Abstract
Objective:
The transition from psoriasis (Ps) to psoriatic arthritis (PsA) occurs in 20-30% patients, however, the mechanisms underlying the emergence of musculoskeletal disease are not well understood. Metabolic disease is prevalent in Ps patients, but whether metabolic factors, other than obesity, increase arthritis risk in Ps patients is not known.
Methods:
To characterize the metabolic alterations during the progression of arthritis in Ps patients, we analyzed cross-sectional healthy control (HC) and PsA samples and longitudinal Ps serum samples, before and after PsA onset. Non-targeted metabolomic profiling was performed using liquid chromatography–mass spectrometry (LC-MS).
Results:
We identified several serum metabolites that differed between PsA, Ps patients and HC. Differentially abundant bile acids, purines, pyrimidines, glutathione, lipids and amino acid metabolites were noted in these 3 groups. We also noted differences between Ps patients who progressed (PsP) or did not progress (PsNP) to PsA. Bile acids and butyrate were depressed in PsP compared to PsNP patients and the level of inflammatory lipid mediators increased following PsA diagnosis. In particular, the combination of leukotriene B4 and glycoursodeoxycholic acid sulfate were sensitive and specific predictors of PsA progression
Conclusion:
We observed notable differences in bile acid, purine, lipid, and amino acid-derived metabolites, among the HC, Ps and PsA patients and identified changes during the transition from Ps to PsA. The decreased bile acid, butyrate and elevated guanine levels in Ps patients at risk for PsA were particularly striking and may reflect gut microbial dysbiosis and dysregulated hepatic metabolism leading to altered proliferation of immune cells and enhanced cytokine expression.
Keywords: Psoriatic arthritis, psoriasis, metabolomics, longitudinal cohort, bile acid
INTRODUCTION
Psoriatic arthritis (PsA) is closely associated with psoriasis (Ps), and 20-30% of patients with psoriasis develop PsA (1). Arthritis onset in Ps patients is often characterized by development of fatigue and arthralgia, while other patients develop rapid onset of joint pain and swelling (2). Several studies indicate that aberrant inflammatory pathways are triggered long before symptom onset, and the wide spectrum of presenting symptoms often results in delayed diagnosis, thereby increasing the likelihood of long-term musculoskeletal damage and permanent disability (3). Therefore, early identification of Ps patients at increased risk for PsA holds potential to facilitate prompt therapeutic intervention and improve outcomes. Unfortunately, established serum diagnostic biomarkers of PsA and arthritis risk in Ps patients are not available.
One of the most striking relationships in psoriatic disorders is the strong association between obesity and the PsA development (4). Furthermore, key metabolic diseases are often present in established PsA patients including type 2 diabetes (5), metabolic syndrome (6), and fatty liver (7). Moreover, PsA patients are at increased risk to develop cardiovascular disease compared to controls (8). These epidemiologic associations raise the possibility of an interaction between metabolic and inflammatory pathways that favor the transition to PsA and propel synovial inflammation, enthesitis and joint damage. This potential metabolic-inflammation link (9) may provide opportunities to identify diagnostic and PsA transition biomarkers.
Several reports examined serum and plasma metabolites in Ps, PsA patients, and healthy controls (HC) (10–14). Comparison of Ps and PsA serum or plasma metabolites revealed differences between Ps and PsA patients (10–15). The majority of these prior studies were cross-sectional and a significant proportion of patients were treated with systemic therapies (including biologics) before metabolomic profiling. Importantly, systemic treatments such as anti-TNF agents significantly alter patient metabolic profiles (16) and may obscure interpretation of metabolomic shifts that take place before or after disease onset. To investigate the link between metabolic changes and disease progression in Ps patients, we monitored a longitudinal cohort of biologically naive Ps patients, collected serial serum samples, and evaluated molecular differences in Ps patients who did (PsP) and did not progress to PsA (PsNP) and were not on systemic therapies. To reveal metabolomic changes, we performed non-targeted metabolomic profiling of the collected serum samples using liquid chromatography–mass spectrometry (LC-MS).
MATERIALS AND METHODS
A). Study design and patient cohort
This study was performed at the University of Rochester Medical Center and conducted in compliance with Helsinki principles. The study protocol was approved by the Institutional Review Board (IRB) of the University of Rochester. Serum samples were collected from Ps and PsA patients along with healthy controls (HC) recruited and consented in the dermatology and rheumatology clinics of the University of Rochester Medical Center, Patients were enrolled in the International Psoriatic Arthritis Research Team (IPART) Registry, a longitudinal cohort of Ps and PsA patients. The diagnosis of Ps was confirmed by a dermatologist (Francisco Tausk) and the diagnosis of PsA by a rheumatologist (Christopher Ritchlin) based on Classification Criteria for Psoriatic Arthritis (CASPAR) criteria (17). Psoriasis and PsA patients on systemic agents including Disease Modifying Anti-Rheumatic Drugs (DMARDs) or biologic agents were excluded. PsA patients were enrolled in the IPART Registry before starting systemic therapies and served as cross-sectional controls. Psoriasis patients, not on systemic therapy, were also enrolled in the registry and followed longitudinally for development of PsA based on CASPAR criteria.
Patients in the IPART Registry were contacted by a coordinator every 6 months and completed a questionnaire. If they developed new musculoskeletal pain, they were evaluated by a rheumatologist for development of PsA (CASPAR criteria). They were also instructed to contact coordinators if they developed new joint symptoms. Patients who developed PsA had serum samples collected prior to initiation of systemic therapy which were stored in −80 freezer for subsequent analysis. The average patient follow-up was 4.5 years.
B). Serum metabolomic analysis
Serum samples were collected, centrifuged at 1700g for 10 min, and immediately stored at −80 degree Celsius. To identify critical metabolic differences between Ps, and PsA patients, we performed non-targeted global metabolic profiling of serum samples collected from a subset of PsNP (n=21) and PsA-T (n=34) patients in our biorepository and compared them with serum metabolites of healthy subjects (n=16). Frozen serum sample aliquots were shipped on dry ice for the metabolomic profiling at a facility of Metabolon Inc, Durham, NC, USA. Thereby, samples were analyzed using reverse-phase and Hydrophilic Interaction Liquid Chromatography (HILIC) coupled to Q-Exactive mass spectrometry with the collected serum samples (18).
C). Statistical approach
Statistical analysis was used to identify significantly altered metabolites within the cohorts. The raw metabolomic profiling data without further normalization was obtained from Metabolon Inc. Subsequently; appropriate statistical analysis was performed with the metabolomics data after performing cumulative sum scaling (CSS) normalization. The abundance of specific metabolites present in the serum samples was compared between groups based on the likelihood ratio test and considered statistically significant at a false discovery rate (FDR)-corrected p-value of <0.05, referred to as differentially abundant metabolites (DAM). In-depth analysis was performed to compare two groups (e.g., Ps vs. PsA) or to compare multiple groups (HC, Ps, and PsA), as specified in the text. Data were tested for normal distribution on a per-metabolite basis within each sample group using a Jarque Bera test for skewness and kurtosis. Of 846 metabolites, the number normally distributed within each sample group was: HC: 514, PsNP: 440, PsA: 458, PsP-B: 572, PsP-A: 608. Overall, 61.3% of the data were normally distributed. Thus, non-parametric testing (Mann-Whitney U-test) was performed to determine significance in comparisons between groups. Cluster analysis was based on Ward’s method to create heatmaps (19).
Multivariate analysis was performed using a combination of principal component analysis (PCA) and orthogonal projection to latent structures–discriminant analysis (PLS-DA) (20) using MetaboAnalyst (19). Sparse PLS-DA was conducted following logarithmic transformation (base 10) and pareto scaling, and considering all 846 variables per component.
RESULTS
We recruited 71 subjects in this pilot study. Thirty-four Ps patients were recruited and followed longitudinally for development of arthritis. Twenty-one Ps patients were non-progressors and did not transition to PsA (PsNP) while 13 patients (PsP) developed PsA during a 4-year time period. We examined and collected sera on the PsP patients before (PsP-B) and after (PsP-A) the development of PsA. We enrolled 21 PsA, and 16 healthy patients (HC) as cross-sectional controls. The PsA-T group is comprised of the cross-sectional PsA and PsP-A patients. The PsP-A patients met CASPAR criteria for PsA and the metabolomic profiles of patients in the PsA and PsP-A groups demonstrated overlapping characteristics so they were analyzed together (Supplementary Figure 1). Analysis of the serum samples using LC-MS enabled the quantification of 846 metabolites with adequate coverage. A schematic of the patient cohorts is depicted in Figure 1A and Figure 1B demonstrates patient demographics and phenotypic features. The patients had a wide range of baseline PASI scores (9.8 +/− 9.05) and the 21 PsA patients had polyarthritis at baseline. A wide range of disease duration of PsA was observed in the cross-sectional PsA group and varied from less than one year to 26 years with a median of three years.
Figure 1. Subject cohorts.
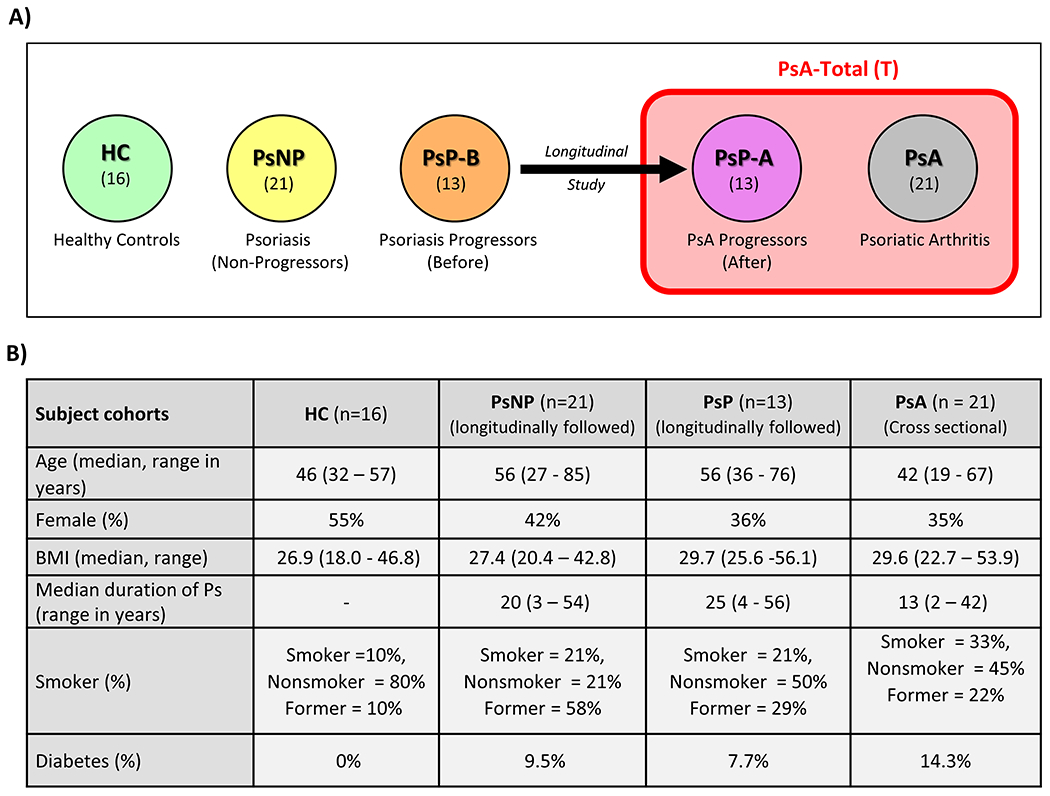
(A) Schematic view of subject cohorts. (B) Clinical and demographic data of study participants.
The characteristics of the PsA cross-sectional and Ps longitudinal patients were consistent across the groups. Group level comparisons were performed to identify differentially abundant metabolites between: a) PsA and Ps, and b) PsP and PsNP (supplementary table 1) over a period of 4 years. Statistical analysis using sparse partial least squares discriminant analysis (sPLDSA) (20) identified metabolites associated with diverse metabolic pathways that differed between PsNP and PsA-T, or PsP and PsNP. We also focused on identifying the major metabolic changes observed during disease progression to PsA.
Metabolic differences between Controls, Ps and PsA patients
Serum metabolites were compared in 34 PsA-T, 21 Ps and 16 HC patients. We identified distinct differences between the 3 groups (Figure 2A). The statistical analysis and subsequent visualization of the metabolomics data in the volcano plot revealed significant differences in metabolite levels between PsNP and PsA-T (Figure 2B). Most notably, the abundance of metabolites related to lipid and bile acid metabolism was altered in the sera of PsA-T patients compared to PsNP. Specifically, a higher level of 12-HHTrE in PsA-T patients (>2-fold, p <0.0001) compared to PsNP and HC. In addition, decreased levels of bile acids were observed in sera from PsA-T compared to PsNP patients. Among these, levels of 3 secondary bile acids, glycoursodeoxycholic acid sulfate, glycodeoxycholate 3-sulfate, and deoxycholic acid 12-sulfate, significantly differed between these 2 groups (all >50% lower in PsA-T compared to PsNP, p <0.001) (Figure 2B). Analysis also revealed higher levels (30 - 70% higher, p <0.01) of metabolites related to nucleic acid (purine) metabolism including guanine, in PsA-T compared to PsNP samples.
Figure 2. Metabolomic differences between healthy control, psoriasis and psoriatic-arthritis patients.
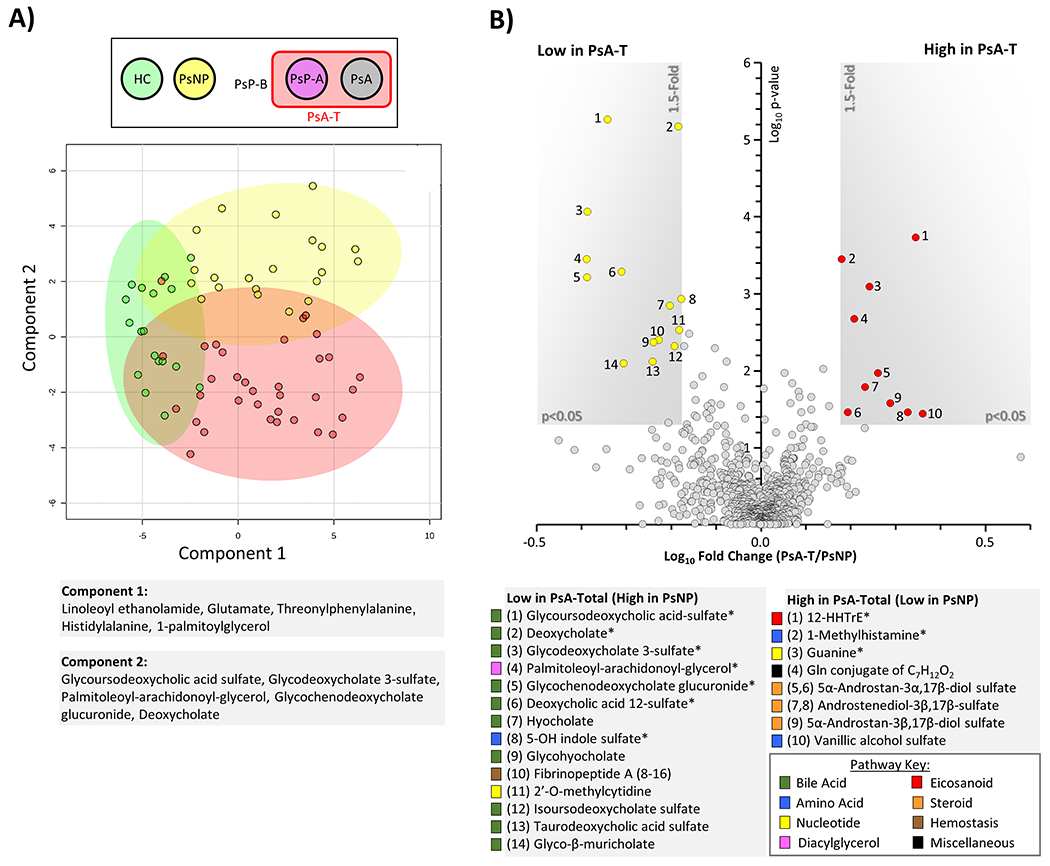
(A): Sparse partial least squares discriminant analysis (sPLDSA) between healthy controls (HC, green), Ps non-progressors (PsNP, yellow), and PsA-total (PsA-T, red) patients. sPLSDA plot from MetaboAnalyst 5 software is shown, with shaded areas representing 95% confidence intervals for each group. Alongside axes are shown the top 5 loadings for each principal component. (B): Volcano plot comparison between PsA-T and PsNP. X-axis shows Log10 of fold change. Y-axis shows −Log10 of p-value (Mann-Whitney U test). Metabolites achieving both statistical significance (p<0.05) and greater than 1.5-fold difference between groups, are shown inside the shaded gray areas, and identified with numbers (see legend below plot). The relevant pathways for individual metabolites are color-coded (see pathway key). Data are means from 21 PsNP and 34 PsA-T individuals. Metabolites marked with an asterisk (*) are significant after FDR correction (Storey q value <0.05).
Metabolic differences between PsNP and PsP
To identify metabolic differences between PsP and PsNP, we analyzed serum samples from 13 PsP-B, and from 21 PsNP patients. sPLSDA revealed significant changes along the component 2 axis for the PsP relative to PsNP (Figure 3), whereas within the PsP group, differences were primarily observed along the component 1 axis. Note the shift in metabolite profiles in the transition to PsA. Further analysis revealed that 6 key metabolites related to bile acid metabolism were significantly lower (p<0.01) in PsP-B compared to PsNP (Figure 4A). Among these metabolites, the secondary bile acid deoxycholate, glycoursodeoxycholic acid sulfate, and deoxycholic acid 12-sulfate, were most differentially abundant (>50% lower in PsP-B, p <0.001). The heat map in Figure 4B shows differences in the metabolite levels which are consistent within the individual patient cohorts. Multiple logistic regression analysis revealed that both leukotriene B4 and glycoursodeoxycholic acid sulfate in log scale were significant predictors for progression to PsA. Higher levels of leukotriene B4 and glycoursodeoxycholic acid sulfate significantly decreased the odds of progressing to PsA. For every one-unit increase in leukotriene B4 levels in log scale, the odds of progressing to PsA decreases 47% (adjusted OR = 0.53, 95% CI: 0.30 – 0.96). Similarly, for every one-unit increase in glycoursodeoxycholic acid sulfate in log scale, the odds of progressing to PsA decrease 95% (adjusted OR = 0.05, 95% CI: 0.01 – 0.49). The ROC curve (Figure 4C) demonstrated both high sensitivity and high specificity for the two biomarkers in predicting progression to PsA with area under the curve equal to 0.9462 (Figure 4C).
Figure 3. Metabolomic differences between non-progressors and progressors.
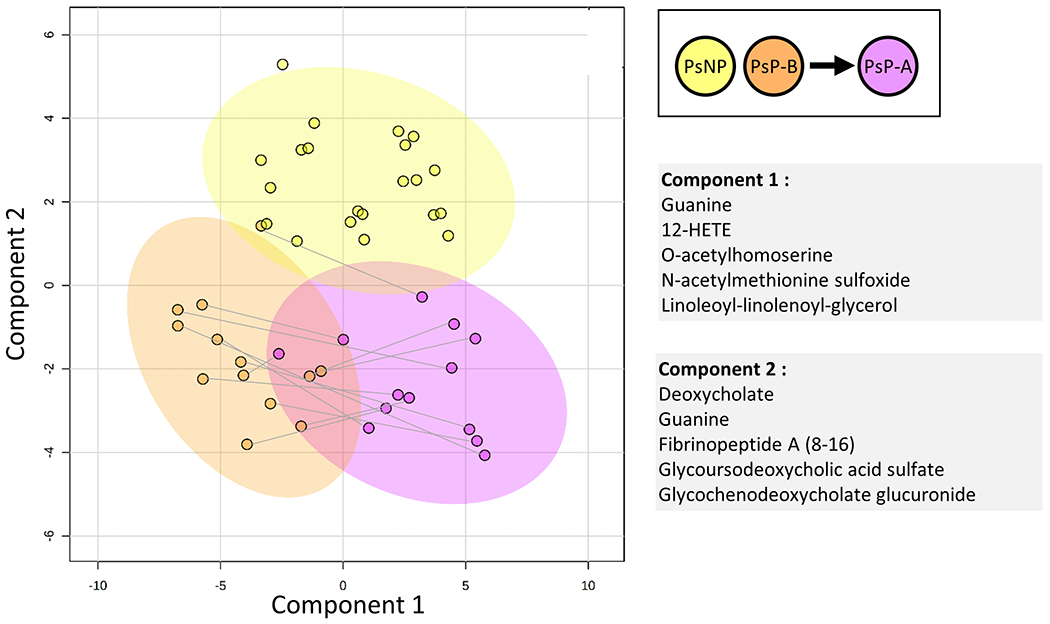
Sparse partial least squares discriminant analysis (sPLDSA) between PsNP (yellow), and PsP-B (orange), and PsP-A (purple) patients. sPLSDA plot from MetaboAnalyst 5 software is shown, with shaded areas representing 95% confidence intervals for each group. Alongside axes are shown the top 5 loadings for each principal component.
Figure 4. Metabolomic differences between PsNP and PsP.
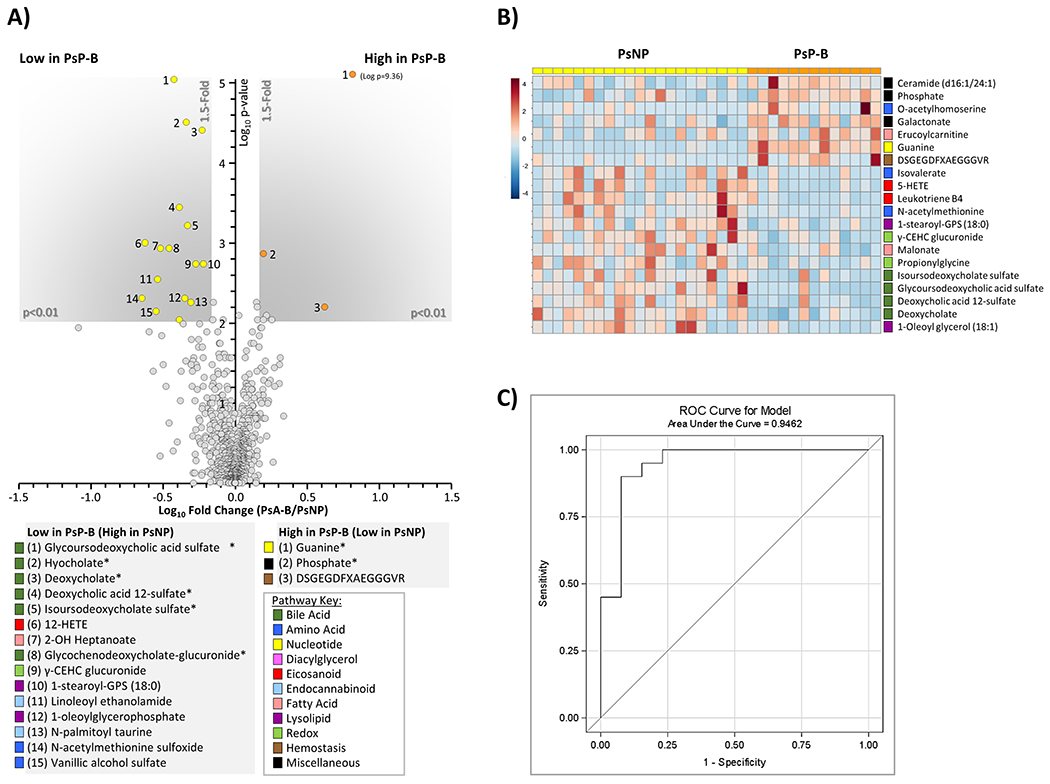
(A): Volcano plot showing metabolites different between PsNP and PsP-B. X-axis shows Log10 of fold change. Y-axis shows −Log10 of p-value (Mann-Whitney U test). Metabolites achieving both statistical significance (p<0.01) and greater than 1.5-fold difference between groups, are shown inside the shaded gray areas, and identified with numbers (see legend below plot). Data are respective means from 21 PsNP and 13 PsP-B samples. Metabolites marked with an asterisk (*) are significant after FDR correction (Storey q value <0.05). The relevant pathways for individual metabolites are color-coded (see pathway key). (B): Cluster heat map (MetaboAnalyst 5.0, Euclidean/Ward algorithm) showing top 20 altered metabolites by t-test. The relevant pathways for individual metabolites are color-coded (see pathway key). (C): ROC curve for the multiple logistic regression model.
The level of bile acids differed significantly between PsNP and PsP-B patients (Figure 5A). In addition, levels of bile acids were lower in PsA-T compared to PsNP. The synthesis of primary bile acids takes place in the liver and secondary bile acid production results from metabolism of primary bile acid metabolites by the gut microbiota as shown in Figure 5B.
Figure 5. Bile acid levels differ significantly between PsNP and PsP patients.
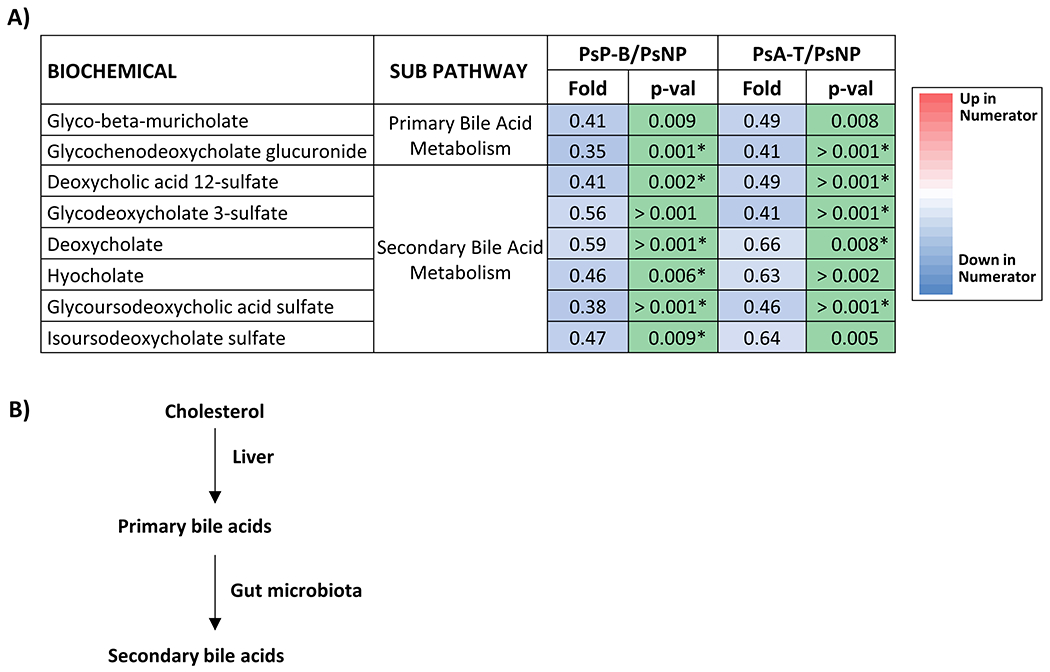
A) Differential abundance of primary and secondary bile acids in samples collected from Ps and PsA patients. Serum samples were analyzed with an unbiased approach and detected primary and secondary bile acid levels were compared between Ps patients who did (PsP, n=13) or did not progress (PsNP, n=21). Samples collected before or after the onset of arthritis from 13 PsP patients were compared with 21 PsNP. Differentially abundant primary and secondary bile acids achieving statistical significance (p<0.05) are shown. In addition, metabolites that are significant after FDR correction (Storey q value <0.05) are further marked with an asterisk (*); B) schematic diagram depicting synthesis of primary and secondary bile acids from cholesterol in the liver and microbial metabolism in the gut.
Metabolic shifts in the patients who progressed to PsA
To identify the major metabolic changes during the transition from Ps to PsA, we compared longitudinal serum samples collected before and after the onset of arthritis in 13 PsP patients. Levels of multiple purines, lipid and amino acid metabolites changed during the transition to PsA (Figures 6A, 6B). The levels of cysteinyl-glycine, guanine, and linoleoyl-linoleoyl glycerol levels (Figure 6A, 6B) significantly decreased (>50%, p <0.001), after the onset of PsA. Linoleoyl glycerol is an endocannabinoid that is transformed by neutrophils and eosinophils to novel lipoxygenase metabolites (21) and cysteinyl-glycine is a dipeptide intermediate in glutathione metabolism (22). In contrast, multiple inflammatory lipid metabolites (4-HDoHE, 12-HETE, 12-HHTrE) were elevated (>2-fold, p<0.01) in the PsP-A patients. In addition, significant increases in several metabolites related to glutathione metabolism were noted after PsA progression, including 5-oxoproline which exhibited a >2-fold increase, p <0.001. Notably, although guanine was elevated prior to PsA progression, the level decreased following PsA onset (Figure 6C). In contrast, guanine levels were comparable between the PsNP and HC (Supplementary Figure 2A). The low level of bile acids in the PsP-B patients remained low following development of PsA. The level of butyrate, a key short chain fatty acid (SCFA) produced during gut flora-mediated fermentation of dietary fibers, was significantly lower (p <0.05) in PsP-B compared to PsNP but increased following the development of PsA (Figure 6D). The levels of butyrate were higher in PsNP compared to the HC and significantly higher compared to the PsP-B group (Supplementary Figure 2B). These data suggest that elevated butyrate in the PsNP group lessen the probability of developing arthritis but further studies are required to support this impression.
Figure 6. Metabolomic differences during longitudinal progression from Ps to PsA.
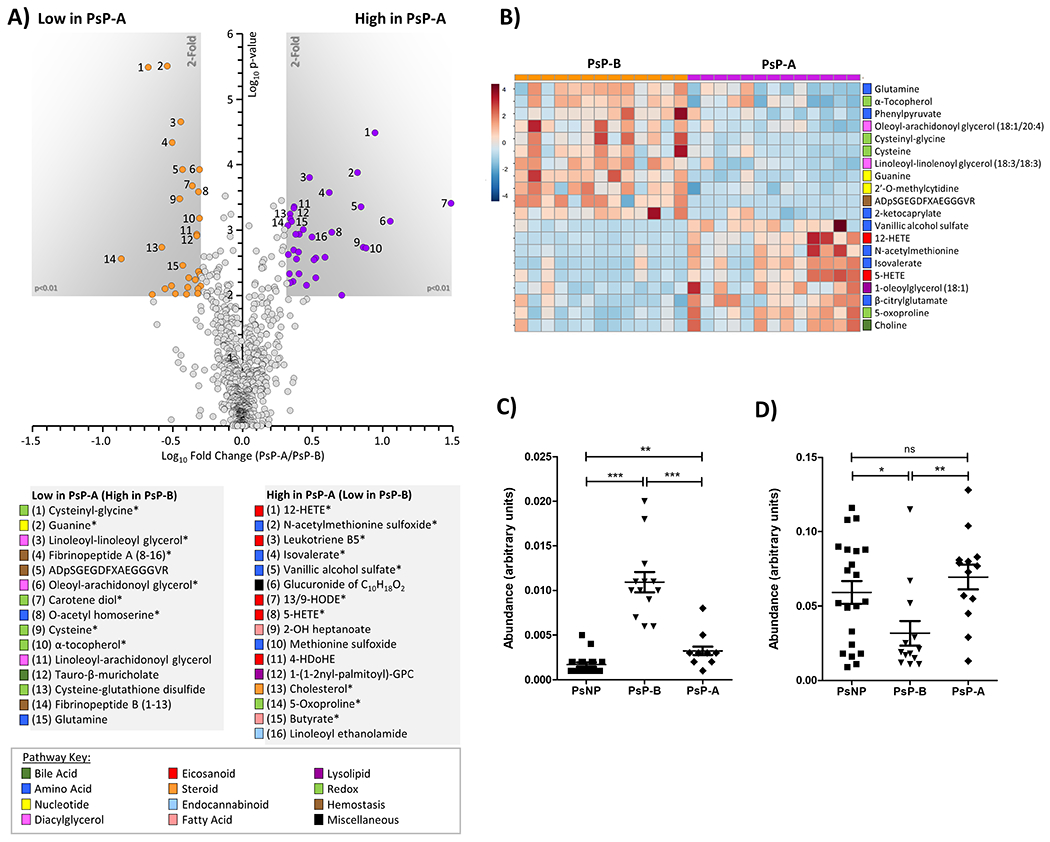
(A): Volcano plot comparing metabolites between PsP-B and PsP-A patients. X-axis shows Log10 of fold change. Y-axis shows −Log10 of p-value (Mann-Whitney U-test). Metabolites achieving both statistical significance (p<0.01) and greater than 1.5-fold difference between groups, are shown inside the shaded gray areas, and identified with numbers (see legend below plot). Data are means from 13 individuals. Metabolites marked with an asterisk (*) are significant after FDR correction (Storey q value <0.01). The relevant pathways for individual metabolites are color-coded (see pathway key). (B): Cluster heat map (MetaboAnalyst 5.0, Euclidean/Ward algorithm) showing top 20 altered metabolites by t-test achieving p<0.05. The relevant pathways for individual metabolites are color-coded (see pathway key). (C and D): Serum guanine (C) and butyrate (D) levels were compared between samples from 13 PsP and 21 PsNP and normalized data presented as a dot plot. ‘***’ indicates p-value less than 0.0001 and ‘*’ indicates p-value less than 0.01.
These findings highlight significant metabolic shifts in the transition to PsA characterized by depressed levels of dipeptides, guanine, bile acids and butyrate and increased levels of inflammatory lipid metabolites and glutathione metabolites following arthritis onset.
DISCUSSION
The circulating metabolome provides a continuous record of the production and consumption of metabolites and reflects the combined input from environmental exposures and the gut microbiota (23, 24). Moreover, metabolite analysis reveals pathways and mechanisms downstream of proteomics and genomics (24). We identified key metabolites that differ in abundance between Ps, PsA patients and controls. We observed a metabolomic shift in bile acids and butyrate in Ps patients who progressed to PsA compared to those who did not progress. These findings provide insights into novel pathways linked to microbial dysbiosis and dysregulated hepatic metabolism with potential to alter the immune response and promote joint inflammation.
Serum primary and secondary bile metabolites were significantly lower in Ps-P compared to PsNP patients and remained depressed following the onset of arthritis. Primary bile acids (cholic and deoxycholic acid) are synthesized in the liver from cholesterol, conjugated to glycine or taurine and secreted in the duodenum (25). The majority of bile acids (95%) are reabsorbed in the ileum and undergo enterohepatic circulation and re-enter the liver via the portal vein (26, 27). The remaining 5% of bile acids reach the colon and are deconjugated and metabolized to secondary bile acids by the resident microbiota. Bile acids are required for the emulsification of lipids, glucose metabolism, host defense and immune homeostasis (28).
Bile acids regulate immunologic responses by multiple mechanisms. Both primary and secondary bile acids activate nuclear and plasma membrane receptors including farnesoid X receptor (FXR) and-protein-coupled bile acid receptor-1 (TGR5) expressed on macrophages, dendritic cells and NKT cells (29). Engagement of these receptors-by bile acids promote an anti-inflammatory response through the suppression of Tumor Necrosis Factor (TNF), Interleukin (IL)-6 and IL-8 and by inhibition of the Nuclear Factor (NF)κB transcription factor (30). Secondary bile acids suppress T-helper 17 (Th17) cell differentiation by direct suppression of RAR-related orphan receptor (ROR)γT and enhance the differentiation of CD4+ lymphocytes to FOXP3+ T regulatory cells (31, 32). Bile acids also modulate the gut microbiome structure by controlling bacterial overgrowth and protecting the epithelial barrier via FXR signaling (33). Thus, bile acids maintain immune homeostasis through regulation of the microbial environment, inhibition of innate immune signaling and suppression of Th17 differentiation.
Decreased serum bile acid levels are associated with several immune-mediated inflammatory diseases, including multiple sclerosis (23), inflammatory bowel disease (34), and Ps (35). We did not observe a significant difference in bile acid levels between Ps patients and HCs, however, primary and secondary bile acids were reported to be lower in Ps patients although levels in those who went on to develop PsA were not examined (35, 36). Based on the observation that rheumatoid arthritis (RA) improved in patients with jaundice (37), RA patients were treated with a short course of intravenous bile acids and experienced significant short-term improvement in arthritis but treatment was complicated by development of phlebitis (38). In a subsequent report, expression of the TGR5 bile acid receptor was significantly lower in RA PBMC compared to healthy controls and levels correlated inversely with the DAS28 outcome measure (30). In preclinical models, therapy with the secondary bile acid, lithocholic acid (LCA), significantly lessened joint inflammation in the collagen-induced arthritis (CIA) model (30), and bile acid treatment suppressed development of psoriasis lesions in the IL-23 minicircle model via inhibition of RORγt and C-C Motif Chemokine Ligand (CCL) 20 expression (39). Thus, accumulating evidence supports a role for disrupted bile acid metabolism in the development of systemic inflammation observed in RA and psoriasis and a number of other immune mediated disorders.
The low serum butyrate levels noted in the Ps-B cohort provide an additional link to dysbiosis in arthritis pathogenesis. Butyrate is a SCFA and the product of gut microbial metabolism that maintains an intact intestinal barrier, inhibits NFκB mediated cytokine release and decreases bacterial translocation (40). Moreover, butyrate treatment of mice prior to the onset of CIA improved intestinal barrier function and significantly decreased the severity of inflammatory arthritis (41). Elevated serum levels of butyrate were associated with non-progression to arthritis in individuals at increased risk (anti-citrullinated antibody positive with musculoskeletal pain) to develop RA (42). Thus, similar to findings in RA, lower levels of serum butyrate noted in the PsP patients may contribute to increased arthritis risk in this population. Interestingly, butyrate levels were depressed in the PsP-B patients but increased after the onset of PsA. The mechanisms underlying this increase are unknown but may represent a homeostatic response to the onset of systemic inflammation following arthritis onset.
Depressed levels of bile acids, particularly primary bile acids may be due to impaired endogenous production of cholesterol, the key precursor. Nonalcoholic fatty liver disease is prevalent in PsA (43) yet levels of primary and secondary bile acids are elevated in this disorder (44). Moreover, none of the 13 patients who developed PsA, had a history of fatty liver, metabolic syndrome or elevated liver function tests prior to arthritis onset. Interactions between cholesterol pathways and immune function are well established and inflammation, via TLR activation, can inhibit cholesterol synthesis (45, 46). Thus, cholesterol biosynthetic pathways can modulate immune function and inflammation can interfere with cholesterol synthesis. Collectively, the lower levels of butyrate and multiple secondary bile acid metabolites in PsP compared to PsNP support the contribution of gut microbiota and related metabolites to arthritis onset. Further studies are required though to determine if the reduced levels of bile acids are also related to altered hepatic synthesis and whether the dysregulation of bile acid and butyrate have a correlative or causative role in the transition to PsA.
Elevated levels of inflammatory and anti-inflammatory lipid metabolites were observed in PsP-A patients, including the docosanoid 4-HDoHE and multiple eicosanoids including 5-HETE, 12-HETE, and 12-HHTrE. It is important to note that Coras et al. performed in-depth lipid profiling and reported elevated levels of many pro and anti-inflammatory eicosanoids in PsA and elevated levels of some eicosanoids correlated with disease severity (10, 15). Although our metabolomics platform was not directed toward in-depth profiling of lipids, our findings from untargeted analysis nevertheless concur with their results (10, 15). The increase of pro-inflammatory lipid metabolites such as leukotriene B4, and leukotriene B5 are indicative of systematic inflammation in PsA patients, while elevated levels of anti-inflammatory eicosanoids such as 5-HETE and 12-HETE likely result from inflammation-triggered responses to minimize tissue damage.
We observed differences in metabolites related to nucleotide metabolism between Ps and PsA patients. The elevated guanine level in Ps patients at risk for PsA was particularly striking, and may reflect altered immune cell proliferation (47) and enhanced cytokine release (48). During the transition from Ps to PsA, we observed a changing pattern of purine and pyrimidine metabolites over time. In particular, guanine levels significantly decreased after the onset of arthritis. This paradoxical observation (increased guanine levels before PsA onset, but decreased after PsA onset) requires further investigation. Published reports of serum or plasma metabolites in Ps and PsA patients showed that despite the apparent overlap in the disease pathology, several metabolic features differ in Ps and PsA patients (11, 13, 15). In particular, distinctive differences in the abundance of key metabolites related to lipid and amino acid metabolism between the Ps and PsA patients were identified (11, 13, 15). However, in these studies, a significant number of PsA patients received systemic treatment before sample collections. Systemic therapies alter metabolic profiles in Ps patients, as demonstrated by a recent study (16).
To our knowledge, our study is the first comprehensive analysis of longitudinal serum metabolites in patients before and after the onset of PsA who are not taking systemic therapies. Despite the advantages of this approach, only 13 patients developed PsA over the course of the study. We are aware that our observations can potentially be confounded by a number of demographic and biological factors. We did match our patients on age and BMI, but we were unable to adjust for other variables such as lipids, physical activity, and hormonal status, which may impact identified metabolites, due to the small sample size. Therefore, although our findings revealed distinctive metabolic alterations during disease progression to PsA, we do not know if these results are characteristic of a larger PsA population, a disease with considerable heterogeneity. The difficulties of recruiting and closely monitoring a large number of Ps patients for the development of PsA over months to years is a significant challenge that will have to be considered in future studies. Furthermore, it is unclear to what degree the changes we observed in the Ps-P cohort are unique to PsA, compared to other forms of inflammatory arthritis such as RA and axial spondyloarthritis (axSpA).
The potential contribution of bile acids and butyrate to RA and other inflammatory disorders reviewed above coupled with reports of altered inflammatory lipid signaling in in both RA (49) and axSpA (50), may limit the utility of such metabolites to serve as a unique PsA biomarker. Nonetheless, our multivariate logistic regression model that included known risk factors for development of PsA revealed that the inclusion of leukotriene B4 and glycoursodeoxycholic acid sulfate improved the sensitivity and specificity of the diagnosis of PsA transition with an AUC of 94%. These findings are very encouraging and point to the possibility of a diagnostic biomarker but require confirmation in a blinded cohorts of psoriasis patients, along with patients at risk for RA and axSpA.
In summary, non-targeted metabolomic analysis of a longitudinal Ps cohort and a cross-sectional group of PsA patients and HC identified key metabolite differences in bile acid, lipid and purine metabolism. Most notable were the bile acid and butyrate signatures in psoriasis patients who went on to develop PsA and the increased level of inflammatory lipid mediators in patients following the diagnosis of PsA. The implications of this study are that the transition from Ps to PsA may be influenced by liver dysfunction and the metabolites produced by commensal gut microbiota and thereby indicate the presence of a metabolic-inflammation axis. Enhanced understanding of the differentially abundant serum metabolites may reveal pivotal disease mechanisms that underlie transition to PsA and biomarkers of arthritis risk in Ps patients.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dustina Holt, Angela Kluzniak, Debbie Campbell, Amy Wielgosz, Amanda Howell, Marc Nuzzo, Samantha Moore and Jennifer Albrecht for their technical assistance in patient selection, sample collection, biobanking of samples in our biorepository, sample retrieving and blinding of samples for unbiased analysis of metabolites in this study. We also want to thank Drs. John Looney and Ben Korman for their informative suggestions.
This work was supported by the funding from National Psoriasis Foundation (PsA Biomarker Grant), AbbVie and UCB Pharmaceuticals through support of the International Psoriatic Arthritis Research Team (IPART) Registry, University of Rochester (Department of Medicine Pilot Grant, and Health Sciences Center for Computational Innovation Pilot Grant), and National Institute of Health (R01-HL071158, R01-AR069000).
Footnotes
All authors declare they have no conflict of interest.
REFERENCES
- 1.Ritchlin CT, Colbert RA, Gladman DD. Psoriatic Arthritis. N Engl J Med. 2017;376(10):957–70. [DOI] [PubMed] [Google Scholar]
- 2.Eder L, Polachek A, Rosen CF, Chandran V, Cook R, Gladman DD. The Development of Psoriatic Arthritis in Patients With Psoriasis Is Preceded by a Period of Nonspecific Musculoskeletal Symptoms: A Prospective Cohort Study. Arthritis Rheumatol. 2017;69(3):622–9. [DOI] [PubMed] [Google Scholar]
- 3.Scher JU, Ogdie A, Merola JF, Ritchlin C. Preventing psoriatic arthritis: focusing on patients with psoriasis at increased risk of transition. Nat Rev Rheumatol. 2019;15(3):153–66. [DOI] [PubMed] [Google Scholar]
- 4.Kumthekar A, Ogdie A. Obesity and Psoriatic Arthritis: A Narrative Review. Rheumatol Ther. 2020;7(3):447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eder L, Chandran V, Cook R, Gladman DD. The Risk of Developing Diabetes Mellitus in Patients with Psoriatic Arthritis: A Cohort Study. J Rheumatol. 2017;44(3):286–91. [DOI] [PubMed] [Google Scholar]
- 6.Haroon M, Gallagher P, Heffernan E, FitzGerald O. High prevalence of metabolic syndrome and of insulin resistance in psoriatic arthritis is associated with the severity of underlying disease. J Rheumatol. 2014;41(7):1357–65. [DOI] [PubMed] [Google Scholar]
- 7.Ortolan A, Lorenzin M, Tadiotto G, Russo FP, Oliviero F, Felicetti M, et al. Metabolic syndrome, non-alcoholic fatty liver disease and liver stiffness in psoriatic arthritis and psoriasis patients. Clin Rheumatol. 2019;38(10):2843–50. [DOI] [PubMed] [Google Scholar]
- 8.Gialouri CG, Fragoulis GE. Cardiovascular disease in Psoriatic arthritis: facts and unmet needs. Rheumatology (Oxford). 2021. [DOI] [PubMed] [Google Scholar]
- 9.Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542(7640):177–85. [DOI] [PubMed] [Google Scholar]
- 10.Coras R, Kavanaugh A, Kluzniak A, Holt D, Weilgosz A, Aaron A, et al. Differences in oxylipin profile in psoriasis versus psoriatic arthritis. Arthritis Res Ther. 2021;23(1):200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armstrong AW, Wu J, Johnson MA, Grapov D, Azizi B, Dhillon J, et al. Metabolomics in psoriatic disease: pilot study reveals metabolite differences in psoriasis and psoriatic arthritis. F1000Res. 2014;3:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koussiouris J, Looby N, Anderson M, Kulasingam V, Chandran V. Metabolomics Studies in Psoriatic Disease: A Review. Metabolites. 2021;11(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Looby N, Roszkowska A, Reyes-Garces N, Yu M, Baczek T, Kulasingam V, et al. Serum metabolic fingerprinting of psoriasis and psoriatic arthritis patients using solid-phase microextraction-liquid chromatography-high-resolution mass spectrometry. Metabolomics. 2021;17(7):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kishikawa T, Arase N, Tsuji S, Maeda Y, Nii T, Hirata J, et al. Large-scale plasma-metabolome analysis identifies potential biomarkers of psoriasis and its clinical subtypes. J Dermatol Sci. 2021;102(2):78–84. [DOI] [PubMed] [Google Scholar]
- 15.Coras R, Kavanaugh A, Boyd T, Huynh Q, Pedersen B, Armando AM, et al. Pro- and anti-inflammatory eicosanoids in psoriatic arthritis. Metabolomics. 2019;15(4):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamleh MA, Snowden SG, Grapov D, Blackburn GJ, Watson DG, Xu N, et al. LC-MS metabolomics of psoriasis patients reveals disease severity-dependent increases in circulating amino acids that are ameliorated by anti-TNFalpha treatment. J Proteome Res. 2015;14(1):557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor W, Gladman D, Helliwell P, Marchesoni A, Mease P, Mielants H, et al. Classification criteria for psoriatic arthritis: development of new criteria from a large international study. Arthritis Rheum. 2006;54(8):2665–73. [DOI] [PubMed] [Google Scholar]
- 18.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 19.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic acids research. 2018;46(W1):W486–W94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee LC, Liong CY, Jemain AA. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: a review of contemporary practice strategies and knowledge gaps. Analyst. 2018;143(15):3526–39. [DOI] [PubMed] [Google Scholar]
- 21.Archambault AS, Tinto F, Dumais E, Rakotoarivelo V, Kostrzewa M, Plante PL, et al. Biosynthesis of the Novel Endogenous 15-Lipoxygenase Metabolites N-13-Hydroxy-octodecadienoyl-ethanolamine and 13-Hydroxy-octodecadienoyl-glycerol by Human Neutrophils and Eosinophils. Cells. 2021;10(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachhawat AK, Yadav S. The glutathione cycle: Glutathione metabolism beyond the gamma-glutamyl cycle. IUBMB Life. 2018;70(7):585–92. [DOI] [PubMed] [Google Scholar]
- 23.Bhargava P, Smith MD, Mische L, Harrington E, Fitzgerald KC, Martin K, et al. Bile acid metabolism is altered in multiple sclerosis and supplementation ameliorates neuroinflammation. J Clin Invest. 2020;130(7):3467–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106(10):3698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019;576(7785):143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann AF, Hagey LR. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell Mol Life Sci. 2008;65(16):2461–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiorucci S, Carino A, Baldoni M, Santucci L, Costanzi E, Graziosi L, et al. Bile Acid Signaling in Inflammatory Bowel Diseases. Dig Dis Sci. 2021;66(3):674–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perino A, Demagny H, Velazquez-Villegas L, Schoonjans K. Molecular Physiology of Bile Acid Signaling in Health, Disease, and Aging. Physiol Rev. 2021;101(2):683–731. [DOI] [PubMed] [Google Scholar]
- 29.Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11(1):55–67. [DOI] [PubMed] [Google Scholar]
- 30.Li ZY, Zhou JJ, Luo CL, Zhang LM. Activation of TGR5 alleviates inflammation in rheumatoid arthritis peripheral blood mononuclear cells and in mice with collagen IIinduced arthritis. Mol Med Rep. 2019;20(5):4540–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581(7809):475–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Hang S, Fang Y, Bae S, Zhang Y, Zhang M, et al. A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe. 2021;29(9):1366–77 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol Aspects Med. 2017;56:54–65. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sorokin AV, Domenichiello AF, Dey AK, Yuan ZX, Goyal A, Rose SM, et al. Bioactive Lipid Mediator Profiles in Human Psoriasis Skin and Blood. J Invest Dermatol. 2018;138(7):1518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Chuan-Jian L, Ling H, Jing-Wen D, Ze-Hui H, Yu-Hong Y, et al. Untargeted serum metabonomics study of psoriasis vulgaris based on ultra-performance liquid chromatography coupled to mass spectrometry. Oncotarget. 2017;8(56):95931–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hench PS. Analgesia accompanying hepatitis and jaundice in cases of arthritis, fibrositis and siatic pain Mayo Clinic Proceedings, 1933;8. [Google Scholar]
- 38.Bruusgaard A, Andersen RB. Abnormal bile acid metabolism in rheumatoid arthritis. Preliminary communication. Dan Med Bull. 1976;23(2):95–8. [PubMed] [Google Scholar]
- 39.Shi Z, Wu X, Santos Rocha C, Rolston M, Garcia-Melchor E, Huynh M, et al. Short-Term Western Diet Intake Promotes IL-23Mediated Skin and Joint Inflammation Accompanied by Changes to the Gut Microbiota in Mice. J Invest Dermatol. 2021;141(7):1780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis K, Lutgendorff F, Phan V, Soderholm JD, Sherman PM, McKay DM. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm Bowel Dis. 2010;16(7):1138–48. [DOI] [PubMed] [Google Scholar]
- 41.Tajik N, Frech M, Schulz O, Schalter F, Lucas S, Azizov V, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun. 2020;11(1):1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinsson K, Durholz K, Schett G, Zaiss MM, Kastbom A. Higher serum levels of short-chain fatty acids are associated with non-progression to arthritis in individuals at increased risk of RA. Ann Rheum Dis. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pakchotanon R, Ye JY, Cook RJ, Chandran V, Gladman DD. Liver Abnormalities in Patients with Psoriatic Arthritis. J Rheumatol. 2020;47(6):847–53. [DOI] [PubMed] [Google Scholar]
- 44.Gottlieb A, Canbay A. Why Bile Acids Are So Important in Non-Alcoholic Fatty Liver Disease (NAFLD) Progression. Cells. 2019;8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGillicuddy FC, de la Llera Moya M, Hinkle CC, Joshi MR, Chiquoine EH, Billheimer JT, et al. Inflammation impairs reverse cholesterol transport in vivo. Circulation. 2009;119(8):1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puleston DJ, Villa M, Pearce EL. Ancillary Activity: Beyond Core Metabolism in Immune Cells. Cell Metab. 2017;26(1):131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rathbone MP, Middlemiss PJ, Gysbers JW, DeForge S, Costello P, Del Maestro RF. Purine nucleosides and nucleotides stimulate proliferation of a wide range of cell types. In Vitro Cell Dev Biol. 1992;28A(7–8):529–36. [DOI] [PubMed] [Google Scholar]
- 48.Lee J, Chuang TH, Redecke V, She L, Pitha PM, Carson DA, et al. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc Natl Acad Sci U S A. 2003;100(11):6646–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sano Y, Toyoshima S, Miki Y, Taketomi Y, Ito M, Lee H, et al. Activation of inflammation and resolution pathways of lipid mediators in synovial fluid from patients with severe rheumatoid arthritis compared with severe osteoarthritis. Asia Pac Allergy. 2020;10(2):e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ou J, Xiao M, Huang Y, Tu L, Chen Z, Cao S, et al. Serum Metabolomics Signatures Associated With Ankylosing Spondylitis and TNF Inhibitor Therapy. Front Immunol. 2021;12:630791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


