Abstract
Critically ill patients infected with SARS-CoV-2 display adaptive immunity, but it is unknown if they develop cross-reactivity to variants of concern (VOCs). We profiled cross-immunity against SARS-CoV-2 VOCs in naturally infected, non-vaccinated, critically ill COVID-19 patients. Wave-1 patients (wild-type infection) were similar in demographics to Wave-3 patients (wild-type/alpha infection), but Wave-3 patients had higher illness severity. Wave-1 patients developed increasing neutralizing antibodies to all variants, as did patients during Wave-3. Wave-3 patients, when compared to Wave-1, developed more robust antibody responses, particularly for wild-type, alpha, beta and delta variants. Within Wave-3, neutralizing antibodies were significantly less to beta and gamma VOCs, as compared to wild-type, alpha and delta. Patients previously diagnosed with cancer or chronic obstructive pulmonary disease had significantly fewer neutralizing antibodies. Naturally infected ICU patients developed adaptive responses to all VOCs, with greater responses in those patients more likely to be infected with the alpha variant, versus wild-type.
Keywords: SARS-CoV-2, COVID-19, Intensive care units, Antibodies, Neutralizing, Adaptive immunity, SARS-CoV-2 alpha variant, SARS-CoV-2 beta variant, SARS-CoV-2 gamma variant, SARS-CoV-2 delta variant
Abbreviations: COVID-19, coronavirus disease 2019; ICU, intensive care unit; WT, wild-type; VOC, variants of concern; RBD, receptor binding domain; ACE2, angiotensin-converting enzyme; REB, research ethics board; P/F, arterial partial pressure to inspired oxygen; MODS, multi-organ dysfunction score; SOFA, sequential organ failure assessment; VTE, venous thromboembolism; MFI, median fluorescence intensity; IQR, interquartile range; ROC, receiver operating characteristic; AUC, area-under-the-curve; COPD, chronic obstructive pulmonary disease
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by SARS-CoV-2; patients are often admitted to the intensive care unit (ICU) for increased monitoring and potential life-saving interventions, where the mortality rate can be high [1]. COVID-19 induces an innate immune response [2,3] that includes increased interferons, tumor necrosis factor, bradykinin, serine proteases, soluble thrombomodulin and clot lysis times [[4], [5], [6], [7], [8]], thereby contributing to microvascular and thrombotic disease [9,10]. A humoral immune response follows the innate reaction in critically ill COVID-19 patients, with robust production of SARS-CoV-2-specific antibodies [11,12].
Since the emergence of wild-type (WT) SARS-CoV-2, multiple variants have evolved that show higher transmissibility and antigenicity and are, therefore, classified as variants of concern (VOCs) [13]. Prominent VOCs are derived from the following lineages: B.1.1.7 (alpha, United Kingdom), B.1.351 (beta, South Africa), P1 (gamma, Brazil) and B.1.617.2 (delta, India) [14]. Increased infectivity of the virus has been attributed to prominent mutations in the N501Y (present in lineages B.1.1.7, B.1.351 and P.1) and the E484K (present in lineages B.1.351 and P.1.) [15]. Mutations have evolved independently and, thus, are strongly believed to have an impact on viral reproductivity rate. The VOC databases and platforms have been reviewed in detail [16].
Viral entry requires host attachment and fusion of SARS-CoV-2 to cellular membranes, promoted by the interaction of the receptor binding domain (RBD), which lies within the S1 subunit of the spike protein (15), and the angiotensin-converting enzyme 2 (ACE2) of the host-cell [17]. ACE2 is a receptor protein, highly expressed in cells of the lung alveoli, small intestine and blood vessels [18], as well as other tissues, and serves as main entrance for cell invasion by the SARS-CoV-2. Neutralizing antibodies bind to the RBD, thereby interfering with the molecular interactions between the virus and the host cell and abolishing viral entry. Mutations within the Spike S1, especially in the RBD domain, are considered a potential threat for generating virus mutants with a higher susceptibility rate and/or an immune escape mechanism. Indeed, mutations in the spike protein are associated with elevated transmissibility rate due to a modified interaction of the mutated protein with the host ACE2 receptor [19].
Measurements of neutralizing antibodies against WT SARS-CoV-2 and its VOCs are important to understand host immunity. Until recently, accurate measurement of neutralizing antibodies had required viral isolates and infectious clones, and needed to be performed under Biosafety Level-3 conditions. Individual immunoassays have now been developed using SARS-CoV-2 RBD recombinant antigens that include mutations present in most widespread variants [20]. Moreover, a multiplex assay for neutralizing antibodies was developed using Luminex beads coupled with Spike S1 protein for the original WT SARS-CoV-2, or one of the four circulating VOCs: alpha; beta; gamma; and delta. To the best of our knowledge, critically ill COVID-19 patients have not been investigated with multiplex technology for cross immunity to the SARS-CoV-2 VOCs. A better understanding of host immunity to SARS-CoV-2 VOCs may influence future care, including patient cohorting, prognostication, therapies and expectant management of late complications [21].
We hypothesized that critically ill COVID-19 patients will demonstrate cross-immunity to SARS-CoV-2 VOCs. Using multiplex technology, the primary aim of this study was to measure neutralizing antibodies to SARS-CoV-2 in naturally infected, non-vaccinated, critically ill patients. Our specific objectives were: (1) to measure and compare the neutralizing potential of human SARS-CoV-2 antibodies against WT and four circulating VOCs; (2) to compare the neutralizing activity kinetics of antibodies in SARS-CoV-2 infected ICU patients from Wave-1 (WT infection) to Wave-3 (mixed WT and alpha variant infection); and (3) to determine potential differences in neutralizing antibody production against SARS-CoV-2 VOCs between critically (ICU) and non-critically (Ward) ill patients.
2. Materials and methods
This study was approved by the Western University, Human Research Ethics Board (REB ID# 1670; issued March 20, 2020) and the Hamilton Integrated Research Ethics Board (REB ID# 10-532; issued October 19, 2020).
2.1. Study participants and clinical data
Patients were enrolled after admission to one of our two participating academic hospitals, London Health Sciences Centre (London, Ontario) and Hamilton General Hospital (Hamilton, Ontario). Informed consent was obtained from all study patients, or when critically ill, their substitute decision makers. COVID-19 was first suspected based on standard hospital screening procedures [22], and then confirmed as part of standard hospital testing by detection of two SARS-CoV-2 viral genes using polymerase chain reaction [23]. Blood sampling was on ICU days 1 and 3. Patient baseline characteristics were recorded on admission and included age, sex, comorbidities, medications, hematologic labs, creatinine, arterial partial pressure to inspired oxygen (P/F) ratio, and chest x-ray findings. We calculated Multiple Organ Dysfunction Score (MODS) [24] and Sequential Organ Failure Assessment (SOFA) score [25] for both Wave-1 and Wave-3 COVID-19 patient groups to enable objective comparison of their illness severity. Both patient groups were characterized as having confirmed or suspected sepsis diagnosis using Sepsis 3.0 criteria [25]. We also recorded clinical interventions received during the observation period including use of antibiotics, anti-viral agents, systemic corticosteroids, vasoactive medications, venous thromboembolism (VTE) prophylaxis, anti-platelet or anti-coagulation treatment, renal replacement therapy, high flow oxygen therapy, and mechanical ventilation (invasive and non-invasive). For comparison to critically ill patients, we also included an age- and sex-matched non-critically ill COVID-19 patient cohort who were admitted to the hospital respiratory ward with moderate disease. All patients were non-vaccinated for SARS-CoV-2.
2.2. Blood draws
Standard operating procedures were used to ensure all samples were treated rapidly and equally (https://translationalresearchcentre.com/) [26,27]. Blood was obtained in the morning from critically ill ICU patients via indwelling catheters using vacuum serum separator tubes and placed immediately on ice. If a venipuncture was required, research blood draws were coordinated with a clinically indicated blood draw. In keeping with accepted research phlebotomy protocols for adult patients, blood draws did not exceed maximal volumes [28]. Once transferred to a negative pressure hood, blood was centrifuged and sera isolated, aliquoted at 250 μL and frozen at −80 °C. All samples remained frozen until use and freeze/thaw cycles were avoided.
2.3. Multiplex neutralizing antibody detection
Neutralizing antibody activity against WT SARS-CoV-2 and four VOCs was determined in human plasma using a multiplexed serological screening kit according to manufacturer’s instructions (Invitrogen™ SARS-CoV-2 Variants Neutralizing Antibody Human 5-Plex ProcartaPlex™ Panel; Cat. No. EPX050-16015-901, which utilized Luminex® xMAP™ fluorescent bead-based technology (Luminex Corp., 12212 Technology Blvd, Austin, TX, 78727, USA)). The assay plate was treated according to the manufacturer’s instructions contained within the ProcartaPlex Kit. Briefly for ProcartaPlex assay setup, 50 μl Luminex Capture Beads (coated with the spike protein) were added per well on microtiter plates, 25 μl prediluted serum sample or positive/negative control were added. After incubation (2 h, and washing steps), the biotinylated detection reagent (biotinylated ACE) was added and incubated for 0.5 h. After washing steps, Streptavidin-PE was added (50 μl), and incubated (0.5 h). After washing steps, the plates are measured on a compatible Luminex® system (Bio-Plex™ 200 system, Bio-Rad Laboratories, 1000 Alfred Nobel Drive, Hercules, CA, 94547, USA).
Data were expressed as the median fluorescence intensity (MFI) of the raw data values, with control values converted to “% Neutralization” by applying the formula: Neutralization (%) = (1 − (MFI of samples/MFI of negative control)) × 100. The cut-off level of 20% was based upon the measurement of 160 healthy control samples in the Neutralizing Antibody ProcartaPlex Panel during development. Since these samples were derived from blood donations before onset of the COVID-19 pandemic from healthy volunteers, no antibodies showing ACE blocking activity due to prior SARS CoV2 infection could be expected in these samples. The background level of these healthy control samples in the assay showed <20% neutralizing activity; thus, when applying this cut-off level, the assay obtained 100% specificity.
2.4. Population statistics
Medians (IQRs) and frequencies (%) were used to report ICU patient baseline characteristics for continuous and categorical variables, respectively; continuous variables were compared using Mann-Whitney U tests (and Wilcoxon matched-pairs signed-rank tests, as appropriate), and categorical variables were compared using Fisher’s exact chi-square tests. For demographic, clinical and scatter plot data, P-values < 0.05 (*) were considered statistically significant, whereas a P < 0.01 was used to control for repeated measures where necessary. Scatter plots were generated, and all analyses were conducted using GraphPad Prism version 9.2.0 for Windows (GraphPad Software, California, USA) and SPSS version 27 (IBM Corp., Armonk, NY, USA).
2.5. Machine learning
A random forest classifier was also trained on the variables to predict Wave-1 versus Wave-3. A random forest is a set of decision trees that can be interrogated to identify the features that have the highest predictive value. To control overfitting, COVID-19 classifiers were trained and tested using a three-fold cross validation with a random forest of 10 trees with a max depth of three. A receiver operating characteristic (ROC) curve with an accompanying area-under-the-curve (AUC) statistic was generated to determine the random forest’s ability to distinguish between outcomes, using the reduced variables where the data were split into training and validation datasets.
3. Results
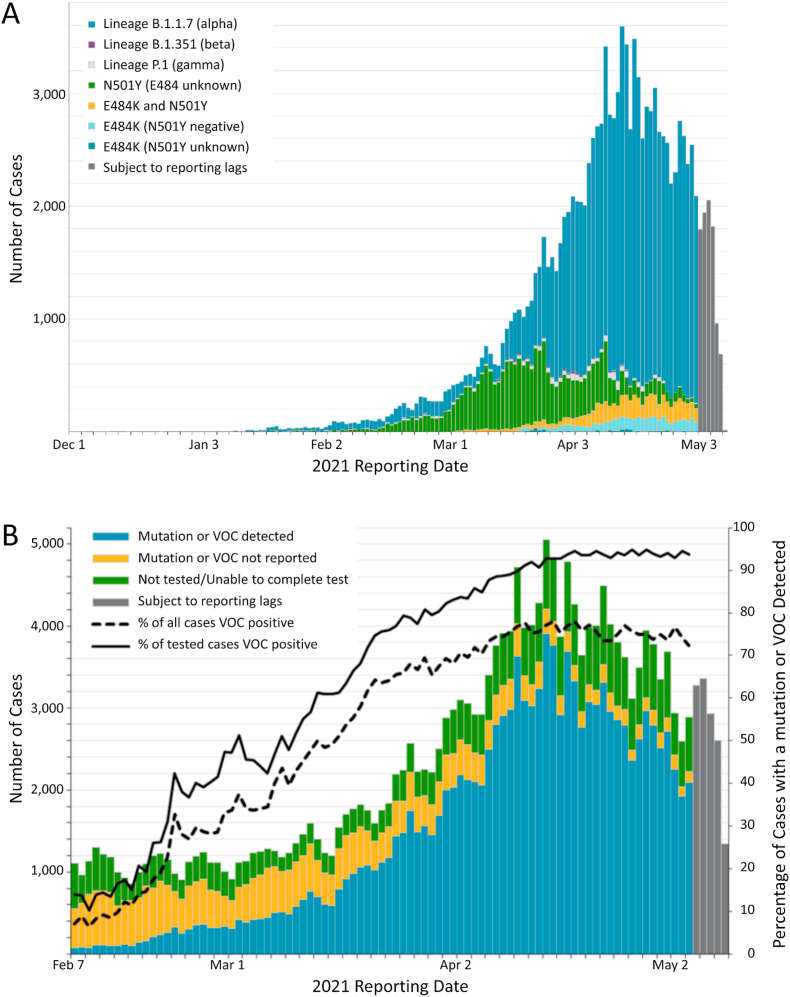
We investigated 14 COVID-19 patients infected during Wave-1 (median years of age = 61.0, IQR = 54.0, 67.0) and 29 COVID-19 patients infected during Wave-3 (median years of age = 62.0, IQR = 53.5, 72.5). Wave-1 patients were infected at the beginning of the pandemic, prior to May 2020, when only the WT virus was circulating in our geographical area (Province of Ontario, Canada). In contrast, Wave-3 patients were reported after December 2020, when the predominate circulating virus was the alpha VOC (Fig. 1A and B). Baseline demographic characteristics, comorbidities, laboratory values and chest x-ray findings are reported in Table 1. Wave-3 COVID-19 patients with either a WT or alpha variant infection had higher MODS (P = 0.032) and SOFA scores (P = 0.004), and were more likely to receive invasive mechanical ventilation (P = 0.008) and to have received steroid treatment (P = 0.001), when compared to Wave-1 COVID-19 patients.
Fig. 1.
Number of confirmed COVID-19 cases with a known variant of concern or mutation detected by date reported to public health unit: Province of Ontario (Canada), December 1, 2020 to May 9, 2021 (“third wave”). (A) Data for cases with a B.1.1.7, B.1.351, and P.1 lineage detected or any of the mutations. Reported date is based on the date the case was reported, not the date that the VOC or mutation was identified. B) Data for all cases with the percentage of cases with a mutation or VOC detected. The denominator includes all confirmed COVID-19 cases, including those that were unable to be tested for VOCs. These data were adapted with the permission of Public Health Ontario. Public Health Ontario assumes no responsibility for the content of any publication resulting from translation/changes/adaptation of Public Health Ontario documents by third parties.
Table 1.
COVID-19 ICU patient admission demographics and clinical data.
| Variable | Wave-1 (n = 14) | Wave-3 (n = 29) | P-value |
|---|---|---|---|
| Age (yrs) | 61 (54, 67) | 62 (53.5, 72.5) | 0.421 |
| Male sex | 6 (43) | 19 (66) | 0.158 |
| Weight (kg) | 90.8 (76.5, 110.5) | 91.0 (77.8, 99.8) | 0.777 |
| Height (cm) | 170 (161, 173) | 170 (160, 176.5) | >0.994 |
| Body Mass Index | 30.5 (27.1, 41.8) | 31.5 (27.1, 39.3) | >0.994 |
| Sepsis 3.0 criteria | 14 (100) | 29 (100) | >0.994 |
| MODS | 4 (3, 6) | 6 (4, 8) | 0.032* |
| SOFA | 5 (2, 9) | 9 (7, 11) | 0.004* |
| Comorbidities | |||
| Diabetes | 5 (36) | 9 (31) | >0.994 |
| Hypertension | 7 (50) | 17 (59) | 0.594 |
| Coronary artery/heart disease | 2 (14) | 5 (17) | >0.994 |
| Chronic/congestive heart failure | 0 (0) | 2 (7) | >0.994 |
| Chronic kidney disease | 2 (14) | 2 (7) | 0.585 |
| Cancer | 2 (14) | 4 (14) | >0.994 |
| COPD | 1 (7) | 2 (7) | >0.994 |
| Laboratories | |||
| Hemoglobin | 122 (102, 135) | 117 (99, 138) | 0.897 |
| White Blood Cell count | 8.5 (6.9, 16.1) | 9.3 (8.0, 11.5) | 0.846 |
| Neutrophils | 7.3 (5.6, 12.6) | 8.1 (5.8, 10.1) | 0.870 |
| Lymphocytes | 0.7 (0.6, 1.0) | 0.7 (0.5, 1.1) | 0.902 |
| Platelets | 206 (134, 294) | 224 (156, 313) | 0.437 |
| Creatinine | 82 (58, 187) | 96 (69, 153) | 0.805 |
| International Normalized Ratio | 1.2 (1.1, 1.3) | 1.1 (1.0, 1.2) | 0.068 |
| Partial Thromboplastin Time | 28 (25, 31) | 27 (24, 31) | 0.499 |
| Lactate | 1.5 (1.0, 2.0) | 1.2 (1.0, 1.7) | 0.305 |
| Pulmonary pathology | |||
| Bilateral pneumonia | 13 (93) | 27 (93) | >0.994 |
| P:F ratio | 107 (66, 162) | 105 (74, 147) | 0.897 |
| Interventions | |||
| High-flow nasal cannula | 8 (57) | 14 (48) | 0.586 |
| Non-invasive mechanical ventilation | 6 (43) | 1 (3) | 0.003* |
| Invasive mechanical ventilation | 10 (71) | 29 (100) | 0.008* |
| Vasoactive medication | 11 (79) | 28 (97) | 0.094 |
| Renal replacement therapy | 2 (14) | 4 (14) | >0.994 |
| Steroids | 3 (21) | 28 (97) | <0.001* |
| Anti-virals | 3 (21) | 2 (7) | 0.309 |
| Antibiotics | 14 (100) | 28 (97) | >0.994 |
| Antiplatelet | 5 (36) | 6 (21) | 0.457 |
| Anticoagulation | 13 (93) | 29 (100) | 0.326 |
| Outcomes | |||
| ICU Length of Stay (days) | 17 (11, 27) | 18 (11, 29) | 0.678 |
| Died | 7 (50) | 12 (41) | 0.594 |
Continuous variables are presented as median, interquartile range (IQR); categorical are variables presented as n (%). *p < 0.05. MODS, Multi-Organ Dysfunction Score; SOFA, Sequential Organ Failure Assessment Score; COPD, Chronic obstructive pulmonary disease; P/F ratio, PaO2/FiO2 ratio.
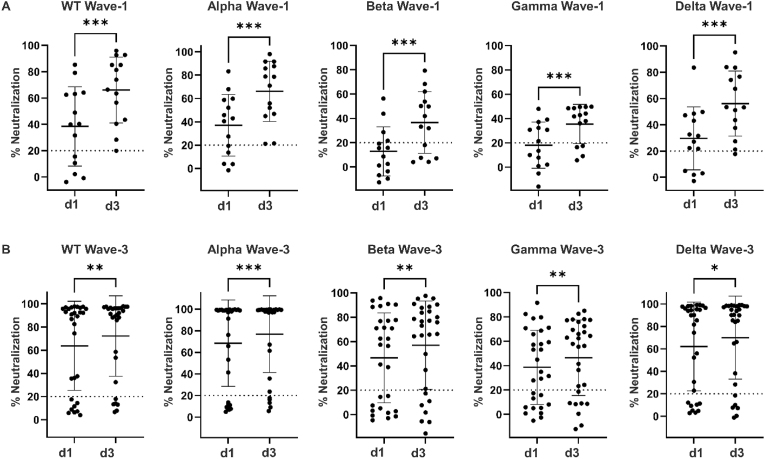
Neutralizing antibodies against WT SARS-CoV-2 and all four VOCs were determined in plasma on ICU day-1 and day-3 for both Wave-1 and Wave-3 COVID-19 patients (Fig. 2A and B). The neutralizing antibody activity against WT SARS-CoV-2 and all four VOCs was significantly increased from ICU day-1 to day-3 during Wave-1 (WT infection; P < 0.001), and to a lesser extent during Wave-3 (WT/alpha infection; P < 0.05 to P < 0.001).
Fig. 2.
Scatter plots comparing neutralizing antibody responses against SARS-CoV-2 WT (WT) and four variants of concern (VOC) compared on ICU days 1 and 3. (A) Neutralizing antibody activity against WT, alpha, beta, gamma and delta variants of Wave-1 COVID-19 subjects (WT infection) compared between ICU day-1 (d1) and day-3 (d3). Statistical significance as indicated (***P < 0.001). (B) Neutralizing antibody activity against WT, alpha, beta, gamma and delta variants of Wave-3 COVID-19 subjects (WT/alpha infection) compared between ICU day-1 (d1) and day-3 (d3). Statistical significance as indicated (*P < 0.05; **P < 0.01; ***P < 0.001).
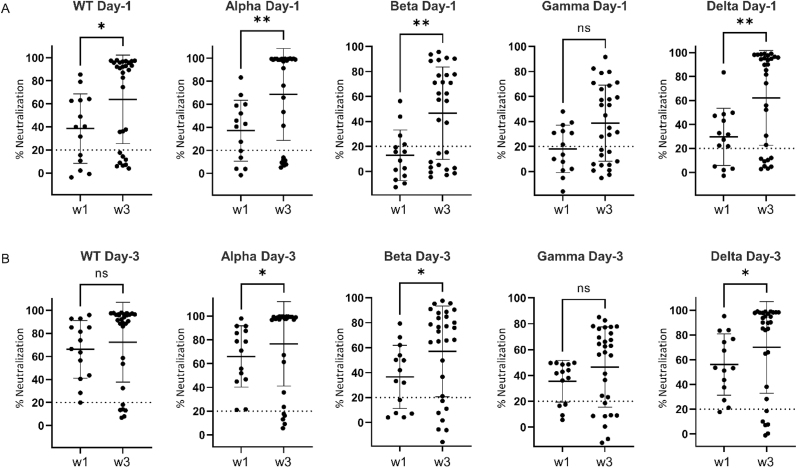
Comparison of neutralization activity between Wave-1 versus Wave-3 COVID-19 patients on ICU day-1 showed significantly higher neutralizing activity against most SARS-CoV-2 VOCs on Wave-3 (P < 0.05 to P < 0.01; Fig. 3A and B). A similar comparison of neutralization activity on ICU day-3 between Wave-1 and Wave-3 COVID-19 patients showed significantly higher neutralizing activity against most SARS-CoV-2 VOCs on Wave-3 (P < 0.05). On both ICU days, the neutralizing activity against the gamma variant was statistically non-significant (P = 0.054). When antibody neutralization of all VOCs (both day-1 and day-3 combined) was compared between Wave-1 (WT infection) and Wave-3 (WT/alpha infection) using feature selection, a good classification accuracy (78%) and a very good AUC (0.84) were generated. Based on the random forest decision trees, a VOC rank order of importance for separating Wave-1 and Wave-3 was identified: alpha (28%) > gamma (23%) > WT (19%) > delta (17%) > beta (13%). Taken together, these data show that COVID-19 patients demonstrated significantly increased potency of neutralizing antibodies against SARS-CoV-2 VOCs on Wave-3 versus Wave-1, with alpha being the most important distinguishing VOC.
Fig. 3.
Scatter plots comparing neutralizing antibody responses against SARS-CoV-2 WT (WT) and four variants of concern (VOC) compared between Wave-1 and Wave-3. (A) Neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on ICU day-1 for COVID-19 subjects comparing Wave-1 (w1; WT infection) to Wave-3 (w3; WT/alpha infection). Statistical significance as indicated (ns, non-significant; *P < 0.05; **P < 0.01). (B) Neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on ICU day-1 for COVID-19 subjects comparing Wave-1 (w1; WT infection) to Wave-3 (w3; WT/alpha infection). Statistical significance as indicated (ns, non-significant; *P < 0.05).
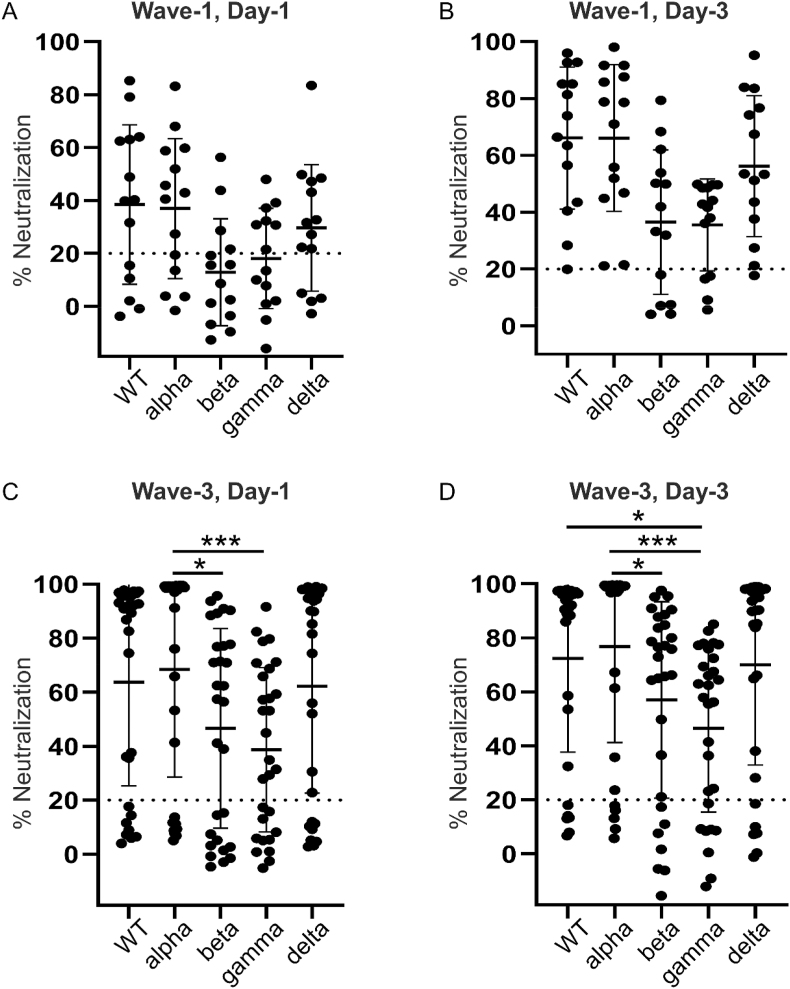
Neutralization activity for all VOCs was examined for the given ICU day and Wave. For Wave-1, there were no significant difference in neutralizing antibodies between variants on either ICU day-1 (Fig. 4A) or day-3 (Fig. 4B). The % neutralization median response for both beta and gamma fell below the cut-off value on Wave-1, Day-1. Analysis of Wave-3 showed significantly greater neutralizing antibodies on ICU day-1 between the alpha variant, as compared to beta (P < 0.05) and gamma (P < 0.001) variants (Fig. 4C); on ICU day-3, neutralizing antibodies to both WT and alpha variants were significantly greater than the gamma variant (P < 0.05 and P < 0.001, respectively), whereas only the alpha variant was significantly greater than the beta variant (P < 0.05) (Fig. 4D).
Fig. 4.
Scatter plots comparing neutralizing antibody responses against SARS-CoV-2 WT (WT) and four variants of concern (VOC) by Wave and ICU day. (A) Comparison of the neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on Wave-1 (WT infection), ICU day-1 for COVID-19 subjects. (B) Comparison of the neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on Wave-1 (WT infection), ICU day-3 for COVID-19 subjects. (C) Comparison of the neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on Wave-3 (WT/alpha infection), ICU day-1 for COVID-19 subjects. Statistical significance as indicated (*P < 0.05; ***P < 0.001). (D) Comparison of the neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on Wave-3 (WT/alpha infection), ICU day-3 for COVID-19 subjects. Statistical significance as indicated (*P < 0.05; ***P < 0.001).
Examination of clinical factors associated with neutralizing antibody responses <20% revealed strong associations between previous diagnoses of cancer (both Wave-1 and Wave-3 patients; P < 0.01) and chronic obstructive pulmonary disease (COPD; Wave 3 patients only, P = 0.002). With regard to outcomes, the degree of neutralizing antibodies was not associated with mortality for either Wave-1 (WT; day-1. P = 0.805; day-3, P = 0.522) or Wave-3 (WT; day-1, P = 0.388, day-3, P = 0.241; alpha; day-1, P = 0.550; day-3, P = 0.298).
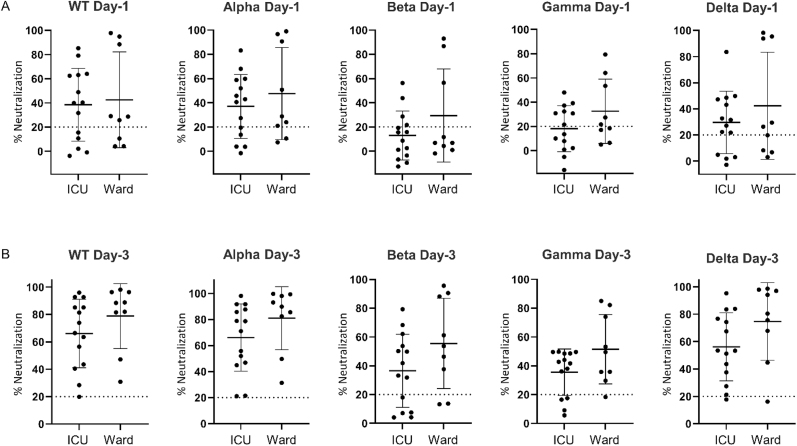
As a final comparison, we investigated SARS-CoV-2 disease severity by comparing Wave-1 ICU patients to age- and sex-matched Ward COVID-19 patients (Table 2). Ward patients were admitted January 2021 to April 2021. Given the admission time period, the Ward patients were considered Wave-3, with a likelihood of either WT or alpha infection (Fig. 1A and B). All Ward patients had bilateral pneumonia and received steroid therapy. Despite a lower disease burden, there were no significant differences in neutralizing antibodies to WT and any VOCs between Wave-1 ICU and Wave-3 Ward patients (Fig. 5A and B). The neutralizing antibody responses in the Ward patients were very similar to Wave-3 ICU patients, as illustrated in Figs. 2B, 4C and D.
Table 2.
COVID-19 ward patient admission demographics and clinical data.
| Variable | Ward Patients (n = 9) |
|---|---|
| Age (yrs), median (IQR) | 60 (41.5, 78.0) |
| Male sex, n (%) | 6 (66.7) |
| Weight (kg), median (IQR) | 83.6 (75.8, 93.4) |
| Height (cm), median (IQR) | 170 (165, 173) |
| Body Mass Index, median (IQR) | 29.2 (23.4, 31.1) |
| Comorbidities, n (%) | |
| Diabetes | 1 (11.1) |
| Hypertension | 3 (33.3) |
| Coronary artery/heart disease | 1 (11.1) |
| Chronic/congestive heart failure | 0 (0) |
| Chronic kidney disease | 0 (0) |
| Cancer | 2 (22.2) |
| COPD | 0 (0) |
| Laboratories, median (IQR) | |
| Hemoglobin | 118 (105, 139) |
| White Blood Cell count | 7.0 (5.3, 10.4) |
| Neutrophils | 5.9 (4.2, 8.1) |
| Lymphocytes | 0.8 (0.5, 1.3) |
| Platelets | 172 (123, 230) |
| Creatinine | 69.0 (57.5, 85.5) |
| International Normalized Ratio | 1.1 (1.1, 1.2) |
| Lactate | 1.6 (1.3, 4.4) |
| Pulmonary pathology, n (%) | |
| Bilateral pneumonia | 9 (100) |
| Interventions, n (%) | |
| High-flow nasal cannula | 6 (66.7) |
| Low-flow nasal cannula | 9 (100) |
| Non-invasive mechanical ventilation | 1 (11.1) |
| Steroids | 9 (100) |
| Anti-virals | 3 (33.3) |
| Antibiotics | 9 (100) |
| Antiplatelet | 3 (33.3) |
| Anticoagulation | 9 (100) |
| Presenting symptoms, n (%) | |
| Fever | 7 (77.8) |
| Cough | 8 (88.9) |
| Anosmia/Ageusia | 2 (22.2) |
| Pharyngitis | 1 (11.1) |
| Headache | 2 (22.2) |
| Myalgias | 5 (55.6) |
| Dyspnea | 9 (100.0) |
| Chest Pain | 2 (22.2) |
| Nausea/Vomiting/Diarrhea | 4 (44.4) |
| Outcomes | |
| Ward Length of Stay (days), median (IQR) | 8.0 (6.0–12.5) |
| Discharged Home, n (%) | 6 (66.7) |
| Died on Ward, n (%) | 2 (22.2) |
| Transferred to ICU, n (%) | 1 (11.1) |
Fig. 5.
Scatter plots comparing neutralizing antibody responses against SARS-CoV-2 WT (WT) and four variants of concern (VOC) compared between ICU (critically ill) and Ward (non-critically ill) patients. (A) Neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on hospital day-1 for COVID-19 subjects comparing critically ill (ICU; WT infection) to non-critically ill (Ward; WT/alpha infection). (B) Neutralizing antibody activity against WT, alpha, beta, gamma and delta variants on hospital day-3 for COVID-19 subjects comparing critically ill (ICU; WT infection) to non-critically ill (Ward; WT/alpha infection).
4. Discussion
In this exploratory study, we demonstrate that critically ill, non-vaccinated, COVID-19 patients exhibited antibody responses to WT and four VOCs that increased over their ICU stay; however, the neutralizing response against SARS-CoV-2 VOCs as determined using a surrogate multiplex neutralization assay was delayed and significantly less in Wave-1 (WT infection) as compared to Wave-3 (mixed WT and alpha infection). Our data suggest that naturally infected ICU survivors have developed humoral responses to all VOCs, albeit variable, with greater responses in those patients more likely to be infected with the alpha variant. Neutralizing antibody responses were insufficient in patients previously diagnosed with either cancer or COPD. No significant differences were seen between ICU (severe COVID-19) and Ward (moderate COVID-19) patients. These data are important to understanding the complexity of the immune response to SARS-CoV-2 during critical illness, and the cross-reactivity expressed for ICU survivors.
SARS-CoV-2 VOCs are emerging due to the high mutation rates associated with coronaviruses [29]. While most mutations appear randomly without impacting the virus, some mutations may lead to increase transmissibility and higher virulence, as well as provide immune escape mechanisms. A comprehensive study on S protein mutations in SARS CoV-2 isolates showed distribution of prominent mutations as basis for setting up a phylogenetic tree of the isolates [30]. Mutations that offer the virus benefit will circulate widely in the population. At the time of our study, four VOCs had been identified and reported world-wide (alpha, beta, gamma and delta). All four VOCs have mutations in the RBD, a region associated with the spike protein [29]. Individuals with neutralizing antibody responses to the WT may not have equivalent humoral responses to new variants [31,32], responses that may be complicated further by critical illness and immunotherapies (e.g., steroids, IL6 inhibitors). Thus, elucidating the neutralizing antibody response in critically ill patients, as we have done in this study, could be important for patient cohorting and potential therapy with either monoclonal antibodies or convalescent plasma.
To measure SARS-CoV-2 neutralizing antibodies, we utilized a novel multiplex assay using Luminex beads coupled with Spike S1 protein for WT or one of the four established VOCs: alpha; beta; gamma; and delta. The protein coupled beads were incubated either with or without (then serving as negative control) patient plasma, followed by an incubation with biotinylated ACE2. The biotinylated ACE2, which binds to the Spike S1 protein on the beads, was determined by measuring median fluorescence intensity with a reporter-dye. The assay was competitive; the negative control without sample gave the highest signal, which was suppressed by interfering neutralizing antibodies present in the plasma sample. Detecting the signal in a multiplex format enabled the direct comparison of neutralizing potential of antibodies towards the original WT, as well as the four VOCs simultaneously, allowing for the study of cross-immunity against SARS-CoV-2 variants.
Our COVID-19 ICU patients were similar to those reported in earlier cohorts [3,[33], [34], [35], [36]] with respect to demographic, comorbidities and clinical presentation. In contrast to Wave-1 patients, Wave-3 COVID-19 patients had higher MODS and SOFA scores, and were more likely to have received invasive mechanical ventilation and steroid treatment. Wave-3 patients were enrolled after December 2020, when the predominate circulating virus in our geographical region was the alpha VOC. Despite reports of the alpha variant being more lethal than the WT [[37], [38], [39]], mortality was not significantly different between Wave-1 and Wave-3, suggesting improved ICU care (e.g., steroid therapy [40]).
Patients admitted to the ICU during Wave-1 developed increasing neutralizing antibodies to all variants with time, as did patients during Wave-3. These data are consistent with our previous reports demonstrating increased WT SARS-CoV-2 antibodies in critically ill ICU patients [11,12]; specifically, we had demonstrated classification accuracy of 96–100% for IgG to the SARS-CoV-2 Spike protein on ICU day-3. Wave-3 patients, when compared to Wave-1, developed more robust antibody responses, particularly for WT, alpha, beta and delta variants. Within Wave-3, neutralizing antibodies were significantly less to beta and gamma VOCs, as compared to WT, alpha and delta. Our data, based on plasma samples from critically ill patients, are similar to reports from both naturally infected non-hospitalized patients and vaccinated patients (BNT162b2) where a robust cross-reactivity to alpha and delta VOCs was reported, and a lesser response reported to beta and gamma VOCs [41,42].
Illness severity did not significantly impact production of neutralizing antibodies to either WT/alpha infection, or via cross-reactivity to the VOCs. Indeed, no significant differences were noted between critically ill (ICU) and non-critically ill (Ward) patients. Mortality was also not related to antibody production, or the lack there of, which is consistent with our previous studies [11,12]. In contrast, using 20% as a threshold for neutralizing antibody specificity, there were significant antibody insufficiencies observed in ICU patients with previous diagnoses of either cancer or COPD. These latter findings are consistent with reports of cancer patients lacking T-cell responses that are associated with an absence of anti-S antibodies following SARS-CoV-2 vaccination [43,44], and the high rates of breakthrough infection in cancer patients despite full vaccination [45]. Humoral immune response to vaccination was also lower in persons with COPD compared to non-COPD controls [46].
Emerging VOCs have potential for increased transmissibility, morbidity and mortality, as well as evading detection with current diagnostics [29,47]. Persistent SARS-CoV-2 infection can result in recurrent deletions in the spike protein as the virus replicates, resulting in less effective neutralizing antibodies to the resultant variants [48]. Three of the VOCs measured here (beta, gamma and delta) are resistant to several monoclonal antibody treatments [49], and both beta and gamma have reduced inhibition by convalescent plasma from immunized patients [[50], [51], [52], [53]]. Our data suggest that patient cohorting based on the four circulating VOCs investigated may not be necessary, unless patients have been previously diagnosed with either cancer or COPD. Moreover, the role of antibody and convalescent plasma therapy may need to be revisited and personalized to the specific VOC infection. For those patients who survive their critical illness, adequate humoral responses were seen with regard to the four circulating VOCs measured here, at least until vaccination was deemed appropriate. Finally, our data also indicate that production of neutralizing antibodies was not reduced by steroid therapy, which was reported to reduce mortality in SARS-CoV-2 patients requiring respiratory support [40], and administered to almost all ICU and ward patients in Wave-3.
Future therapies to reduce ICU mortality may include broadening of the protective immune response through development of variant boosters with antigenic inserts matching the SARS-CoV-2 VOCs of interest [54]; an approach that would require rapid VOC profiling on hospital admission with a multiplex immunoassay, as we have done in this study, followed by immediate boosting. Knowledge on neutralizing antibody responses to specific VOCs in critically ill patients may also guide convalescent plasma therapy, which to date has yielded disappointing results [55], and help personalize VOC-specific neutralizing monoclonal antibody therapy [56]. Finally, ICU mortality may be reduced by targeting the RBD with peptide-based therapies, administering engineered variants of human recombinant soluble ACE2, and/or drug interruption of SARS-CoV-2 endosomal function [47].
As depicted above, we did not find a significant correlation of neutralizing antibodies with morbidity and mortality of the patients; however, a better understanding of the host response may influence vaccination protocols [57], as well as other potential therapies (e.g., hyperbaric oxygen therapy, [58]). Furthermore, a potential association of long-lasting COVID-19 symptoms (Long-COVID) with prior early determination of neutralizing activity may be addressed in forth-coming studies. Indeed, Long-COVID is associated with diffuse and prolonged symptoms after acute SARS-CoV-2 infection, and represents a looming health care crisis with significant strain on health care systems [21].
Our study was not without limitations. First, our patient number was modest and the time points of investigation limited to day-3 of hospitalization. Nonetheless, our study cohorts were representative of SARS-CoV-2 infected patients in many publications, and the day-3 time point has been demonstrated to be a reasonable surrogate for antibody production [11,12]. Second, we cannot be certain that our in vitro testing for neutralizing antibodies accurately reflected individual protection; however, our results are consistent with other reports in non-hospitalized subjects [41,42]. Third, we only investigated four VOCs (alpha, beta, gamma and delta) with other emerging variants on the rise. Importantly, the immunoassay used here can be quickly altered in the future to test for additional neutralizing antibodies (e.g., against omicron). Fourth, while we acknowledge our study is exploratory, it is the first to investigate neutralizing responses in critically ill SARS-CoV-2 patients, and is hypothesis-generating for future studies.
5. Conclusions
In summary, we report that critically ill COVID-19 patients exhibited antibody responses to all VOCs that increase over ICU stay; however, the neutralizing response against SARS-CoV-2 variants was delayed and significantly less in Wave-1 (WT infection) as compared to Wave-3 (WT/alpha infection). Naturally infected ICU patients developed adaptive responses to all VOCs, with greater responses in those patients more likely to be infected with the alpha variant, versus WT. The neutralizing antibody response was not affected by SARS-CoV-2 disease severity, but those patients with a previous diagnosis of either cancer or COPD appeared to have insufficient neutralizing antibody responses. These data may influence future care, including patient cohorting, prognostication, therapies and expectant management of late complications.
Funding
DDF acknowledges funding from Western University (Research), the Departments of Medicine and Pediatrics at Western University, the Lawson Health Research Institute (https://www.lawsonresearch.ca/), the London Health Sciences Foundation (https://lhsf.ca/), the London Community Foundation, and the AMOSO Innovation Fund (INN20-029). PYK was funded by the Department of Medicine, Mid-Career Award, CHY was funded by the Michael DeGroot Fellowship Award, GKJ was funded by the CanVECTOR Studentship Award and the McMaster University group was funded by the Canadian Institutes of Health Research (VR2-172768).
Declaration of competing interest
Some authors are employees of Thermo Fisher Scientific Inc. who manufacture and distribute the anti-SARS-CoV-2 immunoglobulin assays as research use only reagents.
Acknowledgments
We thank the enthusiastic support of the frontline Critical Care Nursing Staff and Research Assistants at London Health Sciences Centre and Hamilton General Hospital.
References
- 1.Armstrong R.A., Kane A.D., Cook T.M. Outcomes from intensive care in patients with COVID-19: a systematic review and meta-analysis of observational studies. Anaesthesia. 2020;75(10):1340–1349. doi: 10.1111/anae.15201. [DOI] [PubMed] [Google Scholar]
- 2.Koyama S., Ishii K.J., Coban C., Akira S. Innate immune response to viral infection. Cytokine. 2008;43(3):336–341. doi: 10.1016/j.cyto.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Iosef C., Martin C.M., Slessarev M., Gillio-Meina C., Cepinskas G., Han V.K.M., et al. COVID-19 plasma proteome reveals novel temporal and cell-specific signatures for disease severity and high-precision disease management. J. Cell Mol. Med. 2022:1–17. doi: 10.1111/jcmm.17622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garvin M.R., Alvarez C., Miller J.I., Prates E.T., Walker A.M., Amos B.K., et al. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;9 doi: 10.7554/eLife.59177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser D.D., Cepinskas G., Slessarev M., Martin C., Daley M., Miller M.R., et al. Inflammation profiling of critically ill coronavirus disease 2019 patients. Crit. Care Explor. 2020;2(6) doi: 10.1097/CCE.0000000000000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill S.E., dos Santos C.C., O’Gorman D.B., Carter D.E., Patterson E.K., Slessarev M., et al. Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive Care Med. Exp. 2020;8(1):75. doi: 10.1186/s40635-020-00361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juneja G.K., Castelo M., Yeh C.H., Cerroni S.E., Hansen B.E., Chessum J.E., et al. Biomarkers of coagulation, endothelial function and fibrinolysis in critically-ill patients with COVID-19: a single-centre prospective longitudinal study. J. Thromb. Haemostasis. 2021;19(6):1546–1557. doi: 10.1111/jth.15327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cani E., Dwivedi D.J., Liaw K.-L., Fraser D.D., Yeh C.H., Martin C., et al. Immunothrombosis biomarkers for distinguishing coronavirus disease 2019 patients from noncoronavirus disease septic patients with pneumonia and for predicting ICU mortality. Crit. Care Explor. 2021;3(12) doi: 10.1097/CCE.0000000000000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser D.D., Patterson E.K., Slessarev M., Gill S.E., Martin C., Daley M., et al. Endothelial injury and glycocalyx degradation in critically ill coronavirus disease 2019 patients: implications for microvascular platelet aggregation. Crit. Care Explor. 2020;2(9) doi: 10.1097/CCE.0000000000000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser D.D., Cepinskas G., Slessarev M., Martin C.M., Daley M., Patel M.A., et al. Critically ill COVID-19 patients exhibit anti-SARS-CoV-2 serological responses. Pathophysiology. 2021;28(2):212–223. doi: 10.3390/pathophysiology28020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser D.D., Cepinskas G., Slessarev M., Martin C.M., Daley M., Patel M.A., et al. Detection and profiling of human coronavirus immunoglobulins in critically ill coronavirus disease 2019 patients. Crit. Care Explor. 2021;3(3) doi: 10.1097/CCE.0000000000000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
- 14.Aleem A., Akbar Samad A.B., Slenker A.K. StatPearls; Treasure Island (FL): 2021. Emerging Variants of SARS-CoV-2 and Novel Therapeutics against Coronavirus (COVID-19) [Google Scholar]
- 15.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahmi M., Kharisma V.D., Ansori A.N.M., Ito M. Retrieval and investigation of data on SARS-CoV-2 and COVID-19 using bioinformatics approach. Adv. Exp. Med. Biol. 2021;1318:839–857. doi: 10.1007/978-3-030-63761-3_47. [DOI] [PubMed] [Google Scholar]
- 17.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SeyedAlinaghi S., Mirzapour P., Dadras O., Pashaei Z., Karimi A., MohsseniPour M., et al. Characterization of SARS-CoV-2 different variants and related morbidity and mortality: a systematic review. Eur. J. Med. Res. 2021;26(1):51. doi: 10.1186/s40001-021-00524-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fraser D.D., Miller M.R., Martin C.M., Slessarev M., Hahn P., Higgins I., et al. Cohort-specific serological recognition of SARS-CoV-2 variant RBD antigens. Ann. Clin. Lab. Sci. 2022;52(4):651–662. [PubMed] [Google Scholar]
- 21.SeyedAlinaghi S., Afsahi A.M., MohsseniPour M., Behnezhad F., Salehi M.A., Barzegary A., et al. Late complications of COVID-19; a systematic review of current evidence. Arch. Acad. Emerg. Med. 2021;9(1):e14. doi: 10.22037/aaem.v9i1.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.https://www.cdc.gov/coronavirus/2019-nCoV/hcp/clinical-criteria.html.
- 23.https://www.fda.gov/media/134922/download.
- 24.Priestap F., Kao R., Martin C.M. External validation of a prognostic model for intensive care unit mortality: a retrospective study using the Ontario Critical Care Information System. Can. J. Anaesth. 2020;67(8):981–991. doi: 10.1007/s12630-020-01686-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brisson A.R., Matsui D., Rieder M.J., Fraser D.D. Translational research in pediatrics: tissue sampling and biobanking. Pediatrics. 2012;129(1):153–162. doi: 10.1542/peds.2011-0134. [DOI] [PubMed] [Google Scholar]
- 27.Gillio-Meina C., Cepinskas G., Cecchini E.L., Fraser D.D. Translational research in pediatrics II: blood collection, processing, shipping, and storage. Pediatrics. 2013;131(4):754–766. doi: 10.1542/peds.2012-1181. [DOI] [PubMed] [Google Scholar]
- 28.Program NHRP . 2009. POLICY: Guidelines for Limits of Blood Drawn for Research Purposes in the Clinical Center M95-9 (Rev) [Google Scholar]
- 29.Thye A.Y., Law J.W., Pusparajah P., Letchumanan V., Chan K.G., Lee L.H. Emerging SARS-CoV-2 variants of concern (VOCs): an impending global crisis. Biomedicines. 2021;9(10) doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nidom R.V., Indrasari S., Normalina I., Nidom A.N., Afifah B., Dewi L., et al. Phylogenetic and full-length genome mutation analysis of SARS-CoV-2 in Indonesia prior to COVID-19 vaccination program in 2021. Bull. Natl. Res. Cent. 2021;45(1):200. doi: 10.1186/s42269-021-00657-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 32.Trombetta C.M., Marchi S., Viviani S., Manenti A., Benincasa L., Ruello A., et al. Serum neutralizing activity against B.1.1.7, B.1.351, and P.1 SARS-CoV-2 variants of concern in hospitalized COVID-19 patients. Viruses. 2021;13(7) doi: 10.3390/v13071347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern. Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhatraju P.K., Ghassemieh B.J., Nichols M., Kim R., Jerome K.R., Nalla A.K., et al. Covid-19 in critically ill patients in the seattle region - case series. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA. 2020;323(16):1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593(7858):270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grint D.J., Wing K., Williamson E., McDonald H.I., Bhaskaran K., Evans D., et al. Case fatality risk of the SARS-CoV-2 variant of concern B.1.1.7 in England, 16 November to 5 February. Euro Surveill. 2021;26(11) doi: 10.2807/1560-7917.ES.2021.26.11.2100256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kow C.S., Merchant H.A., Hasan S.S. Mortality risk in patients infected with SARS-CoV-2 of the lineage B.1.1.7 in the UK. J. Infect. 2021;83(1):e14–e15. doi: 10.1016/j.jinf.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Group R.C., Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., et al. Dexamethasone in hospitalized patients with covid-19. N. Engl. J. Med. 2021;384(8):693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hojjat Jodaylami M., Djaileb A., Ricard P., Lavallee E., Cellier-Goetghebeur S., Parker M.F., et al. Cross-reactivity of antibodies from non-hospitalized COVID-19 positive individuals against the native, B.1.351, B.1.617.2, and P.1 SARS-CoV-2 spike proteins. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-00844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384(15):1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aleman A., Upadhyaya B., Tuballes K., Kappes K., Gleason C.R., Beach K., et al. Variable cellular responses to SARS-CoV-2 in fully vaccinated patients with multiple myeloma. Cancer Cell. 2021;39(11):1442–1444. doi: 10.1016/j.ccell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenberger L.M., Saltzman L.A., Senefeld J.W., Johnson P.W., DeGennaro L.J., Nichols G.L. Antibody response to SARS-CoV-2 vaccines in patients with hematologic malignancies. Cancer Cell. 2021;39(8):1031–1033. doi: 10.1016/j.ccell.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt A.L., Labaki C., Hsu C.Y., Bakouny Z., Balanchivadze N., Berg S.A., et al. COVID-19 vaccination and breakthrough infections in patients with cancer, 33(3) Ann. Oncol. 2021:340–346. doi: 10.1016/j.annonc.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nath K., Viswanathan S., Upham J., Davies J., Towers M., Looke D., et al. Clinical factors associated with the humoral immune response to influenza vaccination in chronic obstructive pulmonary disease. Int. J. Chronic Obstr. Pulm. Dis. 2013;9:51–56. doi: 10.2147/COPD.S53590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khateeb J., Li Y., Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care. 2021;25(1):244. doi: 10.1186/s13054-021-03662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371(6534):1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7) doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med. 2021;27(4):620–621. doi: 10.1038/s41591-021-01270-4. [DOI] [PubMed] [Google Scholar]
- 51.Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184(11):2939–29354 e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184(9):2348–23461 e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184(9):2372–23783 e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bates T.A., McBride S.K., Winders B., Schoen D., Trautmann L., Curlin M.E., et al. Antibody response and variant cross-neutralization after SARS-CoV-2 breakthrough infection. JAMA. 2021;327(2):179–181. doi: 10.1001/jama.2021.22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Begin P., Callum J., Jamula E., Cook R., Heddle N.M., Tinmouth A., et al. Convalescent plasma for hospitalized patients with COVID-19: an open-label, randomized controlled trial. Nat. Med. 2021;27(11):2012–2024. doi: 10.1038/s41591-021-01488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21(6):382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mehraeen E., Dadras O., Afsahi A.M., Karimi A., Pour M.M., Mirzapour P., et al. Vaccines for COVID-19: a systematic review of feasibility and effectiveness. Infect. Disord.: Drug Targets. 2022;22(2) doi: 10.2174/1871526521666210923144837. [DOI] [PubMed] [Google Scholar]
- 58.Oliaei S., SeyedAlinaghi S., Mehrtak M., Karimi A., Noori T., Mirzapour P., et al. The effects of hyperbaric oxygen therapy (HBOT) on coronavirus disease-2019 (COVID-19): a systematic review. Eur. J. Med. Res. 2021;26(1):96. doi: 10.1186/s40001-021-00570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]