KP54/KP13/QRFP/// | 10 | 2.0 |
| VIP and PCAP | VPAC1–2 / PAC1 | 3 | VIP/PACAP1-38/VAPAC1-27/PHI/PHM/GRF | 6 | 2.0 |
| Anaphylatoxin | C3a | 1 | C3a/C5a | 2 | 2.0 |
| Calcium sensing | CaS | 1 | Ca/Mg | 2 | 2.0 |
| Trace amine | TA1–2 | 2 | Tyramine/PEA/octopamine/do pamine/tryptamine | 5 | 2.5 |
| Opioid and opioid like | DOR/KOR/MOR/ORL1 | 4 | Enkephalin/endorphin/dynorphin/orphanin/endomorphin/// | 11 | 2.8 |
| Parathyroid | PTH1–2 | 2 | PTH/PTHrP/TIP39/// | 6 | 3.0 |
| Hormone | |||||
| Bradykinin | B1–2 | 2 | Bradykinin and its variants | 7 | 3.5 |
| Formyl Peptide | FPR1 | 1 | FLMP/cathepsin/annexin/spinorphilin | 4 | 4.0 |
| Bile acid | GPBA | 1 | Lithocholic/deoxycholic/chenodeoxycholic/cholic acids | 4 | 4.0 |
| Cytokine | CCRx | 10 | CCLx/// | 40 | 4.0 |
| Chemokine | CXCRx | 6 | CXCLx/// | 27 | 4.5 |
| Free fattty acid-Long chain | FFA1 | 1 | C:16/C:22/// | 8 | 8.0 |
Non-odorant GPCRs and endogenous ligands identified from the British Journal of Pharmacology Guide to Receptors and Channels, 4th addition, 2009.
Selected receptor variants and endogenous ligands are presented, separated by a front slash (/). The existence of additional, unspecified receptor variants and/or ligands is denoted by three consecutive front slashes (///).
Further to the right along the curve, representative of an increased likelihood of identifying biased, orthosteric ligands, are the cannabinoid (m=2.0) and opioid (m=2.8) receptor families. To the far-right side of the plot in the second peak are families of cytokine (m=4.0) and chemokine receptors (m=4.5), that have relatively greater informational entropy and, consequently retain a higher likelihood of supporting biased, orthosteric ligands. This analysis supports the hypothesis that, for many GPCRs, it may be unproductive to restrict searches for biased compounds to orthosteric sites and more productive to examine allosteric motifs for the determinants of signaling bias.
Allosteric modulation of GPCR signaling
Allosterism was first used to describe the cooperative binding of oxygen molecules to the metalloprotein hemoglobin [18], and today, allosterism is a unifying theme of GPCR regulation. Indeed, multiple allosteric sites are now known to exist within a single receptor [19, 20]. Endogenous GPCR allosteric regulators include, G proteins, β-arrestins, ions (e.g., Zn2+, Na+, and Cl−), aromatic amino acids (e.g., L-phenylalanine, L-tryptophan, and L-tyrosine), lipids, receptor-activity modifying proteins (RAMPs), autoantibodies (particularly in disease states), melanocortin receptor accessory proteins (MRAPs), membrane stretch, and other GPCRs through the formation of homo/heterodimers [21].
Allosteric modulators may enhance or antagonize agonist binding and/or signaling. In the extended operational model of allosterism, these effects are described mathematically by cooperativity parameters α and β (see Box 1) [22, 23]. In this model, α and β values of 1 indicate no effect of the modulator on agonist activities. Intrinsic efficacy is denoted by the transducer parameter τ, with a τ value greater than 0 indicative of intrinsic activity/efficacy. Allosteric ligands fall into general categories based on the magnitude of α and β and exert characteristic effects on agonist dose-response curves (DRCs), see [23–25] for comprehensive reviews of classic allosteric modulators. Positive allosteric modulators (PAMs) increase agonist binding (α>1), signaling efficacy (β>1), or both. Conversely, negative allosteric modulators (NAMs) decrease agonist binding (α<1), signaling efficacy (β<1) or both. All of these so-called ‘pure’ modulators lack intrinsic activity and, therefore, the ability to activate the receptor in the absence of agonist. Modulators with some degree of intrinsic activity are known as ago-allosteric modulators [26]. Because the behavior of a given allosteric modulator at a given receptor can vary depending on the orthosteric ligand used to probe its activity, a phenomenon known as probe dependence, its classification should be placed within the context of the interacting ligand [26].
Box 1. Modeling allosteric modulation.
The actions of systems with allosteric modulators can be described using an extended operational model of allosterism, as previously described by Leach and colleagues [23]:
| Eq. 1 |
In this equation, the functional effect of the agonist and allosteric modulator pair in the system (E), is a function of the maximum functional effect of the system (Em), a binding cooperativity coefficient (α), an efficacy cooperative coefficient (β), the transducer slope parameter for the system (n), and agonist (A) and allosteric modulator (B) concentrations ([A], [B]), equilibrium dissociation constants (KA, KB), and intrinsic efficacies (τA and τB). While practically useful, this model does not weight action at the orthosteric and allosteric sites equally and fails to account for situations in which effects of the agonist and the modulator on the other’s efficacy are not reciprocal.
Here, we present a new variation of this model that better accounts for an equivalence between allosteric and orthosteric sites by the simple addition of a single term. This model makes Eq. 1 invariant to the interchange of agonist A and modulator B by redefining β as (efficacy cooperativity coefficient for the effect of modulator B on agonist A) and introducing the term (highlighted in grey, efficacy cooperativity coefficient for the effect of agonist A on modulator B):
| Eq. 2 |
This implies, mathematically, that orthosteric and allosteric ligands can potentially exert effects of equal or unequal magnitude on each other and on receptor signaling. A demonstration of the curve fits produced by the original extended operational model of allosterism (Eq. 1) and the symmetrical variation (Eq. 2) is provided in Box 1, Figure I.
Biased allosteric modulators (BAMs)
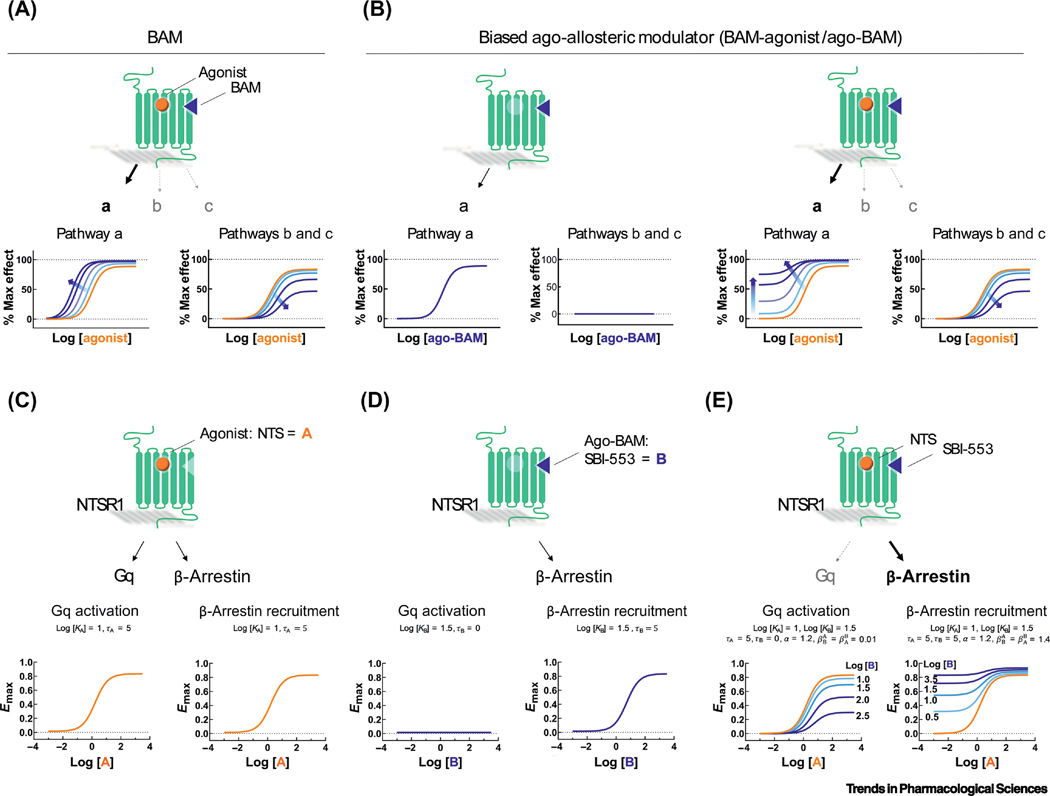
BAMs are set apart from other classes of allosteric modulators by an ability to exert pathway-specific effects. ‘Pure’ BAMs lack intrinsic activity (τ=0) and may or may not alter agonist binding. They selectively enhance signaling through some pathways (i.e., PAM activity; Fig 3A, left) while exerting no effect or antagonizing signaling through another subset of pathways (i.e., neutral allosteric ligand (NAL) or NAM activity; Fig 3A, right). Biased ago-allosteric modulators (ago-BAMs) act both as biasing modulators to exert pathway-specific effects on agonist signaling (Fig 3B, right) and act alone as biased allosteric agonists (Fig 3B, left). That is, τ is greater than 0 for some pathways, but not others.
Figure 3. Effects of BAMs on receptor signaling.
GPCRs (green) are bound by an orthosteric agonist (gold ball) and an allosteric modulator (blue triangle). Binding of the agonist to the receptor results in signaling through multiple pathways - labeled by ‘a’, ‘b’ and ‘c’ (arrow thickness and pathway labels indicate the strength of the signaling outcome). Dose-response curves (below) show the effect of agonist on a given signaling outcome in the presence of increasing allosteric modulator concentrations, indicated by increasing blue color saturation. Newest among the classes of allosteric modulators are the BAM and ago-BAM. BAMs are pure modulators that lack intrinsic activity (τ=0) and may or may not alter the binding affinity (α) of the agonist. Characteristically, BAMs enhance signaling (β>1) through some pathways but exert no effect (β=1) or antagonize signaling (β<1) through other pathway subsets. Ago-BAMs act as biasing modulators to exert pathway-specific effects on agonist signaling and act alone as biased allosteric agonists, that is, τ is greater than 0 for some pathways, but not others. (A) This hypothetical BAM potentiates agonist-induced signaling through pathway ‘a’ while antagonizing agonist-induced signaling through pathways ‘b’ and ‘c’. (B) This hypothetical ago-BAM stimulates signaling through pathway ‘a’ when applied alone but has no effect on pathways ‘b’ or ‘c’. In combination with an orthosteric agonist, this ago-BAM enhances agonist signaling efficacy through pathway ‘a’ while antagonizing signaling through ‘b’ and ‘c’. (C-E) Application of Eq. 2, a symmetrical version of the extended operational model, to the agonist ago-BAM pair of neurotensin (NTS) and SBI-553 at the NTSR1. (C) NTS alone acts as a balanced, full orthosteric agonist, activating both Gαq- and β-arrestin-mediated cellular responses. (D) SBI-553 alone acts as a biased allosteric agonist, activating β-arrestin- but not Gαq-mediated cellular responses. (E) SBI-553 confers β-arrestin bias to NTS by concurrently enhancing NTS-induced β-arrestin recruitment and antagonizing NTS-induced Gαq activation. The qualitative behavior depicted here has been directly observed with the action on SBI-553 on NTSR1 [40, 41].
A growing body of evidence suggests that signaling bias can be achieved with endogenous allosteric modulators. For example, membrane stretch induces β-arrestin-biased allosteric modulation of angiotensin II type 1 receptor [27]. Additionally, homo- and hetero- receptor dimer formation induce biased receptor signaling, as reviewed [28, 29]. The GPCR accessory protein MRAP2 regulates biased signaling and constitutive activity of the ghrelin receptor GHSR1a [30]. Autoantibodies have been identified that confer bias to the calcium sensing receptor [31]. RAMPs can also modulate the signaling preferences of GPCRs, including biasing signaling of the vasoactive intestinal polypeptide 1 receptor [32], calcitonin receptor [33] and adrenomedullin–calcitonin receptor-like receptor [34]. Sodium has been implicated in the regulation of biased signaling by the δ-opioid receptor [35]. The growing list of identified endogenous BAMs suggests biased allosteric modulation is an evolved mechanism for texturizing GPCR signaling.
In addition to endogenous modulators, several classes of xenobiotics have now been identified that can act as biasing modulators, including small molecules, pepducins, and peptides/bitopic ligands [36, 37]. Table 2 lists several GPCRs for which BAMs have been described and the therapeutic arenas in which ligand bias may prove beneficial.
Table 2.
Selected biased allosteric modulators
| Receptor | Receptor Class | Biasing Modulat or Class | Biasing modulator | Agonist | Reported Bias* | Potential Therapeutic Utility | Effect on agonist binding | Intrinsic activity | Allosteric site/Binding determinants | Refs |
|---|---|---|---|---|---|---|---|---|---|---|
| Bias towards G protein coupling or G protein subtype preference | ||||||||||
|
| ||||||||||
| CXCR4 | Class A | Pepducin | ATI 2341 | SDF-1 | Gi>β-arrestin, GRK2, GRK3>> G13 | HIV, leukemias | ND | Gi; minimal β-arrestin, GRK2, GRK3 | ICLs1,2,3 | [52, 53] |
| FP receptor | Class A | Small molecule | PDC113.824 | PGF2α | Gq >G12 | Preterm labor | Increased off-rate of [3H]PGF2α | No | ND | [54] |
| MC3–5R | Class A | Small molecule | Fenoprofen | αMSH | ERK1/2 > cAMP | Inflammatory arthritis, obesity | Did not displace [125I]NDP-MSH | ERK1/2 | ND | [55, 56] |
| FFAR2 | Class A | Small molecule | AZ 1729 | C3 | Gi>Gq/11 | Metabolic disorders | Increased C3 binding; did not fully displace [3H]GLPG0974 | Gi | ND | [57, 58] |
| FSHR | Class A | Small molecule | Thiazolidinone analogs | FSH | Gi>Gs | Infertility | Did not displace [125I]FSH | Gs or Gi or both, analog-specific | TMHs 1,2,3 | [59] |
| δ-opioid receptor | Class A | Small molecule | BMS 986187 | Leu-enkephalin, SNC80 and TAN67 | G protein (general, GTPγS) > β-arrestin2 | Chronic pain, depression | Did not displace [3H]diprenorphine, increased affinity of leu-enkephalin, SNC80, and TAN67 | G protein; minimal β-arrestin2, internalization | ND | [60, 61] |
| Calcium sensing receptor | Class C | Small molecule | AC 265347 | Ca2+o | IP1, ERK1/2 > Ca2+ | Hyper-parathyroidism | ND | IP1; minimal Ca2+ | TMHs | [62, 63] |
| R,R-calcimimetic B | Ca2+o | IP1, ERK1/2 > Ca2+ | ND | IP1; minimal Ca2+ | ND | [62] | ||||
|
| ||||||||||
| Bias towards β-arrestin coupling | ||||||||||
|
| ||||||||||
| NTSR1 | Class A | Small molecule | ML314 | NT, NT8–13 | β-arrestin2 > Gq/Ca2+ | Addiction, psychiatric disorders | Increased [3H]NT binding Bmax and decreased Kd | β-arrestin2, internalization | ND | [38, 39] |
| SBI-553 | NT, NT8–13 | β-arrestin2 > Gq/Ca2+ | Increased [3H]NT binding Bmax and decreased Kd | β-arrestin2, internalization | ND | [40, 41] | ||||
| β2AR | Class A | Pepducin | ICL1–9 | ICYP | β-arrestin > Gs | Myocardial infarction, heart failure | No effect on [125I]ICYP binding | β-arrestin, internalization, ERK1/2 | ICL1 | [46, 47] |
| CB1R | Class A | Small molecule | Org27569 | CP55940 | β-arrestin1 > G protein (general, GTPyS) | Obesity, pain, neurodegenerative disorders | Increased [3H]CP55940 binding | β-arrestin, ERK1/2, internalization | TMHs 3,6, 7; ECLs 2, 3 | [42, 43] |
| Pyrimidinyl biphenylureas | CP55940 | β-arrestin1/2 > G protein | Increased [3H]CP55940 binding, decreased binding of inverse agonist [3H]SR141716A | β-arrestin2, ERK1/2, internalization | ND | [44, 45] | ||||
| CX3CR1 | Class A | Small molecule | AZD8797 | Fractalkine | β-arrestin > G protein (general, GTPyS) | Atherosclerosis, metabolic disorders | Reduced CX3CL1 Bmax, no effect on Kd | minimal Gi | ND | [48] |
ND, not determined; ILC, intracellular loop; TMH, transmembrane helix; ECL, extracellular loop
> indicates an identified bias toward the pathway/effector on the left relative to the pathway/effector on the right.
If there is a unifying theme to be found across these examples, it is one of diversity of action and mechanism. This point is particularly true for the β-arrestin-biased modulators. The most dramatic examples of these are the β-arrestin–biased ago-allosteric modulators of NTSR1, ML314 [38, 39] and SBI-553 [40, 41], and of CB1, ORG27569 [42, 43] and the pyrimidinyl biphenylureas, LDK1285, LDK1288, LDK1305, and PSNCBAM1 [44, 45]. While the β2AR pepducin, ICL1–9, also stimulated receptor β-arrestin recruitment and internalization without activating G protein signaling [46, 47], intrinsic β-arrestin activity is not a unifying characteristic of this class. Indeed, the βarrestin-biased small molecule CX3CR1 allosteric modulator, AZD8797, did not induce β-arrestin recruitment alone [48].
The β-arrestin-biased allosteric modulators to date also exert differential effects on orthosteric ligand binding. ML314 [39], SBI-553 [40], ORG27569 [42] and the pyrimidinyl biphenylureas [44] all induced high-affinity, β-arrestin selective receptor conformations, as evidenced by a concurrent increase in orthosteric ligand affinity in radioligand assays and preferential β-arrestin coupling in recruitment assays. The necessity of β-arrestin to induce this high affinity state is not known. Conversely, the β2AR pepducin ICL1–9 stimulated receptor-β-arrestin interaction without altering agonist binding affinity. Curiously, AZ8797 decreased CXCL1 binding (Bmax) without changing its apparent affinity (Kd) [48].
While the actions of ORG27569 on CB1 and SBI-553 on NTSR1 are similar with respect to G protein antagonism, β-arrestin recruitment, receptor internalization and agonist binding, they also deviate in important ways. SBI-553 biased neurotensin-induced MAPK signaling through β-arrestin but did not increase the pERK1/2 generation when applied alone [40]. These findings suggest that while SBI-553stimulates NTSR1-β-arrestin recruitment, the SBI-553/NTSR1/β-arrestin complex may not be fully signaling competent. The effect of ORG27569 on MAPK signaling is less clear, as different studies have argued that it acts as an allosteric agonist of ERK1/2 signaling via β-arrestin1 [43] or Gi/o [49] or, alternatively, as an allosteric antagonist [50] or an inverse agonist [51]. The β2AR pepducin ICL1–9 stimulated receptor phosphorylation, internalization and β-arrestin-mediated MAPK signaling [47]. In this regard, the pepducin ICL1–9 exhibits effects similar to the β-arrestin-biased β2AR agonist carvedilol [47].
While the actions of BAMs identified to date may be diverse, a shared feature is conferment of a more restricted receptor signaling profile that manifests as pathway-specific efficacy and/or modulation. The ability of BAMs to qualitatively alter receptor signaling highlights the need for an updated allosteric model that gives equal weight to ligands acting at orthosteric and allosteric sites.
A model of allosteric modulation that puts agonist and modulator on equal footing
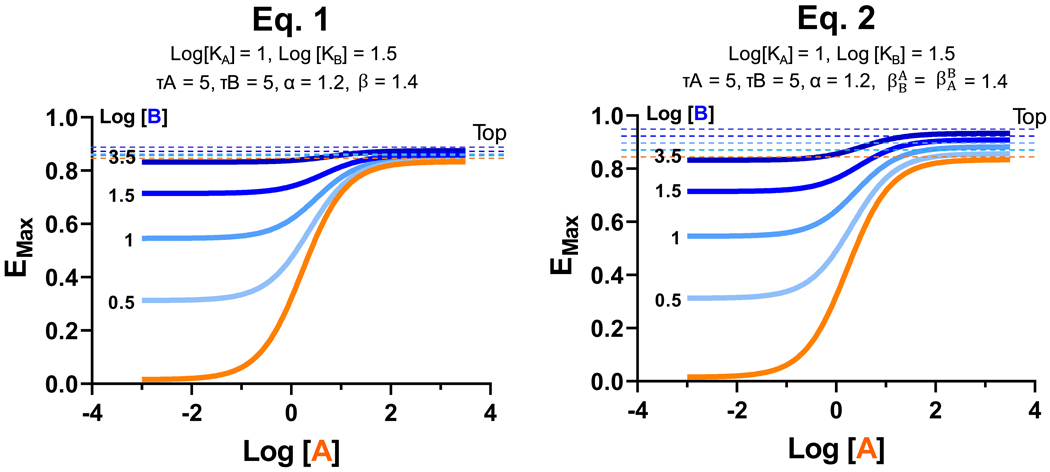
The first modeling equation in Box 1, Eq. 1, extends the operational model from one to two concentration variables, as described [23]. Though more complete than the operational model, it captures orthosteric-allosteric reciprocity in binding cooperativity (α) but not in efficacy cooperativity (β). β is a scaling factor that denotes the magnitude of allosteric effect on orthosteric agonist efficacy and is not reciprocal between orthosteric and allosteric ligands [64]. What is missing from Eq. 1 is a term describing the equivalence of the orthosteric and allosteric sites that makes the model invariant to the interchange of orthosteric and allosteric parameters. This is remedied by the second equation we introduce in Box 1, Eq. 2, that incorporates an added efficacy parameter and its corresponding transducer term . This term forms part of the effective transducer that occurs at very large concentrations of both agonist [A] and modulator [B]. This additional term allows us to resolve the concern raised in Giraldo [64] that in Eq. 1 τB is not included in the expression for Top effect, when it should logically appear. Another advantage of introducing the additional parameter is the ability to explain the effect of the orthosteric agonist on allosteric modulator efficacy in situations where the two effects need to be distinguished (i.e., when the allosteric modulator potentiates the efficacy of the orthosteric agonist, but the orthosteric agonist reduces the efficacy of the allosteric modulator). While the added term may be a noteworthy theoretical improvement, in practice, allosteric signaling parameters are evaluated by fitting systems of equations to functional measurements by nonlinear regression analysis. Absent a means to independently vary the strength of either of the individual transducers τA or τB, it may be difficult to determine both parameters and from Box 1, Eq. 2 without some other underlying assumption, as they only appear together within the sum Thus, while restricting an allosteric analysis to Box 1, Eq. 1 routinely yields good data fits (Box 1, Figure I), it does so by implicitly assuming that does not contribute to the efficacy (is small or 0).
Box 1, Figure I. Optimizing the extended operational model.
While both Eq. 1 (left) and Eq. 2 (right) capture the qualitative effects of positive allosteric modulation, they produce distinct effects on agonist-modulator dose-response curve Top shifts. The most appropriate model may depend on the characteristics of the allosteric modulator, receptor system and assay platform. The horizontal lines represent the Top values of the corresponding colored response curves.
As proof-of-concept, we applied Eq. 2 to the agonist-modulator pair of neurotensin (NTS) and SBI-553 at NTSR1. We recently characterized SBI-553 as a biased ago-allosteric modulator that acts as an ago-PAM for β-arrestin translocation and a PAM-antagonist for Gq signaling [40]. Based on the biology, in this example we assume . As modeled in Figure 3C, NTS is a balanced, full agonist, activating both NTSR1 Gq signaling and stimulating receptor β-arrestin recruitment. SBI-553 alone, in contrast, acts as a biased allosteric agonist, stimulating NTSR1 β-arrestin recruitment without activating the Gq pathways (Figure 3D). That is, SBI-553’s intrinsic activity (τ) is pathway-specific: >0 for β-arrestin recruitment and ≤0 for Gq-associated responses. Both SBI-553’s permissiveness of NT-induced β-arrestin recruitment (Figure 3E, right) and its antagonism of NT-induced Gq signaling (Figure 3E, left) can be modeled and reflect observed behaviors [40, 41]. In addition to pathway-specific intrinsic activity, SBI-553 also exhibits pathway-specific effects on agonist efficacy is ≥ 1 for β-arrestin recruitment and < 1 for Gq signaling. In summary, the symmetrical, extended operational model, Eq. 2, explains the behavior of the NTSR1 BAM SBI-553 [40], and its parent compound ML314 [38, 39].
Why seek biased allosteric modulators of GPCRs as therapeutics?
Despite the historical success of GPCR drug discovery, many GPCR-targeted therapeutics produce severe, dose-limiting side effects. BAMs have therapeutic potential, as they possess the combined positive characteristics of both allosteric and biased ligands. These characteristics together afford an unprecedented degree of temporal-, location-, receptor subtype- and signaling pathway-selectivity. As outlined below, this degree of control may limit off-target side effects, permit the targeting of previously ‘undruggable’ GPCRs and allow for the segregation of on-target therapeutic action from on-target side effects.
Advantages of biased and allosteric drugs
Increased receptor subtype-selectivity. Targeting specific receptors belonging to the same family subtype is often challenging because of the high degree of conservation of their orthosteric binding sites [65, 66]. Allosteric sites are generally less conserved between receptor subtypes and, thus, allow for increased subtype selectivity [67].
Targeting ‘undruggable’ GPCRs. Allosteric modulators provide an opportunity to target GPCRs that were previously intractable to drug discovery efforts because of the unique geometry of the orthosteric binding site or exceptionally high site occupancy by the endogenous ligand[68].
Pathway specificity. As outlined above, biased ligands can selectively engage therapy-relevant signaling pathways. We recently observed that, for the NTSR1, pathway specificity permits the segregation of desired, β-arrestin-mediated anti-addiction actions from unwanted, G protein-mediated hypothermic and hypotensive actions [40].
Preservation of physiological signaling. Allosteric modulators that lack intrinsic activity maintain the natural spatiotemporal “rhythms” of the endogenous ligand, which may limit side effects stemming from disruption of physiological signaling.
Effect Saturation. The dependency of ‘pure’ allosteric modulators on endogenous ligands result in an allosteric “ceiling effect” that increases the likelihood of on-target safety in overdose situations. It is noteworthy, however, that many allosteric ligands have intrinsic activity themselves (i.e., ago-allosteric characteristics, Table 2), which would occlude this benefit.
Combination therapies. New orthosteric-allosteric drug combination treatments are possible, in which a biased allosteric modulator is used to push receptor signaling by an orthosteric drug down select pathways [69].
Disadvantages of biased and allosteric drugs
Loss of benefits of polypharmacology. Recent drug development efforts indicate an increasing focus on target selectivity rather than on polypharmacology [5]. Some of the most efficacious drugs, however, have multiple targets and multiple activities at those targets. By gaining specificity and, potentially, safety, we may be sacrificing efficacy [70].
Less predictable biological effects. Biased agonists activate receptors in a less naturalistic manner that may produce unanticipated on-target effects [71].
Low potency and aqueous solubility. Because allosteric sites can be located at receptor sites that interact with the plasma membrane and allosteric ligands hydrophobic, allosteric modulators may have lower receptor affinities and aqueous solubilities [20].
How can we identify biased and allosteric GPCR ligands?
Binding-based, biased and allosteric drug discovery efforts - even large-scale ones evaluating millions of compounds - have not always been successful [72]. Here, we outline a screening platform based predominantly on multiplexed functional assays that can be complemented by structure-based drug discovery efforts.
Functional screening
High-content screening is particularly well-suited for GPCR drug discovery. Biased ligands can be identified using multiplexed screening platforms that include multiple readouts of G protein- and β-arrestin-dependent activities. Allosteric ligands can be identified by looking for effects of compounds alone and on agonist EC20 function (for PAM activity) or EC80 function (for NAM activity). Using these approaches our group has identified compounds for the NTSR1 with unique pharmacological profiles that would have gone unnoticed if using a ligand-binding or G protein-based platform [38–41]. New platforms involving transforming growth factor-α (TGF-α) shedding responses [73], luciferase complementation [74] and bioluminescence resonance energy transfer (BRET) [75–77] provide a means of directly assessing effector recruitment to the receptor and effector activation, providing a like-for-like comparison of G protein family members, G protein-coupled receptor kinases (GRKs), and β-arrestins. With the inclusion of appropriate reference agonist(s) in the screen, quantitative ‘bias factors’ can be calculated using one of a number of approaches [11]. Functional assays can be followed by saturation radioligand binding assays to confirm target engagement and competition radioligand binding assays to determine effects on agonist affinity (Kd) and site occupancy (Bmax).
Structure-guided drug design
The resolution of the structure of many GPCRs, which permits the use of structure-based, computer-assisted docking, has resulted in the identification of several new allosteric probes [15, 78]. The crystal structures of 44 unique receptors and 205 ligand–receptor complexes now provide a wealth of information to facilitate structure-based drug discovery [5]. Additionally, database and software tools are now available specifically for the identification of allosteric receptor sites and ligands [68]. Technically, the resolution of X-ray structures of receptor-allosteric modulator complexes can be more difficult than those with orthosteric ligands, as allosteric ligands may have relatively weaker affinity and may be highly lipophilic or polyaromatic, limiting their aqueous solubility [20]. At present no GPCR-BAM structures are available, limiting our understanding of non-orthosteric determinants of biased receptor signaling.
Challenges and considerations
Just as biased allosteric modulators possess the combined potential therapeutic advantages of biased and allosteric agents, the search for such compounds is plagued by the challenges associated with identifying both phenomena. While not exhaustive, we are listing some of the more important ones below.
Challenge 1: Ensuring that the screening environment recapitulates the in vivo environment of interest in terms of effector ratios [79, 80].
Challenge 2: Selecting multiple G protein and β-arrestin-associated readouts based on known or anticipated physiological relevance.
Challenge 3: Accounting for temporal and location effects in the determination of signaling bias [9, 81, 82].
Challenge 4: Comparing amplified to linear signaling data in the determination of signaling bias [83].
Challenge 5: Quantifying the bias of ligands that exert differential effects on more than two signaling pathways. Future bias calculations may require multivariate statistics to describe complex ligand activity profiles.
Challenge 6: Accounting for probe dependence of allosteric modulators [67].
Challenge 7: Preparing for species divergence, as allosteric receptor sites are less well-conserved than orthosteric determinants across species [84].
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
GPCRs are the largest family of targets of therapeutic drugs, but many of these medications are plagued by side effects that limit their utility. Classic agonist and antagonist GPCR ligands activate receptor signaling pathways uniformly and bind orthosteric sites that are highly conserved across receptor subtypes, potentially producing on- and off-target side effects. BAMs are an emerging class of GPCR ligands that selectively activate a subset of a receptor’s signaling pathways and bind to less well-conserved, allosteric sites. Biased allosteric modulation is an endogenous mechanism used to fine-tune receptor signaling that may be leveraged to develop more selective therapeutics. Indeed, we recently described a novel β-arrestin ago-BAM of the NTSR1 with desirable pharmacodynamic properties [40]. This lead compound joins a remarkably short list of allosteric xenobiotics known to engender biased receptor signaling, but with a rethinking of GPCR drug development and screening strategies, this should shortly change.
BAMs have the potential to limit both on- and off-target side effects, permit the targeting of previously ‘undruggable’ GPCRs, and avoid adverse effects associated with the dysregulation of endogenous hormone rhythms. Despite these advantages, little has been done practically to develop biased allosteric drugs. Moreover, the diverse mechanisms of action of this new compound class are largely unexplored, presenting exciting new avenues of investigation (see Outstanding Questions). Application of information theory to the ratio of endogenous ligands per GPCR class suggests that, for many GPCRs, orthosteric sites may not have evolved to produce biased receptor signaling. As such, screening for BAMs rather than biased orthosteric agonists may be the more fruitful strategy to identify functionally selective GPCR ligands. The biased pharmacological tool compounds identified in functional and in silico screens will advance our understanding of receptor systems, including linking effector coupling with distinct physiological effects. By arming us with this knowledge and the ability to capitalize on it, BAMs may improve our ability to treat diverse disease conditions. While critical questions remain, for many GPCRs, the future of drug discovery may favor allosterism.
OUTSTANDING QUESTIONS.
What are the structural determinants of biased allosteric modulation?
Are distinct GPCR conformations associated with ‘sets’ of signaling outcomes such that some signaling pathways are inseparable?
Does the promiscuity of GPCRs in terms of G protein coupling in in vitro assays reflect their in vivo signaling patterns?
For how many GPCRs can the effects of various G protein signaling pathways and β-arrestin recruitment be discriminated physiologically?
Will biased and allosteric GPCR ligands make for safer therapeutics?
HIGHLIGHTS.
Biased allosteric modulators (BAMs) are an emerging class of GPCR ligands that can profoundly alter GPCR behavior by exerting pathway-specific effects on receptor signaling.
By increasing receptor subtype and pathway selectivity, BAMs present the opportunity to develop safer, more efficacious therapeutics and target previously ‘undruggable’ GPCRs.
An assessment of the informational entropy inherent to non-odorant GPCR families suggests that, for many receptors, BAMs may be more easily identified than biased, orthosteric ligands.
The actions of BAMs can be described mathematically using extended operational models of allosterism with distinct efficacy and signaling cooperativity parameter values for each individual signaling pathway.
The search for biased and allosteric GPCR ligands is ripe with challenges, but these challenges are surmountable if drug discovery efforts are thoughtfully designed.
The potential therapeutic advantages of BAMs, and promising late-breaking reports on β-arrestin biased modulators that give new-found tangibility to these theoretical advantages, suggest that such drug discovery endeavors will be worthwhile.
ACKNOWLEDGEMENTS
This work was supported, in part, by K99 DA048970, UG3 DA050316 and R37 MH073853.
GLOSSARY
- Agonist:
a ligand that activates a GPCR and increases receptor signaling above basal levels
- Allosteric agonist:
a ligand that activates a GPCR and increases receptor signaling above basal levels following binding at a receptor site distinct from that utilized by the endogenous ligand
- Allosteric binding site:
a topographical receptor site distinct from that at which an endogenous ligand binds
- Allosterism:
an overarching biological phenomenon the describes the reciprocal ability of conformation changes at one site of a macromolecule to induce conformation changes at a topographically distinct site
- Antagonist:
a ligand that blocks activation of a GPCR
- Biased ligand/agonist:
a ligand that activates a GPCR and increases receptor signaling above basal levels for some signaling pathways but not others
- Biased allosteric modulator:
a ligand that binds a receptor site distinct from that at which an endogenous ligand binds, and exerts non-uniform effects on receptor signaling
- Biased signaling:
also referred to as functional selectivity or ligand-directed signaling, refers to signaling through a subset of a receptor’s potential signaling pathways
- Bitopic molecule:
a ligand that binds both the orthosteric binding pocket and an allosteric receptor site
- Effector:
an intracellular protein that physically engages with a GPCR to mediate signaling
- G protein-coupled receptors (GPCRs):
a family of transmembrane receptors that control a wide array of physiological processes and are the targets of many therapeutics
- Information entropy:
a statistic indicating the amount of possible messages or outcomes available to a system
- Off-target side effects:
unwanted drug effects resulting from drug action at a biological target distinct from that producing the desired therapeutic action
- On-target side effects:
unwanted drug effects resulting from drug action at the same biological target that produces the desired therapeutic action
- Operational model of allosterism:
a mathematical model that describes GPCR signaling
- Orthosteric binding site:
physical receptor site through which binding of an endogenous ligand leads to receptor activation
- Pepducin:
a cell-penetrating lipopeptide that modulates GPCR signaling
- Probe dependence:
the magnitude and direction of signaling modulation by an allosteric probe is dependent on the orthosteric ligand with which it is assessed
- Signaling pathways:
defined signaling cascades resulting from activation of effectors. Different signaling pathways can produce distinct physiological effects
- Small molecule:
a low molecular weight (<900 daltons) compound. Can be endogenous (e.g., sugars, amino acids, lipids) or synthetic (e.g., drug candidates)
- System bias:
biased receptor signaling attributable not to the stabilization of a distinct receptor conformation, but rather to the environment in which the receptor is assessed
Footnotes
DISCLAIMER STATEMENT
US Patents 9,868,707 and 10,118,902 relating to the chemistry of ML314, SBI-553, and their derivatives have been issued to the Sanford Burnham Prebys Medical Research Institute and Duke University (M.G.C. and L.S.B.).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lagerstrom MC and Schioth HB, Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov, 2008. 7(4): p. 339–57. [DOI] [PubMed] [Google Scholar]
- 2.Chan HCS, et al. , Enhancing the Signaling of GPCRs via Orthosteric Ions. ACS Cent Sci, 2020. 6(2): p. 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sriram K. and Insel PA, G Protein-Coupled Receptors as Targets for Approved Drugs: How Many Targets and How Many Drugs? Mol Pharmacol, 2018. 93(4): p. 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santos R, et al. , A comprehensive map of molecular drug targets. Nat Rev Drug Discov, 2017. 16(1): p. 19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hauser AS, et al. , Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov, 2017. 16(12): p. 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowden H. and Munro J, Trends in clinical success rates and therapeutic focus. Nat Rev Drug Discov, 2019. 18(7): p. 495–496. [DOI] [PubMed] [Google Scholar]
- 7.Wouters OJ, McKee M, and Luyten J, Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009–2018. JAMA, 2020. 323(9): p. 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JS, Lefkowitz RJ, and Rajagopal S, Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov, 2018. 17(4): p. 243–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein Herenbrink C, et al. , The role of kinetic context in apparent biased agonism at GPCRs. Nat Commun, 2016. 7: p. 10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenakin T. and Christopoulos A, Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat Rev Drug Discov, 2013. 12(3): p. 205–16. [DOI] [PubMed] [Google Scholar]
- 11.Gundry J, et al. , A Practical Guide to Approaching Biased Agonism at G Protein Coupled Receptors. Front Neurosci, 2017. 11: p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajagopal S, Rajagopal K, and Lefkowitz RJ, Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov, 2010. 9(5): p. 373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenakin T, Agonist-receptor efficacy. II. Agonist trafficking of receptor signals. Trends Pharmacol Sci, 1995. 16(7): p. 232–8. [DOI] [PubMed] [Google Scholar]
- 14.Wisler JW, et al. , Recent developments in biased agonism. Curr Opin Cell Biol, 2014. 27: p. 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCorvy JD, et al. , Structure-inspired design of beta-arrestin-biased ligands for aminergic GPCRs. Nat Chem Biol, 2018. 14(2): p. 126–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon CE, A Mathematical Theory of Communication. Bell System Technical Journal, 1948. 27(3): p. 379–423. [Google Scholar]
- 17.Shannon CE, A Mathematical Theory of Communication. Bell System Technical Journal, 1948. 27(4): p. 623–656. [Google Scholar]
- 18.Monod J, Wyman J, and Changeux JP, On the Nature of Allosteric Transitions: A Plausible Model. J Mol Biol, 1965. 12: p. 88–118. [DOI] [PubMed] [Google Scholar]
- 19.Gentry PR, Sexton PM, and Christopoulos A, Novel Allosteric Modulators of G Protein-coupled Receptors. J Biol Chem, 2015. 290(32): p. 19478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Congreve M, Oswald C, and Marshall FH, Applying Structure-Based Drug Design Approaches to Allosteric Modulators of GPCRs. Trends Pharmacol Sci, 2017. 38(9): p. 837–847. [DOI] [PubMed] [Google Scholar]
- 21.van der Westhuizen ET, et al. , Endogenous allosteric modulators of G protein-coupled receptors. J Pharmacol Exp Ther, 2015. 353(2): p. 246–60. [DOI] [PubMed] [Google Scholar]
- 22.Roche D, Gil D, and Giraldo J, Mechanistic analysis of the function of agonists and allosteric modulators: reconciling two-state and operational models. Br J Pharmacol, 2013. 169(6): p. 1189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leach K, Sexton PM, and Christopoulos A, Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol Sci, 2007. 28(8): p. 382–9. [DOI] [PubMed] [Google Scholar]
- 24.Kenakin T, The Quantitative Characterization of Functional Allosteric Effects. Curr Protoc Pharmacol, 2017. 76: p. 9 22 1–9 22 10. [DOI] [PubMed] [Google Scholar]
- 25.Kenakin T. and Strachan RT, PAM-Antagonists: A Better Way to Block Pathological Receptor Signaling? Trends Pharmacol Sci, 2018. 39(8): p. 748–765. [DOI] [PubMed] [Google Scholar]
- 26.Christopoulos A, et al. , International Union of Basic and Clinical Pharmacology. XC. multisite pharmacology: recommendations for the nomenclature of receptor allosterism and allosteric ligands. Pharmacol Rev, 2014. 66(4): p. 918–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W, et al. , Allosteric modulation of beta-arrestin-biased angiotensin II type 1 receptor signaling by membrane stretch. J Biol Chem, 2014. 289(41): p. 28271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goupil E, Laporte SA, and Hebert TE, GPCR heterodimers: asymmetries in ligand binding and signalling output offer new targets for drug discovery. Br J Pharmacol, 2013. 168(5): p. 1101–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith NJ and Milligan G, Allostery at G protein-coupled receptor homo- and heteromers: uncharted pharmacological landscapes. Pharmacol Rev, 2010. 62(4): p. 701–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouault AAJ, et al. , The GPCR accessory protein MRAP2 regulates both biased signaling and constitutive activity of the ghrelin receptor GHSR1a. Sci Signal, 2020. 13(613). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makita N, et al. , An acquired hypocalciuric hypercalcemia autoantibody induces allosteric transition among active human Ca-sensing receptor conformations. Proc Natl Acad Sci U S A, 2007. 104(13): p. 5443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christopoulos A, et al. , Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem, 2003. 278(5): p. 3293–7. [DOI] [PubMed] [Google Scholar]
- 33.Morfis M, et al. , Receptor activity-modifying proteins differentially modulate the G protein-coupling efficiency of amylin receptors. Endocrinology, 2008. 149(11): p. 5423–31. [DOI] [PubMed] [Google Scholar]
- 34.Serafin DS, et al. , Dawn of a New RAMPage. Trends Pharmacol Sci, 2020. 41(4): p. 249–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenalti G, et al. , Molecular control of delta-opioid receptor signalling. Nature, 2014. 506(7487): p. 191–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carr R 3rd and Benovic JL, From biased signalling to polypharmacology: unlocking unique intracellular signalling using pepducins. Biochem Soc Trans, 2016. 44(2): p. 555–61. [DOI] [PubMed] [Google Scholar]
- 37.Carr R 3rd, et al. , Development and characterization of pepducins as Gs-biased allosteric agonists. J Biol Chem, 2014. 289(52): p. 35668–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peddibhotla S, et al. , Discovery of ML314, a Brain Penetrant Non-Peptidic beta-Arrestin Biased Agonist of the Neurotensin NTR1 Receptor. ACS Med Chem Lett, 2013. 4(9): p. 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barak LS, et al. , ML314: A Biased Neurotensin Receptor Ligand for Methamphetamine Abuse. ACS Chem Biol, 2016. 11(7): p. 1880–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slosky LM, et al. , beta-Arrestin-Biased Allosteric Modulator of NTSR1 Selectively Attenuates Addictive Behaviors. Cell, 2020. 181(6): p. 1364–1379 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinkerton AB, et al. , Discovery of beta-Arrestin Biased, Orally Bioavailable, and CNS Penetrant Neurotensin Receptor 1 (NTR1) Allosteric Modulators. J Med Chem, 2019. 62(17): p. 8357–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn KH, Mahmoud MM, and Kendall DA, Allosteric modulator ORG27569 induces CB1 cannabinoid receptor high affinity agonist binding state, receptor internalization, and Gi protein-independent ERK1/2 kinase activation. J Biol Chem, 2012. 287(15): p. 12070–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn KH, et al. , Distinct roles of beta-arrestin 1 and beta-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1). J Biol Chem, 2013. 288(14): p. 9790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jagla CAD, et al. , Pyrimidinyl Biphenylureas Act as Allosteric Modulators to Activate Cannabinoid Receptor 1 and Initiate beta-Arrestin-Dependent Responses. Mol Pharmacol, 2019. 95(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khurana L, et al. , Pyrimidinyl Biphenylureas: Identification of New Lead Compounds as Allosteric Modulators of the Cannabinoid Receptor CB1. J Med Chem, 2017. 60(3): p. 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grisanti LA, et al. , Pepducin-mediated cardioprotection via beta-arrestin-biased beta2-adrenergic receptor-specific signaling. Theranostics, 2018. 8(17): p. 4664–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carr R 3rd, et al. , beta-arrestin-biased signaling through the beta2-adrenergic receptor promotes cardiomyocyte contraction. Proc Natl Acad Sci U S A, 2016. 113(28): p. E4107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cederblad L, et al. , AZD8797 is an allosteric non-competitive modulator of the human CX3CR1 receptor. Biochem J, 2016. 473(5): p. 641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baillie GL, et al. , CB(1) receptor allosteric modulators display both agonist and signaling pathway specificity. Mol Pharmacol, 2013. 83(2): p. 322–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khajehali E, et al. , Biased Agonism and Biased Allosteric Modulation at the CB1 Cannabinoid Receptor. Mol Pharmacol, 2015. 88(2): p. 368–79. [DOI] [PubMed] [Google Scholar]
- 51.Gamage TF, Anderson JC, and Abood ME, CB1 allosteric modulator Org27569 is an antagonist/inverse agonist of ERK1/2 signaling. Cannabis Cannabinoid Res, 2016. 1(1): p. 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quoyer J, et al. , Pepducin targeting the C-X-C chemokine receptor type 4 acts as a biased agonist favoring activation of the inhibitory G protein. Proc Natl Acad Sci U S A, 2013. 110(52): p. E5088–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Planesas JM, et al. , Studying the binding interactions of allosteric agonists and antagonists of the CXCR4 receptor. J Mol Graph Model, 2015. 60: p. 1–14. [DOI] [PubMed] [Google Scholar]
- 54.Goupil E, et al. , A novel biased allosteric compound inhibitor of parturition selectively impedes the prostaglandin F2alpha-mediated Rho/ROCK signaling pathway. J Biol Chem, 2010. 285(33): p. 25624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan XC and Tao YX, Fenoprofen-An Old Drug Rediscovered as a Biased Allosteric Enhancer for Melanocortin Receptors. ACS Chem Neurosci, 2019. 10(3): p. 1066–1074. [DOI] [PubMed] [Google Scholar]
- 56.Montero-Melendez T, et al. , Old drugs with new skills: fenoprofen as an allosteric enhancer at melanocortin receptor 3. Cell Mol Life Sci, 2017. 74(7): p. 1335–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bolognini D, et al. , A Novel Allosteric Activator of Free Fatty Acid 2 Receptor Displays Unique Gi-functional Bias. J Biol Chem, 2016. 291(36): p. 18915–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sergeev E, et al. , A single extracellular amino acid in Free Fatty Acid Receptor 2 defines antagonist species selectivity and G protein selection bias. Sci Rep, 2017. 7(1): p. 13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Landomiel F, et al. , Biased Signaling and Allosteric Modulation at the FSHR. Front Endocrinol (Lausanne), 2019. 10: p. 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanczyk MA, et al. , The delta-opioid receptor positive allosteric modulator BMS 986187 is a G-protein-biased allosteric agonist. Br J Pharmacol, 2019. 176(11): p. 1649–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burford NT, et al. , Discovery, synthesis, and molecular pharmacology of selective positive allosteric modulators of the delta-opioid receptor. J Med Chem, 2015. 58(10): p. 4220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cook AE, et al. , Biased allosteric modulation at the CaS receptor engendered by structurally diverse calcimimetics. Br J Pharmacol, 2015. 172(1): p. 185–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leach K, et al. , Towards a structural understanding of allosteric drugs at the human calcium-sensing receptor. Cell Res, 2016. 26(5): p. 574–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giraldo J, Operational models of allosteric modulation: caution is needed. Trends in Pharmacological Sciences, 2015. 36(1): p. 1–2. [DOI] [PubMed] [Google Scholar]
- 65.Khoury E, Clement S, and Laporte SA, Allosteric and biased g protein-coupled receptor signaling regulation: potentials for new therapeutics. Front Endocrinol (Lausanne), 2014. 5: p. 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burger WAC, et al. , Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors. J Gen Physiol, 2018. 150(10): p. 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conn PJ, Christopoulos A, and Lindsley CW, Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov, 2009. 8(1): p. 41–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu S, et al. , Allosteric Modulator Discovery: From Serendipity to Structure-Based Design. J Med Chem, 2019. 62(14): p. 6405–6421. [DOI] [PubMed] [Google Scholar]
- 69.Ni D, et al. , Combining Allosteric and Orthosteric Drugs to Overcome Drug Resistance. Trends Pharmacol Sci, 2020. 41(5): p. 336–348. [DOI] [PubMed] [Google Scholar]
- 70.Peters JU, Polypharmacology - foe or friend? J Med Chem, 2013. 56(22): p. 8955–71. [DOI] [PubMed] [Google Scholar]
- 71.Luttrell LM, Minireview: More than just a hammer: ligand “bias” and pharmaceutical discovery. Mol Endocrinol, 2014. 28(3): p. 281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conn PJ, et al. , Opportunities and challenges in the discovery of allosteric modulators of GPCRs for treating CNS disorders. Nat Rev Drug Discov, 2014. 13(9): p. 692–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue A, et al. , TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods, 2012. 9(10): p. 1021–9. [DOI] [PubMed] [Google Scholar]
- 74.Inoue A, et al. , Illuminating G-Protein-Coupling Selectivity of GPCRs. Cell, 2019. 177(7): p. 1933–1947 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olsen RHJ, et al. , TRUPATH, an open-source biosensor platform for interrogating the GPCR transducerome. Nat Chem Biol, 2020. 16(8): p. 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Angers S, et al. , Detection of beta(2)-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proceedings of the National Academy of Sciences of the United States of America, 2000. 97(7): p. 3684–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hamdan FF, et al. , High-throughput screening of G protein-coupled receptor antagonists using a bioluminescence resonance energy transfer 1-based beta-arrestin2 recruitment assay. Journal of Biomolecular Screening, 2005. 10(5): p. 463–475. [DOI] [PubMed] [Google Scholar]
- 78.Shoichet BK and Kobilka BK, Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol Sci, 2012. 33(5): p. 268–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Onfroy L, et al. , G protein stoichiometry dictates biased agonism through distinct receptor-G protein partitioning. Sci Rep, 2017. 7(1): p. 7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Urs NM, et al. , Distinct cortical and striatal actions of a beta-arrestin-biased dopamine D2 receptor ligand reveal unique antipsychotic-like properties. Proc Natl Acad Sci U S A, 2016. 113(50): p. E8178–E8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grundmann M. and Kostenis E, Temporal Bias: Time-Encoded Dynamic GPCR Signaling. Trends Pharmacol Sci, 2017. 38(12): p. 1110–1124. [DOI] [PubMed] [Google Scholar]
- 82.Stoeber M, et al. , A Genetically Encoded Biosensor Reveals Location Bias of Opioid Drug Action. Neuron, 2018. 98(5): p. 963–976 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gillis A, et al. , Low intrinsic efficacy for G protein activation can explain the improved side effect profiles of new opioid agonists. Science Signaling, 2020. 13(625). [DOI] [PubMed] [Google Scholar]
- 84.Kennedy JP, et al. , Synthesis and structure-activity relationships of allosteric potentiators of the m(4) muscarinic acetylcholine receptor. ChemMedChem, 2009. 4(10): p. 1600–7. [DOI] [PMC free article] [PubMed] [Google Scholar]