Abstract
Four pneumococcal genes (phtA, phtB, phtD, and phtE) encoding a novel family of homologous proteins (32 to 87% identity) were identified from the Streptococcus pneumoniae genomic sequence. These open reading frames were selected as potential vaccine candidates based upon their possession of hydrophobic leader sequences which presumably target these proteins to the bacterial cell surface. Analysis of the deduced amino acid sequences of these gene products revealed the presence of a histidine triad motif (HxxHxH), termed Pht (pneumococcal histidine triad) that is conserved and repeated several times in each of the four proteins. The four pht genes (phtA, phtB, phtD, and a truncated version of phtE) were expressed in Escherichia coli. A flow cytometry-based assay confirmed that PhtA, PhtB, PhtD and, to a lesser extent, PhtE were detectable on the surface of intact bacteria. Recombinant PhtA, PhtB, and PhtD elicited protection against certain pneumococcal capsular types in a mouse model of systemic disease. These novel pneumococcal antigens may serve as effective vaccines against the most prevalent pneumococcal serotypes.
Streptococcus pneumoniae is a leading bacterial cause of otitis media, meningitis, and pneumonia in the United States and throughout the world (13, 16). Among infants and young children, acute otitis media (AOM) is the most common disease caused by this pathogen (12). It has been estimated that approximately 75% of all children experience at least one episode of AOM and that more than half of these cases are caused by S. pneumoniae (13). S. pneumoniae accounts for nearly 7 million cases of middle ear infections in children under 2 years of age in the United States alone (16). The importance of providing effective prophylactic vaccination for AOM has increased with the emergence of antibiotic-resistant strains of S. pneumoniae (5).
The pneumococcal vaccine in current use is a 23-valent composition of pneumococcal capsular polysaccharides (PS). Whereas this vaccine has been shown to be protective in adults (31), it is of limited use in infants and young children since the latter populations are poor responders to PS antigens (10, 11). To overcome this limitation and to enhance immunogenicity, PS from the predominant pneumococcal serotypes have been conjugated with immunogenic T-cell-dependent carrier proteins, and this vaccine has been approved recently for use in infants (15, 17). While almost 80% of cases of pneumococcal bacteremia and approximately 65% of cases of otitis media (4, 6) in the United States are caused by the seven major serotypes (4, 6B, 9V, 14, 18C, 19F, and 23F) (33) that are included in heptavalent conjugate vaccines, other serotypes that are predominant in other regions of the world are also important in pneumococcal disease (7). Furthermore, the use of vaccines with limited specificity might increase the carriage of pneumococcal serotypes not represented in the formulation (22). Finally, repeated use of the same carrier protein could reduce the immunogenicity of the conjugates (8).
Given these potential limitations associated with conjugated-PS vaccines, much recent attention has been focused on the identification of potential protein-based vaccines. Several pneumococcal proteins, including pneumolysin (18), PsaA (9, 36), PspA (3), and CbpA (M. Dormitzer, T. M. Wizemann, J. E. Adamou, B. Walsh, T. Gayle, S. Koenig, S. Langermann, and J. Johnson, Abstr. 98th Gen. Meet. Am. Soc. Microbiol. 1998, abstr. B-3, p. 56, 1998), also described as PspC (3), are currently being investigated as vaccine targets. Some of these proteins have been shown to be protective in animal models of colonization and systemic disease (25). However, several of these proteins have also been shown to have substantial sequence heterogeneity among different serotypes.
We describe here the characterization of a novel family of cell surface-exposed pneumococcal proteins (the Pht family), including some members that can induce antibodies capable of protecting mice against pneumococcal sepsis and death. These proteins were identified from the S. pneumoniae genome database and were selected based on their putative hydrophobic leader sequences, which are characteristic of proteins exported across the cytoplasmic membrane (14, 26). These novel pneumococcal antigens, either alone or in combination with capsular polysaccharides, could serve as effective vaccines against the most prevalent pneumococcal serotypes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Twenty-three pneumococcal strains, one isolate representative of each of the 23 serotypes contained in multivalent pneumococcal PS vaccines, were obtained from the American Type Culture Collection (ATCC; Manassas, Va.). Additional strains and plasmids used in this study are listed in Table 1. Pneumococcal cultures were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY; Difco, Detroit, Mich.) at 37°C under a 5% CO2 atmosphere or on BBL tryptic soy agar plates containing 5% sheep blood (TSA II; Becton Dickinson, Cockeysville, Md.).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Capsule type | Comment(s) | Sourcea |
|---|---|---|---|
| S. pneumoniae | |||
| WU2 | 3 | Laboratory strain | D. Briles |
| Norway 4 | 4 | Strain used for genomic sequencing | I. Aaberg |
| BG9739 | 4 | Blood isolate from patient with septicemia | D. Briles |
| EF5668 | 4 | Clinical isolate | D. Briles |
| D211 | 4 | Middle ear fluid isolate | R. Dagan |
| D33965 | 4 | Blood isolate | R. Dagan |
| EF6796 | 6A | Blood isolate | D. Briles |
| SJ2 | 6B | Nasal isolate and animal passage strain | P. Flynn |
| D016 | 6B | Middle ear fluid isolate | R. Dagan |
| D17079 | 6B | Blood isolate | R. Dagan |
| D60 | 9V | Middle ear isolate | R. Dagan |
| D3008 | 9V | Cerebral spinal fluid isolate | R. Dagan |
| D17978 | 9V | Blood isolate | R. Dagan |
| SJ9 | 14 | Nasal isolate from patient with pneumonia | P. Flynn |
| D118 | 14 | Middle ear isolate | R. Dagan |
| D105 | 14 | Nasopharyngeal isolate from child with ear infection | R. Dagan |
| D665 | 14 | Blood isolate | R. Dagan |
| SJ12 | 19A | Nasal isolate from patient with fever | P. Flynn |
| SJ1 | 23F | Sputum isolate from patient with cough | P. Flynn |
| SJ6 | 23F | Eye isolate from patient with conjunctivitis | P. Flynn |
| SJ13 | 23F | Nasal isolate from patient with pneumonia | P. Flynn |
| D020 | 23F | Middle ear fluid isolate | R. Dagan |
| D7021 | 23F | Blood isolate | R. Dagan |
| E. coli M15(pREP4) | Host strain for pQE10 | Qiagen | |
| Plasmid pQE10 | E. coli expression vector | Qiagen |
Pneumococcal isolates were obtained from the collections of David Briles (University of Alabama, Birmingham), Ingeborg Aaberge (National Institute of Public Health, Oslo, Norway), Ron Dagan (Seroka Medical Center, Beer-Sheeva, Israel), and Pat Flynn (St. Jude Children's Research Hospital, Memphis, Tenn.) as indicated.
Cloning and expression of pht genes.
The genome of S. pneumoniae (serotype 4 strain; N4) was sequenced by the whole-genome random sequencing method as previously described (T. M. Wizemann et al., submitted for publication). PhtA (originally called Sp36, GenBank accession no. AF291695) was identified from the deduced amino acid sequence of open reading frames (ORFs) predicted by GeneMark software (Gene Pro, Inc., Atlanta, Ga.) by searching for a processing site predicted to be recognized by signal peptidase II (lipoprotein motif) (LxxC; reviewed in reference 35). The ORF encoding PhtA beginning at serine (residue 21) was amplified by PCR using the forward and reverse primer sets indicated in Table 2. The PCR fragment was digested with BamHI and HindIII, ligated into the similarly digested expression vector pQE10 (Qiagen, Chatsworth, Calif.), and transformed into the Escherichia coli host strain M15 (pREP4) (Qiagen). The identity of the cloned DNA fragment was verified by DNA sequencing. The recombinant polyhistidine-tagged PhtA fusion protein (His6PhtA) was purified under denaturing conditions by metal affinity chromatography using Ni-nitrilotriacetic acid resin (Qiagen) as described by the manufacturer, and the denatured protein was dialyzed against phosphate-buffered saline (PBS; pH 7.5) to promote refolding. Purity of the recombinant protein was evaluated by Coomassie blue staining of a sodium dodecyl sulfate (SDS)-polyacrylamide gel. PCR primers sets (Table 2) were used for amplification of phtA (N-terminal half; nucleotides 61 to 1083), phtA (C-terminal half; nucleotides 1159 to 2448), phtB (nucleotides 61 to 2460), phtD (nucleotides 61 to 2520), and phtE (N-terminal half; nucleotides 64 to 1455). Expression and purification procedures were similar to that described above for His6PhtA.
TABLE 2.
Sequence of oligonucleotide primers used to amplify the pht genes from S. pneumoniae N4a
| pht genes | Primerb | Sequence (5′-3′) |
|---|---|---|
| phtA | Primer 1 (F) | ATCGGATCCTTCTTACGAGTTGGGACTGTATCAAGC |
| Primer 2 (R) | AGTCAAGCTTGTTTATTTTTTCCTTACTTACAGATGAAGG | |
| phtB | Primer 3 (F) | TGCCCTAAGTGTTTGTCCCGGGTCCTATGAGCT |
| Primer 4 (R) | AGGTCGAGAAAGCTTTTACTTACTCTCCTTTAATAAAGCCAATAG | |
| phtD | Primer 5 (F) | GCATGCTCCTATGAACTTGGTCGTCA |
| Primer 6 (R) | AAGCTTTTACTATATAGGAGCCGGTTGACT | |
| phtE (N terminal) | Primer 7 (F) | GCATGCGCCCTATGCACTAAACCAGCA |
| Primer 8 (R) | CTGCAGCTAAATGTTTTTTGCGCACCT | |
| phtA (N terminal) | Primer 9 (F) | GGATCCTCTTACGAGTTGGGACTGTATCA |
| Primer 10 (R) | AAGCTTTACCCAATGGTTTGAACGAT | |
| phtA (C terminal) | Primer 11 (F) | GGATCCCTTAAAATAGACTCAAATTCTTCTTTGG |
| Primer 12 (R) | AAGCTTGTTTATTTTTTCCTTACTTACAGATGAA |
Nucleotides underlined in each primer sequence show the positions of the restriction endonuclease sites incorporated to facilitate cloning.
F, forward primer; R, reverse primer.
Southern blot analysis.
Genomic DNA was prepared from each of the 23 pneumococcal strains obtained from the ATCC, as well as from strain SJ2. DNA (ca. 5 μg) was digested with PvuII and BamHI, separated through an 0.8% agarose gel, and transferred to a Hybond-N membrane (Amersham-Pharmacia Biotech, Inc., Piscataway, N.J.). A DNA probe was prepared by amplifying the phtA gene (described above) from strain N4 DNA and labeling the amplified fragment with fluorescein using a random-prime labeling kit (Amersham-Pharmacia). Southern blot analysis was performed under stringent conditions in hybridization buffer (Amersham-Pharmacia) containing 5× standard saline citrate (SSC), 0.5% (wt/vol) blocking agent, 0.1% (wt/vol) SDS, 5% (wt/vol) dextran sulfate, and 100 μg of heterologous DNA per ml at 60°C. Blots were washed with 1× SSC–0.1% (wt/vol) SDS for 15 min at 60°C followed by washing with 0.1× SSC– 0.1% (wt/vol) SDS for 15 min at 60°C. SSC (1×) is composed of 150 mM sodium chloride and 15 mM sodium citrate (pH 7.0). Hybridization signals were detected with an anti-fluorescein antibody conjugated to horseradish peroxidase (HRP) using the ECL kit from Amersham-Pharmacia.
Preparation of whole-cell lysates of S. pneumoniae.
Whole-cell lysates were prepared as previously described (38). The total pneumococcal protein in each lysate was quantitated by the bicinchoninic acid method (BCA Protein Assay Reagent; Pierce, Rockford, Ill.), and lysates were stored at −70°C.
Mouse active immunizations and challenge studies.
C3H/HeJ (C3H) or BALB/cByJ (BALB) mice (6- to 8-week-old females; Jackson Laboratories, Bar Harbor, Maine) were used for protection studies. Animals (10 mice/group) were immunized subcutaneously with recombinant proteins (15 μg in 50 μl of PBS emulsified in 50 μl of complete Freund adjuvant [CFA]). Control mice received PBS with adjuvant alone (sham immunization). Mice were boosted at 3 weeks with an additional 15 μg of protein in incomplete Freund adjuvant (IFA), while the sham group received PBS with IFA. Blood was drawn at week 7, and sera from each group were pooled for analysis by enzyme-linked immunosorbent assay (ELISA). Mice were challenged at week 8 by an intraperitoneal (i.p.) injection of 15 to 100 times the 50% lethal dose (LD50) of the indicated S. pneumoniae strain. The actual number of bacteria injected was determined by plating for CFU. Deaths were recorded daily for 15 days. The virulence of strain SJ2 for mice was enhanced by infecting mice via i.p. injection and isolation of the pneumococci from the peripheral blood.
Generation of rabbit immune sera and passive immunization studies.
New Zealand White rabbits were immunized intradermally with 250 μg of His6PhtA, His6PhtD, or His6PhtE in CFA. On days 29 and 50 the rabbits were boosted with 125 μg of protein in IFA given by s.c. injection. Serum was collected 10 days following the second boost. Rabbit serum was also prepared against a negative control antigen (E. coli, FimC) (20).
For passive immunization studies, 100 μl of rabbit serum (preimmune or hyperimmune) was administered i.p. to C3H or BALB mice 24 h prior to and 4 h following challenge with S. pneumoniae. Mice were monitored for survival as described above for active immunization studies. All animal studies were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at MedImmune, Inc.
Immunoassays. (i) ELISA.
Microtiter wells (Immulon Plate 4; Nunc, Naperville, Ill.) were coated overnight at 4°C with 50 μl of 1 μg of recombinant protein per ml in 0.1 M carbonate buffer (pH 9.6). The plates were washed with PBS containing 0.1% Tween 20 (Sigma Chemical Co.) and blocked for 1 h at room temperature with PBS containing 1% bovine serum albumin (Sigma Chemical Co.) and 1% nonfat dry milk. Twofold serial dilutions of pooled serum were added, and the plates were incubated for 1 h at room temperature. The plates were washed and then incubated with goat anti-mouse immunoglobulin G (IgG) or goat anti-rabbit IgG, both conjugated to HRP (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). After incubation for 1 h, the plates were washed, bound antibody was visualized with 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) diammonium (ABTS) substrate reagent (Kirkegaard & Perry Laboratories), and the absorbance measured at 405 nm. Endpoint titers were determined as the last dilution achieving a twofold increase in signal compared to sera from animals immunized with adjuvant alone.
(ii) SDS-PAGE and immunoblot analysis.
Proteins in whole-cell lysates were separated on an SDS–10% polyacrylamide gel electrophoresis (PAGE) gel (Bio-Rad, Hercules, Calif.) by the method of Laemmli (19) and transferred to nitrocellulose membranes electrophoretically. Membranes were blocked overnight at 4°C in PBS and 0.25% Triton X-100 (PBS-T) containing 5% nonfat dry milk. The membranes were washed two times in PBS-T with 1% nonfat dry milk at room temperature and then incubated for 1 h at room temperature in antiserum diluted to 1:5,000 in wash buffer. The membranes were washed with PBS-T and incubated with HRP-conjugated goat anti-mouse antibody (Amersham-Pharmacia) diluted to 1:5,000 in PBS-T. HRP was detected by exposure to film with ECL reagent (Amersham-Pharmacia). Molecular weight markers (Rainbow markers) were purchased from Amersham-Pharmacia.
Immunoblots of recombinant (His6) Pht proteins were probed with acute or convalescent sera (both diluted to 1:3,000) from patients with culture-confirmed pneumococcal bacteremia. Bound antibody was detected with HRP-conjugated goat anti-human immunoglobulin (Amersham-Pharmacia) and ECL reagent.
(iii) Flow-cytometric analysis of S. pneumoniae.
Mid-log-phase pneumococci were washed once in PBS containing 5% heat-inactivated fetal calf serum (FCS; BioWhittaker, Walkersville, Md.) and adjusted to 5 × 106 bacteria in 0.1 ml. Immune sera (diluted to 1:100) were added, and bacterial cells were held on ice for 1 h. Excess antibody was washed off by centrifugation in 1 ml of PBS containing FCS (wash buffer). Alexa 488-conjugated goat anti-mouse or anti-rabbit immunoglobulin (both from Molecular Probes, Eugene, Oreg.) were added to 1 μg per 5 × 106 bacteria and placed on ice for 30 min. After being washed, the samples were suspended in wash buffer and analyzed with a Becton Dickinson FACStar Plus flow cytometer using Lysys II software for data acquisition and analysis. Antisera to either CbpA or PspA, two known surface-exposed proteins from pneumococci (3, 31), labeled the bacterial cell surface. Antibody to GroEL (Epicentre Technologies, Madison, Wis.), a cytoplasmic protein, did not bind to pneumococci.
Statistics.
A two-sample log-rank test (39) was used to evaluate protection against death in the mouse model of systemic disease.
Nucleotide sequence accession numbers.
The sequence data for phtB, phtD, and phtE have been submitted to the DDBJ, EMBL, and GenBank databases under accession numbers AF318954, AF318955, and AF318956, respectively.
RESULTS
Identification and sequence analysis of the pht gene family.
The deduced amino acid sequence of phtA (Sp36; Wizemann et al., submitted), the first of the pneumococcal histidine triad genes identified in this family, was used to search the S. pneumoniae genome database (http://www.tigr.org/tdb/mdb/mdb.html) (translated in all six ORFs) to identify homologous ORFs. The products of several ORFs showing strong homology to PhtA were identified on four separate DNA fragments. Fragment 1 contained phtA (2,451 bp) and a second incomplete ORF of 920 bp (phtB) located near the end of the fragment. Fragment 2 and fragment 3 contained ORFs of 2,460 bp (phtC) and 2,520 bp (phtD), respectively. Subsequent PCR amplification using primers specific to the 3′ ends of phtA and phtC genes followed by Southern blot analysis revealed that phtB and phtC were fragments of the same gene, subsequently referred to as phtB (data not shown). Fragment 4 contained an ORF of 3,120 bp (phtE) and a second ORF (phtF) that contained several in-frame stop codons that may represent sequencing errors of the unedited DNA sequence or possibly natural mutational events on a pseudogene. The phtF gene was not characterized further. Subsequent analysis indicated that the pht genes are arranged in tandem pairs, with phtD and phtE encoding in the opposite orientation from phtA and phtB (data not shown).
Each of the four Pht ORFs (phtA, phtB, phtD, and phtE) is preceded by a typical ribosome binding site (32). The phtA and phtB genes encode polypeptides of 816 and 819 amino acids with calculated molecular masses of approximately 91.5 and 92.1 kDa, respectively. The phtD and phtE genes encode polypeptides of 839 and 1,039 amino acids with calculated molecular masses of approximately 93.5 and 114.6 kDa, respectively.
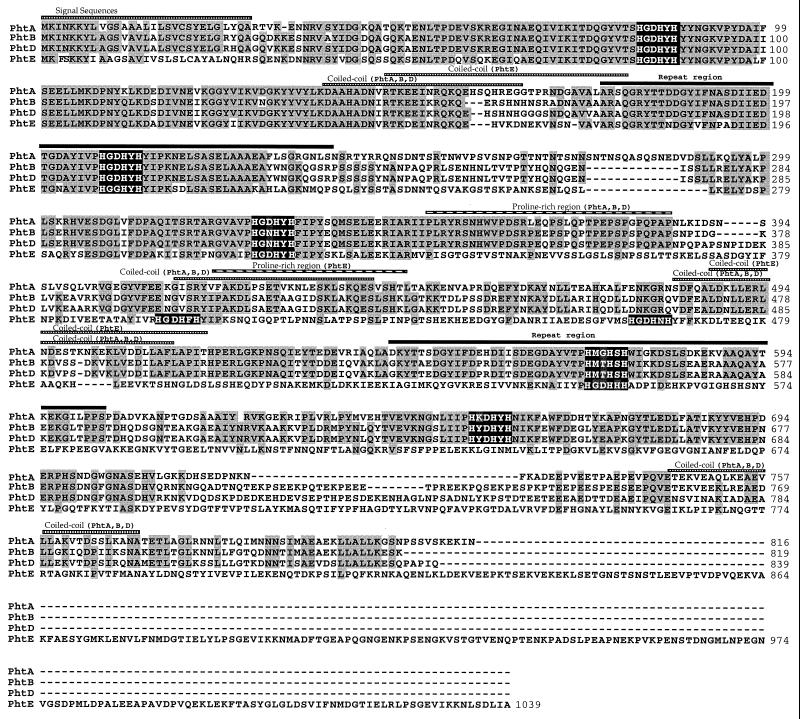
Each gene product contained five (PhtA, PhtB, and PhtD) or six (PhtE) histidine triad motifs (HxxHxH; see Fig. 1), and therefore these proteins were designated Pht (pneumococcal histidine triad) for these characteristic repeats. All four Pht proteins contain segments that are predicted by the Coils algorithm (23) to adopt a coiled-coil conformation, a feature common among gram-positive surface proteins, as well as a proline-rich region (Fig. 1). Two repeats of 61 amino acids each are found within PhtA, PhtB, PhtD, and PhtE (Fig. 1). The N terminus of all four Pht proteins contains a putative sec-dependent hydrophobic leader sequence as predicted by the SignalP algorithm (27) and a signal peptidase II motif (LxxC) (29, 38).
FIG. 1.
Multiple alignment of the Pht protein sequences from S. pneumoniae strain Norway 4. The Pht protein sequences (PhtA, PhtB, PhtD, and PhtE) were aligned using the CLUSTAL algorithm in the DNAStar computer package. Amino acid residues that are conserved in at least three Pht proteins are shaded, and gaps introduced to maximize alignments are indicated by dashed lines. The predicted signal peptides, histidine triad motifs, α-helical coiled-coil regions, proline-rich regions, and 61-amino-acid repeat regions are indicated.
The amino acid sequence identity between the various Pht proteins was determined using the CLUSTAL computer algorithm (DNAStar). PhtA is 61% identical to either PhtB or PhtD. PhtB and PhtD are the most closely related sequences, sharing 87.2% identity. The greatest divergence was seen when we compared the PhtE sequence to PhtA, PhtB, and PhtD proteins (approximately 32% identity). The N-terminal region (amino acids 30 to 350) is highly conserved between the four Pht homologs (>62% identity). Given the genomic organization, The sequence homology, and the conservation of structural features, we propose that all four Pht proteins comprise a single family of pneumococcal proteins.
Southern blot analysis.
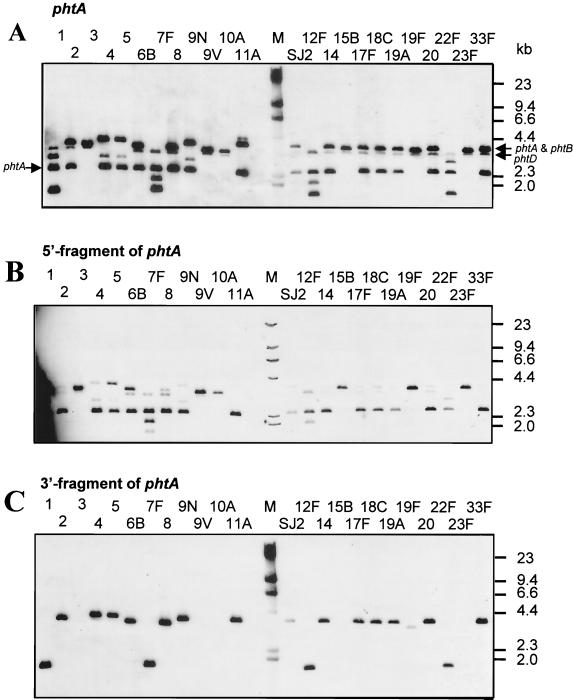
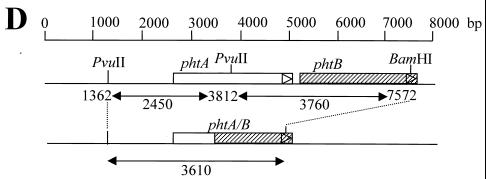
The conservation of the phtA gene among diverse pneumococcal strains was evaluated by Southern blot analysis using genomic DNA isolated from 23 serotypes represented in the current pneumococcal PS vaccine, as well as the SJ2 strain used in our animal model of pneumococcal sepsis (see below). The full-length phtA probe hybridized with DNA of all strains tested (Fig. 2A). For about half of the strains, including the SJ2 strain used in challenge studies, the pattern was that predicted from the sequence of N4 phtA and phtB. A 2,450-bp band representing the 5′ end of phtA, a 3,760-bp band representing the 3′ end of phtA, and nearly all of phtB were detected. A band just below the 3,760-bp band, probably due to hybridization of the phtA probe with the phtD gene, was also detected in most strains. In contrast, for several strains (serotypes 3, 9V, 10A, 15B, 19F, and 23F) the phtA probe hybridized with only a single band, of approximately 3,610 bp (Fig. 2A). The 3,610-bp band was also detected with the probe specific for the 5′ region of phtA (Fig. 2B) but not with the 3′ specific phtA probe (Fig. 2C). The data suggest that in some strains the tandem genes phtA and phtB may have undergone recombination, resulting in the loss of the 3′ region of phtA, the 5′ region of phtB, and the intervening region (Fig. 2D). We have confirmed this hypothesis by sequencing the proposed phtA-phtB hybrid gene from the ATCC pneumococcal strain (serotype 15B) (Fig. 2D). Several strains (serotypes 1, 6B, 12F, and 22F) had additional bands that hybridized with the phtA probe. Whether these bands represent additional pht homologues or are the result of restriction site polymorphism has not yet been determined.
FIG. 2.
Southern blot analysis of the phtA gene. Genomic DNA prepared from 23 strains represented in current pneumococcal PS vaccines, as well as strain SJ2, were digested with BamHI and PvuII. Southern blots were probed with a PCR-generated fragment encompassing the phtA gene (A), the 5′ region (B), or the 3′ region (C) of the phtA gene. Serotypes are identified at the top of each lane. HindIII-digested λ DNA (M) was included, and the molecular weights are shown in kilobases. (D) Schematic representation of the proposed recombination between phtA (open box) and phtB (hatched box) genes of certain strains of S. pneumoniae. The thin line denotes flanking genomic DNA. The phtA-phtB hybrid gene shown is based on the sequence information of pneumococcal strain, ATCC 10354 (serotype 15B). Restriction fragments expected after BamHI and PvuII digestion are shown.
Expression of Pht proteins in S. pneumoniae.
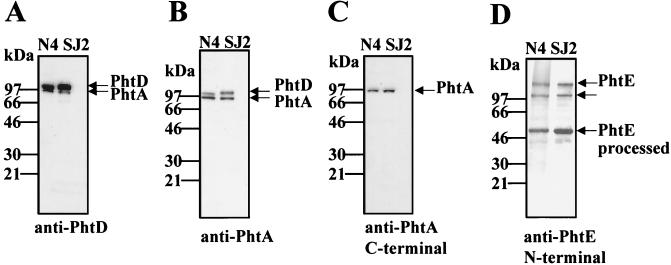
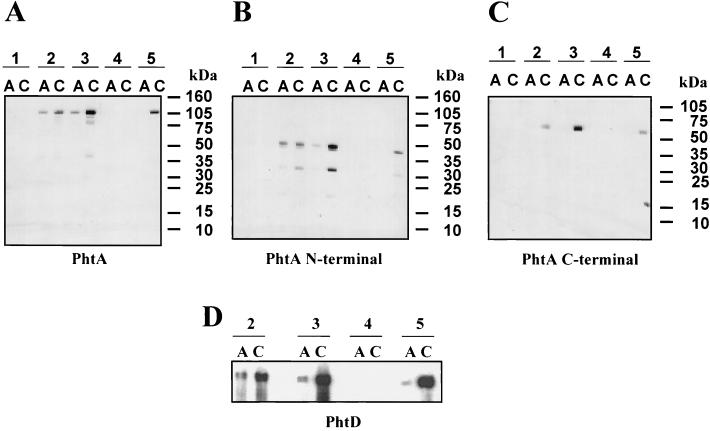
S. pneumoniae strains N4 and SJ2 were evaluated for expression of the Pht proteins by immunoblot analysis of whole-cell lysates using mouse antisera raised against either full-length PhtA, full-length PhtD, or the N-terminal portion of PhtE (Fig. 3). Antisera raised against PhtD detected a predominant protein nearly consistent with its molecular mass (98 kDa; Fig. 3A). A lower-molecular-weight protein of about 96 kDa was also detected. Similarly, antiserum raised against PhtA detected two major proteins with molecular masses of approximately 98 and 96 kDa in both lysates (Fig. 3B). To distinguish which band was PhtA, antisera specific for the N-terminal or C-terminal halves of PhtA were used for immunoblot analysis. Antiserum specific for the more-divergent C-terminal half of PhtA recognized a single band of about 96 kDa, which was nearly consistent with the predicted molecular mass of this protein (Fig. 3C). The reactivity of the antiserum specific for the N-terminal portion of PhtA was similar to that observed with antiserum raised against the full-length molecule (data not shown). Antiserum raised against the N-terminal portion of PhtE detected a predominant band of 52 kDa and two minor bands of about 120 kDa and 98 kDa in both lysates (Fig. 3D). The data suggest that PhtE is rapidly processed from a full-length protein (114.5 kDa) to a smaller, possibly active mature protein (52 kDa). The middle band (98 kDa) may also be a result of PhtE processing or may represent cross-reactivity to PhtD (Fig. 3D).
FIG. 3.
Immunoblot analysis of whole-cell lysates of S. pneumoniae. Whole-cell lysates of S. pneumoniae strains N4 and SJ2 were evaluated by immunoblot analysis with mouse antiserum raised against PhtD (residues 21 to 839) (A), PhtA (residues 21 to 816) (B), the C-terminal half of PhtA (residues 386 to 816) (C), and the N-terminal half of PhtE (residues 21 to 484) (D). The reactive bands are indicated by arrows. The molecular masses (in kilodaltons) of the protein standards are indicated.
Detection of PhtA and PhtD on the bacterial cell surface by flow cytometry.
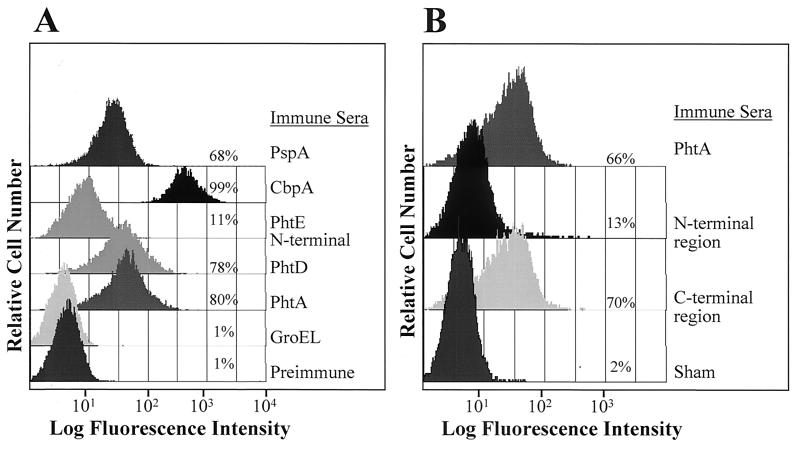
Pneumococci were incubated with hyperimmune rabbit sera raised against full-length PhtA, full-length PhtD, or the N-terminal portion of PhtE, followed by detection with a fluorescently tagged secondary antibody. As shown in Fig. 4A, antisera raised against PhtA or PhtD bound to the cell surface of the N4 strain, as detected by flow cytometry. Weaker labeling of the pneumococcal cell surface was observed with rabbit antiserum raised against the N-terminal portion of PhtE. Antisera specific for the N-terminal or the C-terminal portions of PhtA labeled the pneumococcal cell surface, but the reactivity of antiserum to the C terminus was more pronounced, suggesting that this region of the protein is more accessible to antibody (Fig. 4B). Similar pneumococcal cell surface labeling was observed with strains SJ2 and EF6796, except that antisera specific for the C-terminal portion of PhtA was not detected on the cell surface of strain EF6796, as well as strains WU2 and EF5668 (data not shown). Hyperimmune mouse antisera raised against full-length PhtB also bound to the cell surface of the SJ2 strain (data not shown). Antiserum raised against full-length PhtA bound to the bacterial surface of 98% of the pneumococcal strains tested, including all 23 serotypes represented in the current PS vaccine, 21 primary clinical isolates belonging to 6 additional serotypes (Table 1), and a laboratory strain, WU2 (data not shown). Similar bacterial surface binding was observed with antiserum raised against full-length PhtD (data not shown). Since the rabbit antiserum raised against either full-length PhtA or full-length PhtD were cross-reactive with both molecules (data not shown), it is not possible to determine whether this surface labeling was due to binding of the antisera to PhtA, to PhtD, or to both proteins.
FIG. 4.
Bacterial cell surface reactivity of anti-PhtA and anti-PhtD antisera. The indicated rabbit (A) and mouse (B) immune polyclonal antisera were tested for binding to intact S. pneumoniae strain N4. Bacteria were analyzed by fluorescence-activated cell sorter analysis and compared with samples treated with either rabbit preimmune sera or sera from sham-immunized mice.
Immunization with PhtA, PhtB, or PhtD induces protection against lethal pneumococcal sepsis.
Mice immunized with either the recombinant N-terminal portion of PhtE, the N-terminal or C-terminal portion of PhtA, or full-length PhtA, full-length PhtB, or full-length PhtD all derived from strain N4 generated antibody titers (reciprocal endpoint titers) ranging from 1,024,000 to 4,096,000. Both PhtA and PhtD induced antibodies that protected mice from death following i.p. challenge with either of two heterologous strains, SJ2 (serotype 6B) or EF6796 (serotype 6A) (Table 3). Immunization with either the N-terminal or the C-terminal portion of PhtA also conferred protection against SJ2 challenge, but only the N-terminal half of PhtA induced a protective response against EF6796 challenge. Mice immunized with PhtA or PhtD were not protected against challenge with the homologous and highly virulent strain N4 or with similarly virulent heterologous serotype 1 and 5 strains (data not shown). However, immunization with PhtD protected against challenge with another highly virulent capsular type 4 strain, EF5668, or with the capsular type 3 strain WU2. In contrast, mice immunized with recombinant PhtA were not protected against challenge with either of these two strains. Mice immunized with the N-terminal portion of PhtE were not protected against challenge with strain SJ2, EF6796, WU2, or N4. Immunization with full-length PhtB protected mice against challenge with strain SJ2 (Table 3).
TABLE 3.
Protection of mice immunized with recombinant Pht proteins from heterologous S. pneumoniae challenge
| Immunogen (Pht protein) | No. of mice alive/total no. of mice challenged with:a
|
||||
|---|---|---|---|---|---|
| SJ2 (type 6B) | EF6796 (type 6A) | EF5668 (type 4) | WU2 (type 3) | N4 (type 4) | |
| PhtA | 10/10∗ | 7/10∗ | 0/10 | 1/10 | 1/10 |
| PhtB | 8/10∗ | ND | ND | ND | ND |
| PhtD | 8/10∗ | 9/10∗ | 9/10∗ | 6/10∗ | 0/10 |
| PhtE (N-terminal) | 0/10 | 0/10 | ND | 1/10 | 0/10 |
| PhtA (N-terminal) | 9/10∗ | 5/10∗ | ND | ND | ND |
| PhtA (C-terminal) | 9/10∗ | 2/10 | ND | ND | ND |
| Sham | 0/10 | 1/10 | 0/10 | 1/10 | 0/10 |
Mice were challenged with either strain SJ2 (∼550 CFU; 55 × LD50), EF6796 (∼1,000 CFU; 100 × LD50), EF5668 (∼450 CFU; 90 × LD50), WU2 (∼1,500 CFU; 15 × LD50), or N4 (∼100 CFU; 20 × LD50), and deaths were recorded for up to 15 days. ∗, significantly different (P < 0.05) from sham control group as calculated by the two-sample log-rank test.
ND, not determined.
Passive administration of rabbit hyperimmune serum raised against PhtA (reciprocal endpoint titer of 8,192,000) was also able to protect mice against lethal challenge with strains SJ2 or EF6796 (10 of 10 and 7 of 8 survivors versus 1 of 10 in the control groups, respectively), demonstrating that protection was antibody mediated. For both studies, mice receiving immune serum showed a significant difference in the number of survivors compared to that of the controls (P < 0.005).
Serological responses to PhtA and PhtD during pneumococcal infection in humans.
Paired acute and convalescent sera from five infants with culture-confirmed pneumococcal bacteremia were examined in Western blots for reactivity with recombinant PhtA or PhtD (Fig. 5). Convalescent sera from patients 2, 3, and 5 reacted with both PhtA and PhtD, and this reactivity was more pronounced than the corresponding acute sera (Fig. 5A and D). The difference between the acute and convalescent sera was particularly evident when the sera were tested against the C-terminal half of the protein (Fig. 5C). The sera from patients 1 and 4, which were not reactive with either PhtA or PhtD, also failed to react with other recombinant pneumococcal proteins tested (data not shown). These two patients were younger at the time of diagnosis than the others (2.5 and 3.5 months compared with 4.5 years, 9 months, and 6 months for patients 2, 3, and 5, respectively).
FIG. 5.
Immunogenicity of PhtA and PhtD during pneumococcal infection in humans. The reactivity of sera collected from patients with culture-proven pneumococcal bacteremia (patients 1 and 4, serotype 5; patients 2, 3, and 5, serotypes 1, 12, and 18C, respectively) with PhtA (residues 21 to 816) (A), the N-terminal half of PhtA (residues 21 to 361) (B), the C-terminal half of PhtA (residues 386 to 816) (C), or PhtD (residues 21 to 839) (D) was determined by Western blot analysis. Lanes labeled A (acute) were probed with serum collected shortly after the diagnosis of pneumococcal infection; lanes C (convalescent) were probed with serum collected either 1 month (patients 1, 2, and 3) or 8 days after the first serum collection (patients 4 and 5). The molecular masses (in kilodaltons) of the protein standards are indicated.
DISCUSSION
We have identified a novel family of pneumococcal proteins (the Pht family) from S. pneumoniae which include at least two proteins that are immunologically protective against a subset of pneumococcal isolates in a mouse model of systemic disease. Initially, we found that immunization with either recombinant PhtA or PhtD did not protect mice against challenge with a homologous and highly virulent serotype 4 strain. However, it should be noted that the well-characterized protective antigen PspA derived from the homologous strain also failed to protect mice from death caused by this strain (Wizemann et al., submitted). When mice were challenged with a heterologous serotype 6B isolate, we were able to demonstrate protection with PhtA, PhtB, or PhtD. Similarly, PhtA or PhtD was able to protect mice against challenge with a serotype 6A isolate, but only PhtD was able to protect against a highly virulent serotype 3 strain (WU2) or another capsular type 4 strain, EF5668. We also found that immunization with the C-terminal portion of PhtA did not confer protection against lethal challenge with strain, EF6796. One possible explanation that remains to be tested is that the phtA gene in strain, EF6796, may have undergone recombination with the phtB gene, resulting in the loss of this region of phtA. Consistent with this hypothesis was the observation that specific antiserum to the C-terminal portion of PhtA did not label the pneumococcal cell surface of strain, EF6796.
Pneumococcal isolates vary widely in pathogenicity in mice, and virulence is generally associated with capsular serotype (2). Briles et al. (2) reported that mice infected with capsular serotype 6 isolates took longer to die than those infected with the serotype 1, 4, or 5 isolates. Our data, as well as that of others, suggest that different strains within a given serotype may vary in their degree of virulence in mice and, therefore, affect the ability of a given protein antigen to confer protection in the pneumococcal sepsis model. In this regard, the lack of protection against the homologous strain (N4) following immunization with PhtA or PhtD suggests that there may be other virulence factors contributing to this strains pathogenicity. Alternatively, it is possible that PhtA and/or PhtD may not be expressed in vivo by strain N4.
While the antibodies targeting PhtA, PhtB, or PhtD are protective, the function of these proteins is still unknown. The number of histidine and tyrosine residues in these proteins, including those contained within the histidine triad motif, suggests that they may be involved in metal or nucleoside binding. A number of divalent-metal transporters have been described in gram-positive bacteria, including the adc and psa operons of S. pneumoniae, which were demonstrated to be involved in the transport of zinc (9) and manganese (29), respectively. These transporter systems generally consist of an extracellular metal-binding lipoprotein, a membrane-bound ATP-binding protein, and a permease protein (36). The external metal-binding proteins have been grouped into a separate cluster (cluster 9) of the larger solute-binding protein family (9). The phtD gene is directly downstream from a gene predicted to encode a novel member of the cluster 9 family, whose gene product shares 67% homology with Lmb, a putative laminin-binding protein from Streptococcus agalactiae, encoded by the lmb gene (34). Furthermore, a histidine triad-containing protein is encoded by the gene directly downstream from lmb, originally identified as an unknown gene (GenBank accession no. AF062533). It is possible that the Pht proteins are involved in low-affinity promiscuous metal scavenging prior to transport involving the lipoprotein component. Although phtA and phtB genes are not associated with such a transporter in the genome the PhtA or PhtB proteins could conceivably associate with proteins encoded by one or more such systems located distally on the chromosome. Alternatively, other proteins containing a histidine triad motif, albeit not identical to the motif described here, have been demonstrated to have affinity for nucleosides (21). The residues within and around the first histidine triad bear homology to a conserved motif, VxxHGDxxxxN, found in aminoglycoside phosphotransferases (24). In this regard, a role for these proteins in antibiotic resistance cannot be ruled out. We have also shown that convalescent-phase sera from pneumococcal bacteremia patients recognize PhtA and PhtD, indicating that these proteins are exposed and recognized by the immune system during natural S. pneumoniae infection in humans.
In addition to the data obtained by Southern blot analysis, DNA sequencing of various strains of S. pneumoniae provided further support that phtA and phtD are highly conserved. Comparison of the deduced protein sequences revealed that PhtD shares greater than 95% identity among strains N4 (type 4), EF5668 (type 4), SJ2 (type 6B), and EF6796 (type 6A). Similarly, sequence analysis of phtA from pneumococcal strains ATCC 6301 (type 1), D39 (type 2), N4 (type 4), ATCC 6305 (type 5), and SJ2 (type 6B) indicates that PhtA is also highly conserved, sharing greater than 97% homology among strains (data not shown).
A search of the publicly available microbial genome databases resulted in the identification of two genes from Streptococcus pyogenes that encode predicted proteins containing histidine triad motifs (data not shown). Southern blot analysis of genomic DNA from Staphylococcus aureus probed with a phtA fragment indicated that a homologous gene might be present in this organism (data not shown). The observation that Pht-like proteins are conserved in groups A and B streptococci, and possibly in S. aureus, suggest that these proteins may perform similar functions in these distinct organisms.
All four Pht proteins contain a classical lipoprotein motif (LxxC) (1) within their N-terminal hydrophobic-leader sequences similar to that recognized for processing by signal peptidase II. However, examination of each of the Pht LxxC sequences indicated that the residue preceding the canonical cysteine was either a valine (PhtA, PhtB, and PhtD) or a leucine (PhtE) rather than the usual glycine or alanine found in the majority of bacterial lipoproteins (38). In addition, Triton X-114 phase-partitioning studies followed by immunoblot analysis revealed that PhtA, PhtD, and PhtE proteins were not detected in the detergent phase, in which lipoproteins are typically recovered (data not shown). Further biochemical studies are warranted to address whether Pht proteins are lipoproteins.
In summary, we have identified and characterized a novel family of cell surface-exposed pneumococcal proteins (the Pht family) that elicited protective responses against diverse pneumococcal serotypes. These novel pneumococcal antigens, either alone or in combination with capsular polysaccharides, may serve as effective vaccines that could potentially offer broad coverage against the most prevalent pneumococcal serotypes and enhanced efficacy against mucosal infections.
ACKNOWLEDGMENTS
We thank David Briles, Ingeborg Aeberge, and Pat Flynn for providing the pneumococcal strains used in this work. We thank Gil Choi at Human Genome Sciences, Inc., for providing the phtA DNA construct. We thank Mark Hanson for helpful discussions and critical reading of the manuscript. We gratefully thank the animal facility at MedImmune for assistance in animal work, David Carlin and Harry Yang for assistance with statistical analysis, and Donni Leach for reviewing the manuscript. The contributions of members of the DNA sequencing facility at Human Genome Sciences are also recognized.
REFERENCES
- 1.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J-M, Hakenbeck R, editors. New comprehensive biochemistry. 27. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier Science; 1994. pp. 319–341. [Google Scholar]
- 2.Briles D E, Crain M J, Gray B M, Forman C, Yother J. Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect Immun. 1992;60:111–116. doi: 10.1128/iai.60.1.111-116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briles D E, Hollingshead S K, Swiatlo E, Brooks-Walter A, Szalai A, Virolainen A, McDaniel L S, Benton K A, White P, Prellner K, Hermansson A, Aerts P C, Dijk H V, Crain M J. PspA and PspC: their potential for use as pneumococcal vaccines. Microb Drug Resist. 1997;3:401–408. doi: 10.1089/mdr.1997.3.401. [DOI] [PubMed] [Google Scholar]
- 4.Butler J C, Breiman R F, Lipman H B, Hofmann J, Facklam J. Serotype distribution of Streptococcus pneumoniae infections among preschool children in the United States, 1978–1994: implications for development of a conjugate vaccine. J Infect Dis. 1995;171:885–889. doi: 10.1093/infdis/171.4.885. [DOI] [PubMed] [Google Scholar]
- 5.Butler J C, Hofmann J, Centron M S, Elliot J A, Fracklam R R, Breiman R F the Pneumococcal Sentinel Surveillance Working Group. The continued emergence of drug-resistant Streptococcus pneumoniae in the United States: an update from the Centers for Disease Control and Prevention's Pneumococcal Surveilance System. J Infect Dis. 1996;174:986–993. doi: 10.1093/infdis/174.5.986. [DOI] [PubMed] [Google Scholar]
- 6.Butler J C. Epidemiology of pneumococcal serotypes and conjugate vaccine formulations. Microb Drug Resist. 1997;3:125–129. doi: 10.1089/mdr.1997.3.125. [DOI] [PubMed] [Google Scholar]
- 7.Dagan R, Engelhard D, Piccard E. Epidemiology of invasive childhood pneumococcal infections in Israel. The Israeli Pediatric Bacteremia and Meningitis Group. JAMA. 1992;268:3328–3332. [PubMed] [Google Scholar]
- 8.Dagan R, Eskola J, Leclerc C, Leroy O. Reduced response to multiple vaccines sharing common protein epitopes that are administered simultaneously to infants. Infect Immun. 1998;66:2093–2098. doi: 10.1128/iai.66.5.2093-2098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dintilhac A, Alloing G, Granadel C, Claverys J-P. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol Microbiol. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 10.Douglas R M, Paton J C, Duncan S J, Hansman D J. Antibody response to pneumococcal vaccination in children younger than five years of age. J Infec Dis. 1983;148:131–137. doi: 10.1093/infdis/148.1.131. [DOI] [PubMed] [Google Scholar]
- 11.Douglas R M, Miles H B. Vaccination against Streptococcus pneumoniae in childhood: lack of demonstrable benefit in young Australian children. J Infec Dis. 1984;149:861–869. doi: 10.1093/infdis/149.6.861. [DOI] [PubMed] [Google Scholar]
- 12.Giebink G S. The microbiology of otitis media. Pediatr Infect Dis J. 1989;8:S18–S20. [PubMed] [Google Scholar]
- 13.Gray B M, Dillon H C. Clinical and epidemiological studies of pneumococcal infection in children. Pediatr Infect Dis J. 1986;5:201–207. doi: 10.1097/00006454-198603000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Inouye S, Soberon X, Franceschini T, Nakamura K, Itakura K, Inouye M. Role of positive charge on the amino-terminal region of the signal peptide in protein secretion across the membrane. Proc Natl Acad Sci USA. 1982;79:3438–3441. doi: 10.1073/pnas.79.11.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kädyhty H, Eskola J. New vaccines for the prevention of pneumococcal infections. Emerg Infect Dis. 1996;2:289–295. doi: 10.3201/eid0204.960404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klien J O. The epidemiology of pneumococcal disease in infants and children. Rev Infect Dis. 1981;3:246–253. doi: 10.1093/clinids/3.2.246. [DOI] [PubMed] [Google Scholar]
- 17.Klein D L. Pneumococcal disease and the role of conjugate vaccines. Microb Drug Resist. 1999;5:147–157. doi: 10.1089/mdr.1999.5.147. [DOI] [PubMed] [Google Scholar]
- 18.Kuo J, Douglas M, Ree H K, Lindberg A A. Characterization of a recombinant pneumolysin and its use as a protein carrier for pneumococcal type 18C conjugate vaccines. Infect Immun. 1995;63:2706–2713. doi: 10.1128/iai.63.7.2706-2713.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Langermann S, Palaszynski S, Barnhart M, Augaste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Hal Jones C, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 21.Lima C D, Klein M G, Hendrickson W A. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 22.Lipsitch M. Vaccination against colonizing bacteria with multiple serotypes. Proc Natl Acad Sci USA. 1997;94:6571–6576. doi: 10.1073/pnas.94.12.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 24.Martin P, Jullien E, Courvalin P. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol Microbiol. 1988;2:615–625. doi: 10.1111/j.1365-2958.1988.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 25.McDaniel L S, Sheffield J S, Delucchi P, Briles D E. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect Immun. 1991;59:222–228. doi: 10.1128/iai.59.1.222-228.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munoa F J, Miller K W, Beers R, Graham M, Wu H C. Membrane topology of Escherichia coli prolipoprotein signal peptidase (signal peptidase II) J Biol Chem. 1991;266:17667–17672. [PubMed] [Google Scholar]
- 27.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Novak R, Braun J S, Charpentier E, Tuomanen E. Penicillin tolerance genes of Streptococcus pneumoniae: the ABC-type manganese permease complex Psa. Mol Microbiol. 1998;29:1285–1296. doi: 10.1046/j.1365-2958.1998.01016.x. [DOI] [PubMed] [Google Scholar]
- 29.Pugsley A. The complete secretory pathway in gram-negative bacteria. Microb Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenow C, Ryan P, Weiser J N, Johnson S, Fontan P, Ortqvist A, Masure H R. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol Microbiol. 1997;25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro E D, Berg A T, Austrian R, Schroeder D, Parcells V, Margolis A, Adair R K, Clemmens J D. Protective efficacy of polyvalent pneumococcal polysaccharide vaccine. N Engl J Med. 1991;325:1453–1460. doi: 10.1056/NEJM199111213252101. [DOI] [PubMed] [Google Scholar]
- 32.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triples and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siber G R. Pneumococcal disease: prospects for a new generation of vaccines. Science. 1994;265:1385–1387. doi: 10.1126/science.8073278. [DOI] [PubMed] [Google Scholar]
- 34.Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Schnitzler N, Lutticken R, Podbielski A. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect Immun. 1999;67:871–878. doi: 10.1128/iai.67.2.871-878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutcliff I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talkington D F, Brown B G, Tharpe J A, Koenig A, Russell H. Protection of mice against fatal pneumococcal challenge by immunization with pneumococcal surface adhesin A (PsaA) Microb Pathog. 1996;21:17–22. doi: 10.1006/mpat.1996.0038. [DOI] [PubMed] [Google Scholar]
- 37.Yother J, Handsome G L, Briles D E. Truncated forms of PspA that are secreted from Streptococcus pneumoniae and their use in functional studies and cloning of the pspA gene. J Bacteriol. 1992;174:610–618. doi: 10.1128/jb.174.2.610-618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 39.Zar J H. Biostatistical analysis. 2nd ed. Englewood Cliffs, N.J: Prentice Hall, Inc.; 1984. [Google Scholar]