Abstract
Mitochondria are cellular organelles essential for the function and survival of eukaryotic cells. Nearly all mitochondrial proteins are nuclear-encoded and require mitochondrial import upon their synthesis in the cytosol. Various approaches have been described to study mitochondrial protein import, such as monitoring the entry of radiolabeled proteins into purified mitochondria or quantifying newly synthesized proteins within mitochondria by proteomics. Here, we provide a detailed protocol for a commonly used and straightforward assay that quantitatively examines mitochondrial protein import by monitoring the co-localization of mitochondrially targeted enhanced green fluorescent protein (eGFP) with the mitochondrial fluorescence dye MitoTracker TM Deep Red FM by live cell imaging. We describe the preparation and use of a stable mammalian cell line inducibly expressing a mitochondrial targeting sequence (MTS)-eGFP, followed by quantitative image analysis using an open-source ImageJ-based plugin. This inducible expression system avoids the need for transient transfection while enabling titration of MTS-eGFP expression and thereby avoiding protein folding stress. Overall, the assay provides a simple and robust approach to assess mitochondrial import capacity of cells in various disease-related settings.
This protocol was validated in: Mol Cell (2021), DOI: 10.1016/j.molcel.2021.11.004
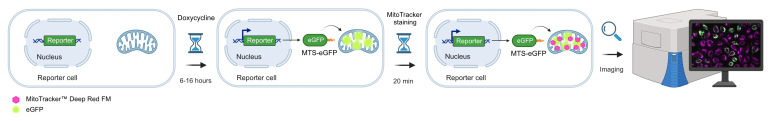
Graphical abstract
Keywords: Mitochondrial protein import , Microscopy , Mitochondria , Protein translocation , Live cell imaging
Background
Mitochondria are crucial for cellular survival. Almost all mitochondrial proteins are encoded in the nuclear genome and proper mitochondrial function and biogenesis rely on mitochondrial protein import ( Dimogkioka et al., 2021 ). Multiple mitochondrial protein import machineries exist; each mediates protein translocation into a distinct mitochondrial sub-compartment ( Schmidt et al., 2010 ). Stress conditions, such as the loss of mitochondrial membrane potential, are known to disturb mitochondrial protein import, and defective import can lead to a series of pathologies, such as neurodegenerative and cardiovascular diseases and cancer ( Geissler et al., 2000 ; Palmer et al., 2021 ).
To investigate mitochondrial protein import, several methods have been established. Radiolabeling of individual mitochondrial proteins followed by incubation with purified mitochondria allows following mitochondrial import of the labeled protein (Murschall et al., 2021; Poveda-Huertes et al., 2021). Besides the technical (e.g., purification of fully active mitochondria) and bureaucratic requirements of performing radioactive lab work, this in vitro approach lacks cellular context and thus does not account for potential contribution of cytosolic factors on protein import. Recently developed proteomics methods allow monitoring the import of newly synthesized proteins into mitochondria on the global level ( Schafer et al., 2022 ). This method is carried out within the cellular context and allows monitoring hundreds of mitochondrial proteins; however, it is a complex proteomics method that requires a mass spectrometry infrastructure not available in most laboratories. Here, we describe a fluorescence microscopy–based approach that allows monitoring mitochondrial import of the previously established mitochondrial targeting sequence (MTS)–enhanced green fluorescent protein (eGFP) reporter protein by live cell imaging ( Chen et al., 2003 ). The reporter consists of eGFP that is selectively targeted to the mitochondrial matrix by means of a N-terminal MTS, which consists of the first 69 amino acids of subunit 9 of the F0-ATPase. Transcription of the reporter gene can be induced with the Tet-On system by treating cells with doxycycline, which allows for a controlled and titratable MTS-eGFP expression. Defective import of the reporter is assessed by its mislocalization to the cytosol, which is monitored by co-staining of mitochondria with MitoTracker TM Deep Red FM. Subsequent co-localization analysis with the ImageJ-implemented Coloc2 plugin allows assessing mitochondrial protein import defects in a quantitative manner.
Materials and Reagents
-
Cultivating cell lines
10 cm tissue culture dish (Sarstedt, catalog number: 83.3902)
6-well cell culture plate (Sarstedt, catalog number: 83.3920.005)
Cell counting slides (dual-chamber) (Bio-Rad, catalog number: 1450015)
0.45 µm filters (Whatman, catalog number: 10462100)
-
HeLa FlpIn TRex cell line (Invitrogen, catalog number: R71407)
Note: This protocol was established for HeLa FlpIn TRex cell lines. It can be transferred to other cell lines that are suitable for microscopy.
HEK 293T cells (human embryonic kidney 293T) (ATCC, catalog number: CRL-3216)
RPMI 1640 medium (+L-Glutamine) (Thermo Fisher Scientific, catalog number: 21875-034)
DMEM medium (+L-Glutamine) (Thermo Fisher Scientific, catalog number 41966029)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, catalog number: 10270-106)
Trypsin-EDTA (0.25%), phenol red (Thermo Fisher Scientific, catalog number: 25200056)
Dulbecco's phosphate buffered saline (PBS) (Thermo Fisher Scientific, catalog number: 14190-169)
Trypan blue solution (Sigma-Aldrich, catalog number: T8154)
-
Stable and inducible MTS-eGFP cell line generation
15 mL tube (Greiner, catalog number: 188271-N)
Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, catalog number: 11668027)
Puromycin (InvivoGen, catalog number: ant-pr-1)
Opti-MEM I (Thermo Fisher Scientific, catalog number: 31985-047)
Polybrene (Sigma-Aldrich, catalog number: TR-1003-G)
pHDM-VSV-G (Addgene, catalog number: 164440); encodes VSV-G protein
pHDM-Hgpm2 (Addgene, catalog number: 164441); encodes codon-optimized HIV gag-pol
pHDM-tatIB (Addgene, catalog number: 164442); encodes HIV-1 Tat accessory protein
pRC-CMV-revIB (Addgene, catalog number: 164443); encodes HIV-1 Rev accessory protein
pLD-puro-MTS-EGFP (Addgene, catalog number: 190270); contains tetracycline/doxycycline-inducible MTS-eGFP and lentiviral packaging signal
-
MTS-eGFP mitochondrial import assay
Cell culture microplate, 96-well, black (Greiner, catalog number: 655090)
MitoTracker TM Deep Red FM (Thermo Fisher Scientific, catalog number: M22426)
Doxycycline (0.25 µg/mL stock in RNAse-free H 2 O) (Sigma-Aldrich, catalog number: D9891-10G)
-
Carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (Sigma-Aldrich, catalog number: C2759)
Note: CCCP is toxic if swallowed. Avoid skin contact and wash hands thoroughly after handling.
Dimethyl sulfoxide for cell culture (DMSO) (VWR, catalog number: A3672.0100)
Gamitrinib-triphenylphosphonium (GTPP) (custom synthesized)
Equipment
-80 °C freezer (Ewald, catalog number: V86-520.1)
TC20 automated cell counter (Bio-Rad, catalog number: 1450102)
CQ-1 confocal quantitative (Yokogawa) or other live cell microscope with 60× magnification (488 nm, 640 nm laser)
Emission filters 525/50 nm for eGFP (CQ-1, Yokogawa)
Emission filters 685/40 nm for MitoTracker TM Deep Red FM (CQ-1, Yokogawa)
Incubator for mammalian cell culture (Thermo Fisher Scientific, catalog number: 16416639)
Software
ImageJ (NIH, version: 1.53e)
Procedure
-
Generation of a stable and inducible MTS-eGFP cell line with lentiviral particles
Note: All steps described in section A have to be conducted in a laboratory approved for lentiviral work.
Seed 3.5 × 10 6 HEK 293T cells in a 10 cm tissue culture dish containing DMEM (v/v) 10% FBS medium to obtain a density of 80% confluence the next day.
Exchange medium with 9 mL of pre-warmed DMEM (v/v) 1% FBS the day after seeding.
Mix plasmids with corresponding ratio (5 μg total DNA for 10 cm culture dish) of pHDM-VSV-G (ratio: 2, 0.4 µg), pHDM-Hgpm2 (ratio: 1, 0.2 µg), pHDM-tatIB (ratio: 1, 0.2 µg), pRC-CMV-revIB (ratio: 1, 0.2 µg), and pLD-puro-MTS-EGFP (ratio: 20, 4 µg) with 500 µL of Opti-MEM I in tube number 1.
-
Mix 15 µL of lipofectamine 2000 transfection reagent with 500 µL of Opti-MEM I in tube number 2.
Note: Instead of Lipofectamine 2000, other transfection reagents can be used.
Add DNA mixture from tube number 1 to tube number 2, mix, and incubate for 10 min at room temperature.
Add mix to the cells in a dropwise manner to reach 10 mL in total.
Exchange medium with 10 mL of pre-warmed DMEM (v/v) 10% FBS after 6 h.
Harvest lentiviral particles after 48 h by collecting medium.
-
Spin the collected supernatant at 2000 × g for 3 min to pellet transferred HEK 293T cells. Transfer the supernatant containing lentiviral particles to a new tube.
Note: Alternatively, 0.45 µm filters can be used to remove transferred HEK 293T cells.
Store the lentiviral particle containing supernatant at 4 °C for up to one week or at -80 °C for longer storage.
Seed 2 × 10 6 HeLa FlpIn TRex cells in a 10 cm tissue culture dish containing RPMI (v/v) 10% FBS medium to obtain a density of 60% confluence the next day.
-
Add 0.8–1 mL of supernatant containing lentiviral particles obtained in step A9 per 10 cm dish with polybrene (final concentration of 8 µg/mL) the day after seeding.
Note: Do not remove the culture medium before adding the supernatant containing lentiviral particles. Also, it is not necessary to determine the concentration of lentiviral particles in the supernatant.
-
Incubate cells with lentiviral particles for 48 h.
Note: Exchange the medium with fresh, pre-warmed RPMI (v/v) 10% FBS medium during the incubation period if the cells show signs of stress.
Start selection of lentivirus-infected HeLa FlpIn TRex cells by puromycin addition (final concentration of 1 µg/mL) to the RPMI (v/v) 10% FBS medium, 48 h after transduction.
-
Select for lentivirus-infected cells for a total of 11 days and by a minimum of three passages in RPMI (v/v) 10% FBS medium containing 1 µg/mL puromycin.
Note: The generated cell line is stable, can be frozen in RPMI (v/v) 10% FBS medium containing 10% DMSO (v/v) at -80 °C and stored ≤ -150 °C. Cells can be cultured in regular RPMI (v/v) 10% FBS medium after completing puromycin selection for 11 days.
-
Cell seeding
-
Aspirate medium from a 10 cm tissue culture dish containing HeLa FlpIn TRex cells carrying the genomically integrated MTS-eGFP reporter grown in RPMI (v/v) 10% FBS at 60%–80% confluence.
Note: This step is performed with cells obtained in the previous step, i.e., after 11 days of puromycin selection.
Wash cell layer once with 5 mL of room temperature PBS and aspirate.
Add 1 mL of 0.25% trypsin to cell layer and incubate the plate for 5 min in the incubator.
Add 9 mL of RPMI (v/v) 10% FBS to stop tryptic reaction and resuspend cells by pipetting up and down repeatedly.
Collect cell suspension in a 15 mL tube and pellet cells by centrifugation at 800 × g for 3 min.
Aspirate supernatant and resuspend cells in 5 mL of RPMI (v/v) 10% FBS by pipetting up and down repeatedly.
Transfer 5 µL of cell suspension to a 1.5 mL tube and mix with 5 µL of trypan blue solution.
Transfer 8.5 µL of stained cell suspension to a counting slide and count viable cells.
Seed 10–20,000 viable cells per well of a black 96-well cell culture microplate in RPMI (v/v) 10% FBS and grow cells overnight in the incubator.
-
-
Treatment, induction of MTS-eGFP expression, and mitochondrial staining
-
Prepare treatment solution containing the compound or vehicle at the appropriate concentration and 0.25 µg/mL doxycycline to induce MTS-eGFP expression in pre-warmed RPMI (v/v) 10% FBS.
Note: According to Moullan et al. (2015), the used doxycycline concentration should not affect mitochondrial function in HeLa cells.
-
Start treatment and doxycycline induction by aspirating the growth medium and adding the treatment medium to the cells in the 96-well plate.
Note: The protocol was tested for 6 h of treatment with 10 µM CCCP, a mitochondrial uncoupler, and 10 µM GTPP, a mitochondrial chaperone inhibitor, both of which rapidly reduce mitochondrial protein import and might be used as positive controls ( Michaelis et al., 2022 ). DMSO was used as vehicle control.
Incubate cells in the incubator during treatment.
Aspirate treatment medium and stain cells with 50 nM MitoTracker TM Deep Red FM in pre-warmed RPMI (v/v) 10% FBS for 20 min in the incubator.
Aspirate medium, wash cells once with PBS, and add fresh RPMI (v/v) 10% FBS.
-
-
Live cell imaging
Transfer the 96-well plate with cells to the Yokogawa CQ-1 microscope chamber pre-equilibrated to 37 °C, 5% CO 2 .
-
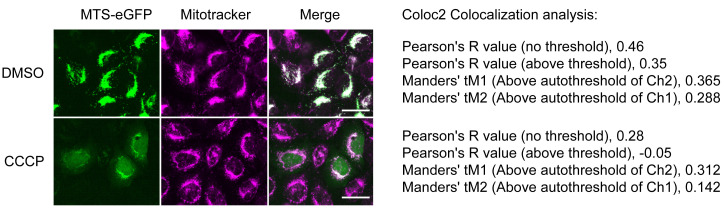
Acquire live cell images of eGFP (488 nm excitation, 525/50 nm emission) and MitoTracker TM Deep Red FM (640 nm excitation, 685/40 nm emission) of at least 100 cells per replicate at 60× magnification with automated focus (example images shown in Figure 1 ).
Notes:
Adjust the laser power and exposure times (maximum 500 ms) to obtain non-saturated images.
It is recommended to acquire z-stacks to obtain in-focus images of as many cells as possible per field of view. Z-stacks are typically acquired with 6–10 slides within 3–5 µm radius around the central focus.
Figure 1. Live cell images of MTS-eGFP and MitoTracker TM Deep Red FM upon DMSO and CCCP treatment.
HeLa FlpIn TRex cells carrying MTS-eGFP were treated for 6 h with doxycycline and 10 µM CCCP or 1 µL/mL DMSO as vehicle control (left). Scale bar represents 25 µm. Results of correlation-based co-localization analysis using ImageJ Coloc2 are shown (right).
Data analysis
Use ImageJ ( Schneider et al., 2012 ) and the implemented Coloc2 plugin for co-localization analysis.
Open images of eGFP and MitoTracker TM Deep Red FM of the same field of view in ImageJ and start Coloc2, following Analyze – Colocalization - Coloc2.
-
Select the images to be analyzed in Channel 1 and 2; select Bisection as Threshold regression and Manders’ correlation in the Coloc2 selection window ( Figure 2 ).
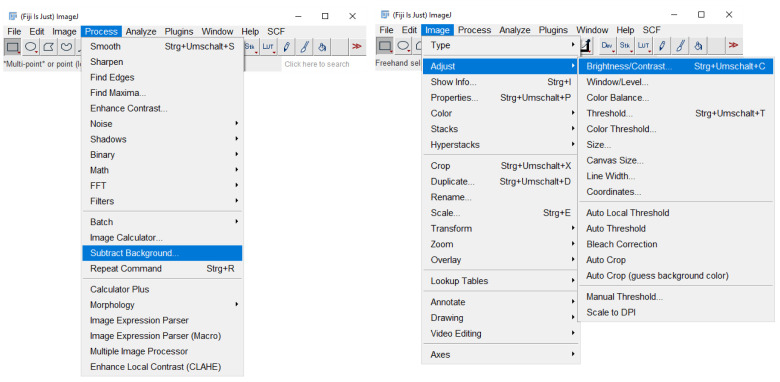
Note: Coloc2 also calculates the Pearson’s coefficient, which is based on another correlation-based co-localization method. The coefficients range from -1 to +1 for Pearson’s and from 0 to 1 for Manders’. The values correlate with the confidence of co-localization. For visualization purposes, perform background subtraction on individual image files using the background subtraction feature in ImageJ and adjust brightness of images to comparable intensities ( Figure 3 ).
Figure 2. Coloc2 selection window.
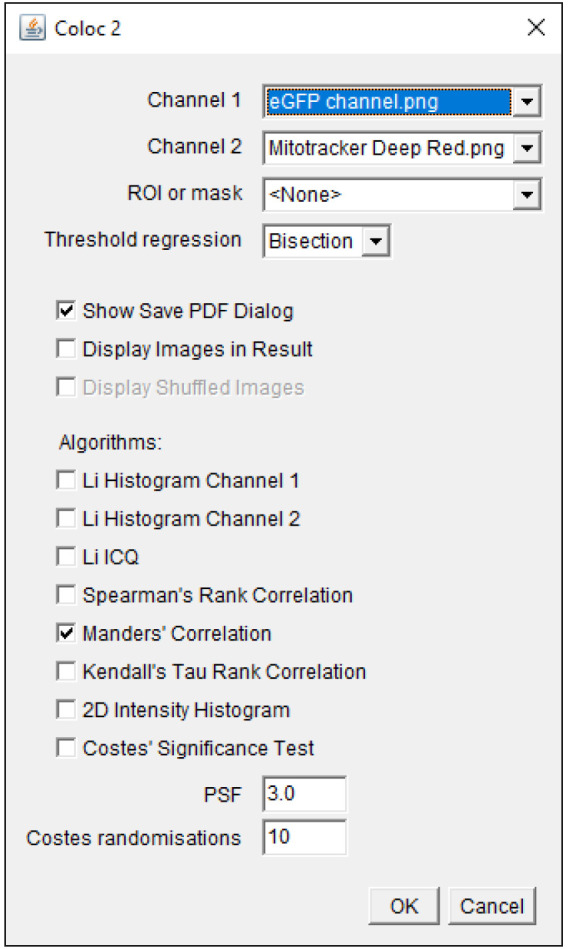
Images for co-localization analysis are selected in Channels 1 and 2. The parameters used for co-localization analysis are displayed.
Figure 3. Image adjustment by background subtraction, brightness, and contrast adjustment in ImageJ.
This figure demonstrates how to access the implemented features for figure preparation.
Notes
This assay can also be combined with siRNA knock downs. For this, MTS-eGFP expression is induced by doxycycline treatment after 2–4 days of siRNA transfection.
Acknowledgments
The MTS-eGFP protein import assay was adapted from Chen et al. (2003). Plasmid lentiviral destination (pLD) was established as described previously by Mak et al. (2010). C.M. acknowledges support from the Deutsche Forschungsgemeinschaft (DFG) SFB 1177 (subproject D08) Project-ID 259130777, Emmy Noether Programm Project ID 390339347, and Excellence Strategy Program of the DFG (Exc 2026), and the European Research Council (ERC) Starting Grant 803565. The protocol was established as part of the following research papers: Schafer et al. (2022) and Michaelis et al. (2022). The graphical abstract was created with BioRender.com.
Competing interests
The authors declare that they have no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Chen H. , Detmer S. A. , Ewald A. J. , Griffin E. E. , Fraser S. E. and Chan D. C. ( 2003 . ). Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development . J Cell Biol 160 ( 2 ): 189 - 200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dimogkioka A. R. , Lees J. , Lacko E. and Tokatlidis K. ( 2021 . ). Protein import in mitochondria biogenesis: guided by targeting signals and sustained by dedicated chaperones . RSC Adv 11 ( 51 ): 32476 - 32493 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Geissler A. , Krimmer T. , Bomer U. , Guiard B. , Rassow J. and Pfanner N. ( 2000 . ). Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b(2) modulates the deltapsi-dependence of translocation of the matrix-targeting sequence . Mol Biol Cell 11 ( 11 ): 3977 - 3991 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mak A. B. , Ni Z. , Hewel J. A. , Chen G. I. , Zhong G. , Karamboulas K. , Blakely K. , Smiley S. , Marcon E. , Roudeva D. , et al. .( 2010 . ). A lentiviral functional proteomics approach identifies chromatin remodeling complexes important for the induction of pluripotency . Mol Cell Proteomics 9 ( 5 ): 811 - 823 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michaelis J. B. , Brunstein M. E. , Bozkurt S. , Alves L. , Wegner M. , Kaulich M. , Pohl C. and Munch C. ( 2022 . ). Protein import motor complex reacts to mitochondrial misfolding by reducing protein import and activating mitophagy . Nat Commun 13 ( 1 ): 5164 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moullan N. , Mouchiroud L. , Wang X. , Ryu D. , Williams E. G. , Mottis A. , Jovaisaite V. , Frochaux M. V. , Quiros P. M. , Deplancke B. , et al. .( 2015 . ). Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research . Cell Rep 10 ( 10 ): 1681 - 1691 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murschall L. M. , Peker E. , MacVicar T. , Langer T. and Riemer J. ( 2021 . ). Protein Import Assay into Mitochondria Isolated from Human Cells . Bio Protoc 11 ( 12 ): e4057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palmer C. S. , Anderson A. J. and Stojanovski D. ( 2021 . ). Mitochondrial protein import dysfunction: mitochondrial disease, neurodegenerative disease and cancer . FEBS Lett 595 ( 8 ): 1107 - 1131 . [DOI] [PubMed] [Google Scholar]
- 9. Poveda-Huertes D. , Taskin A. A. , Dhaouadi I. , Myketin L. , Marada A. , Habernig L. , Buttner S. and Vogtle F. N. ( 2021 . ). Increased mitochondrial protein import and cardiolipin remodelling upon early mtUPR . PLoS Genet 17 ( 7 ): e1009664 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schafer J. A. , Bozkurt S. , Michaelis J. B. , Klann K. and Munch C. ( 2022 . ). Global mitochondrial protein import proteomics reveal distinct regulation by translation and translocation machinery . Mol Cell 82 ( 2 ): 435 - 446 e437 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmidt O. , Pfanner N. and Meisinger C. ( 2010 . ). Mitochondrial protein import: from proteomics to functional mechanisms . Nat Rev Mol Cell Biol 11 ( 9 ): 655 - 667 . [DOI] [PubMed] [Google Scholar]
- 12. Schneider C. A. , Rasband W. S. and Eliceiri K. W. ( 2012 . ). NIH Image to ImageJ: 25 years of image analysis . Nat Methods 9 ( 7 ): 671 - 675 . [DOI] [PMC free article] [PubMed] [Google Scholar]